Abstract
The concept that macrophages play an active role in inflammatory responses began its development in the late 1800s with the now iconic studies by Elie Metchnikoff using starfish larvae and Daphnia [reviewed in Kaufmann SHE: Nat Immunol 2008;9:705–712 and Cavaillon JM: J Leukoc Biol 2011;90:413–424]. Based on his observation of the phagocyte response to a foreign body (rose thorn) and yeast, he proposed that phagocytes acted in host defense and were active participants in the inflammatory process. Flash forward more than 100 years and we find that these basic tenets hold true. However, it is now appreciated that macrophages come in many different flavors and can adopt a variety of nuanced phenotypes depending on the tissue environment in which the macrophage is found. In this brief review, we discuss the role of one type of macrophage termed the alternatively activated macrophage (AAM), also known as the M2 type of macrophage, in regulating allergic lung inflammation and asthma. Recent studies using mouse models of allergic lung inflammation and samples from human asthma patients contribute to the emerging concept that AAMs are not just bystanders of the interleukin (IL)-4- and IL-13-rich environment found in allergic asthma but are also active players in orchestrating allergic lung disease.
Keywords: Interleukin-4, Interleukin-13, Macrophages, Asthma
Introduction
Asthma is a complex disease characterized by chronic inflammation of the respiratory tract leading to airway hyperreactivity and obstruction [1]. Asthma can be classified as atopic (allergic) or nonatopic (nonallergic). The prevalence of atopy and asthma has increased dramatically in adults and children over the past few decades, with estimates that as many as 300 million people are affected by this disease worldwide [2]. There is a disparity in the prevalence of asthma across the world, with developed countries having higher rates of asthma. These differences may be attributable to differences in genetic makeup, lifestyle and environmental factors.
Exposure to allergens, tobacco smoke or other molecules can trigger narrowing of airways and excessive production of mucus by lung epithelial cells. Patients then develop intermittent episodes of wheezing, shortness of breath and coughing [1]. Repeated exposures can lead to airway remodeling. There are rodent models of asthma that mimic many of the pathologies observed in humans, including allergic lung inflammation. Extensive studies in mouse models have shown that inhaled allergens activate epithelial cells, dendritic cells, macrophages, basophils, mast cells and eosinophils. This causes priming of allergen-specific CD4+ T helper cells (TH2 cells), which then produce cytokines such as interleukin-4 (IL-4), IL-13 and IL-5. These cytokines act on multiple cell types to initiate and propagate the characteristic symptoms of asthma mentioned above.
Alternatively Activated Macrophages
Cells of the monocyte-macrophage lineage are able to adopt markedly distinct morphological and functional characteristics depending on the nature of their extracellular environment [3,4,5,6,7,8]. In the presence of IFNγ and lipopolysaccharide, macrophages adopt a potent proinflammatory phenotype and acquire the ability to efficiently kill intracellular bacteria [reviewed in [6,7]]. These cells are characterized by the expression of inducible nitric oxide synthase and the classic proinflammatory cytokines and chemokines (IL-1β, IL-12, TNFα, IP10). Since these were the first type of activated macrophages to be described, they were termed ‘lassically activated’ macrophages (CAM) or M1. These CAM are found under TH1 conditions during immune responses. However, under TH2 conditions in the presence of IL-4 or IL-13, macrophages adopt a different or ‘alternative’ phenotype. These cells, termed alternatively activated macrophages (AAMs) or M2, were first described in 1992 [9,10]. These cells do not express potent proinflammatory mediators; in fact, the expression of inflammatory cytokines is suppressed. Instead, these AAMs express a different set of genes including increased levels of the macrophage mannose receptor (mrc1), arginase 1 (Arg1), resistin-like molecule-α/found in inflammatory zone 1 (FIZZ1; Retnla), chitinase and chitinase-like family members such as acidic mammalian chitinase (AMCase; Chia) and YM1/2 (Chi3L3/4) and a number of fibrotic factors [7,11,12,13]. Furthermore, these AAMs can fuse to form multinucleated giant cells that act to sequester and degrade foreign materials [14]. In the presence of immune complexes, macrophages become regulatory cells producing high levels of IL-10 [6]. During complex inflammatory responses in vivo, there may be a mixture of these stimulatory agents, leading to the presence of macrophages with mixed phenotypes [6].
In addition to being phenotypically different from other types of macrophages, AAMs are functionally distinct. Several reports have shown that AAMs protect the host against parasitic pathogens [15,16] and also decrease inflammation at the site of injury, reducing the pathology observed in response to inhaled pathogens [17,18]. They also play an important role in wound healing and tissue repair processes. However, AAMs are substantially less effective at killing intracellular bacteria than CAM, a phenotype taken advantage of by Francisella tularensis to enhance its survival [17].
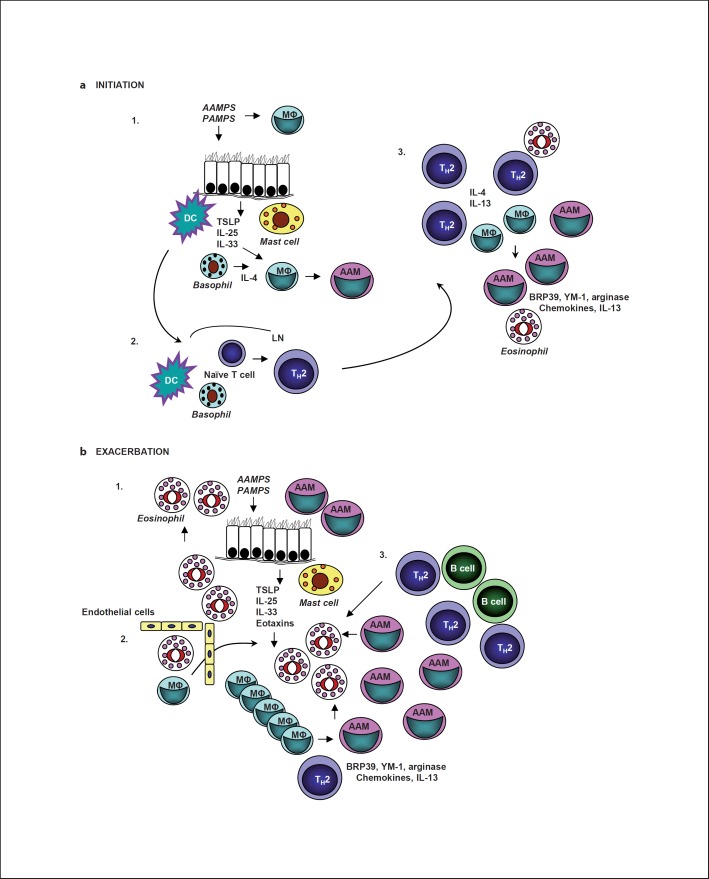
During an immune response, both IL-4 and IL-13 can be made by a number of different cell types. While it is well known that IL-4 and IL-13 are produced by antigen-specific TH2 lymphocytes, they can also be produced by cells of the innate immune system [19]. Pathogen-associated molecular patterns (PAMPS) and patterns found in innocuous allergens (allergen-associated molecular patterns, AAMPS) activate cells through pattern recognition receptors (PRRs). Stimulation of PRRs on epithelial cells or alveolar macrophages can induce the production of innate cytokines such as IL-25, IL-33 and thymic stromal lymphopoietin (TSLP) [20]. Mast cells, basophils, eosinophils, macrophages and the innate lymphoid-like cell (or nuocyte) have been shown to produce IL-4 or IL-13 after stimulation by PAMPS, AAMPS or IL-33 [17,18,19,20,21]. Early production of IL-4 and IL-13 by innate cells is thought to be important for the initiation phase of the TH2-dominant allergic inflammatory response (fig. 1a).
