Abstract
Antimicrobial peptides are important for a healthy host-microbe homeostasis. In infections characterized by low levels of the human cathelicidin, LL-37, induction of its expression increases clearance of pathogens. Our aim was to discover signaling pathways and compounds capable of affecting the expression of LL-37. We recently observed a synergistic induction of LL-37 expression by stimulating the colonic epithelial cell-line HT-29 with lactose and phenylbutyrate (PBA). Here, we studied regulatory circuits mediating this synergism in HT-29 cells stimulated with lactose (60 g/l) and PBA (2 mM) for 24 h by using mass spectrometry and pathway analyses. Selected pathways were evaluated for their involvement in LL-37 regulation in a CAMP gene-luciferase reporter system. Three pathways were examined in detail: thyroid hormone receptor and retinoid X receptor (TR/RXR) activation, eicosanoid signaling and steroid biosynthesis. Induced expression of LL-37 was observed upon stimulation with triiodothyronine (T3, 2.5 nM-1 µM for 3-30 h) and thyroxine (T4, 2.5-10 nM for 24 h). Furthermore, the synergism of lactose and PBA was reduced in cells coincubated with inhibitors of phospholipase A2, cyclooxygenase 2 or HMG-CoA reductase. Based on these results, we conclude that proteomics and pathway analyses are valuable tools for dissecting the regulatory networks involved in LL-37 expression.
Key Words: Antimicrobial peptides, Host defense, LL-37, Proteomics, Pathway analysis
Introduction
Regulation of innate immunity is important for human health and disease. Several defects in this regulation have been linked to susceptibility to infections [1]. At the epithelial interface, where host/microbe interactions first occur, the antimicrobial peptides (AMPs), crucial effectors of innate immunity, are expressed. In vivo AMPs are important in the defense against a range of pathogenic microbes, but also for immunomodulation, wound-healing and regulation of the microflora [2, 3, 4]. Dysregulation of AMP expression has been linked to diseases such as psoriasis, Crohn's disease, shigellosis and tuberculosis [5, 6]. Several compounds that induce the expression of the most common human AMPs, the defensins and cathelicidins, have been discovered. Some of these compounds are potential drug candidates that can be utilized, either alone or as adjunctive drugs to conventional antibiotics for the treatment of infections [7, 8, 9, 10, 11]. These studies indicate that regulation of AMPs such as the cathelicidins are important for front-line defenses against infection and for host-bacterial homeostasis. Although the antimicrobial and immunomodulatory effects of the human cathelicidin, LL-37, are well established, the regulation of its gene remains elusive. However, known transcription factors regulating the CAMP gene encoding LL-37 are VDR, PU.1 and C/EBP epsilon [12, 13, 14]. It has also been demonstrated that 1,25-dihydroxyvitamin D3, lithocholic acid and butyrate/phenylbutyrate (PBA) are inducers of the CAMP gene [5]. In addition, we have recently shown that lactose enhances the expression of LL-37 in colonic and monocytic cell lines [11]. We also showed a synergistic induction of CAMP expression by treating HT-29 cells with a combination of lactose and PBA. In this study, we examined regulatory circuits involved in CAMP gene induction using lactose and/or PBA as stimulants in HT-29 cells. By applying high-throughput label-free quantification mass spectrometry coupled with pathway analyses, several putative regulatory networks involved in CAMP expression were revealed. We assessed the pathways for their involvement in the regulation of the CAMP gene further in experiments using the reporter cell line MN8CampLuc [15]. The combination of the two methods is a novel approach and provides a tool to resolve the regulation of antimicrobial peptides.
Materials and Methods
Chemicals
Simvastatin, pravastatin, lactose, methyl arachidonyl fluorophosphonate (MAFP), acetylsalicylic acid (ASA), mevalonate, cholesterol, triiodothyronine/thyroxine (T3/T4) and all-trans-retinoic acid (ATRA) were purchased from Sigma-Aldrich (St. Louis, Mo., USA). Simvastatin hydroxy acid was obtained from Abcam (Cambridge, UK). Etoricoxib and PBA were from Santa Cruz Biotechnology (Dallas, Tex., USA) and MK-886 from Cayman Chemical (Ann Arbor, Mich., USA). All compounds were diluted in DMSO except for PBA and lactose which were diluted in phosphate-buffered saline (PBS) and ASA which was diluted in ethanol. The final concentration of solvent never exceeded 1% and did not markedly affect LL-37 expression.
Cell Culture and Sample Preparation
HT-29 cells were obtained from the American Type Culture Collection: HTB-38 (Rockville, Md., USA). The cells were cultured in RPMI1640 (Sigma-Aldrich) supplemented with 10% fetal calf serum (FCS, Invitrogen, Carlsbad, Calif., USA), 25 mM HEPES (Invitrogen), 500 µg/ml streptomycin, 500 units/ml penicillin (Invitrogen), 2 mML-glutamine (Sigma-Aldrich) at 37°C and in 5% CO2. At 85–95% confluence, cells were washed with PBS (Invitrogen) and culture media without FCS was applied. Cells were then stimulated with vehicle, 60 g/l lactose and/or 2 mM PBA. After 48 h of stimulation, cells were harvested by trypsinization and collected by centrifugation at 3,500 g for 10 min. The cells were then resuspended in lysis buffer containing 8 M urea, 75 mM NaCl, 50 mM Tris, 10 mM sodium pyrophosphate, pH 8.2 and Complete Mini protease inhibitor cocktail (Roche, Basel, Switzerland). Cell lysates were prepared by sonication on ice for 3 min using cycles of a 6-second pulse at 30% amplitude followed by a 4-second pause. Cell debris was pelleted by centrifugation at 20,000 g for 20 min at 8°C and cell supernatants were stored at −80°C until used. Protein concentration of cell lysate supernatants was measured using the BCA protein assay kit (Thermo Fisher Scientific, Waltham, Mass., USA) and samples were stored at −80°C until used.
Mass Spectrometry and Pathway Analysis
For mass spectrometric analysis, 10 µg protein/cell lysate was utilized. Samples were analyzed in collaboration with Proteomics Karolinska (PK/KI). Proteins in the samples were subjected to reduction and alkylation and cleavage by trypsin in accordance with Cederlund et al. [16]. Tryptic digests were enriched using stage tips (Thermo Fisher Scientific) prior to mass spectrometric analyses. Spectra were recorded utilizing a nanoLC-Q Exactive Orbitrap MS/MS (Thermo Fisher Scientific) and the spectra were analyzed using Mascot (Matrix Science, London, UK; version 2.3.0) and searched against SwissProt_2011.08 (Species: Homo sapiens, 20245 entries). In Mascot, the fixed modifications were defined as carbamidomethylation of cysteines and variable modifications were defined as oxidation of methionine and deamidation of asparagine. Mass tolerances were set to 0.050 Da and 10.0 PPM for ion mass fragments and parent ion tolerance, respectively. For validation of protein identities, the software Scaffold (version 3.3.3, Proteome Software Inc., Portland, Oreg., USA) was utilized. The identity threshold for peptides and proteins was set at ≥95.0% probability. Protein identities and their spectrum intensities were quantified using in-house developed quantitation software (PK/KI) [17] and protein quantities of treated cells were normalized to those of vehicle-treated cells (control). The normalized quantities were further analyzed through the use of IPA (Ingenuity® Systems, www.ingenuity.com). Proteomic data from treated HT-29 cells were analyzed using IPA and protein identifiers from the dataset were mapped onto identifiers in the Ingenuity Knowledge Base. Regulatory and metabolic networks were generated by Ingenuity and ranked in accordance to their significance, as judged by a right-tailed Fisher's exact test, where p < 0.05 was considered significant. In addition, the activation states of transcription factors were also calculated and presented as z-scores [18].
