Abstract
The TUC (TOAD-64/Ulip/CRMP) proteins are homologs of UNC-33, a protein that is required for axon extension and guidance inCaenorhabditis elegans. The TUC proteins are expressed in newly born neurons in the developing nervous system and have been implicated in semaphorin signaling and neuronal polarity. Here, we identify several new variants of the TUC family, each of which is expressed during distinct periods of neural development. We cloned and characterized TUC-4b, a variant of TUC-4a that includes a unique N-terminal extension. The functional relevance of this N-terminal domain is demonstrated by the finding that overexpression of TUC-4b, but not TUC-4a, results in increased neurite length and branching. Furthermore, whereas TUC-4a is expressed throughout life, TUC-4b is expressed exclusively during embryonic development. TUC-4b is localized to SV2 (synaptic vesicle protein 2)-positive vesicles in the central domain of the growth cone, suggesting a potential role in growth cone vesicle transport. Furthermore, TUC-4b interacts with the SH3A (Src homology 3A) domain of intersectin, an endocytic–exocytic adaptor protein. Together, these data suggest that TUC-4b can regulate neurite extension and branching through a mechanism that may involve membrane transport in the growth cone.
Keywords: growth cones, neurite outgrowth, growth cone vesicles, intersectin, CRMP, Ulip, TUC, unc-33
Introduction
The TUC (TOAD-64/Ulip/CRMP) family of proteins has been implicated in axon guidance and outgrowth. The members of this family have been identified previously as TOAD-64 (turned on after division, 64 kDa), Ulip (UNC-33 like protein), CRMP (collapsin response mediator protein), and DRP (dihydropyrimidinase related protein) (Table1) (Quinn et al., 1999). Four isoforms (TUC-1 through TUC-4) that share 70–80% amino acid identity currently comprise the TUC family (Wang and Strittmatter, 1997; Byk et al., 1998;Quinn et al., 1999). TUC-4 (Ulip-1) was identified as a protein that is regulated during neuronal differentiation in the cerebral cortex and in neuronal cell lines in response to NGF or retinoic acid (Minturn et al., 1995b; Byk et al., 1996; Gaetano et al., 1997). TUC-2 (CRMP-62) was identified in an expression screen for proteins that mediate the response to semaphorin-3A (Goshima et al., 1995). Two additional members of this family, TUC-1 and TUC-3, were identified by homology to TUC-2 and TUC-4 (Wang and Strittmatter, 1996). Indirect evidence for a role of the TUCs in axon outgrowth comes from the observation that TUC-4 is expressed in growth cones and is upregulated in motor neurons during axonal regeneration after sciatic nerve lesion (Minturn et al., 1995a). A role for TUC-2 in axon guidance is suggested by the observation that antibodies to CRMP-62 (TUC-2) can inhibit semaphorin-3A-mediated growth cone collapse (Goshima et al., 1995). TUC-2 may also function in the development of neuronal polarity, because its overexpression can lead to the formation of extra axons in cultured hippocampal neurons (Inagaki et al., 2001).
Table 1.
TUC nomenclature
| TUC-1 | TUC-2 | TUC-3 | TUC-4 |
|---|---|---|---|
| CRMP-1 | CRMP-2 | CRMP-3 | CRMP-4 |
| Ulip-3 | CRMP-62 | Ulip-4 | Ulip-1 |
| DRP-1 | Ulip-2 | DRP-3 | |
| DRP-2 | |||
| TOAD-64 |
The TUC (TOAD/Ulip/CRMP) nomenclature has been proposed to unify discussion of this family.
The TUCs are homologous to a protein encoded by unc-33, a gene required for normal axon guidance in Caenorhabditis elegans. In unc-33 mutants, some axons show errors in their trajectories, whereas others follow the correct trajectory but terminate before reaching their normal targets, suggesting thatunc-33 functions in both guidance and outgrowth (Hedgecock et al., 1985; Siddiqui and Culotti, 1991). The gene unc-33encodes three transcripts that are translated into proteins of 55, 72, and 90 kDa. The four original members of the TUC family correspond to the transcript that encodes the 55 kDa isoform of UNC-33. Analysis of the mn260 allele of unc-33 has indicated that the larger transcripts are necessary for the function of the unc-33gene. In mn260, the transcripts that encode the 72 and 90 kDa isoforms of UNC-33 are disrupted by a frame-shift mutation, leaving the transcript that encodes the 55 kDa isoform intact. The phenotype of animals that carry mn260 alleles is only slightly weaker than in animals with null alleles, suggesting that the transcript that encodes the 55 kDa isoform of UNC-33 has little function in the absence of the larger transcripts and that the key functions of the UNC-33 are mediated by the larger isoforms (Li et al., 1992).
The observation that the 72 and 90 kDa isoforms of UNC-33 are essential for the function of the unc-33 gene has led us to ask whether larger transcripts of the TUC family might also exist. Here, we demonstrate the presence of 75 kDa isoforms of TUC-1, TUC-2, and TUC-4. We cloned and characterized one of these isoforms, TUC-4b. TUC-4b is enriched in growth cones, in which it colocalizes with SV2 (synaptic vesicle protein 2)-positive vesicles in the central region of the growth cone. We further show that TUC-4b binds to the SH3A (Src homology 3A) domain of intersectin, a multifunctional adaptor protein that plays a role in membrane transport and neurite outgrowth. Finally, we show that overexpression of TUC-4b, but not TUC-4a, results in increased neurite extension and branching.
Materials and Methods
Antibodies. Antibodies to Rab5 and SV2 were obtained from Transduction Laboratories (Lexington, KY) and the Developmental Studies Hybridoma Bank (University of Iowa, Iowa City, IA), respectively. The antibody to TUC-4 has been described previously as antibody-25 (Minturn et al., 1995a). The antibody to TUC-2 was a gift from Dr. Yasuo Ihara (University of Tokyo, Tokyo, Japan) and has been described previously as C4G (Gu and Ihara, 2000). Polyclonal antibodies to TUC-4b, TUC-1, and TUC-3 were produced at Zymed (San Francisco, CA) by immunizing rabbits with the following peptides: TUC4b, (C)RPGTTDQVPRQKYG; TUC-1, (CGGGGG)NTYLQKPSQ; and TUC-3, (C)PRWHESTKE. Note that residues between parentheses are not part of the TUC sequences but were added for stability or to allow coupling to the KLH carrier. The polyclonal pan-TUC antibody was prepared at Pocono Rabbit Farm & Laboratory (Canadensis, PA) by immunizing rabbits with the thyroglobulin-conjugated peptide: IVNDDQSFYADIYMEDGLIKQIG. Each polyclonal antibody was affinity purified with its respective peptide at Zymed.
cDNA constructs. The full-length TUC-4b clone was generated by reverse transcription (RT)-PCR on RNA prepared from embryonic day 18 (E18) rat brain. The PCR was performed using Pfu polymerase (Stratagene, La Jolla, CA) and the following primers: GCCGCTGTCGCTTGAACC and GAGGGCTTAACTCAGGGATGTG. Single nucleotide overhangs were added to the blunt PCR product by incubation withTaq polymerase, and the resulting product was ligated into the pcDNA3.1/V5His TOPO vector. Note that a stop codon was included in the PCR primer, such that the V5/His tag was not used. The sequence of the TUC-4b insert was confirmed by DNA sequencing. Clones for TUC-1a, TUC-2a, TUC-3a, and TUC-4a were PCR amplified from a neonatal rat hippocampus cDNA library and subcloned into the pcDNA3.1/V5/His vector. The preparation of cDNAs encoding the SH3 domains of intersectin has been described previously (Yamabhai et al., 1998).
Western blot analysis of brains. Brains were dissected from Sprague Dawley rats at the following ages: E12, E15, E18, postnatal day 1 (P1), P7, P14, P21, and adult. Brains were homogenized in 10 mm HEPES with Cθmplete protease inhibitors (Roche Products, Hertforshire, UK). Triton X-100 was added to 1% final concentration, the samples were incubated at 4°C for 20 min and centrifuged at 12,000 × g for 45 min, and the supernatants were prepared for Western blot analysis.
Glutathione S-transferase binding assay. Fusion proteins between each SH3 domain and glutathioneS-transferase (GST) were prepared as described previously (Yamabhai et al., 1998). Binding assays with E18 brain were conducted as described previously (Yamabhai et al., 1998). For binding assays with transfected cell lysates, HEK293 cells were transfected with Lipofectamine (Invitrogen, Gaithersburg, MD) and DNA encoding either TUC-4a or TUC-4b. Twenty four hours after transfection, the cells were lysed in 10 mm HEPES with Cθmplete protease inhibitors (Roche Products). Triton X-100 was added to 1% final concentration, and the samples were rocked at 4°C for 20 min and centrifuged at 12,000 ×g for 45 min. Transfected cell lysate was diluted 1:10 with lysate from untransfected HEK293 cells. One milligram of the diluted lysate was incubated with GST-SH3A immobilized on glutathione-Sepharose (Amersham Biosciences, Arlington Heights, IL) at 4°C for 4 hr. Afterward, the Sepharose was washed three times with 10 mm HEPES with 1% Triton X-100. The bound proteins were eluted by boiling in loading buffer and prepared for Western blot analysis.