Fig. 1.
Possible roles for AAMs in asthma initiation and exacerbation. a Initiation. In step 1, molecular patterns found in pathogens (PAMPS) or allergens (AAMPS) stimulate PRRs expressed on lung epithelial cells and alveolar macrophages. These cells then make cytokines, including IL-25, IL-33 and TSLP, that stimulate cells of the innate immune system. These cytokines can activate dendritic cells (DC) and stimulate granuloctyes (mast cells and basophils) and innate lymphoid cells (not shown) to produce IL-4 or IL-13. IL-4 or IL-13 in combination with IL-33 drive AAM differentiation. In step 2, activated dendritic cells migrate to the draining lymph node (LN) and with the help of an IL-4-producing cell such as a basophil stimulate naive CD4+ T cells to become TH2 effectors or memory cells. In step 3, TH2 effectors migrate to the site of inflammation producing high levels of IL-4, IL-5 and IL-13. These cytokines mediate mucus secretion, AHR, chemokine production, eosinophilic infiltration and further AAM differentiation. b Exacerbation. In step 1, PAMPS or AAMPS stimulate PRRs expressed on lung epithelial cells and alveolar macrophages, leading to production of IL-25, IL-33 and TSLP. These cytokines stimulate granuloctyes (mast cells, basophils, eosinophils) and innate lymphoid cells (not shown) to produce IL-4 or IL-13. IL-4 or IL-13 in combination with IL-33 drive additional AAM differentiation. In step 2, lung epithelial cells and AAMs produce chemokines and other factors that recruit more inflammatory cells from the blood including eosinophils and monocytes. Increased numbers of macrophages in the lung tissue may also come from proliferation of interstitial macrophages as has been noted during parasitic infections [86]. In step 3, allergen-specific memory TH2 cells migrate to the site of inflammation, producing high levels of IL-4, IL-5 and IL-13. These cytokines mediate mucus secretion, AHR, chemokine production, eosinophilic infiltration and further AAM differentiation, setting up a positive amplification loop. MΦ = Macrophage.
The biologic responses to IL-4 and IL-13, including the alternative activation of macrophages, are mediated by high-affinity cell surface receptor complexes consisting of a 140-kDa, high-affinity binding chain (IL-4 receptor α, IL-4Rα) and the common γ-chain (γc), which is also a component of the receptors for IL-2, IL-7, IL-9, IL-15 and IL-21 [22,23]. This complex is called the type I IL-4R. The IL-4Rα can also pair with the IL-13Rα1 chain, forming the type II receptor [24]. IL-4 can bind to and activate both receptor types, while IL-13 only engages the type II receptor. Macrophages express both types of receptors and thus will respond to both IL-4 and IL-13 [25]. Once the cytokines bind to their respective receptor, they activate two different signaling pathways, the signal transducer and activator of transcription (STAT) 6 and insulin receptor substrate (IRS) 2 pathways [22]. STAT6 is necessary for the alternative activation of macrophages in response to IL-4 or IL-13 in vitro [26]. Further studies have shown that AAM gene/protein expression in vivo is also dependent on the IL-4Rα chain and STAT6. Loss of STAT6 signaling caused a significant reduction in Retnla and Chi3L3 mRNA levels in bleomycin-induced lung fibrosis and allergic peritonitis models, respectively [27,28]. Using a murine model of asthma, we found that the number of AAMs recruited to the lungs in mice lacking STAT6 or IL-4Rα was much lower when compared to wild-type (WT) mice [29]. It has also been demonstrated that promoter regions of the Arg1, Retnla and Chi3L3 genes have consensus STAT6-binding sites [28,30,31]. Binding of both STAT6 and CCAAT enhancer-binding protein to the promoter was required for optimal Retnla and Arg1 expression [30,31].
While IL-4 and IL-13 produce many similar biological effects on a variety of cell types due to the sharing of the IL-4Rα chain and the STAT6 pathway, responses to these cytokines can differ in potency and their relative contribution to the different phases of an allergic immune response. We compared IL-4 and IL-13 signaling in primary bone marrow (BM)-derived macrophages (BMMs) from WT or γc−/− mice in vitro [25]. We found that IL-4 was able to strongly activate the phosphorylation of IRS2 in WT but not γc−/− macrophages. The IRS2 from IL-4-treated WT BMMs, but not from γc−/− BMMs, was able to coprecipitate with growth factor receptor-bound protein 2 and strongly recruit the p85 subunit of phosphatidylinositol 3′ kinase (PI-3K). The phosphorylation of STAT6 was induced equally by IL-4 and IL-13 in both types of macrophages. IL-13 failed to substantially induce IRS2 phosphorylation in either. Taken together, these data demonstrate that IL-4 signaling through the type I receptor is more efficient at inducing the activation of the IRS2 pathway and that this function is mediated via the γc. We further analyzed whether the signaling differences had an impact on biological responses to IL-4 and IL-13 in macrophages. Using primary BMMs derived from WT mice, we found that IL-4 is dramatically more potent than IL-13 in inducing a subset of AAM genes (Arg1, Retnla, Chi3L3), even using concentrations of cytokine that elicited similar levels of STAT6 activation. IRS2 activation leads to the activation of PI-3K, suggesting that PI-3K is important for AAM differentiation. Indeed, it was shown that inhibitors of PI-3K suppressed alternative activation and that loss of Src homology 2 domain-containing inositol phosphatase (SHIP), a phosphatidylinositol phosphatase, led to enhanced AAM gene expression in vitro [32,33]. Furthermore, SHIP−/− mice had increased numbers of AAMs in vivo with accumulation of YM1 crystals [33]. Taken together, these results indicate that while AAM gene expression is STAT6 dependent, efficient expression may also be influenced by additional signaling pathways.
Intriguingly, mutation of an immunoreceptor tyrosine-based inhibition motif in the IL-4Rα chain shown to interact with Src homology 2 domain-containing protein tyrosine phosphatase (SHP)-1, SHP-2 and SHIP resulted in increased sensitivity of macrophages to IL-13-mediated AAM activation [34]. The Y709 (WT) BMMs treated with IL-4 led to significantly higher expression of AAM genes (Arg1, Chi3L3) and also Ccl11 than IL-13. Mutation of Y709 residue to F709 resulted in enhanced STAT6 phosphorylation and led to a dramatic amplification of Arg1, Chi3L3 and Ccl11 genes in response to IL-13, while leaving the IL-4-induced responses intact or slightly enhanced [34]. The authors proposed that differential recruitment of SHP-1 by the type I and type II receptors may be the reason for the disproportionate increase in AAM activation induced by IL-13.
The relative role of IL-4 and IL-13 in AAM formation was also evaluated in vivo. Analyses of AAM gene expression in IL-13Rα1−/− mice have indicated that several AAM genes were differentially regulated by IL-4 and IL-13. DNA microarray studies revealed that allergen and IL-4 stimulation of WT or IL-13Rα1−/− mice induced similar Retnla (FIZZ1) expression, but induction of Chia (AMCase) was completely dependent on IL-13Rα1 (type II receptor) [35]. Interestingly, allergen-induced Arg1 expression required the type II receptor, but IL-4-induced Arg1 expression did not. Thus, it appears that IL-4 utilizes both the type I and type II receptors to induce these genes in the lung.
