Abstract
Objective:
To obtain the comprehensive transcriptome profile of human citrulline-specific B cells from patients diagnosed with rheumatoid arthritis (RA).
Methods:
Citrulline and hemagglutinin (HA) specific B cells were flow-sorted using peptide-streptavidin conjugates from peripheral blood of RA patients and healthy individuals respectively. Transcriptome profile of the sorted cells was obtained by RNA-sequencing, and expression of key protein molecules was evaluated by aptamer-based SOMAscan assay and flow cytometry. The ability of these proteins to effect differentiation of osteoclasts and proliferation and migration of synoviocytes was examined by in vitro functional assays.
Results:
Citrulline-specific B cells, in comparison to citrulline-negative B cells differentially express IL15RA, and genes related to protein citrullination and cyclic AMP signaling. By analyzing an independent cohort of CCP seropositive RA patients, we demonstrate that: (1) the expression of IL15Rα protein is enriched in citrulline-specific B cells within RA patients, and (2) surprisingly, all B cells from RA patients were capable of producing epidermal growth factor ligand, amphiregulin (AREG). AREG directly led to increased migration and proliferation of fibroblast-like synoviocytes (FLS), and in combination with anti-citrullinated protein antibodies (ACPA) led to the increased differentiation of osteoclasts.
Conclusion:
To the best of our knowledge, this is the first study documenting the whole transcriptome profile of autoreactive B cells in any autoimmune disease. Our data identifies several genes and pathways that may be targeted by repurposing several FDA- approved drugs and can serve as the foundation for the comparative assessment of B-cell profiles in other autoimmune diseases.
Introduction.
The identification of citrullination, the post-translational modification of arginine residues to citrulline in proteins, catalyzed by the enzymes peptidyl arginine deiminases (PADs), as one of the key factors mediating the breach in tolerance and eliciting anti-citrullinated protein antibody (ACPA) responses, has been a major milestone for RA1,2. The appearance of ACPA in circulation precedes the onset of clinical disease, and ACPA positivity has a sensitivity of 60–70%, and a specificity of greater than 90%, for the diagnosis of RA1. ACPA have shown to trigger human immune effector functions including activation of the complement system and the ability to engage activating Fcγ receptors3. Lastly, it has been demonstrated that ACPA can catalyze bone erosion commonly seen in RA patients4.
Although the indispensable role of B cells in autoantibody production is well recognized, their autoantibody-independent contributions are not as well-defined. Systemic depletion of B cells using rituximab that targets B cells expressing human CD20, a phenotypic cell surface marker, has been shown to be effective clinically for the treatment of a subset of RA patients5. While studies have supported the idea that patients with higher autoantibody titers are likely to respond to rituximab6, the clinical efficacy of rituximab treatment is not necessarily correlated with a decrease in autoantibody titers7. This, in turn, implies a more expanded role for B cells in autoimmune pathogenicity beyond antibody production. Preclinical and clinical data have suggested many different functions of B cells in RA including a role as antigen presenting cells (APCs)8; direct secretion of pro-inflammatory cytokines like tumor necrosis factor (TNF) and interleukin-6 (IL-6)9–11 supporting the organization of tertiary lymphoid tissues within the inflamed synovium8,12; and impacting bone homeostasis through the secretion of receptor activator of nuclear factor kappa-B ligand (RANKL)13. Despite these efforts, a direct interrogation of the roles of the autoreactive B cell compartment, and how they differ from other B cells from the same donor or from B cells from healthy individuals, has not been accomplished.
One of the major challenges in profiling autoreactive B cells within humans is the low frequency of these cells in peripheral blood (< 0.1 % of all B cells in circulation)14. We developed and validated a flow-cytometry based assay for the reliable detection of cyclic citrullinated peptide (CCP) specific B cells ex vivo. We then performed RNA-sequencing (RNA-seq) on these small numbers of cells and the study design was to compare the whole transcriptome profile of CCP-specific B cells (RA-CCPPOS) with that of CCP-negative B cells (RA-CCPNEG) from the same donor and hemagglutinin-specific (HAPOS) B cells from healthy individuals elicited upon vaccination with seasonal influenza virus. We performed a two-pronged comparison to identify key genes expressed on all B cells from RA patients presumably induced by the systemic pro-inflammatory environment prevailing in these patients, as well as those that were only restricted to autoreactive B cells. Our data identified a novel role of B cells as an activator of the epidermal growth factor (EGF) pathways by secretion of EGF receptor ligand amphiregulin (AREG) under pro-inflammatory conditions, and also identified IL15Rα, as a specific biomarker of RA-CCPPOS B cells. Overall, our data suggest that besides being a source of autoantibodies, B cells may also play direct roles in the inflammatory cascades and osteoclastogenesis in RA. Significantly, while some of the identified genes and pathways are the targets of approved therapies for RA, our data also illustrates that drugs approved for other indications might be useful in targeting the RA-CCPPOS B cell compartment in RA. More broadly, to the best of our knowledge, this is the first comprehensive study on the transcriptome profile of human RA-CCPPOS B cells in autoimmune disease and will serve as a resource to further investigate the role of B cells in autoimmunity.
Material and methods:
Patients.
Blood (15 ml) was aspirated in heparin vacutainer tubes (BD Biosciences) from seropositive RA patients after informed consent under IRB approved protocols at BCM, UAB and UH. Patient information is summarized in Table S.1. All RA patients met 1987 ARA (now ACR) classification criteria for RA and were confirmed to be CCP-seropositive.
Flow-sorting of CCP-specific B cells from patients’ blood.
Peripheral blood mononuclear cells (PBMC) isolated from the blood of RA patients by density centrifugation, were blocked with PBS-5% human serum for 20 min at 4°C. All subsequent staining steps were performed in PBS containing 2% serum. PBMCs were stained for B-cell markers with fluorochrome-labeled antibodies, anti-CD19, IgM, and IgD together with T-cell marker using anti-CD3 antibody (Biolegend) according to manufacturer’s instructions. Biotinylated-cyclic citrulline peptide I (CCP; Anaspec), a surrogate peptide to capture anti-citrullinated protein antibody, was then added (1 µg/million PBMCs) followed by labeling with fluorochrome-labeled streptavidin (1 µg/ml). Subsequently, biotinylated-cyclic arginine peptide (CAP; Anaspec) was also added (1 µg/million PBMCs) as a control and labeled with fluorochrome-labeled streptavidin (1 µg/ml), to exclude B-cell cross-reactive to both antigens. Further, CD19POSIgM/IgDNEG B cells (IgG/IgAPOS) were gated and the cell populations CCPPOSCAPNEG and CCPNEGCAPNEG were sorted for validation of cell purity. A total of 125–360 RA-CCPPOS B cells were obtained of 10–15 million PBMCs from each patient. The sorted cells were then grown in 96-well tissue culture plates with 1×105/well of 3T3 fibroblast cells secreting msCD40L (NIH AIDS reagent program), 100 ng/ml of IL-21 (Peprotech) and 5 µg/ml of anti-APO1 antibody (eBiosciences) for a period of 14 days with half media change once after 7 days. Thereafter, cell purity was assessed by validating the specificity of IgG from the culture supernatants against CCP.
Preparation of cDNA library and RNA sequencing.
