Abstract
Commissural neurons in the spinal cord project their axons through the floor plate using a number of molecular interactions, such as netrins and their receptor DCC (deleted in colorectal cancer). However, the molecular cascades that control differentiation of commissural neurons are less characterized. A homeobox gene,MBH1 (mammalian BarH1) was expressed specifically in a subset of dorsal cells in the developing spinal cord. Transgenic mice that carried lacZ andMBH1-flanking genome sequences demonstrated thatMBH1 was expressed by commissural neurons. To analyze the function of MBH1, we established an in vivo electroporation method for the transfer of DNA into the mouse spinal cord. Ectopic expression of MBH1 drove dorsal cells into the fate of commissural neurons with concomitant expression of TAG-1 (transiently expressed axonal surface glycoprotein 1) and DCC. Cells ectopically expressing MBH1 migrated to the deep dorsal horn, in which endogenousMBH1-positive cells accumulated. These results suggest that MBH1 functions upstream of TAG-1 and DCC and is involved in the fate determination of commissural neurons in the spinal cord.
Keywords: MBH1, homeobox, homeodomain, in vivo electroporation, TAG-1, DCC
Introduction
Commissural neurons in both vertebrates and invertebrates transfer information from one side of their bodies to the other through the midline. Molecular mechanisms regulating axon guidance of these neurons have been characterized extensively (Tessier-Lavigne and Goodman, 1996; Mueller, 1999;Kaprielian et al., 2001). Netrins and DCC (deleted in colorectal cancer) play a pivotal role in axon guidance. Some commissural neurons are generated in the developing dorsal spinal cord, in which domains of progenitor cells are specified by helix-loop-helix (HLH) transcription factors (Gowan et al., 2001). The domains are initially established by TGF-β-like signals (Liem et al., 1997) and produce several cell types, which are defined by combinatorial expression of homeobox genes (Lee and Jessell, 1999; Gross et al., 2002; Muller et al., 2002). The most dorsal cell type, dI1 (D1), is generated by an HLH factor, MATH1 (mouse atonal homolog 1). dI1 cells and a subset of commissural neurons are lost in MATH1 knock-out mice (Bermingham et al., 2001; Gowan et al., 2001), whereas ectopic expression ofMATH1 increases the number of dI1 cells (Gowan et al., 2001). However, the molecular cascades that form the link between the generation of the cells and their migration–axon guidance remain to be determined.
Bar-class homeobox (BarH) genes function in the development of various organs. Drosophila BarH genes control the development of the retina (Higashijima et al., 1992a) and peripheral nervous system (Higashijima et al., 1992b). A mammalianBarH gene, MBH1, is expressed at early stages of neurogenesis and is a potential regulator of neural HLH genes in the diencephalon (Saito et al., 1998). Outside of the diencephalon,MBH1 is expressed by postmitotic neurons in the midbrain, hindbrain, spinal cord, and retina (Saito et al., 1998, 2000). Another mammalian BarH gene, MBH2/Barhl1, is also expressed in the spinal cord (Bulfone et al., 2000; Saito et al., 2000) and suggested to be a downstream gene of MATH1 (Bermingham et al., 2001). Xenopus BarH genes, XBH1 andXBH2, which are orthologs of MBH1 andMBH2, respectively, show distinct expression patterns (Patterson et al., 2000). Expression patterns of MBH1 andMBH2 are similar but not identical (Saito et al., 2000), suggesting that expression of the two genes may be controlled by different mechanisms.
In this paper, we made transgenic mice carrying lacZ with the MBH1-flanking sequences and examined the cell types ofMBH1-expressing cells in the developing mouse spinal cord. The function of MBH1 has been revealed using the in vivo electroporation method.
Materials and Methods
Generation and analysis of transgenic mice. TheMBH1-flanking sequences were obtained by screening a 129/SvJ mouse genomic library (Stratagene, La Jolla, CA) using the entire sequence of the MBH1 cDNA (GenBank accession numberAB004056) as a probe. We made a construct that carried thelacZ-coding region from BGZA (Yee and Rigby, 1993; Helms et al., 2000) between the 1 kb 5′ and 2.5 kb 3′ sequences flanking theMBH1-coding region. BGZA was a gift from Dr. J. Johnson (University of Texas Southwestern Medical Center, Dallas, TX). Transgenic mice were generated by standard procedures (Hogan et al., 1986) using fertilized eggs from B6C3F1 (C57BL/6 × C3H) crosses. Staged transgenic embryos were dissected in cold PBS and fixed in 4% paraformaldehyde. Whole mount 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside staining of the embryos was performed as described previously (Verma-Kurvari et al., 1996). Other constructs that carried longerMBH1-flanking sequences also demonstrated the samelacZ expression pattern as the above construct, recapitulating endogenous MBH1 expression. After staining, the embryos were postfixed and embedded in paraffin. Microtome sections (7 μm) were stained with Nuclear Fast Red (Vector Laboratories, Burlingame, CA). For immunostaining, fixed embryos were embedded in OCT compound and sliced at 14 μm using a cryostat.
