Abstract
Several microtubule binding proteins, including CLIP-170 (cytoplasmic linker protein-170), CLIP-115, and EB1 (end-binding protein 1), have been shown to associate specifically with the ends of growing microtubules in non-neuronal cells, thereby regulating microtubule dynamics and the binding of microtubules to protein complexes, organelles, and membranes. When fused to GFP (green fluorescent protein), these proteins, which collectively are called +TIPs (plus end tracking proteins), also serve as powerful markers for visualizing microtubule growth events. Here we demonstrate that endogenous +TIPs are present at distal ends of microtubules in fixed neurons. Using EB3-GFP as a marker of microtubule growth in live cells, we subsequently analyze microtubule dynamics in neurons. Our results indicate that microtubules grow slower in neurons than in glia and COS-1 cells. The average speed and length of EB3-GFP movements are comparable in cell bodies, dendrites, axons, and growth cones. In the proximal region of differentiated dendrites ∼65% of EB3-GFP movements are directed toward the distal end, whereas 35% are directed toward the cell body. In more distal dendritic regions and in axons most EB3-GFP dots move toward the growth cone. This difference in directionality of EB3-GFP movements in dendrites and axons reflects the highly specific microtubule organization in neurons. Together, these results suggest that local microtubule polymerization contributes to the formation of the microtubule network in all neuronal compartments. We propose that similar mechanisms underlie the specific association of CLIPs and EB1-related proteins with the ends of growing microtubules in non-neuronal and neuronal cells.
Keywords: microtubules, microtubule dynamics, microtubule plus end tracking proteins, cytoskeleton, neurons, neuronal differentiation
Introduction
Neurons are signaling cells with a unique polarized composition, which is paralleled by a highly specific microtubule (MT) network organization. For example, MTs are organized nonuniformly in differentiated dendrites, with dynamic (plus) ends pointing both distally and toward the cell body (Baas et al., 1988). In axons, on the other hand, MTs are organized unidirectionally, with all plus ends pointing toward the growth cone. This organization is different from that of other cell types in which minus ends of MTs often are, for example, embedded in the MT organizing center (MTOC), whereas plus ends explore the cytoplasm. In view of these differences and the fact that the distance between cell body and neuronal periphery can be quite extensive, it is logical to assume that the localization and transport of membrane-bound organelles over MT tracks, the formation and maintenance of the neuronal MT cytoskeleton itself as well as the transport of cytoskeletal elements might be organized in a unique manner.
The dynamic properties of MTs are regulated by a large number of cellular factors, including MT-associated proteins (MAPs) and MT motors. Recently, a novel type of microtubule binding has attracted considerable interest, both with respect to the regulation of MT dynamics and intracellular membrane transport over MTs. Live imaging studies have shown that an increasing number of MT regulatory proteins from various organisms specifically associates with the distal ends of growing MTs. This characteristic behavior was documented first for CLIP-170 (cytoplasmic linker protein-170) (Perez et al., 1999); it was explained by a mechanism called “treadmilling” or “plus end tracking,” and MT plus end-binding proteins that show this behavior have since been called “plus end-tracking proteins” or +TIPs (Schuyler and Pellman, 2001). Treadmilling involves the incorporation of +TIPs into growing MT ends either by copolymerization with tubulin or by recognition of a specific feature of the MT distal end and subsequent release from an older, more proximal part of the MT. The kinetics of association and release may be regulated by post-translational modifications of the +TIP, for example, phosphorylation (Rickard and Kreis, 1991; Hoogenraad et al., 2000;Tirnauer et al., 2002; Vaughan et al., 2002). In addition to CLIP-170, CLIP-115, and CLIP-associated proteins or CLASPs (Akhmanova et al., 2001), LIS1 (Han et al., 2001; Coquelle et al., 2002), EB1 (end-binding protein 1) and its associated protein APC (Mimori-Kiyosue et al., 2000a,b) as well as components of the dynein/dynactin motor complex (Han et al., 2001; Vaughan et al., 2002) appear to undergo treadmilling behavior. Besides their role in the regulation of MT dynamic events, some of these +TIPs also may exert an influence on intracellular transport of vesicular cargo via their control over MT tip association of the dynein/dynactin complex (Valetti et al., 1999; Vaughan et al., 1999).