Luciferase Reporter Assay
HT-29 cells that have been stably transfected with a linearized construct consisting of 3,000 base pairs of the CAMP promoter and the CAMP gene fused to the luciferase gene LUC2 were used to confirm potential regulatory pathways with relevant agonist or antagonists. The reporter cell line, denoted MN8CampLuc, produces a fusion protein of hCAP-18, the precursor protein of LL-37 and luciferase (proLL37-luciferase) and responds similarly to stimulants known to modulate the expression of the endogenous CAMP gene in the parental cell line [15]. MN8CampLuc cells were seeded into 96-well plates and grown to 85–95% confluence in culture media (as formulated in the ‘Cell cultures’ section) containing 10% FCS and hygromycin before stimulations. Stimulants were administered in culture media without hygromycin and were incubated for 24 h. The luciferase assay was performed using the Luciferase assay kit (Promega, Fitchburg, Wis., USA) in accordance with the manufacturer's instructions.
Real-Time Reverse Transcription PCR
Total RNA was extracted from cells using the Isolate RNA mini kit (Bioline, London, UK) in accordance with manufacturer's instructions. One microgram of total RNA was used for the synthesis of cDNA utilizing the iScript cDNA synthesis kit (Bio-Rad, Hercules, Calif., USA). Reverse transcription was performed using the CFX96 Real-Time system (Bio-Rad) and iQ SYBR Green supermix (Bio-Rad) with primers specific for the CAMP gene: forward 5′-TCACCAGAGGATTGTGACTTCAA; and reverse 5′-TGAGGGTCACTGTCCCCATAC. The results were normalized to total RNA quantity and presented as relative expression of CAMP in treated cells compared to vehicle control-treated cells.
Statistical Analyses
Data are presented as means and standard deviation (SD) or standard error of the mean (SEM). Statistical differences between groups were evaluated using a Student two-tailed t test (if samples were normally distributed) or by nonparametric Mann-Whitney test with a 95% confidence interval if samples were not normally distributed, as judged by a Shapiro-Wilk normality test. p < 0.05 was considered significant (*** p ≤ 0.001, ** p ≤ 0.005, * p ≤ 0.05).
Results
Proteomic and Pathway Analyses of Cell Lysates
We previously described that lactose and PBA synergistically induce the expression of LL-37 in HT-29 cells [11]. To elucidate the regulatory pathways behind this observation, we performed proteomic analyses of stimulated HT-29 cells. The analyses were performed on lysates of HT-29 cells treated with vehicle, 60 g/l lactose, 2 mM PBA and 2 mM PBA in combination with 60 g/l lactose (PBA/lactose) for 48 h. As a control, CAMP induction was confirmed on mRNA level after stimulation with lactose or PBA or a combination of these. Cell lysates from these cells were subsequently used for proteomic analyses by label-free quantification mass spectrometry. This yielded on average 1,529 protein identities and abundances per sample. Of these identified proteins, 1,323 were present in all samples and thus chosen for pathway analysis (fig. 1). By comparing the relative spectrum intensities of the proteins from treated cells to those in the vehicle control cells and mapping these results to the pathways present in the Ingenuity Knowledge Base, a number of metabolic and regulatory pathways including transcription factors were activated (table 1, 2). There were pronounced differences in the activation pattern of signaling and metabolic pathways between cells stimulated with lactose or PBA. However, we could not detect any differentially regulated metabolic pathways in cells treated with a combination of lactose and PBA compared to those stimulated with only PBA or lactose (table 1). In contrast, there were several differences in transcription factor pattern of cells treated with PBA/lactose to those stimulated with PBA or lactose (table 2). In cells treated with PBA/lactose, pathway analyses indicated that the transcription factors SREBF1, RXRA, estrogen receptor alpha (ESR1), peroxisome proliferator-activated receptor gamma (PPARG), SREBF2, NR3C2 and HNF4α were activated. By comparing the results of the pathway analyses from cells stimulated with PBA/lactose with the results from cells treated with PBA or lactose, a unique activation of RXRA and an enhanced activation (increase in z-score) of ESR1, NR3C2, HNF4α dimer and PPARG in PBA/lactose-stimulated cells was observed (table 2).
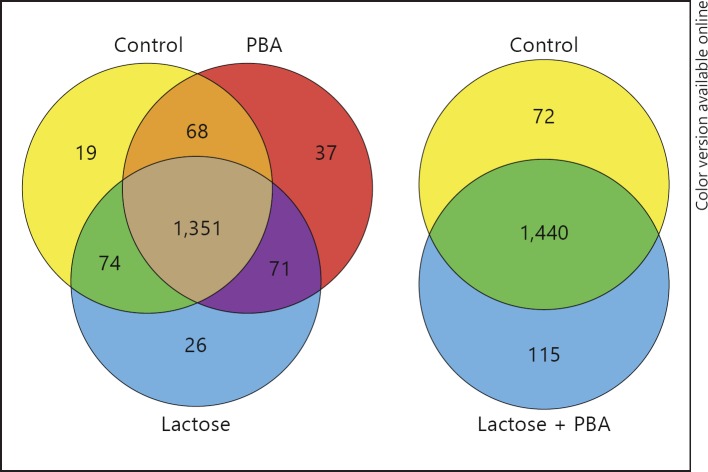
Fig. 1.
Venn diagrams of proteins identified in the proteomic analysis of stimulated HT-29 cell lysates. Protein cell lysates of HT-29 cells stimulated for 24 h with or without 2 mM PBA and 60 g/l lactose, alone or in combination, were analyzed using label-free quantitation with a Q Exactive MS/MS. In cells stimulated with vehicle (Control), 1,512 proteins were detected, in lactose 1,522, in PBA 1,527 and in cells treated with a combination of PBA and lactose 1,555. Numbers indicate the number of unique proteins identified in the sample.
Table 1.
Activated metabolic and regulatory pathways
| Pathway | Lactose | PBA | PBA/lactose |
|---|---|---|---|
| EIF4 signaling* | + | – | + |
| Glycolysis | + | – | + |
| Citrate cycle | – | + | + |
| mTOR signaling* | + | – | + |
| Pentose phosphate pathway | + | – | + |
| Propionate metabolism | – | + | + |
| Butyrate metabolism | – | + | + |
| Eicosanoid signaling* | – | + | + |
| Biosynthesis of steroids* | – | + | + |
| TR/RXR activation* | + | – | + |
Pathways indicated as activated in HT-29 cells treated for 24 h with either lactose (60 g/l), PBA (2 mM) or PBA/lactose in combination, as based on the Ingenuity® pathway analysis. * Pathways selected for further analyses.
