Abstract
Bromodomain and extraterminal domain proteins, especially bromodomain-containing protein 4 (Brd4), have recently emerged as therapeutic targets for several cancers, although the role and mechanism of Brd4 in glioblastoma multiforme (GBM) are unclear. In this study, we aimed to explore the underlying mechanisms of the anti-tumor effects of Brd4 and the bromodomain inhibitor JQ1 on glioma stem cells (GSCs). In vitro, JQ1 and small interfering RNAs targeting Brd4 (siBrd4) inhibited the proliferation and self-renewal of GSCs. In vivo, JQ1 significantly inhibited the growth of xenograft GSCs tumors. The RNA-seq analysis revealed that the PI3K-AKT pathway played an important role in GBM. Vascular endothelial growth factor (VEGF) and VEGF receptor 2 phosphorylation was downregulated by exposure to JQ1 in GSCs, thereby reducing PI3K and AKT activity. In addition, treatment with JQ1 inhibited MMP expression, thereby inhibiting degradation of the extracellular matrix by MMP and angiogenesis in GBM tumors. Suppression of AKT phosphorylation inhibited the expression of the retinoblastoma/E2F1 complex, resulting in cell cycle arrest. In addition, treatment with siBrd4 or JQ1 induced apoptosis by activating AKT downstream target genes involved in apoptosis. In conclusion, these results suggest that Brd4 has great potential as a therapeutic target, and JQ1 has notable anti-tumor effects against GBM which may be mediated via the VEGF/PI3K/AKT signaling pathway.
Keywords: Brd4, GSCs, JQ1, epigenetics, VEGF/PI3K/AKT pathway
Introduction
Glioblastoma multiforme (GBM) is the most common and aggressive form of primary malignant brain tumor (1-3). Although multiple treatments have emerged in recent years, such as surgery, radiotherapy, and chemotherapy, the overall survival of patients with glioblastoma has not changed significantly (4,5). Glioma stem cells (GSCs) are increasingly recognized as a driving force supporting glioma development, resistance to treatment, and malignant recurrence (6). GSCs possess self-renewal capacity and multipotency, and can induce tumorigenesis (7). The elucidation of the molecular mechanism controlling GSCs may provide novel information for development of targeted treatment for GBM (3).
Bromodomain and extraterminal domain (BET) proteins are epigenetic readers that regulate a variety of important biologic processes, such as transcriptional regulation, chromatin remodeling, and DNA damage repair (7-9). BET proteins were revealed to have oncogenic effects for the first time in NUT midline carcinoma (10). By screening a library of small hairpin RNAs targeting known chromatin regulators in acute myeloid leukemia (AML), a subsequent study identified bromodomain-containing protein 4 (Brd4) as an important factor in the maintenance of AML (11). Recently, BET proteins, especially Brd4, have emerged as valuable therapeutic targets for treating several cancers, such as breast, prostate, and liver cancers (12-14). These findings indicated Brd4 could be a therapeutic target for treating GBM.
The small-molecule BET inhibitor JQ1 can competitively bind to acetylated lysine residues in the BET bromodomain to prevent binding of the BET protein with chromatin (15). BET bromodomain inhibitors can inhibit the transcription of c-Myc, and thus, there are potential targets for the clinical treatment of Myc-driven cancer, including GBM (16,17). In several cancers, JQ1 has notable anti-inflammatory and anti-tumor effects in vitro and in vivo (18,19). Although some studies have examined Brd4, its role in GBM tumorigenesis is yet to be determined, and the potential of JQ1 for treating GBM is largely unexplored.
To resolve these uncertainties, we investigated the effects of Brd4 in GSCs using JQ1 or small interfering RNAs (siRNAs) in vitro and in vivo. Transcriptome sequencing analysis of JQ1-treated GSCs revealed that Brd4 is closely associated with the PI3K-AKT pathway. Furthermore, our findings revealed the mechanism by which the vascular endothelial growth factor (VEGF)/PI3K/AKT axis regulates cell cycle progression and apoptosis of GSCs, which would in part explain the involvement of Brd4 in the anti-tumor effects of JQ1. We aimed to confirm the role of Brd4 in the tumorigenesis and progression of GBM, and reveal the underlying molecular mechanisms. Furthermore, our study identified JQ1 as a novel promising inhibitor for treating GBM.
Materials and methods
Cell culture and transfection
Primary murine glioma stem cells (CSC2078 and CSC1589) were isolated from human glial fibrillary acidic protein (hGFAP)-Cre+ p53L/L phosphatase tensin homolog (Pten)L/+ mice as previously described (20); cell isolation was not conducted in the present study. The numbers of GSCs were encoded as the same as the code number of transgenic mice. The hGFAP-Cre transgene to delete p53 alone or in combination with Pten in all CNS lineages using conditional p53 and Pten alleles as previously described (20). Mice (n=42, 15-40 weeks old, 20-22 g, Dana-Farber Cancer Institute) were interbred and maintained on FvB/C57Bl6 hybrid background. 73% of hGFAP-Cre+; P53lox/lox; Ptenlox/+ mice (between ages 15-40 weeks) presented with acute-onset neurological symptoms in clinical analysis. All 42 neurologically symptomatic mice harbored malignant gliomas were classified as anaplastic astrocytomas [World Health Organization (WHO) III, n=28, 66%] or GBM (WHO IV, n=14, 34%). In this study, all manipulations were performed with IACUC approval (20). Patient-derived glioma stem cells (GSCs) TS543 were a gift from Dr Cameron W. Brenner (Memorial Sloan Kettering Cancer Center) (21). CSC2078 and CSC1589 were cultured in neurobasal media (Dulbecco's Modified Eagle's medium/F12, HyClone; GE Healthcare Life Sciences) with epidermal growth factor (20 ng/ml, cat. no. 315-09, PeproTech, Inc.) and basic fibroblast growth factor (10 ng/ml, cat. no. 450-33, PeproTech, Inc.) or differentiation medium (neurobasal media with 1% FBS). TS543 cells were cultured in NeuroCult NS-A proliferation media (human; Stem Cell Technologies) supplemented with epidermal growth factor (20 ng/ml, cat. no. GMP100-15, PeproTech, Inc.) and basic fibroblast growth factor (10 ng/ml, cat. no. 100-18B, PeproTech, Inc.). Cells were cultured at 37°C in a 5% CO2.
CSC2078 cells were seeded into a 6-well plate at 2×105 cells per well overnight. The following day, CSC2078 were transfected with three independent Brd4-specific siRNAs (20 µM; siBrd4s-475, siBrd4s-1648, siBrd4s-3820) and a negative control siRNAs (20 µM) for 48 h. siRNAs was purchased from Gene Pharma and the sequences were described in Table SI. The transfection agent INTERFERin (Thermo Fisher Scientific, Inc.) was used according to the manufacturer's protocol. The efficiency of transfection was determined by western blotting.
Cell viability, cell proliferation, and self-renewal assay
To determine the dose response to JQ1, cells were seeded into a 96-well plate in triplicate at the density of 10,000 cells per well and adhered overnight. Cells were treated with serial dilutions of JQ1 at 37°C for 24 or 48 h. JQ1 was added by 2-fold serial dilutions (the concentration of JQ1 for CSC2078 and TS543: 0.05, 0.1, 0.2, 0.4 and 0.8 µM; the concentration of JQ1 for CSC1589: 0.0625, 0.125, 0.25, 0.5 and 1 µM). CSC2078 cells transfected with siRNAs for 24, 48 and 72 h. The Cell Counting Kit-8 (CCK-8) solution (10 µl, MedChem Express) was added to each well of the plate. Then, the plate was incubated at 37°C for 4 h. The absorbance was measured at 450 nm using a microplate reader. The Cell Counting Kit-8 (cat. no. HY-K0301) and JQ-1 (cat. no. HY-13030) was purchased from MedChem Express.
For the soft agar colony assay, 2,500 cells were plated in the upper layer for each well in 6-well plate in triplicate. The concentration of the lower layers of agar was 0.6% and the upper layers were 0.4%. The agar was mixed with the neurobasal medium or NeuroCult NS-A proliferation media at a ratio of 1:1. The number of colonies in five random microscopic fields were counted after 10-15 days of seeding GSCs and staining by 0.1% crystal violet for 1 h at room temperature. Images were captured using a light microscope at ×400 magnification in five random microscopic fields (BX71 Olympus Corporation).
Self-renewal capacity was measured by neural sphere formation assays. The cells were cultured into 6-well plates (100/well) in triplicate and treated with JQ1 (0.1, 0.2, 0.25 and 0.5 µM) for 24 h and transfected with siRNAs for 48 h at 37°C. The neural spheres were cultured in NSC proliferation media (HyClone; GE Healthcare Life Sciences) containing growth factors (epidermal growth factor, 20 ng/ml; basic fibroblast growth factor, 10 ng/ml) for 10 days at 37°C. After 10 days, neurospheres containing >50 cells were scored (22). The number of neural spheres in each well was counted under an IX71 light microscope (Olympus Corporation) at ×100 and 200 magnification. Data were presented as the percentage of sphere-forming cells relative to control.
Cell-cycle distribution
Cells were seeded in 6-well plates (1×105 cells/well) and treated with JQ1 (0.05, 0.1, 0.125, 0.2, 0.25 and 0.5 µM) and/or linifanib (5 µM, cat. no. HY-50751, MedChem Express) for 24 h. CSC2078 cells were transfected with siRNAs for 48 h. We fixed cells with 1 ml 70% ethanol overnight at 4°C. The collected cells were detected using a propidium iodide (PI)/RNase Staining Solution (cat. no. CY2001-P Sungene Biotech) according to the manufacturer's protocols. The cell cycle was analyzed using a flow cytometer (BD Biosciences) and ModFit LT 5.0 (Verity Software House).
Annexin V apoptosis assay, terminal deoxynucleotidyl-transferase-mediated dUTP nick end labeling (TUNEL) assay
Cells were seeded in 6-well plates overnight at a density of 1×105 cells/well. Then, cells were treated with JQ1 and/or linifanib for 24 h and were transfected with siRNAs for 48 h. The vehicle-, JQ1/linifanib and JQ1 + Linifanib-treated cells were stained with Annexin V-fluorescein isothiocyanate and PI according to the instructions provided with the Apoptosis Detection Kit (cat. no. WLA001, Wanleibio Co., Ltd.). The vehicle-treated cells were treated with dimethyl sulfoxide (DMSO; maximum final concentration of 0.01%). The apoptosis rate was calculated as the sum of early apoptotic and late apoptotic cells Apoptosis was analyzed using flow cytometry.
A TUNEL assay was conducted using the DeadEnd™ Colorimetric TUNEL System (Promega Coporation) and DeadEnd™ Fluorometric TUNEL System (Promega Corporation). The DeadEnd™ Colorimetric TUNEL System was used to detect the apoptosis of CSC2078 treated with JQ1. Cells were treated with JQ1, or vehicle (DMSO at a maximum final concentration of 0.01%) for 24 h. The DeadEnd™ Fluorometric TUNEL System was used to detect the apoptosis of tumors of CSC2078-subcutaneous xenograft mice as described below. The TUNEL assay was performed according to the manufacturer's instructions. Images of cells and tumor tissues were captured using an Olympus BX53 microscope (light and fluorescence) at ×400 magnification. For quantitation, the TUNEL-positive cells were counted in five random area images for each sample using ImageJ 1.46r (National Institutes of Health).