Immunofluorescence. Dorsal root ganglia (DRGs) have large growth cones that are amenable to immunocytochemical analyses of subcellular structure. DRGs were dissected from the lumbar enlargement of E8 chick embryos. DRGs were placed on a laminin-coated coverslip and grown for 12–16 hr in F-12 media with 10% FBS and 5 ng/ml 7S NGF. Cultures were then fixed with 3.7% paraformaldehyde (PFA)–sucrose for 30 min at room temperature and permeabilized with 0.2% Triton X-100 for 3 min. Each coverslip was incubated with the appropriate primary antibodies, followed by the secondary antibodies, and then mounted on glass slides for observation with a Nikon (Tokyo, Japan) PCM 2000 confocal microscope.
Neurite outgrowth assays. Cortical neurons can be readily transfected with foreign genes. E18 cortical cultures from Sprague Dawley rats were dissociated as described previously (Threadgill et al., 1997). After dissociation, cortical cells were plated on poly-l-lysine- and laminin-coated glass coverslips at 100,000 cells per coverslip. The culture media consisted of the following: neurobasal media (Invitrogen), 5% FBS (Hyclone, Logan, UT), B27 supplement (Invitrogen), penicillin–streptomycin, l-glutamine, and sodium pyruvate. After incubation for 24 hr, the cultures were transfected with a modified calcium phosphate technique (Threadgill et al., 1997). For each coverslip, 1 μg of DNA encoding green fluorescent protein (GFP) was combined with 2 μg of DNA encoding TUC-4, TUC-4b, or the empty PRK5 vector. Cells were incubated for an additional 48 hr after transfection, fixed with PFA–sucrose for 30 min at room temperature, and processed for immunofluorescence to GFP. The transfection efficiency varied between 2 and 5%. Immunostaining of cultures with an antibody to a region common to TUC-4a and TUC-4b revealed that each of these proteins was substantially overexpressed in transfected (GFP-positive) cells. Images of transfected neurons were then analyzed to measure the length of the longest neurite for each neuron, as well as the number of branch points contained by each neuron. Digital camera lucida drawings were prepared using a SPOT 2 digital camera (Diagnostic Instruments, Sterling Heights, MI) and Adobe Photoshop 5 (Adobe Systems, San Jose, CA).
Results
Identification of TUC protein variants
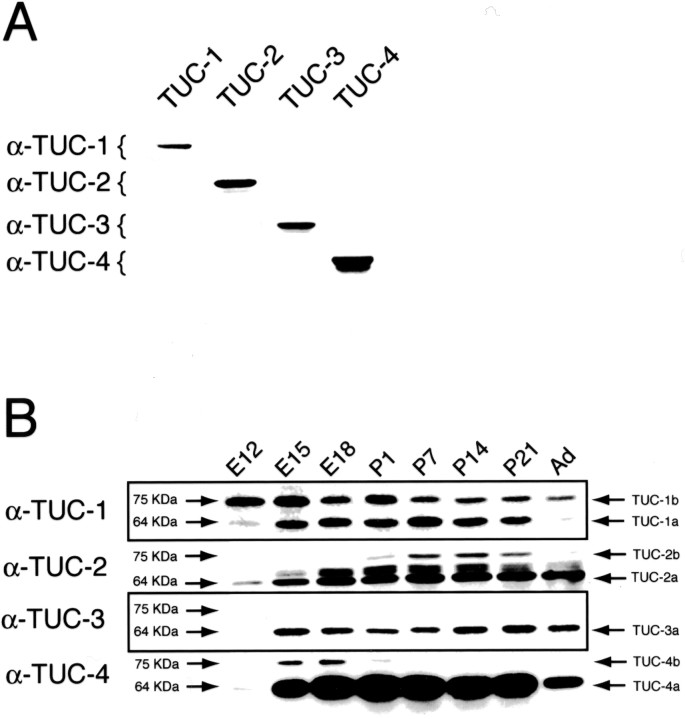
Previous studies have identified cDNAs encoding four isoforms of the TUC family (TUC-1, TUC-2, TUC-3, and TUC-4), each of which encode a protein with a predicted molecular mass of 64 kDa (Wang and Strittmatter, 1996). To identify variants of these proteins, we developed a panel of antibodies specific for each TUC isoform. The specificity of the antibodies was demonstrated by immunoblotting lysates from HEK293 cells transfected with a cDNA for each of the TUC isoforms. Each antibody recognized its target isoform but did not recognize any of the other three isoforms (Fig.1A).
Fig. 1.
Isoform-specific TUC antibodies recognize 64 and 75 kDa proteins. A, Each TUC isoform-specific antibody was tested by immunoblotting lysates from HEK293 cells transfected with cDNA encoding each of the TUC proteins. Each antibody recognized its target isoform and did not cross react with the other isoforms.B, Brain homogenates from rats from E12 to adult (Ad) were immunoblotted with each TUC isoform-specific antibody. Whereas the antibody to TUC-3 recognized only a single 64 kDa protein, each of the other antibodies recognized proteins at 64 and 75 kDa. The antibody to TUC-2 also recognized two proteins at ∼68 kDa.
We next used the isoform-specific antibodies to determine whether any of the four isoforms might have additional variants. Because previous studies have shown that the TUC proteins are developmentally regulated (Minturn et al., 1995a,b; Byk et al., 1996, 1998; Wang and Strittmatter, 1996; Gaetano et al., 1997; Kamata et al., 1998), rat brain lysates from animals at a range of developmental stages were examined (Fig. 1B). The antibody to TUC-3 detected only one protein at 64 kDa, whereas the antibodies to TUC-1, TUC-2, and TUC-4 detected more than a single protein (Fig. 1B). The TUC-1 and TUC-4 antibodies recognized the predicted 64 kDa proteins and, in addition, 75 kDa proteins. The TUC-2 antibody recognized 64 and 75 kDa proteins and detected a protein doublet of ∼68 kDa. These studies suggest that the 75 kDa isoforms are variants of the 64 kDa proteins. We named the 75 kDa forms TUC-1b, TUC-2b, and TUC-4b. We propose that each of the 64 kDa forms, which correspond to the previously cloned TUC cDNAs, be called TUC-1a, TUC-2a, TUC-3a, and TUC-4a (Fig. 1B). This experiment also permitted us to determine the time course of expression of the immunoreactive proteins. Each isoform showed a unique pattern of developmental regulation (Fig. 1B). TUC-1b was detected earlier than the other isoforms, at rat E12, an age at which neurogenesis is just beginning in the brain. Whereas both forms of TUC-1 and TUC-4, as well as TUC-2b, were downregulated in the adult, the TUC-3a and TUC-2a proteins continued to be expressed at high levels in the adult brain. TUC-2b and TUC-4b exhibited the most restricted developmental expression. TUC-4b reached peak levels between E15 and E18, was barely detectable by P1, and was below the limit of detection at later ages. TUC-2b was only weakly expressed at P1, reached its peak expression level between P7 and P14, and declined to a low level by P21.
Cloning of TUC-4b
To determine whether the 75 kDa forms of the TUC proteins were the products of alternate transcripts, we searched the expressed sequence tag (EST) database for cDNAs that matched the TUC cDNAs and also contained additional sequence. We identified a series of overlapping ESTs from embryonic rat libraries that could be assembled into a single putative cDNA with an open reading frame that included a 3′ region that was identical to TUC-4a and a unique 5′ region. The expression of this transcript in rat brain was confirmed by RT-PCR on RNA from E18 rat brain, using primers designed to amplify the full translated region of the transcript (data not shown). As described above, we named this new isoform TUC-4b. The nucleotide sequence of TUC-4b (Fig.2A) predicts a protein of 683 aminoacids with a molecular mass of 74.4 kDa. The first 126 amino acids comprise a unique N-terminal extension, whereas amino acids 127–683 are identical to amino acids 14–570 of TUC-4a (Fig. 2B). Amino acids 1–13 of TUC-4a, which are absent from TUC-4b, correspond to the translated region of TUC-4a exon 1 (Matsuo et al., 2000), indicating that TUC-4b arises from the use of an alternate promoter. To confirm that the predicted TUC-4b protein is expressed in developing rat brain, we generated an antibody to a peptide in the N-terminal, TUC-4b-specific part of the protein (Fig. 2B). The TUC-4b antibody recognized a 75 kDa protein in E18 rat brain lysate that comigrated with the 75 kDa protein recognized by the antibody to TUC-4 (Fig. 2C). As expected, the TUC-4b antibody did not show any immunoreactivity at the position of the 64 kDa TUC-4a protein.
