ABSTRACT
There is currently no diagnostic modality for early-stage pancreatic cancer. Given that adjuvant therapies require further development, the overall survival of pancreatic cancer remains unsatisfactory. Circular RNAs (circRNAs) are a class of noncoding RNAs that play an important role in the progression of many diseases including cancer. CircRNAs mainly bind to microRNAs as microRNA sponges to restore the expression of targeted genes and regulate tumor invasion, metastasis, proliferation, and apoptosis. CircRNAs also play roles in the diagnosis and targeted therapy of tumors. Studies on the mechanisms of action of circRNAs in pancreatic cancer are still in their infancy, but it is anticipated that this field will gradually advance. In this review, we provide a brief introduction to circRNAs from four perspectives: biogenesis, functions, and mechanisms of action, tumor therapy with circRNAs, and circRNAs’ roles in pancreatic cancer.
KEYWORDS: Circular RNA, pancreatic cancer, biogenesis, biomarker, targeted therapy, immunotherapy
1. Introduction
Pancreatic cancer remains one of the cancer types with the most dismal prognosis. This is associated with its concealed onset, early invasion and metastasis, limited surgical options, and unresponsiveness to adjuvant therapy, which all lead to an extremely low 5-year survival rate of 8%. When distant metastasis has occurred, this rate drops to just 3%.1,2 Despite unrelenting research efforts, the morbidity, and mortality of pancreatic cancer continue to rise, and its incidence in younger patients is increasing.3 At present, the major obstacle for treating pancreatic cancer is the lack of specific biomarkers and therapeutic targets, but circular RNAs (circRNAs) may offer a route to overcome this.
CircRNAs are a type of covalent closed circular noncoding RNAs that function similarly to other noncoding RNAs (e.g., miRNA, lncRNA, snRNA, piRNA, and siRNA). Although circRNAs cannot encode proteins, they can regulate the transcription and post-transcriptional modification of genes.4 Unlike their linear counterparts, the 3′ and 5′ ends of circRNAs are joined in a loop, so they lack a 5′ cap and a 3′ poly(A) tail. This makes them less susceptible to degradation by ribonucleases so circRNAs can maintain stable intracellular expression.5 In 1976, Sanger et al. first discovered that some plant viroids are composed of single-stranded circular closed RNA. Three years later, aided by electron microscopy, circular RNA was found in eukaryotic cells.6,7 Subsequently, in animal cells, satellite viruses of hepatitis B virus and hepatitis D virus were also observed to be composed of circRNA.8 However, owing to its low expression, circRNA was considered to be a by-product of accidental abnormal cleavage of precursor mRNAs (pre-mRNAs), which accumulates as “rubbish” in cells; its role in regulating the transcription of genes was also ignored, which resulted in a lack of investigations into circular RNA for decades.9 However, with the development of increasingly sophisticated RNA purification technology, high-throughput RNA sequencing technology, bioinformatic analysis, and various circRNA research tools, more than 30,000 circRNAs have now been predicted, indicating their ubiquity.10 However, these circular RNAs still need to be clearly identified.11
CircRNAs, unlike their linear counterparts, are derived from the noncanonical splicing of a pre-mRNA transcript, also called back-splicing. In this process, a receptor upstream of the RNA sequence binds to a downstream donor to form a loop structure.12–14 Although the length of circRNAs varies greatly, exonic circRNAs and intronic circRNAs, which account for a large proportion of circRNAs, are usually shorter than 200 nucleotides, and can even be shorter than 100.15 Owing to their unique structure, circRNAs are very stable and evolutionarily highly conserved in mammals. Moreover, their expression is tissue-specific and time-specific to some extent.16–18 There is increasing evidence that circRNAs are closely related to autophagy, apoptosis, the cell cycle, and proliferation, suggesting their potential role in various diseases, such as those of the nervous system and cardiovascular system, as well as cancer.19–23 Because of the significant correlation between circRNA expression and clinicopathological factors, and their widespread expression in body fluids including blood, saliva, urine, and gastric juice, the identification of differentially expressed circulating circRNAs for the early diagnosis and prognostic prediction of tumors is not only minimally invasive but also inexpensive.24–27 CircRNAs mainly influence genes that modulate tumor growth, metastasis, proliferation, and chemoresistance through the classical circRNA–miRNA–mRNA axis, including in lung cancer,28,29 colorectal cancer,30,31 gastric cancer,32,33 liver cancer,34,35 and breast cancer.36,37 Therefore, circRNAs may be used as therapeutic targets for cancer. This review briefly discusses the biogenesis and mechanisms of action of circRNAs, as well as their roles in tumor diagnosis and targeted therapy, and finally introduces related research on circRNAs in pancreatic cancer.
2. Classification and biogenesis of circRNAs
Although most circRNAs are only composed of exons encoding protein genes, they generally cannot encode proteins.17 Other circRNAs are composed of components (not exons) alone or in combination with exons, such as introns, noncoding regions, 3′-UTR, 5′-UTR, intergenic regions.38 In this way, circRNAs are divided into exonic circRNAs (EcircRNAs), intronic circRNAs (CiRNAs), and exon–intron circRNAs (intron-retaining circRNAs or EIciRNAs) based on their composition. In addition, recently, Gao et al. used a circRNA identification tool, CIRI, to reveal a class of integrated circRNAs that are spliced from two intronic circRNA fragments, called intergenic circRNAs;39 these are a kind of noncoding circRNA.
2.1. Biogenesis and regulation of exonic circRNAs
Approximately 80% of identified circRNAs belong to exonic circRNAs, which are almost always present in the cytoplasm. The classical function of circRNAs as miRNA sponges mainly involves this kind of circRNA. Jeck and colleagues reported that exonic circRNAs escape the nucleus during cell mitosis, but the specific mechanism behind their transport has remained unclear.40 However, recent studies reported that whether circRNAs are exported from or retained in the nucleus depends on their size, although this hypothesis contradicts the hypothesis that introns are strictly retained in the cell nucleus.41,42 The detailed mechanisms of circRNA export thus require further exploration. In canonical splicing, introns in pre-mRNAs are removed by selective splicing, and the remaining exons are ligated in tandem to form linear mRNAs. However, the generation of exonic circRNAs is different which is called back-splicing. In this process, a highly conserved receptor and donor of a pre-mRNA sequence join together to form a new circRNA. Back-splicing can be divided into three types: 1) lariat-driven circularization (exon skipping), 2) intron complementary pairing-driven cyclization, and 3) resplicing-driven circularization43 (Figure 1).
Figure 1.