Evidence suggests that there is cross-talk between the IL-4/IL-13 pathways and other signaling pathways in macrophages. Apart from STAT6 and CCAAT enhancer-binding protein, other transcription factors such as peroxisome proliferator-activated receptors δ and γ are required for maturation of AAMs and maximal gene expression [36,37]. IL-33, a member of the IL-1 family of cytokines, can also enhance IL-13-induced alternative activation of macrophages in an allergen-induced asthma model in mice [38]. These authors also showed that IL-33 amplified Arg1,Chi3L3 and chemokine production by AAMs in vitro [38].
Role of AAMs in Asthma Pathogenesis
Over the past few years, the contribution of AAMs to TH2 inflammation has been evaluated. Many studies have indicated a role for AAMs in the immune response to helminth infections [39,40,41]. The functions of AAMs in mouse models and in human asthma are just now being explored. Recent advances in this field suggest that AAMs play an active role in exacerbating TH2 responses, inducing eosinophilic inflammation, tissue remodeling and fibrosis, and airway hyperresponsiveness (AHR). However, their role in the initiation and establishment of allergic lung inflammation is less clear. In this section, we focus on our current understanding of how AAMs may participate in asthma pathogenesis.
Studies in parasite infection models were the first to suggest an active role for AAMs in a TH2-driven inflammatory response. Heligmosomoides polygyrus infection of mice elicited a TH2 response, and AAMs accumulated at the site of inflammation in response to IL-4 [39]. The presence of AAMs also correlated with recruitment of eosinophils to the lung and peritoneal cavity in mice infected with Nippostrongylus brasiliensis[40]. A reduction in macrophage numbers by clodronate depletion resulted in reduced eosinophilia in both models. Several strategies to delete the IL-4Rα simultaneously on monocytic and granulocytic cells also suggested a role for AAMs in mediating eosinophilic inflammation in parasite infection models [41].
AAM formation during allergic lung inflammation and the role of AAMs in this response is still unclear. Several studies demonstrated a positive correlation between the presence of macrophages and enhanced eosinophilic inflammation using allergic models in mouse and man [42,43,44,45,46,47,48]. A recent study showed that AAMs contribute to the enhanced asthma response observed in female mice as compared to male mice [45]. Female mice had greater numbers of AAMs in lung tissue than male mice and showed enhanced allergic lung inflammation. Adoptive transfer of in vitro differentiated AAMs by intranasal instillation enhanced allergic lung inflammation observed in male mice. In support of these results, we have found that adoptive transfer of IL-4Rα+/+ BMMs by intraperitoneal injection, but not IL-4Rα−/− macrophages, was sufficient to enhance TH2-dependent, ovalbumin-induced allergic lung inflammation [49]. A separate study analyzing the impact of IL-33 on AAM differentiation in vivo also established a correlation between AAMs and eosinophilia in a mouse model of allergic lung inflammation [44]. Furthermore, infection of mice with Sendai virus was shown to induce production of IL-13 by natural killer T (NKT) cells and by alveolar and interstitial macrophages [43]. This IL-13 then stimulated AAM differentiation in the lung; the AAMs were detected up to 49 days following infection. The increase in AAMs was associated with enhanced mucus production and AHR.
Studies in patients also suggest a role for AAMs in human asthma. Asthmatics have greater numbers of macrophages in their lungs as compared to healthy controls [48,50]. Furthermore, asthmatics have increased levels of chitinase family members in their blood and lung lavage fluid [51]. Interestingly, single-nucleotide polymorphisms found in Chi3L1 (YKL40), Chia (AMCase) and mrc1 (mannose receptor) have been associated with asthma [52,53,54]. Peripheral blood monocytes isolated from children undergoing viral-induced asthma exacerbations had elevated expression of AAM signature genes, including mrc1 encoding the mannose receptor [46]. Kim et al. [43] reported increased numbers of IL-13+ macrophages in bronchoalveolar lavage samples from asthma patients as compared to healthy controls. Most recently, immunohistochemistry of biopsy specimens taken from the lungs of asthma patients and healthy controls demonstrated an increase in AAMs (defined by expression of mannose receptor, CD206 and stabilin-1) in the lungs of asthmatic patients [48]. The percentage of AAMs in the lung tissue correlated with peak expiratory flow variation, suggesting that AAMs may regulate asthma severity. Taken together, results from mouse models of allergic lung inflammation and studies in human asthma patients show that AAMs are important, active contributors to TH2-driven inflammation and are not just bystander cells responding to the TH2 cytokines.
While the evidence for an active role for AAMs in exacerbation of TH2-driven inflammation is mounting, a potential role of AAMs in the initiation phase is less clear. The innate defense system in the lung includes several macrophage populations that reside in distinct environments [55]. The alveolar macrophage is located outside of the epithelial barrier in the airway lumen and regularly comes in direct contact with inhaled agents. Tissue resident macrophages in the lung parenchyma, including interstitial macrophages, are located on the basolateral side of the epithelial barrier. During inflammatory responses, additional monocytes can be recruited from the circulation into the lung tissue and airways. It is not known which type of macrophage actually becomes an AAM in models of lung inflammation, although they all have the potential to respond to IL-4 and IL-13.
Alveolar macrophages are thought to have a somewhat distinct phenotype. They have elevated phagocytic capacity and lower expression of MHC class II molecules, CD80 and CD86 as compared to other macrophage populations [55]. Using strategies to deplete alveolar macrophages and adoptive transfer approaches, several groups have shown that alveolar macrophages can be suppressive for allergic lung inflammation [56]. Interstitial macrophages have also been shown to suppress allergic airway responses to harmless allergens by producing IL-10 and inhibiting dendritic cell activation and migration to lymph nodes [57]. However, it is possible that certain types of PAMPS or AAMPS can shift this suppressive activity towards a promoting activity (fig. 1a).
Several respiratory pathogens and allergens have been shown to induce the alternative activation of alveolar macrophages to AAMs, including respiratory syncytial virus [18], Sendai virus [43], Aspergillus fumigatus[58], influenza [59] and house dust mites [60]. These agents also act on epithelial cells via PRRs to induce production of IL-25, IL-33 and TSLP [20]. These cytokines then act on cells of the innate immune system, leading to the activation of dendritic cells and the production of IL-4 or IL-13 by mast cells, basophils, NKT cells or the newest kid on the block, the so-called innate lymphoid cells [43,61,62,63]. Regardless of the source, the IL-4/IL-13 could then further drive alveolar and interstitial macrophages to become AAMs. While the contribution of AAMs in the initiation phase has not yet been established, several murine studies provided evidence that AAMs can participate in the initiation/sensitization phase by enhancing activation of dendritic cells [45,60]. The activated dendritic cells would then migrate to the draining lymph node where they would present allergen to naive CD4+ T cells in a manner that would drive differentiation to the TH2 type. Upon further allergen exposure, the TH2 cells would populate the lung tissue, inducing a further increase in AAMs and stimulating eosinophilic inflammation, thereby establishing a type 2 inflammatory environment in the lung.
As described above, a role for AAMs in asthma exacerbations is supported by studies in mice and men (fig. 1b). Re-exposure to allergens after initiating and establishing the allergic inflammatory phenotype would lead to IgE receptor cross-linkage on mast cells and activation of TH2 memory cells. Enhanced production of IL-4, IL-5 and IL-13 would lead to enhanced mucus hypersecretion, AHR and AAM differentiation. Lung epithelial cells and AAMs would produce chemokines and other factors that would recruit more inflammatory cells from the blood, including eosinophils and monocytes, setting up an amplification cycle between BM-derived cells and the lung [47]. Children undergoing viral-induced asthma exacerbations had increased numbers of peripheral blood mononuclear cells with an AAM gene signature [46]. Two studies using viral infection of mice lend credence to this model. Sendai virus infection induced the production of IL-13 by NKT cells, which induced AAM differentiation; the IL-13+ AAMs were found in the lungs long after viral infection [43]. Influenza infection induced the production of IL-33, which acted on innate lymphoid cells in the lung to rapidly produce IL-13, thus stimulating mucus secretion by epithelial cells and AHR [64]; influenza infection also stimulated AAM differentiation of alveolar macrophages [59]. Taken together, these results suggest that repeated allergen sensitization and respiratory virus infection can set off an amplification cycle including AAMs that leads to the exacerbation of asthma symptoms.