Fig. 2.
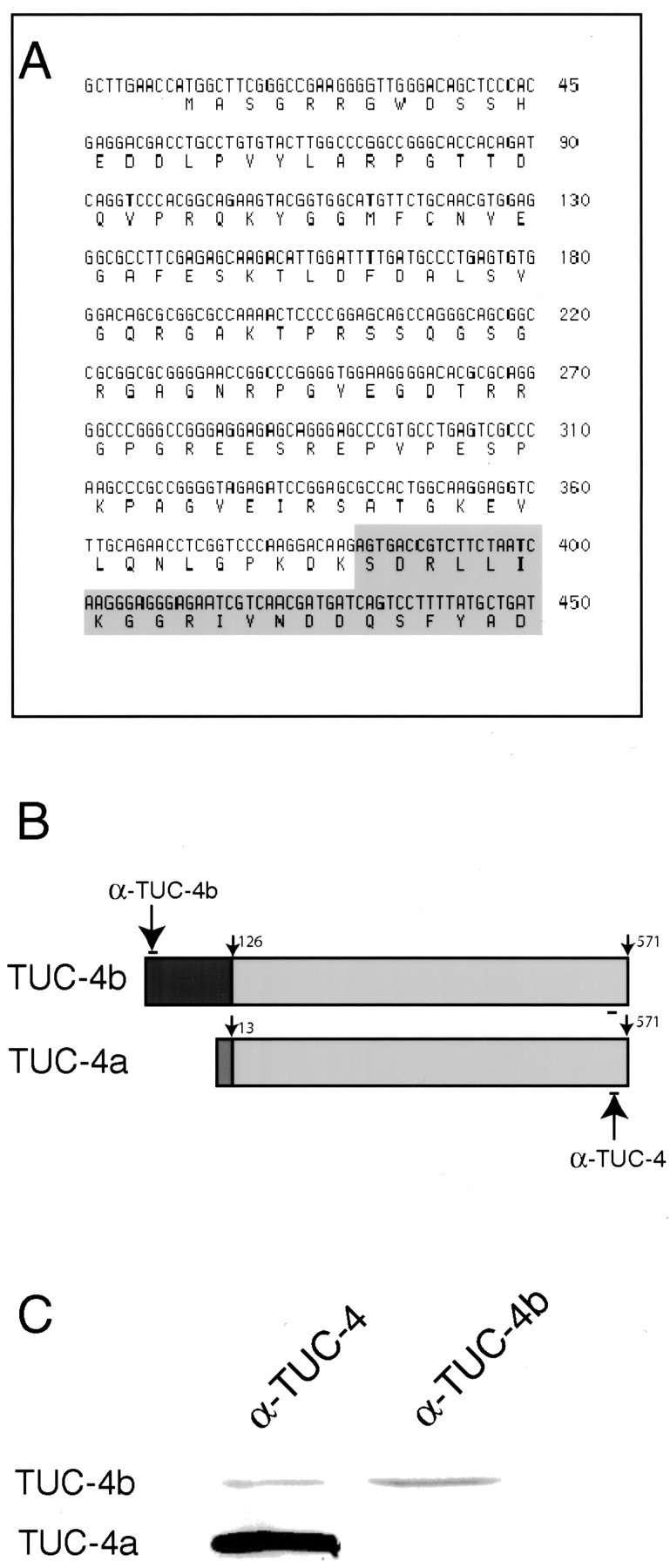
Identification of TUC-4b as a variant of TUC-4. A, Nucleotide sequence encoding the unique N-terminal extension of TUC-4b. The predicted amino acid sequence of the TUC-4b N-terminal extension is shown below the nucleotide sequence. The shaded area represents the beginning of the region of sequence identity with TUC-4a. The full nucleotide sequence of TUC-4b can be found in GenBank (accession number AF398465).B, In TUC-4b, the first 13 amino acids of TUC-4a, which are derived from exon 1, are replaced by a unique 126 amino acid sequence. The TUC-4 antibody (αTUC-4) recognizes an epitope that is present at the C-terminal end of TUC-4a and TUC-4b. The TUC-4b antibody (αTUC-4b) recognizes an epitope that is unique to TUC-4b.C, Western blot of E18 rat brain lysate with the TUC-4 and TUC-4b antibodies. The TUC-4 antibody recognizes proteins at 64 and 75 kDa, whereas the TUC-4b antibody only recognizes a 75 kDa protein.
TUC-4b is expressed in the developing nervous system
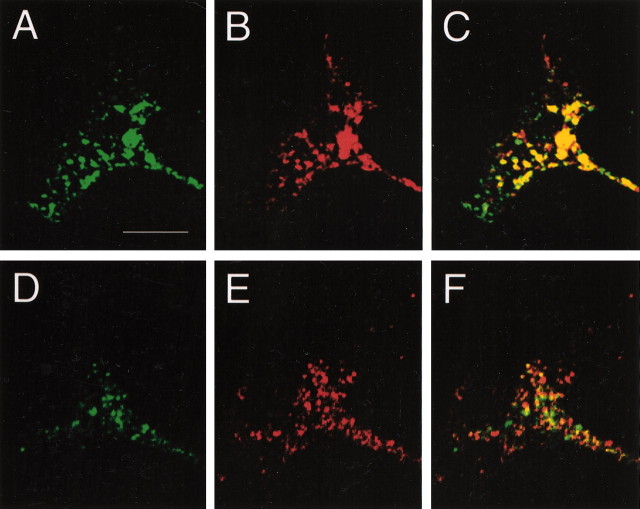
Immunostaining of E17 rat embryos revealed that TUC-4b is expressed in the CNS and PNS. Weak immunoreactivity was observed in the ventricular zone, with stronger immunoreactivity in the developing cortical plate (Fig.3A). TUC-4b was also expressed at high levels in the dorsal root ganglia (Fig. 3B). TUC-4b was expressed throughout the spinal cord with the highest level in the ventral horn (Fig. 3B). Expression of TUC-4b was also observed in the trigeminal ganglia (Fig.3C). To study the subcellular localization of TUC-4b, immunocytochemistry was performed on cultured chick DRG explants. Immunoblotting analysis indicated that the expression levels of TUC-4a and TUC-4b in DRGs is similar to their expression levels in brain (data not shown). Immunostaining with an antibody specific for TUC-4b revealed that the distribution of TUC-4b was highly restricted, with expression concentrated in the growth cones (Fig.4A–C). Immunostaining with an antibody that detects both TUC-4a and TUC-4b demonstrated that TUC-4 is expressed at high levels along neurites and throughout the growth cones (Fig. 4D–F). Although this antibody is not specific for TUC-4a, the restricted pattern of TUC-4b staining allows one to infer that TUC-4a is distributed throughout the neurite and growth cone. Within the growth cone, TUC-4b was restricted to punctae in the central region (Fig. 4G), as revealed by double staining of actin with phalloidin (Fig.4H,I), whereas TUC-4a was expressed throughout the growth cone, extending into the filopodia (Fig. 4J–L).
Fig. 3.