In vivo electroporation. Exo utero surgery and electroporation were performed as described previously (Saito and Nakatsuji, 2001). pCAG-EYFP (enhanced yellow fluorescent protein), which carried EYFP downstream of a CAG promoter (Saito and Nakatsuji, 2001), was used as a control. To express bothEYFP and MBH1 in the same cells, pEYFP-MBH1 was constructed by inserting the MBH1-coding region downstream of the second CAG promoter of pCAG-EYFP-CAG (Saito and Nakatsuji, 2001). One microliter of DNA solution (140 nm) in PBS was injected into the central canal of the spinal cord. Five electric pulses at 22 V were delivered to the spinal cord by holding embryos with forceps-type platinum electrodes. Two kinds of electrodes, half-ring type (see Fig. 2A) and round-plate type with a 3 mm diameter (http://www.frontier.kyoto-u.ac.jp/rc01/in_vivo_electroporation.html), were used for gene transfer into the whole and a part of the spinal cord, respectively. The electric pulses were obtained from an electroporator, CUY21EDIT (Nepa Gene, Ichikawa, Japan). Survival and EYFP-positive (EYFP+) rates, which were calculated from surviving embryos/operated and EYFP+ spinal cords/surviving embryos, were 56.7 ± 4.6 and 79.9 ± 2.7%, respectively. For functional analysis of genes, each result was confirmed by using another independently isolated clone with the same structure.
Fig. 2.
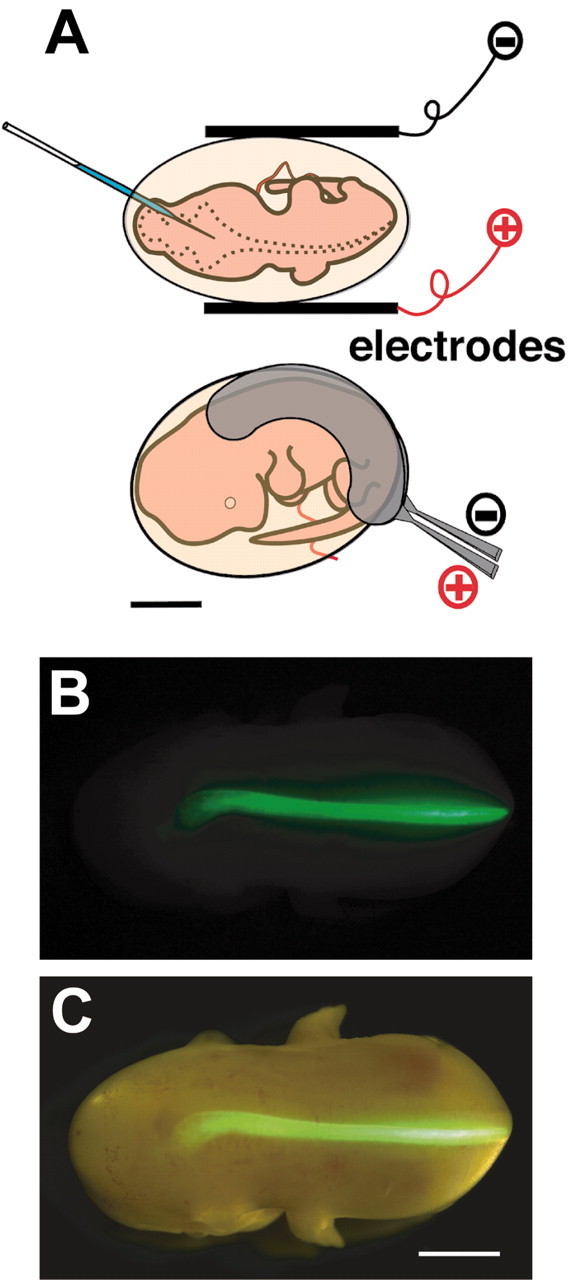
DNA transfer into the mouse spinal cord byin vivo electroporation. A, Schematic representation of DNA injection and electrodes. EYFP was introduced into the E11.5 spinal cord. B, Dorsal view of an embryo at E13.5, 2 d after electroporation. C, Semi-illuminated view of B. EYFP was expressed in only one side of the spinal cord closer to the anode. Scale bars: 2 mm.
In situ hybridization and immunohistochemistry. In situ hybridization was performed as described previously (Saito et al., 1996). Antisense RNA probes were synthesized from plasmids carrying mouse cDNA clones: pMH4–1 for MBH1 and generous gifts from Dr. T. Jessell (Columbia University, New York, NY) forLH2B (a LIM homeobox gene), Dr. R. Kageyama (Kyoto University, Kyoto, Japan) for MATH1, and Dr. Q. Ma (Harvard Medical School, Boston, MA) for Ngn2 (neurogenin 2). Frozen sections were incubated with the following primary antibodies: 4D7 [anti-TAG-1 (transiently expressed axonal surface glycoprotein 1)], AF5 (anti-DCC; Calbiochem, La Jolla, CA), 40.2D6 [anti-Isl1 (Islet-1)], 4F2 [anti-Lim1/2 (a homeodomain protein)], rabbit polyclonal L1 (anti-LH2A/B, gift from Dr. Jessell), goat anti-β-galactosidase (β-gal) (Biogenesis, Kingston, NH), and rabbit anti-GFP (Molecular Probes, Eugene, OR). 4D7, 40.2D6, and 4F2 were obtained from the Developmental Studies Hybridoma Bank (University of Iowa, Iowa City, IA). Signals were visualized by the following secondary antibodies: donkey anti-rabbit IgG, anti-mouse IgG, or anti-mouse IgM conjugated with Cy3 or FITC (Jackson ImmunoResearch, West Grove, PA); and donkey anti-goat IgG conjugated with Alexa Fluor 488 (Molecular Probes). Immunofluorescent studies were performed as described previously (Saito and Nakatsuji, 2001).