Specific Contribution of AAM Products to Allergic Lung Inflammation
An important question arising from these studies is how the AAMs actively enhance the inflammatory response. IL-4/IL-13-stimulated mouse and human macrophages make a number of factors that may elicit effector functions, leading to exacerbation of symptoms. Production of IL-13 by AAMs would clearly elicit enhanced effector responses. However, the precise contribution of the many AAM products to allergic inflammation is not clear.
Arginase Family of Proteins
Arginase is an enzyme that competes with inducible nitric oxide synthase for L-arginine and converts it into ornithine and polyamines, in contrast to inducible nitric oxide synthase, which induces nitric oxide production [65]. There are two main isoforms of arginase, Arg1 and Arg2. Only Arg1 has STAT6 response elements in its promoter region and can respond to TH2 cytokines [66]. However, both Arg1 and Arg2 mRNA have been reported to be upregulated during asthma. Arg1 was STAT6 dependent but Arg2 was not [67]. Arg1 is mainly induced in murine macrophages. Although homologs exist, this protein is not expressed by human AAMs [68].
Arginase expression has been detected in several mouse models of helminth infections as well as asthma [[11,65,67], reviewed in [69]]. Several reports suggest that Arg1-induced production of proline and polyamines leads to collagen synthesis and development of fibrosis [70]. These processes may be harmful in the context of asthma and allergic inflammation but are required for wound healing [65]. Arg1 also helps protect against helminth infections by facilitating parasite killing and reducing pathology associated with TH2 inflammation [71,72]. Arg1 expression in macrophages suppresses TH2 type inflammation and fibrosis in Schistosoma-infected Arg1 conditional knockout mice [72].
Several groups have investigated the role of arginase in allergic lung inflammation with contrasting results. It is known that Arg1 is expressed in the lungs of asthmatic patients as well as mice [73]. Arg1 blockade using specific inhibitors led to reduced AHR and decreased airway eosinophilia, goblet cell hyperplasia, AHR and lung fibrosis [73,74] in a mouse and guinea pig model of chronic asthma, respectively. However, since Arg1 can be expressed by macrophages as well as epithelial cells, it was not clear which of these cell types was contributing to the asthma phenotype. Using a BM chimera approach, Niese et al. [75] reported that BM cells were the predominant source of Arg1. In spite of this, Arg1 expression by BM cells was not required to induce airway inflammation, AHR or collagen deposition.
FIZZ/Resistin-Like Molecule Family of Proteins
FIZZ proteins are a family of proteins secreted by AAMs [reviewed in [69]]. They are also named resistin-like molecules because of the sequence similarity with resistin, a protein that is secreted by adipocytes. Four different subtypes of FIZZ proteins have been characterized, FIZZ1–4. All these proteins have highly conserved C-terminal cysteine residues, which determine their tertiary structure. Murine FIZZ1, which is secreted by both AAM and epithelial cells, does not have a homolog in humans. However, human resistin is more closely related to mouse FIZZ1 than to mouse resistin. FIZZ2 is present in both mice and humans and is mainly expressed in the gut and lung [31,76]. Much less is known about FIZZ3 and FIZZ4. FIZZ1 and FIZZ3 are also hypothesized to play a role in insulin resistance and type 2 diabetes.
Of the four isoforms, FIZZ1 is most widely expressed. FIZZ1 was originally discovered in bronchoalveolar lavage fluid in a mouse model of asthma [77]. Elevated levels of FIZZ1 mRNA have since been detected in parasite infection models [69], allergic lung inflammation [28,29,77], allergic peritonitis [28], bleomycin-induced lung fibrosis [27] and hypoxia-induced pulmonary hypertension [78]. FIZZ1 has been reported to induce smooth muscle proliferation and actin and collagen deposition, causing fibrosis [79]. Similar to Arg1, FIZZ1 also been shown to have a protective role in helminth-induced immunopathology [80]. FIZZ1−/− mice infected with either Schistosoma mansoni or N. brasiliensis displayed significantly increased granulomatous inflammation together with fibrosis in the liver and severe lung pathology, respectively [80]. The contribution of FIZZ1 to asthma is still unclear. Our studies have shown that IL-4 induces robust induction of FIZZ1 mRNA in macrophages in vitro [25], but FIZZ1 protein expression in these cells was absent in vivo in a mouse model of asthma [29].
Chitinase and Chitinase-Like Mammalian Proteins
In addition to the arginase and FIZZ family members, AAMs also produce chitinases and chitinase-like proteins. Chitin, a polymer of N-acetlyglucosamine, is present in helminths, fungi, crustaceans and other lower organisms and is the substrate for the enzyme chitinase [81]. One such protein is AMCase, which is expressed in the stomach and to a lesser extent in the lung. Other members of this family in rodents include YM1, YM2 and breast regression protein (BRP)-39. These proteins lack chitinase activity. YM1/2 lack human homologs [68]; however, AMCase and YKL-40 (the human equivalent of BRP-39) are expressed by human cells [51,82]. A recent study suggests that human chitotriosidase 1 represents the human counterpart of mouse YM1/2 in chronic obstructive lung disease [83].
Induction of chitinase-like mammalian proteins is a characteristic feature of helminth infection as well as asthma and other inflammatory diseases [16,81]. It is hypothesized that chitinases bind and degrade chitin present in the cuticles of helminths. Although YM1 lacks chitinase function, it can bind to saccharides with a free amine group (such as heparin or N-glucosamine). Therefore, it has been postulated that YM1 may be involved in worm egg degradation and tissue repair. Chitinases have also been implicated in asthma. Zhu et al. [82] demonstrated that AMCase expression was greatly enhanced upon ovalbumin sensitization and challenge in mice and also in asthma patients. In both species, epithelial cells as well as macrophages expressed AMCase mRNA and protein. The authors further showed that administration of anti-AMCase antibodies to antigen-challenged or IL-13 transgenic mice led to a significant decrease in pulmonary inflammation, eosinophilia and AHR. Moreover, AMCase blockade also reduced secretion of the IL-13-induced chemokines CCL2 [monocyte chemotactic protein (MCP)-1], CCL8 (MCP-2) and CCL4 (macrophage inflammatory protein-1β) in the lung.
Another chitinase-like protein, BRP-39, was found to be highly expressed in alveolar macrophages and epithelial cells in a murine model of allergic lung inflammation [60]. Mice deficient in BRP-39 elicited reduced antigen-specific TH2 responses including IL-13-induced tissue inflammation and fibrosis. In addition, recombinant BRP-39 was found to directly regulate alternative activation of macrophages [60]. Stimulation of peritoneal and alveolar macrophages with recombinant BRP-39 in vitro led to enhanced Arg1 activity and mannose receptor expression. The contribution of YM1 in asthma is still unclear. YM1 can recruit eosinophils to the site of inflammation and has also been implicated in tissue remodeling and fibrosis [81]. Furthermore, YM1/2 has been shown to enhance TH2 cytokine production by T cells [84]. Allergen sensitization and challenge upregulated YM1 protein levels in both epithelial cells and macrophages in WT mice, which was absent in mice deficient in STAT6 [29].