Total RNA was obtained by sorting antigen-specific B cells (350–1,000 total cells) ex vivo directly into the 100 μl of cell lysis buffer provided in the RNA isolation kit (Macherey-Nagel, RNA-XS). Further, cDNA libraries were synthesized using the commercially available SMART-Seq Ultra Low Input RNA kit (Clontech), as per the manufacturer’s protocols. After preparation of cDNA libraries, they were first tagmented and then barcoded by indexing primers using the Nextra XT kit (Illumina). The libraries were pooled and a 76bp paired-end sequencing was performed on an Illumina HiSeq3000 sequencer to yield a minimum of 17.4 million reads per library (range = 17.4 – 37.3 million).
RNA-sequencing data accession number in Gene Expression Omnibus (GEO): GSE99006
Detailed methods on RNA-seq bioinformatics, ACPA purification, FLS and osteoclastogenesis assays, SOMAmer assays are described in the supplemental information.
Results.
Flow-sorting of antigen-specific B cells.
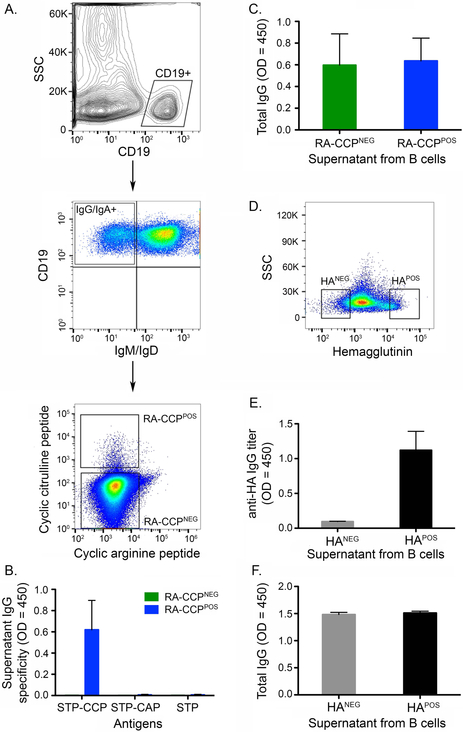
We developed a dual-labeling, flow sorting method using both cyclic citrullinated (CCP) and cyclic arginine peptides (CAP) to isolate RA-CCPPOS B cells. In order to verify the purity of our sorting method, an equal number of cells within the CCPPOSCAPNEG (hereafter referred to as RA-CCPPOS B cells), CCPNEGCAPPOS and CCPNEGCAPNEG (hereafter referred to as RA-CCPNEG) populations (Fig. 1A) were sorted in 96 well plates and grown in vitro for 14 days. The purity of our sorting strategy was validated by testing the supernatants after in vitro culture, which confirmed that only the immunoglobulins secreted in B-cell culture established from the RA-CCPPOS B cell population demonstrated a specific reactivity towards the CCP but not towards streptavidin or control cyclic arginine peptide (Fig. 1B-C). After validation of our sorting strategy, a total of 350–1000 RA-CCPPOS B cells (0.01 – 0.1 %) from the blood of four RA patients were used directly for the preparation of cDNA libraries ex vivo to ensure minimal perturbations to the transcriptional profile (Table S.1). Both RA-CCPPOS and RA-CCPNEG B cells were confirmed to be predominantly of the memory phenotype based on the surface expression of CD27 and IgD (Fig. S.1A).
Figure 1. Isolation of an enriched population of RA-CCPPOS and HA-specific B cells.
A. Representative flow plots depicting the sorting strategy of RA-CCPPOS and RA-CCPNEG B cells. Cells were first gated as CD19POSIgM/IgDNEG B cells (IgG/IgAPOS), thereafter, RA-CCPPOS B cells were flow sorted as CCPPOSCAPNEG and RA-CCPNEG cells were sorted as CCPNEGCAPNEG B-cell population. B. ELISA on supernatants, tested for antigen specificity of RA-CCPPOS and RA-CCPNEG B cells, expanded and differentiated in vitro (n=3). C. ELISA on supernatants, measuring total Ig from RA-CCPPOS and RA-CCPNEG B cells, expanded and differentiated in vitro (n = 3). D. Representative flow plot showing isolation of HAPOS and HANEG B cells, sorted with a similar gating strategy as described in panel A. E. ELISA on supernatants, tested for (E) HA reactivity and (F) total Ig from HANEG and HAPOS B-cell populations (n = 4). Error bars in ELISA results indicate standard error of the mean. STP – Streptavidin, Ig – Immunoglobulin, CCP – Cyclic citrullinated peptide, CAP – Cyclic arginine peptide.
In order to have a comparative analysis of B-cell transcriptome profile during autoimmunity versus normal immune response to vaccination, HA-specific B cells (hereafter referred to as HAPOS B cells) were isolated from blood of four healthy individuals vaccinated with the seasonal influenza vaccine. Our ability to enrich for HAPOS B cells was validated by the same three step procedure used for RA-CCPPOS B cells: (a) antigen labeling and flow-sorting a total of 3500 HAPOS and HANEG cells from PBMCs of these vaccinated donors, (b) in vitro expansion and differentiation, and (c) ELISA testing for HA-reactivity on the culture supernatants (Fig. 1D-F). Similar to the B cells from RA patients, HAPOS B cells from healthy individuals also displayed a CD27+ memory phenotype (Fig. S.1B). We did not observe a significant difference in the frequency of memory B cells between different samples of RA-CCPPOS, RA-CCPNEG, and HAPOS B cells (Fig. S.1C). Subsequent to validation, 1000–2000 HAPOS B cells from the same four donors were used to construct cDNA libraries ex vivo for RNA-sequencing (RNA-seq). In order to ensure that the differences in the gene expression profile of RA-CCPPOS B cells was not due to the composition of different isotypes of B cells (IgG vs IgA), we analyzed our RNA-seq data for transcripts associated with IgG and IgA molecules, and confirmed that no significant differences were observed between RA-CCPPOS, RA-CCPNEG, and HAPOS B cells (Fig. S.1D).
Transcriptome analysis revealed that RA-CCPPOS, RA-CCPNEG, and HAPOS B cells could be distinguished based on the differentially expressed genes.