Results
MBH1 expression marked a subset of cells in the spinal cord
Expression of MBH1 was detected in the mouse dorsal spinal cord at embryonic day 10.5 (E10.5) (Fig.1A). At E11.5, the expression expanded ventrally to the deep dorsal horn (Fig.1C). Later than E12.5, the expression was mainly restricted to the deep dorsal horn (Fig. 1E). The pattern of the expression during development resembled that of ventral migration of some dorsal neurons (Leber and Sanes, 1995), suggesting thatMBH1 was expressed by these migrating neurons. A stream of cells between the deep dorsal horn and the floor plate also expressedMBH1 at E12.5 (Fig. 1E,arrows). MBH1 expression in the ventral spinal cord became confined to a group of cells dorsolateral to the floor plate at later stages (see Fig. 4B), suggesting thatMBH1+ cells in the stream are under ventromedial migration at E12.5.
Fig. 1.
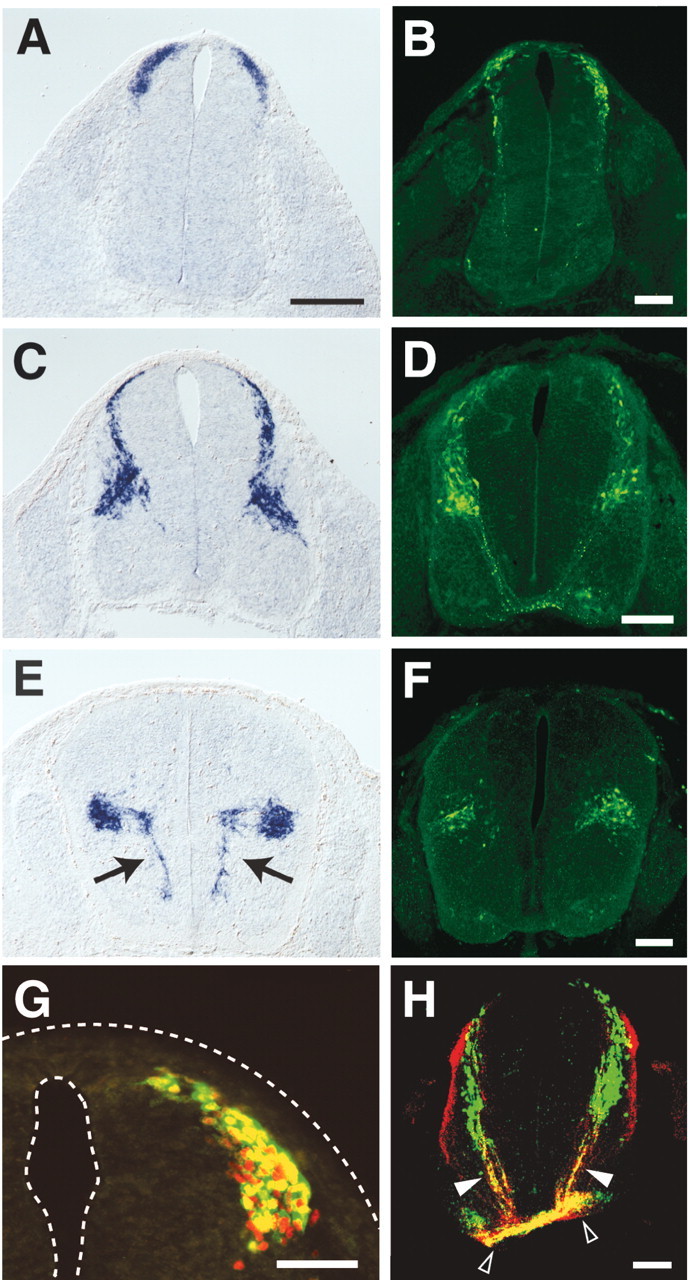
MBH1 expression in the developing mouse spinal cord. Transverse sections at brachial levels of embryos at E10.5 (A, B), E11.5 (C,D) and E12.5 (E, F) were hybridized with antisense cRNA probes for MBH1(A, C, E) and immunostained using an anti-β-gal antibody (B,D, F). Embryos carrying theMBH1/lacZ transgene were used (B, D, F–H).Arrows indicate streams ofMBH1+ cells between the deep dorsal horn and the floor plate. Control hybridization using a sense-strand probe of MBH1 gave no specific signals (data not shown). Similar expression patterns of MBH1 were observed from cervical to lumbar levels. G, Transverse section of the E10.5 spinal cord was stained with antibodies against the β-gal (green) and LH2A/B proteins (red). H, Double-label immunostaining with the anti-β-gal (green) and anti-TAG-1 (red) antibodies of the E11.5 spinal cord. Filled and open arrowheadsindicate β-gal+ axons and ventral funiculi, respectively. Scale bars: (in A) A,C, E, 200 μm; B,D, F–H, 100 μm.
Fig. 4.
