Abstract
Infections exert important evolutionary pressures shaping the human genome, especially on genes involved in host defense. A crucial step for host defense is recognition of pathogens by pattern recognition receptors on innate immune cells, among which Toll-like receptor 4 (TLR4) is one of the best known. Genetic variation in TLR4 (Asp299Gly, Thr399Ile) has been recently described. Haplotype frequencies of these polymorphisms differ among African, Asian and European populations, suggesting evolutionary pressures exerted by local infections. The TLR4 299Gly/399Ile haplotype, characteristic mainly of European populations, has relatively high frequency in the Iberian peninsula. This region is also described as refuge area during the last glacial maximum 20,000 years ago, from which repopulation of Europe took place. We speculate that a genetic bottleneck in the Iberian peninsula could have promoted the increased frequency of this haplotype by genetic drift. This hypothesis is supported by three arguments: (1) the West-East gradient of prevalence in the haplotype among European populations; (2) ancient DNA from Neolithic burials in the Iberian peninsula, dated 6,600–4,500 years before present, confirmed the relatively high frequency of this haplotype in the region, and (3) no functional differences between this haplotype and wild-type TLR4 have been found. In contrast, the disappearance of the 299Gly/399Thr haplotype in Europe is most likely due to negative selection due to sepsis. In conclusion, differences in distribution of TLR4 polymorphisms Asp299Gly and Thr399Ile in European populations are most likely due to a combination of population migration events combined with selection due to sepsis.
Key Words: TLR4, Europe, Sepsis, Neolithic
Introduction
Genes encoding proteins important for host defense against infections are constantly exposed to pressures from infectious diseases, suggesting that pathogenic microorganisms are one of the most important evolutionary selective forces shaping our genome during human history [1, 2]. Selection among immunity genes, both positive and balancing natural selection, is relatively more commonly found compared to other gene classes [1]. The first step of the host response during infection is represented by the recognition of conserved structures of bacteria, viruses, fungi and protozoa by pattern recognition receptors (PRRs) [3]. Several classes of PRRs have been described, among which the most studied are the Toll-like receptors (TLRs) [4]. The central role of TLRs for the host defense against infections is suggested by their largely unchanged repertoire in all animal phyla (consisting of six major TLR classes) [5], and by their slow rate of evolution, most likely due to selection for conservation of function [5, 6]. In addition to these phylogenetic data, evidence has been provided that in Homo sapiens, host defense genes, and among them PRRs, are associated with recent selection events [7, 8].
TLR4, encoded by the gene with the same name located on chromosome 9, is the main receptor for the lipopolysaccharide component of Gram-negative bacteria. It also recognizes other pathogen-associated molecular patterns from mycobacteria, fungi, viruses and protozoa such as malaria [9, 10, 11, 12, 13]. More than 35 TLR4 polymorphisms have been described, but most of them are in introns or do not lead to changes in the structure of the molecule [14]. However, two nonsynonymous single nucleotide polymorphisms (SNPs) located in the extracellular leucine-rich repeat domain of TLR4 may result in differential ligand recognition: an A/G transition at SNP rs4986790 (896A/G) that causes an Asp/Gly amino acid change at position 299 of the molecule, and a C/T transition at SNP rs4986791 (1196C/T) that causes a Thr/Ile amino acid change at position 399 (fig. 1a). These mutations affect the ligand-binding region (Asp299Gly) of TLR4 and the co-receptor binding region (Thr399Ile) of the receptor, respectively [14]. It has been suggested that these TLR4 polymorphisms have important functional consequences related to the induction of pro- and anti-inflammatory cytokine production, and that they modulate the systemic inflammatory response syndrome in septic shock [15] and influence susceptibility to Gram-negative infections [16].
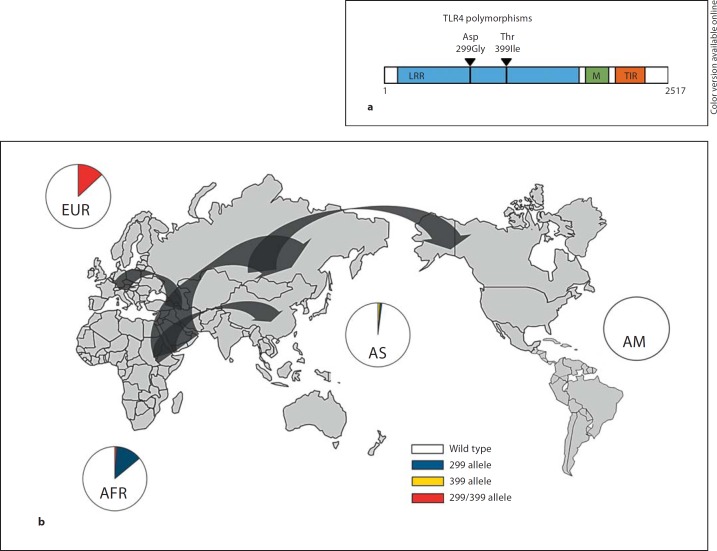
Fig. 1.