Hoechst 33342 staining
CSC1589 cells were treated with JQ1 for 24 h. Then, the CSC1589 were fixed with 4% paraformal-dehyde for 5 min. Then, CSC1589 were stained with 1 mg/ml Hoechst (Beyotime Biotechnology Inc., Nantong, China) for 10 min at room temperature in the dark. Finally, the cells were washed three times with PBS and imaged by Olympus BX53 microscope (fluorescence) at ×400 magnification.
RNA isolation, reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA of CSC2078 and TS543 cells was extracted using TRIzol (Thermo Fisher Scientific, Inc.) according to the manufacturer's protocols. Following isolation, total RNA was reverse transcribed using the Revert Aid First Strand cDNA Synthesis Kit (cat. no. K1622; Thermo Fisher Scientific, Inc.). Briefly, RNA (2 µg) was incubated for 60 min at 42°C and for 5 sec at 70°C. qPCR was conducted using the SYBR® Premix Ex Taq™ II (Tli RNaseH Plus, cat. no. RR820A, Takara Bio, Inc.) according to the manufacturer's protocols. The thermocycling conditions were as follows: 95°C initial denaturation for 30 sec; followed by 40 cycles of denaturation at 95°C for 5 sec, and annealing and extension at 60°C for 30 sec) using CFX96 (Bio-Rad Laboratories, Inc.). After being normalized to the expression levels of GADPH, the relative mRNA expression levels were determined by the 2−ΔΔCq method (23). The primers used are described in Table SII.
Protein extraction and western blotting
GSCs were lysed in phenylmethylsulfonyl fluoride and radioimmunopre-cipitation assay lysis buffer (1:100, Wuhan Boster Biological Technology, Ltd.), and the collected lysates were treated by sonication at 4°C, then centrifuged at 12,000 × g at 4°C for 15 min. The supernatants were used as cell lysates. The protein concentration was analyzed by a BCA protein assay kit (cat. no. PP1001, Bioteke Corporation). Samples containing 30 µg of protein were separated by SDS-PAGE (10, 12 and 15%) and transferred onto polyvinylidene difluoride membranes which were probed with antibodies. Prior, the membranes were blocked with 5% non-fat milk in Tris-buffered saline containing 0.05% Tween-20 at room temperature for 1 h. Subsequently, the membranes were incubated overnight at 4°C with the following antibodies (1:1,000 dilutions) against BRD4 antibody [EPR5150(2); ab128874) from Abcam; phosphorylated (phosphor)-AKT (Ser473, cat no. 4060) and phospho-retinoblastoma (Rb; Ser807/811, cat. no. 9308S) from Cell Signaling Technology, Inc.; total AKT (cat. no. 10176-2-AP), c-Myc (cat. no. 10828-1-AP), Bcl2-associated X protein (BAX; cat. no. 50599-2-lg), BCL2 (cat. no. 12789-1-AP), caspase 3/p17/p19 (cat. no. 19677-1-AP), P27 (cat. no. 25614-1-AP), E2F1 (cat. no. 12171-1-AP), matrix metalloproteinase 2 (MMP2; cat. no. 10373-2-AP), MMP9 (cat. no. 10375-2-AP) and Actin antibody from ProteinTech Group, Inc.; CCND1 (A11022) from ABclonal Biotech Co., Ltd.; proliferating cell nuclear antigen (PCNA; PC10, SC-56) from Santa Cruz Biotechnology, Inc.; Rb (G3-245) from BD Pharmingen (BD Biosciences); VEGF (cat. no. 05-443) from Upstate Biotechnology; Histone H2A.X (D17A3) XP (H2AX, cat. no. 7631), phospho-Histone H2A.X (Ser139, 20E3, γH2AX; cat. no. 9718), VEGFR2 (cat. no. 55B11) and phospho-VEGF receptor 2 (Tyr1175, cat. no. 19A10) from Cell Signaling Technology, Inc. AKT (phospho Thr308) from Immunoway Biotechnology. The secondary antibodies, horseradish peroxidase (HRP)-conjugated Affinipure Goat Anti-Mouse IgG (H+L) (cat. no. SA00001-1; 1:2,000) and HRP-conjugated Affinipure Goat Anti-Rabbit IgG (H+L) (cat. no. SA00001-2, 1:2,000) antibodies were purchased from ProteinTech Group, Inc. After visualization using the enhanced chemiluminescence (Thermo Fisher Scientific, Inc.), the density of the bands was analyzed using image analysis software (Image J 1,46r).
Immunofluorescence
CSC1589 cells were cultured in 12-well plates at density of 1×105 cells/well and treated with JQ1 for 0.125 or 0.25 µM. Single cell suspensions were seeded on coverslips pre-coated with fibronectin (1 µg/ml, cat. no. F1141-5MG, Sigma-Aldrich; Merck KGaA) and ornithine (50 mg/ml, P3655-50MG, Sigma-Aldrich; Merck KGaA) overnight. CSC1589 cells were fixed with 4% paraformaldehyde at room temperature for 10-15 min and permeabilized with 0.3% Triton X-100 (Sigma-Aldrich; Merck KGaA) in PBS for 5 min. Then, potential non-specific binding sites were blocked with antibody dilution buffer [10% fetal bovine serum and (Sigma-Aldrich; Merck KGaA) and 1% bovine serum albumin (Gentihold) in PBS]. Sections was incubated with an antibody against Nestin (ab6320, 1:50; Abcam) overnight at 4°C. After washing the sections with PBS for 5 min, secondary antibodies were applied [Donkey anti-Mouse IgG (H+L) ReadyProbes™ Secondary Antibody, Alexa Fluor® 488, R37114, 1:5,000; Invitrogen; Thermo Fisher Scientific, Inc.] for 30 min at room temperature before mounting in water soluble mounting medium (Wuhan Boster Biological Technology, Ltd.) with DAPI (room temperature, 2 min). After the final washing step by PBS, the glass coverslips were mounted upside-down and visualized with an Olympus BX53 microscope at ×400 magnification.
RNA-seq analysis
Total RNA of CSC2078 cells was isolated using the TRIzol, after which the quality, concentration and integrity were determined using a NanoDrop spectrophotometer (NanoDrop Technologies; Thermo Scientific Fisher, Inc.). RNA (3 µg) was used for the RNA sample preparations. Sequencing libraries were generated using the TruSeq RNA Sample Preparation Kit (Illumina, Inc.). Briefly, mRNA was purified from total RNA using poly-T oligoattached magnetic beads. Fragmentation was carried out using divalent cations under elevated temperature (94°C, 5 min) in an Illumina proprietary fragmentation buffer. First strand cDNA was synthesized using random oligonucleotides and SuperScript II (25°C for 10 min; 42°C for 15 min; 70°C for 15 min, Illumine, Inc.) reverse transcriptase. Then, the second strand cDNA synthesis was performed by DNA Polymerase I and RNase H (16°C for 1 h, Illumine, Inc.). Remaining overhangs were converted into blunt ends via exonuclease/polymerase activities and the enzymes were removed. After adenylation of the 3′ends of the DNA fragments, Illumina PE adapter oligonucleotides were ligated to prepare for hybridization. To select cDNA fragments of the preferred 200 bp in length, the library fragments were purified using the AMPure XP system (Beckman Coulter, Inc.). DNA fragments with ligated adaptor molecules on both ends were selectively enriched using Illumina PCR Primer Cocktail (Illumina, Inc.) in a 15 cycle PCR reaction (Applied Biosystems; Thermo Fisher Scientific, Inc.). The thermocycling conditions were as follows: 98°C initial denaturation for 30 sec; followed 98°C denaturation for 10 sec, and 60°C annealing for 30 sec; and 72°C extension for 30 sec. Products were purified (AMPure XP system) and quantified using the Agilent high sensitivity DNA assay on a Bioanalyzer 2100 system (Agilent Technologies, Inc.). The sequencing library was then sequenced on a Hiseq platform (Illumina, Inc.) by Shanghai Personal Biotechnology Co. Ltd.
Gene ontology (GO) and Kyoto encyclopedia of genes and genomes (KEGG) pathway analysis
We used HTSeq(0.9.1) statistics (https://htseq.readthedocs.io/en/release_0.11.1/history.html#version-0-9-1) to compare the Read Count values on each gene as the original expression of the gene, and then used fragments per kilobase of transcript per million to standardize the expression. Then we used DESeq (1.30.0) (24) to analyze the genes of difference expression with screened conditions as follows: Expression difference multiple |log2FoldChange| >1, significance, P<0.05. In addition, we used R language Pheatmap (1.0.8) software package (https://cran.r-project.org/web/packages/pheatmap/index.html) to perform bi-directional clustering analysis of all different genes of samples. We mapped all the genes to terms in the GO database and calculated the numbers of differentially enriched genes in each term. Based on the whole genome, terms with significant enrichment of differentially enriched genes were calculated by hypergeometric distribution. We counted the number of differentially expressed genes following KEGG pathway enrichment analysis (https://www.genome.jp/kegg/); the metabolic pathways and signaling pathways that differentially expressed genes mainly participate in were identified.
Animal studies
BALB/c male nude mice (age, 5-8 weeks) were obtained from Beijing Vital River Laboratory Animal Technology. A total of 10 mice each weighing 18-22 g were used. Mice were housed under a 12 h light/dark cycle in an air-conditioned room at 22±2°C with free access to food and water. In xenograft implantation experiments, nude mice were injected subcutaneously in the right flank with CSC2078 cells (1×107) suspended in 100 µl PBS. After 2 days, nude mice of the JQ1-treated group were administered intraperitoneally with JQ1 (50 mg/kg/day) for 17 days. DMSO-containing JQ1 was diluted with physiological saline to obtain a final solution of 5 mg/ml in JQ1-treated groups. Mice of the control group were injected an equal amount of physiological saline containing 5% DMSO per day. We measured the weight and tumor size every day. Tumor size was measured with calipers. The tumor volume was calculated with the following formula: V=(length) 2× (width)/2. The humane endpoints were judged by mouse activity assessment (hunching, stationary, poor grooming and ruffling) or the mouse weight loss (>20% of total body weight). No animals were sacrificed due to meeting these endpoints; all mice were sacrificed via carbon dioxide after 17 days. The 10 mice were anesthetized by an intraperitoneal injection of 10% chloral hydrate at a dosage of 300 mg/kg body weight prior to carbon dioxide euthanasia; 100% carbon dioxide was applied 10-30% volume per minute and the mice were euthanized after exposure to carbon dioxide for 5-6 min. Mice death was confirmed when no spontaneous breathing for 2-3 min and no blinking reflex were observed. No secondary method was applied to verify mice death. After the mice were euthanized, tumor tissues were extracted were divided into two parts: One part was frozen at -80°C for protein and RNA extraction, and the other part was fixed in 4% paraformaldehyde (room temperature, 24 h) for immunofluorescent staining. The splanchnic tissues (heart, liver, spleen, lung and kidney) were excised for hemoxylin and eosin (H&E) staining. The largest tumor in the control group had a diameter of 13 mm and the JQ1-treated group had a diameter of 7 mm. Animal experiments were approved by the Animal Ethics Committee of Jilin University [approval no. 2018;(54)], and by the Institutional Committee for the Care and Use of Laboratory Animals of the Experimental Animal Center of Jilin University.