All live imaging studies so far have documented the behavior of +TIPs, fused to GFP (green fluorescent protein) (GFP+TIPs), in non-neuronal cells. Because the growth rates of MTs as measured with various GFP+TIPs are similar to values obtained after the injection of fluorescently labeled tubulin (see Komarova et al., 2002a), the dynamic behavior of MTs is not altered significantly by low expression levels of the +TIPs. Thus GFP+TIPs appear to be excellent tools for studying MT growth dynamics in living cells. Using these markers in neurons, we have addressed the important question of whether changes in the MT network during neuronal differentiation are reflected by changes in MT polymerization dynamics in different neuronal compartments. We first show that endogenous CLIPs and EB1-related proteins are present at the distal ends of neuronal MTs. Using EB3-GFP as a marker, we subsequently highlight polymerizing MTs in two different types of cultured neurons. Our results support the conclusion that local MT polymerization events occur throughout neuronal differentiation. We propose that similar mechanisms control the association of +TIPs with MT distal ends in neurons and non-neuronal cells.
Materials and Methods
Antibodies and immunocytochemistry. For immunocytochemistry the cultured neurons were washed in PBS and fixed first in freshly prepared −20°C methanol/1 mm EGTA, followed by a 2% paraformaldehyde fixation at room temperature (20 min for each step). Subsequent antibody incubation and washing steps have been described, as have the CLIP-115- and CLIP-170-specific antisera (numbers 2221, 2238, and 2360) (Hoogenraad et al., 2000; Coquelle et al., 2002). A combination of these sera was used, each in a 1:1000 dilution. For EB3 the novel polyclonal antibodies (number 02-1005-07) were raised in rabbit, as described previously (Hoogenraad et al., 2000), using glutathione S-transferase-EB3 (GST-EB3) as an antigen. Monoclonal antibodies against EB1 (Transduction Laboratories, Lexington, KY), actin (Chemicon, Temecula, CA), acetylated α-tubulin, tyrosine α-tubulin, and β-tubulin (Sigma, St. Louis, MO) were used in a dilution of 1:100. FITC-conjugated goat anti-rabbit (1:100; Nordic Laboratories, Tilburg, The Netherlands) and Alexa 594-conjugated goat anti-mouse (1:500; Molecular Probes, Eugene, OR) were used as secondary antibodies. Signals were captured as described previously (Hoogenraad et al., 2000; Akhmanova et al., 2001), using either a Leica DMRBE microscope with a 100× oil immersion lens (numerical aperture, 1.3) and a Hamamatsu C4880 camera or a Zeiss (Jena, Germany) LSM510 confocal microscope with a 63× oil immersion lens (numerical aperture, 1.4). EB3 and EB1 fluorescent intensities at plus ends were analyzed with Image-Pro Plus, version 4.5 (Media Cybernetics, Silver Spring, MD). After subtraction of the background a threshold was put in the red fluorescent channel to select the plus ends. Then the red and green intensities were measured at the same spot. Small spots (≤0.1 μm2) and large spots (>1 μm2) were discarded from the analysis. The confocal images representing EB3-tubulin costaining (see Fig.2H,I) were reconstructed with a Quick Maximum Likelihood Estimation (Huygens2 pro, Scientific Volume Imaging, Hilversum, The Netherlands) that is based on a theoretical point spread function. Different color channels were analyzed with different point spread functions. After analysis the channels were merged again.