Chemokines
In addition to these products, AAMs make a number of chemokines. IL-4/IL-13 stimulation of macrophages induces the production of eotaxin 1 and 2 (CCL11 and CCL24), which recruit eosinophils [85]. AAMs also produce CCL2 (MCP-1), CCL22 (macrophage-derived chemokine) and CCL17 (thymus and activation-regulated chemokine), which recruit macrophages and TH2 cells [7]. IL-4 stimulation of AAM has been shown to enhance the production of macrophage inflammatory protein-1α (CCL3), a granulocyte chemokine [85]. In humans, IL-4 stimulates production of CCL13, CCL14, CCL17, CCL18, CCL22 and CCL24 [reviewed in [7]]. These chemokines attract cells that express chemokine receptors CCR1–4 to the site of inflammation. Thus, AAMs produce a number of chemokines capable of enhancing recruitment of TH2 cells, eosinophils and macrophages to the lung.
Conclusion
The diverse states of macrophage activation play important roles in immune responses to pathogens and other agents. The chronic activation of macrophages has been linked to a variety of inflammatory diseases. Within the site of inflammation, macrophages can produce a range of cytokines, chemokines, growth factors and proteolytic enzymes that may participate in either tissue damage or tissue repair. The AAMs and their secreted products are induced by a number of pathogens and allergens. The presence of these AAMs in human airways and lungs has been correlated with asthma severity. Furthermore, in mouse models of allergic lung inflammation, AAMs have been shown to actively contribute to disease severity. Several reports further show that viral infection induces AAMs in the lung, which could impact on the subsequent enhanced responsiveness to allergens. Thus, AAMs and their products represent potential therapeutic targets for the treatment of asthma.
Disclosure Statement
This work was supported by US Public Health Service grant AI038985 to A.D.K.
References
- 1.Busse WW, Rosenwasser LJ. Mechanisms of asthma. J Allergy Clin Immunol. 2003;111((3 suppl)):S799–S804. doi: 10.1067/mai.2003.158. [DOI] [PubMed] [Google Scholar]
- 2.Global Initiative for Asthma: Global strategy for asthma management and prevention 2010 http://www.ginasthma.com. [Google Scholar]
- 3.Kaufmann SHE. Immunology's foundation: the 100 year anniversary of the Nobel prize to Paul Erlich and Elie Metchnikoff. Nat Immunol. 2008;9:705–712. doi: 10.1038/ni0708-705. [DOI] [PubMed] [Google Scholar]
- 4.Cavaillon JM. The historical milestones in the understanding of leukoctye biology initiated by Elie Metchnikoff. J Leukoc Biol. 2011;90:413–424. doi: 10.1189/jlb.0211094. [DOI] [PubMed] [Google Scholar]
- 5.Stout RD, Suttles J. Functional plasticity of macrophages: reversible adaptation to changing microenvironments. J Leukoc Biol. 2004;76:509–513. doi: 10.1189/jlb.0504272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat Rev Immunol. 2008;8:958–969. doi: 10.1038/nri2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martinez FO, Helming L, Gordon S. Alternative activation of macrophages: an immunologic functional perspective. Annu Rev Immunol. 2009;27:451–483. doi: 10.1146/annurev.immunol.021908.132532. [DOI] [PubMed] [Google Scholar]
- 8.Gordon S, Martinez FO. Alternative activation of macrophages: mechanism and function. Immunity. 2010;32:593–604. doi: 10.1016/j.immuni.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 9.Stein M, Keshav S, Harris N, Gordon S. IL-4 potently enhances murine macrophage mannose receptor activity: a marker of alternative immunologic macrophage activation. J Exp Med. 1992;176:287–292. doi: 10.1084/jem.176.1.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chensue SW, Terebuh PD, Warmington KS, Hershey SD, Evanoff HL, Kunkel SL, Higashi GI. Role of IL-4 and IFNγ in Schistosoma mansoni egg-induced hypersensitivity granuloma formation: orchestration, relative contribution, and relationship to macrophage function. J Immunol. 1992;148:900–914. [PubMed] [Google Scholar]
- 11.Loke P, Nair MG, Parkinson J, Guilano D, Blaxter M, Allen JE. IL-4 dependent alternatively activated macrophages have a distinct in vivo gene expression phenotype. BMC Immunol. 2002;3:7–17. doi: 10.1186/1471-2172-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nair MG, Cochrane DW, Allen JE. Macrophages in chronic type 2 inflammation have a novel phenotype characterized by the abundant expression of Ym1 and Fizz1 that can be partly replicated in vivo. Immunol Lett. 2003;85:173–177. doi: 10.1016/s0165-2478(02)00225-0. [DOI] [PubMed] [Google Scholar]
- 13.Sandler NG, Mentink-Kane MM, Cheever AW, Wynn TA. Global gene expression profiles during acute pathogen-induced pulmonary inflammation reveal divergent roles for Th1 and Th2 responses in tissue repair. J Immunol. 2003;171:3655–3667. doi: 10.4049/jimmunol.171.7.3655. [DOI] [PubMed] [Google Scholar]
- 14.McNally AK, Anderson JM, Interleukin-4 induces foreign body giant cells from human monocytes/macrophages Differential lymphokine regulation of macrophage fusion leads to morphological variants of multinucleated giant cells. Am J Pathol. 1995;147:1487–1499. [PMC free article] [PubMed] [Google Scholar]
- 15.Loke P, Gallagher I, Nair MG, Zang X, Brombacher F, Mohrs M, Allison JP, Allen JE. Alternative activation is an innate response to injury that requires CD4+ T-cells to be sustained during chronic infection. J Immunol. 2007;179:3926–3936. doi: 10.4049/jimmunol.179.6.3926. [DOI] [PubMed] [Google Scholar]
- 16.Anthony RM, Rutitzky LI, Urban JF, Jr, Stadecker MJ, Gause WC. Protective immune mechanisms in helminth infection. Nat Rev Immunol. 2007;7:975–987. doi: 10.1038/nri2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shirey KA, Cole LE, Keegan AD, Elkins KE, Vogel SN. Francisella tularensis LVS induces macrophage alternative activation as a survival mechanism. J Immunol. 2008;181:4159–4167. doi: 10.4049/jimmunol.181.6.4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shirey KA, Pletneva LM, Puche AC, Keegan AD, Prince GA, Blanco JCG, Vogel SN. Control of RSV-induced lung injury by alternatively activated macrophages is IL-4R alpha-, TLR4-, and IFN-beta-dependent. Mucosal Immunol. 2010;3:291–300. doi: 10.1038/mi.2010.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gessner A, Mohrs K, Mohrs M. Mast cells, basophils, and eosinophils acquire constitutive IL-4 and IL-13 transcripts during lineage differentiation that are sufficient for rapid cytokine production. J Immunol. 2005;174:1063–1072. doi: 10.4049/jimmunol.174.2.1063. [DOI] [PubMed] [Google Scholar]