Immunohistochemistry
Animals of the vehicle- and JQ1-treated CSC2078 xenograft model groups were sacrificed and the tumors were rinsed in PBS, followed by fixation with 4% paraformaldehyde for 24 h at room temperature. The tumors were cut into five-µm thick sections and mounted on poly-D-lysine-coated slides. The sections were then deparaffinized with xylene and rehydrated via alcohol, followed by 100% alcohol for 5 min, 95% for alcohol for 5 min, 75% alcohol for 5 min and 50% alcohol for 5 min. Then, the sections were washed with PBS (1X) for 5 min twice at room temperature. Antigen retrieval was performed by application of citrate buffer pH 6.00 for 20 min. The slides were incubated with primary antibodies overnight at 4°C, against c-Myc (cat. no. 10828-1-AP, 1:100), BAX (cat. no. 50599-2-lg, 1:100), BCL2 (cat. no. 12789-1-AP, 1:100), MMP2 (cat. no. 10373-2-AP, 1:100) and MMP9 (cat. no. 10375-2-AP, 1:100) from ProteinTech Group, In. Subsequently, HRP secondary antibodies were applied for 40 min at 37°C (SV0002; Wuhan Boster Biological Technology, Ltd.) according to the manufacturer's protocols. DAB (DA1010 Beijing Solarbio Science & Technology Co., Ltd.) was applied at room temperature for 15 min and analysis was performed using a BX53 microscope (Olympus Corporation).
Statistical analyses
Statistical analyses were performed with Microsoft Excel (Microsoft Corporation) and GraphPad Prism 6 software (GraphPad Software, Inc.). Comparison of the two sets of data was using an unpaired Student's t-test. For comparison of more than two sets, one-way ANOVA with a Newman-Keuls post-hoc test was conducted. All quantifications were performed with at least three independent experiments. Data are expressed as the mean ± standard error of the mean. P<0.05 was considered to indicate a statistically significant difference.
Results
Brd4 is a potential therapeutic target of GBM
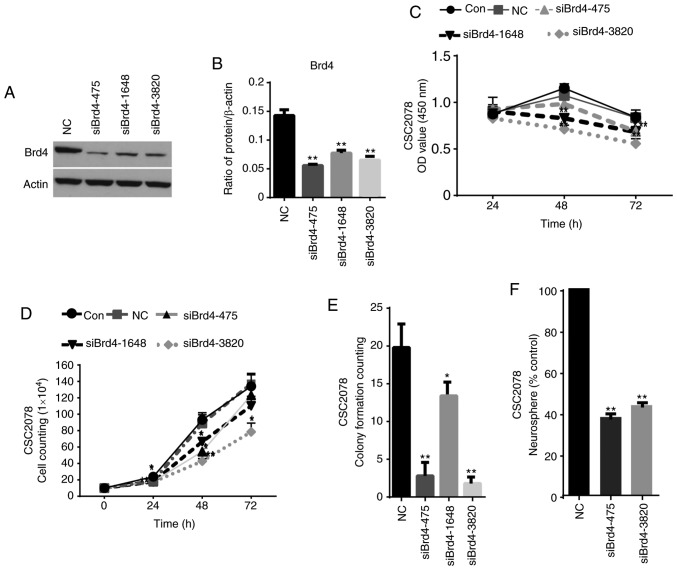
To evaluate the role of epigenetic genes in GSCs, we acquired GSCs from hGFAP-Cre+ p53L/L PtenL/+ mice (20). We used siRNAs to silence the expression of Brd4 to detect the effects of Brd4 on the activity, proliferation and self-renewal ability of GSCs. Then, western blotting was performed to detect the protein expression of Brd4 in CSC2078 transfected with the Brd4-specific siRNAs. Compared with the control group, we found that the expression of Brd4 was significantly inhibited in the CSC2078 cells treated with siBrd4s, especially siBrd4s-475 and siBrd4s-3820. This demonstrated the effectiveness of siRNAs targeting Brd4 (Fig. 1A and B). We next explored the effects of silencing Brd4 on the proliferation and self-renewal of GSCs. We detected the cell viability of CSC2078 transfected with siBrd4s using a CCK-8 assay. The results showed that silencing of Brd4 significantly reduced cell viability (Fig. 1C). A cell growth curve and a soft agar colony formation assays demonstrated that the three independent Brd4 siRNAs had significant anti-proliferative effects on CSC2078 cells compared with the control (Figs. 1D and E, and S2A). Then, we conducted neural sphere formation assays to detect the effects of the most effective siRNAs on the self-renewal ability of CSC2078 cells. The results revealed that the number of neural spheres in the siBrd4 groups were significantly lower than the control group (Fig. 1F). The results illustrated that siRNAs targeting Brd4 effectively inhibited the self-renewal ability of CSC2078 cells (Figs. 1F and S2E). Taken together, these results suggested that Brd4 had important functions in mediating an anti-tumor effect on GSCs, and that Brd4 could be a potential therapeutic target of in ablating GSCs.
Figure 1.
Comprehensive analyses identify Brd4 as a therapeutic target in glioblastoma multiforme. (A and B) Western blotting analysis of Brd4 expression in CSC2078 treated with siRNAs. (C) CSC2078 cells were infected with si-NC or siBrd4 (20 µM). Cell viability was detected by the CCK-8 assay. Error bars represent standard error of the mean. (D) Cell proliferation was assessed by cell growth counting assay. Error bars represent standard error of the mean. (E) Soft agar assay for colony formation of CSC2078 treated with negative control siRNAs or siBrd4 (mean ± standard error of the mean of five fields). (F) Sphere-forming ability of CSC2078 transfected with si-NC or siBrd4s treatment were measured (mean ± standard error of the mean of five fields). *P<0.05 and **P<0.01 vs. Con or NC. Brd4, bromodomain-containing protein 4; Con, control; NC, negative control; si, siRNA, small interfering RNA.
JQ1 suppresses the proliferation and self-renewal of GSCs
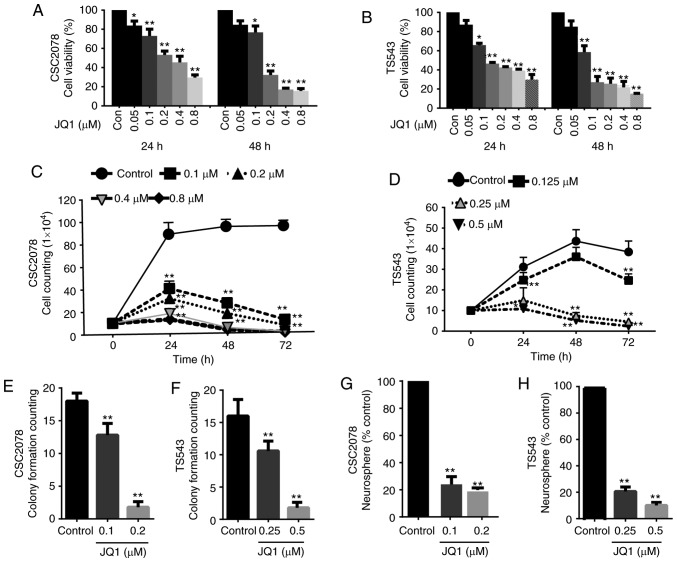
To explore the effects of JQ1 on tumorigenic properties, we assessed cell proliferation and self-renewal in TS543, CSC2078 and CSC1589. CCK-8 assays revealed that cell viability was significantly reduced compared with the control after 24 and 48 h of treatment with JQ1 in a concentration-dependent manner (Figs. 2A and B, and S1A). Next, we analyzed cell proliferation using cell growth curves following 3 days of treatment of CSC2078, TS543 and CSC1589 with JQ1. JQ1 significantly inhibited the proliferation of all three GSC lines compared with the control (Figs. 2C and D, and S1B). In addition, JQ1 significantly reduced the clone formation capacity of GSCs compared with the control. Specifically, the number of cell clones was significantly lower after 10 days of treatment with JQ1. This further supported that JQ1 effectively inhibited GSCs proliferation (Figs. 2E and F, S1C and D, and S2A). In addition, neural sphere formation assays revealed that neural sphere numbers were significantly decreased by treatment with various concentrations of JQ1 for 10 days compared with the control (Figs. 2G and H, and S2B-D).
Figure 2.
Treatment with JQ1 reduces the proliferation and self-renewal of glioma stem cells. (A and B) Cell viability of CSC2078 and TS543 was detected by a Cell Counting Kit-8 assay. CSC2078 and TS543 cells were treated with JQ1 for 24 and 48 h (error bars represent the standard error of the mean). (C and D) Cell proliferation of CSC2078 and TS543 cells following treatment with JQ1 was assessed by a cell growth counting assay (error bars represent the standard error of the mean). (E and F) Soft agar assay for the colony formation of CSC2078 and TS543 cells treated with JQ1 for 10 days (means ± represent the standard error of the mean of five fields). (G and H) Sphere-forming ability of CSC2078 and TS543 cells treated with JQ1 for 10 days were measured (means ± standard error of the mean of five fields). *P<0.05, **P<0.01 vs. control.
These results demonstrated that the treatment of GSCs with JQ1 has similar effects as silencing of Brd4. The anti-neoplastic effects of JQ1 effectively inhibited the proliferation and self-renewal of GSCs.
JQ1 and siBrd4 inhibits cell proliferation by inducing G1 arrest and apoptosis of GSCs
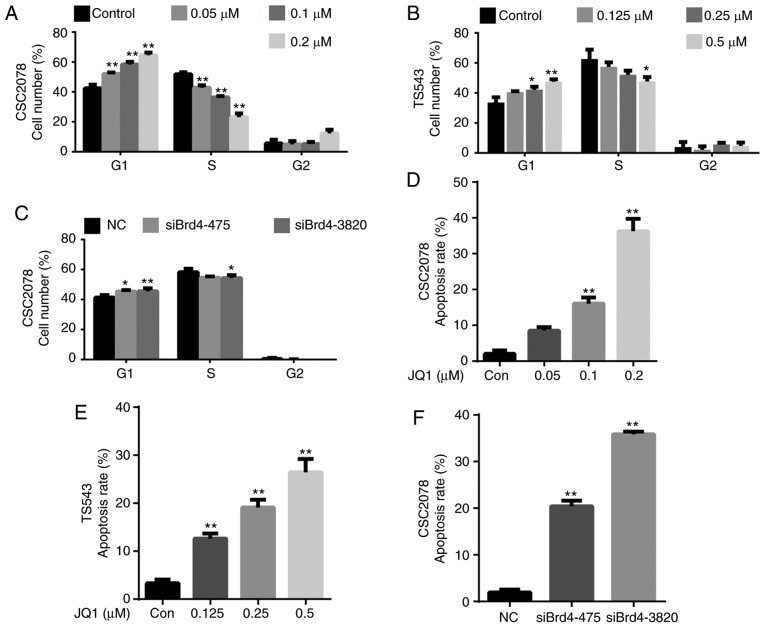
To further explore the mechanisms by which JQ1 and siBrd4 inhibit cell proliferation, we evaluated their effects on the cell cycle and apoptosis of GSCs. We analyzed the cell cycle via flow cytometry of CSC2078 and TS543 cells treated with JQ1 for 24 h, and CSC2078 transfected with siBrd4 for 48 h. We found that JQ1 significantly increased the percentage of cells in G1 and reduced that in S phase compared with the control; notable effects on the percentage of cells in G2/M phase were observed (Figs. 3A and B, and S3). We observed a similar G1/S cell cycle arrest effect in CSC2078 transfected with siBrd4 (Figs. 3C and S3). Collectively, these data suggested that JQ1 impeded cell cycle progression.