Table 2.
Transcription factors activated in HT-29 cells treated with lactose, PBA or PBA/lactose
| Transcription regulator | Predicted activation state | Regulation z-score | p-value of overlap |
|---|---|---|---|
| Transcription factors activated in cells stimulated with PBA/lactose | |||
| PPARG* | activated | 4.1 | 8.58E-16 |
| SREBF1* | activated | 2.9 | 7.43E-10 |
| RXRA* | activated | 2.8 | 3.98E-03 |
| ESR1* | activated | 2.4 | 5.67E-02 |
| PPARA* | activated | 2.3 | 1.20E-18 |
| SREBF2 | activated | 2.3 | 4.18E-08 |
| NR3C2 | activated | 2.1 | 1.05E-02 |
| HNF4α dimer | activated | 2 | 1.11E-02 |
| Transcription factors activated in cells stimulated with lactose | |||
| SREBF1 | activated | 2.8 | 7.43E-10 |
| SREBF2 | activated | 2.6 | 4.18E-08 |
| STAT5A | activated | 2.5 | 2.32E-02 |
| HIF1A* | activated | 2.3 | 9.53E-05 |
| PPARG | activated | 2.1 | 8.58E-16 |
| ESR1 | activated | 2 | 1.60E-02 |
| Transcription factors activated in cells stimulated with PBA | |||
| PPARA | activated | 3.2 | 1.20E-18 |
| ESRRA | activated | 2.4 | 6.87E-08 |
| SIRT1 | activated | 2 | 2.60E-02 |
Transcription factors indicated as activated by the Ingenuity pathway analysis in HT-29 cells treated with either lactose, PBA or PBA/lactose in combination. A positive z-score indicates a measure of consistency of a gene expression pattern with the activation of a given transcription factor.
Transcription factors selected for further analyses.
However, since we focused only on pathways pertaining to the regulation of the CAMP gene, we chose, after initial screening based on bibliographic and preliminary experimental data of the highest-ranked regulatory networks (literature searches for pathways connected to innate immune regulation, feasibility of interference with specific pathways and pilot experiments performed using the reporter cell line MN8CampLuc [15], data not shown), to examine a subset of pathways activated in cells stimulated with PBA/lactose (table 1, 2: indicated by asterisks). Out of eleven pathways examined in the reporter cell line MN8CampLuc [15], three pathways were deemed as interesting based on an altered expression of the reporter protein proLL37-luciferase. These pathways were thyroid hormone receptor and retinoid X receptor (TR/RXR) activation, eicosanoid signaling and steroid biosynthesis and were selected for further analyses (fig. 2, 3, 4). In order to evaluate the validity of the pathway analyses, activated pathways were analyzed in the reporter system by incubation with agonists or antagonists of key proteins of the respective pathways.
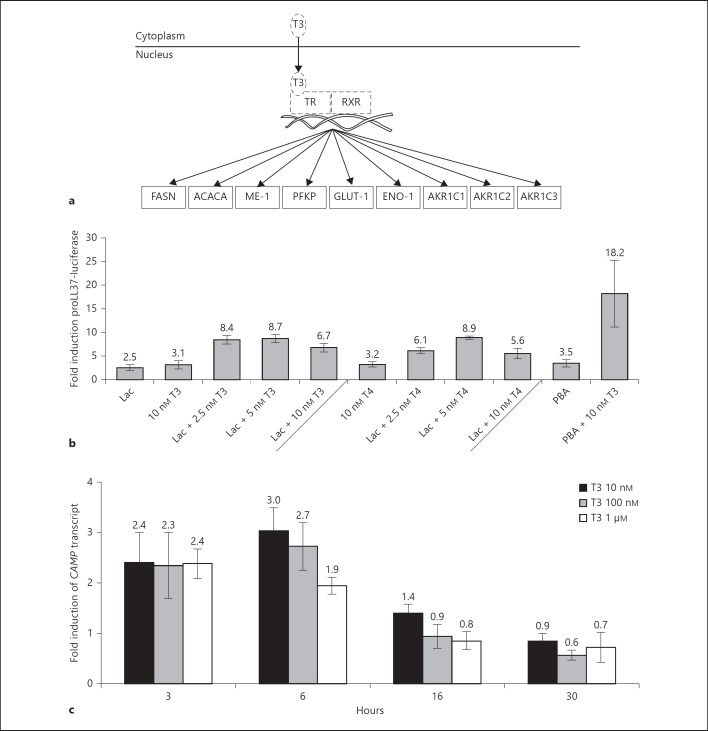
Fig. 2.
a Proteins involved in TR/RXR activation detected as induced by the proteomic analysis after stimulation of HT-29 cells. Solid lines around proteins indicate those that were identified and induced in HT-29 cells treated with lactose or PBA/lactose. Hatched lines indicate proteins or hormones not detected in the proteomic analysis. b Screen of the MN8CampLuc cell line with compounds involved in TR/RXR signaling. Incubation for 24 h of MN8CampLuc cells with thyroid hormones (i.e. T3 or T4, concentrations are indicated in the graph) together with either 60 g/l lactose (Lac) or 2 mM PBA induces the expression of proLL37-luciferase. The results are shown as fold induction relative to vehicle control and the data shows the mean and SD of at least 4 independent experiments performed in duplicate. c Kinetic analysis of CAMP transcript expression in parental HT-29 cells treated with thyroid hormone. Incubation of HT-29 cells with 10 nM, 100 nM and 1 µM T3 for 3, 6, 16 and 30 h, respectively. Data depict fold induction related to vehicle control, as measured by RT-PCR, and show the mean and SD of at least 3 independent experiments performed in triplicate.
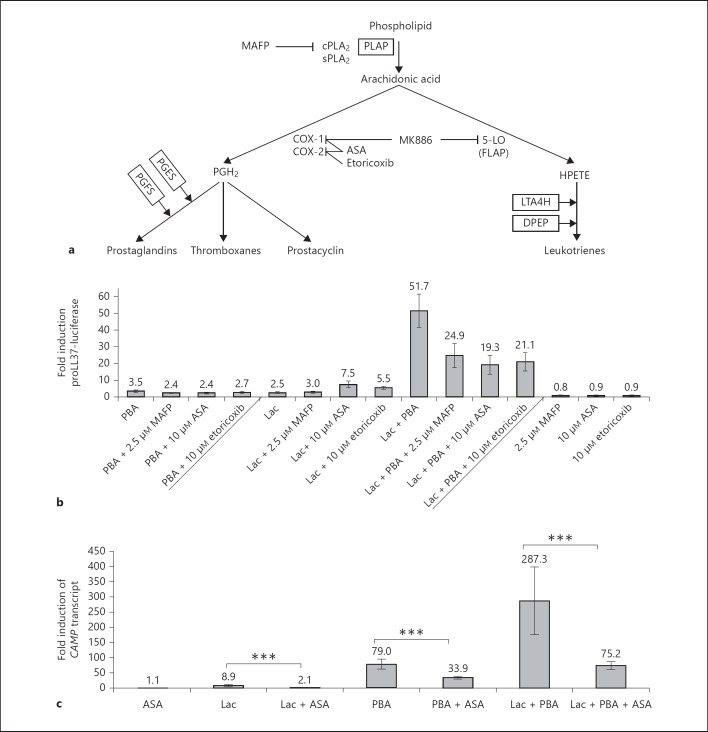
Fig. 3.