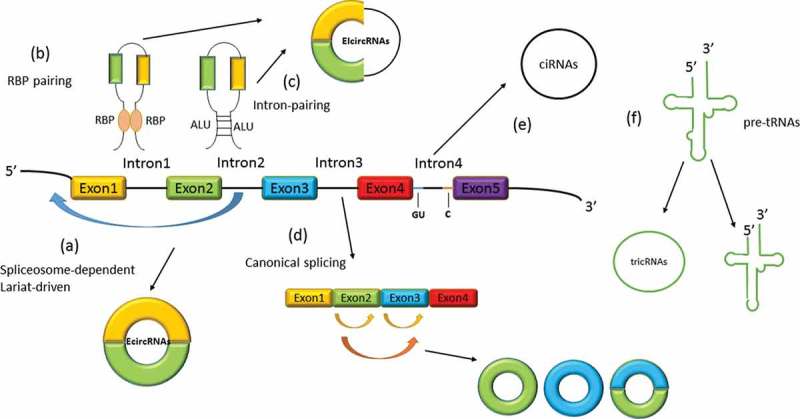
Biogenesis of circRNAs. (a) Spliceosome-dependent lariat-driven circularization: The spliceosome joins the downstream 5′ donor and upstream 3′ receptor together, from which back-splicing proceeds, which leads to the creation of EcircRNAs or EIcircRNAs. (b) RBPs can form a bridge-like connection through binding to flanking introns, which induces the formation of EcircRNAs. (c) Intron complementary pairing-driven circularization: Flanking introns with a content-specific sequence such as Alu element facilitate the release of EcircRNAs. (d) Resplicing-driven circularization: Mature exonic linear RNA may once again undergo self-back-splicing, which produces EcircRNAs consisting of one or more exons. (e) In flanking introns, sequences including a GU-rich element and a C-rich element are apt to form lariats and then form ciRNAs through circularization. (f) Introns in pre-tRNAs can separate and cyclize to form tricRNAs. This figure is adapted from Xu et al.44
Lariat-driven circularization refers to the formation of a lariat-like structure by a partial sequence of a pre-mRNA, which allows the upstream receptor and downstream donor that are far apart on the chromosome to be spatially close together. When coming into proximity of each other, the two can bind together via the action of a spliceosome with a stable 3′-5′ phosphodiester bond; the exons and introns in-between are spliced to form an intermediate exon–intron circRNA.45 One or more intra-lariat splicing events can occur in exon–intron circRNAs, and new exon–intron circRNAs and intron lariats can form.46
Intron complementary pairing-driven circularization is the first established and most common model of the formation of circRNAs. Different from lariat-driven circularization, intron complementary pairing-driven circularization is induced by the reverse complementary sequence pairing on the flanking introns to approximate the spatial distance between the splicing sites.47 Interestingly, most of the flanking intron complementary sequences that are capable of mediating circRNA production contain specific complementary repeat short sequences. Among them, introns with complementary Alu elements are more likely to induce circularization of exons than any other complementary intron components.11 A flanking intron comprising a complimentary repeat short sequence of about 30–40 nt is sufficient to mediate base pairing and subsequent RNA circularization. However, under specific conditions, intron base pairing increases the stability of linear RNA and inhibits RNA circularization.48 Since most intron reverse-complementing pairing sequences contain Alu elements, this feature can help us identify the circularization site of pre-mRNAs and also identify circRNAs.
Resplicing-driven circularization refers to the process of back-splicing of mature linear exonic mRNA. During this process, the resulting circRNAs can only be exonic circRNAs, including one or more exons.43
The generation of circRNA is modulated by a series of RNA-binding proteins (RBPs). Quaking (QKI),49 Muscleblind (MBL),45 and Fused-in Sarcoma (FUS)50 are positive regulators that upregulate the rate of circRNA formation. Taking MBL proteins as an example, two MBL proteins can bind to the MBL binding sites located on the flanking intron of the circMBL pre-mRNA, and dimerize to form a bridge-like junction. The RNA sequence between MBL dimer forms a lariat-like structure, which causes the receptor and the donor to be spatially close to each other; this, in turn, induces RNA back-splicing, hinders linear canonical splicing, and promotes the production of circMBL.45 Besides, ADAR1 and DHX9 can participate in circRNA negative regulation through the aforementioned intron pairing-driven circularization. Adenosine-to-inosine (A-to-I) RNA editing of double-stranded RNA-specific adenosine deaminase (ADAR) is thought to be associated with reduced efficiency of circRNA generation. Studies have shown that ADAR can bind to Alu elements responsible for intron pairing, causing annealing and deformation, resulting in an inability to perform reverse complementation pairing and dysfunction of back-splicing.51 Ivanov et al. found that the knockout of ADAR in vitro resulted in the twofold upregulation of expression of 84 circular RNAs and the downregulated expression of their linear counterparts.52 DHX9 is an important cofactor in the process of ADAR regulation of circRNA production.53 Intriguingly, RBPs can regulate the formation of circRNAs in both directions. Some studies have shown that, although FUS positively regulates the production of some circRNAs, it has the opposite effect in the production of others.50
2.2. Biogenesis of intronic circRNAs
In both canonical splicing and back-splicing, the introns between exons are usually debranched and degraded. However, under certain circumstances, introns can avoid debranching and form a stable ring structure. There are major differences between the biogenesis of intronic circRNAs and the biogenesis of exonic ones. The generation of stable intronic circRNAs is mainly dependent on a 7-nt GU-rich element near the 5′ splicing site and an 11-nt C-rich element near the debranching site.38 Therefore, the splicing sites of intronic circRNAs can be predicted based on these two elements. An in-depth study on the details of intron-driven circularization revealed that this process is divided into two steps. First, the 2′-OH on the branching point adenylate (bpA) attacks the 5′ splicing site to produce a 3′-OH in the 5′ exon. Next, the newly generated 3′-OH attacks the 3′ splicing end to generate an intron lariat and a new transcript consisting of selectively combined exons.48 Subsequently, this newly generated intron lariat undergoes 3′ tail degradation, eventually forming a mature intronic circRNA.38 Distinct from exonic circRNAs, intronic circRNAs are stably expressed in the nucleus and modulate gene expression. Intronic circRNAs can be degraded by RNA debranching enzymes due to the existence of a specific 2′-5′ linkage.38 Recent studies have found that, in the process of tRNA formation, introns in the pre-tRNA experience back-splicing, forming a special intronic circRNA called tricRNA.54
Although the efficacy of circRNA generation is much lower than that of their linear counterparts, circRNAs are abundant in cells due to their resistance against ribonuclease degradation. However, an excess of circRNAs in cells may be cytotoxic.55 In this context, the following question can be posed: How do circRNAs get transported out of cells? Studies have shown that exosomes and cellular microbubbles may be “vehicles” for transporting excess circRNA out of cells.55 Studies on exosomes have also shown that circRNAs are more abundant than linear RNAs in exosomes, which also supports this hypothesis.56
3. Underlying functions of circRNAs
3.1. Circrnas as miRNA sponges
The most prominent function of exonic circRNA in the cytoplasm is as an miRNA sponge. It is well known that miRNAs bind to the 3′-UTR of mRNAs and inhibit the expression of target genes. ceRNAs such as circRNAs generally contain one or more miRNA response elements (MREs), and MREs can bind to corresponding miRNAs to induce resumption of the expression of miRNA downstream genes.57 Given the diverse functions of miRNA-targeted genes, circRNAs also exert versatile effects through acting as miRNA sponges. In tumor progression, according to their particular functions, circRNAs can be divided into tumor-promoting and tumor-suppressing types.