a Genomic structure of the TLR4 gene with the position of Asp299Gly and Thr399Ile indicated. LRR = Leucine-rich repeat domain; M = membrane domain; TIR = Toll/IL-1 receptor domain. b World distribution of the TLR4 haplotypes in human populations. Circles indicate average values of heterozygous and homozygous genotype frequencies combined per geographical region (white, wild-type; blue, 299Gly; yellow, 399Ile; red, 299Gly/399Ile; colors in online version only). Exact frequency data are listed in online suppl. table S1. The 299Gly haplotype is concentrated in Africa, and the 299Gly/399Ile haplotype is concentrated in Europe. Arrows indicate the main out-of-Africa migration routes of modern Homo sapiens. EUR = Europe, data based on populations from Spain (n = 107), the Netherlands (n = 209), Germany (n = 632), Romania (n = 102), Greece (n = 162); AS = Asia, data based on populations from China (n = 100), Indonesia (n = 98) and Papua New Guinea (n = 49); AM = indigenous American populations, data based on a population from Surinam (n = 99); AFR = Africa, data based on populations from Mali (n = 484), Cameroon (n = 142), Tanzania (n = 121), Sudan (n = 101). Adapted from Ferwerda et al. [19].
Due to the modulation of cytokine production that may ensue in individuals bearing these TLR4 polymorphisms, it has been proposed that TLR4 Asp299Gly and Thr399Ile SNPs influence susceptibility to both infectious and inflammatory diseases [15, 17], and that they may be therefore one factor explaining morbidity patterns in different populations. In the present review we present an overview of the information pertaining to the prevalence of TLR4 polymorphisms in various populations, and we propose a model explaining the distribution of TLR4 polymorphisms in modern human populations on the European continent.
Prevalence of TLR4 Polymorphisms in Different Human Populations
The prevalence of TLR4 alleles varies greatly between different populations around the world (fig. 1b; online suppl. table S1, www.karger.com/doi/10.1159/000329492). Based on the presence of Asp299Gly and Thr399Ile TLR4 polymorphisms in populations from both Africa and Europe, it is likely that these polymorphisms are ancient and occurred more than 65,000 years ago in Africa, before the migration of Homo sapiens out of Africa and the separation of the populations from these two continents [18]. Important differences have been described in the prevalence of TLR4 alleles in various populations, possibly depending on local infectious pressures and demographic history. The Asp299Gly SNP has a high prevalence in sub-Saharan Africa, and it has been proposed to have protective effects against mortality from malaria, although the precise mechanisms of how this protection is mediated is not yet known [19]. However, the TLR4 haplotype containing solely this mutation seems to have disappeared from East Asians and Americans. In contrast, Asp299Gly has been found to be present in co-segregation with Thr399Ile, and this haplotype has a high prevalence especially in European populations [19, 20].
The precise mechanisms that have led to these differences are still under investigation. On the one hand, evolutionary processes driven by local infectious pressures could be one factor shaping the genetics of TLR4 and the prevalence of the TLR4 polymorphisms. On the other hand, neutral processes such as population migration and genetic drift may also have played an important role. One finding that may support this latter scenario is that of recent genetic studies showing a low number of fixed or nearly fixed genetic differences between human populations. This pattern indicates that the selective pressures encountered by the different populations are weak enough to allow nonselective forces such as genetic bottlenecks and population history (including migration routes) to influence the genetic make-up [21]. Genetic bottlenecks are encountered in certain periods or places during history when a population has gone through a significant decrease in number of individuals, either due to unfavorable conditions (for example, climatic change and epidemics of disease), or during migration of a small population to a new unpopulated area. It is well known that population bottlenecks strongly increase genetic drift.
It is likely that both selective (infectious) pressures, demographic history and genetic drift have played an important role in the distribution of TLR4 polymorphisms on the different continents. The main events of the colonization of the world by the modern humans are represented by the exit out of Africa some 65,000 years ago, with subsequent complete colonization of Australia (50,000 years ago), Asia (40,000 years ago), Europe (35,000 years ago) and the Americas (15,000 years ago) [22]. The relatively high prevalence of the haplotype containing the Asp299Gly allele in sub-Saharan Africa suggests a possible beneficial role of this variant allele. Indeed, we have shown that individuals bearing this allele display a more efficient production of pro-inflammatory cytokines upon challenge with bacterial stimuli, and children bearing the polymorphism are protected against cerebral malaria [19]. On the other hand, the demographic history has also likely influenced the distribution of TLR4 polymorphisms: for example, indigenous populations of South America, living in hot climates for a relatively short period of time (last 10,000 years at most), originate from human populations that lived in the cold climates of North-East Asia for much longer before that, and these Asian populations lack any TLR4 polymorphisms [19]. The variant TLR4 Asp299Gly genotype that protects against malaria has been probably lost during the 25,000-year immigration to the Americas across the cold climates of North-East Asia, climates in which the TLR4 299Gly allele was likely deleterious due to increased mortality from septic shock [15] and possibly respiratory syncytial virus [23].