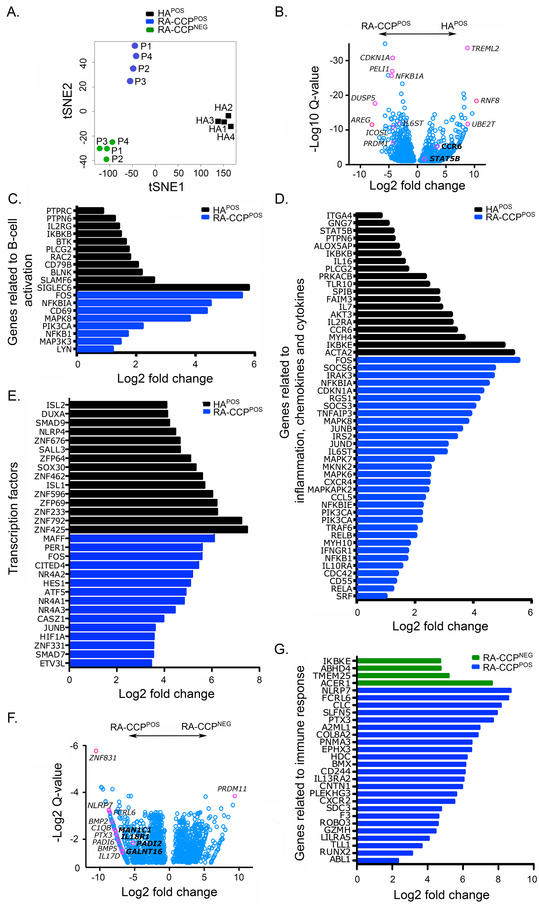
The cDNA libraries generated ex vivo from 12 samples (4 paired RA-CCPPOS B cells and RA-CCPNEG populations, and 4 HAPOS B cell populations) were barcoded, pooled and sequenced using 76bp paired-ends to yield a minimum of 17 million reads per library. After validation of the RNA-seq populations (Fig. S.2), differential analyses using the DEseq package revealed that 1658 genes (false discovery rate, FDR < 0.1) were differentially expressed in RA-CCPPOS B cells in comparison to the HAPOS B cells, and 431 genes were identified as differentially expressed genes (DEGs) in comparison to the RA-CCPNEG B cells (Table S.3 and S.4). We utilized t-distributed stochastic neighbor embedding (t-SNE), to demonstrate that these identified DEGs could clearly resolve the three distinct cellular populations 15 (Fig. 2A). As expected, both the number of identified DEGs and their relative change in expression was lower in comparing the RA-CCPPOS and RA-CCPNEG B cells from the same donors, in contrast to comparing the RA-CCPPOS B cells to the HAPOS B cells (Table S.3 and S.4). We evaluated the statistical power of our study to differentiate the RA-CCPPOS and RA-CCPNEG B cell subsets using powsimR, and estimated it to be 0.86±0.18 (mean±SEM, based on 100 simulations).
Figure 2. RA-CCPPOS, RA-CCPNEG, and HAPOS B cells can be differentiated based on DEGs.
A. t-Distributed Stochastic Neighbor Embedding (t-SNE) visualization of sample relationships using the differentially expressed genes. B. Volcano plots generated using fold change and q-value of DEGs shows enrichment of genes related autoimmunity and inflammation in RA-CCPPOS B cells in comparison to HAPOS B cells. (C-E). Gene ontology plots indicating fold change in expression of genes related to B-cell activation, inflammation, chemokines and cytokines, and top fifteen transcription factors that are differentially enriched in RA-CCPPOS versus HAPOS B cells. F. Volcano plot generated using fold change and q-value of DEGs between RA-CCPPOS B cells and RA-CCPNEG B cells from the same donors. G. Gene ontology plot depicting fold change in expression of genes related to immune responses that are highly expressed in RA-CCPPOS B cells in comparison to RA-CCPNEG B cells from the same donors.
Comparisons between RA-CCPPOS and HAPOS B cells.
A number of candidate DEGs that are well validated in autoimmunity including the cyclin kinase p21 (CDKN1A)16, ubiquitin ligase PelI117, and the costimulatory molecule ICOSLG18 were identified. The epidermal growth factor ligand, amphiregulin (AREG) was identified as the transcript with the largest change in expression (Fig. 2B). By classifying these same DEGs into the Gene Ontology (GO) categories, we grouped them as being related to B-cell activation, genes related to inflammation, and transcription factors. We observed differences in the expression of genes related to B-cell activation (Fig. 2C) with increased expression of kinases like phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha (PI3KCA)19 and mitogen-activated protein kinases (MAPK7/MAPK8) known to cause autoimmunity under deregulated activation20. In addition, several genes related to inflammation including signaling molecules like TNFAIP3 and IL6ST, and T-cell recruiting chemokine-like CCL5 and chemokine receptor CXCR4 were upregulated in RA-CCPPOS B cells (Fig. 2D)21,22.
Analysis of the differentially expressed transcription factors (TFs) revealed upregulation of positive regulatory domain I-binding factor 1 (PRDM1) and ETS variant 3 (ETV3 and ETV3L) within RA-CCPPOS B cells; these TFs are known to inhibit c-Myc and consequently cell growth23, while at the same time promoting B-cell differentiation and immunoglobulin (Ig) secretion24. Similarly, the small MAF family TFs (MAFF and MAFG) that can directly impact Ig secretion were enriched in these RA-CCPPOS B cells (Fig. 2E)25.
We also sought to determine if there was a B-cell specific change in expression of genes known to be high-risk loci in RA, documented through large-scale genetic efforts26. Out of the known RA-specific loci, 9 genes were also identified as DEGs (SH2B3, CCR6, ILF3, TXNDC11, PTPRC, PRDM1, TNFAIP3, TRAF6, and LBH) [Fig. S.3].
Comparisons between RA-CCPPOS and RA-CCPNEG B cells.
The candidate DEGs upregulated within the RA-CCPPOS population in comparison to RA-CCPNEG B cells within these same donors included: pentraxin PTX3 (Fig. 2F, G) that recognizes pathogen-associated molecular patterns (PAMP) and its binding partner in the complement cascade (C1QB)27,28; the bone morphogenetic proteins (BMP2 and BMP5); and the peptidyl arginine deaminase citrullinating enzymes (PADI2 and PADI6)29 [Fig. 2F]. Additionally, several immune-related transcripts like the inflammasome-associated protein NLRP730 and the Fc receptor-like protein FCRL631 were also upregulated within RA-CCPPOS B cells (Fig. 2F, G).
Cytokine and signaling pathways enriched in RA-CCPPOS B cells.
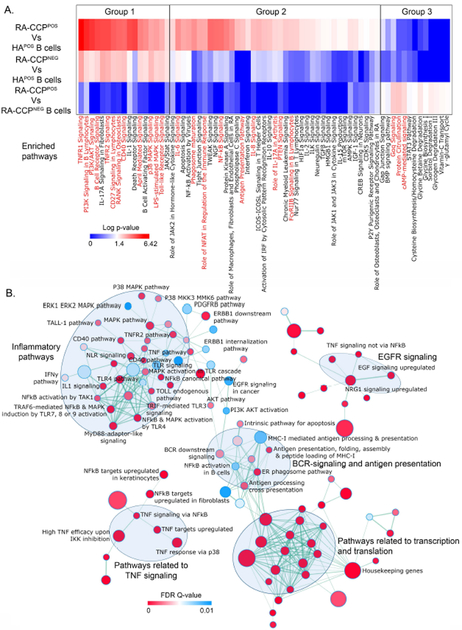
The differentially expressed pathways were obtained by matching the expression dataset using Ingenuity pathway analysis (IPA), filtering the pathways directly related to immune cells and their functions, and visualized as a heatmap by using the log p-values (Fig. 3A). In order to facilitate comparisons, we identified three groups of pathways that were enriched: both in the RA-CCPPOS and RA-CCPNEG B-cell populations in comparison to the HAPOS B cells (group 1), only in the RA-CCPPOS population but not the RA-CCPNEG or HAPOS B cells (group 2), and only in the RA-CCPPOS population in comparison to the RA-CCPNEG B cells (group 3) [Fig. 3A]. The group 1 pathways comprised of B-cell activation pathways (B-cell receptor signaling, PI3K/Akt, and p38 MAPK), toll-like receptor-based activation (TLR signaling and LPS-stimulated MAPK pathways), and the TNF superfamily ligand-mediated signaling (CD40, APRIL signaling) and their receptor signaling pathways (TNFR1, TNFR2, CD27, RANK, and CD40). The group 2 pathways comprised of inflammatory cytokine signaling (IL6, IL8, and IL-17A) and RA-specific signatures (role of macrophages, endothelial cells and osteoclasts in rheumatoid arthritis); B-cell activation/homeostasis (PTEN, NF-κB, and NFAT activation) and function (phagosome maturation, FcγRIIb signaling, and antigen presentation). Finally, group 3 showed enrichment in the pathways for protein citrullination, G-protein coupled receptor signaling (Gαq signaling) and cyclic AMP (cAMP) signaling (Fig. 3A). Overall, these data suggest that while TNF signaling has a global impact on all B cells from RA patients (group 1), the RA-CCPPOS B cells in comparison to the RA-CCPNEG B cells demonstrate a potential role in protein citrullination and effector functionality.