ciRS-7 (a circRNA sponge of miR-7) and circSRY are two typical miRNA sponges. ciRS-7 contains more than 70 miR-7 binding sites and can bind to miR-7 via Argonaute (AGO).4 miR-7 has a wide range of functions and is involved in multiple signaling pathways. Therefore, many tumors can be regulated through the ciRS-7–miR-7 axis.58 However, miR-671 can bind to ciRS-7, leading to its linearization followed by its degradation mediated by AGO-2, leading to the release of miR-7.59 Many other circRNAs can also act as miRNA sponges, and even interact with long noncoding RNAs (lncRNAs).60 However, Guo and Militello et al. indicated that owing to their low expression and short length, some circRNAs cannot act as miRNA sponges, suggesting that miRNA sponge activity may not be shared among all circRNAs.61
3.2. Circrnas as protein sponges
Besides acting as miRNA sponges, circRNAs can bind to proteins to regulate the behavior of cells. The aforementioned RNA-binding protein MBL can increase the rate of circMBL formation by binding to the pre-MBL mRNA. circMBL can bind MBL and induce the latter’s dysfunction, thereby reducing its own expression level, maintaining a dynamic balance.62 It was also reported that circ-Amotl1 can bind to PDK1 and AKT1 in cardiomyocytes, leading to AKT1 phosphorylation for transport into the nucleus to protect the myocardium from damage.45 It was also discovered that circ-Amotl1 can bind to STAT3 and c-Myc and transfer them into the nucleus to promote cell proliferation, invasion, and tumorigenesis.37 Therefore, circ-Amotl1 can be used as a target in targeted therapy. Moreover, some circRNAs can serve as “scaffolds” for the binding of two different proteins, mediating their interaction. For example, circFOXO3 binds to CDK2 and p21 complex to form a triplet, which accelerates the dissociation of p21 and CDK2, phosphorylates cyclinA and cyclinE, and promotes cell cycle transition.22 In addition, p53 can also bind to murine double minute (MDM2) through circFOXO3 to promote its own degradation.21
3.3. Translation into proteins
Owing to their lack of a 5′ cap and a 3′ poly(A) tail, circRNAs can generally not be translated into proteins, unlike their linear counterparts. However, in vitro studies have revealed that some circRNAs containing an open reading frame (ORF) and an internal ribosome entry site (IRES) element possess the ability to bind endogenous ribosomes and be translated into proteins.63 The recruitment of YTHDF3 and EIf4G2 by the m6A modification site of circRNAs is associated with the potential of circRNAs to be translated into proteins.64 The classical example of a circRNA that can be translated into a protein is in the hepatitis D virus. When the hepatitis D virus infects host cells, the circRNA in it is translated into D-hepatic antigens.8 Moreover, circFBXW7 and circSHPRH containing IRES can encode FBXW7-185aa and SHPRH-146aa, respectively. Knocking out the IRES sequence can lead to the inactivation of these circRNAs in glioma and downregulate the expression of both FBXW7-185aa and SHPRH-146aa.65,66 However, it remains controversial whether circRNA-encoded proteins can be used as tumor therapeutic targets or tumor biomarkers.66
3.4. Transcription modulator
Exon–intron circRNAs can form RNA–RNA junctions with U1 small nuclear ribonucleoprotein (U1 snRNP) to activate the RNA pol II complex and facilitate the expression of paternal genes.67 However, intronic circRNAs, such as circANKRD52 and circ sirt7, can interact with RNA pol II complex to hamper the expression of paternal genes.38 A coding gene can form a linear RNA through canonical splicing transcription or a circRNA through noncanonical splicing. Because of their requirement for common exons, the above two options compete in terms of achieving their expression, leading to a dynamic balance between them.11 Some exonic circRNAs also contain promoters of linear RNAs. Therefore, increased expression of circRNAs can disable the translation of related linear RNAs; HIPK2/3 expression has been reported as a good example of this.11
4. Roles of circRNAs in cancer therapy
4.1. Circrnas as biomarkers for diagnosis and prognosis
Owing to them lacking open 3′ and 5′ ends, circRNAs are not easily degraded by ribonucleases and exonucleases, so they can stably and widely exist in cells, even in body fluids; this facilitates their simple and early detection. Moreover, the fact that circRNAs are evolutionarily conserved and exhibit tissue-specific expression supports their use as tumor biomarkers.
Bachmayr-Heyda et al. asserted that, in highly proliferating cells, the variety of circRNAs is reduced compared with that in cells with a low rate of proliferation. In line with this, the abundance of circRNAs in tumor tissues should be significantly reduced relative to that in adjacent normal tissues.68 They subsequently confirmed this in a study on colorectal cancer.23 In addition to profiles of circRNA abundance, certain aberrantly expressed circRNAs can often be used for the early diagnosis of cancer, with sufficient efficacy to challenge conventional tumor biomarkers. Hsa_circ_0000190 is downregulated in gastric cancer and its diagnostic specificity and sensitivity are even better than those of carcinoembryonic antigen (CEA) and carbohydrate antigen (CA19-9).69 High expression of ciRS-7 in hepatocellular carcinoma has been reported to be correlated with microvascular metastasis and alpha fetoprotein (AFP) levels in hepatocellular carcinoma.70 Given the stability of circRNA and its positive correlation with AFP, the combination of AFP and ciRS-7 could further improve the sensitivity and specificity of liver cancer diagnosis. CEA is a conventional tumor biomarker for colorectal cancer, but most patients in whom CEA positivity has developed have already reached the terminal stage of this disease. Dou et al. found that, in KRAS-mutated colorectal cancer, specific circRNAs are downregulated and others are upregulated compared with their levels in paracarcinoma tissue, suggesting a potential link between circRNAs and colorectal cancer gene mutations.71 However, biopsy for examination is invasive. Since Memczak et al. discovered that blood contains enough circRNAs to be detected, circRNAs have been widely found to be stably expressed in blood, saliva, urine, and gastric juice, among other body fluids.26 In view of the minimal invasiveness of liquid biopsy, circRNAs have gradually gained clinical value as tumor biomarkers. Moreover, circRNA expression in exosomes secreted by tumor cells was found to be at least double that in normal cell exosomes, with them acting to regulate the behavior of other tumor cells.72 This activity may be due to the higher proliferative capacity of tumor cells and their increased production of circRNAs, so more exosomes are required to transport circRNAs out of cells. Considering the difference in the expression of circRNAs between tumor exosomes and normal exosomes, analysis of the differentially expressed circRNAs in exosomes can also be used for the early diagnosis of tumors. Besides, the specific fusion circRNAs encoded by fusion genes can also identify cancers. For instance, fcircM1 produced by the MLL/AF9 fusion gene in leukemia and circRNA produced by the EML4-ALK fusion gene in non-small-cell lung cancer can be used not only as biomarkers for diagnosis but also as targets for tumor therapy.29,73
CircRNAs can also predict tumor stage and prognosis. For example, downregulation of hsa_circ_0000745 was discovered in cancerous tissues and plasma of patients with gastric cancer; its level in gastric cancer tissues was found to be closely related to tumor differentiation, and its plasma level in patients was related to the TNM stage of gastric cancer.74 Moreover, hsa_circ_0000520 was found to be aberrantly decreased in gastric cancer tissues and plasma, the level of which was also positively correlated with the TNM stage of gastric cancer; furthermore, its expression level in plasma was positively correlated with the CEA level.75 Furthermore, the expression level of hsa_circ_002059 in patients with gastric cancer was significantly lower postoperatively than that before surgery, while its expression level was significantly correlated with distant metastasis, TNM stage, gender, and age.76 CircCCDC66 is also highly expressed in colorectal cancer, and a higher circCCDC66 level was shown to be linked to a worse prognosis.31 Finally, in liver cancer, the overexpression of hsa_circ_0005075 was revealed to be closely related to clinicopathological factors such as tumor size.77
4.2. Circrnas in cancer treatment
The function of circRNAs as miRNA sponges can affect a variety of tumor progression-related signaling pathways. The basic strategy for targeted therapy with circRNAs is to inhibit the expression of tumor-promoting circRNAs by gene knockout, antisense oligonucleotides, and small interfering RNAs (siRNAs), or to transfect artificial tumor-suppressing circRNAs. siRNAs can bind to circRNA-specific back-splicing sites and specific sequences on precursor mRNAs, such as upstream receptor, downstream donor, and Alu element, to cause dysfunction of RNA lariat formation. Besides, the employment of CRISPR/Cas9 technology to knock out tumor-promoting circRNA-expressing genes is also a promising approach.78 Moreover, administering tumor-suppressing circRNAs exogenously or synthesizing circRNAs specific for tumor-promoting miRNAs also has clinical value. For example, Liu et al. constructed an miRNA sponge targeting miR-210 and miR-183–96-182 by inserting multiple miRNA binding sites into the gene, successfully inhibiting the growth and invasiveness of bladder cancer cells, and promoting their apoptosis.79
4.3. Circrnas and tumor immunotherapy
Via their functions as miRNA and protein sponges, circRNAs can bind to miRNAs and proteins involved in modulating tumor immunity. For example, hsa_circ_0020397 was discovered to bind to miR-138 and indirectly promoted the expression of its targets, including PD-L1, which binds PD-1 and hence mediates tumor immune escape.80 Moreover, circFOXO3 can compete with p53 for binding to MDM2, preventing p53’s degradation; this is important as p53 plays a key role in inducing immune responses.81 Abnormal circRNAs can be found in tumor cells due to gene mutations and chromosomal variations. Owing to their stability and heterogeneity, these circRNAs can be used as tumor antigens to induce immune responses. Studies have shown that abnormal circRNAs produced in tumor cells can be secreted by exosomes or vesicles and presented to immunocytes, such as Tregs, to stimulate immune responses.82 Additionally, proteins produced by the translation of circRNAs can also elicit immune responses. Moreover, in exosomes, circRNAs can coexist with miRNAs or mRNAs. Therefore, circRNAs may bind to miRNAs or mRNAs to stabilize them and inhibit their degradation, rather than directly regulate immune responses themselves. After reaching their targeted immunocytes, circRNAs release miRNAs or mRNAs to regulate immunocyte function. CircRNAs can regulate tumor immune responses through various mechanisms, as described above. However, further study of the roles of circRNAs in tumor immune responses and immune escape is still warranted.