The Asp299Gly and Thr399Ile TLR4 Alleles Are Co-Segregated in Europe
The TLR4 diversity in populations of the European continent, as in populations elsewhere, presents a particular profile. Interestingly, all studies to date have shown a very high level of co-segregation between the Asp299Gly and Thr399Ile polymorphisms in the European population, while the haplotypes containing either of the two polymorphisms alone are very rare, if present at all [19, 20]. This is in fact not excessively surprising, if we have in mind the hierarchical temporal origin of the mutations (that is, the 399Ile mutation occurred in a 299Gly allele) and assume that each site cannot mutate twice and that recombination is limited, because the rate is low or there has not been too much time for a substantial recombination to happen. In this case, even if the recombination rate for the region is higher (6 cM/Mb) than the genome average (1 cM/Mb), the distance between both SNPs is not too long, so the effective recombination rate between the SNPs is proportional to the distance. Of course, even if the recombination rate is very low, after the passing of a substantial amount of time, recombinations can occur. The question has been posed whether the prevalence of the Asp299Gly and Thr399Ile alleles (and especially of the haplotype containing both mutations) in the Eurasian landmass is mainly influenced by genetic drift and demographic history, as previously proposed [19], or whether yet to be described selective forces are at work.
One approach to assess whether infections exerted selective pressure on the TLR4 variants is to investigate these polymorphisms in populations of different ethnic origins that for a long period of time lived in the same geographical location and under the same infectious pressure. One would expect that in case of strong infectious pressure in populations of sufficient size, the prevalence of ancient polymorphisms such as the TLR4 SNPs would become similar in the populations, irrespective of their ethnicity. We have recently performed an investigation of various polymorphisms and TLR4 haplotypes in 15 populations from Iran, a country ideal for such a study, considering its rich ethnic diversity and its key location on the routes of migration during the out-of-Africa human migration. In contrast to the homogeneity of the TLR4 polymorphisms in other populations such as those from Africa or Europe, the Iranian subpopulations display a broad heterogeneity of TLR4 Asp299Gly and Thr399Ile, with all combinations of the TLR4 SNPs possible being present in close geographical proximity [Ioana et al., in submission]. These findings suggest a weak or absent selection pressure on TLR4 polymorphisms in the Middle East, supporting the hypothesis of genetic drift as the major force driving the prevalence of TLR4 heterogeneity in Eurasian populations.
In contrast to Iran, however, European populations are much more homogenous in terms of TLR4 polymorphisms, the only variant haplotype being that in which Asp299Gly and Thr399Ile are co-segregated. Functional analysis of this haplotype did not show a major difference in the response of the variant TLR4 to pathogens, when compared with the wild-type haplotype [19, 24]. On the one hand, these data, corroborated with the lack of evolutionary pressures on TLR4 SNPs suggested in Middle Eastern populations (see above), suggest that the distribution of TLR4 polymorphisms in the European population is most likely the result of demography and genetic drift, rather than selection. On the other hand, the presence of just one variant TLR4 haplotype suggests that this distribution may be the result of a population bottleneck that promoted the increase in the prevalence of one variant haplotype in the ensuing population(s).
It is difficult at present to demonstrate that this scenario has indeed been the mechanism explaining the distribution of TLR4 polymorphisms across Europe. However, there are indirect arguments that support the hypothesis that the distribution of the TLR4 299Gly/399Ile haplotype across Europe may be the result of a population bottleneck and subsequent genetic drift occurring in the Iberian refuge during the last glacial maximum (LGM) approximately 20,000 years ago.
TLR4 299Gly/399Ile Haplotype in the LGM Iberian Refuge
Distribution of TLR4 SNP Prevalence in Various Populations across the Continent
Europe has been colonized by modern humans through several waves of migrants coming from the Middle East during the Upper Paleolithic and the Neolithic [25]. Based on genetic data on both mitochondrial and Y-chromosome DNA, the extant genetic pool of Europeans is believed to consist up to 80% of lineages from the Upper Paleolithic and only 20% from new arrivals during the Neolithic [26, 27]. In addition, a key event in the population history of Europe has been caused by the harsh climatic changes occurring during the LGM.