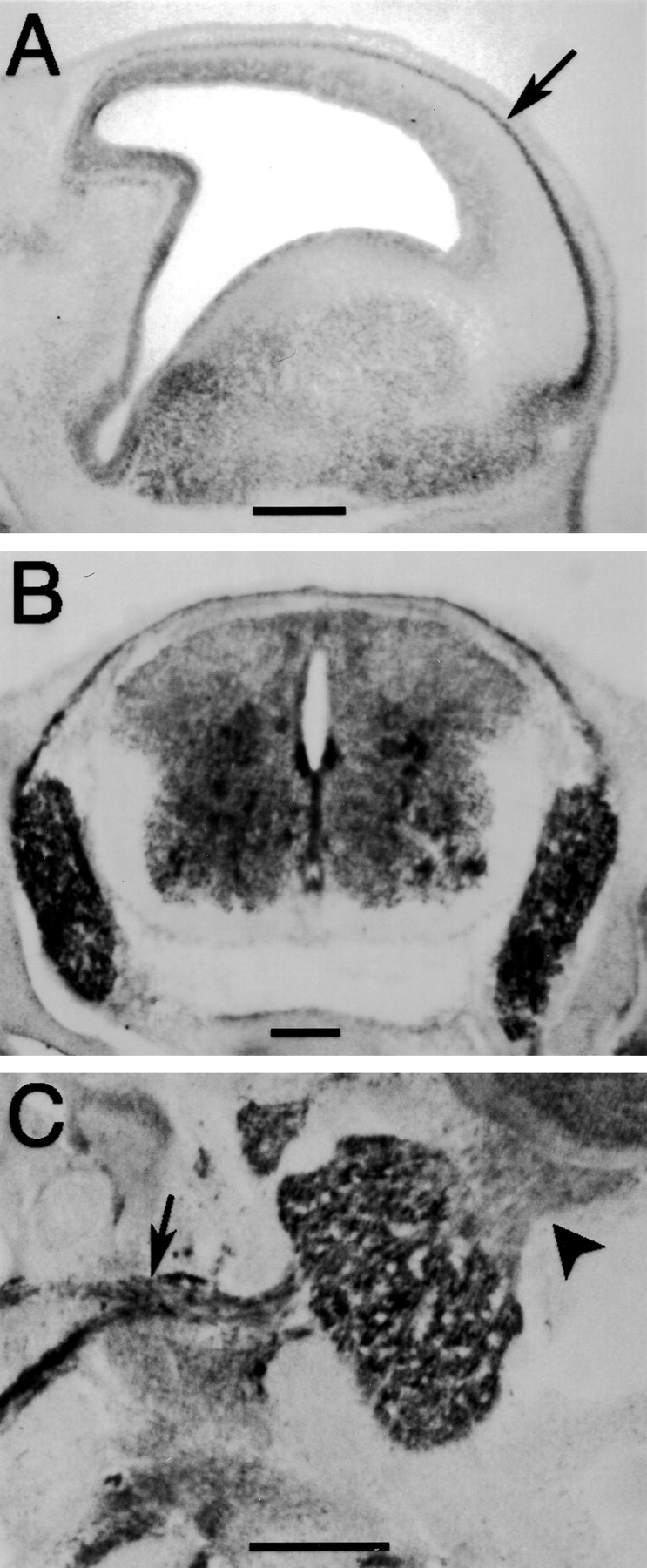
TUC-4b is expressed in the CNS and PNS of E17 rat embryos. A, Sagittal section through the telencephlon stained with the antibody to TUC-4b. TUC-4b was expressed at low levels in the ventricular zone and at higher levels in the cortical plate (arrow). Scale bar, 500 μm. B, Coronal section of E17 spinal cord stained with an antibody to TUC-4b. TUC-4b was expressed in the dorsal root ganglia and in the spinal cord. Scale bar, 200 μm. C, Sagittal section through the trigeminal ganglia. TUC-4b was detected in the trigeminal ganglia and in its peripheral (arrow) and central (arrowhead) processes. Scale bar, 200 μm.
Fig. 4.
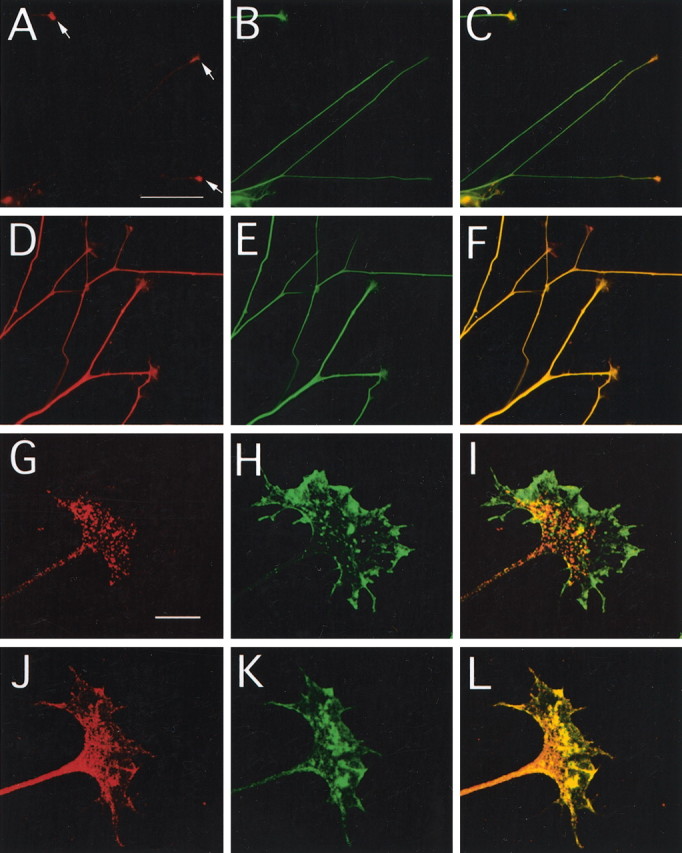
TUC-4b is enriched in the central domain of growth cones from DRG neurons. A–F, DRG explant cultures were stained with antibodies to TUC-4b (A), TUC-4 (D), or tubulin (B,E). The merged images are shown inC and F. Scale bar, 100 μm. TUC-4b immunoreactivity was weak in neurites and strong in the growth cones (see arrows in A). TUC-4 immunoreactivity was strong throughout the neurite and growth cones.G–L, High-power images of growth cones stained with antibodies to TUC-4b (G), TUC-4 (J), or phalloidin (H,K). The merged images are shown inI and L. Scale bar, 10 μm. TUC-4b immunoreactivity was localized to punctate structures in the central domain of the growth cone. TUC-4 immunoreactivity was distributed throughout the growth cone, extending into the filopodia.
TUC-4b is localized to growth cone vesicles
The central domain of the growth cone contains many large vesicles, some endocytic (Bunge, 1977; Cheng and Reese, 1985, 1987;Dailey and Bridgman, 1993) and others exocytic (Cheng and Reese, 1987;Pfenninger and Friedman, 1993; Zakharenko and Popov, 1998). To further define the intracellular localization of TUC-4b, we performed colocalization experiments with markers of endocytic and exocytic membranes. TUC-4b partially colocalized with SV2, a synaptic vesicle marker that is thought to regulate exocytosis (Fig.5A–C). In contrast, TUC-4b did not colocalize with Rab5, a marker of early endosomes (Fig.5D–F). Colocalization between TUC-4b and SV2 indicates that TUC-4b is associated with vesicles in the central domain of the growth cone. Because SV2 is a marker of exocytic vesicles in neurons, it is likely that these vesicles are part of the exocytic pathway in the growth cone.
Fig. 5.
TUC-4b is localized to vesicles in the growth cone. A–C, DRG explant cultures were colabeled with antibodies to TUC-4b (A) and SV2 (B), and the merged image is seen inC. D–F DRG explant cultures were colabeled with antibodies to TUC-4b (D) and Rab5 (E), and the merged image is seen inF. Scale bar, 10 μm.
TUC-4b interacts with the SH3A domain of intersectin
Intersectin is a scaffolding protein that has five SH3 domains (SH3A–SH3E), which are sites for protein–protein interactions (Roos and Kelly, 1998; Yamabhai et al., 1998; Sengar et al., 1999). Through its protein interactions, intersectin has been demonstrated to function in endocytosis (Yamabhai et al., 1998; Simpson et al., 1999) and the regulation of actin dynamics (Hussain et al., 2001), and it has also been implicated in exocytosis (Okamoto et al., 1999). Like TUC-4b, intersectin is enriched in growth cones and is also distributed in a punctate pattern (Tong et al., 2000). Moreover, a recent study from our laboratory has demonstrated that intersectin is highly expressed in growth cones of developing neurons, in which it appears to function in the regulation of neurite branching (Quinn et al., 2001). During the course of our studies on intersectin, we performed binding assays with extracts from E18 rat brains using GST fusion proteins encoding each of the five SH3 domains of the protein. Proteins that bound to each of the five SH3A domains were analyzed by SDS-PAGE (Fig.6A). As reported previously, dynamin bound to the SH3A, SH3C, and SH3E domains and was visible as a 110 kDa band on a Coomassie blue-stained gel (Yamabhai et al., 1998). In addition, an unknown, 65 kDa protein bound to the SH3A domain and, to a lesser degree, to the SH3E domain. The 65 kDa protein was not readily detected in binding assays from adult brain (data not shown).
Fig. 6.
