Abstract
MASH1, a basic helix-loop-helix transcription factor, is widely expressed by neuronal progenitors in the CNS and PNS, suggesting that it plays a role in the development of many neural regions. However, in mice lacking a functional Mash1 gene, major alterations have been reported in only a few neuronal populations; among these is a generalized loss of olfactory receptor neurons of the olfactory epithelium. Here, we use a transgenic reporter mouse line, in which the cell bodies and growing axons of subsets of central and peripheral neurons are marked by expression of a tau-lacZ reporter gene (the Tattler-4 allele), to look both more broadly and deeply at defects in the nervous system of Mash1−/−mice. In addition to the expected lack of olfactory receptor neurons in the main olfactory epithelium, developingMash1−/−;Tattler-4+/− mice exhibited reductions in neuronal cell number in the vomeronasal organ and in the olfactory bulb; the morphology of the rostral migratory stream, which gives rise to olfactory bulb interneurons, was also abnormal. Further examination of cell proliferation, cell death, and cell type-specific markers inMash1−/− animals uncovered parallels between the main olfactory epithelium and the vomeronasal organ in the regulation of sensory neuron development. Interestingly, this analysis also revealed that, in the olfactory epithelium of Mash1−/− animals, there is an overproduction of proliferating cells that co-express markers of both neuronal progenitors and supporting cells. This finding suggests that olfactory receptor neurons and olfactory epithelium supporting cells may share a common progenitor, and that expression ofMash1 may be an important factor in determining whether these progenitors ultimately generate neurons or glia.
Keywords: bHLH transcription factor, olfactory epithelium, olfactory receptor neuron, transgenic mouse, olfactory bulb, rostral migratory stream, subventricular zone, vomeronasal organ, neural progenitor, lineage, supporting cell, granule cell
Introduction
Basic helix-loop-helix (bHLH) transcription factors appear to play a conserved role in determining neuronal fate during development (Brunet and Ghysen, 1999; Guillemot, 1999). In Drosophila, proneural genes such asachaete, scute, and atonal instruct neuronal fate determination (Jan and Jan, 1994), and at least some homologs of these genes play analogous roles in vertebrates. For example, loss of function studies in mice have shown thatMash1, Ngn1, and Ngn2 are required for the development of specific subsets of neurons (Guillemot et al., 1993;Fode et al., 1998; Ma et al., 1998).
Mash1, a homolog of achaete and scute, is expressed by neuronal progenitors in the developing PNS and CNS. In the CNS, Mash1 is expressed in the developing telencephalon, including ventricular zone (VZ) of the developing olfactory bulb (OB) and ganglionic eminences, diencephalon, midbrain, spinal cord, and retina (Guillemot and Joyner, 1993; Ma et al., 1997; Horton et al., 1999). Mash1 is also expressed in the PNS, including the olfactory epithelium (OE) and the sympathetic, parasympathetic, and enteric nervous systems (Guillemot and Joyner, 1993; Gordon et al., 1995; Blaugrund et al., 1996; Ma et al., 1997). Although this widespread expression suggests that Mash1 plays a role in the development of many neural regions, the initial analysis of gene-targeted Mash1−/− mice detected deficits only in olfactory receptor neurons (ORNs) and neurons of the autonomic nervous system (Guillemot et al., 1993). Subsequent studies demonstrated additional and more subtle roles for Mash1, often acting in concert with other transcriptional regulators, in regulating cell fate and neuronal differentiation in various CNS regions (Casarosa et al., 1999; Horton et al., 1999; Torii et al., 1999; Tomita et al., 2000;Hatakeyama et al., 2001; Marquardt et al., 2001; Nieto et al., 2001). These findings, and the discovery of thalamocortical axon pathfinding defects resulting from loss of Mash1 (Tuttle et al., 1999), suggest that other neural deficits are likely to be present inMash1−/− animals.
To look systematically for additional phenotypes in Mash1−/−animals, as well as other neurodevelopmental mutants, we developed a transgenic mouse line (Tattler-4) that allows rapid visualization of subsets of neurons and their axons during development. Tattler-4 mice were generated using a promoter fragment from theTα1 α-tubulin gene to express atau-lacZ fusion gene. The transgene is expressed in cell bodies and axons of subsets of PNS and CNS neurons during terminal neuronal differentiation and initial axon outgrowth. By breeding theMash1 null allele onto Tattler-4 and examining the primary olfactory pathway with β-galactosidase histochemistry, we were able to identify abnormalities in Mash1−/− animals not only in OE, but also in the vomeronasal organ (VNO), OB, and rostral migratory stream (RMS), a structure containing neural progenitors that give rise to OB interneurons. Analysis of gene expression in the developing OE and VNO of Mash1−/− animals uncovered parallels in genetic regulation of sensory neuron development between these two tissues and provided clues that ORNs and OE supporting cells share a common progenitor.
Materials and Methods
Transgenic mice. TheTα1:tau-lacZ transgene was generated by ligating a 5.5 kb XhoI/SpeI fragment containing the tau-lacZ fusion and SV40 splice site and poly(A) addition sequence (Callahan and Thomas, 1994) downstream of a 1.1 kb BssHII/XbaI fragment of theTα1 α-tubulin promoter (Gloster et al., 1994) (see Fig. 1A). To generate transgenic mice, the Tα1:tau-lacZ construct was linearized with AscI/PmeI and injected into pronuclei of fertilized mouse ova (CB6 F2) using standard techniques (Hogan, 1994) in the University of California, Irvine, Transgenic Mouse Facility. Transgenic offspring were identified by PCR of tail DNA and bred against CD-1 mice (Charles River Laboratories, Wilmington, MA) to generate four independent transgenic mouse lines, named Tα1-tubulin tau-lacZ expressing reporter (Tattler) 1–4. Tattler-4 mice, used in this study, were bred for more than eight generations on a CD-1 background. Mash1+/– mice were a generous gift from F. Guillemot (IGBMC, Strasbourg, France) (Guillemot et al., 1993) and were maintained on a CD-1 background, where the OE phenotype is fully penetrant (Cau et al., 1997; this study).Tattler-4+/+;Mash1+/– animals were mated withMash1+/– females to generateTattler-4+/–;Mash1+/+ and Tattler-4+/–;Mash1−/−littermate embryos for analysis.
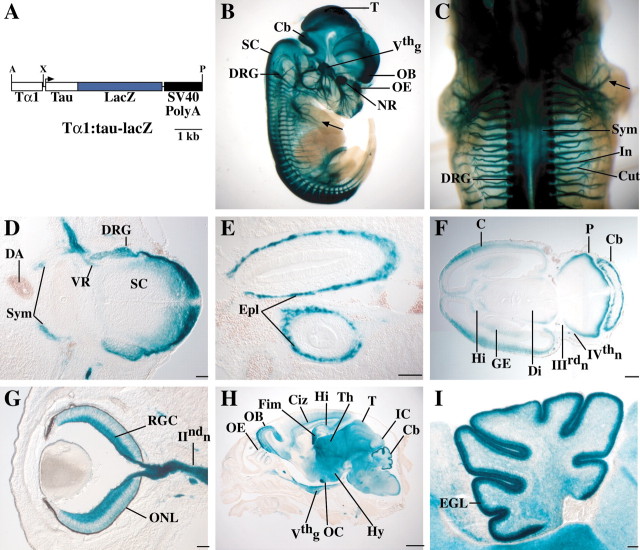
Fig. 1.
A, Diagram of theTα1:tau-lacZ transgenic construct used to generate Tattler-4 mice. The tau-lacZ reporter fusion is expressed under the control of theTα1 α-tubulinpromoter. A polyadenylation signal from SV40 is included in the construct. A, AscI; X,XhoI; P, PmeI.B-I, Tα1:tau-lacZexpression in Tattler-4 transgenic mice. Expression was visualized by X-gal staining, as detailed in Materials and Methods. B, Sagittal view of E13.5 Tattler-4 transgenic embryo stained with X-gal. Staining is present in the neural retina (NR), olfactory epithelium (OE), olfactory bulb (OB), tectum (T), cerebellum (Cb), dorsal spinal cord (SC), trigeminal ganglion (Vthg), and dorsal root ganglia (DRG). Sensory nerves in the limb (arrow) are also stained. C, Dorsal view of E13.5 embryo. The sympathetic chain (Sym) underlying the spinal cord, dorsal root ganglia (DRG), cutaneous (Cut), and intercostal (In) spinal nerves are stained. D, Transverse section through the spinal cord of an E13.5 embryo. Staining is present in the dorsal spinal cord (SC) and dorsal root ganglia (DRG) as well as in the sympathetic chain (Sym). Staining in the ventral root (VR) likely represents preganglionic sympathetic axons, because the ventral horn motor neurons are not stained. The dorsal aorta (DA) is labeled for reference (dorsal is at the right). E, Horizontal section through the abdomen of an E13.5 embryo. Staining is present in the enteric plexus (Epl) of the developing gut.F, Horizontal section through the head of an E13.5 embryo stained in whole mount. Staining is present at the margin of the cerebral cortex (C), pons (P), cerebellum (Cb), diencephalon (Di), ganglionic eminence (GE), and hippocampus (Hi). The oculomotor (IIIrdn) and trochlear (IVthn) cranial nerves are also stained. G, Horizontal section through the eye of an E16.5 embryo. Prominent staining is present in the retinal ganglion cells (RGC) and their axons in the optic nerve (IIndn). Some staining in the developing outer nuclear layer (ONL) is also apparent. H, Parasagittal section through the head of a P0 animal. Staining is present in the OE,OB, intermediate zone of the cerebral cortex (Ciz), hippocampus (Hi), fimbria (Fim), thalamus (Th), optic chiasm (OC), hypothalamus (Hy), tectum (T), inferior colliculus (IC), cerebellum (Cb), and trigeminal ganglion (Vthg).I, Parasagittal section through the cerebellum of a P0 Tattler-4 mouse. Staining is present in cells of the external granule layer (EGL). Scale bars: D,E, G, I, 100 μm;F, 300 μm; H, 1 mm.
Genotype was determined by PCR (30 cycles, annealing temperature 58°C) of tail or yolk sac DNA using oligonucelotide primers (Invitrogen, San Diego, CA) specific for lacZfor Tattler-4 animals or for Mash1 and neo to distinguish Mash1+/+, +/−, −/−genotypes (lacZ, GenBank accession number V00296; LacZ-Forward, 5′-TGATGAAAGCTGGCTACAT-3′; LacZ-Reverse, 5′-ACCACCGCACGATAGAGATT-3′; Mash1, GenBank U68534; Mash1-Forward, 5′-CCAACTGGTTCTGAGGAC-3′; Mash1-Reverse, 5′-CCCATTTGACGTAGTTGG-3′; neo, GenBank U43611; Neo-Forward, 5′-GATCTCCTGTCATCTCACCT-3′; Neo-Reverse, 5′-ATGGGTCACGACGAGATCCT-3′).
In situ hybridization, immunohistochemistry, and terminal deoxynucleotidyl transferase-mediated biotinylated UTP nick end labeling staining. One hour before they were killed, timed-pregnant female mice were injected intraperitoneally with 50 μg/gm body weight of 5-bromo-2′-deoxyuridine (BrdU) (5 mg/ml in 0.9% saline, 0.007N NaOH). Pregnant dams were killed, and embryos were dissected and fixed overnight in 4% paraformaldehyde in 0.02m NaPO4, 0.15m NaCl, pH 7.5. Embryos were washed in PBS, cryoprotected in 30% sucrose/PBS, and sectioned at 12 or 20 μm on a cryostat. Sections were collected on Superfrost/Plus slides (Fisher Scientific, Houston, TX) and stored at −80°C until use. For staging of embryos, 12:00 P.M. on the day a vaginal plug was detected was designated embryonic day (E) 0.5.