The early Paleolithic populations of Northern and Central Europe became extinct or retreated to the South, in several refuges in the Iberian, Italian and Balkan peninsula, a scenario supported by both archeological [28] and genetic [29] evidence (fig. 2a). After the retreat of the ice, an important population expansion from the Iberian and Balkan refuges, followed by the repopulation of Europe, has been proposed [29]. Several studies on mitochondrial DNA have confirmed the Upper Paleolithic Iberian/Franco-Cantabrian refuge as a major source for the European gene pool [30, 31]. Other authors have disputed the exclusive role of the Franco-Cantabrian refuge as an origin of the South-Western recolonization of Europe, suggesting different local expansions with low effects on continental Europe [32, 33].
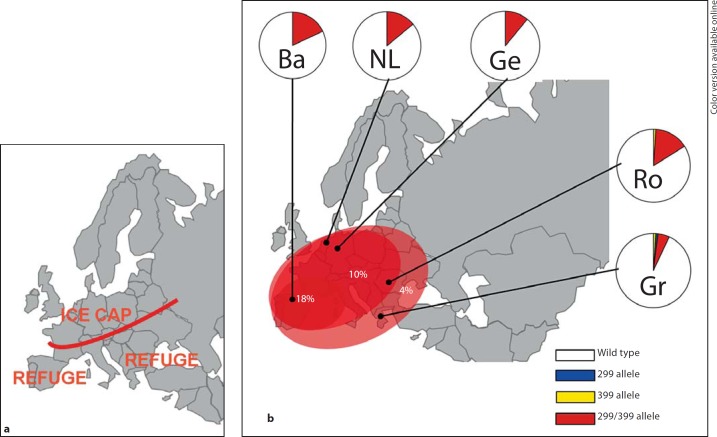
Fig. 2.
aGeographical representation of the last glacial maximum era about 20,000 years BP with indicated the position of the ice cap and the refuge areas in the Iberian, Italian and Balkan peninsula. b Distribution of the TLR4 haplotypes in European human populations at present. Circles indicate average values of heterozygous and homozygous genotype frequencies combined per geographical region (white, wild-type; blue, 299Gly; yellow, 399Ile; red, 299Gly/399Ile; colors in online version only). Exact frequency data are listed in online suppl. table S1. The 299Gly/399Ile haplotype has a frequency of 18% in the Iberian peninsula which is diluted to 10% and 4% in Central European and South-Eastern European populations, respectively. Ba = Basque, Spain (n = 107); NL = The Netherlands (n = 209); Ge = Germany (n = 632); Ro = Romania (n = 102); Gr = Greece (n = 162).
TLR4 polymorphisms have been most likely brought into Europe already in the Upper Paleolithic by the early modern human colonists. This assumption is based on the fact that the African population has a low 2–3% prevalence of the TLR4 299Gly/399Ile co-segregated haplotype, and this most likely traveled with the first wave of migration out of Africa. However, the strong increase in the prevalence in later European populations may have had two sources: on the one hand, an increase in the prevalence in the Middle East and subsequent spread in Europe, a scenario that is, however, not supported by any evidence, or a later increase in prevalence in Europe. One scenario for this latter hypothesis is that the TLR4 299Gly/399Ile haplotype increased in frequency during the LGM in one of the South-European refuges as a result of the population bottleneck. The decreasing West-East gradient of TLR4 299Gly/399Ile prevalence in European populations may support this second scenario: the high frequency of 18% of individuals bearing at least one allele of the TLR4 299Gly/399Ile haplotype in Basque Country is diluted to 10% in Central European populations and down to 4% in South-Eastern European populations in Greece (fig. 2b; online suppl. table S1) [19], although one has to be careful with clines observed for alleles with relatively low frequencies [34]. This raises the possibility that the prevalence of the variant TLR4 haplotype increased in the LGM Iberian refuge, from which it spread throughout Europe during the recolonization of the continent, similar to the spread of several major lineages of mitochondrial DNA such as haplogroup V and H [28, 30].