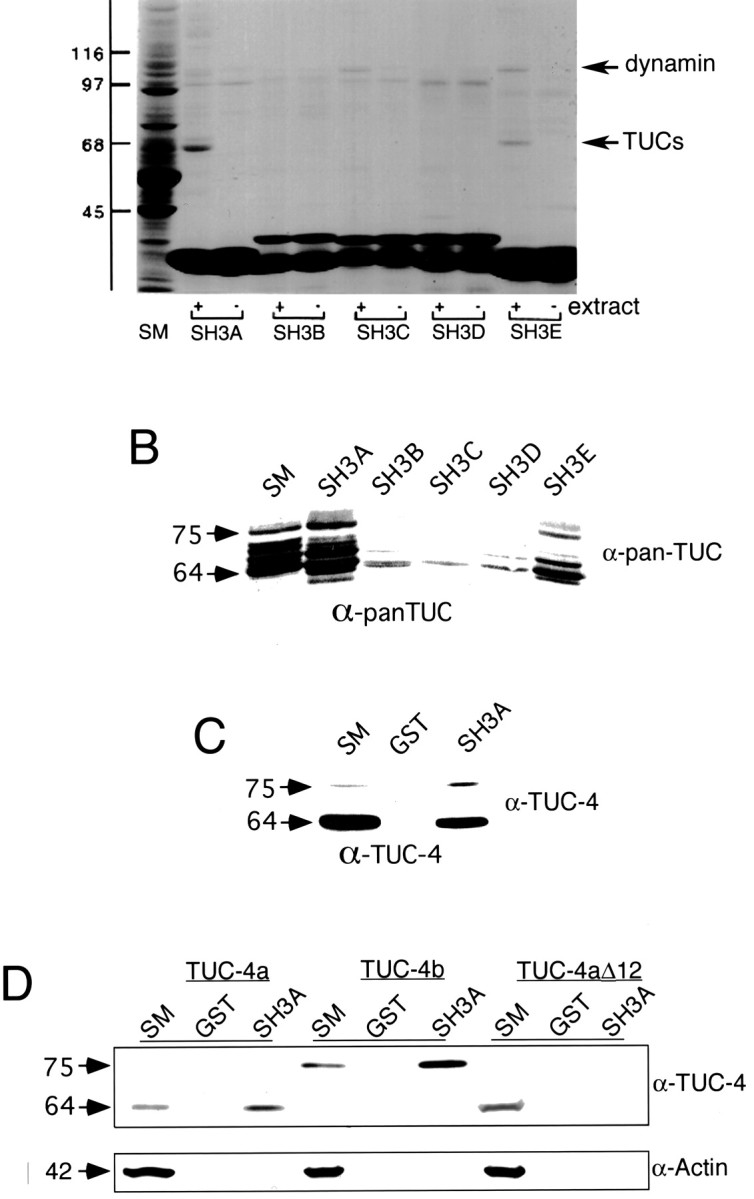
The TUC family interacts with the SH3A domain of intersectin. A, GST fusion proteins encoding the individual SH3 domains of intersectin (SH3A–SH3E) were precoupled to glutathione-Sepharose and incubated with Triton X-100-soluble proteins from E18 rat brains (+ extract) or with buffer alone (− extract). The beads were washed, and specifically bound material was eluted and processed for SDS-PAGE along with an aliquot of the soluble extract [starting material (SM)] equivalent to of the protein added to the beads. The gels were stained with Coomassie blue to reveal the full protein profile. An unknown protein (TUCs) bound to the SH3A and SH3E domains and was later determined to belong to the TUC family. The strong bands below the 45 kDa marker are the GST fusion proteins that elute from the beads. The band at 110 kDa represents dynamin as indicated and as described previously (Yamabhai et al., 1998). The band at 97 kDa is an Escherichia coli-derived protein, as evidenced by its presence in the no-extract lanes. B, Binding assays were conducted with each of the five SH3 domains of intersectin, and bound proteins were immunoblotted with the pan-TUC antibody. Ten percent of the starting material (SM) was included on each gel for reference. The TUC family bound to the SH3A domain and to a lesser degree the SH3E domain but not to the SH3B, SH3C, or SH3D domains.C, Proteins that bound to the SH3A domain were immunoblotted with the TUC-4 antibody, which recognizes both TUC-4a and TUC-4b. Whereas TUC-4a bound at levels <10% of the starting material (SM), TUC-4b bound at levels substantially >10% of the starting material. D, Binding assays were conducted with the SH3A domain and lysates from cells that were transfected with either TUC-4a or TUC-4b, or TUC-4aΔ12. Bound fractions were immunoblotted with an antibody that recognizes a region in common between TUC-4a and TUC-4b. To assess possible nonspecific binding, the bound fractions were also immunoblotted with an antibody against actin. Whereas TUC-4a was slightly enriched relative to 10% of the starting material (SM), TUC-4b was highly enriched relative to 10% of the starting material. Neither TUC-4aΔ12 nor actin bound to the SH3A domain.
N-terminal sequence analysis suggested that the 65 kDa protein might belong to the TUC family (data not shown). We performed an immunochemical analysis of the proteins that bound to each of the SH3 domains, using an antibody that recognizes all of the TUC isoforms (Fig. 6B). Members of the TUC family bound to the SH3A domain and to a lesser degree the SH3E domain but showed little or no interaction with the SH3B, SH3C, and SH3D domains (Fig. 6B). Many of the TUC-immunoreactive bands observed in the starting material bound to the SH3A domain; however, a 75 kDa protein bound with apparently higher affinity than other proteins, because it demonstrated the highest degree of enrichment in the bound fractions relative to the starting material. To determine whether the 75 kDa protein corresponds to TUC-4b, proteins bound by the SH3A domain were immunoblotted with the antibody that recognizes both TUC-4a and TUC-4b (Fig. 6C). Whereas there was less TUC-4a in the bound fraction, the 75 kDa TUC-4b band was enriched in the bound fraction, relative to the starting material (Fig.6C). To confirm an interaction between TUC-4b and the SH3A domain of intersectin, we tested the binding of recombinant TUC-4a and TUC-4b to SH3A (Fig. 6D). The amount of TUC-4a that was eluted from the SH3A affinity matrix corresponded to a slight enrichment relative to the starting material. In contrast, TUC-4b was highly enriched in the SH3A-bound fraction relative to the starting material. To determine whether the increased SH3A binding capacity of TUC-4b was the result of sequences lost or sequence gained relative to TUC-4a, we created a TUC-4aΔ12 protein. TUC-4aΔ12 consists of TUC-4a with the first 12 amino acids deleted and replaced by a single methionine. TUC-4aΔ12 did not bind to the SH3A domain, suggesting that the first 12 amino acids of TUC-4a are required for binding to the SH3A domain. Because these amino acids do not contain any proline-rich sequences, it is likely that their requirement for SH3A binding is indirect.
TUC-4b stimulates neurite outgrowth
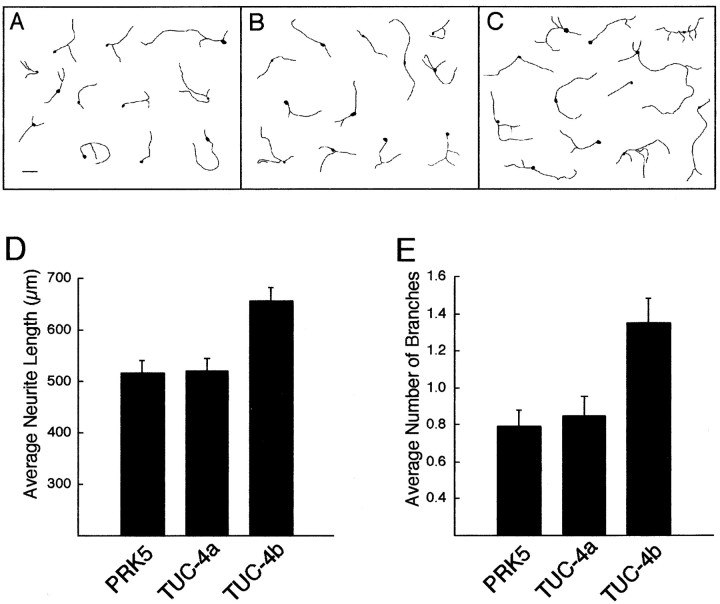
The marked enrichment of TUC-4b binding to the SH3A domain, compared with TUC-4a and TUC-4aΔ12, led us to ask whether TUC-4b might play a role in neurite outgrowth. Primary neurons from E18 rat cerebral cortex were cotransfected with GFP and TUC-4a, TUC-4b, or the empty PRK5 vector as a control after 1 d in culture. The cultures were incubated for an additional 48 hr and then fixed and stained with an antibody to GFP. In some experiments, the neurons were stained with an antibody to TUC-4, confirming that both TUC-4a and TUC-4b were overexpressed in the GFP-positive neurons (data not shown). Figure7 shows camera lucida drawings of representative examples of neurons cotransfected with GFP and PRK5 (Fig. 7A), TUC-4a (Fig. 7B), and TUC-4b (Fig.7C). The number of branches and the length of the longest neurite were determined for each of 100 GFP-positive neurons. Neurons transfected with TUC-4a showed no change in the length of the longest neurite (Fig. 7D) or in the number of branches (Fig.7E) when compared with control-transfected neurons. In contrast, neurons transfected with TUC-4b showed a 71% increase in the number of branches (Fig. 7E) and a 27% increase in the length of the longest neurite (Fig. 7D).