Figure 3.
Inhibition of Brd4 by JQ1 or siRNAs induced cell cycle arrest and apoptosis in glioma stem cells. (A-C) CSC2078 and TS543 were treated with JQ1 for 24 h. CSC2078 cells were transfected with si-NC or siBrd4 for 48 h. Cell cycle was analyzed by flow cytometry. Quantitation of the cell cycle of CSC2078 and TS543. Data are representative of three independent experiments (error bars represent standard error of the mean). (D-F) CSC2078 and TS543 cells were treated with JQ1 for 24 h. CSC2078 cells were transfected with siRNAs for 48 h. Apoptosis was analyzed by flow cytometry. Quantitation of the apoptosis rate of CSC2078 and TS543. Data are representative of three independent experiments (error bars represent standard error of the mean). *P<0.05, **P<0.01 vs. control or NC. Brd4, bromodomain-containing protein 4; Con, control; NC, negative control; si, siRNA, small interfering RNA.
Treatment with JQ1 and siBrd4s also induced apoptosis in GSCs. We assessed the apoptosis of CSC2078 and TS543 cells via flow cytometry using Annexin V/PI staining. The number of apoptotic cells was significantly increased in both cells lines treated with JQ1 compared with the control (Figs. 3D and E, S4, S7 and S8A-D). Similarly, silencing of Brd4 using siRNAs significantly induced the apoptosis of CSC2078 cells compared with the control (Figs. 3F, S4, and S8E-G). Consistent with the aforementioned results, Hoechst 33342 staining illustrated that the number of apoptotic CSC1589 cells was significantly with JQ1 treatment than in the control group (Fig. S5A and B). The results of TUNEL staining also confirmed the ability of JQ1 to promote the apoptosis of CSC2078 cells (Fig. S5C and D). These data suggested that JQ1 reduces the viability and inhibits the proliferation of GSCs by inducing cell cycle arrest and apoptosis.
Downregulation of Brd4 by JQ1 can potentially promote GSC differentiation
GO enrichment analysis revealed that treatment of CSC2078 with JQ1 induced multiple changes of biological activities, among which 'cell differentiation' was particularly prominent (Fig. S6A). RNA-seq analysis of differentially expressed genes (DEGs) illustrated that the neural stem cell (NSC) markers Nestin and ciliary neurotrophic factor were significantly downregulated in the JQ1-treated groups (Fig. S6B). We detected the expression of Nestin via immuno-fluorescence staining of CSC1589 cells. The results indicated that protein expression levels of Nestin were significantly downregulated in JQ1-treated cells (Fig. S6C and D). Then, we detected the mRNA expression of Nestin by RT-qPCR in CSC2078. The results indicated that the mRNA expression levels of Nestin were significantly downregulated after treatment with JQ1 for 24 h (Fig. S6E). In addition, the astrocyte marker S-100β was upregulated, whereas the oligodendrocyte marker platelet-derived growth factor α (PDGFα) was strongly downregulated (Fig. S6B). The neuronal precursor marker class III b-tubulin was markedly upregulated in JQ1-treated cells. The results revealed that JQ1 can inhibit the expression of NSC markers and promote GSC differentiation to astrocytes under proliferative conditions (Fig. S6B).
PI3K/AKT signaling pathway plays an important role in mediating the anti-tumor effect of JQ1
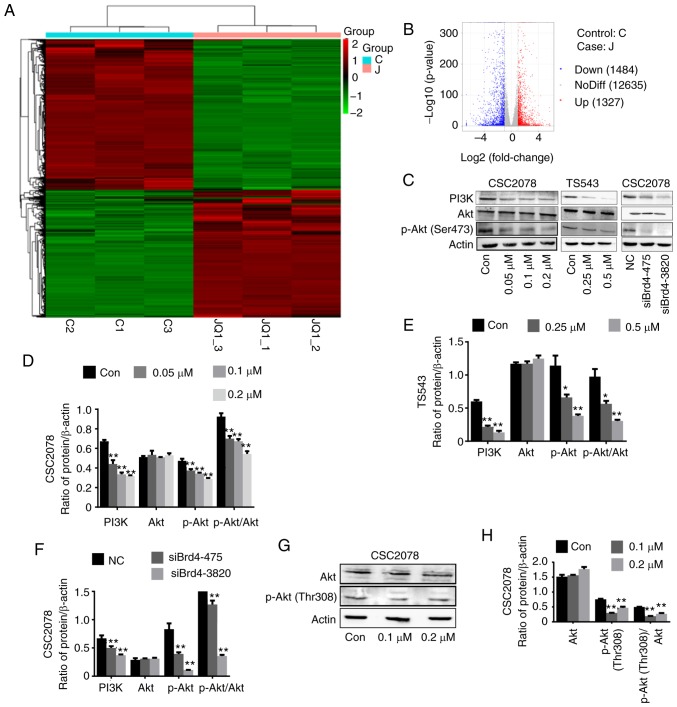
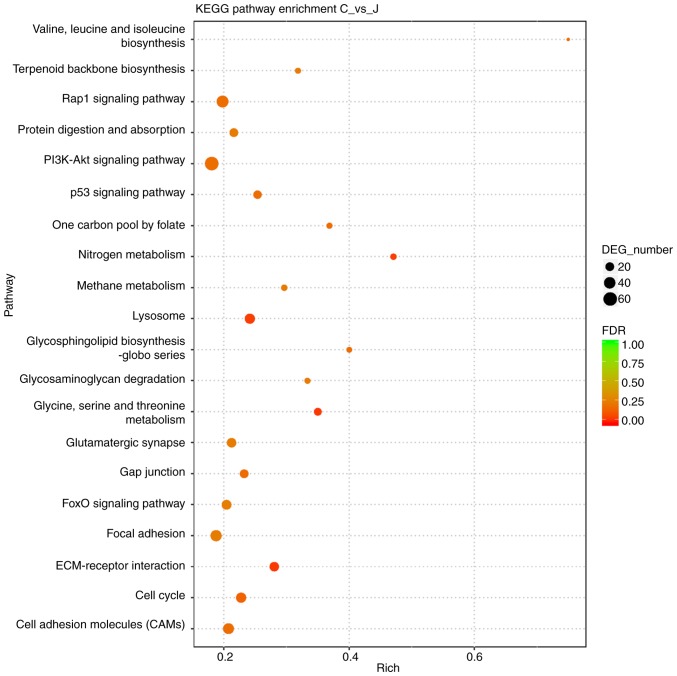
To further explore the mechanism by which Brd4 regulates the growth of GSCs, we performed RNA-seq comparisons of the CSC2078 cells treated with JQ1 for 24 h. The results revealed that 1,327 and 1,484 genes were upregulated and downregulated, respectively (Fig. 4A and B). KEGG is a collection of databases containing genomes, biological pathways, diseases. Compared with the whole genomic background, these databases can identify pathways of significant enrichment between candidate target genes (25). The top 20 pathways of candidate target gene enrichment were shown in the enrich scatter diagram, and enrichment factors, q-value and gene number were used to report KEGG enrichment degree. Our results showed that PI3K/AKT signaling pathway was significantly enriched and the q-value of this pathway was close to zero; the DEG numbers were higher (Fig. 5). Therefore, we used western blotting to investigate the relationship between Brd4 and the PI3K/AKT signal pathway. The results demonstrated that inhibition of Brd4 using siRNAs or JQ1 inhibited PI3K and phospho-AKT (Ser473 or Thr308); thus, we speculated that Brd4 regulates GSC growth through PI3K/AKT signaling (Fig. 4C-H).
Figure 4.
PI3K/AKT signaling pathway serves an important role in glioblastoma multiforme. (A) The cluster heat map of expression patterns of upregulated and downregulated genes of CSC2078 after treatment with JQ1 as detected by RNA-Seq (upregulated, red; downregulated, green). (B) Different gene number of CSC2078 after JQ1 treatment vs. control. (C-F) Western blotting analysis expression of PI3K, AKT and p-AKT (Ser473) in control, JQ1-treated group and siBrd4 group (error bars represent standard error of the mean). (G and H) Western blotting analysis expression of p-AKT (Thr308) in CSC2078 cells treated with JQ1 (error bars represent standard error of the mean), *P<0.05, **P<0.01 vs. control. P, phosphorylated; siBrd4, small interfering RNA against bromodomain-containing protein 4.
Figure 5.
KEGG pathway categories of DEGs in CSC2078 from RNA-Seq analysis of gene expression changes in CSC2078 cells treated with JQ1 for 24 h. The 20 most significantly enriched KEGG pathways of CSC2078 after treatment with JQ1. The x-axis represented KEGG enrichment scores and the y-axis represented pathway terms. The colors of circle indicated FDR and the size of circle indicated the numbers of DEGs. Large red circles indicated that the enrichment of the pathway was higher and the number of DEG number was larger in the pathway. DEGs, differentially expressed genes; FDR, false discovery rate; KEGG, Kyoto Encyclopedia of Genes and Genomes.
JQ1 exerts anti-tumor effects in GSCs through the VEGF/PI3K/AKT signaling pathway
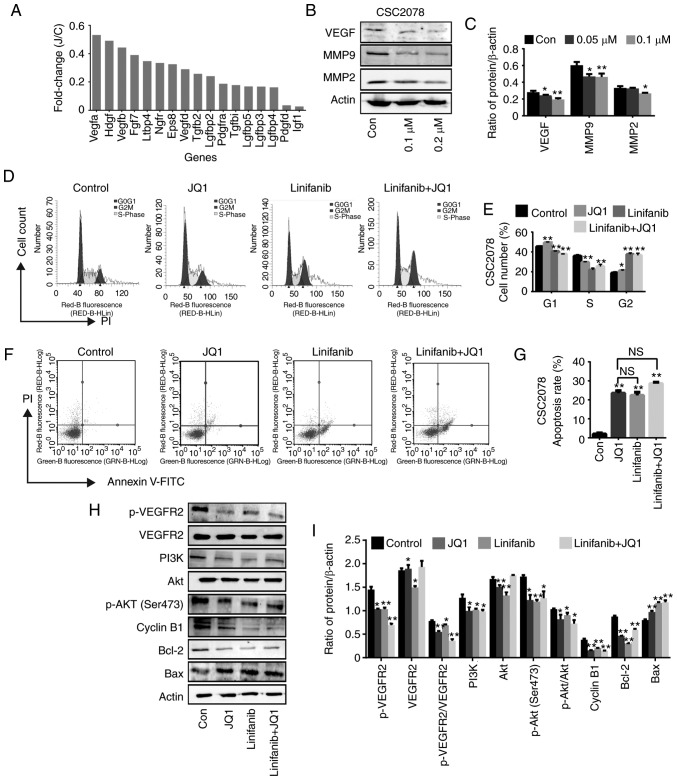
It has been reported that PI3K signaling is activated by various growth factors, including VEGF (26). We mainly analyzed the downregulated growth factors in CSC2078 cells treated with JQ1 from the results of RNA-seq analysis It was found that VEGF was inhibited to the highest degree (Fig. 6A). VEGF is released by cancer cells to induce tumor angiogenesis and can be secreted from the extracellular matrix (ECM) by MMP9 to promotes tumor growth (27). Thus, we used western blotting to detect the expression of VEGF and MMP in CSC2078 cells treated with JQ1. The results demonstrated that the expression of VEGF, MMP2 and MMP9 were significantly inhibited by JQ1 (Fig. 6B and C). Therefore, we preliminarily speculated that JQ1 exhibits anti-tumor effects via the VEGF/PI3K/AKT signaling pathway. In addition, we proposed that MMP2 and MMP9 proteins downstream of VEGF can be downregulated by JQ1, controlling the angiogenesis and metastasis of GBM and inhibiting tumor development.