TLR4 Polymorphism in Ancient DNA Samples from the Iberian Peninsula
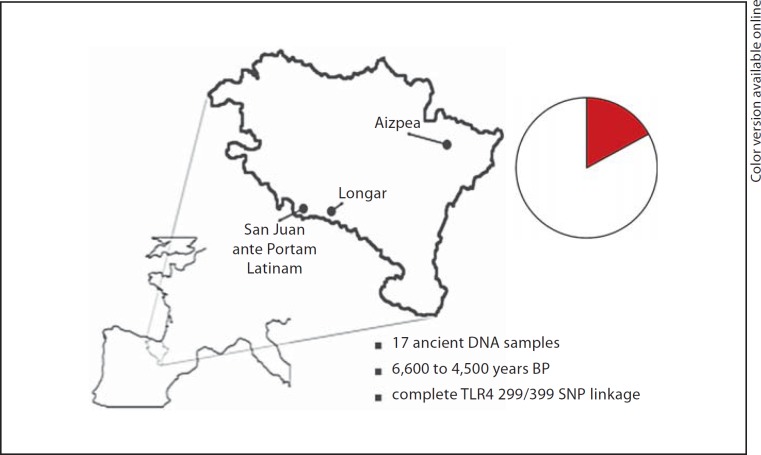
In order to provide additional arguments for this hypothesis, we have investigated the presence of TLR4 polymorphisms in ancient samples from the Iberian peninsula. We have investigated 17 prehistoric samples, 12 from the site of San Juan ante Portam Latinam, which is located in the province of Araba (Basque Country, Spain), and four additional samples that came from the site of Longar, a site located in the South of the province of Navarre (Spain) (fig. 3). 14C dating of human bone remains from these sites date the San Juan ante Portam Latinam site at approximately 5,000 years before present (BP), and that of Longar site at approximately 4,500 years BP. One additional sample corresponds to the remains of a woman from the Mesolithic site of Aizpea (Navarre, Spain), which is dated at approximately 6,600 years BP. A detailed description of the methodology and of the extensive measures of avoiding contamination with modern DNA is presented in the supplementary information.
Fig. 3.
Neolithic burial locations in the Basque Country. Circle indicates the frequency of individuals bearing the 299G/399I haplotype (red in the online version) in the Neolithic population from the Iberian peninsula.
When TLR4 polymorphisms were assessed in these ancient DNA samples, we have identified two individuals heterozygous and one individual homozygous for the 299Gly/399Ile TLR4 haplotype, corresponding to 17% of individuals bearing at least one allele of the 299Gly/399Ile haplotype, very close to the prevalence of the polymorphism in the modern Basque population. In contrast, none of these ancient DNA samples showed the presence of either Asp299Gly or Thr399Ile SNPs alone. One has to concede that no definitive arguments can be provided that the DNA samples analyzed here mirror exactly the Upper Paleolithic populations of the Iberian refuge. However, this presumption is likely, because the samples analyzed here are from the beginning of the Neolithic period when agriculture had just been brought to the Iberian peninsula, and the Basque population is known to have the lowest prevalence of Neolithic genetic lineages [29]. The high prevalence of the co-segregated TLR4 299Gly/399Ile haplotype in ancient DNA from the Iberian peninsula, especially reflected by the identification of an individual homozygous for this haplotype, supports the hypothesis of a population bottleneck in the Iberian isolated populations during the LGM as the source of the high prevalence of this TLR4 haplotype in Western European populations.
Conclusions
The overview data presented here show the variable distribution of TLR4 polymorphisms among populations from the various continents. While there is little doubt that PRRs have evolved under selection driven by infections, the prevalence of polymorphisms in these receptors, such as the Asp299Gly and Thr399Ile TLR4 polymorphisms, depends on both their functional consequences for the function of the molecule, and on the history of the populations in which they are tested. In case of some TLR4 haplotypes, selection due to infectious pressures seems to have played an important role (the presence of Asp299Gly SNP in sub-Saharan Africa due to protection from malaria), while in Europe the distribution of TLR4 polymorphisms seems to have been shaped mainly by population migration and genetic drift. One plausible explanation for the high prevalence of the TLR4 299Gly/399Ile haplotype in European populations might be the genetic drift occurring in the isolated Upper Paleolithic populations in the Iberian refuge during the last LGM.
Are there consequences of these differences in TLR4 polymorphisms for the modern human populations? Although TLR4 has evolved as a pathogen receptor, a strong body of evidence accumulated during the past decade which supports its involvement in inflammatory and autoimmune processes as well. TLR4 can bind endogenous ligands (such as heat-shock proteins) that function as danger-associated molecular patterns and can thus initiate sterile inflammation [35]. In this manner, the host defense mechanisms that have beneficial effects during infection can be hijacked to exert deleterious effects of exaggerated inflammatory or autoimmune reactions [36]. In this respect, it has been recently shown that TLR4 polymorphisms can increase the severity of atopy in patients with asthma [37, 38], or susceptibility to rheumatoid arthritis [17] in European populations, and it can be hypothesized that similar processes are important for other autoimmune diseases as well. Thus, evolutionary processes that occurred long ago during the evolutionary history of Homo sapiens still influence the susceptibility to disease in modern human populations.
Disclosure Statement
None of the authors declare any conflict of interest regarding this paper.
Supplementary Material
Supplementary data
Acknowledgements
This work was partly supported by a Vici grant of the Netherlands Organization for Scientific Research (to M.G.N.), and by grants UPV05/150 (University of the Basque Country), GIC07/43 (Basque Government), GCL-2007-65515 (Spanish Ministry of Science and Innovation), IT453-07-GIG07/43 and IT542-10-CIG100/46 (Basque Government to Research groups of the Basque University System) to C.R. M.I. was supported by the Sectoral Operational Programme Human Resources Development (SOP HRD), financed from the European Social Fund and by the Romanian Government under the contract number POSDRU/89/1.5/S/64109.