For in situ hybridization, sections were fixed onto slides in 4% paraformaldehyde/PBS, washed in PBS, incubated with proteinase K (25 μg/ml, 10–15 min), refixed in 4% paraformaldehyde/PBS, acetylated (0.25% acetic anhydride in 0.1 mtriethanolamine, pH 8.0), and hybridized at 60°C in 50% formamide, 5× SSC, 300 μg/ml Yeast tRNA, 100 μg/ml heparin, 1× Denhardt's, 0.1% Tween 20, 0.1% CHAPS, and 5 mm EDTA containing 100 ng/ml probe. Unbound probe was removed by washing (0.2× SSC, 60°C), and probes were detected using alkaline phosphatase-conjugated sheep anti-DIG antibodies (1:2000) (Roche Molecular Biochemicals, Indianapolis, IN) and visualized using 5-bromo-4-chloro-3-indolyl-phosphate (BCIP)/4-nitroblue tetrazolium chloride (NBT) as substrate. Slides were dehydrated and mounted in Pro-Texx (Lerner Laboratories, Pittsburgh, PA) before viewing. The probes that were used were as follows: 375 bp fragment of mouse Mash1 coding region, 2.0 kb fragment of mouse Mash1 gene including coding region and 3′UTR [Clone 1 in Guillemot and Joyner (1993)], 1.2 kb fragment of rat ngn1 gene (Ma et al., 1996), 391 bp fragment of mouseNcam coding region (Barthels et al., 1987), 1.7 kb fragment of the mouse Trp2 coding region (Vannier et al., 1999), and 155 bp fragment of the mouse reelin coding region (D'Arcangelo et al., 1995). The steel probe consisted of an 879 bp fragment of the mouse steel coding region and 3′UTR (GenBank M57647) (bp 386–1265), generated by RT-PCR of mouse E12.5 head total RNA. The PCR product was cloned into the pCR2.1 vector (Invitrogen), and its identity was confirmed by sequencing. The Gad67 probe consisted of an 867 bp fragment of the mouse Gad67 3′UTR (GenBank NM008077) (bp 1987–2854), generated by RT-PCR of mouse P1 brain total RNA. The PCR product was cloned into pBluescript (Stratagene, La Jolla, CA), and its identity was confirmed by sequencing.
For BrdU immunostaining, sections were permeabilized in 0.1% Triton X-100 in PBS (30 min, room temperature), treated with 2N HCl (1 hr, 37°C), neutralized in HBSS (Invitrogen), blocked in 0.1% Triton X-100, 10% bovine calf serum (BCS) (HyClone, Logan, UT) in PBS, incubated with anti-BrdU antibody (clone BU1/75) (1:1000 in 10% BCS in PBS; overnight, 4°C) (Harlan Sera-Lab, Sussex, UK), and visualized with Texas Red-conjugated goat anti-rat IgG (1:50) (Jackson ImmunoResearch, West Grove, PA). Immunostaining with monoclonal anti-neuron-specific nuclear protein (NeuN) (1:500 dilution) (Chemicon, Temecula, CA) was performed in the same manner but without acid treatment, and primary antibody was detected using Texas Red-conjugated goat anti-mouse IgG1 (1:50) (Southern Biotechnology, Birmingham, AL). For immunostaining with 12F8 monoclonal anti-polysialic acid (PSA) neural cell adhesion molecule (NCAM) (Carl Lagenaur, University of Pittsburgh), hybridoma supernatant was applied overnight at 4°C and detected using Texas Red-conjugated goat anti-rat IgM (1:50) (Jackson ImmunoResearch).
Terminal deoxynucleotidyl transferase-mediated biotinylated UTP nick end labeling (TUNEL) staining to detect DNA fragmentation in situ was performed as described previously (Holcomb et al., 1995), using Texas Red-conjugated NeutrAvidin (Molecular Probes, Eugene, OR) to detect incorporated Biotin-16-dUTP (Roche Molecular Biochemicals).
β-galactosidase histochemistry. E13.5 whole-mount staining was performed as described previously (Murray et al., 2000). For older embryos, tissues were dissected and fixed by immersion in 2 mm MgCl2, 4% paraformaldehyde in 0.02 mNaPO4, 0.15 m NaCl, pH 7.5, for 2–4 hr at room temperature, washed three times 5 min in 2 mm MgCl2 in PBS at room temperature, cryoprotected in 30% sucrose, 2 mmMgCl2 in PBS, and sectioned at 30 μm on a cryostat. Sections were collected on gelatin-coated slides, postfixed in 2 mm MgCl2, 0.5% glutaraldehyde, in PBS for 15 min, and permeabilized in 2 mm MgCl2, 0.1% Triton X-100, 0.01% deoxycholate, in PBS for 10 min. Sections were stained overnight in 1 mg/ml 5-bromo-4-chloro-3-indolyl β-d-galactopyranoside (X-gal), 5 mmK3Fe(CN)6, 5 mmK4Fe(CN)6, 2 mm MgCl2, 0.1% Triton X-100, 0.01% deoxycholate, in PBS at 37°C, and then dehydrated and mounted in Pro-Texx.
Results
Subsets of neurons in the PNS and CNS are marked in Tattler-4 transgenic mice
To study the connectivity of developing neuronal populations in the embryonic mouse nervous system, we generated transgenic mice that express an axon-targeted reporter gene in differentiating neurons. A 1.1 kb fragment of 5′ regulatory sequence from theTα1 tubulin gene was used to drive expression of a reporter construct (tau-lacZ), which consisted of a fragment of the bovine gene encoding the microtubule-associated protein, tau, fused to the bacterial β-galactosidase gene (lacZ) (Callahan and Thomas, 1994; Gloster et al., 1994). The resulting construct (Tα1:tau-lacZ) (Fig.1A) was used to generate four independent transgenic mouse lines, which were namedTα1-tubulin tau-lacZ expressing reporter (Tattler) 1–4. The Tattler-4 strain had the most extensive pattern of expression and was chosen for further analysis.
To reveal the overall expression pattern ofTα1:tau-lacZ expression in Tattler-4 mice, transgenic embryos were fixed and stained with X-gal as whole mounts at day 13.5 of gestation (E13.5). As shown in Figure 1, B andC, X-gal staining was specific to neurons and was present in both neuronal cell bodies and axons in a subset of neural structures in both the CNS and PNS. In the CNS, prominent staining was observed in the retina, olfactory bulbs, telencephalon, diencephalon, midbrain, hindbrain, and spinal cord (Fig. 1B). In addition, cranial sensory ganglia and nerves, including the trigeminal, oculomotor, and trochlear nerves, were labeled by X-gal staining (Fig.1B). In the PNS, expression was seen in the dorsal root ganglia and cutaneous and intercostal spinal nerves (Fig.1C). Sections made from E13.5 X-gal-stained whole mounts revealed that both sympathetic nerve fibers (Fig. 1D) and the enteric plexus were labeled (Fig. 1E). These sections also revealed that staining of the dorsal spinal cord could be attributed to labeled fibers of the dorsal root entry zone; developing motoneurons and interneurons were not labeled (Fig.1D). Brain sections from E13.5 whole mounts showed X-gal staining at the margins of structures such as cortex, diencephalon, ganglionic eminence, and pons, but not in the respective ventricular zones (Fig. 1F). This suggests that the transgene is expressed primarily by postmitotic neurons rather than neuronal progenitors and is consistent with what is known about time of onset of terminal neuronal differentiation in these regions (Angevine, 1970; Pierce, 1973; Nornes and Carry, 1978; McConnell, 1981). Because previous studies have shown that the Tα1 tubulin promoter fragment that we used drives reporter gene expression in embryonic neurons at the time of terminal differentiation and initial axon outgrowth (Gloster et al., 1994, 1999), these observations suggested that X-gal staining in Tattler-4 mice reveals the axons and, to a lesser extent, cell bodies, of newly differentiating neurons.
Examination of Tattler-4 embryos at older ages revealed high levels of reporter expression in distinct subsets of neurons in different regions of the CNS. For example, in the developing neural retina at E16.5, retinal ganglion cells and their axons were darkly stained; light staining was also observed in cells of the developing outer nuclear layer (ONL) (Fig. 1G). At postnatal day (P) 0, staining could be seen in a subset of cells and their axons in the cerebral cortex, hippocampus, fimbria, and striatum; fiber tracts in the diencephalon and midbrain were also labeled (Fig.1H). Especially prominent staining was observed in the ORNs of the OE and associated olfactory nerve, the glomeruli and mitral/tufted cell layer of the olfactory bulb; and cells of the external granule layer, but not Purkinje cells, in the cerebellum (Fig.1H,I). Expression of theTα1:tau-lacZ transgene in only a subset of CNS and PNS neurons can likely be attributed to sensitivity of theTα1-tubulin promoter fragment to the site of transgene integration; other investigators have generated transgenic lines using this promoter to drive a nuclear lacZ reporter gene, and the subset of expressing neurons varied in different lines (Gloster et al., 1994). Because expression ofTα1:tau-lacZ by specific populations of neurons in Tattler-4 mice is a stable characteristic of all mice in this transgenic line, it provides a useful tool for selectively examining the behavior of these cells as development of the nervous system proceeds.
Tα1:tau-lacZ reporter gene expression reveals unexpected changes in the olfactory bulb and rostral migratory stream of Mash1−/− mice
Because the Tα1:tau-lacZ reporter gene is expressed strongly in several structures of the primary olfactory pathway in Tattler-4 mice, we used these animals to help us examine development of this pathway in animals in which the gene encoding the basic helix-loop-helix transcription factor, MASH1, has been disrupted by homologous recombination. Mash1−/− mice die at birth and have been demonstrated previously to have a profound reduction in the number of ORNs in the OE lining the nasal cavity (Guillemot et al., 1993). Because Mash1 is known to be expressed in the developing telencephalon, including the OB (Guillemot and Joyner, 1993;Sommer et al., 1996), we hypothesized that other defects might be present in the primary olfactory pathway of Mash1−/− mice. To test this possibility, we bred the Mash1 knock-out allele onto the Tattler-4 background and compared olfactory structures in Mash1−/− animals and their wild-type littermates.
We first examined mice around the time of birth (E18.5/P0), becauseMash1−/− animals only survive to this age. At E18.5, X-gal staining of Mash1+/+;Tattler-4+/− mice revealed expression of tau-lacZ in ORN cell bodies within the OE and in ORN axons projecting to the OB (Fig.2A). Also labeled by X-gal staining were cells in the mitral/tufted cell layer of the OB; these are the cell types onto which ORNs synapse. However, cells in the developing OB granular layer, which contains primarily granule cell interneurons, were not stained (Fig. 2A) (Farbman, 1992). In contrast to the situation in wild-type embryos, the OE ofMash1−/−;Tattler-4+/− embryos was devoid of X-gal staining and was much thinner than normal, and ORN axons were virtually absent (Fig. 2B). Thus, the Tattler-4allele clearly revealed the deficit in ORN development known to occur in the absence of Mash1 function.
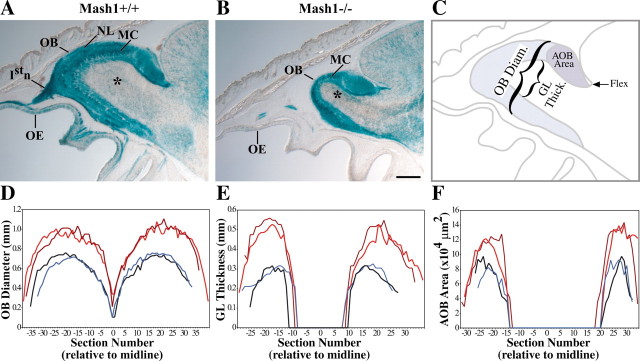
Fig. 2.