Figure 6.
JQ1 induces cycle arrest and apoptosis of glioma stem cells through the VEGF/PI3K/AKT signaling pathway. (A) The mRNA expression of genes related growth factors in CSC2078 expressed as fold change. (B and C) Western blotting analysis of VEGF, MMP9 and MMP2 expression in CSC2078 treated with JQ1 (error bars represent standard error of the mean). (D and E) CSC2078 cells were treated with JQ1 and or linifanib for 24 h. JQ1=100 nM, linifanib=5 µM. The cell cycle was analyzed by flow cytometry. Quantitation of the cell cycle. Data are representative of three independent experiments (error bars represent standard error of the mean). (F and G) CSC2078 cells were treated with JQ1 and or linifanib for 24 h. JQ1=100 nM, linifanib=5 µM. Effects of CSC2078 apoptosis were detected by flow cytometry. Quantitation of the apoptosis rate of CSC2078. Data are representative of three independent experiments. (Error bars represent standard error of the mean). (H and I) Western blotting analysis of p-VEGFR2, VEGFR2, PI3K, Akt, p-Akt (Ser473), CyclinB1, Bcl-2 and Bax in control, JQ1 and/or linifanib treated groups. JQ1=100 nM, linifanib=5 µM. Densitometry was performed and fold change of protein expression was presented (error bars represent standard error of the mean). *P<0.05, **P<0.01 vs. Con. Bax, Bcl2-associated X protein; Con, control; J/C, JQ1/Con; MMP, matrix metalloproteinase; p, phosphorylated; VEGFR2, vascular endothelial growth factor; VEGFR2, receptor 2.
In order to further verify that JQ1 plays an anti-tumor role in GSCs through the VEGF/PI3K/AKT signaling pathway, we selected the VEGR2 inhibitor linifanib for further experiments. Linifanib is a novel tyrosine-kinase inhibitor, which selectively targets VEGFR and PDGFR, and has low off-target inhibitory activity (28). We analyzed the cell cycle via flow cytometry of CSC2078 treated with JQ1 and/or linifanib for 24 h. We found that JQ1, linifanib alone or in combination induced the cell cycle arrest of CSC2078 (Fig. 6D and E). In addition, treatment with JQ1 and/or Linifanib significantly induced CSC2078 cell apoptosis. Importantly, the apoptosis rate in JQ1-treated group was not significant different compared with that linifanib-treated group or the combination group (Figs. 6F and G, and S9). Consistent with our observations, there was a reduction in the levels of phospho-VEGFR2, PI3K, phospho-AKT (Ser473), Cyclin B1 and Bcl-2 in CSC2078 treatment with JQ1, Linifanib or in combination for 24 h. However, the expression of Bax was increased and VEGFR2 and AKT was not significantly altered (Fig. 6H and I). Compared with treatment with each agent alone, co-treatment with JQ1 and linifanib did not cause greater attenuation of PI3K/AKT activity and further induce cell cycle arrest/apoptosis. These results further indicated that JQ1 exerts anti-tumor effects on GSCs through the VEGF/PI3K/AKT signaling pathway.
JQ1 and siBrd4 induces cycle arrest and apoptosis of GSCs through the VEGF/PI3K/AKT signaling pathway
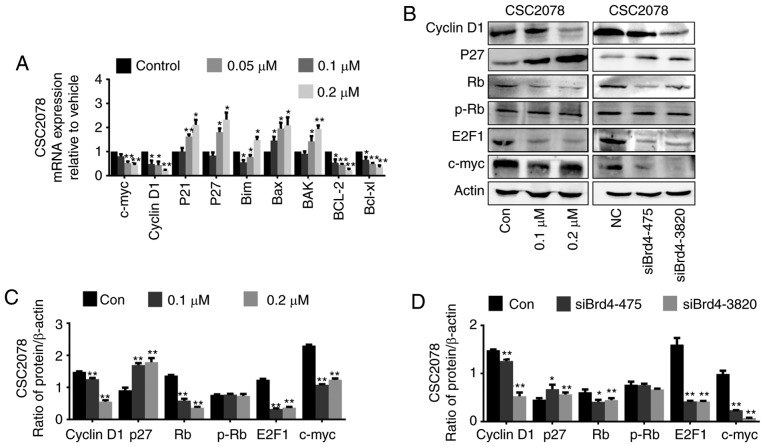
The cell cycle was significantly enriched in KEGG pathway enrichment analysis. To further investigate the mechanism by which Brd4 regulates cell cycle progression in GSCs, we used RT-qPCR to detect cell cycle-related genes, namely c-Myc, cyclin D1, p21, and p27, in CSC2078 and TS543 cells treated with JQ1. We found that the mRNA expression levels of the c-Myc and CyclinD1 were significantly inhibited, while P21 and P27 were significantly upregulated (Figs. 7A and 8A). Then, we detected the protein levels of cell cycle proteins including Cyclin D1, CDK inhibitor (CDKI) p27, Rb, phospho-Rb, E2F1 and c-Myc after treatment CSC2078 with JQ1, or siBrd4 by western blotting. We found that the protein expression of the c-Myc and CyclinD1 was significantly inhibited, while P27 was significantly upregulated. JQ1 or siBrd4 reduced the expression of Rb protein, but the expression level of p-Rb protein did not significantly change; thus, Rb protein was not activated. Therefore, E2F1 cannot be released from the RB/E2F1 complex to promote transcription of downstream genes. The Rb/E2F1 complex was deactivated resulting in a cycle arrest in GSCs treated with JQ1 or siRNAs (Fig. 7B-D).
Figure 7.
Inhibition of the vascular endothelial growth factor/PI3K/AKT signaling pathway induces cell cycle arrest in glioma stem cells. (A) Reverse transcription-quantitative polymerase chain reaction of c-Myc, cyclinD1, P21, P27, Bim, BAX, Bak, Bcl-2 and Bcl-xl in CSC2078 treated with JQ1 for 24 h (error bars represent standard error of the mean). (B) Western blotting analysis of cyclinD1, P27, Rb, p-Rb, E2F1 and c-Myc in CSC2078 treated by JQ1 or siBrd4. (C and D) Quantification (error bars represent standard error of the mean). *P<0.05, **P<0.01 vs. Control. p, phosphorylated Rb, retinoblastoma; siBrd4, small interfering RNA against bromodomain-containing protein 4.
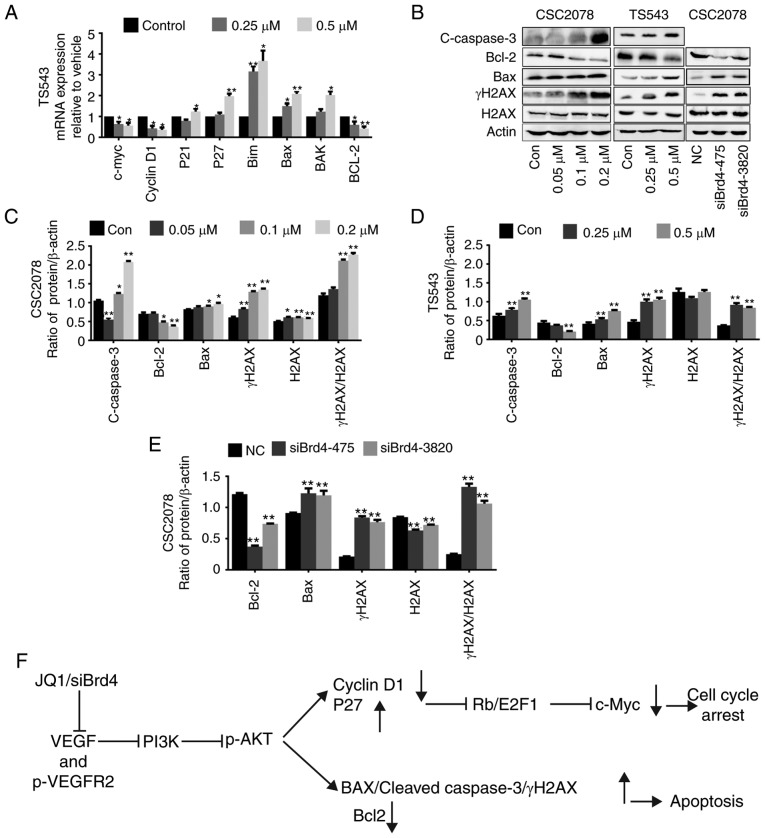
Figure 8.
Inhibition of VEGF/PI3K/AKT signaling pathway induces apoptosis in GSCs. (A) Reverse transcription-quantitative polymerase chain reaction of c-Myc, cyclinD1, P21, P27, Bim, BAX, Bak and Bcl-2 in TS543 treated with JQ1 for 24 h (error bars represent standard error of the mean). (B) Western blotting analysis of Bcl-2, BAX, cleaved-caspase 3, γH2AX and H2AX in GSCs treated with JQ1 or siBrd4. (C-E) Quantification (error bars represent standard error of the mean). (F) Proposed schematic model of VEGF/PI3K/AKT signaling pathway in GSCs. *P<0.05, **P<0.01 vs. Con. BAX, Bcl-2-associated X protein; Bak, Bcl2 antagonist/killer 1; Con, control; GSCs, glioma stem cells; H2AX, H2A histone family member X; siBrd4, small interfering RNA against bromodomain-containing protein 4; VEGF, vascular endothelial growth factor.