5. Circrnas in pancreatic cancer
Although intense investigations of the roles of circRNAs in tumor progression have been performed, only a few studies have focused on circRNAs in pancreatic cancer. Such studies have only been initiated in recent years, particularly in China. Although the study of circRNAs in pancreatic cancer is still in its infancy, we here summarize the roles of circRNAs in pancreatic cancer, especially the involvement of specific types of circRNA in pancreatic cancer progression, targeted therapy, and drug resistance (Table 1), with the aim of drawing attention to this promising field.
Table 1.
CircRNAs and pancreatic cancer.
| CircRNAs | Expression | Function | Refs |
|---|---|---|---|
| hsa_circ_0006988 | Up | Biomarker | 83 |
| hsa_circ_0000977 | Up | MiR-874-3p sponge | 84 |
| hsa_circ_0006215 | Up | MiR-378a-3p sponge | 85 |
| circ_0007534 | Up | MiR-625 and miR-892b sponge 90 | 86 |
| hsa_circ_0001649 | Down | Biomarker and target | 87 |
| hsa_circ_0036627 | Up | MiR-338 sponge | 88 |
| circIRAS | Up | MiR-122 sponge | 89 |
| circ_100782 | Up | MiR-124 sponge | 90 |
| circRHOT1 | Up | miR-26b,miR-125a,miR-330 and miR-382 sponge | 91 |
| ciRS-7 | Up | MiR-7 sponge | 92 |
| circZMYM2 | Up | MiR-335-5p sponge | 93 |
| circ_0030235 | Up | MiR-1253 and MiR-1294 sponge | 94 |
Most of the circRNAs in pancreatic cancers that have been studied or are now under investigation were first identified by Li et al.95 They analyzed six pairs of pancreatic cancer and paracancerous tissues and found that the circRNA expression profiles in these two tissues were significantly different; they also evaluated another 20 pairs of tissue samples for further confirmation. These abnormally expressed circRNAs may become tumor biomarkers and therapeutic targets of pancreatic cancer. These results were registered in the Gene Expression Omnibus (GEO; No. GSE69362).
Since then, the results of Li et al. have become the cornerstone for further research on the mechanisms of action of circRNAs in the progression and chemoresistance of pancreatic cancer.95,96 For example, inspired by the research of Li et al., Sun and colleagues found that hsa_circ_0006988 has potential as a tumor biomarker for pancreatic cancer, as revealed by an analysis using circRNA research tools, such as circBase and cric2Traits. Hsa_circ_0006988 was significantly elevated in pancreatic cancer tissues and plasma. Its expression level in pancreatic cancer tissues was also found to be closely related to tumor blood vessel infiltration and lymphatic metastasis, and its expression in plasma was significantly correlated with CA19-9 level, N stage, blood vessel infiltration, and lymphatic metastasis. In addition, hsa_circ_0006988 combined with CA19-9 was also found to have higher sensitivity and specificity for the early diagnosis of pancreatic cancer.83 The role of the hsa_circ_0006215–miR-378a-3p–SERPINA4 signaling axis in the progression of pancreatic cancer was also demonstrated, for the first time in pancreatic cancer, they demonstrated that circRNAs can function as miRNA sponges and modulate the expression of tumor-related genes.84 Wang’s team also analyzed the circRNA microarray expression profile in pancreatic cancer, and obtained results similar to those of Li et al. The results of this study are also registered in GEO (No. GSE79634).85 On this basis, hsa_circ_0000977 was discovered to be aberrantly expressed in pancreatic cancer tissues, and was shown to restore PLK1 through binding to miR-874-3p, which silenced the function of PLK1; moreover, PLK1 overexpression was shown to be associated with poor prognosis in numerous cancers.97 Meng et al. also discovered that upregulated circ_0007534-facilitated cell proliferation and invasion, and inhibited apoptosis by acting as a sponge towards miR-625 and miR-892b.86 Moreover, Li’s team discovered that circRHOT1 was elevated in pancreatic cancer and facilitated cell proliferation, invasion, and metastasis by binding miR-26b, miR-125a, miR-330, and miR-382.91 The aforementioned relationship between ciRS-7 and miR-7 in pancreatic cancer progression was also confirmed to occur through promoting the expression of EGFR and STAT3.92 Then, circZMYM2/miR-335-5p/JMJD2C and circ_0030235/miR-1253 or miR-1294 axis were also identified as the miRNA sponges function of circRNAs.93,94 Unlike the aforementioned increase in the expression of circRNAs in pancreatic cancer, hsa_circ_0001649 was found to be downregulated in pancreatic cancer tissue and cell lines, and its low expression is often accompanied by advanced tumor staging and poor tissue grading. However, the exogenous administration of hsa_circ_0001649 can suppress the proliferative capacity of pancreatic cancer cell lines and induce apoptosis, suggesting that it can be used as an exogenous anticancer agent.87 As mentioned earlier, exosomes secreted by tumor cells can act as messengers that transmit signals between cells. Research has shown that plasma exosomes in patients with pancreatic cancer are rich in circ-PDE8A and are associated with tumor progression and prognosis. Moreover, an in-depth study revealed that circ-PDE8A can activate the MACC/MET/ERK signaling pathway through competitive binding to miR-338 to facilitate tumor progression.88 The same team also observed that circ-IARS contained in plasma exosomes of patients with pancreatic cancer can be carried to human microvascular endothelial cells, upregulating F-actin expression and improving local cell adhesion by upregulating RhoA and RhoA-GTP levels through binding to miR-122; this, in turn, increases endothelial cell monolayer permeability, promoting tumor cell invasion and metastasis.89 CircRNAs may also play an important role in chemoresistance in pancreatic cancer. Ding et al. confirmed that there was a difference between the expression profiles of circRNAs in gemcitabine-resistant pancreatic cancer cell lines and in normal pancreatic cancer cell lines; they also suggested that circRNAs may play a role in pancreatic cancer chemoresistance by acting as miRNA sponges affecting the MRPK and mTOR signaling pathway.98 At the same time, Huang et al. constructed a PANC-1-GR gemcitabine-resistant cell line and analyzed the difference in the expression profiles of circRNAs between PANC-1-GR and PANC-1.The two circRNAs (chr14: 101402109–101464448+, chr4: 52729603–52780244+) with the most significant differences between the two groups were identified, and it was found that silencing their expression restored the sensitivity of pancreatic cancer resistant cell lines, while their overexpression weakened this sensitivity.99 Another study showed that circ_100782 is upregulated in pancreatic cancer and increases the expression of downstream IL-6R and STAT3 by binding to miR-124, thereby promoting the proliferation of the pancreatic cancer cell line BxPC-3.90 The upregulation of STAT3 can mediate tumor immune escape. Therefore, circ_100782 may play a role in tumor immunity of pancreatic cancer, although the specific mechanisms involved in this remain to be explored.100
6. Conclusions and future prospects
Because of a dearth of specific targeting molecules, preoperative neoadjuvant chemotherapy and adjuvant therapy after radical surgery cannot significantly improve the long-term survival rate of patients, which is the major bottleneck in the treatment of pancreatic cancer. Studies have shown that the interactions between various molecules in the microenvironment of pancreatic cancer tumors, as well as pancreatic cancer stromal cells and tumor cells, play crucial roles in the progression of this disease. By continuing the search for new molecular targets that are specific to pancreatic cancer, versatile molecular targets could become the main focus of research to overcome pancreatic cancer.