Alterations in the OE and OB of Mash1−/−mice. A, B, E18.5 littermate embryos heterozygous for the Tattler-4 reporter allele and Mash1+/+ (A) orMash1−/− (B) were sectioned at 30 μm in the sagittal plane and stained with X-gal. Anterior is on the left; dorsal is at the top. The olfactory epithelium (OE), mitral/tufted cell layer (MC), outer nerve layer (NL), olfactory nerve (Istn), and granular layer (asterisk) are shown. Scale bar, 300 μm. C, Diagram to illustrate the measurements taken to quantify changes in OB and AOB size. OB diameter and granular layer (GL) thickness were measured in the plane perpendicular to the cribriform plate, at the point midway between the anterior tip of the OB and the flexure at the junction of the anterior border of the cerebral cortex and the dorsal surface of the OB (labeled brackets). OB diameter was measured as the distance between the dorsal and ventral surfaces of the OB; GL thickness was measured as the distance across the unstained granular layer in X-gal-stained tissue. AOB area (light blue shading) was measured using NIH Image 1.61. D, Decreased OB diameter inMash1−/− embryos. To quantify changes in the size of the OB, serial 30-μm-thick sagittal sections through the entire extent of both OBs were measured in two Mash1−/−;Tattler-4+/−and two Mash1+/+;Tattler-4+/− animals at E18.5. A total of 67–80 measurements were taken per animal and plotted relative to the distance of the measured section from the midline.Mash1−/− embryos (blue lines) and wild-type embryos (red lines) are shown.E, Decreased GL thickness in Mash1−/−embryos. A total of 32–50 measurements were taken per animal and plotted as in D. F, Reduced AOB area in Mash1−/− embryos. A total of 24–35 measurements were taken per animal and plotted as in D.
X-gal staining of Mash1−/−;Tattler-4+/− embryos also revealed that the OBs of these animals were reduced in size compared with their wild-type littermates (Fig. 2B). In addition to a size reduction caused by absence of the olfactory nerve layer of the bulb (expected because of the lack of ingrowing ORN axons), the granular layer of the OB appeared to be greatly reduced in size in Mash1−/−;Tattler-4+/− embryos relative toMash1+/+;Tattler-4+/− littermates (Fig.2A,B, asterisks). Moreover, the accessory olfactory bulb (AOB) (the synaptic target of vomeronasal sensory neurons) (Halpern, 1987) also appeared to be smaller in Mash1−/−;Tattler-4+/− embryos.
To quantify these changes, we measured the diameter of the OB, the thickness of the granular layer (the cell layer containing developing granule interneurons) within the OB, and the area of the AOB through the entire extent of both bulbs in twoMash1−/−;Tattler-4+/− and twoMash1+/+;Tattler-4+/− littermate embryos (Fig.2C). The data, shown in Figure 2D-F, demonstrate clear reductions in size of both the OB and the AOB inMash1 mutant animals. The diameter of the OB ofMash1−/−;Tattler-4+/− animals was reduced by 27% relative to their Mash1+/+;Tattler-4+/− littermates, and the thickness of the granular layer was reduced by 45% inMash1 mutants relative to wild-type littermates. The average area of the AOB also showed a large reduction, being decreased by 30% in Mash1−/−;Tattler-4+/− animals relative to theirMash1+/+;Tattler-4+/− littermates. To confirm that the reduction in the size of the OB in Mash1−/−;Tattler-4+/−animals is not caused by the Tattler-4 allele, the diameter of the OB was measured in Mash1−/− animals and their wild-type littermates maintained on a CD-1 background. A similar decrease in OB diameter (29%) in Mash1−/− animals relative to wild types was observed (wild type, 0.91 ± 0.03 mm;Mash1−/−, 0.65 ± 0.01 mm, for three animals of each genotype; more than eight sections measured per animal).
Granule cell interneurons of the OB originate in the subventricular zone of the lateral ventricles and migrate to the OB in a structure known as the RMS (Luskin, 1993; Lois and Alvarez-Buylla, 1994). The Tattler-4 reporter allele is expressed by cells of the RMS, allowing this structure to be visualized easily in X-gal-stained sections of normal and Mash1−/− animals on theTattler-4 background. In sagittal sections through the brains of wild-type animals at E18.5, the RMS could be seen as a stream of cells, many of which were stained with X-gal, extending from the anterior limit of the lateral ventricle to the caudal boundary of the OB (Fig. 3A). Interestingly, in Mash1−/−;Tattler-4+/− embryos, the RMS was present, but its shape was different, being noticeably thicker inMash1−/−;Tattler-4+/− animals than in their wild-type littermates (Fig. 3B). Moreover, the density of X-gal-stained cells in the RMS of Mash1−/−;Tattler-4+/−animals was consistently greater than what was observed in wild types (Fig. 3, compare C, D). Because induced absence of the highly sialylated form of NCAM is associated with similar morphological changes in the RMS, as well as reduced OB size (Bruses and Rutishauser, 2001), we used a monoclonal antibody specific for the PSA moieties on the “embryonic” form of NCAM to stain sections through the RMS in E17.5 Mash1−/− andMash1+/+ littermates (Chung et al., 1991). As shown in Figure 3E, in wild-type embryos, essentially the entire forebrain and OB are stained with the 12F8 monoclonal anti-PSA-NCAM. The cell-dense RMS and ventricular zone of the OB (visualized with a nuclear stain in Fig. 3G) are also immunopositive for PSA-NCAM, although as described previously, staining is less intense than in the surrounding brain regions (Chung et al., 1991). InMash1−/− embryos, the level of anti-PSA-NCAM immunoreactivity observed in the RMS and OB ventricular zone was similar to that in wild types (Fig.3F,H). Thus, there is no obvious change in expression of PSA-NCAM in the absence ofMash1 function.
Fig. 3.
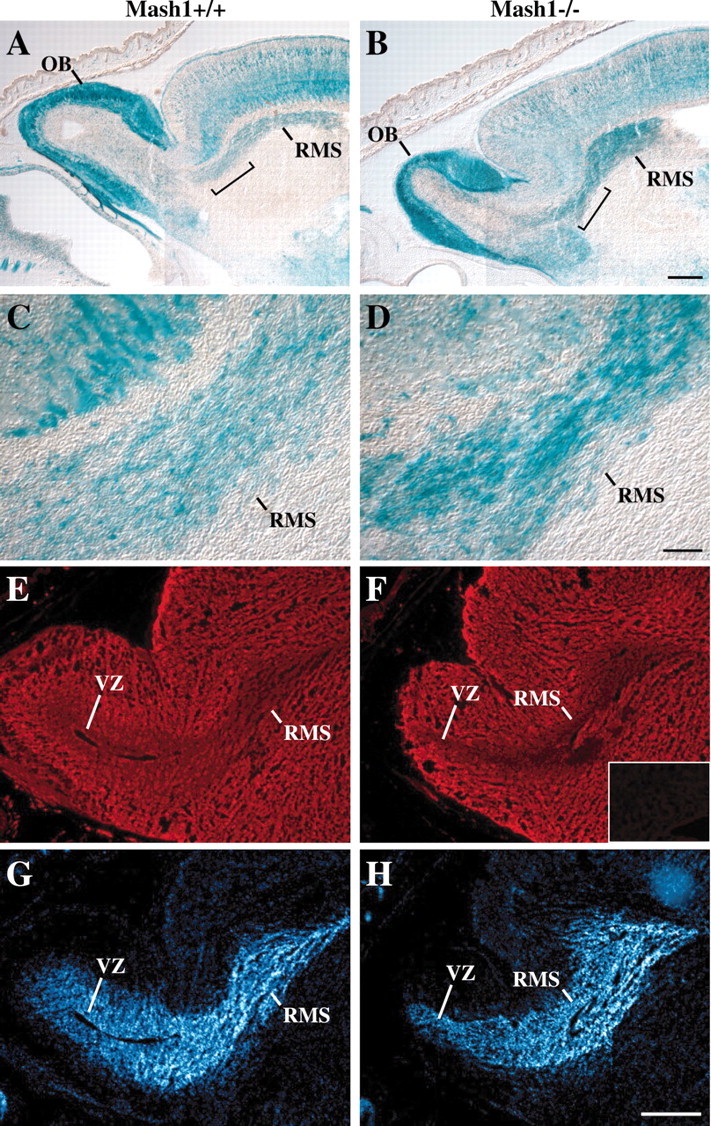
Comparison of the rostral migratory stream inMash1−/− and Mash1+/+ embryos.A–D, Thirty micrometer sagittal sections through the head of an E18.5 Mash1+/+;Tattler-4+/− embryo (A, C) and aMash1−/−;Tattler-4+/− littermate (B,D) stained with X-gal. C,D, Higher power images of the regionsbracketed in A and B.E–H, Twelve micrometer sagittal sections through the head of an E17.5 Mash1+/+ embryo (E,G) and a Mash1−/− littermate (F, H) processed for PSA-NCAM immunostaining (monoclonal anti-PSA-NCAM 12F8) (Chung et al., 1991) (E, F) or nuclear DNA staining (bisbenzimide H 33258 counterstain of same sections) (G,H). Inset in Fshows a negative control (no primary antibody) section of forebrain, including a portion of the lateral ventricle, for comparison. Rostral migratory stream (RMS) and ventricular zone (VZ) are shown. Scale bars: (in B)A, B, 250 μm; (in D)C, D, 50 μm; (inH) E–H, 300 μm.
Many of the neuronal cells migrating in the RMS are known to be progenitors of OB granule cells; these progenitors are unusual in that they express markers characteristic of differentiated neurons (e.g., neuronal tubulins) while still continuing to proliferate (Luskin, 1998). Granule cell progenitors follow a stereotyped migratory route into the OB: they first migrate tangentially into the OB along the ventricular zone and then disperse radially into the surrounding granular layer (Alvarez-Buylla, 1997; Luskin, 1998). To determine whether this pattern of cell migration is disrupted in Mash1−/−animals, E17.5 embryos were given a pulse of BrdU to label migratory progenitors and then killed 1 hr later, and their brains were fixed and processed for BrdU immunoreactivity. As shown in Figure4, A and B, the patterns of dispersal of BrdU-labeled cells were very different inMash1−/− embryos and their wild-type littermates. In wild-type embryos, many BrdU+ cells could be seen outside of the ventricular zone, dispersed within the granular layer (Fig.4A). In contrast, BrdU+ cells in the OB ofMash1−/− animals were restricted to the ventricular zone, which itself appeared thinner than that of wild types (Fig.4B). These observations were confirmed by counting the number of BrdU-labeled cells in three concentric bands surrounding the ventricle of the OB in these sections (Fig. 4C). InMash1+/+ embryos, many cells were found to have migrated radially out of the ventricular zone and into the surrounding granular layer (i.e., out of Band I into Bands II and III), whereas inMash1−/− animals, virtually all BrdU-positive cells in the OB were found within the ventricular zone (Band I). This observation suggested that, in Mash1−/− animals, granule cell progenitors may fail to migrate out of the RMS and into the developing granular layer of the OB, resulting in a deficit in differentiated granule cells and a marked reduction in OB size.
Fig. 4.
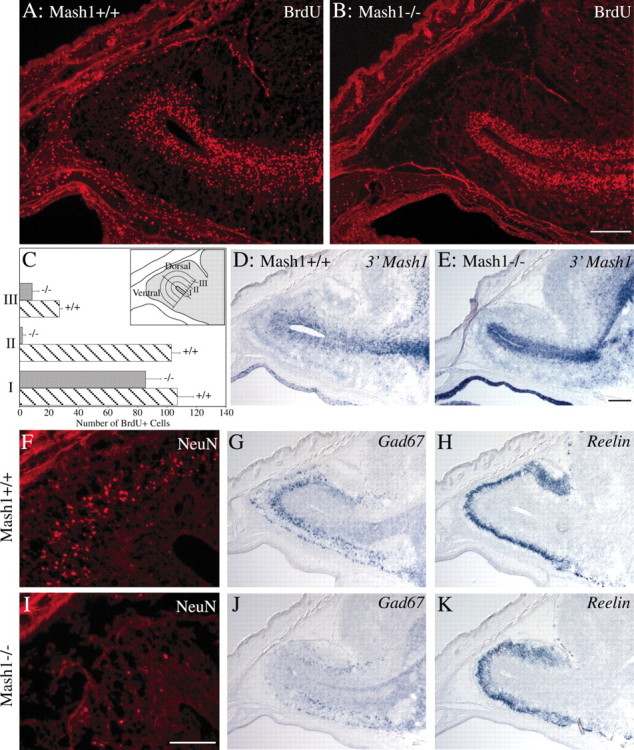
Alterations in RMS, VZ, and granular layer of the OB in Mash1−/− embryos. A,B, D–K, Sagittal sections, 12 μm (A, B, F,I) and 20 μm (D,E, G, H, J,K), through the heads of E17.5Mash1+/+ embryos (A, D,F–H) and Mash1−/− littermates (B, E, I–K) were processed for BrdU or NeuN immunostaining or in situhybridization for 3′ Mash1 UTR, Gad67, orReelin as indicated. Scale bars: (in B)A, B, 200 μm; (inI) F, I, 100 μm; (in E) D, E,G, H, J, K, 200 μm. C, Quantification of BrdU-incorporating cells in the ventricular zone and granular layer of the OB in Mash1−/− embryos (gray bars) and wild-type (striped bars) littermates. The total number of BrdU+ cells was counted in a series of three 83-μm-wide (approximate width of the ventricular zone) bands proceeding dorsally or ventrally from the OB ventricular surface (inset). Values: Band I, 107 (range, ±11) BrdU+ cells (wild type) and 85.5 (range, ±10.5) BrdU+ cells (Mash1−/−); Band II, 103 (range, ±6) BrdU+ cells (wild type) and 2 (range, ±2) BrdU+ cells (Mash1−/−); Band III, 27 (range, ±2) BrdU+ cells (wild type) and 8.5 (range, +4.5) BrdU+ cells (Mash1−/−).