a Proteins involved in eicosanoid signaling detected as induced by the proteomic analysis after stimulation of HT-29 cells. Closed lines around proteins indicate those that were identified and induced in HT-29 cells treated with PBA or PBA/lactose. b Screen of the MN8CampLuc cell line using compounds involved in eicosanoid signaling. Incubation for 24 h of MN8CampLuc cells with inhibitors to key enzymes of the COX-dependent eicosanoid biosynthesis pathway (MAFP to cPLA2, ASA to COX-1 and 2, etoricoxib to COX-2). The results are shown as fold induction relative to vehicle control and the data shows the mean and SD of at least four independent experiments performed in duplicate. c CAMP in parental HT-29 cells after stimulation with lactose and/or PBA or in combination with ASA. HT-29 cells were incubated for 24 h with lactose, PBA or PBA/lactose separately or together with 10 µM ASA an antagonist to COX-1 and 2. The results are shown as fold induction relative to vehicle control and the data show the mean and SEM of at least 3 independent experiments performed in triplicate. *** p ≤ 0.001.
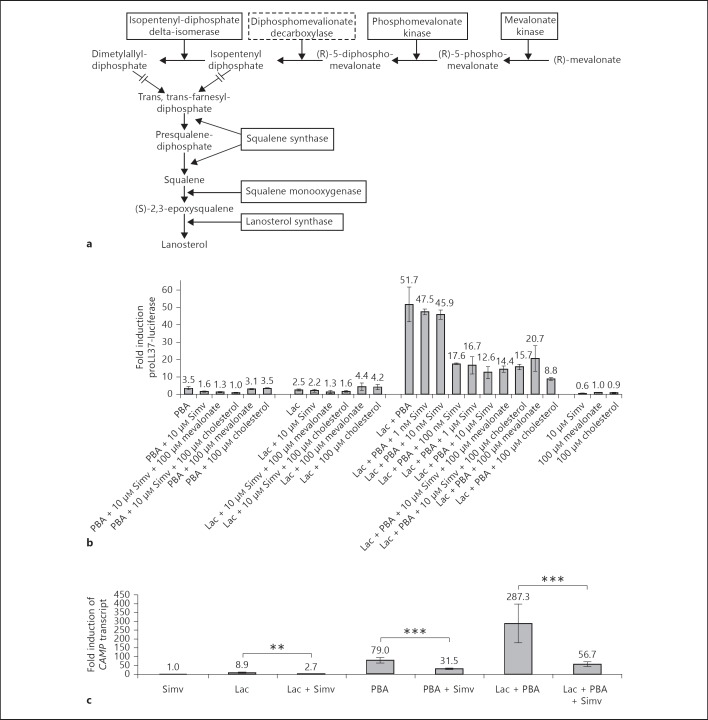
Fig. 4.
a Proteins involved in steroid biosynthesis detected as induced by the proteomic analysis in stimulated HT-29 cells. Closed lines around proteins indicate enzymes that were identified and induced in HT-29 cells treated with lactose or PBA/lactose. Hatched lines indicate proteins not detected in the proteomic analysis. b Screen of the MN8CampLuc cell line using compounds affecting the steroid biosynthesis pathway. Incubation for 24 h of MN8CampLuc cells with an inhibitor to HMG-CoA reductase [simvastatin (Simv) 10 µM] results in an inhibition of the PBA/lactose-mediated synergism on proLL37-luciferase expression. No rescuing effect was observed by administration of either mevalonate or cholesterol in cells incubated with simvastatin. The results are shown as fold induction relative to vehicle control and the data show the mean and SD of at least 4 independent experiments performed in duplicate. c The level of CAMP transcripts in parental HT-29 cells after stimulation with inhibitors of steroid biosynthesis. HT-29 cells incubated with lactose, and/or PBA or PBA/lactose with 10 µM simvastatin (Simv) for 24 h. The results are shown as fold induction relative to vehicle control and the data show the mean and SEM of at least 3 independent experiments performed in triplicate. ** p ≤ 0.005; *** p ≤ 0.001.
Thyroid Hormone Receptor/Retinoid X Receptor Activation
In cells treated with 60 g/l lactose, 2 mM PBA or a combination of PBA/lactose, pathway analyses indicated an increased activity in TR/RXR signaling since a number of proteins known to be regulated by TR/RXR were induced in the cells (table 1). These proteins were fatty acid synthase (FASN), acetyl-CoA carboxylase 1 (ACACA), NADP-dependent malic enzyme (ME-1), 6-phosphofructokinase type C (PFKP), glucose transporter type 1 (GLUT-1), α-enolase (ENO-1) and aldo-keto reductase family 1 members C1, 2 and 3 (AKR1C1-3) (fig. 2a), as determined by using IPA (Ingenuity). To confirm the involvement of the TR/RXR pathway, the reporter cell line MN8CampLuc was stimulated with the TR ligands T3 and T4 [19] and the RXR ligand ATRA. In these stimulations we sought to evaluate if these ligands could invoke a similar response of CAMP induction as observed by stimulations with lactose or PBA in the parental cell line HT-29.
A 3.1-fold increase in the fusion protein proLL37-luciferase expression was observed after 24 h of stimulation with 10 nM T3, a 3.2-fold induction with 10 nM T4 (fig. 2b) and 1.4-fold with 500 nM ATRA (online suppl. fig. 1; for all online suppl. material, see www.karger.com/doi/10.1159/000355931). In cells stimulated with a combination of T3 and ATRA, the expression of proLL37-luciferase was additively increased to 5.5-fold (online suppl. fig. 1). When stimulating cells with a combination of 60 g/l lactose and T3 or T4, proLL37-luciferase induction was observed at all concentrations of T3 and T4 tested with an optimal concentration of 5 nM for both (8.7- and 8.9-fold, respectively). Interestingly, 2 mM of PBA exhibited a synergistic effect in the induction of proLL37-luciferase when it was combined with 10 nM of T3 (18.2-fold) (fig. 2b). The induction of CAMP transcripts was observed in the parental HT-29 cell line when cells were stimulated with varying concentrations of T3. The highest induction of CAMP was observed in cells stimulated with 10 nM T3 (fig. 2c). This induction of CAMP was observed already after 3 h (2.4-fold, 10 nM T3) with a maximal induction of CAMP at 6 h (3-fold, 10 nM T3). Interestingly, the inducing effect of T3 was down to basal expression at 16 h (0.9 and 0.8-fold, for 100 nM T3 and 1 µM T3, respectively). In conjunction with these results, we found TRIAC, T4 and liothyronine to upregulate proLL37-luciferase in the screening of the commercially available Prestwick Chemical Library as described by Nylen et al. [15].