Fig. 7.
Overexpression of TUC-4b increases neurite outgrowth. A–C, Digital camera lucida drawings illustrating representative examples of E18 cortical neurons cotransfected with GFP and PRK-5 control vector (A), TUC-4a (B), or TUC-4b (C). Scale bar, 100 μm.D, Graph representing the average length of the longest neurite for neurons transfected with PRK5 (n = 88), TUC-4a (n = 90), or TUC-4b (n = 101). Transfection of neurons with TUC-4b increased neurite length by 27% relative to neurons transfected with PRK5, a statistically significant result (p < 0.05; unpairedt test). E, Graph representing the average number of branch points for each neuron transfected with PRK5 (n = 91), TUC-4a (n = 83), or TUC-4b (n = 103). Transfection of neurons with TUC-4b increased neurite branching by 71% relative to neurons transfected with PRK5, a statistically significant result (p < 0.05; unpaired ttest).
Discussion
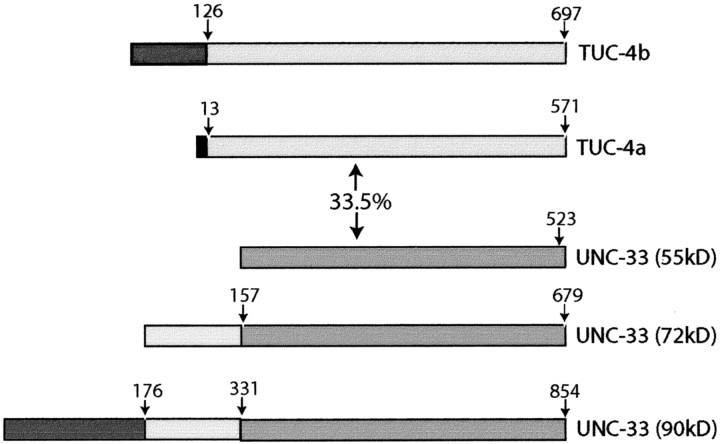
Over the last several years, a family of proteins homologous to the C. elegans unc-33 gene products have been identified in rodents, human, and chick and named the TUC (TOAD-64/Ulip/CRMP) family of proteins (Quinn et al., 1999). Mutations in unc-33 give rise to disruptions in axonal pathways and a severely uncoordinated phenotype (Hedgecock et al., 1985; Siddiqui and Culotti, 1991). On the basis of homology to unc-33, but little direct experimental evidence, it has been suggested that the TUC proteins play a role in axon growth and guidance. The unc-33gene produces proteins of 55, 72, and 90 kDa (Li et al., 1992); the previously identified TUC proteins correspond to the 55 kDa UNC-33 protein (Fig. 8). Analysis of mutations in unc-33 indicate that the 55 kDa protein has minimal function in the absence of the two larger proteins, suggesting that larger isoforms of the TUC family might, similarly, have significant roles in axon growth and guidance in vertebrates. Here, we demonstrate the existence of several new members of the TUC family. We cloned and characterized TUC-4b, a 75 kDa variant of TUC-4a. TUC-4b consists of a core region that is identical to amino acid numbers 13–571 of TUC-4a, with the first 12 amino acids of TUC-4a being replaced by a unique 125 amino acid N-terminal extension. The core region that is shared between TUC-4a and TUC-4b is 33.5% identical to the 55 kDa isoform of UNC-33 (Fig. 8). Whereas TUC-4a is distributed throughout neurons and their axons, TUC-4b is highly expressed specifically in the growth cone, in which it is localized to vesicles in the central domain of the growth cone. Although previous studies have implicated the TUC proteins in semaphorin signal transduction and the establishment of neuronal polarity, here we present direct evidence for a role of TUC-4b in neurite growth by showing that overexpression of TUC-4b, but not TUC-4a, leads to increased neurite extension and branching.
Fig. 8.
The TUC family is based on a TUC-4 core region that is 33.5% identical to the C. elegans 55 kDa isoform of UNC-33. TUC-4a consists of the core region plus a 12 amino acid N-terminal extension. TUC-4b consists of the core region plus a 126 amino acid extension. The TUC-4a and TUC-4b N-terminal extensions are unique and do not have any homology to UNC-33 or any other known proteins. The 55 kDa isoform of UNC-33 represents a core region that is 33.5% identical to the TUC-4 core region. The 72 kDa isoform of UNC-33 consists of the UNC-33 core region plus a 156 amino acid N-terminal extension. The 90 kDa UNC-33 isoform consists of the entire sequence of the 72 kDa isoform plus an additional 175 amino acid N-terminal extension.
The N-terminal region of the TUC family members may mediate distinct functions
Antibodies specific for each of the four previously identified 64 kDa TUC isoforms reveal the existence of 75 kDa variants of TUC-1, TUC-2, and TUC-4, and, in addition, 68 kDa variants of TUC-2. We cloned a transcript for TUC-4b, the 75 kDa variant of TUC-4a, and obtained partial sequence of the 75 kDa TUC-1b variant (data not shown). Sequence analysis indicates that, like TUC-4b, TUC-1b comprises an N-terminal extension that replaces the first 13 amino acids of TUC-1a. It is reasonable to predict an analogous structure for TUC-2b, but we have not yet identified clones encoding this variant. The structure of the additional, 68 kDa variants of TUC-2 has not yet been determined; they could arise from alternate splicing or from posttranslational processing. The existence of multiple versions of the TUC protein, all based on the 55 kDa core region defined by UNC-33, begins to suggest a common theme in which the N-terminal extensions may specify unique functions of each TUC variant. It is important to note that our ability to detect variants immunochemically is limited by whether a particular epitope is shared among variants, leaving open the possible expression of additional variants beyond those identified here.
TUC-4b may function with intersectin to regulate neurite outgrowth
TUC-4b interacts with the SH3A domain of intersectin, a protein that can regulate membrane dynamics and neurite outgrowth. Intersectin contains two Eps15-homology domains, a central helical domain, and five SH3 domains (SH3A–SH3E) (Yamabhai et al., 1998; Sengar et al., 1999). The helical domain interacts with SNAP-25 (synaptosome-associated protein of 25 kDa), suggesting a function in exocytosis (Okamoto et al., 1999). The SH3A, SH3C, and SH3E domains interact with dynamin and synaptojanin, both of which are involved in endocytosis (Yamabhai et al., 1998). Disruption of the interactions between the SH3A domain and its ligands, but not any of the other four SH3 domains, results in an increase in neurite branching in cortical neurons (Quinn et al., 2001). Here, we report that overexpression of TUC-4b, in the same experimental system, results in increased neurite branching. Therefore, it is possible that TUC-4b may regulate neurite branching through its interaction with the SH3A domain of intersectin. However, we cannot rule out the function of mSos, another SH3A domain-specific intersectin-binding partner (Tong et al., 2000), in the role of the SH3A domain in neurite branching.
TUC-4b may regulate vesicle function in the growth cone
The localization of TUC-4b to SV2-positive vesicles suggests that TUC-4b might be associated with vesicles in the exocytic pathway of the growth cone. SV2 is a transmembrane glycoprotein that localizes to secretory vesicles in endocrine cells and in neurons (Buckley and Kelly, 1985). In mature neurons, SV2 is localized to presynaptic vesicles. In developing neurons, it is localized to growth cone vesicles (Buckley and Kelly, 1985). The function of SV2 has been best studied in chromaffin cells, in which it regulates exocytosis but is not required for exocytic fusion in itself. Chromaffin cells that encode a mutant form of SV2 have fewer readily releasable vesicles and are deficient in SNARE (soluble N-ethylmaleimide-sensitive factor attached protein receptor) complexes, suggesting that SV2 may regulate the progression of vesicles to a fusion-competent state (Xu and Bajjalieh, 2001). Although the role of SV2 in the growth cone has not been studied, it is likely that it also functions to regulate exocytosis in the growth cone. The overlapping localization of SV2 and TUC-4b would be consistent with TUC-4b participating in the exocytic pathway of the growth cone. To test this hypothesis, it will be necessary to develop methods to determine the effect of TUC-4b loss of function on vesicle transport in the growth cone.