To investigate this idea, we performed in situ hybridization experiments to determine whether, in Mash1−/− animals, the BrdU+ cells apparently “trapped” in the RMS and VZ of the OB are progenitors that would normally express the Mash1 gene. To do this, we took advantage of the fact that the noncoding second exon of the Mash1 gene was still present in the targeting vector used to generate the Mash1−/− animals that were used in our study (Guillemot et al., 1993). It has been shown previously that transcripts continue to be made from the disrupted Mash1allele in CNS neural progenitors present in Mash1−/−animals (Horton et al., 1999). This finding suggested that a probe that includes the Mash1 3′UTR could be used to identify cells that express Mash1 transcripts in Mash1−/−animals, as well as normal Mash1-expressing progenitors in wild types. When we performed in situ hybridization experiments using such a probe (3′ Mash1 probe), we found that the pattern of hybridization in the RMS/VZ and developing granular layer of the OB showed close correspondence to the pattern of anti-BrdU immunoreactivity (Fig. 4D,E). Cells positive for the 3′ Mash1 probe were present in the VZ and dispersed within the developing OB granular layer in wild-type animals (Fig. 4D), whereas they were restricted to the VZ in the OB of Mash1−/− animals (Fig. 4E). This finding suggested that the cells to which the Mash1 3′ probe hybridized are neuronal progenitors that would normally expressMash1 and migrate out of the RMS/VZ and into the granular layer of the OB. However, in Mash1−/− animals, these cells are unable to migrate and contribute to OB development, apparently because of a defect resulting from lack of Mash1function.
To further investigate the role of Mash1 in the development of intrinsic OB neuronal cell types, we performed in situhybridization experiments using specific markers to compare the relative sizes of differentiated OB cell populations inMash1−/− animals and their wild-type littermates. Differentiated granule cells were detected using a monoclonal antibody to the neuronal nuclear marker NeuN (Mullen et al., 1992) and a cRNA probe for Gad67 (Bulfone et al., 1998), whereas OB mitral cells were identified using a probe for Reelin (D'Arcangelo et al., 1995). As shown in Figure 4, the number of NeuN+ andGad67+ cells was drastically reduced in the granular layer of Mash1−/− OB (Fig.4I,J) compared with wild type (Fig. 4F,G), indicating that differentiated granule cells are greatly reduced in number inMash1−/− OB. In contrast, there was no apparent decrease in the number of cells expressing the mitral cell markerReelin in Mash1−/− OB (Fig. 4, compareH, K).
Together, these data indicate that in Mash1−/− animals, OB granule cell progenitors appear to be generated but are unable to migrate out of the RMS and VZ into the developing granular layer, resulting in a dramatic decrease in granule cell number and a marked reduction in OB size.
Mash1 is required for sensory neuron development in the vomeronasal organ
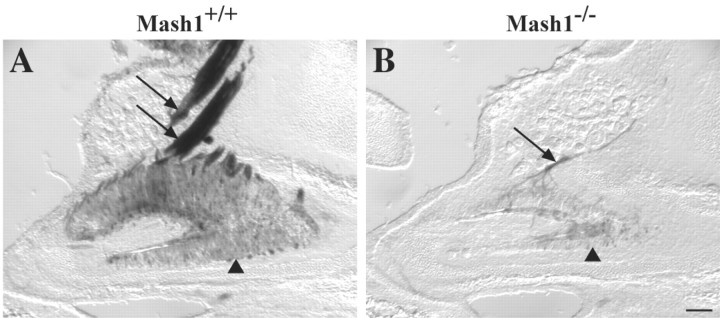
Although detection of most odors is mediated by ORNs in the main OE, pheromone detection is mediated by sensory neurons of the VNO, a tube-shaped sensory epithelium that lies within the ventral portion of the nasal septum (Halpern, 1987). Like the main OE, the VNO is derived from the olfactory placode (Farbman, 1992). Although ORN development in the main OE is Mash1 dependent (Cau et al., 1997), a recent report has suggested that genesis of sensory neurons in the VNO does not require Mash1 (Cau et al., 2002); however, that study examined only early development (E10.5–12.5). Given that much of the neurogenesis in the VNO is known to occur late during fetal development and in the early postnatal period (for review, see Halpern, 1987), it seemed possible that a requirement for Mash1 function might not yet be evident in the VNO by E12.5. To resolve this question, we examined the VNO in Mash1−/−;Tattler-4+/− embryos around the time of birth (E18.5). In wild-type (Mash1+/+;Tattler-4+/−) animals, theTα1:tau-lacZ transgene is expressed by VNO sensory neurons, and the vomeronasal nerve connecting the VNO to the AOB is heavily labeled by X-gal staining (Fig.5A). However, inMash1−/− animals, the epithelium is much thinner than in wild-type animals, and contains almost no X-gal-stained neurons (Fig.5B). In addition, the size of the vomeronasal nerve is reduced dramatically. These results indicate that most of the sensory neurons of the VNO depend on Mash1 for their proper development. Moreover, they suggest that the decrease in size of the AOB in Mash1−/− animals, shown in Figure 2, is likely to be caused, at least in part, by the loss of sensory afferents from the VNO.
Fig. 5.
Alterations in the VNO of Mash1−/−mice. A, B, Sagittal sections through the VNO of an E18.5 Mash1+/+;Tattler-4+/−embryo (A) and aMash1−/−;Tattler-4+/− littermate (B) stained with X-gal. Anterior is on theleft, and dorsal is at the top.A, Sensory neurons in the VNO epithelium (arrowhead) stain with X-gal as do the vomeronasal nerves (arrows). B, The number of sensory neurons and the size of the VNO (arrowhead) as well as the size of the vomeronasal nerve (arrow) is reduced in the Mash1−/−;Tattler-4+/− littermate. Scale bar, 100 μm.
To determine whether other aspects of VNO sensory neuron development show similarities to ORN development in the main OE, we examined the VNO for expression of markers characteristic of different cell stages in the ORN developmental pathway (Fig.6). ORN development appears to involve three stages of proliferating neuronal progenitor cells, all of which are present in the OE at E14.5–15.5: a neuronal stem cell [defined functionally, because no definitive marker for it has been identified (Mumm et al., 1996)] gives rise to neuronal progenitors that expressMash1 (Fig. 6A) (Gordon et al., 1995).Mash1-expressing progenitors then give rise to the immediate neuronal precursors (INPs) of ORNs (Calof and Chikaraishi, 1989). INPs do not express Mash1, but instead expressNeurogenin1 (Ngn1), another proneural gene homolog encoding a bHLH transcription factor (Fig.6B) (Cau et al., 1997; Calof et al., 1998, 2002; Wu et al., 2003). The progeny of INPs rapidly differentiate into ORNs and express NCAM (Fig. 6C) (Calof and Chikaraishi, 1989; DeHamer et al., 1994; Calof et al., 1998). In Mash1−/−animals, development of ORNs ceases early in this pathway, and as a consequence expression of the INP- and neuron-specific genes,Ngn1 and Ncam, is drastically reduced in the OE (Fig. 6E,F) (Cau et al., 1997).
Fig. 6.
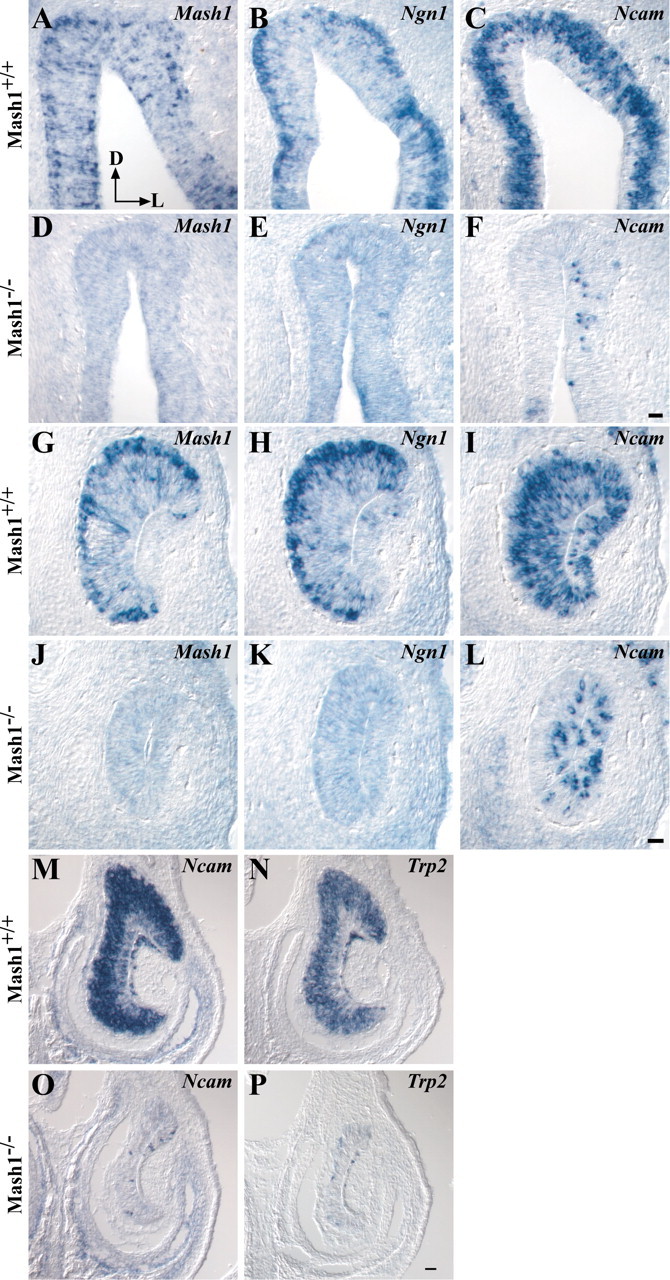
Gene expression in the OE and VNO ofMash1−/− embryos. A–P, Coronal sections of E14.5 wild-type OE (A–C) and VNO (G–I) or Mash1−/− littermate OE (D–F) and VNO (J–L) and E17.5 wild-type (M, N) orMash1−/− littermate VNO (O,P) were processed for in situhybridization as described in Materials and Methods. Dorsal is at thetop, and lateral is on the right(A, arrows). Scale bar: (inF) A–F, 20 μm; (in L) G–L, 20 μm; (in P)M–P, 50 μm.
