Abstract
Paneth cell α-defensins are antimicrobial peptides involved in the control of the intestinal microbiota and immunological homeostasis. In mice, they are encoded by multiple, highly homologous genes (Defa). The transcriptional activity of ileal Defa genes was studied in response to pharmacological and genetic perturbations of the intestinal environment of C57BL/6 mice. Defa gene transcription was sensitive to oral antibiotic administration suggesting that commensal microbes regulate Defa expression. Ileal microbiota analysis showed that decreased transcription of Defa genes correlated with depletion of Lactobacillus. Defa expression was partially restored in vivo by lactobacillus administration to antibiotic-treated mice. Defa transcripts were less abundant in ex vivo, microbiota-free intestinal explants but recovered after explant exposure to UV-killed bacteria, Toll-like receptor (TLR)-2 or TLR4 agonists. Genetic deficiency of several TLRs or MyD88 led to dramatic drops in Defa transcription in vivo. These results show that Paneth cell Defa genes are regulated by commensal bacteria through TLR-MyD88 signaling and provide a further understanding of the dysregulation of intestinal homeostasis that occurs as a result of imbalances in the populations of commensal bacteria.
Key Words: Defensins, Toll-like receptors, Intestinal microbiota, Paneth cells, Antimicrobial peptides, Cryptdins
Introduction
Together with other antimicrobials, enteric α-defensins are secreted into the lumen of the mammalian small intestine [1]. They are thought to defend against pathogenic insults as suggested by in vivo studies, where gain of a human defensin 5 (HD5) in a transgenic mouse model improved protection from enteric Escherichia coli and Salmonella infections whereas deficiency of Mmp7 (the enzyme responsible for processing cryptdin precursors to their mature form) correlated with increased susceptibility to those infections [2, 3]. In addition, recent evidence indicates that enteric α-defensins participate in the control of intestinal homeostasis by modifying the commensal microbial composition and in turn, influencing the immunological landscape of the intestinal mucosa [4].
In the healthy adult gut, enteric α-defensins are produced exclusively by Paneth cells in the crypts of the small intestine [1]. In mice, enteric α-defensins, the cryptdins, are encoded by a group of highly homologous genes (Defa), located on the defensin gene cluster on chromosome 8. The repertoire of C57BL/6 mice includes more than 20 potentially active Defa genes and numerous pseudogenes [5, 6]. In addition, the C57BL/6 Defa genotype is different from that of other laboratory mouse strains; more than 10 transcriptionally active genes identified in other mouse lines have not been found in C57BL/6 and several genes are unique to this mouse strain [5, 6].
Expression analyses of Defa genes have been of limited use in understanding α-defensin biology in mouse models of enteric diseases. A major problem is the high gene sequence homology, which makes PCR primer design challenging and clouds the validity of microarray data. This fact, combined with the incomplete knowledge of the strain diversity in the Defa gene pool, has led to the generation of a significant amount of data that require reinterpretation in light of the recent C57BL/6 Defa genotyping [5, 6]. Other than an involvement of the β-catenin pathway [7, 8], very little is known about the control of intestinal α-defensin expression. Previous studies on the role of bacteria in cryptdin synthesis have produced conflicting results [9, 10, 11, 12]. Here, we studied the transcriptional activity and regulation of ileal Defa genes in adult C57BL/6 mice by qPCR. Two major, unique aspects of this work are the generation and validation of primers specific for the Defa gene repertoire of this mouse strain, and the analysis of the majority of the known genes of C57BL/6 mice. We found significant variation in the steady-state Defa gene expression, which was responsive to antibiotic-induced perturbations of the intestinal environment. Most interestingly, Defa transcriptional activity correlated to specific changes in the composition of the intestinal microbiota and was affected by genetic deficiencies in the TLR-MyD88 signaling pathway. Collectively, these findings show that Paneth cell Defa gene expression is controlled by the intestinal commensal microbiota through the TLR-MyD88 pathway.
Materials and Methods
Animals
Eight- to twelve-week-old, wild-type C57BL/6 and congenic sex- and age-matched MyD88–/–, Tlr2–/–, Tlr4–/–, Tlr2–/–/ Tlr4–/–, Tlr9–/– or Nod2–/– mice were raised in the same room in the animal facility of the Child and Family Research Institute. All knockout mouse strains were backcrossed onto a C57BL/6 genetic background between 6 and 10 times and housed in the same room as wild-type C57BL/6 for a minimum of 3 years. Animal protocols for this study were approved by the University of British Columbia's Animal Care Committee. Antibiotics were administered in the drinking water for 4 days at 450 mg/l streptomycin or 750 mg/l metronidazole. Mice were sacrificed and tissue samples of the terminal ileum were taken approximately 1 inch from the ileocecal junction for RNA purification, analysis of microbiota and microscopy.
Ileal Explants
Ileal explants were prepared essentially as described [13]. Briefly, the distal region of the ileum was resected and flushed with sterile PBS. The tissue was opened longitudinally and washed repeatedly with sterile PBS to eliminate the remaining intestinal content and immediately placed in DMEM/10% FCS/100 µg/ml streptomycin/100 U/ml penicillin. The tissues were divided into 2-mm-long sections in sterile conditions and incubated at 37°C in humidified 5% CO2 atmosphere. After 1 h, the media were replaced and the samples were incubated for an additional 2 h in the same medium. Thereafter, the explants were transferred to fresh media containing E. coli lipopolysaccharide (LPS) (Sigma), Pam2CSK4, Pam3CSK4 (InvivoGene, http://www.invivogen.com/, cat. No. tlrl-pm2s and tlrl-pms) or UV-killed Lactobacillus johnsonii, Lactobacillus reuteri or E. coli (all previously isolated from the intestinal commensal population of experimental animals) and incubated for 16 h at 37°C in a humidified 5% CO2 atmosphere.