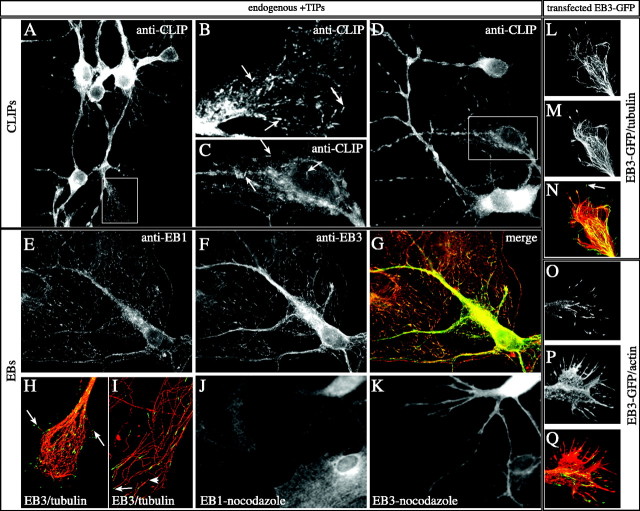
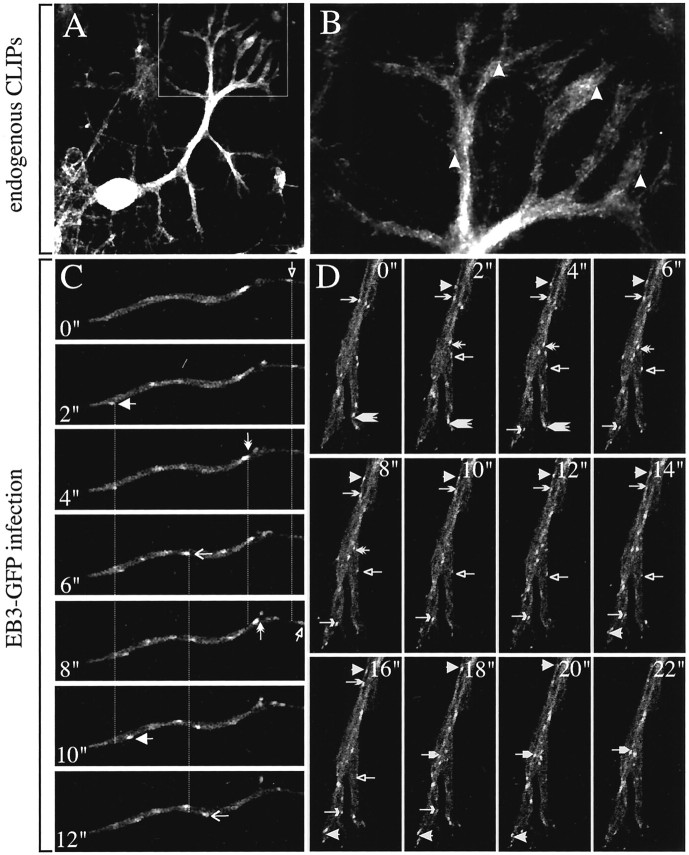
Fig. 2.
+TIPs in hippocampal neurons. A–K, Hippocampal neurons of 2–6 d in culture were fixed and incubated with a mixture of antibodies recognizing both CLIP-115 and CLIP-170 (A–D), with an antibody mixture recognizing EB1 and EB3 (E–G), with a mixture recognizing EB3 and β-tubulin (H, I), or with the single EB1 (J) and EB3 (K) antibodies.B (growth cone) and C (neurites) are magnifications of the rectangles in A andD, respectively. Examples of comet-like CLIP dashes are indicated by arrows. In G the merged image of EB1 (E) and EB3 (F) staining is shown, in which EB1 is inred and EB3 is in green. Clear comet-like labeling is visible with both antibodies in the neuron and in the two glial cells. In H (neuronal growth cone) andI (cell periphery of a glial cell) deconvolution of confocal images is used to demonstrate clearly the EB3 localization to MT distal ends. The large arrows in H andI indicate clear examples of such EB3 localization. Thesmall arrow in I points toward a MT end, which is not labeled by EB3. Given that the other end of this small MT is stained, we presume that this end represents a MT minus end. The cell periphery of the glial cell in I is particular in that it contains several of such “free” MTs. In Jand K nocodazole was added to the culture medium before fixation of the cells. Note that the typical comet-like staining of EB1 and EB3 has vanished after this brief treatment. L–Q, Hippocampal neurons, transfected with EB3-GFP, were fixed 3–4 d after plating and stained with cytoskeletal markers. Costaining of EB3-GFP (L) in a growth cone with antibodies against tyrosinated α-tubulin (M) reveals that the GFP signals are located at the tip of MTs (the arrowindicates an example of such a tip in the merged image inN). Costaining of EB3-GFP (O) in a transfected neuron with antibodies against actin, followed by Alexa 594-conjugated secondary antibody incubation (P), reveals that EB3-GFP-positive MT plus ends are present in the growth cone and sometimes are detected in actin-rich filopodial extensions (see merged image inQ).