References
- 1.Barreiro LB, Quintana-Murci L. From evolutionary genetics to human immunology: how selection shapes host defence genes. Nat Rev Genet. 2010;11:17–30. doi: 10.1038/nrg2698. [DOI] [PubMed] [Google Scholar]
- 2.Ferrer-Admetlla A, Bosch E, Sikora M, Marques-Bonet T, Ramirez-Soriano A, Muntasell A, Navarro A, Lazarus R, Calafell F, Bertranpetit J, Casals F. Balancing selection is the main force shaping the evolution of innate immunity genes. J Immunol. 2008;181:1315–1322. doi: 10.4049/jimmunol.181.2.1315. [DOI] [PubMed] [Google Scholar]
- 3.Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 4.Akira S, Takeda K, Kaisho T. Toll-like receptors: critical proteins linking innate and acquired immunity. Nat Immunol. 2001;2:675–680. doi: 10.1038/90609. [DOI] [PubMed] [Google Scholar]
- 5.Roach JC, Glusman G, Rowen L, Kaur A, Purcell MK, Smith KD, Hood LE, Aderem A. The evolution of vertebrate Toll-like receptors. Proc Natl Acad Sci USA. 2005;102:9577–9582. doi: 10.1073/pnas.0502272102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leulier F, Lemaitre B. Toll-like receptors – taking an evolutionary approach. Nat Rev Genet. 2008;9:165–178. doi: 10.1038/nrg2303. [DOI] [PubMed] [Google Scholar]
- 7.Barreiro LB, Ben-Ali M, Quach H, Laval G, Patin E, Pickrell JK, Bouchier C, Tichit M, Neyrolles O, Gicquel B, Kidd JR, Kidd KK, Alcais A, Ragimbeau J, Pellegrini S, Abel L, Casanova JL, Quintana-Murci L. Evolutionary dynamics of human Toll-like receptors and their different contributions to host defense. PLoS Genet. 2009;5:e1000562. doi: 10.1371/journal.pgen.1000562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sabeti PC, Schaffner SF, Fry B, Lohmueller J, Varilly P, Shamovsky O, Palma A, Mikkelsen TS, Altshuler D, Lander ES. Positive natural selection in the human lineage. Science. 2006;312:1614–1620. doi: 10.1126/science.1124309. [DOI] [PubMed] [Google Scholar]
- 9.Krishnegowda G, Hajjar AM, Zhu J, Douglass EJ, Uematsu S, Akira S, Woods AS, Gowda DC. Induction of proinflammatory responses in macrophages by the glycosylphosphatidylinositols of Plasmodium falciparum: cell signaling receptors, glycosylphosphatidylinositol (GPI) structural requirement, and regulation of GPI activity. J Biol Chem. 2005;280:8606–8616. doi: 10.1074/jbc.M413541200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kurt-Jones EA, Popova L, Kwinn L, Haynes LM, Jones LP, Tripp RA, Walsh EE, Freeman MW, Golenbock DT, Anderson LJ, Finberg RW. Pattern recognition receptors TLR4 and CD14 mediate response to respiratory syncytial virus. Nat Immunol. 2000;1:398–401. doi: 10.1038/80833. [DOI] [PubMed] [Google Scholar]
- 11.Lien E, Means TK, Heine H, Yoshimura A, Kusumoto S, Fukase K, Fenton MJ, Oikawa M, Qureshi N, Monks B, Finberg RW, Ingalls RR, Golenbock DT. Toll-like receptor 4 imparts ligand-specific recognition of bacterial lipopolysaccharide. J Clin Invest. 2000;105:497–504. doi: 10.1172/JCI8541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Malley R, Henneke P, Morse SC, Cieslewicz MJ, Lipsitch M, Thompson CM, Kurt-Jones E, Paton JC, Wessels MR, Golenbock DT. Recognition of pneumolysin by Toll-like receptor 4 confers resistance to pneumococcal infection. Proc Natl Acad Sci U S A. 2003;100:1966–1971. doi: 10.1073/pnas.0435928100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weindl G, Naglik JR, Kaesler S, Biedermann T, Hube B, Korting HC, Schaller M. Human epithelial cells establish direct antifungal defense through TLR4-mediated signaling. J Clin Invest. 2007;117:3664–3672. doi: 10.1172/JCI28115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smirnova I, Hamblin MT, McBride C, Beutler B, Di RA. Excess of rare amino acid polymorphisms in the Toll-like receptor 4 in humans. Genetics. 2001;158:1657–1664. doi: 10.1093/genetics/158.4.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lorenz E, Mira JP, Frees KL, Schwartz DA. Relevance of mutations in the TLR4 receptor in patients with Gram-negative septic shock. Arch Intern Med. 2002;162:1028–1032. doi: 10.1001/archinte.162.9.1028. [DOI] [PubMed] [Google Scholar]