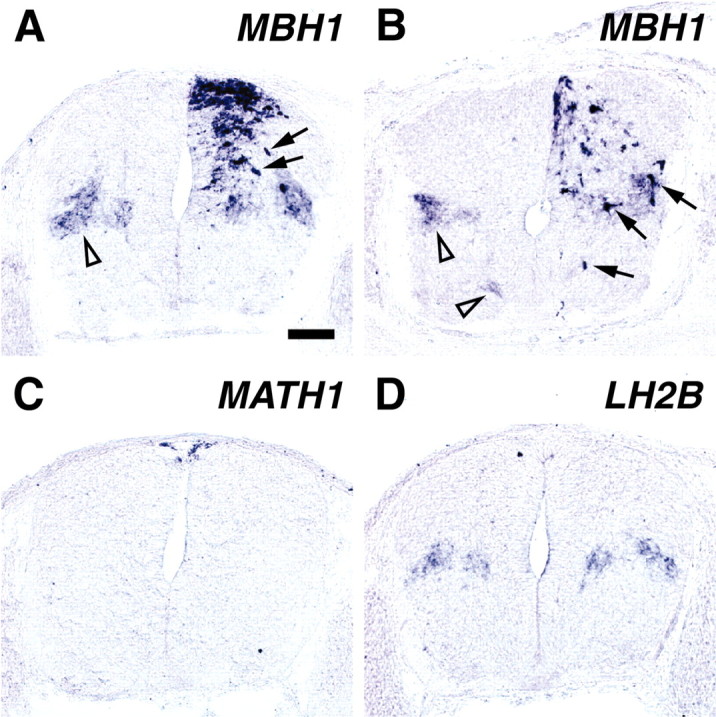
Comparison of MBH1-misexpressing cells with endogenous MBH1+ cells using in situ hybridization. After electroporation at E11.5, transverse sections of the mouse spinal cord were annealed with antisense cRNA probes of MBH1 (A,B), MATH1 (C), and LH2B (D). Right sides were transfected. Embryos were recovered at E12.5 (A,C, D) and at E13.5 (B). Arrowheads andarrows indicate endogenousMBH1+ domains andMBH1-misexpressing cells, respectively. Expression ofMATH1 and LH2B was not upregulated at E13.5 either (data not shown). Transfection with EYFPalone did not affect the expression of the genes (data not shown). Scale bar (in A): A–D, 100 μm.
Characterization of MBH1-expressing cells
To examine which types of cells expressed MBH1, transgenic mice with lacZ under the control of theMBH1-flanking DNA sequences were generated. The transgenic mice expressed the lacZ product β-gal in a pattern recapitulating endogenous MBH1 expression (Fig.1B,D,F). Coexpression of MBH1 and lacZ was confirmed by immunostaining using an anti-MBH1 antibody (data not shown). All β-gal+ cells were labeled with antibodies against the LH2A/B proteins (Fig. 1G), a marker of dI1 cells, but not with antibodies against the Isl1 and Lim1/2 proteins (data not shown), suggesting that MBH1 is expressed by dI1 cells.
Because the β-gal protein spreads throughout the cytoplasm, it enabled us to examine the morphologies ofMBH1+ cells. β-gal+ signals were detected in axons projecting to the floor plate and ventral funiculi (Fig.1H). The β-gal+ axons were labeled with specific markers of commissural neurons, anti-TAG-1 and anti-DCC antibodies (Fig. 1H; data not shown). These results indicate that MBH1 is expressed by commissural neurons.
In vivo electroporation into the spinal cord
To examine the function of MBH1, we established a system for the forced expression of a gene in the mouse spinal cord by modifying our in vivo electroporation method to the brain (Saito and Nakatsuji, 2001). The uterine wall was cut to see embryos clearly, and DNA was injected into the central canal of the spinal cord (Fig. 2). Then electric pulses were applied to the spinal cord using half-ring-type electrodes (Fig.2A). The electrodes helped better survival of embryos by limiting electric pulses mainly onto the spinal cord. After electroporation, a reporter gene, EYFP, was expressed only in one side of the spinal cord (Fig.2B,C).
Ectopic expression of MBH1
Using this system, pEYFP-MBH1, which carried both theEYFP and MBH1 genes downstream of ubiquitous CAG promoters, was introduced into the E11.5 mouse spinal cord. At this stage, DNA will be taken up by cells that are not fated to expressMBH1 for the following two reasons: (1) endogenousMBH1+ cells are away from the ventricle (Fig. 1C), and (2) DNA is transferred to cells adjacent to the ventricle by this method (Saito and Nakatsuji, 2001). DNA will be also introduced mainly into dorsal cells, because the central canal is wider in the dorsal side. Transfection withEYFP alone as a control mostly labeled only dorsal cells, as expected (Fig. 3A). In contrast, more ventral EYFP+ cells were generated by coexpression with MBH1 (Fig. 3B,arrows). The ventral EYFP+cells had morphologies similar to some commissural neurons (Silos-Santiago and Snider, 1992). Transfection of MBH1 also produced more EYFP+ commissural axons (Fig. 3B, arrowhead). The cells transfected withEYFP alone projected ascending EYFP+ axons ipsilaterally (Fig.3C, arrow) but not commissural axons (Fig.3E). In contrast, cells ectopically expressingMBH1 projected ascending commissural axons (Fig.3F, arrowhead). Coexpression of MBH1and EYFP in the same cells was confirmed by immunostaining using the anti-MBH1 antibody (data not shown). There were more TAG-1+ axons observed in theMBH1-transfected side (Fig. 3G). EYFP+ axons were labeled with the anti-TAG-1 antibody (Fig. 3H). Similarly, more DCC+ axons were generated by ectopic expression of MBH1, and the EYFP+ axons were also DCC+ (data not shown). These results indicate that the axons of the cells ectopically expressingMBH1 had acquired molecular identities as commissural axons and that ipsilaterally projecting dorsal neurons were transfated into commissural neurons by misexpression of MBH1.