Recent studies on circRNAs have gradually revealed their various roles in tumor progression. The most common and important function of circRNAs is as miRNA sponges. Through the circRNA–miRNA–mRNA axis, circRNAs can up- or downregulate gene expression and affect tumor progression. The roles of circRNAs in tumor immunity, exocytic transport, as protein sponges, and their translation into functional proteins can also affect tumor progression. These functions of circRNAs, either inhibiting the expression of tumor-promoting circRNAs or exogenously upregulating the expression of tumor-suppressing ones, have been utilized for tumor-targeted therapy.
Studies of circRNAs for the diagnosis and treatment of pancreatic cancer are still in their infancy, but have already made some progress. For example, it has been shown that there is a significant difference in the expression profile of circRNAs in pancreatic cancer tissues compared with that in adjacent tissues, which has also been supported by plasma analyses. Additionally, in-depth studies have demonstrated that circRNAs in pancreatic cancer can modulate the behavior of pancreatic cancer tumors, such as their invasion, metastasis, immune escape, and chemoresistance, through various signaling axes by binding to miRNAs. This suggests that circRNAs play versatile roles in pancreatic cancer; moreover, the identification of these signaling pathways can form the basis of a search for therapeutic targets for pancreatic cancer. Within the field of study of circRNAs in pancreatic cancer, there is still a lack of convictive animal experiments and clinical research to confirm the role of circRNAs in clinical treatment. Therefore, further study on the mechanisms of action of circRNA in pancreatic cancer is still awaited. We believe that, in the near future, circRNAs should benefit the early diagnosis and targeted treatment of pancreatic cancer, given their analytical advantages and an extensive range of functions.
Funding Statement
This work was supported by the China Academy of Medical Sciences Innovation Fund for Medical Sciences [2016-I2M-3-019];Non-profit Central Research Institute Fund of Chinese of Academy of Medical Sciences [2018PT32014].
Acknowledgments
This work was supported by the China Academy of Medical Sciences Innovation Fund for Medical Sciences (CIFMS; Grant No. 2016-I2M-3-019) and the Non-profit Central Research Institute Fund of the Chinese Academy of Medical Sciences (Grant No. 2018PT32014). We thank Liwen Bianji, Edanz Group China (www.liwenbianji.cn/ac), for editing the English text of a draft of this manuscript.
Conflicts of interest
All authors have no conflicts of interest to declare.
References
- 1.Siegel RL, Miller KD, Jemal A.. Cancer statistics,2019. CA Cancer J Clin. 2019;69(1):7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Kamisawa T, Wood LD, Itoi T, Takaori: K. Pancreatic cancer. Lancet. 2016;388(10039):73–85. doi: 10.1016/S0140-6736(16)00141-0. [DOI] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal: A. Cancer statistics. CA Cancer J Clin. 2016;66(1):7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 4.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 5.Qu S, Yang X, Li X, Wang J, Gao Y, Shang R, Sun W, Dou K, Li H. Circular RNA: a new star of noncoding RNAs. Cancer Lett. 2015;365(2):141–148. doi: 10.1016/j.canlet.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 6.Sanger HL, Klotz G, Riesner D, et al. Viroids are singlestranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci USA. 1976;73:3852–3856. doi: 10.1073/pnas.73.8.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hsu MT, Coca-Prados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature. 1979;280(5720):339–340. [DOI] [PubMed] [Google Scholar]
- 8.Kos A, Dijkema R, Arnberg AC, van der Meide PH, Schellekens H. The hepatitis delta (delta) virus possesses a circular RNA. Nature. 1986;323(6088):558–560. doi: 10.1038/323558a0. [DOI] [PubMed] [Google Scholar]
- 9.Cocquerelle C, Mascrez B, Hétuin D, Bailleul B. Mis-splicing yields circular RNA molecules. Faseb J. 1993;7(1):155–160. [DOI] [PubMed] [Google Scholar]
- 10.Caiment F, Gaj S, Claessen S, Kleinjans: J. High-throughput data integration of RNA-miRNA-circRNA reveals novel insights into mechanisms of benzo[a]pyrene-inducedcarcinogenicity. Nucleic Acids Res. 2015;43(5):2525–2534. doi: 10.1093/nar/gkv115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. Circular RNAs are abundant, conserved, and associated with ALU repeats. Rna. 2013;19(2):141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barrett SP, Wang PL, Salzman: J. Circular RNA biogenesis can proceed through an exon-containing lariat precursor. Elife. 2015;4:e07540. doi: 10.7554/eLife.06416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schindewolf C, Braun S, Domdey: H. In vitro generation of a circular exon from a linear pre-mRNA transcript. Nucleic Acids Res. 1996;24(7):1260–1266. doi: 10.1093/nar/24.7.1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Starke S, Jost I, Rossbach O, Schneider T, Schreiner S, Hung LH, Bindereif A. Exon circularization requires canonical splice signals. Cell Rep. 2015;10(1):103–111. doi: 10.1016/j.celrep.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 15.Lasda E, Parker: R. Circular RNAs: diversity of form and function. Rna. 2014;20(12):1829–1842. doi: 10.1261/rna.047126.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. CircularRNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7(2):e30733. doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 18.Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9(9):e1003777. doi: 10.1371/journal.pgen.1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang R, Zhang Y, Han B, Bai Y, Zhou R, Gan G, Chao J, Hu G, Yao H. Circular RNA HIPK2 regulates astrocyte activation via cooperation of autophagy and ER stress by targeting MIR124-2HG. Autophagy. 2017;13(10):1722–1741. doi: 10.1080/15548627.2017.1356975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang J, Wang P, Wan L, Xu S, Pang D. The emergence of noncoding RNAs as Heracles in autophagy. Autophagy. 2017;13(6):1004–1024. doi: 10.1080/15548627.2017.1312041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Du WW, Fang L, Yang W, Wu N, Awan FM, Yang Z, Yang BB. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. 2017;24(2):357–370. doi: 10.1038/cdd.2016.