These results indicate that CAMP expression is regulated by thyroid hormones and that the effect can be enhanced in combination with either lactose or PBA.
Eicosanoid Signaling
In cells stimulated with PBA and PBA/lactose, but not in cells stimulated with lactose separately (table 1), pathway analyses indicated that eicosanoid signaling was activated because a number of proteins involved in this signaling were upregulated. These proteins were phospholipase A-2-activating protein (PLAP), leukotriene A4 hydrolase (LTA4H), prostaglandin E synthase (PGES), prostaglandin F synthase (PGFS) and dipeptidase 1 (DPEP) [20] (fig. 3a). The majority of these proteins are involved in the cyclooxygenase (COX)-dependent pathway of eicosanoid synthesis. Therefore, several pharmacological inhibitors for this pathway were employed in order to elucidate if the PBA/lactose mediated induction of CAMP was dependent on the COX pathway. Cells were stimulated with a combination of PBA/lactose and MAFP, a selective and irreversible pharmacological inhibitor of both calcium-dependent and calcium-independent cytosolic phospholipase A2 (cPLA2) [21]. This resulted in a significant reduction of proLL37-luciferase expression by 52% (from 51.7-fold to 24.9-fold; p < 0.001) (fig. 3b). Similarly, a 59% inhibition of the synergism was achieved by incubation of PBA/lactose-stimulated cells with the selective COX-2 inhibitor etoricoxib (p = 0.001) [22]. Incubation of PBA/lactose-stimulated cells with ASA, an inhibitor of both COX-1 and COX-2 [22], resulted in a 63% reduction of proLL37-luciferase expression (p = 0.001; fig. 3b). To further dissect the influence of eicosanoid signaling on the PBA/lactose-mediated induction of LL-37 expression in MN8CampLuc, cells were stimulated with either lactose, PBA or PBA/lactose together with MAFP, etoricoxib or ASA. The lactose-mediated induction of proLL37-luciferase was not significantly affected by 10 µM MAFP (p = 0.247). Notably, we observed a significant induction of the fusion protein expression in the reporter system with incubation of cells with lactose and 10 µM ASA or 10 µM etoricoxib in combination (both p < 0.001) compared to stimulations with lactose alone (fig. 3b). In comparison to cells incubated with only PBA, there was a significant reduction in the expression of LL-37 in the reporter system when stimulating cells with a combination of PBA with the eicosanoid inhibitors ASA (p = 0.009), etoricoxib (p = 0.028) or MAFP (p = 0.001) (fig. 3b). However, in the parental HT-29 cells incubated with lactose, PBA or PBA/lactose all showed a significant decrease in the expression of CAMP transcript when incubated with 10 µM ASA relative to cells not stimulated with ASA (fig. 3c).
The proteomic analyses also indicated that LTA4H and DPEP of the 5-lipoxygenase (5-LO)-dependent branch of eicosanoid synthesis were induced in cell lysates from cells stimulated with PBA and PBA/lactose. Hence, MK-886 an antagonist to 5-LO-activating protein (FLAP) was utilized to inhibit the 5-LO pathway of leukotriene biosynthesis [23]. As opposed to cells stimulated with PBA and 1 µM MK-886, cells incubated with lactose and MK-886 showed a significant (p = 0.04) reduction in fusion protein expression in the reporter assay. There was no significant reduction of the synergistic effect of PBA/lactose when incubating cells with MK-886 (p = 0.590; online suppl. fig. 2).
Taken together, these results indicate that the synergism between lactose and PBA on CAMP gene induction is dependent on functional COX-2 eicosanoid signaling.
Biosynthesis of Steroids
In cells stimulated with lactose or PBA/lactose, a number of enzymes involved in the biosynthesis of steroids were induced (table 1). These were mevalonate kinase, phosphomevalonate kinase, isopentenyl-diphosphate delta-isomerase, squalene synthase, squalene monooxygenase and lanosterol synthase (fig. 4a). To evaluate if this pathway contributes to the observed CAMP gene induction in cells stimulated with lactose and PBA/lactose, cells were incubated with simvastatin, a 3-hydroxy-3-methylglutaryl coenzyme A (HMG-CoA) reductase inhibitor. Incubation of cells with 100 nM of simvastatin resulted in a 66% reduction of the PBA/lactose-mediated induction of proLL37-luciferase, with a further reduction to 76% at 10 µM (fig. 4b). In addition, simvastatin hydroxy acid and pravastatin, a hydrophilic statin that does not need activation to exert its effect, showed similar reducing effects on the synergistic induction of proLL37-luciferase by lactose and PBA (online suppl. fig. 3). However, no rescue of the observed reduction of proLL37-luciferase by simvastatin was achieved with either 100 µM mevalonate or 100 µM cholesterol (fig. 4b). Of the cells treated with simvastatin together with lactose (p = 0.205) or PBA, only PBA induction was significantly affected (p = 0.003). Notably, the addition of either mevalonate or cholesterol did not rescue the simvastatin-mediated reduction of proLL37-luciferase expression in cells treated with PBA (fig. 4b).
A significant reduction of CAMP transcripts was also observed in parental HT-29 cells incubated with either lactose, PBA or PBA/lactose together with 10 µM simvastatin compared to cells stimulated only with vehicle (fig. 4c).
Taken together, these results indicate that the synergism between lactose and PBA in the induction of CAMP is blocked by statin treatment in a mevalonate- and cholesterol-independent manner.
Discussion
Biochemical Analyses of Metabolic and Regulatory Pathways
We used proteomics combined with bioinformatic pathway analyses to dissect the regulatory pathways involved in the induction of the CAMP gene by lactose and/or PBA. This is a novel approach to resolve the complex regulation of cathelicidin expression. The initial outcome of this method yielded eleven intracellular signaling pathways and transcription factors characterized for their importance for AMP expression in innate immunity.
As a validation of the methodology, we could observe that pathways previously reported to be activated in PBA-stimulated or lactose-stimulated cells were also activated in our samples. For example, we observed that the incubation of cells with lactose led to an increase of enzymes involved in glycolysis and the pentose phosphate pathway, and the PBA biosynthesis of steroids and butyrate/propionate metabolism was activated in cells in response to PBA treatment [24, 25].
Notably, we could not detect any additionally activated metabolic pathways in cells treated with the PBA/lactose combination to those activated in cells treated with lactose or PBA separately. The lack of such pathways may be the result of the limited proteomic dataset or an actual physiological situation. However, we could observe differences in the activation states of specific transcription factors. The main differences between PBA/lactose-stimulated cells and those stimulated with PBA or lactose were the activation of the transcription factor retinoid X receptor alpha (RXRA) and a further increase in the activation of ESR1 and PPARG, indicating their involvement in cathelicidin regulation. RXRA forms a heterodimer with VDR which is a known activator of CAMP gene expression. In addition, RXRA is a partner for PPARG. A link between LL-37 expression and estrogen signaling has recently been proposed [26].