In the VNO at E14.5, Mash1 is expressed predominantly by cells in the basal region, with scattered Mash1+ cells located more apically in the epithelium (Fig. 6G). This pattern is similar to the pattern of Mash1 expression in the main OE (Fig. 6A) and is consistent with what is known concerning the location of proliferating progenitors in VNO (Weiler et al., 1999). Ngn1 is also expressed in the basal progenitor cell region of the VNO (Fig. 6H), and the neuronal marker Ncam is expressed throughout the sensory neuron-containing layers (Fig. 6I). To determine whether this is indicative of a developmental hierarchy of gene expression in the VNO sensory neuron lineage similar to that of ORNs in the main OE, we asked whether expression of Ngn1 andNcam in the VNO is also dependent on Mash1function. Ngn1 is essentially absent in the VNO ofMash1−/− embryos at E14.5 (Fig. 6K), consistent with the idea that Ngn1 expression in the VNO isMash1-dependent. In addition, a dramatic reduction in the number of Ncam-expressing neurons was also apparent in the VNO of Mash1−/− embryos (Fig. 6L). By E17.5, near the time of birth, only a few scattered sensory neurons remain in the VNO of Mash1−/− animals, as indicated by the decrease of hybridization for probes to either Ncam or the VNO neuron-specific channel, Trp2 (Fig.6M-P) (Liman et al., 1999). Altogether, these findings demonstrate that Mash1 function is required for the normal development of sensory neurons in the VNO, as it is in the main OE. In addition, they suggest that similar hierarchies of gene expression regulate neuronal differentiation in these two sensory epithelia.
Proliferating neural progenitors are present in the olfactory epithelium and vomeronasal organ of Mash1−/−embryos
The failure of sensory neurons to differentiate in the VNO and OE in the absence of Mash1 function raises questions about the fate of neural progenitor cells in these structures. Because the OE and VNO are thinner and have many fewer neurons in Mash1−/−animals (Fig. 6), we hypothesized that progenitor cells might also be absent. To determine whether this was the case, we injected pregnant dams (day 14.5 of gestation) with BrdU and fixed embryos 1 hr later to identify cells in S phase. In wild-type embryos at E14.5, BrdU-labeled cells are located in the basal and apical layers of the OE and in the basal two-thirds of the VNO, regions in which proliferating progenitor cells are known to be located in both of these structures (Fig.7A,B) (Smart, 1971; Cuschieri and Bannister, 1975). We also found numerous BrdU+ cells present in both OE and VNO of E14.5 Mash1−/−embryos (Fig. 7C,D). Interestingly, BrdU+ cells in the OE and VNO of Mash1−/− embryos are altered in their relative locations within the epithelia compared with wild types. Rather than being localized to the basal and apical compartments of the epithelia, BrdU+ cells are present throughout the apical–basal extent of the epithelia in Mash1−/− OE and VNO. In addition, the shape of BrdU-incorporating nuclei differs in embryos of the two genotypes. In Mash1−/− animals, the nuclei are larger and spindle-shaped, rather than round or oval in shape, as they are in wild-type embryos (Fig. 7C,D).
Fig. 7.
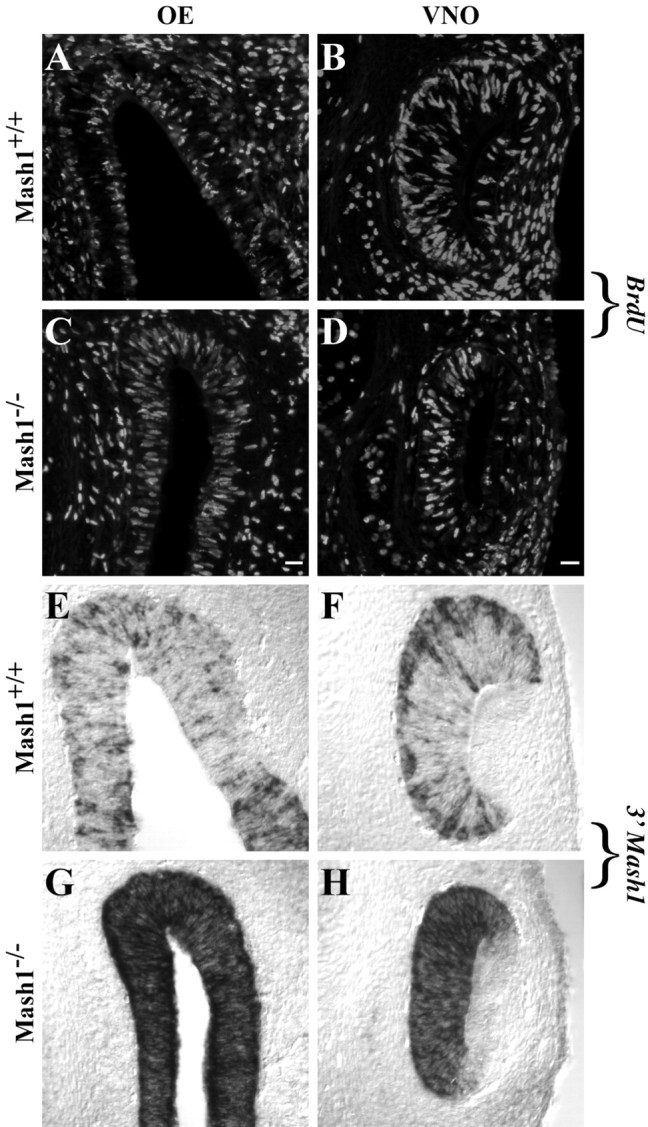
. BrdU incorporation andMash1 expression in the OE and VNO of Mash1−/−embryos. A–H, E14.5 wild-type (A, B, E,F) and Mash1−/−(C, D, G,H) littermates were sectioned coronally and processed for BrdU immunohistochemistry (A–D) orin situ hybridization with the probe for the 3′Mash1 UTR (E–H). Scale bar: (inC) A, C, E,G, 20 μm; (in D)B, D, F, H, 20 μm.
To determine whether the number of proliferating cells is altered inMash1−/− OE and VNO, we performed cell counts in six to eight sections from two animals of each genotype. In wild-type OE, there were 572 (range, ±85) BrdU+ cells per millimeter versus 376 (range, ±2) BrdU+ cells per millimeter OE in Mash1−/−animals. Because the OE is thinner in Mash1−/− mice, we also calculated the density of BrdU+ cells per unit area, and these numbers were very similar: 80 (range, ±25) BrdU+ cells/10,000 μm2 in wild-type OE versus 81 (range, ±6) BrdU+ cells/10,000 μm2 inMash1−/− OE. Because the VNO is a circular structure in cross section, it is not possible to accurately count the number of BrdU+ cells per unit length, so only the density of BrdU+ cells per unit area was obtained for this structure: 234 (range, ±5) BrdU+ cells/25,000 μm2 in wild-type VNO and 201 (range, ±15) BrdU+ cells/25,000 μm2in Mash1−/− VNO. Thus, the density of proliferating cells is very similar in both the OE and VNO of wild-type andMash1−/− animals.
To determine whether the proliferating cells in the OE and VNO of mutant embryos might be neural progenitors, we used the 3′ Mash1 in situ hybridization probe to detect cells expressing mutantMash1 transcripts (i.e., presumptive neural progenitors). The pattern of hybridization in the OE and VNO of wild-type embryos observed with this probe was identical to that seen in earlier experiments using a probe containing only the Mash1 coding region (compare Fig. 7E,F with Fig.6A,G). In Mash1−/−embryos, however, the 3′ Mash1 probe hybridized to the vast majority of cells in OE and VNO, and BrdU-incorporating cells (Fig. 7, compare G, H with C,D) were among the expressing cells in both structures. These findings indicate that the proliferating cells present in mutant OE and VNO are capable of expressing Mash1, and so in this respect they have at least one characteristic of sensory neuron progenitors. However, these cells fail to express normal Mash1 orNgn1 transcripts, and for the most part they fail to give rise to sensory neurons in either the OE or VNO (Fig. 6) (Guillemot et al., 1993; Gordon et al., 1995; Cau et al., 1997), indicating that they have lost the ability to give rise to neurons.
3′ Mash1-expressing cells in Mash1−/−olfactory epithelium express a sustentacular cell marker
What then is the fate of 3′ Mash1-expressing proliferating cells in Mash1−/− OE and VNO? To determine whether these cells have characteristics of other cell types in these epithelia, we first used a commercial antiserum to keratins to mark horizontal basal cells (Calof and Chikaraishi, 1989); no difference was observed in the pattern of staining between wild-type and mutant OE and VNO (data not shown). The Steel gene has been shown to be expressed by supporting cells (sustentacular cells) of the OE, and a previous study noted that Steel-expressing cells are still present in the OE of newborn Mash1−/− animals (Guillemot et al., 1993). We generated a Steel probe by RT-PCR (see Materials and Methods) and performed in situ hybridization experiments on the OE and VNO of E14.5 and E17.5 Mash1−/−embryos and wild-type littermates. The results are shown in Figure8. In wild-type OE at both ages,Steel expression is evident in the apical cytoplasm of sustentacular cells, which are arrayed in a single layer immediately above Ncam-expressing ORNs in the epithelium (Fig.8A,B,G,H).InMash1−/− OE, in contrast, Steel appears to be expressed by the majority of cells throughout the basal–apical extent of the epithelium, whereas almost no Ncam-positive ORNs are present (Fig.8D,E,J,K). (Steel is not expressed in either wild-type orMash1−/− VNO; data not shown.) Thus, there appeared to be many more Steel-expressing cells in Mash1−/−than in wild-type OE, and the position of these cells overlapped with those incorporating BrdU and expressing the 3′ Mash1UTR (compare Fig. 7C with Fig.8E,F,K,L). To determine the extent of this overlap, we counted the percentage of cells expressing each marker in Mash1−/− OE at E14.5: 84.6% (range, ±3.4%) of cells in the mutant OE expressSteel, and 96% (range, ±0.4%) of cells in mutant OE also label with the 3′ Mash1 probe, so it must be the case that most of the cells in the mutant OE express both markers. Thus, in the absence of Mash1 function, the OE becomes populated by proliferating cells that have characteristics of both neuronal progenitors (expression of the 3′ Mash1 UTR) and supporting (sustentacular) cells (expression of Steel).
Fig. 8.
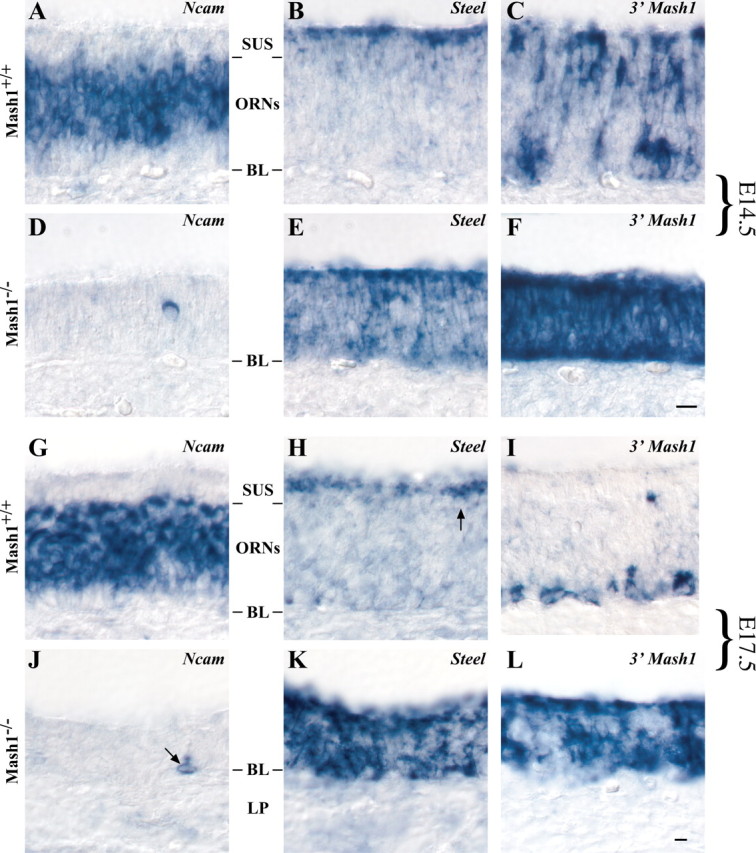
. Ncam,Steel, and 3′ Mash1 expression in wild-type and Mash1−/− embryos. A–L, E14.5 (A–F) and E17.5 (G–L) embryos were sectioned in the coronal plane and processed for in situ hybridization. Sustentacular cell layer (SUS), olfactory receptor neuron layer (ORNs), basal lamina (BL), and lamina propria (LP) are shown. Scale bar: (inF) A–F, 10 μm; (in L) G-L, 10 μm.