- 20.Gregory LG, Lloyd CM. Orchestrating house dust mite-associated allergy in the lung. Trends Immunol. 2011;32:402–411. doi: 10.1016/j.it.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barlow JL, McKenzie AN. Nuocytes: expanding the innate cell repertoire in type-2 immunity. J Leukoc Biol. 2011;90:867–874. doi: 10.1189/jlb.0311160. [DOI] [PubMed] [Google Scholar]
- 22.Nelms K, Keegan AD, Zamorano J, Ryan JJ, Paul WE. The IL-4 receptor: signaling mechanisms and biologic functions. Annu Rev Immunol. 1999;17:701–738. doi: 10.1146/annurev.immunol.17.1.701. [DOI] [PubMed] [Google Scholar]
- 23.Leonard WJ. The molecular basis of X-linked severe combined immunodeficiency: defective cytokine receptor signaling. Annu Rev Med. 1996;47:229–239. doi: 10.1146/annurev.med.47.1.229. [DOI] [PubMed] [Google Scholar]
- 24.Callard RE, Matthews DJ, Hibbert L. IL-4 and IL-13 receptors: are they one and the same? Immunol Today. 1996;17:108–110. doi: 10.1016/0167-5699(96)80600-1. [DOI] [PubMed] [Google Scholar]
- 25.Heller NM, Qi X, Juntilla I, Shirey KA, Vogel SN, Paul WE, Keegan AD. Type I IL-4Rs selectively activate IRS-2 to induce target gene expression in macrophages. Sci Signal. 2008;1:ra17. doi: 10.1126/scisignal.1164795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moreno JM, Mikailenko I, Tondravi M, Keegan AD. IL-4 promotes the formation of functional multi-nucleated giant cells from macrophage precursors by a STAT6-dependent mechanism. J Leukoc Biol. 2007;82:1542–1553. doi: 10.1189/jlb.0107058. [DOI] [PubMed] [Google Scholar]
- 27.Liu T, Jin H, Ullenbruch M, Hu B, Hashimoto N, Moore B, McKenzie A, Lukacs NW, Phan SH. Regulation of found in inflammatory zone 1 expression in bleomycin-induced lung fibrosis: role of IL-4/IL-13 and mediation via STAT-6. J Immunol. 2004;173:3425–3431. doi: 10.4049/jimmunol.173.5.3425. [DOI] [PubMed] [Google Scholar]
- 28.Welch JS, Escoubet-Lozach L, Sykes DB, Liddiard K, Greaves DR, Glass DK. TH2 cytokines and allergic challenge induce Ym1 expression in macrophages by a STAT6-dependent mechanism. J Biol Chem. 2002;277:42821–42829. doi: 10.1074/jbc.M205873200. [DOI] [PubMed] [Google Scholar]
- 29.Dasgupta P, Chapoval SP, Smith EP, Keegan AD. Transfer of in vivo primed transgenic T cells supports allergic lung inflammation and FIZZ1 and Ym1 production in an IL-4Rα and STAT6 dependent manner. BMC Immunol. 2011;12:60. doi: 10.1186/1471-2172-12-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gray MF, Poljakovic M, Kepka-Lenhart D, Morris SM Jr. Induction of arginase I transcription by IL-4 requires a composite DNA response element for STAT6 and C/EBPbeta. Gene. 2005;353:98–106. doi: 10.1016/j.gene.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 31.Stutz AM, Pickart LA, Trifilieff A, Baumruker T, Prieschl-Strassmayr E, Woisetschlager M. The Th2 cell cytokines IL-4 and IL-13 regulate found in inflammatory zone 1/resistin-like molecule alpha gene expression by a STAT6 and CCAAT/enhancer-binding protein-dependent mechanism. J Immunol. 2003;170:1789–1796. doi: 10.4049/jimmunol.170.4.1789. [DOI] [PubMed] [Google Scholar]
- 32.Weisser SB, McLarren KW, Voglmaier N, van Netten-Thomas CJ, Antov A, Flavell RA, Sly LM. Alternative activation of macrophages by IL-4 requires SHIP degradation. Eur J Immunol. 2011;41:1742–1753. doi: 10.1002/eji.201041105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rauh MJ, Ho V, Pereira C, Sham A, Sly LM, Lam V, Huxham L, Minchinton AI, Mui A, Krystal G. SHIP represses the generation of alternatively-activated macrophages. Immunity. 2005;23:361–374. doi: 10.1016/j.immuni.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 34.Tachdjian R, Al Khatib S, Schwinglshackl A, Kim HS, Chen A, Blasioli J, Mathias C, Kim HY, Umetsu DT, Oettgen HC, Chatila TA. In vivo regulation of the allergic response by the IL-4 receptor alpha chain immunoreceptor tyrosine-based inhibitory motif. J Allergy Clin Immunol. 2010;125:1128–1136. doi: 10.1016/j.jaci.2010.01.054. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Munitz A, Brandt EB, Mingler M, Finkelman FD, Rothenberg ME. Distinct roles for IL-13 and IL-4 via IL-13 receptor alpha1 and the type II IL-4 receptor in asthma pathogenesis. Proc Natl Acad Sci USA. 2008;105:7240–7245. doi: 10.1073/pnas.0802465105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Odegaard JI, Ricardo-Gonzalez RR, Red Eagle A, Vats D, Morel CR, Goforth MH, Subramanian V, Mukundan L, Ferrante AW, Chawla A. Alternative M2 activation of Kupffer cells by PPARdelta ameliorates obesity-induced insulin resistance. Cell Metab. 2008;7:496–507. doi: 10.1016/j.cmet.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Odegaard JI, Ricardo-Gonzalez RR, Goforth MH, Morel CR, Subramanian V, Mukundan L, Red Eagle A, Vats D, Brombacher F, Ferrante AW, Chawla A. Macrophage-specific PPARgamma controls alternative activation and improves insulin resistance. Nature. 2007;447:1116–1120. doi: 10.1038/nature05894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kurowska-Stolarska M, Stolarski B, Kewin P, Murphy G, Corrigan CJ, Ying S, Pitman N, Mirchandani A, Rana B, van Rooijen N, Shepherd M, McSharry C, McInnes IB, Xu D, Liew FY. IL-33 amplifies the polarization of alternatively activated macrophages that contribute to airway inflammation. J Immunol. 2009;183:6469–6477. doi: 10.4049/jimmunol.0901575. [DOI] [PubMed] [Google Scholar]
- 39.Anthony RM, Urban JF Jr, Alem F, Hamed HA, Rozo CT, Boucher JL, Van Rooijen N, Gause WC. Memory TH2 cells induce alternatively activated macrophages to mediate protection against nematode parasites. Nat Med. 2006;12:955–960. doi: 10.1038/nm1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Voehringer D, van Rooijen N, Locksley RM. Eosinophils develop in distinct stages and are recruited to peripheral sites by alternatively activated macrophages. J Leukoc Biol. 2007;81:1434–1444. doi: 10.1189/jlb.1106686. [DOI] [PubMed] [Google Scholar]
- 41.Brombacher F, Arendse B, Peterson R, Holscher A, Holscher C. Analyzing classical and alternative macrophage activation in macrophage/neutrophil-specific IL-4Rα-deficient mice. Methods Mol Biol. 2009;531:225–252. doi: 10.1007/978-1-59745-396-7_15. [DOI] [PubMed] [Google Scholar]
- 42.Kelly-Welch AE, Melo MEF, Ford A, Noben-Trauth N, Smith E, Haudenschild C, Keegan AD. Complex role of the IL-4Rα in a murine model of airway inflammation: expression of the IL-4Rα on nonlymphoid cells of bone marrow origin contributes to severity of inflammation. J Immunol. 2004;172:4545–4555. doi: 10.4049/jimmunol.172.7.4545. [DOI] [PubMed] [Google Scholar]