Of eleven metabolic and regulatory pathways detected as activated by PBA/lactose stimulation we chose to examine three, i.e. TR/RXR activation, eicosanoid signaling and steroid biosynthesis (table 1, 2: marked with asterisks). Using specific agonists and antagonists, these pathways were systematically investigated for their involvement in the expression of the CAMP gene.
Thyroid Hormone Receptor/Retinoid X Receptor Activation
We observed that supplementation of culture media with thyroid hormones T4 or T3 could induce the CAMP gene (fig. 2b, c). Thyroid hormones are commonly associated with the regulation of metabolism; however, evidence has been accumulated that they confer additional immunomodulatory functions (review: [19]). In addition, it has been reported that children resistant to thyroid hormone show an increased incidence of respiratory tract infections [27]. In this study, we observed an induction of LL-37 expression in response to thyroid hormone supplementation in colonic epithelial cells. Adding the hormonally inactive form T4 also exerted induction of the fusion protein proLL37-luciferase. This effect would be explained either by T4 acting as a weak ligand to TRs [28] or through the conversion of T4 to T3 by deiodinases expressed in the colonic cells [29]. The observed induction of thyroid hormones on proLL37-luciferase expression as well as a synergism with T3 and lactose or PBA was not investigated for a direct causal effect. However, there are putative binding sites of TR, the T3 response element (TRE, sequence: AGGTCA) [30], present in the promoter of the CAMP gene at 2,100 and 1,830 basepairs upstream of the CAMP start site. This indicates that there may be a direct effect of thyroid hormones on CAMP gene expression. One possible relation of thyroid hormones to the induction of CAMP gene expression is that enterocyte differentiation is driven by these hormones as well as by short-chain fatty acids such as the CAMP gene-inducer butyrate [31, 32, 33]. The induction of the CAMP gene by T3 may, in fact, be a result of an altered differentiation state in the cells. In addition, the association of thyroid hormones to immunity is established and there are reports on thyroid hormones acting as modulators of immune cells, specifically natural killer cells [19]. Our results demonstrate that thyroid hormones can also modulate the expression of immune genes in epithelial cells, which lends further credibility to an immunomodulatory role for thyroid hormones.
In many cases, transcription of early-response and immune genes is tightly regulated or the mRNA is highly destabilized, resulting in a destruction of mRNA signal after induction [34]. Luciferase protein has a half-life of approximately 3 h (promega.com) which might be further extended in the proLL37-luciferase fusion protein. It is also known that protein translation may lag behind the transcriptional induction by several hours, and this could explain why the induction of transcript is lost at the time point where we observe induction in the luciferase assay.
The indicated activation of RXR and the fact that this transcription factor forms a heterodimer with VDR motivated the stimulation of cells with the RXR ligand ATRA. By this stimulation we observed a weak but significant induction of the CAMP gene (p = 0.010; online suppl. fig. 1). This result, together with the indication of an activated state of RXR, suggests the involvement in the regulation of CAMP gene. An additional role of RXR is by pairing with the thyroid hormone receptor, and hence TR activation of target genes in the absence of its ligand is inhibited [35]. Indeed, we observed an additive induction of reporter protein expression in the reporter assay when stimulating with a combination of TR (T3) and RXR (ATRA) ligands. Furthermore, ATRA exhibits several additional functions and can act as a ligand of the retinoic acid receptor and RAR-related orphan receptor beta [36].
Eicosanoid Signaling
We observed that the PBA/lactose effect on proLL37-luciferase expression can be inhibited by antagonists for key enzymes in the signaling of eicosanoid production. There is accumulating evidence that there are regulatory circuits in cells that connect the expression of AMPs to the production of eicosanoids. For example, it has been shown that LTB4 can induce the release of LL-37 from neutrophils and, conversely, LL-37 can stimulate the translocation of 5-LO and thus increase the production of LTB4 [37]. By inhibiting key enzymes of the eicosanoid biosynthesis, we could show that the inducing effect of PBA and lactose was inhibited by inhibiting COX-2. It has been reported that colon cancer cell lines may respond with apoptosis when challenged with COX-2 inhibitors [38]. However, in these studies, the incubation time was 72 h and the concentrations of the COX-2 inhibitor were in the millimolar range. In agreement with Chen et al. [39],we did not observe an decrease in cell vitality after 24 h of stimulation with the COX-2 inhibitor, compared to cells stimulated with vehicle control (data not shown). Taken together, our observations extend previous observations of the crosstalk of eicosanoids and LL-37 [37].
Steroid Biosynthesis
We observed an inhibitory effect on the PBA/lactose-mediated induction on CAMP gene expression by HMG-CoA reductase inhibitors (pravastatin, simvastatin and simvastatin hydroxy acid). Statins inhibit the enzyme HMG-CoA reductase, catalyzing the conversion of 3-hydroxy-3-methylglutaryl-CoA to mevalonate, resulting in a reduction also in the downstream products of steroid biosynthesis. However, we could not observe a rescuing effect of the inhibition of the PBA/lactose on the fusion protein proLL37-luciferase induction by supplementation of exogenous mevalonate or cholesterol, suggesting a cholesterol-independent effect of statins on CAMP gene expression. Although further experiments are needed for exclusion of a cholesterol-independent effect on LL-37 expression, there are reports on cholesterol-independent effects of statins, e.g. reduction of inflammation and oxidative stress and the improvement of endothelial function [40]. In the case of induction by lactose and the inhibition thereof by simvastatin, there are discrepancies between the different methods used, e.g. RT-PCR versus the luciferase assay. As discussed earlier, we believe this to be dependent on the reporter protein turnover and the stability of the mRNA. Greater reduction probably occurs over time rather than at the specific time point of 24 h.
Concluding Remarks
We used label-free detection mass spectrometry coupled with a bioinformatic tool in order to study the regulatory pathways involved in PBA and/or lactose induction of the CAMP gene. We identified three novel pathways (TR/RXR activation, eicosanoid and steroid biosynthesis) involved in the regulation of the CAMP gene. T3, a hormone and a drug used for the treatment of hypothyroidism, was shown to be a potent inducer of LL-37 in the presence of PBA/lactose. Thus, it is likely that an intact thyroid axis is required for the optimal expression and release of LL-37, especially in the gut where lactose and butyrate, an analogue of PBA, are commonly present. This finding could have implications for gastrointestinal immunity among patients with a dysfunctional thyroid gland. In contrast, common drugs such as COX-inhibitors and statins were shown to decrease LL-37 expression. This could possibly lead to a diminished inflammatory reaction or to an immunodeficient state with low LL-37 expression. Taken together, our novel experimental approach has revealed three pathways for LL-37 expression, which provide a framework for further functional studies of the relevance of AMPs in health and disease.
Supplementary Material
Supplementary data
Acknowledgements
The authors are supported by the Swedish Research Council (56X-1217-01-6 and 20854, a Linneus grant CERIC), The Swedish Strategic Foundation: RBd08-0014, Cancerfonden: CAN 2011/559, Karolinska Institutet, the Icelandic Centre for Research (RANNIS) and the University of Iceland Research Fund. Gudmundur H. Gudmundsson is a visiting scientist at Karolinska Institutet supported by the Wenner-Gren Foundation. We would like to extend our sincere gratitude to Marie Ståhlberg and Dr. Dorothea Rutishauser at PK/KI, Karolinska Institutet for their invaluable help in sample preparation, mass spectrometric analyses and expertise in the interpretation of mass spectrometric data.