Fig. 3.
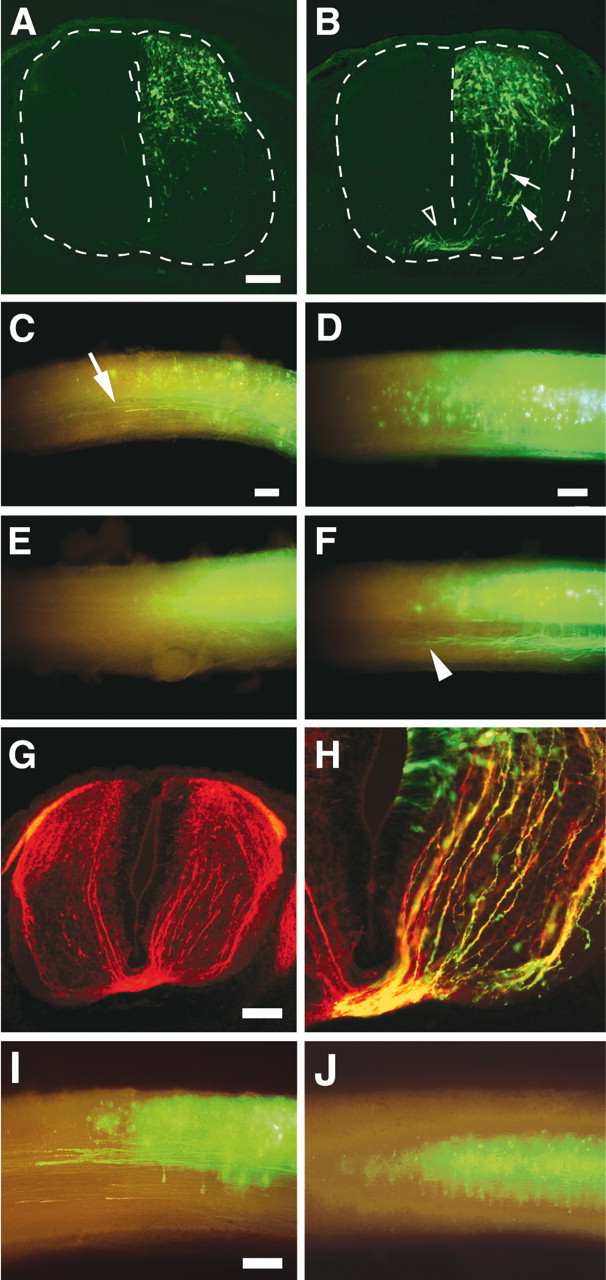
Generation of commissural neurons by ectopic expression of MBH1. E11.5 mouse spinal cords were transfected with EYFP alone (A,C, E, I) and bothEYFP and MBH1 (B,D, F–H, J). Right sides of sections were closer to the anode and transfected with these genes (A, B, G,H). A, B, Transverse sections of the spinal cord at brachial levels, 2 d after electroporation. Arrows andarrowhead indicate EYFP+ cells with morphologies similar to migratory commissural neurons and commissural axons, respectively. Similar patterns of EYFP+ cells and axons were observed through all axial levels of the spinal cord in all electroporated EYFP+ embryos (n = 15 for EYFP alone;n = 25 for coexpression of EYFP andMBH1). Lateral (C, D,I, J) and ventral (E, F) views of the spinal cord electroporated at lumbar levels, 3 (C–F) and 4 (I,J) d after electroporation. Rostral is to theleft. Arrow and arrowheadindicate EYFP+ ipsilaterally projecting and commissural axons, respectively. G, H, Transverse section of the E12.5 spinal cord, 1 d after transfection with EYFP and MBH1. The section was stained with antibodies against TAG-1 (red) and GFP (green). Misexpression of eitherLH2B, a LIM homeobox gene, or PHD1, a paired-like homeobox gene expressed in the dorsal spinal cord (Saito et al., 1996), did not generate more commissural axons. Scale bars: (inA) A, B, 100 μm; (inC) C, E, 200 μm; (inD) D, F, 200 μm;G, 100 μm; (in I)I, J, 200 μm.
Migration patterns of MBH1-misexpressing cells
Four days after electroporation, cells expressingEYFP alone remained in the dorsal spinal cord (Fig.3I). In contrast, most ofMBH1-misexpressing cells were observed in the middle of the spinal cord (Fig. 3J), suggesting thatMBH1-misexpressing cells may have migrated from the dorsal spinal cord. To compare MBH1-misexpressing cells with endogenous MBH1+ cells, in situ hybridization was performed (Fig.4). At E12.5, 1 d after electroporation, more cells expressing MBH1 were detected in the dorsal area of the MBH1-transfected side (Fig.4A). Expression levels of MBH1 were higher in the MBH1-misexpressing cells than those of endogenousMBH1, reflecting a strong activity of the CAG promoter. Those cells appeared to migrate toward the deep dorsal horn, whereas the endogenous MBH1+ cells had already settled in the deep dorsal horn at this stage (Fig.4A, arrowhead). Two days after electroporation, many MBH1-misexpressing cells settled down in the deep dorsal horn, in which the endogenousMBH1+ cells accumulated (Fig.4B). A minor population of theMBH1-misexpressing cells was detected in the ventral spinal cord as well as the endogenous MBH1+cells. These results suggest that the cells ectopically expressingMBH1 migrate to endogenousMBH1+ domains.