Increased apoptosis in olfactory epithelium and vomeronasal organ of Mash1−/− embryos
Despite the high level of proliferation in Mash1−/−OE and VNO (Fig. 7), by the time of birth these epithelia are much thinner than those of wild-type animals (Figs. 2, 5). This observation suggests that the proliferating progenitors present in Mash1−/−epithelia do not survive, but instead may be dying at an abnormally high rate. We and others have observed previously that there is an increased level of apoptotic death in cells of Mash1−/−OE at E13.5–15.5 (Calof et al., 1996b; Cau et al., 1997). To determine whether cells in Mash1−/− VNO also display abnormal levels of apoptosis, we performed TUNEL assays onMash1−/− VNO at E14.5 (Holcomb et al., 1995) (OE was also assessed as a positive control). The results are shown in Figure9. In wild-type epithelia, almost no TUNEL+ cells can be observed, whereas numerous TUNEL+ cells are present in both OE and VNO of Mash1−/− animals. Counting the number of TUNEL+ nuclei confirmed the dramatic increase in the number of apoptotic cells in Mash1−/− VNO (e.g., the number of TUNEL+ cells was increased more than fivefold in Mash1−/−VNO) (Fig. 9F). Thus, despite being able to proliferate and being capable of expressing markers of both neuronal progenitor cells (Mash1) and sustentacular cells (Steel), many cells in the Mash1−/− VNO and OE are unable to survive.
Fig. 9.
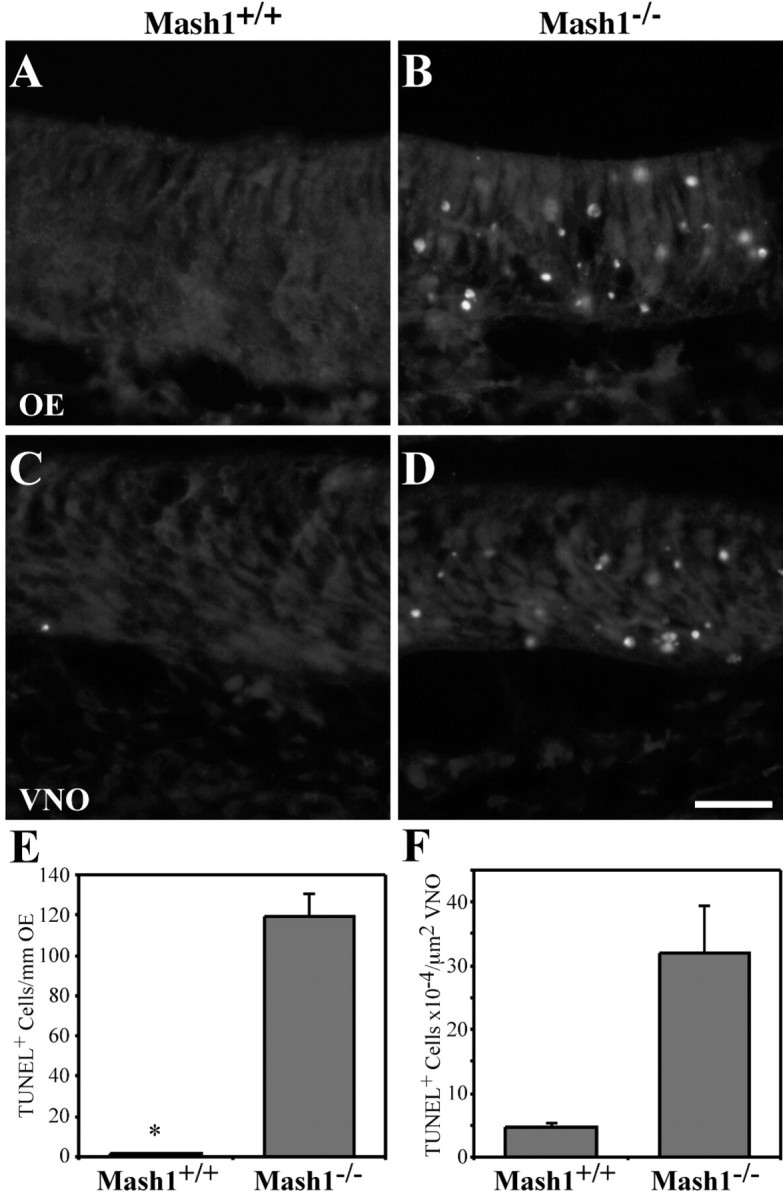
Apoptosis in the OE and VNO of Mash1−/−embryos. A–D, E14.5 wild-type (A, C) and Mash1−/−(B, D) littermates were sectioned horizontally at 12 μm and processed for TUNEL labeling as described previously (Holcomb et al., 1995). Scale bar, 20 μm.E, The number of TUNEL+ cells per millimeter of OE was counted in a minimum of 20 fields representing at least 5.2 mm of OE in one animal of each genotype. Values are 1.8 (±0.53 SEM) TUNEL+ cells per millimeter OE for wild-type and 119.8 (± 10.87 SEM) TUNEL+ cells per millimeter OE for Mash1−/−. F, The number of TUNEL+ cells per square micrometers of VNO was counted in a minimum of eight fields representing at least 115,000 μm2 of VNO. Values are 4.64 × 10−4 (±0.75 SEM) TUNEL+ cells per square micrometer VNO for wild-type and 31.8 × 10−4(±7.57) TUNEL+ cells per square micrometer VNO forMash1−/−.
Discussion
Loss of Mash1 function leads to abnormalities of olfactory bulb progenitors and granule cells
One of the more striking effects in the CNS that we observed inMash1−/− mice was a marked decrease in OB size, evident in both the main OB and the AOB (Fig. 2). Although some reduction in size of the main OB could be expected from the diminished number of ingrowing ORN and VNO axons (Figs. 2, 5), our observations clearly show that much of the reduction occurs in a layer of interneurons, the OB granule cells (Fig. 2E). These neurons derive from progenitors that are produced in the subventricular zone of the lateral ventricle, migrate through the RMS into the VZ of the OB, and finally disperse into the OB granule layer.
The fact that Mash1 is expressed in the VZ of the OB (Fig.4) (Guillemot and Joyner, 1993; Sommer et al., 1996), as well as the VZ and subventricular zone of the ganglionic eminences (Casarosa et al., 1999; Horton et al., 1999), which contribute cells to the RMS (Wichterle et al., 1999), suggests that loss of Mash1function directly affects OB granule neurons and/or their precursors. The alternate possibility—that reduced afferent (ORN) input to the OB in Mash1−/− mice indirectly affects granule cell number—is plausible, given data that naris occlusion in early postnatal rodents can cause granule cell apoptosis in the ipsilateral OB (Frazier and Brunjes, 1988; Frazier-Cierpial and Brunjes, 1989;Najbauer and Leon, 1995; Fiske and Brunjes, 2001). However, the time course of deafferentation-induced granule cell apoptosis is likely to be too slow (∼10 d) (Petreanu and Alvarez-Buylla, 2002) to explain the abnormalities in Mash1−/− OB at E18.5, just 3–4 d after the initial generation of granule neurons (Hinds, 1968).
Interestingly, in Mash1−/−;Tattler-4+/− animals, the RMS is noticeably abnormal (Fig. 3A,B), being both shorter and thicker along the anterior-posterior axis and exhibiting stronger X-gal staining (i.e., increased density of neuronal cells) (Fig. 3C,D). These changes are reminiscent of mice in which the polysialylated form of NCAM has been rendered nonfunctional by gene inactivation or enzymatic treatment (Cremer et al., 1994; Ono et al., 1994). In such animals, interference with neuronal cell migration into the OB causes an accumulation of cells in the RMS and a reduction in OB size (Bruses and Rutishauser, 2001). If loss of Mash1 function also results in a migration defect in the RMS, then the fact that Mash1−/− animals exhibit normal expression of PSA-NCAM (Fig.3E,F) indicates that the cause of this defect would have to be different. It has previously been reported that ventral forebrain neuronal progenitors migrate abnormally in Mash1−/− embryos, and this has been linked to premature neuronal differentiation (Horton et al., 1999). The possibility that OB granule neurons also differentiate prematurely is certainly consistent with the observed increased in X-gal staining in the RMS ofMash1−/−;Tattler-4+/− animals, as theTα1:tau-lacZ transgene is selectively turned on during neuronal differentiation (Fig. 1) (Gloster et al., 1999).
Neurogenesis in the vomeronasal organ: parallels with the olfactory epithelium
Like the main OE, the VNO derives from the olfactory placode, maintains a neuroepithelial structure, projects to the OB, and uses a family of specialized seven-transmembrane odorant receptors to transduce olfactory signals (Halpern, 1987; Dulac, 2000). Despite these similarities, Cau et al. (2002) recently reported only a modest reduction in neuron number in the VNO of Mash1−/− animals at E12.5 and concluded that the generation of VNO neurons, unlike those of the main OE, must depend to a large extent on a factor other thanMash1. Our observations here support a different view. At E14.5, we observed a substantially reduced number of neurons in theMash1−/− VNO, and by E17.5 a profound reduction was obvious (Fig. 6). These data suggest that although the earliest neurogenesis in the VNO may be Mash1-independent, most of the later production of neurons, known to occur late in fetal development and in the early postnatal period (Halpern, 1987), requiresMash1 function. Interestingly, in the main OE it is also the case that the very earliest generated neurons (those born by E9.5) are relatively Mash1 independent, whereas those produced later require Mash1 (Cau et al., 1997). Thus, our results suggest that the molecular details of neurogenesis in the VNO and OE may be more similar than suspected previously. Such a view is supported further by our finding in the E14.5 VNO that Mash1 is required for expression not just of neuron-specific markers (Ncam, Trp2), but also of Ngn1, a gene that, in the main OE, marks a progenitor cell stage interposed betweenMash1+ cells and neurons (Cau et al., 1997; Calof et al., 2002).
Do supporting cells and sensory neurons of the olfactory epithelium share a common progenitor?
Despite the deficit in neuron number in the OE and VNO ofMash1−/− embryos as early as E14.5 (Fig. 6), we found that many cells in these tissues are proliferating and most express theMash1 3′ UTR (Fig. 7). The observation that theMash1 promoter is active in so many more cells inMash1−/− OE and VNO than in wild types suggests that far fewer cells in developing OE and VNO normally express Mash1transcripts than are competent to do so. It also supports the findings of Horton and colleagues (1999), whose studies of gene expression in the ventral forebrain of Mash1−/− embryos implied that MASH1 negatively regulates its own expression (Horton et al., 1999). Those authors argued that, in Mash1 nulls, absence of MASH1 protein (required for Notch-Delta signaling) results in a breakdown of the lateral inhibition that is necessary for correct specification of neuronal progenitors from a larger field of competent cells (Lewis, 1996). The idea that Mash1 functions in the OE through a Notch signaling pathway is supported by the finding that activation of the expression of several genes in this pathway fails to occur inMash1−/− OE (Cau et al., 2000, 2002). Altogether, these observations support a model in which the OE and VNO inMash1−/− embryos are populated by early proliferating progenitor cells that transcribe (aberrant) Mash1transcripts, but lacking Mash1 function, fail to differentiate properly and subsequently undergo apoptosis (Fig. 9).
Interestingly, most (perhaps all) of these presumed early progenitors in Mash1−/− OE express Steel, a marker of supporting (sustentacular) cells (Fig. 8), together with aberrantMash1 transcripts (Figs. 7, 8). This suggests that most of these cells are developing along a sustentacular cell pathway. They also exhibit the elongated nuclei characteristic of sustentacular cells (Fig. 7) (Smart, 1971; Cuschieri and Bannister, 1975). The obvious implication is that, early in embryonic development, the OE may contain bipotential progenitors that subsequently become restricted to a neuronal (ORN) or glial (sustentacular) fate, and that Mash1function is required for the neuronal determination event. Indeed, in the inner ear, another placode-derived sensory epithelium, it has been shown that sensory and supporting cells share a common progenitor (Corwin and Cotanche, 1988; Ryals and Rubel, 1988; Fekete et al., 1998). Moreover, in other areas of the nervous system, bHLH transcription factors have been shown to act to promote neuronal, and inhibit glial, fate determination (Tomita et al., 2000; Morrison, 2001;Nieto et al., 2001).