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44(6):2846–2858. doi: 10.1093/nar/gkw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bachmayr-Heyda A, Reiner AT, Auer K, Sukhbaatar N, Aust S, Bachleitner-Hofmann T, Mesteri I, Grunt TW, Zeillinger R, Pils D. Correlation of circular RNA abundance with proliferation—exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues. Sci Rep. 2015;5:8057. doi: 10.1038/srep08057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, Chen D, Gu J, He X, Huang S. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25:981–984. doi: 10.1038/cr.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bahn JH, Zhang Q, Li F, Chan TM, Lin X, Kim Y, Wong DT, Xiao X. The landscape of microRNA,Piwi-interacting RNA, and circular RNA in human saliva. Clin Chem. 2015;61:221–230. doi: 10.1373/clinchem.2014.230433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Memczak S, Papavasileiou P, Peters O, Rajewsky N. Identification and characterization of circular RNAs as a new class of putative biomarkers in human blood. PLoS One. 2015;10:e0141214. doi: 10.1371/journal.pone.0141214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim KM, Abdelmohsen K, Mustapic M, Kapogiannis D, Gorospe M. RNA in extracellular vesicles. Wiley Interdiscip Rev RNA. 2017;8:e1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qiu M, Xia W, Chen R, Wang S, Xu Y, Ma Z, Xu W, Zhang E, Wang J, Fang T, et al. The circular RNA circPRKCI promotes tumor growth in lung adenocarcinoma. Cancer Res. 2018;78(11):2839–2851. doi: 10.1158/0008-5472.CAN-17-2808. [DOI] [PubMed] [Google Scholar]
- 29.Tan S, Gou Q, Pu W, Guo C, Yang Y, Wu K, Liu Y, Liu L, Wei YQ, Peng Y. Circular RNA F-circEA produced from EML4-ALK fusion gene as a novel liquid biopsy biomarker for non-small cell lung cancer. Cell Res. 2018;28(6):693–695. doi: 10.1038/s41422-018-0033-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zeng K, Chen X, Xu M, Liu X, Hu X, Xu T, Sun H, Pan Y, He B. Wang: circHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell Death Dis. 2018;9(4):417. doi: 10.1038/s41419-018-1111-y. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Hsiao KY, Lin YC, Gupta SK, Chang N, Yen L, Sun HS, Tsai SJ. Noncoding effects of circular RNA CCDC66 promote Colon Cancer growth and metastasis. Cancer Res. 2017;77(9):2339–2350. doi: 10.1158/0008-5472.CAN-16-1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang J, Liu H, Hou L, Wang G, Zhang R, Huang Y, Chen X, Zhu J. Circular RNA_LARP4 inhibits cell proliferation and invasion of gastric cancer by sponging miR-424-5p and regulating LATS1 expression. Mol Cancer. 2017;16(1):151. doi: 10.1186/s12943-017-0719-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen J, Li Y, Zheng Q, Bao C, He J, Chen B, Lyu D, Zheng B, Xu Y, Long Z, et al. Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer. Cancer Lett. 2017;388:208–219. doi: 10.1016/j.canlet.2016.12.006. [DOI] [PubMed] [Google Scholar]
- 34.Chen G, Shi Y, Liu M, Sun J. circHIPK3 regulates cell proliferation and migration by sponging miR-124 and regulating AQP3 expression in hepatocellular carcinoma. Cell Death Dis. 2018;9(2):175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Han D, Li J, Wang H, Su X, Hou J, Gu Y, Qian C, Lin Y, Liu X, Huang M, et al. 2017. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology. 66(4):1151–1164. [DOI] [PubMed] [Google Scholar]
- 36.He R, Liu P, Xie X, Zhou Y, Liao Q, Xiong W, Li X, Li G, Zeng Z, Tang H. circGFRA1 and GFRA1 act as ceRNAs in triple negative breast cancer by regulating miR-34a. J Exp Clin Cancer Res. 2017;36(1):145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang Q, Du WW, Wu N, Yang W, Awan FM, Fang L, Ma J, Li X, Zeng Y, Yang Z, et al. A circular RNA promotes tumorigenesis by inducing c-myc nuclear translocation. Cell Death Differ. 2017;24(9):1609–1620. doi: 10.1038/cdd.2017.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, Zhu S, Yang L, Chen LL. Circular intronic long noncoding RNAs. Mol Cell. 2013;51(6):792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 39.Gao Y, Wang J, Zhao F. CIRI: an efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol. 2015;16:4. doi: 10.1186/s13059-015-0667-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32(5):453–461. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang C, Liang D, Tatomer DC, Wilusz JE. A length-dependent evolutionarily conserved pathway controls nuclear export of circular RNAs. Genes Dev. 2018;32(9–10):639–644. doi: 10.1101/gad.314856.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wan Y, Hopper AK. Size matters: conserved proteins function in length-dependent nuclear export of circular RNAs. Genes Dev. 2018;32(9–10):600–601. doi: 10.1101/gad.316216.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kameyama T, Suzuki H, Mayeda: A. Re-splicing of mature mRNA in cancer cells promotes activation of distant weak alternative splice sites. Nucleic Acids Res. 2012;40(16):7896–7906. doi: 10.1093/nar/gks520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cui X, Wang J, Guo Z, Li M, Li M, Liu S, Liu H, Li W, Yin X, Tao J, et al. Emergingfunction and potential diagnostic value of circular RNAs in cancer. Mol Cancer. 2018;17(1):123. doi: 10.1186/s12943-018-0877-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, Kadener S. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56(1):55–66. [DOI] [PubMed] [Google Scholar]
- 46.Zaphiropoulos PG. Circular RNAs from transcripts of the rat cytochrome P450 2C24 gene: correlation with exon skipping. Proc Natl Acad Sci USA. 1996;93(13):6536–6541. doi: 10.1073/pnas.93.13.6536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L. Complementary sequence-mediated exon circularization. Cell. 2014;159:134–147. doi: 10.1016/j.cell.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 48.Liang D, Wilusz JE. Short intronic repeat sequences facilitate circular RNA production. Genes Dev. 2014;28(20):2233–2247. doi: 10.1101/gad.251926.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA, Goodall GJ. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160(6):1125–1134. doi: 10.1016/j.cell.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 50.Errichelli L, Dini Modigliani S, Laneve P, Colantoni A, Legnini I, Capauto D, Rosa A, De Santis R, Scarfo R, Peruzzi G, et al. FUS affects circular RNA expression in murine embryonic stem cell-derived motor neurons. Nat Commun. 2017;8:14741. doi: 10.1038/ncomms14741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Levanon EY, Eisenberg E, Yelin R, Nemzer S, Hallegger M, Shemesh R, Fligelman ZY, Shoshan A, Pollock SR, Sztybel D, et al. Systematic identifcation of abundant A-to-I editing sites in the human transcriptome. Nat Biotechnol. 2004;22(8):1001–1005. doi: 10.1038/nbt996. [DOI] [PubMed] [Google Scholar]