As AKT can activate apoptosis in many types of cancer, we investigated the effect of Brd4 on apoptotic genes in GSCs. We used RT-qPCR to verify the effects of JQ1 and siBrd4 on important apoptotic genes in PI3K/AKT signaling, including Bim, BAX, Bak, Bcl-2, and Bcl-xl in CSC2078 and/or TS543 cells (Figs. 7A and 8A). We also investigated the effects of JQ1 or siBrd4 on apoptotic proteins, including BAX, Bcl-2, and cleaved caspase 3 in GSCs via western blotting. We found that the protein expression of cleaved caspase 3 and Bax were significantly upregulated, while Bcl-2 was inhibited. In addition, we found that the protein expression of γH2AX were significantly upregulated. JQ1 or siBrd4 caused DNA damage as indicated by the ratio of γH2AX/H2AX, which further promoted the apoptosis of GSCs. From our results, the trend was consistent between JQ1- and siBrd4s-treated cells. The results indicated that siBrd4 or JQ1 could exhibit pro-apoptotic effects through the VEGF/PI3K/AKT signal axis in GSCs (Fig. 8B-E).
JQ1 has anti-tumor activity in a glioblastoma tumor xenograft model
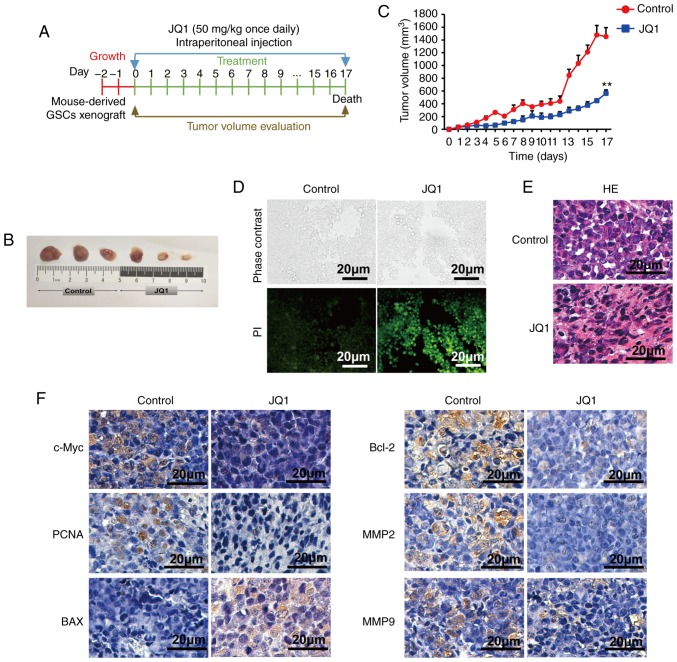
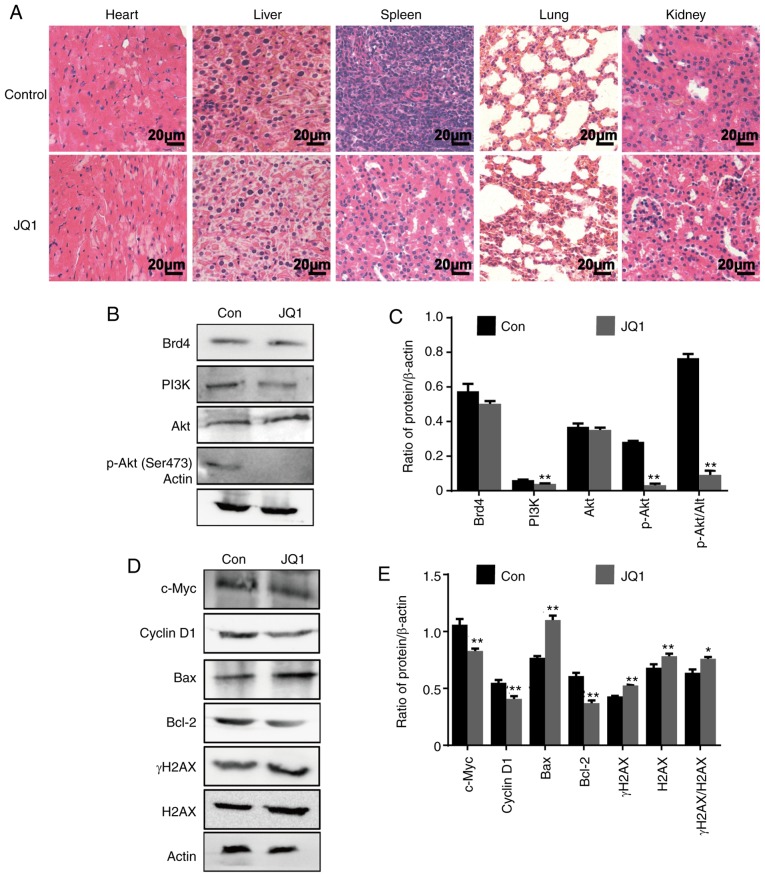
In addition, we assessed the therapeutic effects of JQ1 on a glioblastoma tumor xenograft model. Nude mice bearing CSC2078 subcutaneous tumors were treated with 50 mg/kg of JQ1 via an intraperitoneal injection once daily and sacrificed on the 17th day (Fig. 9A). The results demonstrated that the tumor volume was significantly smaller in the JQ1-treated group than that in the control group (Fig. 9B and C). Apoptosis in tumor tissues obtained from control and JQ1-treated mice was detected using TUNEL staining. The number of apoptotic cells was markedly increased in JQ1-treated tumors (Fig. 9D). The results of H&E staining indicated that tumor tissues displayed obvious nuclear shrinkage in JQ1-treatment group (Fig. 9E). The results were consistent with the in vitro findings. Immunohistochemical staining revealed that c-Myc, PCNA, Bcl-2, MMP2 and MMP9 expression was decreased in tumor tissues from JQ1-treated mice, whereas BAX expression was elevated (Fig. 9F). To detect the toxicity of JQ1 in mice, H&E staining was performed in the organs of mice. There were no obvious histopathological findings in the JQ1-treated mice, as shown in Fig. 10A. Compared with the findings in control tumors, the protein expression levels of Brd4 and AKT were markedly unchanged in JQ1-treated tumors, whereas PI3K and phosphor-AKT (Ser473) was downregulated. The results were consistent with the in vitro findings (Fig. 10B and C). We observed significant reductions in c-Myc, Cyclin D1, and Bcl-2 levels in JQ1-treated mice compared with the control. By contrast, BAX and γH2AX expression was significantly increased (Fig. 10D and E). Together, these data suggested that JQ1 could effectively inhibit tumorigenesis and the development of GBM. The therapeutic effects of JQ1 may warrant a clinical trial.
Figure 9.
Effects of JQ1 treatment on survival in glioblastoma multiforme. (A) The schematic showed the formation protocol of CSC2078 subcutaneous xenograft in nude mice for JQ1 or control experiment. (B) The images of tumor tissues resected from the control and JQ1 treatment groups. (C) Tumor volume quantification for CSC2078 xenografts in mice (n=5 mice for treatment group and control, error bars represent standard error of the mean). **P<0.01 vs. control. (D) Terminal deoxynucleotidyl-transferase-mediated dUTP nick end labeling staining of apoptotic cells in tumor samples as described in (B). Green, positive apoptosis cells. Scale bar=20 µm. (E) H&E staining of tumor tissues. Scale bar=20 µm. (F) Intratumoral molecular changes of tumor samples were detected using immunohistochemistry analysis. Scale bar=20 µm. BAX, Bcl-2-associated X protein; MMP, matrix metalloproteinase; PCNA, proliferating cell nuclear antigen.
Figure 10.
JQ1 has notable anti-tumor effects on CSC2078 subcutaneous xenograft mice with low toxicity. (A) H&E staining of the heart, liver, spleen, lung and kidney tissues. Scale bar=20 µm. (B and C) Western blotting analysis proteins expression of Brd4, PI3K, AKT and P-AKT (Ser473), in tumor tissues (Error bars represent standard error of the mean). (D and E) Western blotting analysis of c-Myc, Cyclin D1, BAX, Bcl-2, γH2AX and H2AX expression in tumor tissues (error bars represent standard error of the mean). *P<0.05, **P<0.01 vs. Con. Con, control; p, phosphorylated; BAX, Bcl-2-associated X protein; Brd4, bromodomain-containing protein 4; H2AX, H2A histone family member X.
Discussion
Previous reports and the current study have demonstrated that Brd4 is of great value as a therapeutic target for GBM (22,29,30). Therefore, therapies targeting Brd4 may aid the development of more effective treatment options for improving quality of life and prolonging the survival of patients with GBM (31).
Previous studies illustrated that epigenetic abnormalities were widespread in glioma; thus, epigenetic analysis might be critical for developing more effective treatment strategies for GBM (32,33). The epigenetic reader Brd4 has emerged as a therapeutic target for many cancers. Brd4 is an important therapeutic target for NUT midline cancer and hematopoietic diseases, and encouraging results have been obtained (11,34,35). Research on Brd4 as a drug target for hepatocarcinoma, breast cancer, and pancreatic cancer has become more extensive in past decade (14,36,37). To date, few studies have explored the role of Brd4 as a drug target for glioma cells, especially GSCs. GBM is a highly heterogeneous tumor; this heterogeneity is dominated by the presence of GSCs (7). Most importantly, the reason to study GSCs is that they have shown to be highly tumorigenic in vivo, and exhibited marked resistance to conventional chemotherapy and radiotherapy (38,39). In addition, GSCs are present throughout the tumor and can migrate along white matter pathways, often evading even gross-total resection, which provides a possibility for the recurrence of GBM (40). Over the past decade, the body of research regarding GSCs has indicated that highly resistant and tumorigenic sub-populations are maintained in specific microenvironmental niches, including the vascular niche (41). GBM is one of the tumors with the highest degree of vascularization in solid tumors (42). Microvascular hyperplasia has been considered as an important feature of the initiation and development of GBM (42). GSCs highly promotes angiogen-esis and the expression of VEGF, attracting endothelial cells to the tumor and driving neovascular growth (41,43). Therefore, using GSCs to perform experiments may increase the value of our results.
JQ1 and Brd4 have co-crystal structures, and thus, JQ1 has high affinity and specificity for Brd4 (44). In our experiments, we reported that the anti-tumor effects of JQ1 are consistent with those of siBrd4. Importantly, the results of RNA-seq experiments indicated that JQ1 selected a few closely related targets and affected only a few genes. JQ1 exhibits a small number of off-target effects, improving its efficacy as an epigenetic therapy (45). In addition, JQ1 has excellent pharmacokinetic properties including 49% oral bioavailability and a strong ability to cross the blood-brain barrier, which provides a basis for its use in treating GBM (46). Previous experiments focused on the inhibitory effect of JQ1 on the proliferation of tumor cells, and JQ1 also has a certain potential to promote cell differentiation (47). In our study, we found that JQ1 can inhibit the expression of NSC markers Nestin and ciliary neurotrophic factor and promote GSC differentiation to astrocytes under proliferative conditions. The role of JQ1 in promoting GSCs needs further study.