Expression Analysis
Sections and ileal explants were collected in RNA Later (Qiagen) and total RNA was isolated using the RNeasy kit (Qiagen). cDNA was prepared from 1 µg of RNA using the Quantitech Reverse Transcription kit (Qiagen). Quantitative PCRs (qPCRs) were done on an Applied Biosystems 7500 using the DyNamo SYBR Green PCR Kit from New England Biolabs. The qPCR primers and conditions for the Defa genes are shown in table 1, the primers for the housekeeping gene 36B4 [ribosomal protein, large, P0 (Rplp0), NM 007475.5] were TCTGGAGGGTGTCCGCAAC (forward) and CTTGACCTTTTCAGTAAGTGG (reverse); the primers for matrix metalloproteinase 7 (Mmp7) were CACTCTAGGTCATGCCTTCGC (forward) and GGTGGCAGCAAACAGGAAGTTC (reverse). Efficiencies for the 36B4 PCRs were 1.9055 at 58°C, 1.9333 at 60°C and 1.8544 at 62°C. qPCR specificity was verified by cloning of PCR products using TOPO TA 4.0 vector and E. coli TOP 10 chemically competent cells (Invitrogen, Carlsbad, Calif., USA) and sequencing of 10–20 independent clones. Relative expression data were generated using the ddCt method, corrected for primer efficiencies according to Pfaffl et al. [14]. Relative expressions from intestinal explants were normalized against the relative expression of villin (expressed exclusively in the epithelial cell layer) as an indication of epithelial survival. The data were normalized for each sample using the formula Rel Exp(gene of interest)/Rel Exp(villin).
Table 1.
The gene composition of each amplification group as defined in this work
| Genes included in group | Group name | Remarks |
|---|---|---|
| Defa3 and OTTMUSG00000019892 | Defa3 | Gene duplication, identical genes and protein products |
| Defa5-related genes* | Defa5 | Four genes differing in one or two nucleotides in the pre- or pro- regions but producing identical mature proteins, identical PCR products |
| Defa20** and OTTMUSG00000019860 | Defa20 | Gene duplication, identical genes and protein products |
| Defa21 and Defa22 | Defa21/22 | Two different genes, amplified together, different PCR products |
| Defa23 and OTTMUSG00000019762 | Defa23 | Gene duplication, identical genes and protein products |
| Defa24 and OTTMUSG00000019742 | Defa24 | Two different genes, amplified together, identical PCR products |
| Defa25 and OTTMUSG00000019857 | Defa25 | TATA box-less promoters |
| Defa26 and OTTMUSG00000019784 | Defa26 | Two different genes, identical mature proteins, identical PCR products |
| Defa4 | Defa4 | Gene not present in C57BL/6 mice |
With the exception of Defa4, gene ID numbers are from the Vertebrate Genome Annotation (VEGA) database (http://vega.sanger.ac.uk/index.html) following Amid et al. [5].
The VEGA database ID numbers are Defa3 (OTTMUSG00000019782), Defa20 (OTTMUSG00000019856), Defa21 (OTTMUSG00000019489), Defa22 (OTTMUSG00000019763), Defa23 (OTTMUSG00000019488), Defa24 (OTTMUSG-00000019980), Defa25 (OTTMUSG00000019700) and Defa26 (OTTMUSG00000019889). Defa4 is NCBI NM_010039.
Defa5-related genes include OTTMUSG00000018258, OTTMUSG00000019785, OTTMUSG00000018259 and OTTMUSG-00000019924.
The names of the Defa genes as used here correspond to and substitute former designations such as Cryp (e.g. Defa3 is Cr3 or Cryp3, or Defcr3) and so on, with the exception of Defa20, which has been also called Cr4-C57B6 or Crp4 (B6a) [10, 34] and Defa21/22, which have been called Crp4 (B6b) [34].
Microbial Analysis
Total bacteria were enumerated by SYBR green DNA staining. Ileal contents were homogenized in 1 ml of PBS and then fixed in 3.7% formalin. Fixed samples were diluted and applied to 0.2 µm Anodisc 25 filters (Whatman International Ltd.), then stained with SYBR green (Invitrogen), diluted 1:400 in PBS, for 15 min. Filters were mounted on glass slides with 30 µl of antifade solution. Images were collected on an Olympus IX81 microscope from 3 randomly chosen fields for each sample and cell counts were averaged. Microbial composition was assessed in tissue samples collected from the ileum by terminal restriction fragment length polymorphism analysis (T-RFLP) and cloning and sequencing. Total DNA was isolated from ileal tissue using the QIAamp DNA mini kit (Qiagen) according to the manufacturer's instructions, with the addition of a 60-second bead-beating step at level 5 on a FastPrep-24 (MP Biomedicals, Solon, Ohio, USA). Bacterial 16S rRNA genes were PCR amplified using broad-range primers 8F [15] 5′-end-labeled with 6-FAM, and 926r [16] and subjected to digestion with HaeIII. Fragment length determination of 6-FAM-labeled products was performed on an ABI 3730 capillary sequencer (Applied Biosystems) and electrophoregrams were processed using GeneMarker (State College, Pa., USA). For the phylogenetic identification of terminal restriction fragments of interest, pooled PCR-amplified products from each treatment group were cloned into the pCR® 4 TOPO vector and transformed into E. coli TOP10 chemically competent cells (Invitrogen) and 64 clones per treatment were sequenced. Sequences were classified using the naïve Bayesian rRNA classifier in Ribosomal Database Project [17]. Selective culture of lactic acid bacteria was performed on Rogosa agar under anaerobic conditions.
Microscopy
Tissue sections were prepared and stained with hematoxylin-eosin (HE) by WaxIt Inc. (Vancouver, B.C., Canada). HE images were taken in an Olympus VS110 using the VS-ASW software. Immunofluorescent Ab localization was performed on paraffin-embedded tissue sections (5 µm) after paraffin removal with xylene and rehydration by sequential immersion in 100, 95, and 75% ethanol and water. Antigen retrieval was performed in 10 mM citric acid pH 6.0 at 100°C for 30 min. Immunostaining was performed using a rabbit anti-human lysozyme (Dako A0099, 1:600). Fluorescence was visualized using a Zeiss AxioImager 2 and processed with the AxioImager software.
Data Analysis
Data were analyzed with GraphPad Prism version 4.0 (GraphPad Software Inc., San Diego, Calif., USA). Statistical comparisons were done with Mann-Whitney A tests with 95% confidence intervals. Sequence alignment and the phylogenetic tree were done using ClustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/).