References
- 1.Netea MG, Wijmenga C, O'Neill LA. Genetic variation in Toll-like receptors and disease susceptibility. Nat Immunol. 2012;13:535–542. doi: 10.1038/ni.2284. [DOI] [PubMed] [Google Scholar]
- 2.Salzman NH, Hung K, Haribhai D, Chu H, Karlsson-Sjoberg J, Amir E, Teggatz P, Barman M, Hayward M, Eastwood D, Stoel M, Zhou Y, Sodergren E, Weinstock GM, Bevins CL, Williams CB, Bos NA. Enteric defensins are essential regulators of intestinal microbial ecology. Nat Immunol. 2010;11:76–83. doi: 10.1038/ni.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bowdish DM, Davidson DJ, Lau YE, Lee K, Scott MG, Hancock RE. Impact of ll-37 on anti-infective immunity. J Leukoc Biol. 2005;77:451–459. doi: 10.1189/jlb.0704380. [DOI] [PubMed] [Google Scholar]
- 4.Heilborn JD, Nilsson MF, Kratz G, Weber G, Sorensen O, Borregaard N, Stahle-Backdahl M. The cathelicidin anti-microbial peptide ll-37 is involved in re-epithelialization of human skin wounds and is lacking in chronic ulcer epithelium. J Invest Dermatol. 2003;120:379–389. doi: 10.1046/j.1523-1747.2003.12069.x. [DOI] [PubMed] [Google Scholar]
- 5.van der Does AM, Bergman P, Agerberth B, Lindbom L. Induction of the human cathelicidin LL-37 as a novel treatment against bacterial infections. J Leukocyte Biol. 2012;92:735–742. doi: 10.1189/jlb.0412178. [DOI] [PubMed] [Google Scholar]
- 6.da Silva FP, Machado MCC. Antimicrobial peptides: clinical relevance and therapeutic implications. Peptides. 2012;36:308–314. doi: 10.1016/j.peptides.2012.05.014. [DOI] [PubMed] [Google Scholar]
- 7.Raqib R, Sarker P, Bergman P, Ara G, Lindh M, Sack DA, Nasirul Islam KM, Gudmundsson GH, Andersson J, Agerberth B. Improved outcome in shigellosis associated with butyrate induction of an endogenous peptide antibiotic. Proc Natl Acad Sci USA. 2006;103:9178–9183. doi: 10.1073/pnas.0602888103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sarker P, Ahmed S, Tiash S, Rekha RS, Stromberg R, Andersson J, Bergman P, Gudmundsson GH, Agerberth B, Raqib R. Phenylbutyrate counteracts shigella mediated downregulation of cathelicidin in rabbit lung and intestinal epithelia: a potential therapeutic strategy. PLoS One. 2011;6:e20637. doi: 10.1371/journal.pone.0020637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu PT, Stenger S, Tang DH, Modlin RL. Cutting edge: vitamin D-mediated human antimicrobial activity against mycobacterium tuberculosis is dependent on the induction of cathelicidin. J Immunol. 2007;179:2060–2063. doi: 10.4049/jimmunol.179.4.2060. [DOI] [PubMed] [Google Scholar]
- 10.Martineau AR, Timms PM, Bothamley GH, Hanifa Y, Islam K, Claxton AP, Packe GE, Moore-Gillon JC, Darmalingam M, Davidson RN, Milburn HJ, Baker LV, Barker RD, Woodward NJ, Venton TR, Barnes KE, Mullett CJ, Coussens AK, Rutterford CM, Mein CA, Davies GR, Wilkinson RJ, Nikolayevskyy V, Drobniewski FA, Eldridge SM, Griffiths CJ. High-dose vitamin D(3) during intensive-phase antimicrobial treatment of pulmonary tuberculosis: a double-blind randomised controlled trial. Lancet. 2011;377:242–250. doi: 10.1016/S0140-6736(10)61889-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cederlund A, Kai-Larsen Y, Printz G, Yoshio H, Alvelius G, Lagercrantz H, Stromberg R, Jornvall H, Gudmundsson GH, Agerberth B. Lactose in human breast milk an inducer of innate immunity with implications for a role in intestinal homeostasis. PLoS One. 2013;8:e53876. doi: 10.1371/journal.pone.0053876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Termen S, Tollin M, Rodriguez E, Sveinsdottir SH, Johannesson B, Cederlund A, Sjovall J, Agerberth B, Gudmundsson GH. Pu.1 and bacterial metabolites regulate the human gene camp encoding antimicrobial peptide LL-37 in colon epithelial cells. Mol Immunol. 2008;45:3947–3955. doi: 10.1016/j.molimm.2008.06.020. [DOI] [PubMed] [Google Scholar]
- 13.Wang TT, Nestel FP, Bourdeau V, Nagai Y, Wang Q, Liao J, Tavera-Mendoza L, Lin R, Hanrahan JW, Mader S, White JH. Cutting edge: 1,25-dihydroxyvitamin D3 is a direct inducer of antimicrobial peptide gene expression. J Immunol. 2004;173:2909–2912. doi: 10.4049/jimmunol.173.5.2909. [DOI] [PubMed] [Google Scholar]
- 14.Verbeek W, Lekstrom-Himes J, Park DJ, Dang PM, Vuong PT, Kawano S, Babior BM, Xanthopoulos K, Koeffler HP. Myeloid transcription factor C/EBPepsilon is involved in the positive regulation of lactoferrin gene expression in neutrophils. Blood. 1999;94:3141–3150. [PubMed] [Google Scholar]
- 15.Nylen F, Miraglia E, Cederlund A, Ottosson H, Stromberg R, Gudmundsson GH, Agerberth B. Boosting innate immunity: development and validation of a cell-based screening assay to identify LL-37 inducers. Innate Immun. 2013 doi: 10.1177/1753425913493338. E-pub ahead of print. [DOI] [PubMed] [Google Scholar]
- 16.Cederlund A, Agerberth B, Bergman P. Specificity in killing pathogens is mediated by distinct repertoires of human neutrophil peptides. J Innate Immun. 2010;2:508–521. doi: 10.1159/000317665. [DOI] [PubMed] [Google Scholar]
- 17.Lyutvinskiy Y, Yang H, Rutishauser D, Zubarev RA. In silico instrumental response correction improves precision of label-free proteomics and accuracy of proteomics-based predictive models. Mol Cell Proteomics. 2013;12:2324–2331. doi: 10.1074/mcp.O112.023804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rajagopalan D, Agarwal P. Inferring pathways from gene lists using a literature-derived network of biological relationships. Bioinformatics. 2005;21:788–793. doi: 10.1093/bioinformatics/bti069. [DOI] [PubMed] [Google Scholar]