Figure 3. Enrichment maps of pathways enriched in RA-CCPPOS B cells as compared to HAPOS B cells.
A. Heat map generated using log-transformed p-values of DE-pathways from IPA indicated enrichment of pro-inflammatory pathways in RA-CCPPOS B cells as compared to HAPOS B cells and upregulation of protein citrullination in RA-CCPPOS B cells versus RA-CCPNEG B cells. Highlighted pathways are marked in red. B. The GSEA-derived enriched C2 curated pathways in RA-CCPPOS B cells were plotted using the enrichment map application in Cytoscape using a cutoff FDR q-value = 0.01 and a p-value = 0.005. Nodes (circles colored red and blue) represent pathways and the edges (green lines) represents overlapping gene among pathways. The size of nodes represents the number of genes enriched within the pathway and the thickness of edges represents the number of overlapping genes. The color of nodes was adjusted to an FDR q value range from 0 – 0.01. Clusters of pathways are labeled as groups with a similar theme. All pathways represented here are enriched in RA-CCPPOS B cell populations.
Based on the substantially larger number of pathways and greater changes in expression, identified in the RA-CCPPOS B cells in comparison to the HAPOS B cells by IPA (groups 1 and 2 combined), we performed gene-set enrichment analyses (GSEA) comparing these two populations. We interrogated the changes in these populations against the Molecular Signatures Database (Hallmark and C2 curated gene sets). As shown in Fig. 3B, five major clusters of pathways were significantly upregulated (FDR q-value < 0.1) in the RA-CCPPOS B cells: transcription and translation; B-cell receptor signaling; EGFR signaling; TNF signaling; and inflammatory cytokines and chemokines.
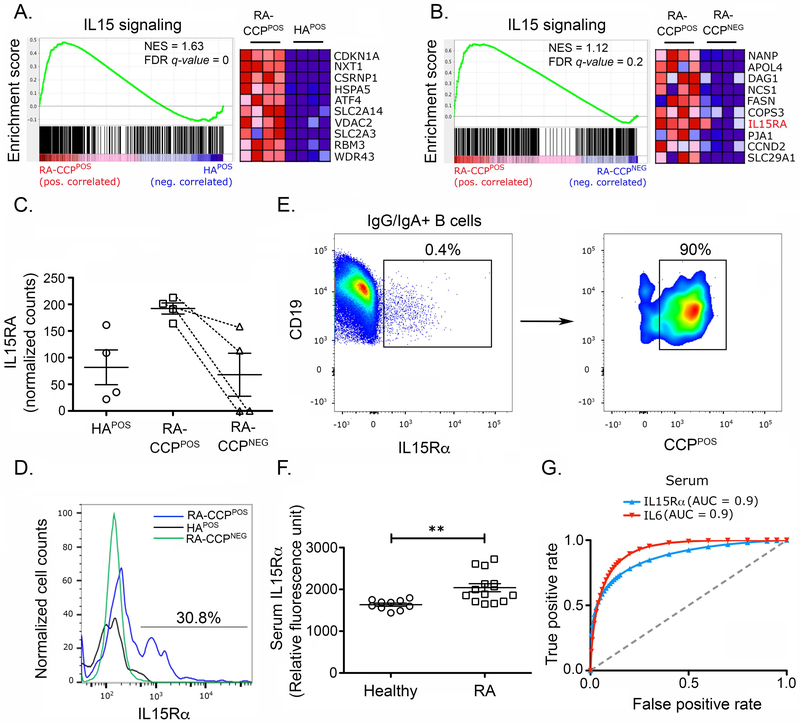
IL5RA expression is enriched in RA-CCPPOS B cells within RA patients.
As outlined above, there was an abundance of cytokine and inflammation-related pathways, enriched in RA-CCPPOS B cells. We focused our attention on IL15 signaling and performed enrichment analyses comparing our different B cell populations against known human IL15 mediated signaling. RA-CCPPOS B cells showed a specific enrichment in IL15 mediated signaling in comparison to either the HAPOS B cells or the RA-CCPNEG B cells (Fig. 4A-B). We next examined the relative abundance of both IL15 and its private receptor, IL15RA. Indeed, IL15RA (Fig. 4C) transcripts were upregulated within RA-CCPPOS B cells relative to either RA-CCPNEG or HAPOS B cells. However, a significant difference was not observed in the expression of IL15 (Fig. S.4).
Figure 4. IL15Rα and IL15 signaling are enriched in RA-CCPPOS B cells.
A. GSEA plots of IL15 signaling in A. RA-CCPPOS versus HAPOS B cells, and B. RA-CCPPOS versus RA-CCPNEG B cells, with heatmap of top ten genes enriched in the pathway using normalized transcript levels. C. Normalized transcript count of IL15RA in RA-CCPPOS, RA-CCPNEG, and HAPOS B cells. Although non-parametric tests demonstrate significant differences between RA-CCPPOS B cells and other two populations, since these would not account for multiple hypothesis testing, we have not shown these results. D. Representative flow-cytometric plot of IL15Rα expression on RA-CCPPOS, RA-CCPNEG, and HAPOS B cells (n = 4). E. IL15Rα expression on RA-CCPPOS B cells when gated on IL15Rα positive IgG/IgA B cells (n = 3). Cells sequentially gated as CD19+ IgM/IgD- (IgG/IgAPOS) IL15RαPOS CCPPOS. F. Serum concentration of IL15Rα in RA patients (n = 11) and healthy individuals (n = 10) evaluated by SOMAscan. Error bars indicate standard error of the mean. G. Receiver operating characteristic curve (ROC) indicating the predictive value of IL15Rα and IL-6 for RA. * p-value < 0.05, ** p-value < 0.01 * p-value < 0.05. NES – Normalized enrichment score; FDR – False discovery rate. Statistical analysis in panel F is by Mann-Whitney U test.
In order to validate the elevated expression of IL15Rα, we utilized an independent cohort of CCP-seropositive RA patients (Table S.1) and performed ex vivo surface staining on B cells by flow cytometry, using the same gating strategy as described earlier (Fig. 1, S.1). Consistent with the RNA-seq results, there was a distinct subpopulation of RA-CCPPOS B cells expressing IL15Rα in all of the patients tested while these were clearly absent in either the RA-CCPNEG or HAPOS B cell populations (Fig. 4D, Fig. S.5). Interestingly, within RA patients, 75– 90% of IL15Rα expressing IgG/IgA B cells, were identified to be RA-CCPPOS B cells, thus indicating that IL15Rα expression was significantly enriched in this population (Fig. 4E).