- 43.Kim EY, Battaile JT, Patel AC, You Y, Agapov E, Holtzman MJ. Persistent activation of an innate immune response translates respiratory viral infection into chronic lung disease. Nat Med. 2008;14:633–640. doi: 10.1038/nm1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kurowska-Stolarska, M, Stolarski B, Kewin P, Murphy G, Corrigan CJ, Ying S, Pitman N, Mirchandani A, Rana B, van Rooijen N, Shepherd M, McSharry C, McInnes IB, Xu D, Liew FY. IL-33 amplifies the polarization of alternatively activated macrophages that contribute to airway inflammation. J Immunol. 2009;183:6469–6477. doi: 10.4049/jimmunol.0901575. [DOI] [PubMed] [Google Scholar]
- 45.Melgert BN, Oriss TB, Qi Z, Dixon-McCarthy B, Geerlings M, Hylkema MN, Ray A, Macrophages Regulators of sex differences in asthma? Am J Respir Cell Mol Biol. 2009;42:595–603. doi: 10.1165/rcmb.2009-0016OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Subrata LS, Bizzintino J, Mamessier E, Bosco A, McKenna KL, Wikstrom ME, Goldblatt J, Sly PD, Hales BJ, Thomas WR, Laing IA, LeSouëf PN, Holt PG. Interactions between innate antiviral and atopic immunoinflammatory pathways precipitate and sustain asthma exacerbations in children. J Immunol. 2009;183:2793–2800. doi: 10.4049/jimmunol.0900695. [DOI] [PubMed] [Google Scholar]
- 47.Holt PG, Strickland DH. Interactions between innate and adaptive immunity in asthma pathogenesis: new perspectives from studies on acute exacerbations. J Allergy Clin Immunol. 2010;125:963–972. doi: 10.1016/j.jaci.2010.02.011. [DOI] [PubMed] [Google Scholar]
- 48.Melgert BN, Hacken NH, Rutgers B, Timens W, Postma DS, Hylkema MN. More alternative activation of macrophages in lungs of asthmatic patients. J Allergy Clin Immunol. 2011;127:831–833. doi: 10.1016/j.jaci.2010.10.045. [DOI] [PubMed] [Google Scholar]
- 49.Ford AQ, Dasgupta P, Smith EP, Mikhailenko I, Noben-Trauth N, Keegan AD. Adoptive transfer of IL-4Rα+ macrophages is sufficient to support Th2-driven alternative macrophage activation and to enhance eosinophilic inflammation in a mouse model of allergic lung inflammation. BMC Immunol. 2012;13:6. doi: 10.1186/1471-2172-13-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Viksman MY, Bochner BS, Peebles RS, Schleimer RP, Liu MC. Expression of activation markers on alveolar macrophages in allergic asthmatics after endobronchial or whole-lung allergen challenge. Clin Immunol. 2002;104:77–85. doi: 10.1006/clim.2002.5233. [DOI] [PubMed] [Google Scholar]
- 51.Chupp GL, Lee CG, Jarjour N, Shim YM, Holm CT, He S, Dziura JD, Reed J, Coyle AJ, Kiener P, Cullen M, Grandsaigne M, Dombret MC, Aubier M, Pretolani M, Elias JA. A chitinase-like protein in the lung and circulation of patients with severe asthma. N Engl J Med. 2007;357:2016–2027. doi: 10.1056/NEJMoa073600. [DOI] [PubMed] [Google Scholar]
- 52.Ober C, Tan Z, Sun Y, Possick JD, Pan L, Nicolae R, Radford S, Parry RR, Heinzmann A, Deichmann KA, Lester LA, Gern JE, Lemanske RF, Jr, Nicolae DL, Elias JA, Chupp GL. Effect of variation in CHI3L1 on serum YKL-40 level, risk of asthma, and lung function. N Engl J Med. 2008;358:1682–1691. doi: 10.1056/NEJMoa0708801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bierbaum S, Nickel R, Koch A, Lau S, Deichmann KA, Wahn U, Superti-Furga A, Heinzmann A. Polymorphisms and haplotypes of acid mammalian chitinase are associated with bronchial asthma. Am J Respir Crit Care Med. 2005;172:1505–1509. doi: 10.1164/rccm.200506-890OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hattori T, Konno S, Hizawa N, Isada A, Takahashi A, Shimizu K, Shimizu K, Gao P, Beaty TH, Barnes KC, Huang SK, Nishimura M. Genetic variations in the mannose receptor gene (MRC1) are associated with asthma in two independent populations. Immunogenetics. 2009;61:731–738. doi: 10.1007/s00251-009-0403-x. [DOI] [PubMed] [Google Scholar]
- 55.Lohmann-Matthes ML, Steinmüller C, Franke-Ullmann G. Pulmonary macrophages. Eur Respir J. 1994;7:1678–1689. [PubMed] [Google Scholar]
- 56.Careau E, Bissonnette EY. Adoptive transfer of alveolar macrophages abrogates bronchial hyperresponsiveness. Am J Respir Cell Mol Biol. 2004;31:22–27. doi: 10.1165/rcmb.2003-0229OC. [DOI] [PubMed] [Google Scholar]
- 57.Bedoret D, Wallemacq H, Marichal T, Desmet C, Quesada Calvo F, Henry E, Closset R, Dewals B, Thielen C, Gustin P, de Leval L, Van Rooijen N, Le Moine A, Vanderplasschen A, Cataldo D, Drion PV, Moser M, Lekeux P, Bureau F. Lung interstitial macrophages alter dendritic cell functions to prevent airway allergy in mice. J Clin Invest. 2009;119:3723–3738. doi: 10.1172/JCI39717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bhatia S, Fei M, Yarlagadda M, Qi Z, Akira S, Saijo S, Iwakura Y, van Rooijen N, Gibson GA, St Croix CM, Ray A, Ray P. Rapid host defense against Aspergillus fumigatus involves alveolar macrophages with a predominance of alternatively activated phenotype. PLoS One. 2011;6:e15943. doi: 10.1371/journal.pone.0015943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen WH, Toapanta FR, Shirey KA, Zhang L, Giannelou A, Page C, Frieman MB, Vogel SN, Cross AS. Potential role for alternatively activated macrophages in the secondary bacterial infection during recovery from influenza. Immunol Lett. 2012;141:227–234. doi: 10.1016/j.imlet.2011.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee CG, Hartl D, Lee GR, Koller B, Matsuura H, Da Silva CA, Sohn MH, Cohn L, Homer RJ, Kozhich AA, Humbles A, Kearley J, Coyle A, Chupp G, Reed J, Flavell RA, Elias JA. Role of breast regression protein 39 (BRP-39)/chitinase 3-like-1 in Th2 and IL-13-induced tissue responses and apoptosis. J Exp Med. 2009;206:1149–1166. doi: 10.1084/jem.20081271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hammad H, Lambrecht BN. Dendritic cells and airway epithelial cells at the interface between innate and adaptive immune responses. Allergy. 2011;66:579–587. doi: 10.1111/j.1398-9995.2010.02528.x. [DOI] [PubMed] [Google Scholar]
- 62.Barrett NA, Austen KF. Innate cells and TH2 immunity in airway inflammation. Immunity. 2009;31:425–437. doi: 10.1016/j.immuni.2009.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wills-Karp M, Finkelman FD. Innate lymphoid cells wield a double-edged sword. Nat Immunol. 2011;12:1025–1027. doi: 10.1038/ni.2142. [DOI] [PubMed] [Google Scholar]