Results
Differential Expression of Defa Genes in the Terminal Ileum of C57BL/6 Mice
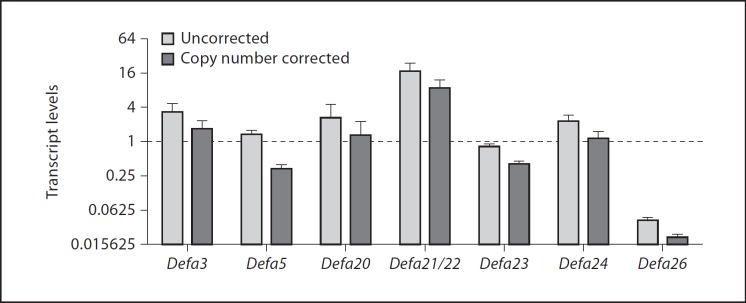
A careful analysis of the published work on cryptdin gene expression reveals that (1) frequently, primer sets designed to amplify one particular Defa gene are potentially able to cross-amplify others; this problem is often aggravated by failure to report the PCR conditions and lack of product sequence verification, (2) in vivo expression data for genes not yet found in C57BL/6 mice are often provided in studies using this mouse strain (e.g. Defa1, Defa4 and Defa10), probably as a result of the primer design problem mentioned above, (3) in most studies, only one or two Defa genes are analyzed; however, generalized conclusions about Defa expression are made. To avoid these inconsistencies, we designed a novel set of primers for the detection of relative amounts of multiple Defa transcripts by qPCR. The high homology of the Defa genes prevented the generation of individual primer pairs; instead, the genes were clustered in subgroups based on coding sequence homology and phylogenetic distance of their promoters (online suppl. fig. 1; for all online supplementary material see www.karger.com?doi=10.1159/000341630). Six subgroups were formed including two genes each, coding for identical or quasi-identical mature protein products (table 2). An additional subgroup was created with the four Defa5-related genes and another one with Defa4, a gene cloned from 129SvJ mice, which has not yet been found in C57BL/6 mice. PCR conditions for specific amplification (table 1) were carefully developed, and were validated by sequencing of cloned PCR products generated with each primer mix. Specific amplification of expected targets was obtained and verified for all primer mixes. Expression of Defa3, Defa5, Defa20, Defa21, Defa22, Defa23, Defa24 and Defa26 was confirmed in C57BL/6 mice (fig. 1) whereas transcripts for Defa25 were undetectable, consistent with the absence of a TATA box in its promoter [5]. Defa4 transcripts were not detected in C57BL/6 but were readily detected in BalbC and 129SvJ mice (online suppl. fig. 2). The results in figure 1 show that the homeostatic transcript levels of independent Defa genes are dissimilar in vivo.
Table 2.
The primers set for the C57BL/6 Defa genes qPCR
| Gene group | Primers sequences | Product length, bp | qPCR annealing, ° C |
qPCR efficiency |
|---|---|---|---|---|
| Defa3 | F: GAGAGATCTGGTATGCTATTG R: AGCAGAGTGTGTACATTAAATG |
98 | 62 | 1.956 |
| Defa5 | F: TCAAAAAAGCTGATATGCTATTG R: AGCTACAGCAGAATACGAAAG |
106 | 58 | 1.982 |
| Defa20 | F: GAGAGATCTGGTATGCTATTG R: AGAACAAAAGTCGTCCTGAG |
86 | 62 | 2.125 |
| Defa21/22 | F: GAGAGATCTGATCTGCCTTTG R: CAGCGCAAAAAAGGTCCTGC |
87 | 64 | 2.079 |
| Defa23 | F: GAGAGATCTGGTATGCTATTG R: AGCAGAGCGTGTATATTAAATG |
98 | 62 | 1.376 |
| Defa24 | F: GAGAGATCTGGTATGCTATTG R: AGCAGAGCATGTACAATAAATG |
98 | 62 | 1.556 |
| Defa25 | F: GTGAAGATCTGATATGCTATTG R: ACCAGAGCATGTACATTAAATG |
99 | 58 | 1.980 |
| Defa26 | F: ATTGTAGAAAAAGAGGCTGTAC R: AGCAGAGTGTGTACATTAAATG |
81 | 62 | 1.802 |
| Defa4* | F: TGAGAGGTTTGTTATGCTATTG R: AGCAGTACAAAAATCGTATTCC |
90 | 60 | 1.992 |
Reaction cycling conditions were: one enzyme activation step at 95°C for 10 min followed by 40 cycles of 95° C, 30 s, annealing for 30 s at the specified temperature and extension at 72° C for another 30 s. Fluorescence readings were taken during the extension time.
Not present in C57BL/6 mice.
Fig. 1.
Paneth cell Defa genes are not equally expressed. Ileal transcript levels of Defa genes in C57BL/6 mice, relative to the transcript levels of the Rplp0 gene (defined as 1, dashed line). Open bars represent the overall relative expression for each subgroup. Solid bars represent relative expression of each individual gene within a subgroup (this was arbitrarily calculated by dividing the overall relative expression of the subgroup by the number of genes within the subgroup; this correction assumes equal expression between the members of each independent subgroup; 2 genes for Defa3, 20, 21/22, 23, 24 and 26, and 4 genes within the Defa5 group). Data shown are medians and ranges from the average of two qPCRs; n = 5 animals per group.
Oral Antibiotic Treatments Modify the Defa Expression Pattern
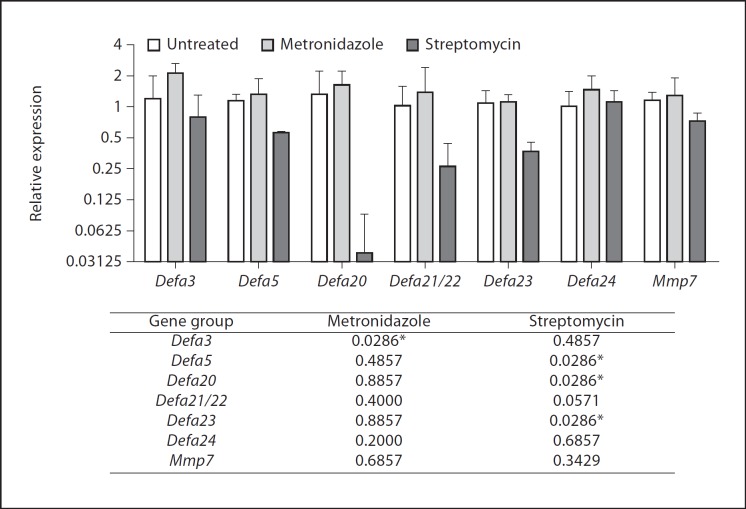
Clinical use of antibiotics is frequently associated with the expansion of opportunistic pathogens, such as vancomycin-resistant enterococcus [18] and Clostridium difficile[19]. This is thought to result from alterations of the commensal intestinal microbiota and perturbations of mucosal immune function such as disrupted mucus layer and imbalances in immune cell populations [reviewed in [20]]. To determine the susceptibility of Defa expression to antibiotic treatment, independent groups of mice from the same supplier (Jackson) and same housing facility were administered streptomycin or metronidazole, which are known to target distinct bacterial groups (aerobes and anaerobes, respectively). Oral streptomycin treatment for 4 days resulted in decreased expression of several Defa genes (fig. 2) whereas metronidazole induced moderate expression increases that reached statistical significance only for Defa3 (fig. 2). Expression of Mmp7 (which in intestinal epithelial cells is stimulated by bacteria [21]) was unaffected in response to antibiotic administration (fig. 2). Interruption of streptomycin treatment led to a gradual increase in the Defa gene transcripts that were the most clearly affected by treatment (online suppl. fig. 3).