The existence of a common ORN–sustentacular lineage has been suggested by some, but not all, studies. For example, fate maps of single cells in Xenopus olfactory placode demonstrated a common progenitor for ORNs and sustentacular cells, at least in early development (Burd et al., 1994). When adult rat OE was lesioned with methyl bromide (which kills both neurons and sustentacular cells) and allowed to regenerate, retroviral lineage mapping suggested the existence of common ORN–sustentacular progenitors (Huard et al., 1998). In contrast, in animals lesioned by olfactory bulbectomy (which kills ORNs but not sustentacular cells), proliferation of cells that express Mash1 and give rise to neurons increases greatly, but proliferation of cells that become sustentacular cells does not (Gordon et al., 1995; Calof et al., 1996a). Moreover, retroviral lineage analysis of OE in unlesioned postnatal rats failed to find any evidence for a common ORN–sustentacular lineage (Caggiano et al., 1994).
Because sustentacular cells become a self-renewing population shortly after their appearance in the OE, at ∼E13.5 (Smart, 1971; Cuschieri and Bannister, 1975; Weiler and Farbman, 1998), one explanation for these data is that the common ORN–sustentacular progenitor generates sustentacular cells only when “needed” to do so. Intriguingly, in the OE there is strong evidence that neuronal production is repressed by feedback signals from ORNs (Mumm et al., 1996; Wu et al., 2003). Perhaps combinations of feedback signals from both ORNs and sustentacular cells cooperatively tell bipotential progenitors not only when to proliferate, but also what cell types to make.
Footnotes
This work was supported by National Institutes of Health Grants DC03583 to A.L.C., NS26862 to A.D.L., HD38761 to A.D.L. and A.L.C., and the March of Dimes Birth Defects Foundation (FY00-660 to A.L.C.). We thank Carl Lagenaur for 12F8 anti-PSA-NCAM monoclonal antibody, Tom Curran for reelin probe, and Shimako Kawauchi for help within situs.
Correspondence should be addressed to Anne L. Calof, Department of Anatomy and Neurobiology, 364 Med Surge II, University of California, Irvine, Irvine, CA 92697-1275. E-mail:alcalof@uci.edu.
References
- 1.Alvarez-Buylla A. Mechanism of migration of olfactory bulb interneurons. Semin Cell Dev Biol. 1997;8:207–213. doi: 10.1006/scdb.1996.0134. [DOI] [PubMed] [Google Scholar]
- 2.Angevine JB., Jr Time of neuron origin in the diencephalon of the mouse. An autoradiographic study. J Comp Neurol. 1970;139:129–187. doi: 10.1002/cne.901390202. [DOI] [PubMed] [Google Scholar]
- 3.Barthels D, Santoni MJ, Wille W, Ruppert C, Chaix JC, Hirsch MR, Fontecilla-Camps JC, Goridis C. Isolation and nucleotide sequence of mouse NCAM cDNA that codes for a Mr 79,000 polypeptide without a membrane-spanning region. EMBO J. 1987;6:907–914. doi: 10.1002/j.1460-2075.1987.tb04837.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blaugrund E, Pham TD, Tennyson VM, Lo L, Sommer L, Anderson DJ, Gershon MD. Distinct subpopulations of enteric neuronal progenitors defined by time of development, sympathoadrenal lineage markers and Mash-1-dependence. Development. 1996;122:309–320. doi: 10.1242/dev.122.1.309. [DOI] [PubMed] [Google Scholar]
- 5.Brunet JF, Ghysen A. Deconstructing cell determination: proneural genes and neuronal identity. BioEssays. 1999;21:313–318. doi: 10.1002/(SICI)1521-1878(199904)21:4<313::AID-BIES7>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 6.Bruses JL, Rutishauser U. Roles, regulation, and mechanism of polysialic acid function during neural development. Biochimie. 2001;83:635–643. doi: 10.1016/s0300-9084(01)01293-7. [DOI] [PubMed] [Google Scholar]
- 7.Bulfone A, Wang F, Hevner R, Anderson S, Cutforth T, Chen S, Meneses J, Pedersen R, Axel R, Rubenstein JL. An olfactory sensory map develops in the absence of normal projection neurons or GABAergic interneurons. Neuron. 1998;21:1273–1282. doi: 10.1016/s0896-6273(00)80647-9. [DOI] [PubMed] [Google Scholar]
- 8.Burd GD, Collazo A, Fraser SE. Cell lineage in the formation and regeneration of the olfactory placodes. Soc Neurosci Abstr. 1994;20:1275. [Google Scholar]
- 9.Caggiano M, Kauer JS, Hunter DD. Globose basal cells are neuronal progenitors in the olfactory epithelium: a lineage analysis using a replication-incompetent retrovirus. Neuron. 1994;13:339–352. doi: 10.1016/0896-6273(94)90351-4. [DOI] [PubMed] [Google Scholar]
- 10.Callahan CA, Thomas JB. Tau-beta-galactosidase, an axon-targeted fusion protein. Proc Natl Acad Sci USA. 1994;91:5972–5976. doi: 10.1073/pnas.91.13.5972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Calof AL, Chikaraishi DM. Analysis of neurogenesis in a mammalian neuroepithelium: proliferation and differentiation of an olfactory neuron precursor in vitro. Neuron. 1989;3:115–127. doi: 10.1016/0896-6273(89)90120-7. [DOI] [PubMed] [Google Scholar]
- 12.Calof AL, Hagiwara N, Holcomb JD, Mumm JS, Shou J. Neurogenesis and cell death in olfactory epithelium. J Neurobiol. 1996a;30:67–81. doi: 10.1002/(SICI)1097-4695(199605)30:1<67::AID-NEU7>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 13.Calof AL, Holcomb JD, Mumm JS, Haglwara N, Tran P, Smith KM, Shelton D. Factors affecting neuronal birth and death in the mammalian olfactory epithelium. Ciba Found Symp. 1996b;196:188–205. doi: 10.1002/9780470514863.ch13. [DOI] [PubMed] [Google Scholar]
- 14.Calof AL, Mumm JS, Rim PC, Shou J. The neuronal stem cell of the olfactory epithelium. J Neurobiol. 1998;36:190–205. doi: 10.1002/(sici)1097-4695(199808)36:2<190::aid-neu7>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 15.Calof AL, Bonnin A, Crocker C, Kawauchi S, Murray RC, Shou J, Wu H-H. Progenitor cells of the olfactory receptor neuron lineage. Microsc Res Tech. 2002;58:176–188. doi: 10.1002/jemt.10147. [DOI] [PubMed] [Google Scholar]
- 16.Casarosa S, Fode C, Guillemot F. Mash1 regulates neurogenesis in the ventral telencephalon. Development. 1999;126:525–534. doi: 10.1242/dev.126.3.525. [DOI] [PubMed] [Google Scholar]
- 17.Cau E, Gradwohl G, Fode C, Guillemot F. Mash1 activates a cascade of bHLH regulators in olfactory neuron progenitors. Development. 1997;124:1611–1621. doi: 10.1242/dev.124.8.1611. [DOI] [PubMed] [Google Scholar]
- 18.Cau E, Gradwohl G, Casarosa S, Kageyama R, Guillemot F. Hes genes regulate sequential stages of neurogenesis in the olfactory epithelium. Development. 2000;127:2323–2332. doi: 10.1242/dev.127.11.2323. [DOI] [PubMed] [Google Scholar]
- 19.Cau E, Casarosa S, Guillemot F. Mash1 and Ngn1 control distinct steps of determination and differentiation in the olfactory sensory neuron lineage. Development. 2002;129:1871–1880. doi: 10.1242/dev.129.8.1871. [DOI] [PubMed] [Google Scholar]
- 20.Chung WW, Lagenaur CF, Yan YM, Lund JS. Developmental expression of neural cell adhesion molecules in the mouse neocortex and olfactory bulb. J Comp Neurol. 1991;314:290–305. doi: 10.1002/cne.903140207. [DOI] [PubMed] [Google Scholar]
- 21.Corwin JT, Cotanche DA. Regeneration of sensory hair cells after acoustic trauma. Science. 1988;240:1772–1774. doi: 10.1126/science.3381100. [DOI] [PubMed] [Google Scholar]
- 22.Cremer H, Lange R, Christoph A, Plomann M, Vopper G, Roes J, Brown R, Baldwin S, Kraemer P, Scheff S, Barthels D, Rajewsky K, Wille W. Inactivation of the N-CAM gene in mice results in size reduction of the olfactory bulb and deficits in spatial learning. Nature. 1994;367:455–459. doi: 10.1038/367455a0. [DOI] [PubMed] [Google Scholar]
- 23.Cuschieri A, Bannister LH. The development of the olfactory mucosa in the mouse: light microscopy. J Anat. 1975;119:277–286. [PMC free article] [PubMed] [Google Scholar]
- 24.D'Arcangelo G, Miao GG, Chen SC, Soares HD, Morgan JI, Curran T. A protein related to extracellular matrix proteins deleted in the mouse mutant reeler. Nature. 1995;374:719–723. doi: 10.1038/374719a0. [DOI] [PubMed] [Google Scholar]
- 25.DeHamer MK, Guevara JL, Hannon K, Olwin BB, Calof AL. Genesis of olfactory receptor neurons in vitro: regulation of progenitor cell divisions by fibroblast growth factors. Neuron. 1994;13:1083–1097. doi: 10.1016/0896-6273(94)90047-7. [DOI] [PubMed] [Google Scholar]
- 26.Dulac C. Sensory coding of pheromone signals in mammals. Curr Opin Neurobiol. 2000;10:511–518. doi: 10.1016/s0959-4388(00)00121-5. [DOI] [PubMed] [Google Scholar]
- 27.Farbman AI. Cell biology of olfaction. Cambridge UP; New York: 1992. [Google Scholar]
- 28.Fekete DM, Muthukumar S, Karagogeos D. Hair cells and supporting cells share a common progenitor in the avian inner ear. J Neurosci. 1998;18:7811–7821. doi: 10.1523/JNEUROSCI.18-19-07811.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fiske BK, Brunjes PC. Cell death in the developing and sensory-deprived rat olfactory bulb. J Comp Neurol. 2001;431:311–319. [PubMed] [Google Scholar]
- 30.Fode C, Gradwohl G, Morin X, Dierich A, LeMeur M, Goridis C, Guillemot F. The bHLH protein NEUROGENIN 2 is a determination factor for epibranchial placode-derived sensory neurons. Neuron. 1998;20:483–494. doi: 10.1016/s0896-6273(00)80989-7. [DOI] [PubMed] [Google Scholar]
- 31.Frazier LL, Brunjes PC. Unilateral odor deprivation: early postnatal changes in olfactory bulb cell density and number. J Comp Neurol. 1988;269:355–370. doi: 10.1002/cne.902690304. [DOI] [PubMed] [Google Scholar]
- 32.Frazier-Cierpial L, Brunjes PC. Early postnatal cellular proliferation and survival in the olfactory bulb and rostral migratory stream of normal and unilaterally odor-deprived rats. J Comp Neurol. 1989;289:481–492. doi: 10.1002/cne.902890312. [DOI] [PubMed] [Google Scholar]