Since IL15Rα can also be converted to the soluble form by proteolytic cleavage by TNF-alpha converting enzyme (TACE)32, we next investigated the concentration of soluble IL15Rα (sIL15Rα) in RA patient’s cohorts. Although elevated concentrations of sIL-15Rα in the synovium are known to be associated with increased disease activity, there are no known reports that have documented elevated sIL15Rα in the serum of seropositive RA patients33. To systematically validate a large number of candidate proteins within cohorts of RA patient’s sera, including sIL15Rα, we took advantage of the aptamer-based SOMAscan assay that can detect analytes at picomolar concentrations by utilizing very small volumes of biological samples (Fig. S.6)34. In a cohort of CCP seropositive RA patients (11 samples) compared to healthy donors (10 samples), we observed significantly higher concentrations of both IL-6 and IL-8 but not in a large panel of other soluble analytes including IL-1β or IL-17 (Fig. S.7). When we evaluated sIL15Rα, we observed significantly higher concentrations in the sera of RA patients in comparison to healthy controls (Fig. 4F). Lastly, we also looked at the predictive value of serum sIL15Rα determined by the SOMAscan assay and found that sIL15Rα displays both high sensitivity and specificity for RA patients in comparison to healthy controls (Fig. 4G). We confirmed the statistical power of our cohort, by estimating the power for sIL15Rα (0.88) using pROC.
EGFR pathways and molecular validation of AREG in RA B cells.
AREG is a member of the epidermal growth factor family of ligands that signal through the EGFR receptors. It has been previously documented that TNF signaling in conjunction with IL-1β signaling can lead to the up regulation of AREG32,35. Our data support a pivotal role for AREG in RA-CCPPOS B cells (Fig. 5A), and the downstream target pathway, EGF-mediated signaling (Fig. 5B). In order to validate our findings, we directly interrogated AREG expression within individual B cells on an independent cohort of seropositive RA patients by flow cytometry (Table S.1). As expected, we did not observe expression of AREG on RA-CCPPOS B cells ex vivo (Fig. S.8) since AREG is secreted as soluble protein upon cleavage by the TNF-α-converting enzyme (TACE)32. In order to determine if AREG expression can be induced in vitro in B cells, we sought to mimic the nature of help afforded by T helper cells in vivo in RA36. Accordingly, populations of RA-CCPPOS, RA-CCPNEG, and HAPOS B cells were flow sorted and incubated with both soluble CD40L and IL-21 for 14 days. Under these conditions, both RA-CCPPOS and RA-CCPNEG B cells showed induction of AREG in >80 % of cells (Fig. 5C), and this was consistent with our RNA-seq data on these populations (Fig. 5A). A tendency towards higher expression was observed when RA-CCPPOS and HAPOS B cells were compared for AREG expression but owing to the high variance in frequency of AREG expressing HAPOS B cell populations this change was not significant. Taken together, these findings suggest that under polarizing conditions, at least in vitro, B cells from RA patients act as a source of AREG, a molecule with a known ability to have a global impact on multiple cell types.
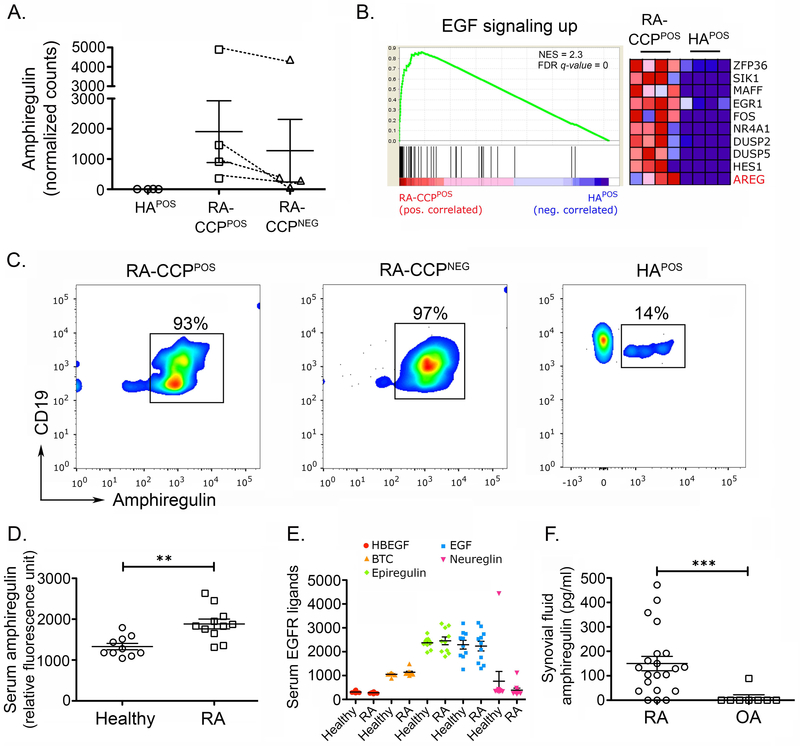
Figure 5. Amphiregulin and associated pathways are enriched in RA-CCPPOS B cells.
A. Normalized transcript count of AREG in RA-CCPPOS, RA-CCPNEG and HAPOS B cells. Although non-parametric tests demonstrate significant differences between RA-CCPPOS B cells and HAPOS B cells, since these would not account for multiple hypothesis testing, we have not shown these results. B. GSEA plot of epidermal growth factor pathway with heat map indicating normalized transcript levels of top ten genes in the pathway for each sample. C. Flow-cytometric evaluation of AREG expression on RA-CCPPOS, RA-CCPNEG, and HAPOS B cells after in vitro expansion (n = 4). D. Serum concentration of AREG in RA patients (n = 11) and healthy individuals (n = 10) evaluated by SOMAscan. Statistical power of this assay was estimated to be 0.98. E. Serum concentrations of all other EGF ligands identified by SOMAscan aptamer assay. F. ELISA on synovial fluid (SF) from patients with RA (n = 21) as compared to control osteoarthritis (OA) [n = 8]. * p-value < 0.05, ** p-value < 0.01. Statistical analysis in panels D and F is by Mann-Whitney U test, and in panel E is by Kruskal-Wallis test. Error bars represent standard error of mean.
AREG expression is elevated in serum and synovial fluid of RA patients.
Among the different EGF ligands that are known to induce proliferation, growth, and differentiation by signaling through the EGFR receptors37, AREG plays a unique role in its ability to induce both cell proliferation and cellular differentiation upon receptor binding38. Although other reports have documented serum AREG levels in RA, these data are contradictory39–41. We evaluated the concentrations of AREG and the other EGF ligands in the sera of CCP-seropositive RA patients with the SOMAscan array, as outlined above. In this cohort of samples, we observed a significantly higher concentration of AREG (Fig. 5D) but none of the other EGF ligands like heparin-binding EGF-like growth factor (HBEGF), betacellulin (BTC), epiregulin, EGF and neuregulin (Fig. 5E). We also observed a high sensitivity and specificity of serum AREG in predicting the occurrence of RA (Fig. S.9). The estimated statistical power of our cohort for AREG = 0.98. Since RA is a disease with localized inflammation, we further evaluated the abundance of AREG in the synovial fluid (SF) from CCP-seropositive RA patients (21 samples) and compared it with the SF from the non-inflammatory osteoarthritis (OA, 8 samples). Our ELISA results showed that the levels of AREG were significantly higher in RA as compared to osteoarthritis (OA) patients, wherein only one patient’s SF tested positive for AREG (Fig. 5F).