Fig. 2.
Transcript levels of Defa and Mmp7 genes in the ileum of antibiotic-treated mice, relative to untreated animals. The graph shows medians and ranges, the table shows the Mann-Whitney test p values with respect to the untreated controls; * p < 0.05: significant differences. Results shown are one representative experiment of three; n = 4 females per group.
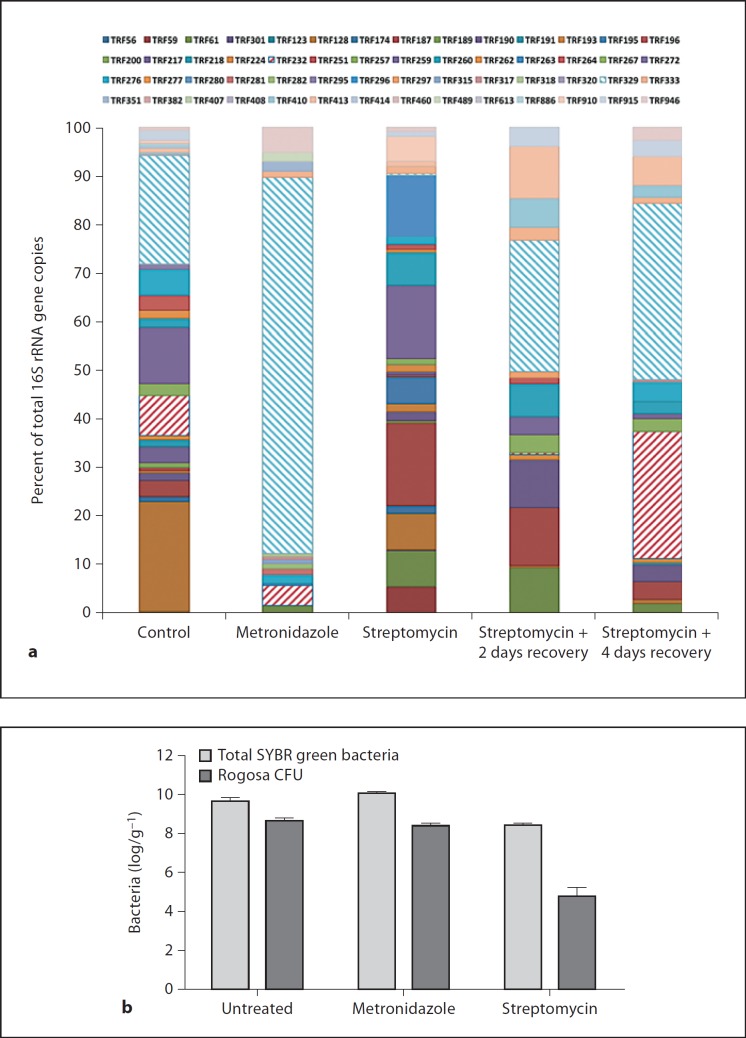
To identify changes in bacterial groups that correlated with changes in Defa expression, the ileal microbial populations of untreated and antibiotic-treated mice were quantified and characterized by T-RFLP and cloning/sequencing of 16S rRNA genes (fig. 3a). Streptomycin reduced total luminal bacteria in the ileum (fig. 3b) whereas metronidazole depleted specific bacterial populations. Based on the respective effects of these antibiotics on Defa expression, we aimed to identify candidate microbes that were depleted by streptomycin, unaffected by metronidazole and that recovered after streptomycin removal. Only two TRFs, TRF232 and TRF329, fit these criteria. Clone libraries revealed that TRF232 represents a yet uncultured Turicibacter (accession No. EF406422) whereas TRF329 represents members of the genus Lactobacillus. The clone library generated from metronidazole-treated mice was nearly exclusively represented by sequences >99% identical to Lactobacillus johnsonii and Lactobacillus reuteri. Changes in the lactobacillus populations of streptomycin-treated mice were confirmed by plating/isolation on Rogosa agar (fig. 3b) and identification by sequencing of the 16S rRNA gene. These results show that the presence of lactobacilli associates with homeostatic Defa transcriptional activity whereas lactobacillus depletion by antibiotic treatment correlates with a drop in ileal Defa gene expression.
Fig. 3.
Changes in microbial populations associated with antibiotic treatment. a Average HaeIII-digested T-RFLP profiles of ileal 16S rRNA gene populations of mice untreated, treated with metronidazole or streptomycin, and 2 or 4 days after suspension of streptomycin treatment. Each bacterial phylotype/TRF peak is represented by a different color; peaks of major interest are marked by hatched bars (TRF232 red and TRF329 blue). * p < 0.05: significant differences. b Total (SYBR staining) and culturable (Rogosa agar) bacteria in ileal contents, data are means ± SE.