As a drug target for tumor treatment, Brd4 is involved in many signaling pathways, including the Wnt, Hedgehog, and PI3K-AKT pathways (48-50). There are two reasons why we selected the PI3K/AKT pathway for further research in our study. Firstly, the PI3K/AKT signaling pathway plays an important role in the regulation of signal transduction (51). The PI3K/AKT signaling pathway mediates various biological processes such as cell proliferation, apoptosis, metabolism and angiogenesis in GBM (52,53). Over the last few decades, it has been recognized that this signaling pathway is frequently activated by genetic and epigenetic alterations in malignant brain tumors, including GBM. Hyper-activation of the PI3K/AKT signaling pathway is closely related to rapid growth, tumor progression and multidrug resistance of GBM cells (52). Secondly, in addition to the PI3K/AKT signaling pathway, there are the Rap1, P53 and FoxO signaling pathways. The enrichment factors and false discovery rate values of these signaling pathways are not significantly superior to the PI3K/AKT signaling pathway. However, the PI3K/AKT signaling pathway has the most significant DEG numbers in these signaling pathways. In conclusion, the PI3K/AKT signaling pathway was selected for further analysis in our study. AKT mainly has two important phosphorylation sites, Thr308 and Ser473; serine 473 phosphorylation is necessary for the full activation of AKT (54,55). It has been reported that phosphorylation of T308 only partially activates AKT, but it is not yet clear whether it is necessary prior to phosphorylation of S473 (55). Secondly, it has reported that elevated AKT activation in human cancers can result from enhanced activation phosphorylation of AKT on the Ser473 site (56). Therefore, in the present study, we chose phospho-AKT (Ser473) as the PI3K substrate for research. AKT is an important mediator of VEGF. PI3K/AKT signaling mediates the VEGF-induced angiogenic stimulation of endothelial cells (57). As important components of the PI3K/AKT signaling pathway, AKT isoforms play key roles in several cellular processes including anti-apoptosis, proliferation, growth, DNA repair, transport, metabolism, angiogenesis, and stem cell self-renewal (58,59). In this article, we reported the novel VEGF/PI3K/AKT signaling axis that serves an important role in GSCs. The experimental results of this study indicated that JQ1 inhibits VEGF and phospho-VEGFR2 expression, thereby reducing the expression of PI3K downstream of VEGF and inhibiting the activity of phosphor-AKT (Ser473 or Thr308). In addition, although the mechanism is unclear, MMP protein expression was significantly decreased by JQ1, preventing VEGF secreting from the ECM to promote angiogenesis and tumor growth (27). Additionally, treatment with JQ1 or siBrd4 caused corresponding changes in apoptosis- and cell cycle-related genes downstream of AKT. The Rb protein binds to and regulates members of the E2F family. E2F1 is a transcriptional activator of E2F family members that positively regulates the transition from G1 phase to S phase (60-62). Active cyclin D/CDK complexes phosphorylate the Rb protein, and phosphorylated Rb is unable to interact with E2F. Therefore, E2F is activated, permitting it to promote the transcription of genes necessary for entry into S phase (63). By contrast, in our study, because AKT phosphorylation at Ser473 was inhibited, the expression of the downstream target gene cyclin D1 was decreased, and that of CDKIs (P21 and P27) was increased. Thus, the Rb protein was not phosphorylated, and E2F1 cannot be released from the Rb/E2F1 complex to facilitate transcription of downstream genes including c-Myc, leading to cell cycle arrest. In addition, after Brd4 silencing, the expression of apoptosis-related genes downstream of AKT, such as cleaved caspase 3, BAX, and Bcl-2, was affected. Inhibition of Brd4 by siRNAs or JQ1 treatment resulted in increased DNA damage, which further promoted apoptosis in GSCs.
The role of Brd4 in GBM has attracted our attention, but there were some limitations in our study. In the future, further experiments are needed to silence or overexpress the genes involved in the pathway to confirm the results and obtain further insight. JQ1 also has great potential for promoting the differentiation of GSCs, making further research valuable. Despite its limitations, this study identified the epigenetic protein Brd4 as a promising target for the treatment of GBM and proposed the feasibility of treating GBM using JQ1. In summary, our experiments identified promising prospects of epigenetic-based targeted therapeutic approaches for GBM, and these findings will may facilitate the rapid clinical evaluation of this new strategy.
Supplementary Data
Acknowledgments
Not applicable.
Abbreviations
- GBM
glioblastoma multiforme
- Brd4
bromodomain-containing protein 4
- BET
bromodomain and extraterminal domain
- GSCs
glioma stem cells
- VEGF
vascular endothelial growth factor
- MMP
matrix metalloproteinase protein
- ECM
extracellular matrix
Funding
The present study was funded by the National Natural Science Foundation of China (grant nos. 81773217 and 81472344) and Jilin Provincial Education Department (grant no. Jijiaokehezi [2016]455) and Jilin University Bethune Plan B Projects (grant no. 2015220) and Jilin Provincial Research Foundation for the Development of Science and Technology Projects (grant no. 20190701065GH) and Project funded by China Postdoctoral Science Foundation (grant no. 2018M631885).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
NW, BG, LZ and YL contributed to the conception and design, acquisition of data, analysis and interpretation of data. HZ contributed to the conception and revised the manuscript critically. LX, QW and DW performed molecular analyses and statistical tests. HL, XC and SZ contributed to analysis and interpretation of data. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
Animal experiments were approved by the Animal Ethics Committee of Jilin University [approval no. 2018;(54)], and by the Institutional Committee for the Care and Use of Laboratory Animals of the Experimental Animal Center of Jilin University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Appin CL, Brat DJ. Molecular genetics of gliomas. Cancer J. 2014;20:66–72. doi: 10.1097/PPO.0000000000000020. [DOI] [PubMed] [Google Scholar]
- 2.Aldape K, Zadeh G, Mansouri S, Reifenberger G, von Deimling A. Glioblastoma: Pathology, molecular mechanisms and markers. Acta Neuropathol. 2015;129:829–848. doi: 10.1007/s00401-015-1432-1. [DOI] [PubMed] [Google Scholar]
- 3.Auffinger B, Spencer D, Pytel P, Ahmed AU, Lesniak MS. The role of glioma stem cells in chemotherapy resistance and glioblastoma multiforme recurrence. Expert Rev Neurother. 2015;15:741–752. doi: 10.1586/14737175.2015.1051968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dawson MA, Kouzarides T. Cancer epigenetics: From mechanism to therapy. Cell. 2012;150:12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 5.Balça-Silva J, Matias D, Carmo AD, Sarmento-Ribeiro AB, Lopes MC, Moura-Neto V. Cellular and molecular mechanisms of glioblastoma malignancy: Implications in resistance and therapeutic strategies. Semin Cancer Biol. 2018 Sep 25; doi: 10.1016/j.semcancer.2018.09.007. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 6.Ahmed AU, Auffinger B, Lesniak MS. Understanding glioma stem cells: Rationale, clinical relevance and therapeutic strategies. Expert Rev Neurother. 2013;13:545–555. doi: 10.1586/ern.13.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu W, Ma Q, Wong K, Li W, Ohgi K, Zhang J, Aggarwal A, Rosenfeld MG. Brd4 and JMJD6-associated anti-pause enhancers in regulation of transcriptional pause release. Cell. 2013;155:1581–1595. doi: 10.1016/j.cell.2013.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu T, Kamikawa YF, Donohoe ME. Brd4's bromodomains mediate histone H3 acetylation and chromatin remodeling in pluripotent cells through P300 and Brg1. Cell Rep. 2018;25:1756–1771. doi: 10.1016/j.celrep.2018.10.003. [DOI] [PubMed] [Google Scholar]
- 9.Donati B, Lorenzini E, Ciarrocchi A. BRD4 and cancer: Going beyond transcriptional regulation. Mol Cancer. 2018;17:164. doi: 10.1186/s12943-018-0915-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.French CA, Ramirez CL, Kolmakova J, Hickman TT, Cameron MJ, Thyne ME, Kutok JL, Toretsky JA, Tadavarthy AK, Kees UR, et al. BRD-NUT oncoproteins: A family of closely related nuclear proteins that block epithelial differentiation and maintain the growth of carcinoma cells. Oncogene. 2008;27:2237–2242. doi: 10.1038/sj.onc.1210852. [DOI] [PubMed] [Google Scholar]
- 11.Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, Magoon D, Qi J, Blatt K, Wunderlich M, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi J, Wang Y, Zeng L, Wu Y, Deng J, Zhang Q, Lin Y, Li J, Kang T, Tao M, et al. Disrupting the interaction of BRD4 with diacetylated Twist suppresses tumorigenesis in basal-like breast cancer. Cancer Cell. 2014;25:210–225. doi: 10.1016/j.ccr.2014.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Asangani IA, Dommeti VL, Wang X, Malik R, Cieslik M, Yang R, Escara-Wilke J, Wilder-Romans K, Dhanireddy S, Engelke C, et al. Therapeutic targeting of BET bromodomain proteins in castration-resistant prostate cancer. Nature. 2014;510:278–282. doi: 10.1038/nature13229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou J, Li W, Guo J, Li G, Chen F, Zhou J. Downregulation of miR-329 promotes cell invasion by regulating BRD4 and predicts poor prognosis in hepatocellular carcinoma. Tumour Biol. 2016;37:3561–3569. doi: 10.1007/s13277-015-4109-4. [DOI] [PubMed] [Google Scholar]
- 15.Leal AS, Williams CR, Royce DB, Pioli PA, Sporn MB, Liby KT. Bromodomain inhibitors, JQ1 and I-BET 762, as potential therapies for pancreatic cancer. Cancer Lett. 2017;394:76–87. doi: 10.1016/j.canlet.2017.02.021. [DOI] [PubMed] [Google Scholar]