- 52.Ivanov A, Memczak S, Wyler E, Torti F, Porath HT, Orejuela MR, Piechotta M, Levanon EY, M L, Dieterich C, et al. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 2015;10(2):170–177. doi: 10.1016/j.celrep.2014.12.019. [DOI] [PubMed] [Google Scholar]
- 53.Aktaş T, Avşar Ilık İ, Maticzka D, Bhardwaj V, Pessoa Rodrigues C, Mittler G, Manke T, Backofen R, Akhtar A. DHX9 suppresses RNA processing defects originating from the Alu invasion of the human genome. Nature. 2017;544(7648):115–119. doi: 10.1038/nature21715. [DOI] [PubMed] [Google Scholar]
- 54.Noto JJ, Schmidt CA, Matera AG. Engineering and expressing circular RNAs via tRNA splicing. RNA Biol. 2017;14(8):978–984. doi: 10.1080/15476286.2017.1317911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huang G, Li S, Yang N, Zou Y, Zheng D, Xiao T. Recent progress in circular RNAs in human cancers. Cancer Lett. 2017;404:8–18. doi: 10.1016/j.canlet.2017.07.002. [DOI] [PubMed] [Google Scholar]
- 56.Lasda E, Parker R. Circular RNAs co-precipitate with extracellular vesicles: a possible mechanism for circRNA clearance. PLoS One. 2016;11(2):e01484077. doi: 10.1371/journal.pone.0148407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu J, Liu T, Wang X,He A.. Circles reshaping the RNA world: from waste to treasure. Mol Cancer. 2017;16(1):58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Salzman J. Circular RNA expression: its potential regulation and function. Trends Genet. 2016;32(5):309–316. doi: 10.1016/j.tig.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hansen TB, Wiklund ED, Bramsen JB, Villadsen SB, Statham AL, Clark SJ, Kjems J. miRNAdependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. Embo J. 2011;30(21):4414–4422. doi: 10.1038/emboj.2011.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kleaveland B, Shi CY, Stefano J, Bartel DP. A network of noncoding regulatory RNAs acts in the mammalian brain. Cell. 2018;174(2):350–362.e17. doi: 10.1016/j.cell.2018.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, Luo Y, Lyu D, Li Y, Shi G, et al. Circular RNA profling reveals an abundant circHIPK3 that regulates cell growthby sponging multiple miRNAs. Nat Commun. 2016;7:11215. doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zeng Y, Du WW, Wu Y, Yang Z, Awan FM, Li X, Yang W, Zhang C, Yang Q, Yee A, et al. A circular RNA binds to and activates AKT phosphorylation and nuclear localization reducing apoptosis and enhancing cardiac repair. Theranostics. 2017;7(16):3842–3855. doi: 10.7150/thno.19764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen CY, Sarnow P. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science. 1995;268(5209):415–417. [DOI] [PubMed] [Google Scholar]
- 64.Yang Y, Fan X, Mao M, et al. 2017. Extensive translation of circular RNAs driven by N(6)-methyladenosine[J]. Cell Res. 27(5):626–641. doi: 10.1038/cr.2017.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang Y, Gao X, Zhang M, Yan S, Sun C, Xiao F, Huang N, Yang X, Zhao K, Zhou H, et al. Novel Role of FBXW7 circular RNA in repressing glioma tumorigenesis. J Natl Cancer Inst. 2018;110(3):304–315. doi: 10.1093/jnci/djx166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang M, Huang N, Yang X, Luo J, Yan S, Xiao F, Chen W, Gao X, Zhao K, Zhou H, et al. A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene. 2018;37(13):1805–1814. doi: 10.1038/s41388-017-0019-9. [DOI] [PubMed] [Google Scholar]
- 67.Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Zhong G, Yu B, Hu W, Dai L, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22(3):256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 68.Drucker E, Krapfenbauer K. Pitfalls and limitations in translation from biomarker discovery to clinical utility in predictive and personalised medicine. Epma J. 2013;4(1):7. doi: 10.1186/1878-5085-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen S, Li T, Zhao Q, Xiao B, Guo J. Using circular RNA hsa_circ_0000190 as a new biomarker in the diagnosis of gastric cancer. Clin Chim Acta. 2017;466:167–171. doi: 10.1016/j.cca.2017.01.025. [DOI] [PubMed] [Google Scholar]
- 70.Xu L, Zhang M, Zheng X, Yi P, Lan C, Xu M. The circular RNA ciRS-7 (Cdr1as) acts as a risk factor of hepatic microvascular invasion in hepatocellular carcinoma. J Cancer Res Clin Oncol. 2017;143(1):17–27. doi: 10.1007/s00432-016-2256-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dou Y, Cha DJ, Franklin JL, Higginbotham JN, Jeppesen DK, Weaver AM, Prasad N, Levy S, Coffey RJ, Jg P, et al. Circular RNAs are down-regulated in KRAS mutant colon cancer cells and can be transferred to exosomes. Sci Rep. 2016;6:37982. doi: 10.1038/srep37982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mercer TR, Dinger ME, Mattick: JS. Long noncoding RNAs: insights into functions. Nat Rev Genet. 2009;10(3):155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 73.Guarnerio J, Bezzi M, Jeong JC, Paffenholz SV, Berry K, Naldini MM, Lo-Coco F, Tay Y, Beck AH, Pandolfi PP. Oncogenic role of fusion-circRNAs derived from cancer-associated chromosomal translocations. Cell. 2016;165(2):289–302. doi: 10.1016/j.cell.2016.03.020. [DOI] [PubMed] [Google Scholar]
- 74.Huang M, He YR, Liang LC, Huang Q, Zhu ZQ. Circular RNA hsa_circ_0000745 may serve as a diagnostic marker for gastric cancer. World J Gastroenterol. 2017;23(34):6330–6338. doi: 10.3748/wjg.v23.i34.6330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sun H, Tang W, Rong D, Jin H, Fu K, Zhang W, Liu Z, Cao H, Cao X. Hsa_circ_0000520, a potential new circular RNA biomarker, is involved in gastric carcinoma. Cancer Biomark. 2018;21(2):299–306. doi: 10.3233/CBM-170379. [DOI] [PubMed] [Google Scholar]
- 76.Li P, Chen S, Chen H, Mo X, Li T, Shao Y, Xiao B, Guo J. Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clin Chim Acta. 2015;444:132–136. doi: 10.1016/j.cca.2015.02.018. [DOI] [PubMed] [Google Scholar]