GFP–fusion constructs. The GFP-CLIP-115 andGFP-CLIP-170 constructs have been described (Perez et al., 1999; Hoogenraad et al., 2000; Akhmanova et al., 2001). Mutant GFP-CLIP-170 (called GFP-CLIP-170ΔHind), lacking the C-terminal domain and part of the coiled-coil region of rat CLIP-170, was generated by HindIII/BamHI digestion of GFP-CLIP-170 in the pEGFP vector, removal of the insert containing the 3′ CLIP-170 sequence, and religation of blunted ends. For cloning ofEB1, EB2, and EB3 cDNAs gene-specific primers were designed, with restriction sites for in-frame cloning after PCR amplification into pEGFP-N1(Clontech, Palo Alto, CA). Primer sequences were based on the following accession numbers: AW106491 (Image clone 2225780, mouse EB1); AW231083 (Image clone 2644901, mouseEB2 or RP1); AA2892 (Image clone 714028; humanEB3 cDNA). Western blot analysis of COS-1 cells transfected with the different cDNAs (Hoogenraad et al., 2000) shows that all EB-GFP fusions are produced with an expected molecular weight (see Fig.1).
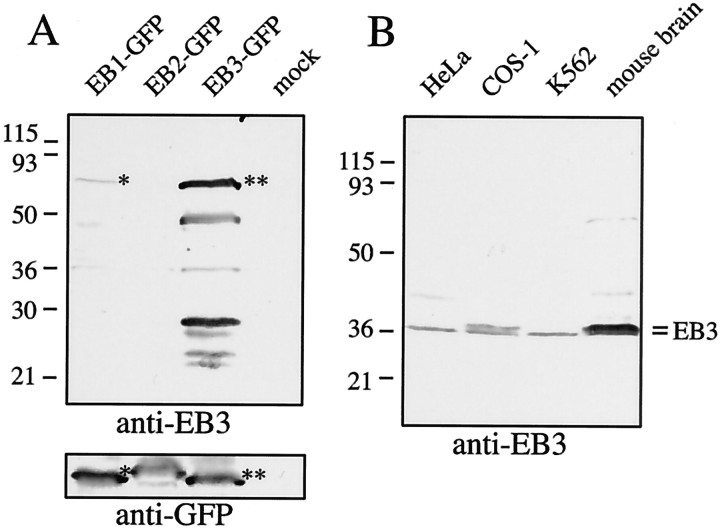
Fig. 1.
Characterization of anti-EB3 antiserum. Novel anti-EB3 antibodies were raised in rabbit against GST-EB3 and characterized on Western blot. A, COS-1 cells were mock transfected or transfected with EB1, EB2, and EB3 fused to GFP. Thebottom panel (Western blot with anti-GFP antiserum) demonstrates that each fusion protein is expressed at a similar level. The top panel (Western blot with the novel anti-EB3 antiserum) shows that anti-EB3 antibodies recognize full-length EB3-GFP (marked by a double asterisk). EB1-GFP (marked by asingle asterisk) also is weakly recognized, whereas EB2-GFP is not. B, Equal amounts of whole-cell protein extract from HeLa, COS-1, and K562 cells and from mouse brain are analyzed. Two isoforms of EB3, which are expressed more abundantly in brain extracts, are recognized by the novel anti-EB3 antiserum.