- 16.Agnese DM, Calvano JE, Hahm SJ, Coyle SM, Corbett SA, Calvano SE, Lowry SF. Human toll-like receptor 4 mutations but not CD14 polymorphisms are associated with an increased risk of Gram-negative infections. J Infect Dis. 2002;186:1522–1525. doi: 10.1086/344893. [DOI] [PubMed] [Google Scholar]
- 17.Radstake TR, Franke B, Hanssen S, Netea MG, Welsing P, Barrera P, Joosten LA, van Riel PL, van den Berg WB. The Toll-like receptor 4 Asp299Gly functional variant is associated with decreased rheumatoid arthritis disease susceptibility but does not influence disease severity and/or outcome. Arthritis Rheum. 2004;50:999–1001. doi: 10.1002/art.20114. [DOI] [PubMed] [Google Scholar]
- 18.Chiaroni J, Underhill PA, Cavalli-Sforza LL. Y chromosome diversity, human expansion, drift, and cultural evolution. Proc Natl Acad Sci USA. 2009;106:20174–20179. doi: 10.1073/pnas.0910803106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferwerda B, McCall MB, Alonso S, Giamarellos-Bourboulis EJ, Mouktaroudi M, Izagirre N, Syafruddin D, Kibiki G, Cristea T, Hijmans A, Hamann L, Israel S, ElGhazali G, Troye-Blomberg M, Kumpf O, Maiga B, Dolo A, Doumbo O, Hermsen CC, Stalenhoef AF, van CR, Brunner HG, Oh DY, Schumann RR, de la RC, Sauerwein R, Kullberg BJ, van d, Netea MG. TLR4 polymorphisms, infectious diseases, and evolutionary pressure during migration of modern humans. Proc Natl Acad Sci USA. 2007;104:16645–16650. doi: 10.1073/pnas.0704828104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ferwerda B, McCall MB, Verheijen K, Kullberg BJ, van d, Netea MG. Functional consequences of Toll-like receptor 4 polymorphisms. Mol Med. 2008;14:346–352. doi: 10.2119/2007-00135.Ferwerda. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coop G, Pickrell JK, Novembre J, Kudaravalli S, Li J, Absher D, Myers RM, Cavalli-Sforza LL, Feldman MW, Pritchard JK. The role of geography in human adaptation. PLoS Genet. 2009;5:e1000500. doi: 10.1371/journal.pgen.1000500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li JZ, Absher DM, Tang H, Southwick AM, Casto AM, Ramachandran S, Cann HM, Barsh GS, Feldman M, Cavalli-Sforza LL, Myers RM. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- 23.Awomoyi AA, Rallabhandi P, Pollin TI, Lorenz E, Sztein MB, Boukhvalova MS, Hemming VG, Blanco JC, Vogel SN. Association of TLR4 polymorphisms with symptomatic respiratory syncytial virus infection in high-risk infants and young children. J Immunol. 2007;179:3171–3177. doi: 10.4049/jimmunol.179.5.3171. [DOI] [PubMed] [Google Scholar]
- 24.Erridge C, Stewart J, Poxton IR. Monocytes heterozygous for the Asp299Gly and Thr399Ile mutations in the Toll-like receptor 4 gene show no deficit in lipopolysaccharide signalling. J Exp Med. 2003;197:1787–1791. doi: 10.1084/jem.20022078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Richards M, Macaulay V, Hickey E, Vega E, Sykes B, Guida V, Rengo C, Sellitto D, Cruciani F, Kivisild T, Villems R, Thomas M, Rychkov S, Rychkov O, Rychkov Y, Golge M, Dimitrov D, Hill E, Bradley D, Romano V, Cali F, Vona G, Demaine A, Papiha S, Triantaphyllidis C, Stefanescu G, Hatina J, Belledi M, Di RA, Novelletto A, Oppenheim A, Norby S, Al-Zaheri N, Santachiara-Benerecetti S, Scozari R, Torroni A, Bandelt HJ. Tracing European founder lineages in the Near Eastern mtDNA pool. Am J Hum Genet. 2000;67:1251–1276. [PMC free article] [PubMed] [Google Scholar]
- 26.Richards M, Corte-Real H, Forster P, Macaulay V, Wilkinson-Herbots H, Demaine A, Papiha S, Hedges R, Bandelt HJ, Sykes B. Paleolithic and neolithic lineages in the European mitochondrial gene pool. Am J Hum Genet. 1996;59:185–203. [PMC free article] [PubMed] [Google Scholar]