Next we examined whether the misexpression of MBH1 affects other genes. Expression of MATH1 and LH2B, which are related to the differentiation of commissural neurons, was not upregulated (Fig. 4C,D; data not shown). Furthermore, no increase of LH2A/B+ cells was detected using the anti-LH2A/B antibody (data not shown). These findings suggest that MATH1 and LH2A/B are not downstream of MBH1.
Discussion
Commissural neuron differentiation by MBH1
The results obtained by the ectopic expression of MBH1suggest that MBH1 regulates at least three aspects of the differentiation of commissural neurons. Expression of TAG-1 and DCC, which are markers of commissural neurons, were induced byMBH1. Because DCC is a receptor of netrins, the ectopic expression of DCC may be responsible for axon projection of theMBH1-misexpressing cells to the floor plate. Their axons elongated along the floor plate after crossing it, as do endogenous commissural neurons. The MBH1-misexpressing cells appeared to migrate to the endogenous MBH1+domains. These findings suggest that several genes involved in the differentiation of commissural neurons are regulated downstream ofMBH1.
There are several types of commissural neurons at various dorsoventral domains in the spinal cord. Only two domains wereMBH1+, showing that MBH1is expressed by a subset of commissural neurons. The expression ofMBH1 at E10.5, which was restricted to the dorsal edge of the spinal cord, was similar to that of MATH1, but the expression of MATH1 was limited to the ventricular zone and detected at E9.5 (Helms and Johnson, 1998; data not shown), earlier than that of MBH1. All β-gal+cells of the MBH1/lacZ transgenic embryos expressed LH2A/B, which is a marker of dI1 cells and expressed downstream of MATH1. These results indicate that MBH1 is expressed in a lineage of cells that have expressed MATH1. All LH2A/B+ cells, however, appeared not to be β-gal+, suggesting that MBH1is expressed in a subset of LH2A/B+ cells. The expression patterns of MBH1 were similar to those ofLH2B in the spinal cord (Fig. 4; data not shown), suggesting that MBH1 may be expressed in the same lineage of cells that express LH2B.
Misexpression of MBH1 generated more commissural neurons without induction of LH2A/B, suggesting that LH2A/B may not exert the same function as MBH1 in the differentiation of commissural neurons. This was confirmed by ectopic expression of LH2B in the spinal cord, which did not produce more commissural neurons (data not shown). On the other hand, misexpression of MATH1 generated more commissural neurons (data not shown), suggesting thatMATH1 is upstream of MBH1. At E11.5, the domain of MBH1 expression closely resembled the β-gal+ domain of transgenic mice that carried lacZ under the control of MATH1-flanking sequences (Helms and Johnson, 1998). Moreover, the 3′MBH1-flanking sequence used for the transgenic mice in this study contained an E-box (CAGCTG), which could bind the MATH1 protein (Akazawa et al., 1995; Helms et al., 2000). These findings suggest thatMBH1 may be a downstream target of the MATH1 protein.
A recent report has shown that excess commissural neurons were generated in Lbx1 (a Ladybird-like homeobox gene 1) mutant mice because of mis-specification of dorsal interneurons (Gross et al., 2002). This is similar to our results from the misexpression of MBH1. However, expression of Isl1 and Lim1/2, which are affected in the mutant mice, were not perturbed by the misexpression of MBH1(data not shown). This result suggests that MBH1 generates ectopic commissural neurons independently of a transcriptional cascade exerted in the Lbx1 mutant mice.
Regulation of cell migration by MBH1
The transgenic mice carrying lacZ with theMBH1-flanking sequences visualizedMBH1+ cells. At E10.5 and E11.5, the stages when the MBH1+ cells were located between the dorsal edge and the deep dorsal horn in the spinal cord, they showed morphologies typical of some migratory neurons (unipolar with leading processes) (Leber and Sanes, 1995). Together with expression patterns of MBH1, this suggests thatMBH1 is expressed during the migration of commissural neurons. The endogenous MBH1+ cells migrated to the deep dorsal horn along the marginal zone of the spinal cord. In contrast, MBH1-misexpressing cells appeared not to follow the same route as the endogenousMBH1+ cells but rather to take a direct shortcut route from their birthplaces in the ventricular zone to the deep dorsal horn. These observations suggest that MBH1may instruct the cells where to migrate, responding to an extracellular factor in the spinal cord. The factor may be released from the deep dorsal horn to attract the cells or may exclude the cells from the dorsolateral region of the spinal cord.
MBH1 was also expressed by granule cells during their migration in the developing cerebellum (Saito et al., 2000).MATH1, TAG-1, and DCC are all expressed in the developing cerebellum as well (Yamamoto et al., 1990; Akazawa et al., 1995;Livesey and Hunt, 1997), suggesting that there is a common cascade of genes between commissural neurons and the cerebellum.