- 33.Gloster A, Wu W, Speelman A, Weiss S, Causing C, Pozniak C, Reynolds B, Chang E, Toma JG, Miller FD. The T α 1 α-tubulin promoter specifies gene expression as a function of neuronal growth and regeneration in transgenic mice. J Neurosci. 1994;14:7319–7330. doi: 10.1523/JNEUROSCI.14-12-07319.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gloster A, El-Bizri H, Bamji SX, Rogers D, Miller FD. Early induction of Talpha1 alpha-tubulin transcription in neurons of the developing nervous system. J Comp Neurol. 1999;405:45–60. doi: 10.1002/(sici)1096-9861(19990301)405:1<45::aid-cne4>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 35.Gordon MK, Mumm JS, Davis RA, Holcomb JD, Calof AL. Dynamics of MASH1 expression in vitro and in vivo suggest a non-stem cell site of MASH1 action in the olfactory receptor neuron lineage. Mol Cell Neurosci. 1995;6:363–379. doi: 10.1006/mcne.1995.1028. [DOI] [PubMed] [Google Scholar]
- 36.Guillemot F. Vertebrate bHLH genes and the determination of neuronal fates. Exp Cell Res. 1999;253:357–364. doi: 10.1006/excr.1999.4717. [DOI] [PubMed] [Google Scholar]
- 37.Guillemot F, Joyner AL. Dynamic expression of the murine Achaete-Scute homologue Mash-1 in the developing nervous system. Mech Dev. 1993;42:171–185. doi: 10.1016/0925-4773(93)90006-j. [DOI] [PubMed] [Google Scholar]
- 38.Guillemot F, Lo LC, Johnson JE, Auerbach A, Anderson DJ, Joyner AL. Mammalian achaete-scute homolog 1 is required for the early development of olfactory and autonomic neurons. Cell. 1993;75:463–476. doi: 10.1016/0092-8674(93)90381-y. [DOI] [PubMed] [Google Scholar]
- 39.Halpern M. The organization and function of the vomeronasal system. Annu Rev Neurosci. 1987;10:325–362. doi: 10.1146/annurev.ne.10.030187.001545. [DOI] [PubMed] [Google Scholar]
- 40.Hatakeyama J, Tomita K, Inoue T, Kageyama R. Roles of homeobox and bHLH genes in specification of a retinal cell type. Development. 2001;128:1313–1322. doi: 10.1242/dev.128.8.1313. [DOI] [PubMed] [Google Scholar]
- 41.Hinds JW. Autoradiographic study of histogenesis in the mouse olfactory bulb. I. Time of origin of neurons and neuroglia. J Comp Neurol. 1968;134:287–304. doi: 10.1002/cne.901340304. [DOI] [PubMed] [Google Scholar]
- 42.Hogan B. Manipulating the mouse embryo: a laboratory manual, Ed 2. Cold Spring Harbor Laboratory; Plainview, NY: 1994. [Google Scholar]
- 43.Holcomb JD, Mumm JS, Calof AL. Apoptosis in the neuronal lineage of the mouse olfactory epithelium: regulation in vivo and in vitro. Dev Biol. 1995;172:307–323. doi: 10.1006/dbio.1995.0025. [DOI] [PubMed] [Google Scholar]
- 44.Horton S, Meredith A, Richardson JA, Johnson JE. Correct coordination of neuronal differentiation events in ventral forebrain requires the bHLH factor MASH1. Mol Cell Neurosci. 1999;14:355–369. doi: 10.1006/mcne.1999.0791. [DOI] [PubMed] [Google Scholar]
- 45.Huard JM, Youngentob SL, Goldstein BJ, Luskin MB, Schwob JE. Adult olfactory epithelium contains multipotent progenitors that give rise to neurons and non-neural cells. J Comp Neurol. 1998;400:469–486. [PubMed] [Google Scholar]
- 46.Jan YN, Jan LY. Genetic control of cell fate specification in Drosophila peripheral nervous system. Annu Rev Genet. 1994;28:373–393. doi: 10.1146/annurev.ge.28.120194.002105. [DOI] [PubMed] [Google Scholar]
- 47.Lewis J. Neurogenic genes and vertebrate neurogenesis. Curr Opin Neurobiol. 1996;6:3–10. doi: 10.1016/s0959-4388(96)80002-x. [DOI] [PubMed] [Google Scholar]
- 48.Liman ER, Corey DP, Dulac C. TRP2: a candidate transduction channel for mammalian pheromone sensory signaling. Proc Natl Acad Sci USA. 1999;96:5791–5796. doi: 10.1073/pnas.96.10.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lois C, Alvarez-Buylla A. Long-distance neuronal migration in the adult mammalian brain. Science. 1994;264:1145–1148. doi: 10.1126/science.8178174. [DOI] [PubMed] [Google Scholar]
- 50.Luskin MB. Restricted proliferation and migration of postnatally generated neurons derived from the forebrain subventricular zone. Neuron. 1993;11:173–189. doi: 10.1016/0896-6273(93)90281-u. [DOI] [PubMed] [Google Scholar]
- 51.Luskin MB. Neuroblasts of the postnatal mammalian forebrain: their phenotype and fate. J Neurobiol. 1998;36:221–233. [PubMed] [Google Scholar]
- 52.Ma Q, Kintner C, Anderson DJ. Identification of neurogenin, a vertebrate neuronal determination gene. Cell. 1996;87:43–52. doi: 10.1016/s0092-8674(00)81321-5. [DOI] [PubMed] [Google Scholar]
- 53.Ma Q, Sommer L, Cserjesi P, Anderson DJ. Mash1 and neurogenin1 expression patterns define complementary domains of neuroepithelium in the developing CNS and are correlated with regions expressing notch ligands. J Neurosci. 1997;17:3644–3652. doi: 10.1523/JNEUROSCI.17-10-03644.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ma Q, Chen Z, del Barco Barrantes I, de la Pompa JL, Anderson DJ. Neurogenin1 is essential for the determination of neuronal precursors for proximal cranial sensory ganglia. Neuron. 1998;20:469–482. doi: 10.1016/s0896-6273(00)80988-5. [DOI] [PubMed] [Google Scholar]
- 55.Marquardt T, Ashery-Padan R, Andrejewski N, Scardigli R, Guillemot F, Gruss P. Pax6 is required for the multipotent state of retinal progenitor cells. Cell. 2001;105:43–55. doi: 10.1016/s0092-8674(01)00295-1. [DOI] [PubMed] [Google Scholar]
- 56.McConnell JA. Identification of early neurons in the brainstem and spinal cord. II. An autoradiographic study in the mouse. J Comp Neurol. 1981;200:273–288. doi: 10.1002/cne.902000207. [DOI] [PubMed] [Google Scholar]
- 57.Morrison SJ. Neuronal differentiation: proneural genes inhibit gliogenesis. Curr Biol. 2001;11:R349–351. doi: 10.1016/s0960-9822(01)00191-9. [DOI] [PubMed] [Google Scholar]
- 58.Mullen RJ, Buck CR, Smith AM. NeuN, a neuronal specific nuclear protein in vertebrates. Development. 1992;116:201–211. doi: 10.1242/dev.116.1.201. [DOI] [PubMed] [Google Scholar]
- 59.Mumm JS, Shou J, Calof AL. Colony-forming progenitors from mouse olfactory epithelium: evidence for feedback regulation of neuron production. Proc Natl Acad Sci USA. 1996;93:11167–11172. doi: 10.1073/pnas.93.20.11167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Murray RC, Tapscott SJ, Petersen JW, Calof AL, McCormick MB. A fragment of the Neurogenin1 gene confers regulated expression of a reporter gene in vitro and in vivo. Dev Dyn. 2000;218:189–194. doi: 10.1002/(SICI)1097-0177(200005)218:1<189::AID-DVDY16>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 61.Najbauer J, Leon M. Olfactory experience modulated apoptosis in the developing olfactory bulb. Brain Res. 1995;674:245–251. doi: 10.1016/0006-8993(94)01448-q. [DOI] [PubMed] [Google Scholar]
- 62.Nieto M, Schuurmans C, Britz O, Guillemot F. Neural bHLH genes control the neuronal versus glial fate decision in cortical progenitors. Neuron. 2001;29:401–413. doi: 10.1016/s0896-6273(01)00214-8. [DOI] [PubMed] [Google Scholar]
- 63.Nornes HO, Carry M. Neurogenesis in spinal cord of mouse: an autoradiographic analysis. Brain Res. 1978;159:1–6. doi: 10.1016/0006-8993(78)90105-1. [DOI] [PubMed] [Google Scholar]
- 64.Ono K, Tomasiewicz H, Magnuson T, Rutishauser U. N-CAM mutation inhibits tangential neuronal migration and is phenocopied by enzymatic removal of polysialic acid. Neuron. 1994;13:595–609. doi: 10.1016/0896-6273(94)90028-0. [DOI] [PubMed] [Google Scholar]
- 65.Petreanu L, Alvarez-Buylla A. Maturation and death of adult-born olfactory bulb granule neurons: role of olfaction. J Neurosci. 2002;22:6106–6113. doi: 10.1523/JNEUROSCI.22-14-06106.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pierce ET. Time of origin of neurons in the brain stem of the mouse. Prog Brain Res. 1973;40:53–65. doi: 10.1016/S0079-6123(08)60679-2. [DOI] [PubMed] [Google Scholar]
- 67.Ryals BM, Rubel EW. Hair cell regeneration after acoustic trauma in adult Coturnix quail. Science. 1988;240:1774–1776. doi: 10.1126/science.3381101. [DOI] [PubMed] [Google Scholar]
- 68.Smart IH. Location and orientation of mitotic figures in the developing mouse olfactory epithelium. J Anat. 1971;109:243–251. [PMC free article] [PubMed] [Google Scholar]
- 69.Sommer L, Ma Q, Anderson DJ. Neurogenins, a novel family of atonal-related bHLH transcription factors, are putative mammalian neuronal determination genes that reveal progenitor cell heterogeneity in the developing CNS and PNS. Mol Cell Neurosci. 1996;8:221–241. doi: 10.1006/mcne.1996.0060. [DOI] [PubMed] [Google Scholar]
- 70.Tomita K, Moriyoshi K, Nakanishi S, Guillemot F, Kageyama R. Mammalian achaete-scute and atonal homologs regulate neuronal versus glial fate determination in the central nervous system. EMBO J. 2000;19:5460–5472. doi: 10.1093/emboj/19.20.5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Torii M, Matsuzaki F, Osumi N, Kaibuchi K, Nakamura S, Casarosa S, Guillemot F, Nakafuku M. Transcription factors Mash-1 and Prox-1 delineate early steps in differentiation of neural stem cells in the developing central nervous system. Development. 1999;126:443–456. doi: 10.1242/dev.126.3.443. [DOI] [PubMed] [Google Scholar]
- 72.Tuttle R, Nakagawa Y, Johnson JE, O'Leary DD. Defects in thalamocortical axon pathfinding correlate with altered cell domains in Mash-1-deficient mice. Development. 1999;126:1903–1916. doi: 10.1242/dev.126.9.1903. [DOI] [PubMed] [Google Scholar]
- 73.Vannier B, Peyton M, Boulay G, Brown D, Qin N, Jiang M, Zhu X, Birnbaumer L. Mouse trp2, the homologue of the human trpc2 pseudogene, encodes mTrp2, a store depletion-activated capacitative Ca2+ entry channel. Proc Natl Acad Sci USA. 1999;96:2060–2064. doi: 10.1073/pnas.96.5.2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Weiler E, Farbman AI. Supporting cell proliferation in the olfactory epithelium decreases postnatally. Glia. 1998;22:315–328. doi: 10.1002/(sici)1098-1136(199804)22:4<315::aid-glia1>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 75.Weiler E, McCulloch MA, Farbman AI. Proliferation in the vomeronasal organ of the rat during postnatal development. Eur J Neurosci. 1999;11:700–711. doi: 10.1046/j.1460-9568.1999.00476.x. [DOI] [PubMed] [Google Scholar]
- 76.Wichterle H, Garcia-Verdugo JM, Herrera DG, Alvarez-Buylla A. Young neurons from medial ganglionic eminence disperse in adult and embryonic brain. Nat Neurosci. 1999;2:461–466. doi: 10.1038/8131. [DOI] [PubMed] [Google Scholar]
- 77.Wu H-H, Ivkovic S, Murray RC, Jaramillo S, Lyons KM, Johnson JE, Calof AL. Autoregulation of neurogenesis by GDF11. Neuron. 2003;37:197–207. doi: 10.1016/s0896-6273(02)01172-8. [DOI] [PubMed] [Google Scholar]