Cell culture and protein expression. COS 1 cells were cultured and transfected as described previously (Akhmanova et al., 2001). Mouse hippocampal neurons were isolated from embryonic day 17 (E17) embryos and cultured on the basis of published procedures (Dotti et al., 1988; de Hoop et al., 1998). Neurons were electroporated immediately after isolation, using highly purified DNA (EndoFree Maxi Prep, Qiagen, Hilden, Germany). Thus hippocampi were dissected from fetal brains, treated with 0.25% trypsin for 15 min at 37°C, washed in Ca/Mg-free HBSS, and dissociated by repeated passage through a constricted Pasteur pipette. Directly after isolation 25 μg of plasmid DNA was added to the suspension of neurons in HBSS (0.4 ml), and neurons were electroporated in a Bio-Rad (Hercules, CA) Gene Pulser at 850 V, 25 mF, and 200 Ω (time constant, 0.8 sec). Subsequently, neurons were plated on 22 mm poly-l-lysine-coated coverslips. Plating medium contained 2 mm sodium butyrate for 17–24 hr to enhance the expression of cytomegalovirus CMV promoter-driven genes. After this time the coverslips with the neuronal cultures were placed upside down in 3.5 cm dishes containing N2 medium (de Hoop et al., 1998) conditioned by a confluent monolayer of astroglial cells. Small paraffin droplets on the coverslips prevented the neurons from making contact with the glia.
Mouse Purkinje cell (PC) neurons were isolated from E18 FVB/N embryos. Cerebella were washed twice in HBSS and then treated with 0.5% Trypsin-EDTA for 13–15 min at 37°C. After centrifugation the cerebella were washed with growth medium, i.e., basal modified Eagle's (Invitrogen, San Diego, CA), containing 10% horse serum (Invitrogen), 10 μg/ml gentamycin, 0.5% glucose (Sigma), 1 mm sodium pyruvate (Sigma), and 1% N2 supplements (Invitrogen). Cells were triturated with fresh growth medium, filtered through a 70 μm nylon cell strainer, and plated in 1 ml aliquots (1 × 106 cells/ml) onto four-well LabTek II chambered cover glasses (Nalge Nunc, Rochester, NY) coated overnight with poly-l-ornithine (500 μg/ml; Sigma). Neuronally enhanced cultures were obtained by replacing one-half of the medium at 2 d in vitro (2 DIV) and then twice per week with serum-free culture medium containing 2% B27 supplement (Invitrogen).
To express foreign proteins in PC neurons, we used Semliki Forest virus (SFV)-mediated gene delivery (Ehrengruber et al., 1999; Lundstrom et al., 2001a,b). The EB3-GFP construct was cloned into the pSFV2 vector according to the manufacturer's instructions (Invitrogen). Constructs were packaged into SFV replicons, using coelectroporation of helper and vector RNA into baby hamster kidney-21 cells. Cultured PCs (or hippocampal neurons) were infected between 10 and 17 DIV by the addition of 1 and 5 μl of SFV infectious replicons to the cultures.
Live cell imaging and analysis of GFP movements. Cells were analyzed at 37°C on a Zeiss LSM510 confocal laser-scanning microscope as described previously (Akhmanova et al., 2001). In most experiments the optical slice (z-dimension) was set to 1 μm. Other settings that were used (e.g., laser intensity and gain value) differed slightly in the various experiments and were adapted to obtain optimal signal-to-noise ratios. Nocodazole (Sigma) and taxol (Molecular Probes) were added at 0.1 or 10 μm final concentrations. Images of GFP+TIP movements in transfected cells were acquired every 1–3.5 sec. Image capture time was typically <1 sec in COS-1 cells. In contrast, capture times for neuronal imaging were in the range of 1–3.5 sec because of the lower signals that were present at MT tips in these cells. Images were recorded and movies were assembled by using LSM510 software.