- 19.De Vito P, Incerpi S, Pedersen JZ, Luly P, Davis FB, Davis PJ. Thyroid hormones as modulators of immune activities at the cellular level. Thyroid. 2011;21:879–890. doi: 10.1089/thy.2010.0429. [DOI] [PubMed] [Google Scholar]
- 20.Funk CD. Prostaglandins and leukotrienes: advances in eicosanoid biology. Science. 2001;294:1871–1875. doi: 10.1126/science.294.5548.1871. [DOI] [PubMed] [Google Scholar]
- 21.Lio YC, Reynolds LJ, Balsinde J, Dennis EA. Irreversible inhibition of Ca(2+)-independent phospholipase A2 by methyl arachidonyl fluorophosphonate. Biochim Biophys Acta. 1996;1302:55–60. doi: 10.1016/0005-2760(96)00002-1. [DOI] [PubMed] [Google Scholar]
- 22.Dallob A, Hawkey CJ, Greenberg H, Wight N, De Schepper P, Waldman S, Wong P, DeTora L, Gertz B, Agrawal N, Wagner J, Gottesdiener K. Characterization of etoricoxib, a novel, selective COX-2 inhibitor. J Clin Pharmacol. 2003;43:573–585. [PubMed] [Google Scholar]
- 23.Dixon RA, Diehl RE, Opas E, Rands E, Vickers PJ, Evans JF, Gillard JW, Miller DK. Requirement of a 5-lipoxygenase-activating protein for leukotriene synthesis. Nature. 1990;343:282–284. doi: 10.1038/343282a0. [DOI] [PubMed] [Google Scholar]
- 24.Wright JM, Zeitlin PL, Cebotaru L, Guggino SE, Guggino WB. Gene expression profile analysis of 4-phenylbutyrate treatment of IB3-1 bronchial epithelial cell line demonstrates a major influence on heat-shock proteins. Physiol Genomics. 2004;16:204–211. doi: 10.1152/physiolgenomics.00160.2003. [DOI] [PubMed] [Google Scholar]
- 25.EMBL-EBI: Rex - retrieval of experiments 2686, 2012, 2012
- 26.Luthje P, Brauner H, Ramos NL, Ovregaard A, Glaser R, Hirschberg AL, Aspenstrom P, Brauner A. Estrogen supports urothelial defense mechanisms. Sci Transl Med. 2013;5:190ra180. doi: 10.1126/scitranslmed.3005574. [DOI] [PubMed] [Google Scholar]
- 27.Brucker-Davis F, Skarulis MC, Grace MB, Benichou J, Hauser P, Wiggs E, Weintraub BD. Genetic and clinical features of 42 kindreds with resistance to thyroid hormone. The National Institutes of Health Prospective Study. Ann Intern Med. 1995;123:572–583. doi: 10.7326/0003-4819-123-8-199510150-00002. [DOI] [PubMed] [Google Scholar]
- 28.Sandler B, Webb P, Apriletti JW, Huber BR, Togashi M, Cunha Lima ST, Juric S, Nilsson S, Wagner R, Fletterick RJ, Baxter JD. Thyroxine-thyroid hormone receptor interactions. J Biol Chem. 2004;279:55801–55808. doi: 10.1074/jbc.M410124200. [DOI] [PubMed] [Google Scholar]
- 29.Lee JK, Gordon PR, Stall GM, Gilchrest BA, Kaplan MM. Phenolic and tyrosyl ring iodothyronine deiodination by the caco-2 human colon carcinoma cell line. Metabolism. 1989;38:1154–1161. doi: 10.1016/0026-0495(89)90151-0. [DOI] [PubMed] [Google Scholar]
- 30.Velasco LF, Togashi M, Walfish PG, Pessanha RP, Moura FN, Barra GB, Nguyen P, Rebong R, Yuan C, Simeoni LA, Ribeiro RC, Baxter JD, Webb P, Neves FA. Thyroid hormone response element organization dictates the composition of active receptor. J Biol Chem. 2007;282:12458–12466. doi: 10.1074/jbc.M610700200. [DOI] [PubMed] [Google Scholar]
- 31.Meng S, Wu JT, Archer SY, Hodin RA. Short-chain fatty acids and thyroid hormone interact in regulating enterocyte gene transcription. Surgery. 1999;126:293–298. [PubMed] [Google Scholar]
- 32.Tanaka Y, Bush KK, Klauck TM, Higgins PJ. Enhancement of butyrate-induced differentiation of HT-29 human colon carcinoma cells by 1,25-dihydroxyvitamin D3. Biochem Pharmacol. 1989;38:3859–3865. doi: 10.1016/0006-2952(89)90596-0. [DOI] [PubMed] [Google Scholar]
- 33.Lu X, Walker T, MacManus JP, Seligy VL. Differentiation of HT-29 human colonic adenocarcinoma cells correlates with increased expression of mitochondrial RNA: effects of trehalose on cell growth and maturation. Cancer Res. 1992;52:3718–3725. [PubMed] [Google Scholar]
- 34.Schott J, Stoecklin G. Networks controlling mRNA decay in the immune system. Wiley Interdiscip Rev RNA. 2010;1:432–456. doi: 10.1002/wrna.13. [DOI] [PubMed] [Google Scholar]
- 35.Li D, Yamada T, Wang F, Vulin AI, Samuels HH. Novel roles of retinoid X receptor (RXR) and RXR ligand in dynamically modulating the activity of the thyroid hormone receptor/RXR heterodimer. J Biol Chem. 2004;279:7427–7437. doi: 10.1074/jbc.M311596200. [DOI] [PubMed] [Google Scholar]
- 36.Stehlin-Gaon C, Willmann D, Zeyer D, Sanglier S, Van Dorsselaer A, Renaud JP, Moras D, Schule R. All-trans retinoic acid is a ligand for the orphan nuclear receptor ROR beta. Nat Struct Biol. 2003;10:820–825. doi: 10.1038/nsb979. [DOI] [PubMed] [Google Scholar]
- 37.Wan M, Sabirsh A, Wetterholm A, Agerberth B, Haeggstrom JZ. Leukotriene b4 triggers release of the cathelicidin LL-37 from human neutrophils: novel lipid-peptide interactions in innate immune responses. FASEB J. 2007;21:2897–2905. doi: 10.1096/fj.06-7974com. [DOI] [PubMed] [Google Scholar]
- 38.Grosch S, Maier TJ, Schiffmann S, Geisslinger G. Cyclooxygenase-2 (COX-2)-independent anticarcinogenic effects of selective COX-2 inhibitors. J Natl Cancer Inst. 2006;98:736–747. doi: 10.1093/jnci/djj206. [DOI] [PubMed] [Google Scholar]
- 39.Chen WS, Wei SJ, Liu JM, Hsiao M, Kou-Lin J, Yang WK. Tumor invasiveness and liver metastasis of colon cancer cells correlated with cyclooxygenase-2 (COX-2) expression and inhibited by a COX-2-selective inhibitor, etodolac. Int J Cancer. 2001;91:894–899. doi: 10.1002/1097-0215(200102)9999:9999<894::aid-ijc1146>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 40.Liao JK. Clinical implications for statin pleiotropy. Curr Opin Lipidol. 2005;16:624–629. doi: 10.1097/01.mol.0000191913.16321.60. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data