- 16.Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, Kastritis E, Gilpatrick T, Paranal RM, Qi J, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bandopadhayay P, Bergthold G, Nguyen B, Schubert S, Gholamin S, Tang Y, Bolin S, Schumacher SE, Zeid R, Masoud S, et al. BET bromodomain inhibition of MYC-amplified medulloblastoma. Clin Cancer Res. 2014;20:912–925. doi: 10.1158/1078-0432.CCR-13-2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Das A, Chai JC, Yang CS, Lee YS, Das ND, Jung KH, Chai YG. Dual transcriptome sequencing reveals resistance of TLR4 ligand-activated bone marrow-derived macrophages to inflammation mediated by the BET inhibitor JQ1. Sci Rep. 2015;5:16932. doi: 10.1038/srep16932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shao Q, Kannan A, Lin Z, Stack BC, Jr, Suen JY, Gao L. BET protein inhibitor JQ1 attenuates Myc-amplified MCC tumor growth in vivo. Cancer Res. 2014;74:7090–7102. doi: 10.1158/0008-5472.CAN-14-0305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng H, Ying H, Yan H, Kimmelman AC, Hiller DJ, Chen AJ, Perry SR, Tonon G, Chu GC, Ding Z, et al. p53 and Pten control neural and glioma stem/progenitor cell renewal and differentiation. Nature. 2008;455:1129–1133. doi: 10.1038/nature07443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang R, Vider J, Kovar JL, Olive DM, Mellinghoff IK, Mayer-Kuckuk P, Kircher MF, Blasberg RG. Integrin avβ3-targeted IRDye 800CW near-infrared imaging of glioblastoma. Clin Cancer Res. 2012;18:5731–5740. doi: 10.1158/1078-0432.CCR-12-0374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheng Z, Gong Y, Ma Y, Lu K, Lu X, Pierce LA, Thompson RC, Muller S, Knapp S, Wang J. Inhibition of BET bromodomain targets genetically diverse glioblastoma. Clin Cancer Res. 2013;19:1748–1759. doi: 10.1158/1078-0432.CCR-12-3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, Tanabe M. Data, information, knowledge and principle: Back to metabolism in KEGG. Nucleic Acids Res. 2014;42(Database Issue):D199–D205. doi: 10.1093/nar/gkt1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ola R, Dubrac A, Han J, Zhang F, Fang JS, Larrivée B, Lee M, Urarte AA, Kraehling JR, Genet G, et al. PI3 kinase inhibition improves vascular malformations in mouse models of hereditary haemorrhagic telangiectasia. Nat Commun. 2016;7:13650. doi: 10.1038/ncomms13650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee SH, Jeong D, Han YS, Baek MJ. Pivotal role of vascular endothelial growth factor pathway in tumor angiogenesis. Ann Surg Treat Res. 2015;89:1–8. doi: 10.4174/astr.2015.89.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aversa C, Leone F, Zucchini G, Serini G, Geuna E, Milani A, Valdembri D, Martinello R, Montemurro F. Linifanib: Current status and future potential in cancer therapy. Expert Rev Anticancer Ther. 2015;15:677–687. doi: 10.1586/14737140.2015.1042369. [DOI] [PubMed] [Google Scholar]
- 29.Pastori C, Daniel M, Penas C, Volmar CH, Johnstone AL, Brothers SP, Graham RM, Allen B, Sarkaria JN, Komotar RJ, et al. BET bromodomain proteins are required for glioblastoma cell proliferation. Epigenetics. 2014;9:611–620. doi: 10.4161/epi.27906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ishida CT, Zhang Y, Bianchetti E, Shu C, Nguyen TTT, Kleiner G, Sanchez-Quintero MJ, Quinzii CM, Westhoff MA, Karpel-Massler G, et al. Metabolic reprogramming by Dual AKT/ERK inhibition through imipridones elicits unique vulnerabilities in glioblastoma. Clin Cancer Res. 2018;24:5392–5406. doi: 10.1158/1078-0432.CCR-18-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Segatto M, Fittipaldi R, Pin F, Sartori R, Dae Ko K, Zare H, Fenizia C, Zanchettin G, Pierobon ES, Hatakeyama S, et al. Epigenetic targeting of bromodomain protein BRD4 counteracts cancer cachexia and prolongs survival. Nat Commun. 2017;8:1707. doi: 10.1038/s41467-017-01645-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dunn GP, Rinne ML, Wykosky J, Genovese G, Quayle SN, Dunn IF, Agarwalla PK, Chheda MG, Campos B, Wang A, et al. Emerging insights into the molecular and cellular basis of glio-blastoma. Genes Dev. 2012;26:756–784. doi: 10.1101/gad.187922.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kondo Y, Katsushima K, Ohka F, Natsume A, Shinjo K. Epigenetic dysregulation in glioma. Cancer Sci. 2014;105:363–369. doi: 10.1111/cas.12379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.French CA, Miyoshi I, Kubonishi I, Grier HE, Perez-Atayde AR, Fletcher JA. BRD4-NUT fusion oncogene: A novel mechanism in aggressive carcinoma. Cancer Res. 2003;63:304–307. [PubMed] [Google Scholar]
- 35.Roe JS, Vakoc CR. The essential transcriptional function of BRD4 in acute myeloid leukemia. Cold Spring Harb Symp Quant Biol. 2016;81:61–66. doi: 10.1101/sqb.2016.81.031039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Andrieu G, Tran AH, Strissel KJ, Denis GV. BRD4 regulates breast cancer dissemination through Jagged1/Notch1 signaling. Cancer Res. 2016;76:6555–6557. doi: 10.1158/0008-5472.CAN-16-0559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sahai V, Kumar K, Knab LM, Chow CR, Raza SS, Bentrem DJ, Ebine K, Munshi HG. BET bromodomain inhibitors block growth of pancreatic cancer cells in three-dimensional collagen. Mol Cancer Ther. 2014;13:1907–1917. doi: 10.1158/1535-7163.MCT-13-0925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bayin NS, Modrek AS, Placantonakis DG. Glioblastoma stem cells: Molecular characteristics and therapeutic implications. World J Stem Cells. 2014;6:230–238. doi: 10.4252/wjsc.v6.i2.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aum DJ, Kim DH, Beaumont TL, Leuthardt EC, Dunn GP, Kim AH. Molecular and cellular heterogeneity: The hallmark of glioblastoma. Neurosurg Focus. 2014;37:E11. doi: 10.3171/2014.9.FOCUS14521. [DOI] [PubMed] [Google Scholar]
- 40.Ropolo M, Daga A, Griffero F, Foresta M, Casartelli G, Zunino A, Poggi A, Cappelli E, Zona G, Spaziante R, et al. Comparative analysis of DNA repair in stem and nonstem glioma cell cultures. Mol Cancer Res. 2009;7:383–392. doi: 10.1158/1541-7786.MCR-08-0409. [DOI] [PubMed] [Google Scholar]
- 41.Thomas TM, Yu JS. Metabolic regulation of glioma stem-like cells in the tumor micro-environment. Cancer Lett. 2017;408:174–181. doi: 10.1016/j.canlet.2017.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carmeliet P, Jain RK. Angiogenesis in cancer and other diseases. Nature. 2000;407:249–257. doi: 10.1038/35025220. [DOI] [PubMed] [Google Scholar]
- 43.Jain RK, di Tomaso E, Duda DG, Loeffler JS, Sorensen AG, Batchelor TT. Angiogenesis in brain tumours. Nat Rev Neurosci. 2007;8:610–622. doi: 10.1038/nrn2175. [DOI] [PubMed] [Google Scholar]
- 44.Shi J, Vakoc CR. The mechanisms behind the therapeutic activity of BET bromodomain inhibition. Mol Cell. 2014;54:728–736. doi: 10.1016/j.molcel.2014.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dawson MA, Kouzarides T, Huntly BJ. Targeting epigenetic readers in cancer. N Engl J Med. 2012;367:647–657. doi: 10.1056/NEJMra1112635. [DOI] [PubMed] [Google Scholar]
- 46.Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee S, Rellinger EJ, Kim KW, Craig BT, Romain CV, Qiao J, Chung DH. Bromodomain and extraterminal inhibition blocks tumor progression and promotes differentiation in neuroblastoma. Surgery. 2015;158:819–826. doi: 10.1016/j.surg.2015.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Engelke CG, Chinnaiyan AM. aBETting therapeutic resistance by Wnt signaling. Cell Res. 2015;25:1187–1188. doi: 10.1038/cr.2015.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tang Y, Gholamin S, Schubert S, Willardson MI, Lee A, Bandopadhayay P, Bergthold G, Masoud S, Nguyen B, Vue N, et al. Epigenetic targeting of Hedgehog pathway transcriptional output through BET bromodomain inhibition. Nat Med. 2014;20:732–740. doi: 10.1038/nm.3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chautard E, Ouédraogo ZG, Biau J, Verrelle P. Role of Akt in human malignant glioma: From oncogenesis to tumor aggressiveness. J Neurooncol. 2014;117:205–215. doi: 10.1007/s11060-014-1382-9. [DOI] [PubMed] [Google Scholar]
- 51.McCubrey JA, Steelman LS, Chappell WH, Abrams SL, Franklin RA, Montalto G, Cervello M, Libra M, Candido S, Malaponte G, et al. Ras/Raf/MEK/ERK and PI3K/PTEN/Akt/mTOR cascade inhibitors: How mutations can result in therapy resistance and how to overcome resistance. Oncotarget. 2012;3:1068–1111. doi: 10.18632/oncotarget.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao HF, Wang J, Shao W, Wu CP, Chen ZP, To ST, Li WP. Recent advances in the use of PI3K inhibitors for glioblastoma multiforme: Current preclinical and clinical development. Mol Cancer. 2017;16:100. doi: 10.1186/s12943-017-0670-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lv D, Jia F, Hou Y, Sang Y, Alvarez AA, Zhang W, Gao WQ, Hu B, Cheng SY, Ge J, et al. Histone acetyltransferase KAT6A upregulates PI3K/AKT signaling through TRIM24 binding. Cancer Res. 2017;77:6190–6201. doi: 10.1158/0008-5472.CAN-17-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Freudlsperger C, Horn D, Weißfuß S, Weichert W, Weber KJ, Saure D, Sharma S, Dyckhoff G, Grabe N, Plinkert P, et al. Phosphorylation of AKT(Ser473) serves as an independent prognostic marker for radiosensitivity in advanced head and neck squamous cell carcinoma. Int J Cancer. 2015;136:2775–2785. doi: 10.1002/ijc.29328. [DOI] [PubMed] [Google Scholar]
- 55.Yung HW, Charnock-Jones DS, Burton GJ. Regulation of AKT phosphorylation at Ser473 and Thr308 by endoplasmic reticulum stress modulates substrate specificity in a severity dependent manner. PLoS One. 2011;6:e17894. doi: 10.1371/journal.pone.0017894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liao Y, Hung MC. Physiological regulation of Akt activity and stability. Am J Transl Res. 2010;2:19–42. [PMC free article] [PubMed] [Google Scholar]
- 57.Li F, Sawada J, Komatsu M. R-Ras-Akt axis induces endothelial lumenogenesis and regulates the patency of regenerating vasculature. Nat Commun. 2017;8:1720. doi: 10.1038/s41467-017-01865-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu Q, Turner KM, Alfred Yung WK, Chen K, Zhang W. Role of AKT signaling in DNA repair and clinical response to cancer therapy. Neuro Oncol. 2014;16:1313–1323. doi: 10.1093/neuonc/nou058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Khan KH, Yap TA, Yan L, Cunningham D. Targeting the PI3K-AKT-mTOR signaling network in cancer. Chin J Cancer. 2013;32:253–265. doi: 10.5732/cjc.013.10057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sun H, Chang Y, Schweers B, Dyer MA, Zhang X, Hayward SW, Goodrich DW. An E2F binding-deficient Rb1 protein partially rescues developmental defects associated with Rb1 nullizygosity. Mol Cell Biol. 2006;26:1527–1537. doi: 10.1128/MCB.26.4.1527-1537.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jiang H, Martin V, Gomez-Manzano C, Johnson DG, Alonso M, White E, Xu J, McDonnell TJ, Shinojima N, Fueyo J. The RB-E2F1 pathway regulates autophagy. Cancer Res. 2010;70:7882–7893. doi: 10.1158/0008-5472.CAN-10-1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chang MM, Lai MS, Hong SY, Pan BS, Huang H, Yang SH, Wu CC, Sun HS, Chuang JI, Wang CY, Huang BM. FGF9/FGFR2 increase cell proliferation by activating ERK1/2, Rb/E2F1, and cell cycle pathways in mouse Leydig tumor cells. Cancer Sci. 2018;109:3503–3518. doi: 10.1111/cas.13793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sammons SL, Topping DL, Blackwell KL. HR+, HER2-advanced breast cancer and CDK4/6 inhibitors: Mode of action, clinical activity, and safety profiles. Curr Cancer Drug Targets. 2017;17:637–649. doi: 10.2174/1568009617666170330120452. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.