Various functions of BarH genes
Some commissural neurons are generated downstream ofNgn2 (Simmons et al., 2001). We showed that forced expression of MBH1 upregulates Ngn2 in P19 cells, reflecting expression patterns of the two genes in the developing diencephalon (Saito et al., 1998). Ngn2 was not activated by ectopic expression of MBH1 in the developing spinal cord (data not shown). MBH1 requires another unknown factor that is transiently expressed in P19 cells to upregulateNgn2 (Saito et al., 1998). The factor may have been absent in the spinal cord at the stage when MBH1 was ectopically expressed. MBH1 was expressed in mitotically active cells in the ventricular zone of the diencephalon, whereas postmitotic cells expressed MBH1 in the spinal cord. MBH1 may have different functions at different stages of neurogenesis. Similarly,Drosophila BarH genes show various functions (Higashijima et al., 1992a,b).
In vivo electroporation in mouse
Both gain- and loss-of-function analyses are essential to establish gene function. Gene knock-out techniques have enabled the loss-of-function analysis of many genes in mouse. On the other hand, gain-of-function approaches have been used extensively in chick. The genes and gene combinations that regulate some stages of development are not exactly the same between chick and mouse. This report demonstrates that the in vivo electroporation technique is a powerful tool to reveal gene function in the mouse. This technique will greatly facilitate functional analyses of genes, because it may also be applied to knock-out and transgenic mice.
Footnotes
This work was supported in part by a grant from the Japan Society for the Promotion of Science and Grants-in-Aids for Scientific Research on Priority Areas–Neural Net Project and –Advanced Brain Science Project from Ministry of Education, Culture, Sports, Science, and Technology of Japan (T.S.). We thank Masuko Tanaka and Junko Kutsuna for their technical assistance and Dr. Takayuki Sakurai for his kind advice for generating transgenic mice. We are grateful to Drs. Thomas Jessell, Jane Johnson, Ryoichiro Kageyama, and Qiufu Ma for antibodies and plasmids. We also acknowledge the Developmental Studies Hybridoma Bank maintained by the University of Iowa (Iowa City, IA) for supply of monoclonal antibodies.
Correspondence should be addressed to Tetsuichiro Saito, Department of Development and Differentiation, Institute for Frontier Medical Sciences, Kyoto University, Shogoin, Sakyo-ku, Kyoto 606-8507. E-mail:tesaito@frontier.kyoto-u.ac.jp.
References
- 1.Akazawa C, Ishibashi M, Shimizu C, Nakanishi S, Kageyama R. A mammalian helix-loop-helix factor structurally related to the product of Drosophila proneural gene atonal is a positive transcriptional regulator expressed in the developing nervous system. J Biol Chem. 1995;270:8730–8733. doi: 10.1074/jbc.270.15.8730. [DOI] [PubMed] [Google Scholar]
- 2.Bermingham NA, Hassan BA, Wang VY, Fernandez M, Banfi S, Bellen HJ, Fritzsch B, Zoghbi HY. Proprioceptor pathway development is dependent on MATH1. Neuron. 2001;30:411–422. doi: 10.1016/s0896-6273(01)00305-1. [DOI] [PubMed] [Google Scholar]
- 3.Bulfone A, Menguzzato E, Broccoli V, Marchitiello A, Gattuso C, Mariani M, Consalez GG, Martinez S, Ballabio A, Banfi S. Barhl1, a gene belonging to a new subfamily of mammalian homeobox genes, is expressed in migrating neurons of the CNS. Hum Mol Genet. 2000;9:1443–1452. doi: 10.1093/hmg/9.9.1443. [DOI] [PubMed] [Google Scholar]
- 4.Gowan K, Helms AW, Hunsaker TL, Collisson T, Ebert PJ, Odom R, Johnson JE. Crossinhibitory activities of Ngn1 and Math1 allow specification of distinct dorsal interneurons. Neuron. 2001;31:219–232. doi: 10.1016/s0896-6273(01)00367-1. [DOI] [PubMed] [Google Scholar]
- 5.Gross MK, Dottori M, Goulding M. Lbx1 specifies somatosensory association interneurons in the dorsal spinal cord. Neuron. 2002;34:535–549. doi: 10.1016/s0896-6273(02)00690-6. [DOI] [PubMed] [Google Scholar]
- 6.Helms AW, Johnson JE. Progenitors of dorsal commissural interneurons are defined by MATH1 expression. Development. 1998;125:919–928. doi: 10.1242/dev.125.5.919. [DOI] [PubMed] [Google Scholar]
- 7.Helms AW, Abney AL, Ben-Arie N, Zoghbi HY, Johnson JE. Autoregulation and multiple enhancers control Math1 expression in the developing nervous system. Development. 2000;127:1185–1196. doi: 10.1242/dev.127.6.1185. [DOI] [PubMed] [Google Scholar]