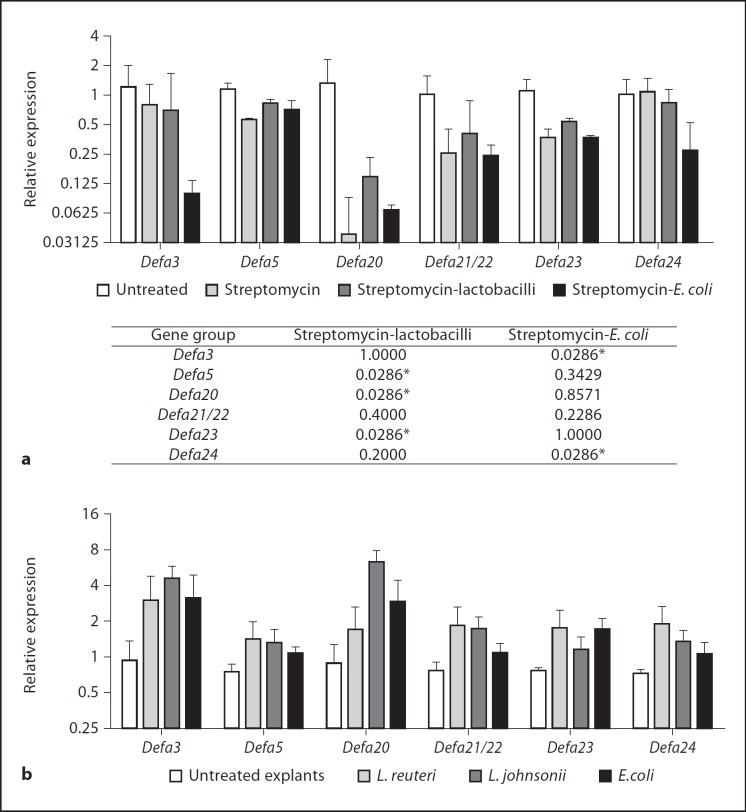
The contribution of lactobacilli to the regulation of Defa expression was confirmed by introducing heat-killed L. johnsonii and L. reuteri to the drinking water of streptomycin-treated mice. The introduction of 108 lactobacilli resulted in a partial rebound in the expression of Defa5, Defa20 and Defa23 whereas a commensal heat-killed E. coli did not have the same effect (fig. 4a). Intriguingly, the expression of Defa3 and Defa24, which reacted rather poorly to streptomycin administration, was significantly reduced by the administration of the heat-killed commensal E. coli. Since the in vivo antibiotic treatments did not completely eliminate the ileal microbiota, we decided to test the effect of lactobacilli and E. coli in cultured intestinal explants. In spite of the limitations inherent in tissue explant culture (such as progressive dying), ileal explants are efficiently devoid of commensals and therefore useful to study their effects on epithelial function. The explants were exposed to UV-killed bacteria for 16 h and Defa expression measured by qPCR. As shown in figure 4b, exposure to bacteria induced the transcription of Defa genes, demonstrating that in this model, bacterial stimulation participates in the control of Defa gene expression.
Fig. 4.
a Oral administration of heat-killed commensal bacteria to streptomycin-treated wild-type C57BL/6 mice partially restores ileal Defa expression in vivo; the data shown are Defa transcript levels relative to untreated animals. The table shows the Mann-Whitney test p values with respect to the streptomycin-treated animals; * p < 0.05: significant differences. b Exposure to commensal bacteria partially restores Defa expression in ileal explants from wild-type C57BL/6 mice. Results shown are relative Defa expressions normalized to relative villin expression for each sample (RelExp Defa/RelExp villin), as a correction for epithelial survival of the individual explants. The graphs show medians and ranges. Results shown are one representative experiment of two; n = 4.
Homeostatic Defa Expression Requires TLR-MyD88 Signaling
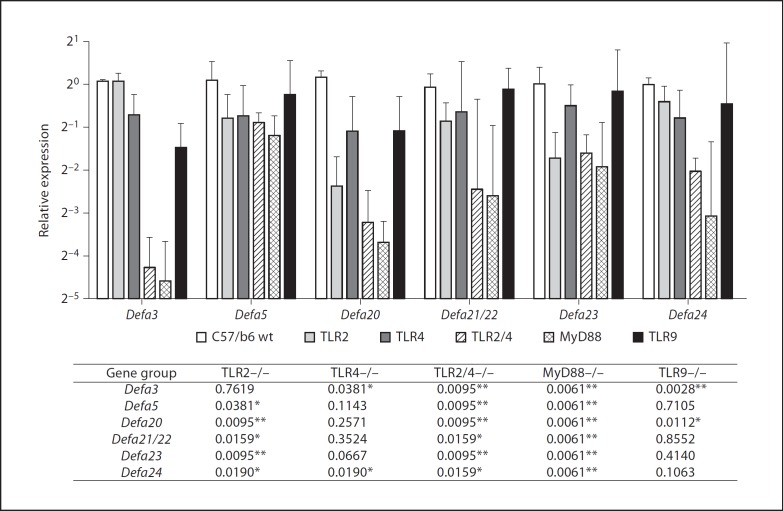
Mice with genetic deficiencies in bacterial sensing (Tlr2–/–, Tlr4–/–, Tlr9–/–, Tlr2/4–/– or MyD88–/–) and C57BL/6 wild-type controls were tested for ileal transcript levels of Defa (all the animals came from colonies housed at the CFRI). As shown in figure 5, mild to significant reductions in expression were observed in Tlr2–/– mice, most notably for Defa20 and Defa23. Tlr4–/– mice showed a more moderate impact, with significant changes only for Defa3 and Defa24 although reductions were also observed for other genes. Defa3 and Defa20 transcription was affected in Tlr9–/– animals, but the rest of the genes were insensitive to TLR9 deficiency. However, the Tlr2–/–/Tlr4–/– double-deficient mice displayed very dramatic reductions in transcript levels for most genes. Deficiency of the downstream adaptor molecule MyD88 also resulted in marked decreases in Defa transcription (fig. 5), in a manner not associated with either gross histological alterations of the intestinal epithelium, reduced number of Paneth cells or reduced levels of lysozyme transcripts (online suppl. fig. 4). Moreover, the expression of Defa20 was measured in a small group of litermates, co-housed MyD88–/– (n = 3) and MyD88–/+ heterozygous (n = 2) mice derived from heterozygous parents to ensure that differences in expression were not a result of separate housing. The observed differences in transcript levels were consistent with mice housed independently, suggesting that disparities in Defa expression are associated with genotype rather than environment (not shown). In general, transcript levels of Defa in MyD88–/– mice were very similar to those of the Tlr2–/–/ Tlr4–/– double knockouts, indicating that the TLR-MyD88 pathway is important for homeostatic expression of Defa genes (fig. 5).
Fig. 5.