- 64.Chang YJ, Kim HY, Albacker LA, Baumgarth N, McKenzie AN, Smith DE, Dekruyff RH, Umetsu DT. Innate lymphoid cells mediate influenza-induced airway hyper-reactivity independently of adaptive immunity. Nat Immunol. 2011;12:631–638. doi: 10.1038/ni.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hesse M, Modolell M, La Flamme AC, Schito M, Fuentes JM, Cheever AW, Pearce EJ, Wynn TA. Differential regulation of nitric oxide synthase-2 and arginase-1 by type 1/type 2 cytokines in vivo: granulomatous pathology is shaped by the pattern of L-arginine metabolism. J Immunol. 2001;167:6533–6544. doi: 10.4049/jimmunol.167.11.6533. [DOI] [PubMed] [Google Scholar]
- 66.Louis CA, Mody V, Henry WL, Jr, Reichner JS, Albina JE. Regulation of arginase isoforms I and II by IL-4 in cultured murine peritoneal macrophages. Am J Physiol. 1999;276:R237–R242. doi: 10.1152/ajpregu.1999.276.1.R237. [DOI] [PubMed] [Google Scholar]
- 67.Zimmermann N, Mishra A, King NE, Fulkerson PC, Doepker MP, Nikolaidis NM, Kindinger LE, Moulton EA, Aronow BJ, Rothenberg ME. Transcript signatures in experimental asthma: identification of STAT6-dependent and -independent pathways. J Immunol. 2004;172:1815–1824. doi: 10.4049/jimmunol.172.3.1815. [DOI] [PubMed] [Google Scholar]
- 68.Raes G, Van den Bergh R, De Baetselier P, Ghassabeh GH, Scotton C, Locati M, Mantovani A, Sozzani S. Arginase-1 and Ym1 are markers for murine, but not human, alternatively activated myeloid cells. J Immunol. 2005;174:6561. doi: 10.4049/jimmunol.174.11.6561. author reply 6561-6562. [DOI] [PubMed] [Google Scholar]
- 69.Nair MG, Guild KJ, Artis D. Novel effector molecules in type 2 inflammation: lessons drawn from helminth infection and allergy. J Immunol. 2006;177:1393–1399. doi: 10.4049/jimmunol.177.3.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wynn TA. Fibrotic disease and the T(H)1/T(H)2 paradigm. Nat Rev Immunol. 2004;4:583–594. doi: 10.1038/nri1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Maizels RM, Balic A, Gomez-Escobar N, Nair M, Taylor MD, Allen JE. Helminth parasites - masters of regulation. Immunol Rev. 2004;201:89–116. doi: 10.1111/j.0105-2896.2004.00191.x. [DOI] [PubMed] [Google Scholar]
- 72.Pesce JT, Ramalingam TR, Mentink-Kane MM, Wilson MS, El Kasmi KC, Smith AM, Thompson RW, Cheever AW, Murray PJ, Wynn TA. Arginase-1-expressing macrophages suppress Th2 cytokine-driven inflammation and fibrosis. PLoS Pathog. 2009;5:e1000371. doi: 10.1371/journal.ppat.1000371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.North ML, Khanna N, Marsden PA, Grasemann H, Scott JA. Functionally important role for arginase 1 in the airway hyperresponsiveness of asthma. Am J Physiol Lung Cell Mol Physiol. 2009;296:L911–L920. doi: 10.1152/ajplung.00025.2009. [DOI] [PubMed] [Google Scholar]
- 74.Maarsingh H, Dekkers BG, Zuidhof AB, Bos IS, Menzen MH, Klein T, Flik G, Zaagsma J, Meurs H. Increased arginase activity contributes to airway remodelling in chronic allergic asthma. Eur Respir J. 2011;38:318–328. doi: 10.1183/09031936.00057710. [DOI] [PubMed] [Google Scholar]
- 75.Niese KA, Collier AR, Hajek AR, Cederbaum SD, O'Brien WE, Wills-Karp M, Rothenberg ME, Zimmermann N. Bone marrow cell derived arginase 1 is the major source of allergen-induced lung arginase but is not required for airway hyperresponsiveness, remodeling and lung inflammatory responses in mice. BMC Immunol. 2009;10:33–38. doi: 10.1186/1471-2172-10-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Artis D, Wang ML, Keilbaugh SA, He W, Brenes M, Swain GP, Knight PA, Donaldson DD, Lazar MA, Miller HR, Schad GA, Scott P, Wu GD. RELMbeta/FIZZ2 is a goblet cell-specific immune-effector molecule in the gastrointestinal tract. Proc Natl Acad Sci USA. 2004;101:13596–13600. doi: 10.1073/pnas.0404034101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Holcomb IN, Kabakoff RC, Chan B, Baker TW, Gurney A, Henzel W, Nelson C, Lowman HB, Wright BD, Skelton NJ, Frantz GD, Tumas DB, Peale FV, Jr, Shelton DL, Hébert CC. FIZZ1, a novel cysteine-rich secreted protein associated with pulmonary inflammation, defines a new gene family. EMBO J. 2000;19:4046–4055. doi: 10.1093/emboj/19.15.4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yamaji-Kegan K, Su Q, Angelini DJ, Myers AC, Cheadle C, Johns RA. Hypoxia-induced mitogenic factor (HIMF/FIZZ1/RELMalpha) increases lung inflammation and activates pulmonary microvascular endothelial cells via an IL-4-dependent mechanism. J Immunol. 2010;185:5539–5548. doi: 10.4049/jimmunol.0904021. [DOI] [PubMed] [Google Scholar]
- 79.Teng X, Li D, Champion HC, Johns RA. FIZZ1/RELMalpha, a novel hypoxia-induced mitogenic factor in lung with vasoconstrictive and angiogenic properties. Circ Res. 2003;92:1065–1067. doi: 10.1161/01.RES.0000073999.07698.33. [DOI] [PubMed] [Google Scholar]
- 80.Pesce JT, Ramalingam TR, Wilson MS, Mentink-Kane MM, Thompson RW, Cheever AW, Urban JF, Jr, Wynn TA. Retnla (relmalpha/fizz1) suppresses helminth-induced Th2-type immunity. PLoS Pathog. 2009;5:e1000393. doi: 10.1371/journal.ppat.1000393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lee CG. Chitin, chitinases, and chitinase-like proteins in allergic inflammation and tissue remodeling. Yonsei Med J. 2009;50:22–30. doi: 10.3349/ymj.2009.50.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhu Z, Zheng T, Homer RJ, Kim YK, Chen NY, Cohn L, Hamid Q, Elias JA. Acidic mammalian chitinase in asthmatic Th2 inflammation and IL-13 pathway activation. Science. 2004;304:1678–1682. doi: 10.1126/science.1095336. [DOI] [PubMed] [Google Scholar]
- 83.Agapov E, Battaile JT, Tidwell R, Hachem R, Patterson GA, Pierce RA, Atkinson JJ, Holtzman MJ. Macrophage chitinase 1 stratifies chronic obstructive lung disease. Am J Respir Cell Mol Biol. 2009;41:379–384. doi: 10.1165/rcmb.2009-0122RC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cai Y, Kumar RK, Zhou J, Foster PS, Webb DC. Ym1/2 promotes Th2 cytokine expression by inhibiting 12/15(S)-lipoxygenase: identification of a novel pathway for regulating allergic inflammation. J Immunol. 2009;182:5393–5399. doi: 10.4049/jimmunol.0803874. [DOI] [PubMed] [Google Scholar]
- 85.Watanabe K, Jose PJ, Rankin SM. Eotaxin-2 generation is differentially regulated by LPS and IL-4 in monocytes and macrophages. J Immunol. 2002;168:1911–1918. doi: 10.4049/jimmunol.168.4.1911. [DOI] [PubMed] [Google Scholar]
- 86.Jenkins SL, Ruckerl D, Cook PC, Jones LH, Finkelman FD, van Rooijen N, MacDonald AS, Allen JE. Local macrophage proliferation, rather than recruitment from the blood, is a signature of TH2 inflammation. Science. 2011;332:1284–1288. doi: 10.1126/science.1204351. [DOI] [PMC free article] [PubMed] [Google Scholar]