Amphiregulin promotes migration and proliferation of fibroblast-like synoviocytes (FLS) in vitro.
One of the hallmarks of RA pathogenesis is the activation of the FLS, which is characterized by a tumor-like aggressive phenotype within the joints42. We tested the ability of AREG to increase the invasiveness of human RA-FLS and observed that AREG promoted the increased migration of FLS in wound healing assays, in comparison to controls (Fig. 6A, B). We confirmed the expression of EGFR on RA-FLS, suggesting that the increased invasiveness is likely due to increased AREG-EGFR signaling (Fig. 6C). We also observed that RA-FLS proliferate more in the presence of AREG in comparison to controls (Fig. 6D). Taken together, our results thus indicate that high-levels of AREG in RA patient’s blood and SF could be responsible for an aggressive phenotype of FLSs in these patients.
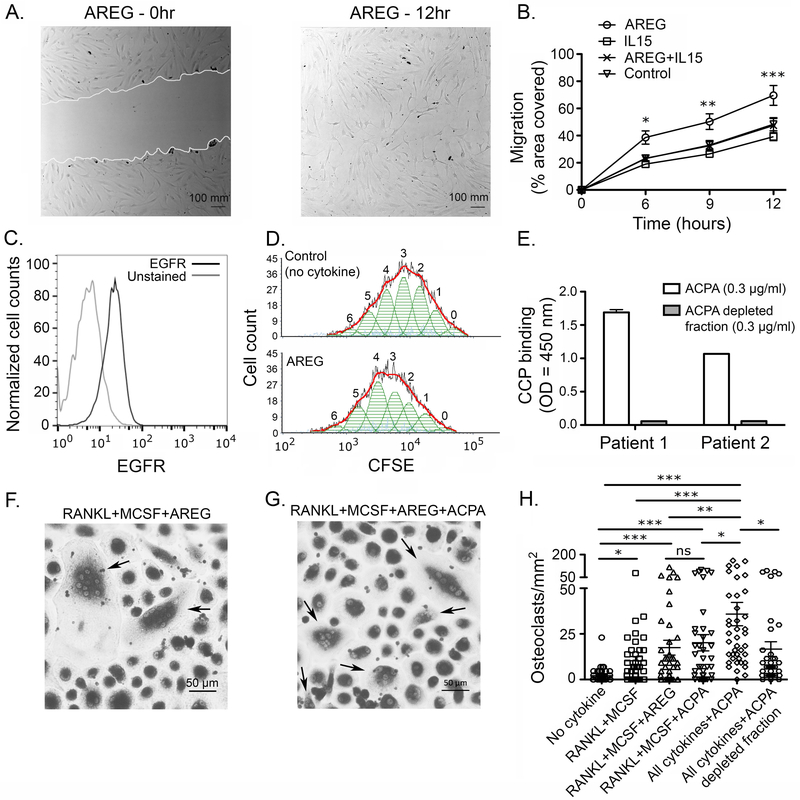
Figure 6. Amphiregulin enhances migration and proliferation of fibroblast-like synoviocytes, and synergizes with ACPA in mediating osteoclast differentiation.
A. Representative scratch assay depicting cell-migration; experiments were performed twice in triplicates on FLS obtained from one RA patient. B. Migration of FLS represented as the scratch area covered. C. Expression of EGFR on FLS; gray line represents unstained cells; replicates = 3. D. The proliferation of FLS measured using CFSE dilution; replicates = 3. E. ELISA to evaluate ACPA enrichment from the plasma of two RA patients against CCP; replicates = 2. (F, G) Representative examples of differentiation of osteoclasts from blood monocytes under two different conditions; arrows indicate differentiated osteoclasts with ≥ 3 nuclei. H. Number of osteoclasts under different culture conditions; ACPA from three RA patients was used with PBMC from three healthy individuals; replicates = 4; ten random images per well were taken at 20x magnification. * p-value < 0.05, ** p-value < 0.01, *** p-value < 0.001. Two-way ANOVA was used in B and Kruskal-Wallis test with Dunn’s comparison was used in H. * in panel B represents significance between AREG and control. Error bars represent standard error of the mean in B, H and standard deviation in E.
Functional synergy between AREG and ACPA in mediating osteoclastogenesis.
The role of AREG in promoting osteolytic activity has been detailed as one of the mediators of bone metastases in breast cancers43. Independently, the role of ACPA in enabling osteoclast differentiation has been described in RA4. We thus interrogated the functional synergy between these molecules in mediating osteoclastogenesis. First, we purified the ACPA antibodies by binding to a CCP-functionalized column (Fig. 6E). Next, we evaluated the ability of each of these molecules either by themselves or in combination to promote the differentiation of multi-nucleated Tartrate-resistant acid phosphatase (TRAP) positive osteoclasts from monocyte precursors. Although AREG by itself did not significantly increase the differentiation of osteoclasts; the number of osteoclasts was observed to be higher in co-cultures with AREG and ACPA in comparison to all other conditions tested (Fig. 6F-G). Our findings thus suggest that AREG can synergize with ACPA in the differentiation of osteoclasts from blood monocytes.
Discussion.
The role of RA-CCPPOS B cells as a source of autoantibodies contributing to disease pathology in RA has been extensively studied but the functional programs that help define the biology of the cell have remained elusive. One of the challenges we encountered was the robust and reliable identification of RA-CCPPOS B cells which account for <0.1% of total peripheral B cells of RA patients14. We successfully designed a sensitive flow sorting method using peptide-streptavidin conjugates, and sorted both the IgGPOS and IgAPOS CCPPOS B cells, owing to the fact that the antibody response against citrullinated proteins in RA comprises both of IgG and IgA type immunoglobulins44.
Based on the RNA-seq data, we validated that the expression of the IL15Rα protein on the cell surface was enriched in the RA-CCPPOS B cell population, and confirmed that increased levels of sIL15Rα can be detected within the blood of an independent cohort of CCP-seropositive RA patients. IL-15 mediated signaling has been targeted in phase I-II clinical trials by using HuMax-IL15 (Table S.2), a human monoclonal antibody that inhibits the bioactivity of IL-15, and has shown modest improvements in disease activity45. Synthetic DMARDs including tofacitinib (Janus kinase inhibitor) inhibit pathways that are also downstream of IL-15Rα derived signaling46. Our data suggest that the targeting of IL15Rα might provide therapeutic benefit in RA either by directly targeting the RA-CCPPOS B cells or by the elimination of sIL15Rα in the blood of RA patients. In this regard, it is worth emphasizing that sIL15Rα is known to increase the potency of IL15 function by 100 fold when complexed together, and enables signaling in cells that might otherwise lack IL15Rα47. As suggested previously, the enrichment of IL15Rα expression within the RA-CCPPOS B cells might be indicative of an altered differentiation state48.