TLR2, TLR4 and MyD88 deficiencies cause a drop in the ileal transcript levels of Defa genes. The graph shows medians and ranges; the table shows the Mann-Whitney test p values with respect to the C57BL/6 wild-type (wt) animals; * p < 0.05 and ** p < 0.01: significant differences. Results shown are the average of two qPCRs, n = 6–9 males per group.
In contrast to TLR deficiency, lack of NOD2 had no significant effect on Defa expression, as compared to sex- and age-matched C57BL/6 wild-type controls from the same supplier and housed in the same facility (online suppl. fig. 5). Different from Defa genes, Reg3β and Reg3γ expression was insensitive to TLR deficiencies although it was decreased in MyD88–/– mice (online suppl. fig. 6). Our results suggest that TLR2- and TLR4-mediated signaling is a fundamental mechanism for the maintenance of homeostatic expression of Defa genes in the intestinal epithelium of adult mice.
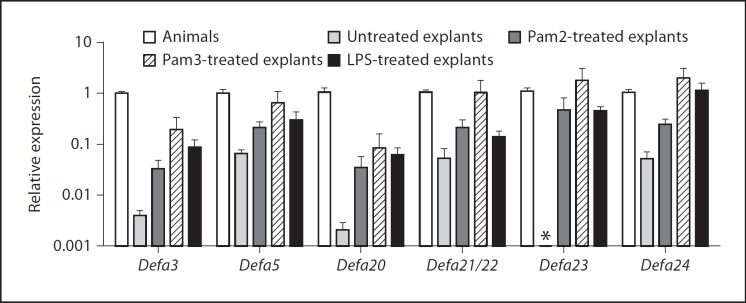
The involvement of TLR2 and TLR4 signaling in Defa expression was verified ex vivo using ileal explants. Ileal sections from C57BL/6 mice were treated in culture with 10 ng/ml of Pam2CSK4 (a Tlr 2/6 heterodimer agonist), 100 ng/ml of Pam3CSK4 (a Tlr 2/1 heterodimer agonist) or 10 ng/ml of E. coli LPS. The results in figure 6 show significant Defa expression drops in untreated, cultured ileal explants with respect to the in vivo levels, which was predicted based on the proposed role of bacteria in Defa regulation. Expression increased 10–1,000-fold after treatment of the explants with TLR2 and TLR4 agonists. Defa20 and Defa23, the genes most sensitive to TLR2 deficiency in vivo, were also the most sensitive to the ex vivo culture conditions (Defa23 transcription became undetectable in untreated explants). In general, stimulation with Pam3CSK4 (the agonist of the TLR2/1 heterodimer) produced the highest increases in Defa expression.
Fig. 6.
Treatment with TLR2 and TLR4 agonists (Pam2CSK4, 10 ng/ml; Pam3CSK4; 100 ng/ml and LPS, 10 ng/ml) restores Defa expression in ex vivo ileal explants from wild-type C57BL/6 mice. Results shown are relative Defa expressions normalized to relative villin expression for each sample (Rel Exp Defa/Rel Exp villin), as a correction for epithelial survival of the individual explants. The asterisk marks the lack of detectable Defa23 transcripts in the untreated explants. Results shown are one representative experiment of two; n = 4 or 5 explants per group. Pam2CSK4 and Pam3CSK4 are synthetic diacylated and triacylated lipopeptide agonists of the TLR2/6 and 2/1 heterodimers, respectively.
Discussion
Paneth cell α-defensins are a major type of enteric antimicrobials. Reduced abundance or loss of function has been associated with Crohn's disease [22, 23], susceptibility to enteric pathogens [2, 3], changes in composition of microbiota and disruption of intestinal immune homeostasis [4]. Their production has been typically considered invariable and independent of bacterial stimulation [24]; however, we observed that transcript levels of several Defa genes are sensitive to antibiotic treatment and correlate with changes in the intestinal microbial composition. Our results strongly associate Lactobacillus with maximal Paneth cell α-defensin expression and show that the commensal population plays a significant role in driving ileal Defa expression. Although it was not tested here, it is quite plausible that other Gram-positive bacteria are also able to induce expression. Observations in clinical specimens from patients with Crohn's disease support the role of commensal bacteria in regulating α-defensin expression since both, alterations in the intestinal microbial composition (including the depletion of lactobacilli) and reduction of α-defensin levels are observed in patients with Crohn's disease [25, 26].
Antibiotic treatments often lead to secondary clinical intestinal complications [18, 20]; it is possible that those arise due (at least in part) to alterations of α-defensin synthesis profile. There is evidence that Paneth cell α-defensins play a role in modulating the intestinal mucosal immune response directly [27, 28] and indirectly [4], and have been shown to inhibit the toxin B of C. difficile[29], a common opportunistic pathogen associated with antibiotic therapy [19]. Thus, reductions in Defa expression have the potential to impact the immunological tone of the gut in a significant way.
In contradiction with previous reports [12], we failed to observe any defect of Defa expression in NOD2-deficient mice. In contrast, the TLR-MyD88 pathway seems important for maximal Defa expression. Out of the TLRs tested, TLR2 was the most critical, being important for expression of most gene subgroups. This is in contrast with the work of Dessein et al. [30], who reported that TLR2 and MyD88 deficiency had no impact on the expression of cryptdins. However, they studied Defa1 and Defa4, two genes that are absent in C57BL/6 mice. In our study, most Defa genes displayed milder expression reductions in single Tlr2–/– or Tlr4–/– mice than in the double Tlr2–/–/Tlr4–/– or the MyD88–/– strains, suggesting a compensatory action between these two TLRs. This is also supported by the responses of the intestinal explants to both TLR2 and TLR4 agonists.
Activation of the β-catenin pathway is so far the only mechanism known to regulate Defa gene expression [7, 8]. Our results demonstrate a role of MyD88 for cryptdins expression in the adult mouse intestine, as previously proposed [31] and provide clear evidence of the involvement of at least TLR2 and TLR4. This expands the known function of MyD88 in driving the expression of Paneth cell antimicrobials [32] and highlights the critical role of the TLR-MyD88 pathway in intestinal homeostasis. The normal expression of lysozyme in TLR- and MyD88-deficient mice suggests that the observed defects in Defa expression are not the result of abnormal Paneth cell development.