Distances traveled by GFP+TIP dashes were measured in different neuronal areas. The velocity of the different +TIP dashes was calculated by dividing the distances traveled by time spent traveling. We included only dashes that could be followed for at least three consecutive frames. To measure the percentage of forward or anterograde EB3-GFP movements (i.e., movements toward the distal end of neurons) and backward or retrograde movements (i.e., toward cell body of neurons), we measured fluorescent displacements occurring within one complete time-lapse movie within selected areas of the neuron (i.e., in different neuronal compartments). In this analysis all movements were included irrespective of how many consecutive frames they lasted. Also, each fluorescent movement within the selected area was traced to determine the average number of consecutive frames that we could follow the displacements. Percentages of forward and backward movement are expressed either relative to the total number of fluorescent movements observed or relative to the number of fluorescent MT distal ends (see Table 2). Because the values that were obtained do not differ considerably, this analysis suggests that the average duration of forward and backward displacements is similar.
Table 2.
Velocity of EB3-GFP dashes in neurons and glia
| Neuronal compartment | Average velocity2-a (number of dashes measured) |
|---|---|
| Hippocampal cultures | |
| Cell body | 0.22 ± 0.06 (64) |
| Neurites | 0.22 ± 0.04 (52) |
| Neurite forward | 0.22 ± 0.06 (91) |
| Neurite backward | 0.21 ± 0.08 (20) |
| Growth cone | 0.20 ± 0.07 (35) |
| Glia | 0.43 ± 0.11 (63) |
| Retracting axons2-b | 0.35 ± 0.06 (37) |
| PC cultures | |
| Dendrite forward | 0.24 ± 0.07 (60) |
| Dendrite backward | 0.26 ± 0.07 (21) |
| Axon forward | 0.21 ± 0.04 (75) |
| Axon backward | 0.24 ± 0.04 (8) |
| Cell body | 0.22 ± 0.05 (20) |
| Golgi cell | 0.28 ± 0.05 (12) |
| Glia | 0.44 ± 0.11 (42) |
μm/sec ± SD.
EB3-GFP velocity derived from infected hippocampal cultures.
Results
+TIP localization in cultured neurons
Several +TIPs have been shown to localize to the ends of a subset of MTs in fixed cultured cells in line with their plus end tracking behavior in live cells. We wanted to analyze whether a similar MT distal end localization is present in neurons, because this has not yet been described. We focused our analysis on CLIP-115 and CLIP-170, EB1 and EB3. Both CLIP-115 (De Zeeuw et al., 1997) and EB3 (Nakagawa et al., 2000) mRNAs are enriched in the CNS, making the analysis of the distribution pattern of these proteins particularly interesting. Antibodies against CLIP-115 and CLIP-170 have been described previously (Hoogenraad et al., 2000), and a commercial monoclonal antiserum against EB1 is available. To obtain EB3 localization data, we raised a novel polyclonal antiserum in rabbit. When tested on Western blot, this anti-EB3 antiserum specifically recognizes EB3-GFP in transfected COS-1 cells (Fig. 1A, lane 3). Transfected EB1-GFP (Fig. 1A, lane 1) also is recognized weakly, whereas EB2-GFP (Fig. 1A, lane 2) is not detected. In extracts from different cell lines and from mouse brain the novel anti-EB3 antiserum mainly recognizes two closely migrating EB3 isoforms of ∼36 kDa (Fig. 1B), which are enriched in the brain extract as compared with the cell line lysates and likely represent the two splice forms of EB3 that have been described previously (Su and Qi, 2001). Together, these results indicate that the novel antiserum is specific for EB3.