- 27.Semino O, Passarino G, Oefner PJ, Lin AA, Arbuzova S, Beckman LE, De BG, Francalacci P, Kouvatsi A, Limborska S, Marcikiae M, Mika A, Mika B, Primorac D, Santachiara-Benerecetti AS, Cavalli-Sforza LL, Underhill PA. The genetic legacy of Paleolithic Homo sapiens sapiens in extant Europeans: a Y chromosome perspective. Science. 2000;290:1155–1159. doi: 10.1126/science.290.5494.1155. [DOI] [PubMed] [Google Scholar]
- 28.Achilli A, Rengo C, Magri C, Battaglia V, Olivieri A, Scozzari R, Cruciani F, Zeviani M, Briem E, Carelli V, Moral P, Dugoujon JM, Roostalu U, Loogvali EL, Kivisild T, Bandelt HJ, Richards M, Villems R, Santachiara-Benerecetti AS, Semino O, Torroni A. The molecular dissection of mtDNA haplogroup H confirms that the Franco-Cantabrian glacial refuge was a major source for the European gene pool. Am J Hum Genet. 2004;75:910–918. doi: 10.1086/425590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Soares P, Achilli A, Semino O, Davies W, Macaulay V, Bandelt HJ, Torroni A, Richards MB. The archaeogenetics of Europe. Curr Biol. 2010;20:R174–R183. doi: 10.1016/j.cub.2009.11.054. [DOI] [PubMed] [Google Scholar]
- 30.Torroni A, Bandelt HJ, Macaulay V, Richards M, Cruciani F, Rengo C, Martinez-Cabrera V, Villems R, Kivisild T, Metspalu E, Parik J, Tolk HV, Tambets K, Forster P, Karger B, Francalacci P, Rudan P, Janicijevic B, Rickards O, Savontaus ML, Huoponen K, Laitinen V, Koivumaki S, Sykes B, Hickey E, Novelletto A, Moral P, Sellitto D, Coppa A, Al-Zaheri N, Santachiara-Benerecetti AS, Semino O, Scozzari R. A signal, from human mtDNA, of postglacial recolonization in Europe. Am J Hum Genet. 2001;69:844–852. doi: 10.1086/323485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pereira L, Richards M, Goios A, Alonso A, Albarran C, Garcia O, Behar DM, Golge M, Hatina J, Al-Gazali L, Bradley DG, Macaulay V, Amorim A. High-resolution mtDNA evidence for the late-glacial resettlement of Europe from an Iberian refugium. Genome Res. 2005;15:19–24. doi: 10.1101/gr.3182305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Izagirre N, de la RC. An mtDNA analysis in ancient Basque populations: implications for haplogroup V as a marker for a major paleolithic expansion from southwestern europe. Am J Hum Genet. 1999;65:199–207. doi: 10.1086/302442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garcia O, Fregel R, Larruga JM, Alvarez V, Yurrebaso I, Cabrera VM, Gonzalez AM. Using mitochondrial DNA to test the hypothesis of a European post-glacial human recolonization from the Franco-Cantabrian refuge. Heredity. 2011;106:37–45. doi: 10.1038/hdy.2010.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Excoffier L, Foll M, Petit R. Genetic consequences of range expansions. Ann Rev Ecol Evol System. 2009;40:481–501. [Google Scholar]
- 35.Beutler B. Microbe sensing, positive feedback loops, and the pathogenesis of inflammatory diseases. Immunol Rev. 2009;227:248–263. doi: 10.1111/j.1600-065X.2008.00733.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Di RA. Population genetics models of common diseases. Curr Opin Genet Dev. 2006;16:630–636. doi: 10.1016/j.gde.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 37.Fageras BM, Hmani-Aifa M, Lindstrom A, Jenmalm MC, Mai XM, Nilsson L, Zdolsek HA, Bjorksten B, Soderkvist P, Vaarala O. A TLR4 polymorphism is associated with asthma and reduced lipopolysaccharide-induced interleukin-12(p70) responses in Swedish children. J Allergy Clin Immunol. 2004;114:561–567. doi: 10.1016/j.jaci.2004.04.050. [DOI] [PubMed] [Google Scholar]
- 38.Yang IA, Barton SJ, Rorke S, Cakebread JA, Keith TP, Clough JB, Holgate ST, Holloway JW. Toll-like receptor 4 polymorphism and severity of atopy in asthmatics. Genes Immun. 2004;5:41–45. doi: 10.1038/sj.gene.6364037. [DOI] [PubMed] [Google Scholar]
- 39.Cooper A, Poinar HN. Ancient DNA: do it right or not at all. Science. 2000;289:1139. doi: 10.1126/science.289.5482.1139b. [DOI] [PubMed] [Google Scholar]
- 40.Hofreiter M, Serre D, Poinar HN, Kuch M, Paabo S. Ancient DNA. Nat Rev Genet. 2001;2:353–359. doi: 10.1038/35072071. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data