Our data for the first time also revealed the expression of EGFR ligand AREG on autoreactive B cells, which was also the most differentially expressed gene in RA-CCPPOS B cells as compared to HAPOS B cells. We confirmed the surface expression of AREG on B cells under inflammatory conditions, identified AREG as a candidate biomarker in CCP-seropositive RA and have evaluated the functional impact of AREG on the cellular effectors of RA. AREG is expressed as a membrane-bound molecule, which is activated upon proteolytic cleavage by TACE32, thereby acting in an autocrine or paracrine manner and influence cell survival, proliferation and motility49.
Contradictory reports exist on the elevated concentrations of AREG in the blood of RA patients, with evidence for both significantly increased concentrations, and no significant differences, in comparison to healthy donors39–41. One confounding factor in these reports is CCP-seropositivity. Consistent with our studies with RA-CCPPOS B cells, our results here demonstrate significant increases in the concentration of AREG in the blood of CCP-seropositive RA patients in comparison to healthy donors. In parallel, immunohistochemistry has confirmed the expression of both AREG and EGFR within the synovial tissue of RA patients32,40. Investigations using a mouse model of inflammation/RA harboring the transgenic IL6 signal transducer (IL6ST) gene has shown the importance of epidermal growth factors including AREG in the pathogenesis of arthritis; wherein targeting AREG either by neutralizing antibodies or shRNA ameliorated the disease severity41.
The induction of AREG expression is governed by the cytokines TGFβ, TNFα, and IL1β32,35, known to be abundant in RA synovial fluid and plasma50 (Fig S.10). Our data illustrates, AREG can directly promote the proliferation and invasiveness of FLS. As outlined by elegant studies in breast cancer metastasis models, AREG is also known to increase the differentiation and activity of osteoclasts from PBMCs in the presence of RANKL and M-CSF43, both of which are abundantly available in the synovial compartment of RA patients. Our in vitro data further advances the ability of AREG to act in concert with ACPA to enhance the differentiation of osteoclasts from blood monocytes. Since our RNA-seq data has outlined a role for both IL6ST and the IL-6 signaling in RA-CCPPOS B cells, this suggests that IL6 signaling might facilitate AREG upregulation. Overall, our results indicate that under proinflammatory conditions all B cells in RA may contribute to the production of AREG irrespective of antigen specificity, which may potentially affect other cells in the joints.
Through this study, we are reporting for the first time a comprehensive transcriptome profile of RA-CCPPOS B cells in RA. To the best of our knowledge, this is also the first report on whole transcriptome profiling of antigen-specific B cells in any human autoimmune disorder. Our results portray B cells as not merely autoantibody producers but as a source of diverse molecules which can influence proliferation, differentiation, and activation of other pathogenic cells types. We anticipate that these data will serve as a foundational data set for investigating multiple hypotheses on the roles of B cells in RA and other autoimmune disorders, and enable drug discovery and validation based on the biology of RA-CCPPOS B cells in RA.
We also recognize that while our report represents an important first step, further studies are required to gain a much deeper understanding of autoreactive B cells in autoimmune biology. Comprehensive studies on the relationships between sIL15Rα and AREG within the synovial compartment, and all of the different cell types that can secrete these molecules, and how they influence the expression and secretion of each other and other signaling cascades, need to be performed. Although direct profiling of synovial B cells can reflect their contribution to RA pathophysiology, obtaining these cells is challenging from a clinical perspective. Arthroscopy is considered an invasive procedure and hence not routinely performed in clinical practice. Similarly, although synovial tissue can be accessed from patients undergoing joint replacement, these patients have end-stage disease not reflective of therapeutically relevant disease. Lastly, although single-cell RNA-seq (scRNA-seq) is better suited for studying moderately or highly expressed transcripts, it would be interesting to perform scRNA-seq to document the heterogeneity in autoimmune B cells, to complement our existing results, especially with synovial B cells.
Supplementary Material
Acknowledgments.
This publication was supported by NIH (R01CA174385), CPRIT (RP130570 and RP180466), MRA Award (509800), Welch Foundation (E1774), Owens Foundation, CDMRP (CA160591), and NSF (1705464) and the UAB RADAR fund. We would like to acknowledge the UH Seq-n-edit core for RNA-seq, Intel for the loan of computing cluster, and the UH Center for Advanced Computing and Data Systems (CACDS) for high-performance computing facilities.
References.
- 1.van der Linden MP et al. Value of anti-modified citrullinated vimentin and third-generation anti-cyclic citrullinated peptide compared with second-generation anti-cyclic citrullinated peptide and rheumatoid factor in predicting disease outcome in undifferentiated arthritis and rheumatoid arthritis. Arthritis Rheum 60, 2232–2241, (2009). [DOI] [PubMed] [Google Scholar]
- 2.Wagner CA et al. Identification of anticitrullinated protein antibody reactivities in a subset of anti-CCP-negative rheumatoid arthritis: association with cigarette smoking and HLA-DRB1 ‘shared epitope’ alleles. Ann Rheum Dis 74, 579–586, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boross P & Verbeek JS The complex role of Fcgamma receptors in the pathology of arthritis. Springer Semin Immunopathol 28, 339–350, (2006). [DOI] [PubMed] [Google Scholar]
- 4.Harre U et al. Induction of osteoclastogenesis and bone loss by human autoantibodies against citrullinated vimentin. J Clin Invest 122, 1791–1802, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dorner T, Radbruch A & Burmester GR B-cell-directed therapies for autoimmune disease. Nat Rev Rheumatol 5, 433–441, (2009). [DOI] [PubMed] [Google Scholar]
- 6.Lal P et al. Inflammation and autoantibody markers identify rheumatoid arthritis patients with enhanced clinical benefit following rituximab treatment. Arthritis Rheum 63, 3681–3691, (2011). [DOI] [PubMed] [Google Scholar]
- 7.Toubi E et al. Changes in macrophage function after rituximab treatment in patients with rheumatoid arthritis. Ann Rheum Dis 66, 818–820, (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takemura S, Klimiuk PA, Braun A, Goronzy JJ & Weyand CM T cell activation in rheumatoid synovium is B cell dependent. J Immunol 167, 4710–4718, (2001). [DOI] [PubMed] [Google Scholar]
- 9.Barr TA et al. B cell depletion therapy ameliorates autoimmune disease through ablation of IL-6-producing B cells. J Exp Med 209, 1001–1010, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McInnes IB & Schett G Cytokines in the pathogenesis of rheumatoid arthritis. Nat Rev Immunol 7, 429–442, (2007). [DOI] [PubMed] [Google Scholar]
- 11.Kalliolias GD & Ivashkiv LB TNF biology, pathogenic mechanisms and emerging therapeutic strategies. Nat Rev Rheumatol 12, 49–62, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takemura S et al. Lymphoid neogenesis in rheumatoid synovitis. J Immunol 167, 1072–1080, (2001). [DOI] [PubMed] [Google Scholar]