- 8.Higashijima S, Kojima T, Michiue T, Ishimaru S, Emori Y, Saigo K. Dual Bar homeo box genes of Drosophila required in two photoreceptor cells, R1 and R6, and primary pigment cells for normal eye development. Genes Dev. 1992a;6:50–60. doi: 10.1101/gad.6.1.50. [DOI] [PubMed] [Google Scholar]
- 9.Higashijima S, Michiue T, Emori Y, Saigo K. Subtype determination of Drosophila embryonic external sensory organs by redundant homeo box genes BarH1 and BarH2. Genes Dev. 1992b;6:1005–1018. doi: 10.1101/gad.6.6.1005. [DOI] [PubMed] [Google Scholar]
- 10.Hogan B, Constantini F, Lacy E. Manipulating the mouse embryo: a laboratory manual, pp 79–203. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1986. [Google Scholar]
- 11.Kaprielian Z, Runko E, Imondi R. Axon guidance at the midline choice point. Dev Dyn. 2001;221:154–181. doi: 10.1002/dvdy.1143. [DOI] [PubMed] [Google Scholar]
- 12.Leber SM, Sanes JR. Migratory paths of neurons and glia in embryonic chick spinal cord. J Neurosci. 1995;15:1236–1248. doi: 10.1523/JNEUROSCI.15-02-01236.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee KJ, Jessell TM. The specification of dorsal cell fates in the vertebrate central nervous system. Annu Rev Neurosci. 1999;22:261–294. doi: 10.1146/annurev.neuro.22.1.261. [DOI] [PubMed] [Google Scholar]
- 14.Liem KF, Jr, Tremml G, Jessell TM. A role for the roof plate and its resident TGFβ-related proteins in neuronal patterning in the dorsal spinal cord. Cell. 1997;91:127–138. doi: 10.1016/s0092-8674(01)80015-5. [DOI] [PubMed] [Google Scholar]
- 15.Livesey FJ, Hunt SP. Netrin and netrin receptor expression in the embryonic mammalian nervous system suggests roles in retinal, striatal, nigral, and cerebellar development. Mol Cell Neurosci. 1997;8:417–429. doi: 10.1006/mcne.1997.0598. [DOI] [PubMed] [Google Scholar]
- 16.Mueller BK. Growth cone guidance: first steps towards a deeper understanding. Annu Rev Neurosci. 1999;22:351–388. doi: 10.1146/annurev.neuro.22.1.351. [DOI] [PubMed] [Google Scholar]
- 17.Muller T, Brohmann H, Pierani A, Heppenstall PA, Lewin GR, Jessell TM, Birchmeier C. The homeodomain factor Lbx1 distinguishes two major programs of neuronal differentiation in the dorsal spinal cord. Neuron. 2002;34:551–562. doi: 10.1016/s0896-6273(02)00689-x. [DOI] [PubMed] [Google Scholar]
- 18.Patterson KD, Cleaver O, Gerber WV, White FG, Krieg PA. Distinct expression patterns for two Xenopus Bar homeobox genes. Dev Genes Evol. 2000;210:140–144. doi: 10.1007/s004270050020. [DOI] [PubMed] [Google Scholar]
- 19.Saito T, Nakatsuji N. Efficient gene transfer into the embryonic mouse brain using in vivo electroporation. Dev Biol. 2001;240:237–246. doi: 10.1006/dbio.2001.0439. [DOI] [PubMed] [Google Scholar]
- 20.Saito T, Lo L, Anderson DJ, Mikoshiba K. Identification of novel paired homeodomain protein related to C. elegans unc-4 as a potential downstream target of MASH1. Dev Biol. 1996;180:143–155. doi: 10.1006/dbio.1996.0291. [DOI] [PubMed] [Google Scholar]
- 21.Saito T, Sawamoto K, Okano H, Anderson DJ, Mikoshiba K. Mammalian BarH homologue is a potential regulator of neural bHLH genes. Dev Biol. 1998;199:216–225. doi: 10.1006/dbio.1998.8889. [DOI] [PubMed] [Google Scholar]
- 22.Saito T, Hama T, Saba R, Nakatsuji N. Mammalian Bar-class homeobox genes. Soc Neurosci Abstr. 2000;26:600.21. [Google Scholar]
- 23.Silos-Santiago I, Snider WD. Development of commissural neurons in the embryonic rat spinal cord. J Comp Neurol. 1992;325:514–526. doi: 10.1002/cne.903250405. [DOI] [PubMed] [Google Scholar]
- 24.Simmons AD, Horton S, Abney AL, Johnson JE. Neurogenin2 expression in ventral and dorsal spinal neural tube progenitor cells is regulated by distinct enhancers. Dev Biol. 2001;229:327–339. doi: 10.1006/dbio.2000.9984. [DOI] [PubMed] [Google Scholar]
- 25.Tessier-Lavigne M, Goodman CS. The molecular biology of axon guidance. Science. 1996;274:1123–1133. doi: 10.1126/science.274.5290.1123. [DOI] [PubMed] [Google Scholar]
- 26.Verma-Kurvari S, Savage T, Gowan K, Johnson JE. Lineage-specific regulation of the neural differentiation gene. MASH1. Dev Biol. 1996;180:605–617. doi: 10.1006/dbio.1996.0332. [DOI] [PubMed] [Google Scholar]
- 27.Yamamoto M, Hassinger L, Crandall JE. Ultrastructural localization of stage-specific neurite-associated proteins in the developing rat cerebral and cerebellar cortices. J Neurocytol. 1990;19:619–627. doi: 10.1007/BF01188031. [DOI] [PubMed] [Google Scholar]
- 28.Yee SP, Rigby PWJ. The regulation of myogenin gene expression during the embryonic development of the mouse. Genes Dev. 1993;7:1277–1289. doi: 10.1101/gad.7.7a.1277. [DOI] [PubMed] [Google Scholar]