It is still unclear how the signaling pathway relevant for Defa expression proceeds downstream of MyD88. We observed that Defa expression was insensitive to NFĸBp50 null deficiency in vivo (not shown) but considering the functional compensation between the Rel family members [33], these results are not conclusive. It should be noted that all the knockout mice tested here are lacking their specified genes in their germ line and are not cell-specific deficient mice. Thus, the dramatic impact of MyD88 deficiency may not reflect the loss of MyD88 signaling within the Paneth cells, but it may instead reflect indirect actions, i.e. the involvement of MyD88 within other cell types, such as macrophages or myofibroblasts resulting in the release of factors that regulate defensin expression by Paneth cells.
In summary, we have presented here novel observations on ileal Defa gene expression in C57BL/6 mice. Future studies are necessary to establish how the observed differences in transcript levels translate into differences in luminal concentrations of active α-defensins and/or intestinal antibacterial activity. We have shown that intestinal commensals are directly linked with α-defensin expression through the TLR-MyD88 pathway. It would be interesting to assess whether the synthesis of a related group of antimicrobial peptides, the cryptdin-related sequences, is subject to similar regulation. As alterations of the intestinal microbial flora and homeostasis are associated with an increasing number of pathological conditions (e.g. inflammatory bowel disease, infection and cancer) our findings bear relevance to the understanding of the molecular mechanisms underlying such conditions.
Supplementary Material
Supplemental Video
Acknowledgements
We would like to thank Rémi Gaudreau for assistance with bright-field microscopy. This work was supported by operating grants to B.B.F. from the Canadian Institutes of Health Research (CIHR) and the Crohn's and Colitis Foundation of Canada (CCFC) to B.B.F. and B.A.V. B.B.F. is the UBC Peter Wall Distinguished Professor and B.A.V. is the Canada Research Chair in Pediatric Gastroenterology (Tier 2) and the Children with Intestinal and Liver Disorders (CHILD) Foundation Research Chair in Pediatric IBD Research. A.M. was a recipient of postdoctoral fellowships from the Michael Smith Foundation for Health Research/Genome BC, and the Natural Sciences and Engineering Research Council of Canada. B.P.W. is a recipient of a fellowship from CIHR in partnership with the Institute of Nutrition, Metabolism and Diabetes; the CIHR Strategy on Patient-Oriented Research; and the Canadian Association of Gastroenterology and its partners, the Crohn's and Colitis Foundation of Canada, Abbott, Ferring Pharmaceuticals, and Merck. M.M. is a recipient of postdoctoral fellowships from the Canadian Association of Gastroenterology/CIHR/CCFC and the Child and Family Research Institute. Present address of A.M.: Department of Microbiology and Infectious Diseases, University of Sherbrooke, Sherbrooke, Qué., Canada.
References
- 1.Porter EM, Bevins CL, Ghosh D, Ganz T. The multifaceted Paneth cell. Cell Mol Life Sci. 2002;59:156–170. doi: 10.1007/s00018-002-8412-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Salzman NH, Ghosh D, Huttner KM, Paterson Y, Bevins CL. Protection against enteric salmonellosis in transgenic mice expressing a human intestinal defensin. Nature. 2003;422:522–526. doi: 10.1038/nature01520. [DOI] [PubMed] [Google Scholar]
- 3.Wilson CL, Ouellette AJ, Satchell DP, Ayabe T, Lopez-Boado YS, Stratman JL, Hultgren SJ, Matrisian LM, Parks WC. Regulation of intestinal alpha-defensin activation by the metalloproteinase matrilysin in innate host defense. Science. 1999;286:113–117. doi: 10.1126/science.286.5437.113. [DOI] [PubMed] [Google Scholar]
- 4.Salzman NH, Hung K, Haribhai D, Chu H, Karlsson-Sjoberg J, Amir E, Teggatz P, Barman M, Hayward M, Eastwood D, Stoel M, Zhou Y, Sodergren E, Weinstock GM, Bevins CL, Williams CB, Bos NA. Enteric defensins are essential regulators of intestinal microbial ecology. Nat Immunol. 2010;11:76–83. doi: 10.1038/ni.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amid C, Rehaume LM, Brown KL, Gilbert JG, Dougan G, Hancock RE, Harrow JL. Manual annotation and analysis of the defensin gene cluster in the c57bl/6j mouse reference genome. BMC Genomics. 2009;10:606. doi: 10.1186/1471-2164-10-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shanahan MT, Tanabe H, Ouellette AJ. Strain-specific polymorphisms in Paneth cell alpha-defensins of C57BL/6 mice and evidence of vestigial myeloid alpha-defensin pseudogenes. Infect Immun. 2011;79:459–473. doi: 10.1128/IAI.00996-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Es JH, Jay P, Gregorieff A, van Gijn ME, Jonkheer S, Hatzis P, Thiele A, van den Born M, Begthel H, Brabletz T, Taketo MM, Clevers H. Wnt signalling induces maturation of Paneth cells in intestinal crypts. Nat Cell Biol. 2005;7:381–386. doi: 10.1038/ncb1240. [DOI] [PubMed] [Google Scholar]
- 8.Andreu P, Colnot S, Godard C, Gad S, Chafey P, Niwa-Kawakita M, Laurent-Puig P, Kahn A, Robine S, Perret C, Romagnolo B. Crypt-restricted proliferation and commitment to the Paneth cell lineage following APC loss in the mouse intestine. Development. 2005;132:1443–1451. doi: 10.1242/dev.01700. [DOI] [PubMed] [Google Scholar]
- 9.Ouellette AJ, Greco RM, James M, Frederick D, Naftilan J, Fallon JT. Developmental regulation of cryptdin, a corticostatin/defensin precursor mRNA in mouse small intestinal crypt epithelium. J Cell Biol. 1989;108:1687–1695. doi: 10.1083/jcb.108.5.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karlsson J, Putsep K, Chu H, Kays RJ, Bevins CL, Andersson M. Regional variations in Paneth cell antimicrobial peptide expression along the mouse intestinal tract. BMC Immunol. 2008;9:37. doi: 10.1186/1471-2172-9-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Putsep K, Axelsson LG, Boman A, Midtvedt T, Normark S, Boman HG, Andersson M. Germ-free and colonized mice generate the same products from enteric prodefensins. J Biol Chem. 2000;275:40478–40482. doi: 10.1074/jbc.M007816200. [DOI] [PubMed] [Google Scholar]