To determine whether endogenous CLIPs associate with the ends of MTs in neurons, we incubated fixed mouse hippocampal cell cultures with antibodies against CLIP-115 or CLIP-170. However, labeling with the individual CLIP antibodies failed to demonstrate a clear plus end staining pattern in neurons, in contrast to the bright staining observed in neighboring glial cells (data not shown). We therefore attempted the staining of neurons with a mixture of the different antibodies. This labeling procedure revealed cytoplasmic labeling, most intense in cell bodies, in addition to a clear staining of comet-like dashes in cell bodies, dendrites, axons, and growth cones (Fig.2; arrows highlight examples of this staining). Double labeling with anti-tubulin antibodies identified these dashes as MT distal ends (data not shown). These data indicate that endogenous CLIPs associate with MT plus ends in cultured hippocampal neurons. However, the labeling is substantially weaker than in non-neuronal cell types, suggesting that less protein is present per MT tip in neurons. This may be the main reason why plus end staining of CLIP-115 in cultured neurons was not observed previously (De Zeeuw et al., 1997).
To analyze whether EB-related proteins also associate with MT distal ends in neuronal cultures, we incubated fixed cells with a mixture of monoclonal antibodies against EB1 and the new polyclonal anti-EB3 antiserum (Fig. 2E–G). In these experiments colocalization is observed of EB1 (Fig. 2E) and EB3 (Fig. 2F) on comet-like dashes in glia and in neurons (merge in Fig. 2G). Because EB1 is a verified marker of growing MT ends in non-neuronal cells (Mimori-Kiyosue et al., 2000b), these data suggest that the same holds true for EB3. Quantification of EB1 and EB3 fluorescent intensity on comets reveals that EB1 signal intensity (expressed in arbitrary units per comet) is similar in glia and neurons (i.e., ∼0.95 AU/comet), whereas EB3 signal increases from ∼0.5 AU in glia to 1 AU in neurons (>1000 comets measured in three glia and four neurons). Although this quantification does not reveal the absolute levels of EB1 or EB3 in neurons and glia, the data suggest that in neurons more EB3 is associated per MT distal end than in glia.
Costaining of EB3 antiserum with monoclonal anti-β-tubulin antibodies reveals that, in those cases in which individual MTs can be distinguished within the dense MT network, the EB3-positive dashes are always located at the ends of MTs, both in neurons (Fig.2H) and glia (Fig. 2I). In the latter example some of the MTs, which are visible, are not embedded in the MTOC. On those MTs EB3 staining is observed only at one of the two ends, which presumably represents the growing end.
Nocodazole and taxol are drugs that interfere with MT dynamics in opposing manners (for review, see Downing, 2000). Nocodazole acts by sequestering tubulin dimers, eventually causing a collapse of the MT network, whereas taxol stabilizes the MT network by specifically binding to MTs. Application of low amounts of both drugs completely abolishes the binding of +TIPs to the ends of growing MTs (Perez et al., 1999; Mimori-Kiyosue et al., 2000b; Akhmanova et al., 2001) despite the fact that MTs are still present and MT-based motors are active. The addition of low concentrations of these drugs to hippocampal cultures leaves the MT network intact, yet both EB1- and EB3-positive comets disappear (Fig. 2J,K; data not shown). Taken together, our results suggest that in neurons, like in non-neuronal cells, CLIPs and EB-related proteins specifically associate with the ends of growing MTs.
Characterization of GFP+TIP behavior in transfected COS-1 cells
Because endogenous +TIP localization (and, by implication, function) is conserved in non-neuronal and neuronal cultures, we next set out to compare GFP+TIP behavior, focusing again on CLIPs and EB1-related proteins. On the basis of published results we inserted the GFP tag at the N terminus of the CLIPs and at the C terminus of the EB1-related proteins (Fig. 3). In the case of GFP-CLIP-170, its dynamic behavior has been described in several reports (Perez et al., 1999; Akhmanova et al., 2001; Komarova et al., 2002a), whereas in the case of EB1-GFP, experiments in liveXenopus A6 cells and human neuroblastoma N2A cells have documented its dynamic behavior (Mimori-Kiyosue et al., 2000b; Morrison et al., 2002). However, neither for GFP-CLIP-115 nor for EB2-GFP and EB3-GFP have live imaging studies yet been reported. We therefore first compared the behavior of all of these proteins in transfected COS-1 cells, which were analyzed under a confocal microscope 20–48 hr after transfection (Fig. 3).