Vesicle transport in the growth cone can regulate axon outgrowth and guidance
Several recent studies have suggested that regulated membrane transport is part of the mechanism that mediates the response to cues that regulate axon outgrowth and guidance, a hypothesis that is strengthened by our finding that TUC-4b is localized to vesicles. Stimulation of growth cones by semaphorins or ephrins induces endocytosis in the growth cone (Fournier et al., 2000; Jurney et al., 2002). However, endocytosis is not stimulated by cytochalasin B, suggesting that endocytosis is not a nonspecific consequence of collapse but rather the specific result of intracellular signaling in response to guidance cues. The small GTPase Rac1 is required for growth cone collapse; however, it does not mediate actin depolymerization during collapse. Rather, Rac1 mediates endocytosis during growth cone collapse (Jin and Strittmatter, 1997; Jurney et al., 2002). Together, these data suggest that endocytosis is part of the biological mechanism that mediates the response to repulsive guidance cues.
Converging lines of evidence indicate that neurite extension is dependent on exocytic addition of membrane to the growth cone plasmalemma. Inhibitors of vesicle fusion proteins, such as TI-VAMP (tetanus neurotoxin-insensitive vesicle-associated membrane protein), SNAP-25, and syntaxin inhibit neurite outgrowth (Osen-Sand et al., 1993; Igarashi et al., 1996; Osen-Sand et al., 1996;Grosse et al., 1999; Martinez-Arca et al., 2001), and overexpression of TI-VAMP or syntaxin increases neurite growth (Hirling et al., 2000;Martinez-Arca et al., 2001), suggesting that exocytosis is a rate-limiting and regulated step in neurite extension. Consistent with these findings, a role for exocytosis in the outgrowth promoting activity of DCC (deleted in colorectal cancer), the netrin receptor, has been described previously (Bouchard et al., 2002). DCC localizes to the plasma membrane and to intracellular vesicles in spinal neurons. Stimulation of spinal neurons with PKA results in exocytosis of the DCC-containing vesicles and enhances neurite outgrowth in an exocytosis-dependent manner. Together, these data suggest that exocytosis is a part of the mechanism that mediates the growth cone response to neurite outgrowth-promoting cues.
In addition to endocytosis and exocytosis, sorting of receptors between membrane compartments also plays a role in mediating the response to guidance cues. In Drosophila, the extracellular protein Slit repels axons from the midline through activation of Robo, the Slit receptor. Axons cross the midline when Comm downregulates Robo. Recent work has indicated that Comm is responsible for the sorting of Robo to endosomes, thereby preventing Robo activation by Slit (Keleman et al., 2002). Receptor sorting in this system further supports a role for vesicle transport in axon guidance.
Conclusions
Despite a large number of studies identifying TUC proteins, the functions of this family remain only partly understood. Previous studies have suggested that the TUC proteins can mediate semaphorin signal transduction, as well as neuronal polarity. Here, we reported for the first time that the TUC family can also regulate neurite outgrowth. This function is mediated through TUC-4b, an N-terminally extended variant of TUC-4a. Although the mechanism of TUC-4b function is not yet known in detail, the localization of TUC-4b to growth cone vesicles and its interaction with intersectin suggest that it may function by affecting membrane dynamics in the growth cone. However, this hypothesized role for TUC-4b in growth cone membrane dynamics will require additional testing. We also identified two other N-terminally extended variants of the TUC proteins that are expressed at distinct stages of neural development, consistent with the range of functions that have been attributed to the TUC family. The further characterization of the family of TUC variants is likely to put us closer to a mechanistic understanding of TUC function. Furthermore, it is important to consider the new variants described here in the interpretation of past studies, most of which have used reagents that cannot distinguish between the 64 and 75 kDa isoforms of the TUC family members.
Footnotes
This research was supported by National Institute of Neurological Disorders and Stroke Grant NS22807 (S.H.) and Canadian Institutes of Health Research (CIHR) Grant MT-15396 (P.S.M.). P.S.M. is a CIHR Investigator. DNA sequencing and oligonucleotide synthesis was performed by the Howard Hughes Medical Institute Biopolymer Laboratory and the W. M. Keck Foundation Biotechnology Resource Laboratory at Yale University. We thank Dr. Yasuo Ihara for the C4G antibody.
Correspondence should be addressed to Christopher C. Quinn at his present address: Department of Pathology, Robert Wood Johnson Medical School, 675 Hoes Lane, Piscataway, NJ 08854-5653. E-mail:quinncc@umdnj.edu.
References
- 1.Bouchard JF, Roux PP, Shekarbi M, Barker PA, Kennedy TE. cAMP elevation rapidly translocates DCC to the plasma membrane and promotes netrin-1-dependent axon extension. Soc Neurosci Abstr. 2002;28:729.14. [Google Scholar]
- 2.Buckley K, Kelly RB. Identification of a transmembrane glycoprotein specific for secretory vesicles of neural and endocrine cells. J Cell Biol. 1985;100:1284–1294. doi: 10.1083/jcb.100.4.1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bunge MB. Initial endocytosis of peroxidase or ferritin by growth cones of cultured nerve cells. J Neurocytol. 1977;6:407–439. doi: 10.1007/BF01178226. [DOI] [PubMed] [Google Scholar]
- 4.Byk T, Dobransky T, Cifuentes-Diaz C, Sobel A. Identification and molecular characterization of Unc-33-like phosphoprotein (Ulip), a putative mammalian homolog of the axonal guidance-associated unc-33 gene product. J Neurosci. 1996;16:688–701. doi: 10.1523/JNEUROSCI.16-02-00688.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Byk T, Ozon S, Sobel A. The Ulip family phosphoproteins—common and specific properties. Eur J Biochem. 1998;254:14–24. doi: 10.1046/j.1432-1327.1998.2540014.x. [DOI] [PubMed] [Google Scholar]
- 6.Cheng TP, Reese TS. Polarized compartmentalization of organelles in growth cones from developing optic tectum. J Cell Biol. 1985;101:1473–1480. doi: 10.1083/jcb.101.4.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng TP, Reese TS. Recycling of plasmalemma in chick tectal growth cones. J Neurosci. 1987;7:1752–1759. doi: 10.1523/JNEUROSCI.07-06-01752.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dailey ME, Bridgman PC. Vacuole dynamics in growth cones: correlated EM and video observations. J Neurosci. 1993;13:3375–3393. doi: 10.1523/JNEUROSCI.13-08-03375.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fournier AE, Nakamura F, Kawamoto S, Goshima Y, Kalb RG, Strittmatter SM. Semaphorin3A enhances endocytosis at sites of receptor-f-actin colocalization during growth cone collapse. J Cell Biol. 2000;149:411–422. doi: 10.1083/jcb.149.2.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gaetano C, Matsuo T, Thiele CJ. Identification and characterization of a retinoic acid-regulated human homologue of the unc-33-like phosphoprotein gene (hUlip) from neuroblastoma cells. J Biol Chem. 1997;272:12195–12201. doi: 10.1074/jbc.272.18.12195. [DOI] [PubMed] [Google Scholar]
- 11.Goshima Y, Nakamura F, Strittmatter P, Strittmatter SM. Collapsin-induced growth cone collapse mediated by an intracellular protein related to UNC-33. Nature. 1995;376:509–514. doi: 10.1038/376509a0. [DOI] [PubMed] [Google Scholar]
- 12.Grosse G, Grosse J, Tapp R, Kuchinke J, Gorsleben M, Fetter I, Hohne-Zell B, Gratzl M, Bergmann M. SNAP-25 requirement for dendritic growth of hippocampal neurons. J Neurosci Res. 1999;56:539–546. doi: 10.1002/(SICI)1097-4547(19990601)56:5<539::AID-JNR9>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 13.Gu Y, Ihara Y. Evidence that collapsin response mediator protein-2 is involved in the dynamics of microtubules. J Biol Chem. 2000;275:17917–17920. doi: 10.1074/jbc.C000179200. [DOI] [PubMed] [Google Scholar]