- 12.Kobayashi KS, Chamaillard M, Ogura Y, Henegariu O, Inohara N, Nunez G, Flavell RA. NOD2-dependent regulation of innate and adaptive immunity in the intestinal tract. Science. 2005;307:731–734. doi: 10.1126/science.1104911. [DOI] [PubMed] [Google Scholar]
- 13.Schmidt DR, Holmstrom SR, Fon Tacer K, Bookout AL, Kliewer SA, Mangelsdorf DJ. Regulation of bile acid synthesis by fat-soluble vitamins A and D. J Biol Chem. 2010;285:14486–14494. doi: 10.1074/jbc.M110.116004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edwards U, Rogall T, Blocker H, Emde M, Bottger EC. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 1989;17:7843–7853. doi: 10.1093/nar/17.19.7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muyzer G, de Waal EC, Uitterlinden AG. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brandl K, Plitas G, Mihu CN, Ubeda C, Jia T, Fleisher M, Schnabl B, DeMatteo RP, Pamer EG. Vancomycin-resistant enterococci exploit antibiotic-induced innate immune deficits. Nature. 2008;455:804–807. doi: 10.1038/nature07250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rupnik M, Wilcox MH, Gerding DN. Clostridium difficile infection: New developments in epidemiology and pathogenesis. Nat Rev Microbiol. 2009;7:526–536. doi: 10.1038/nrmicro2164. [DOI] [PubMed] [Google Scholar]
- 20.Willing BP, Russell SL, Finlay BB. Shifting the balance: antibiotic effects on host-microbiota mutualism. Nat Rev Microbiol. 2011;9:233–243. doi: 10.1038/nrmicro2536. [DOI] [PubMed] [Google Scholar]
- 21.Lopez-Boado YS, Wilson CL, Hooper LV, Gordon JI, Hultgren SJ, Parks WC. Bacterial exposure induces and activates matrilysin in mucosal epithelial cells. J Cell Biol. 2000;148:1305–1315. doi: 10.1083/jcb.148.6.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wehkamp J, Harder J, Weichenthal M, Schwab M, Schaffeler E, Schlee M, Herrlinger KR, Stallmach A, Noack F, Fritz P, Schroder JM, Bevins CL, Fellermann K, Stange EF. NOD2 (CARD15) mutations in Crohn's disease are associated with diminished mucosal alpha-defensin expression. Gut. 2004;53:1658–1664. doi: 10.1136/gut.2003.032805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wehkamp J, Salzman NH, Porter E, Nuding S, Weichenthal M, Petras RE, Shen B, Schaeffeler E, Schwab M, Linzmeier R, Feathers RW, Chu H, Lima H, Jr, Fellermann K, Ganz T, Stange EF, Bevins CL. Reduced paneth cell alpha-defensins in ileal Crohn's disease. Proc Natl Acad Sci USA. 2005;102:18129–18134. doi: 10.1073/pnas.0505256102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bevins CL, Salzman NH. Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat Rev Microbiol. 2011;9:356–368. doi: 10.1038/nrmicro2546. [DOI] [PubMed] [Google Scholar]
- 25.Giaffer MH, Holdsworth CD, Duerden BI. The assessment of faecal flora in patients with inflammatory bowel disease by a simplified bacteriological technique. J Med Microbiol. 1991;35:238–243. doi: 10.1099/00222615-35-4-238. [DOI] [PubMed] [Google Scholar]
- 26.Willing BP, Dicksved J, Halfvarson J, Andersson AF, Lucio M, Zheng Z, Jarnerot G, Tysk C, Jansson JK, Engstrand L. A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology. 2010;139:1844–1854. doi: 10.1053/j.gastro.2010.08.049. [DOI] [PubMed] [Google Scholar]
- 27.Lin PW, Simon PO, Gewirtz AT, Neish AS, Ouellette AJ, Madara JL, Lencer WI. Paneth cell cryptdins act in vitro as apical paracrine regulators of the innate inflammatory response. J Biol Chem. 2004;279:19902–19907. doi: 10.1074/jbc.M311821200. [DOI] [PubMed] [Google Scholar]
- 28.Shi J, Aono S, Lu W, Ouellette AJ, Hu X, Ji Y, Wang L, Lenz S, van Ginkel FW, Liles M, Dykstra C, Morrison EE, Elson CO. A novel role for defensins in intestinal homeostasis: regulation of Il-1beta secretion. J Immunol. 2007;179:1245–1253. doi: 10.4049/jimmunol.179.2.1245. [DOI] [PubMed] [Google Scholar]
- 29.Giesemann T, Guttenberg G, Aktories K. Human alpha-defensins inhibit clostridium difficile toxin B. Gastroenterology. 2008;134:2049–2058. doi: 10.1053/j.gastro.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 30.Dessein R, Gironella M, Vignal C, Peyrin-Biroulet L, Sokol H, Secher T, Lacas-Gervais S, Gratadoux JJ, Lafont F, Dagorn JC, Ryffel B, Akira S, Langella P, Nunez G, Sirard JC, Iovanna J, Simonet M, Chamaillard M. Toll-like receptor 2 is critical for induction of Reg3 beta expression and intestinal clearance of Yersinia pseudotuberculosis. Gut. 2009;58:771–776. doi: 10.1136/gut.2008.168443. [DOI] [PubMed] [Google Scholar]
- 31.Gong J, Xu J, Zhu W, Gao X, Li N, Li J. Epithelial-specific blockade of MyD88-dependent pathway causes spontaneous small intestinal inflammation. Clin Immunol. 2010;136:245–256. doi: 10.1016/j.clim.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 32.Vaishnava S, Behrendt CL, Ismail AS, Eckmann L, Hooper LV. Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proc Natl Acad Sci USA. 2008;105:20858–20863. doi: 10.1073/pnas.0808723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoffmann A, Leung TH, Baltimore D. Genetic analysis of NF-kappab/Rel transcription factors defines functional specificities. EMBO J. 2003;22:5530–5539. doi: 10.1093/emboj/cdg534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shirafuji Y, Tanabe H, Satchell DP, Henschen-Edman A, Wilson CL, Ouellette AJ. Structural determinants of procryptdin recognition and cleavage by matrix metalloproteinase-7. J Biol Chem. 2003;278:7910–7919. doi: 10.1074/jbc.M210600200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Video