- 13.Meednu N et al. Production of RANKL by Memory B Cells: A Link Between B Cells and Bone Erosion in Rheumatoid Arthritis. Arthritis Rheumatol 68, 805–816, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kerkman PF et al. Identification and characterisation of citrullinated antigen-specific B cells in peripheral blood of patients with rheumatoid arthritis. Ann Rheum Dis 75, 1170–1176, (2016). [DOI] [PubMed] [Google Scholar]
- 15.Maaten L v. d. & Hinton, G. Visualizing data using t-SNE. Journal of Machine Learning Research 9, 2579–2605, (2008). [Google Scholar]
- 16.Santiago-Raber ML et al. Role of cyclin kinase inhibitor p21 in systemic autoimmunity. J Immunol 167, 4067–4074, (2001). [DOI] [PubMed] [Google Scholar]
- 17.Chang M et al. The ubiquitin ligase Peli1 negatively regulates T cell activation and prevents autoimmunity. Nat Immunol 12, 1002–1009, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamel KM, Cao Y, Olalekan SA & Finnegan A B cell-specific expression of inducible costimulator ligand is necessary for the induction of arthritis in mice. Arthritis Rheumatol 66, 60–67, (2014). [DOI] [PubMed] [Google Scholar]
- 19.Werner M, Hobeika E & Jumaa H Role of PI3K in the generation and survival of B cells. Immunol Rev 237, 55–71, (2010). [DOI] [PubMed] [Google Scholar]
- 20.Thalhamer T, McGrath MA & Harnett MM MAPKs and their relevance to arthritis and inflammation. Rheumatology (Oxford) 47, 409–414, (2008). [DOI] [PubMed] [Google Scholar]
- 21.Kurko J et al. Genetics of rheumatoid arthritis - a comprehensive review. Clin Rev Allergy Immunol 45, 170–179, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nagafuchi Y et al. Immunophenotyping of rheumatoid arthritis reveals a linkage between HLA-DRB1 genotype, CXCR4 expression on memory CD4(+) T cells, and disease activity. Sci Rep 6, 29338, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin Y, Wong K & Calame K Repression of c-myc transcription by Blimp-1, an inducer of terminal B cell differentiation. Science 276, 596–599, (1997). [DOI] [PubMed] [Google Scholar]
- 24.Nutt SL, Fairfax KA & Kallies A BLIMP1 guides the fate of effector B and T cells. Nat Rev Immunol 7, 923–927, (2007). [DOI] [PubMed] [Google Scholar]
- 25.Muto A et al. Identification of Bach2 as a B-cell-specific partner for small maf proteins that negatively regulate the immunoglobulin heavy chain gene 3’ enhancer. EMBO J 17, 5734–5743, (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eyre S et al. High-density genetic mapping identifies new susceptibility loci for rheumatoid arthritis. Nat Genet 44, 1336–1340, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moalli F et al. Pathogen recognition by the long pentraxin PTX3. J Biomed Biotechnol 2011, 830421, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roumenina LT et al. Interaction of C1q with IgG1, C-reactive protein and pentraxin 3: mutational studies using recombinant globular head modules of human C1q A, B, and C chains. Biochemistry 45, 4093–4104, (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang S & Wang Y Peptidylarginine deiminases in citrullination, gene regulation, health and pathogenesis. Biochim Biophys Acta 1829, 1126–1135, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khare S, Luc N, Dorfleutner A & Stehlik C Inflammasomes and their activation. Crit Rev Immunol 30, 463–487, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schreeder DM et al. Cutting edge: FcR-like 6 is an MHC class II receptor. J Immunol 185, 23–27, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu FL, Wu CC & Chang DM TACE-dependent amphiregulin release is induced by IL-1beta and promotes cell invasion in fibroblast-like synoviocytes in rheumatoid arthritis. Rheumatology (Oxford) 53, 260–269, (2014). [DOI] [PubMed] [Google Scholar]
- 33.Santos Savio A et al. Differential expression of pro-inflammatory cytokines IL-15Ralpha, IL-15, IL-6 and TNFalpha in synovial fluid from rheumatoid arthritis patients. BMC Musculoskelet Disord 16, 51, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Albaba D, Soomro S & Mohan C Aptamer-Based Screens of Human Body Fluids for Biomarkers. Microarrays (Basel) 4, 424–431, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Woodworth CD, McMullin E, Iglesias M & Plowman GD Interleukin 1 alpha and tumor necrosis factor alpha stimulate autocrine amphiregulin expression and proliferation of human papillomavirus-immortalized and carcinoma-derived cervical epithelial cells. Proc Natl Acad Sci U S A 92, 2840–2844, (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rao DA et al. Pathologically expanded peripheral T helper cell subset drives B cells in rheumatoid arthritis. Nature 542, 110–114, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wieduwilt MJ & Moasser MM The epidermal growth factor receptor family: biology driving targeted therapeutics. Cell Mol Life Sci 65, 1566–1584, (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zaiss DM, Gause WC, Osborne LC & Artis D Emerging functions of amphiregulin in orchestrating immunity, inflammation, and tissue repair. Immunity 42, 216–226, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamane S et al. Proinflammatory role of amphiregulin, an epidermal growth factor family member whose expression is augmented in rheumatoid arthritis patients. J Inflamm (Lond) 5, 5, (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Swanson CD et al. Inhibition of epidermal growth factor receptor tyrosine kinase ameliorates collagen-induced arthritis. J Immunol 188, 3513–3521, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harada M et al. Temporal expression of growth factors triggered by epiregulin regulates inflammation development. J Immunol 194, 1039–1046, (2015). [DOI] [PubMed] [Google Scholar]
- 42.Bartok B & Firestein GS Fibroblast-like synoviocytes: key effector cells in rheumatoid arthritis. Immunol Rev 233, 233–255, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bolin C et al. Oncostatin m promotes mammary tumor metastasis to bone and osteolytic bone degradation. Genes Cancer 3, 117–130, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kokkonen H et al. Antibodies of IgG, IgA and IgM isotypes against cyclic citrullinated peptide precede the development of rheumatoid arthritis. Arthritis Res Ther 13, R13, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baslund B et al. Targeting interleukin-15 in patients with rheumatoid arthritis: a proof-of-concept study. Arthritis Rheum 52, 2686–2692, (2005). [DOI] [PubMed] [Google Scholar]
- 46.Waldmann TA The biology of IL-15: implications for cancer therapy and the treatment of autoimmune disorders. J Investig Dermatol Symp Proc 16, S28–30, (2013). [DOI] [PubMed] [Google Scholar]
- 47.Machado Diaz AC et al. Proinflammatory soluble interleukin-15 receptor alpha is increased in rheumatoid arthritis. Arthritis 2012, 943156, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tarte K, Zhan F, De Vos J, Klein B & Shaughnessy J Jr. Gene expression profiling of plasma cells and plasmablasts: toward a better understanding of the late stages of B-cell differentiation. Blood 102, 592–600, (2003). [DOI] [PubMed] [Google Scholar]
- 49.Berasain C & Avila MA Amphiregulin. Semin Cell Dev Biol 28, 31–41, (2014). [DOI] [PubMed] [Google Scholar]
- 50.Tetta C, Camussi G, Modena V, Di Vittorio C & Baglioni C Tumour necrosis factor in serum and synovial fluid of patients with active and severe rheumatoid arthritis. Ann Rheum Dis 49, 665–667, (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vieth B, Ziegenhain C, Parekh S, Enard W & Hellmann I powsimR: power analysis for bulk and single cell RNA-seq experiments. Bioinformatics 33, 3486–3488, (2017). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.