- 77.Shang X, Li G, Liu H, Li T, Liu J, Zhao Q, Wang C. Comprehensive circular RNA profiling reveals that hsa_circ_0005075, a new circular RNA biomarker, is involved in hepatocellular Crcinoma development. Medicine (Baltimore). 2016;95(22):e3811. doi: 10.1097/MD.0000000000004864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Piwecka M, Glazar P, Hernandez-Miranda LR, Memczak S, Wolf SA, Rybak-Wolf A, Filipchyk A, Klironomos F, Cerda Jara CA, Fenske P, et al. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science. 2017;357(6357). pii: eaam8526. doi: 10.1126/science.aam8526. [DOI] [PubMed] [Google Scholar]
- 79.Liu Y, Han Y, Zhang H, Nie L, Jiang Z, Fa P, Gui Y, Cai Z. Synthetic miRNA-mowers targeting miR-183-96-182 cluster or miR-210 inhibit growth and migration and induce apoptosis in bladder cancer cells. PLoS One. 2012;7(12):e52280. doi: 10.1371/journal.pone.0052280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang XL, Xu LL, Wang F. Hsa_circ_0020397 regulates colorectal cancer cell viability, apoptosis and invasion by promoting the expression of the miR-138 targets TERT and PD-L1. Cell Biol Int. 2017;41(9):1056–1064. doi: 10.1002/cbin.10826. [DOI] [PubMed] [Google Scholar]
- 81.Huang Y, Yu P, Li W, Ren G, Roberts AI, Cao W, Zhang X, Su J, Chen X, Chen Q, et al. p53 regulates mesenchymal stem cell-mediated tumor suppression in a tumor microenvironment through immune modulation. Oncogene. 2014;33(29):3830–3838. doi: 10.1038/onc.2013.355. [DOI] [PubMed] [Google Scholar]
- 82.Li P, Liu C, Yu Z, Wu M. New insights into regulatory T cells: exosome-and non-coding RNA-mediated regulation of homeostasis and resident Treg cells. Front Immunol. 2016;7:574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yang F, Liu DY, Guo JT, Ge N, Zhu P, Liu X, Wang S, Wang GX, Sun SY. Circular RNA circ-LDLRAD3 as a biomarker in diagnosis of pancreatic cancer. World J Gastroenterol. 2017;23(47):8345–8354. doi: 10.3748/wjg.v23.i47.8345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhu P, Ge N, Liu D, Yang F, Zhang K, Guo J, Liu X, Wang S, Wang G, Sun S. Preliminary investigation of the function of hsa_circ_0006215 in pancreatic cancer. Oncol Lett. 2018;16(1):603–611. doi: 10.3892/ol.2018.8652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Guo S, Xu X, Ouyang Y, Wang Y, Yang J, Yin L, Ge J, Wang H. Microarray expression profile analysis of circular RNAs in pancreatic cancer. Mol Med Rep. 2018;17(6):7661–7671. doi: 10.3892/mmr.2018.8827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hao L, Rong W, Bai L, Cui H, Zhang S, Li Y, Chen D, Meng X. 2018. Upregulated circular RNA circ_0007534 indicates an unfavorable prognosis in pancreatic ductal adenocarcinoma and regulates cell proliferation, apoptosis, and invasion by sponging miR-625 and miR-892b. J Cell Biochem. DOI: 10.1002/jcb.27658 [DOI] [PubMed] [Google Scholar]
- 87.Jiang Y, Wang T, Yan L, Qu L. A novel prognostic biomarker for pancreatic ductal adenocarcinoma: hsa_circ_0001649. Gene. 2018;675:88–93. doi: 10.1016/j.gene.2018.06.099. [DOI] [PubMed] [Google Scholar]
- 88.Li Z, Yanfang W, Li J, Jiang P, Peng T, Chen K, Zhao X, Zhang Y, Zhen P, Zhu J, et al. Tumor-releasedexosomal circular RNA PDE8A promotes invasive growth via the miR-338/MACC1/MET pathway in pancreatic cancer. Cancer Lett. 2018;432:237–250. doi: 10.1016/j.canlet.2018.04.035. [DOI] [PubMed] [Google Scholar]
- 89.Li J, Li Z, Jiang P, Peng M, Zhang X, Chen K, Liu H, Bi H, Liu X, Li X. Circular RNA IARS (circ-IARS) secreted by pancreatic cancer cells and located within exosomes regulates endothelial monolayer permeability to promote tumor metastasis. J Exp Clin Cancer Res. 2018;37(1):177. doi: 10.1186/s13046-018-0822-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hen G, Shi Y, Zhang Y, Sun J. CircRNA_100782 regulates pancreatic carcinoma proliferation through the IL6-STAT3 pathway. Onco Targets Ther. 2017;10:5783–5794. doi: 10.2147/OTT.S150678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Qu S, Hao X, Song W, Niu K, Yang X, Zhang X, Shang R, Wang Q, Li H, Liu Z. Circular RNA circRHOT1 is upregulated and promotes cell proliferation and invasion in pancreatic cancer. Epigenomics. 2019;11(1):53–63. doi: 10.2217/epi-2018-0051. [DOI] [PubMed] [Google Scholar]
- 92.Liu L, Fb L, Huang M, Xie K, Xie QS, Liu CH, Shen MJ, Huang Q. CircularRNA ciRS-7 promotes the proliferation and metastasis of pancreatic cancer by regulating miR-7-mediated EGFR/STAT3 signaling pathway. Hepatobiliary Pancreat Dis Int. 2019:ii S1499-3872(19)30039-6. doi: 10.1016/j.hbpd.2019.03.003 [DOI] [PubMed] [Google Scholar]
- 93.Liu S, Duan Y, Sun D, Chen X, He X. circZMYM2 Competed Endogenously with miR-335-5p to Regulate JMJD2C in pancreatic cancer. Cell Physiol Biochem. 2018;51(5):2224–2236. doi: 10.1159/000495868. [DOI] [PubMed] [Google Scholar]
- 94.Xu Y, Yao Y, Gao P, Cui Y. Upregulated circular RNA circ_0030235 predicts unfavorable prognosis in pancreatic ductal adenocarcinoma and facilitates cell progression by sponging miR-1253 and miR-1294. Biochem Biophys Res Commun. 2019;509(1):138–142. doi: 10.1016/j.bbrc.2018.12.088. [DOI] [PubMed] [Google Scholar]
- 95.Qu S, Song W, Yang X, Wang J, Zhang R, Zhang Z, Zhang H, Li H. Microarray expression profile of circular RNAs in human pancreatic ductal adenocarcinoma. Genom Data. 2015;5:385–387. doi: 10.1016/j.gdata.2015.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Li H, Hao X, Wang H, Liu Z, He Y, Pu M, Zhang H, Yu H, Duan J, Qu S. Circular RNA expression profile of pancreatic ductal adenocarcinoma revealed by microarray. Cell Physiol Biochem. 2016;40(6):1334–1344. doi: 10.1159/000453186. [DOI] [PubMed] [Google Scholar]
- 97.Huang WJ, Wang Y, Liu S, Yang J, Guo SX, L W, H W, Fan YF. Silencing circular RNA hsa_circ_0000977 suppresses pancreatic ductal adenocarcinoma progression by stimulating miR-874-3p and inhibiting PLK1 expression. Cancer Lett. 2018;422:70–80. doi: 10.1016/j.canlet.2018.02.014. [DOI] [PubMed] [Google Scholar]
- 98.Xu C, Yu Y, Ding F. Microarray analysis of circular RNA expression profiles associated with gemcitabine resistance in pancreatic cancer cells. Oncol Rep. 2018;40(1):395–404. doi: 10.3892/or.2018.6450. [DOI] [PubMed] [Google Scholar]
- 99.Shao F, Huang M, Meng F, Huang Q. Circular RNA signature predicts gemcitabine resistance of pancreatic ductal adenocarcinoma. Front Pharmacol. 2018;9:584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zg Y, Awan FM, Du WW, Zeng Y, Lyu J, Wu, Gupta S, Yang W, Yang BB. The circular RNA interacts with STAT3, increasing its nuclear translocation and wound repair by modulating Dnmt3a and miR-17 function. Mol Ther. 2017;25(9):2062–2074. doi: 10.1016/j.ymthe.2017.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]