- 14.Hedgecock EM, Culotti JG, Thomson JN, Perkins LA. Axonal guidance mutants of Caenorhabditis elegans identified by filling sensory neurons with fluorescein dyes. Dev Biol. 1985;111:158–170. doi: 10.1016/0012-1606(85)90443-9. [DOI] [PubMed] [Google Scholar]
- 15.Hirling H, Steiner P, Chaperon C, Marsault R, Regazzi R, Catsicas S. Syntaxin 13 is a developmentally regulated SNARE involved in neurite outgrowth and endosomal trafficking. Eur J Neurosci. 2000;12:1913–1923. doi: 10.1046/j.1460-9568.2000.00076.x. [DOI] [PubMed] [Google Scholar]
- 16.Hussain NK, Jenna S, Glogauer M, Quinn CC, Wasiak S, Guipponi M, Antonarakis SE, Kay BK, Stossel TP, Lamarche-Vane N, McPherson PS. Endocytic protein intersectin-l regulates actin assembly via Cdc42 and N-wasp. Nat Cell Biol. 2001;3:927–932. doi: 10.1038/ncb1001-927. [DOI] [PubMed] [Google Scholar]
- 17.Igarashi M, Kozaki S, Terakawa S, Kawano S, Ide C, Komiya Y. Growth cone collapse and inhibition of neurite growth by Botulinum neurotoxin C1: a t-SNARE is involved in axonal growth. J Cell Biol. 1996;134:205–215. doi: 10.1083/jcb.134.1.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Inagaki N, Chihara K, Arimura N, Menager C, Kawano Y, Matsuo N, Nishimura T, Amano M, Kaibuchi K. CRMP-2 induces axons in cultured hippocampal neurons. Nat Neurosci. 2001;4:781–782. doi: 10.1038/90476. [DOI] [PubMed] [Google Scholar]
- 19.Jin Z, Strittmatter SM. Rac1 mediates collapsin-1-induced growth cone collapse. J Neurosci. 1997;17:6256–6263. doi: 10.1523/JNEUROSCI.17-16-06256.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jurney WM, Gallo G, Letourneau PC, McLoon SC. Rac1-mediated endocytosis during ephrin-A2- and semaphorin 3A-induced growth cone collapse. J Neurosci. 2002;22:6019–6028. doi: 10.1523/JNEUROSCI.22-14-06019.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kamata T, Subleski M, Hara Y, Yuhki N, Kung H, Copeland NG, Jenkins NA, Yoshimura T, Modi W, Copeland TD. Isolation and characterization of a bovine neural specific protein (CRMP-2) cDNA homologous to unc-33, a C. elegans gene implicated in axonal outgrowth and guidance. Brain Res Mol Brain Res. 1998;54:219–236. doi: 10.1016/s0169-328x(97)00332-x. [DOI] [PubMed] [Google Scholar]
- 22.Keleman K, Rajagopalan S, Cleppien D, Teis D, Paiha K, Huber LA, Technau GM, Dickson BJ. Comm sorts robo to control axon guidance at the Drosophila midline. Cell. 2002;110:415–427. doi: 10.1016/s0092-8674(02)00901-7. [DOI] [PubMed] [Google Scholar]
- 23.Li W, Herman RK, Shaw JE. Analysis of the Caenorhabditis elegans axonal guidance and outgrowth gene unc-33. Genetics. 1992;132:675–689. doi: 10.1093/genetics/132.3.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martinez-Arca S, Coco S, Mainguy G, Schenk U, Alberts P, Bouille P, Mezzina M, Prochiantz A, Matteoli M, Louvard D, Galli T. A common exocytotic mechanism mediates axonal and dendritic outgrowth. J Neurosci. 2001;21:3830–3838. doi: 10.1523/JNEUROSCI.21-11-03830.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matsuo T, Stauffer JK, Walker RL, Meltzer P, Thiele CJ. Structure and promoter analysis of the human unc-33-like phosphoprotein gene: E-box required for maximal expression in neuroblastoma and myoblasts. J Biol Chem. 2000;275:25052. doi: 10.1074/jbc.M001312200. [DOI] [PubMed] [Google Scholar]
- 26.Minturn JE, Fryer HJ, Geschwind DH, Hockfield S. TOAD-64, a gene expressed early in neuronal differentiation in the rat, is related to unc-33, a C. elegans gene involved in axon outgrowth. J Neurosci. 1995a;15:6757–6766. doi: 10.1523/JNEUROSCI.15-10-06757.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Minturn JE, Geschwind DH, Fryer HJ, Hockfield S. Early postmitotic neurons transiently express TOAD-64, a neural specific protein. J Comp Neurol. 1995b;355:369–379. doi: 10.1002/cne.903550304. [DOI] [PubMed] [Google Scholar]
- 28.Okamoto M, Schoch S, Sudhof TC. EHSHI/intersectin, a protein that contains EH and SH3 domains and binds to dynamin and SNAP-25. A protein connection between exocytosis and endocytosis? J Biol Chem. 1999;274:18446–18454. doi: 10.1074/jbc.274.26.18446. [DOI] [PubMed] [Google Scholar]
- 29.Osen-Sand A, Catsicas M, Staple JK, Jones KA, Ayala G, Knowles J, Grenningloh G, Catsicas S. Inhibition of axonal growth by SNAP-25 antisense oligonucleotides in vitro and in vivo. Nature. 1993;364:445–448. doi: 10.1038/364445a0. [DOI] [PubMed] [Google Scholar]
- 30.Osen-Sand A, Staple JK, Naldi E, Schiavo G, Rossetto O, Petitpierre S, Malgaroli A, Montecucco C, Catsicas S. Common and distinct fusion proteins in axonal growth and transmitter release. J Comp Neurol. 1996;367:222–234. doi: 10.1002/(SICI)1096-9861(19960401)367:2<222::AID-CNE5>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 31.Pfenninger KH, Friedman LB. Sites of plasmalemmal expansion in growth cones. Brain Res Dev Brain Res. 1993;71:181–192. doi: 10.1016/0165-3806(93)90170-f. [DOI] [PubMed] [Google Scholar]
- 32.Quinn CC, Gray GE, Hockfield S. A family of proteins implicated in axon guidance and outgrowth. J Neurobiol. 1999;41:158–164. [PubMed] [Google Scholar]
- 33.Quinn CC, Wasiak S, Hussain NK, Kinjo TG, Bell A, Kay BK, Baranes D, Hockfield S, McPherson PS. Intersectin regulates neurite outgrowth. Soc Neurosci Abstr. 2001;27:891.3. [Google Scholar]
- 34.Roos J, Kelly RB. Dap160, a neural-specific Eps15 homology and multiple SH3 domain-containing protein that interacts with Drosophila dynamin. J Biol Chem. 1998;273:19108–19119. doi: 10.1074/jbc.273.30.19108. [DOI] [PubMed] [Google Scholar]
- 35.Sengar AS, Wang W, Bishay J, Cohen S, Egan SE. The EH and SH3 domain Ese proteins regulate endocytosis by linking to dynamin and Eps15. EMBO J. 1999;18:1159–1171. doi: 10.1093/emboj/18.5.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Siddiqui SS, Culotti JG. Examination of neurons in wild type and mutants of Caenorhabditis elegans using antibodies to horseradish peroxidase. J Neurogenet. 1991;7:193–211. doi: 10.3109/01677069109167433. [DOI] [PubMed] [Google Scholar]
- 37.Simpson F, Hussain NK, Qualmann B, Kelly RB, Kay BK, McPherson PS, Schmid SL. SH3-domain-containing proteins function at distinct steps in clathrin-coated vesicle formation. Nat Cell Biol. 1999;1:119–124. doi: 10.1038/10091. [DOI] [PubMed] [Google Scholar]
- 38.Threadgill R, Bobb K, Ghosh A. Regulation of dendritic growth and remodeling by Rho, Rac, and Cdc42. Neuron. 1997;19:625–634. doi: 10.1016/s0896-6273(00)80376-1. [DOI] [PubMed] [Google Scholar]
- 39.Tong XK, Hussain NK, de Heuvel E, Kurakin A, Abi-Jaoude E, Quinn CC, Olson MF, Marais R, Baranes D, Kay BK, McPherson PS. The endocytic protein Intersectin is a major binding partner for the Ras exchange factor mSos1 in rat brain. EMBO J. 2000;19:1263–1271. doi: 10.1093/emboj/19.6.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang LH, Strittmatter SM. A family of rat CRMP genes is differentially expressed in the nervous system. J Neurosci. 1996;16:6197–6207. doi: 10.1523/JNEUROSCI.16-19-06197.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang LH, Strittmatter SM. Brain CRMP forms heterotetramers similar to liver dihydropyrimidinase. J Neurochem. 1997;69:2261–2269. doi: 10.1046/j.1471-4159.1997.69062261.x. [DOI] [PubMed] [Google Scholar]
- 42.Xu T, Bajjalieh SM. SV2 modulates the size of the readily releasable pool of secretory vesicles. Nat Cell Biol. 2001;3:691–698. doi: 10.1038/35087000. [DOI] [PubMed] [Google Scholar]
- 43.Yamabhai M, Hoffman NG, Hardison NL, McPherson PS, Castagnoli L, Cesareni G, Kay BK. Intersectin, a novel adaptor protein with two Eps15 homology and five Src homology 3 domains. J Biol Chem. 1998;273:31401–31407. doi: 10.1074/jbc.273.47.31401. [DOI] [PubMed] [Google Scholar]
- 44.Zakharenko S, Popov S. Dynamics of axonal microtubules regulate the topology of new membrane insertion into the growing neurites. J Cell Biol. 1998;143:1077–1086. doi: 10.1083/jcb.143.4.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]