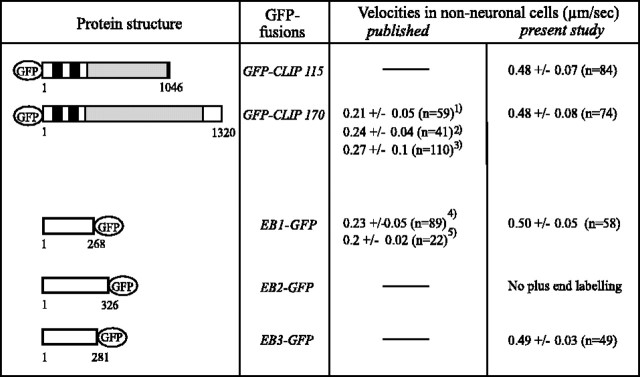
Fig. 3.
Velocities of GFP+TIP fusion proteins in non-neuronal cells. COS-1 cells, expressing the indicated GFP+TIP fusion proteins, were monitored at 37°C on a ZeissLSM510 confocal microscope and analyzed for GFP+TIP velocities. Only cells expressing low levels of the fusion proteins were investigated. For comparison, previously published values are indicated [1Perez et al. (1999); 2Akhmanova et al. (2001); 3Komarova et al. (2002a);4Mimori-Kiyosue et al. (2000); 5Morrison et al. (2002)]. MT-binding domains (black bars) and coiled-coil regions (gray bars) are indicated in the CLIPs. The microtubule-binding motif has not been identified clearly in EB1-related proteins.
For live cell imaging we studied only cells with low expression levels of the GFP fusions, but even under these conditions EB2-GFP binds all along MTs and has no preference for MT distal ends (data not shown). The other GFP+TIPs, however, all move in comet-like dashes, mainly from the presumptive MTOC to the cell periphery, consistent with their preferential association with the growing ends of MTs [see movies 1 (www.eur.nl/fgg/ch1/galjart/jon.html) and 2 (available atwww.jneurosci.org) for GFP-CLIP-115 and EB3-GFP behavior, respectively; the behavior of the other fusion proteins has been published previously and is not shown here]. Costaining with tubulin antibodies in fixed cells reveals that these comets represent MT distal ends, and low doses of nocodazole or taxol abolish GFP+TIPs staining at MT ends in transfected COS-1 cells (data not shown). We conclude that, of the five GFP markers that were analyzed, four (GFP-CLIP-115, GFP-CLIP-170, EB1-GFP, and EB3-GFP) faithfully label the ends of growing MTs in COS-1 cells.
The rates of GFP+TIP movement reported here (Fig. 3) are higher than values published previously (Perez et al., 1999; Mimori-Kiyosue et al., 2000b; Akhmanova et al., 2001; Komarova et al., 2002a; Morrison et al., 2002; Vaughan et al., 2002). This might be attributable to the different cell types used in some of the studies. In addition, we found that slight differences in temperature and/or in culture medium conditions affect these speeds [Fig. 3; compare GFP-CLIP-170 value inAkhmanova et al. (2001) with current value]. A similar result has been obtained by using GFP-p150Glued as a plus end marker (Vaughan et al., 2002). Interestingly, the measured velocities of the different GFP fusion proteins are quite similar in transfected COS-1 cells in the current experiments (Fig. 3) despite the reportedly different functions of CLIPs and EB1-related proteins (Komarova et al., 2002b; Rogers et al., 2002; Tirnauer et al., 2002). These results indicate that at low levels of overexpression these GFP fusion proteins do not affect MT growth dynamics to a great extent.
GFP+TIP expression in mouse hippocampal neurons
Transfection of hippocampal neurons with GFP-CLIP-115, -CLIP-170, -EB1, and -EB3 yields comet-like GFP dashes in cells expressing low amounts of these proteins (Fig. 2L,O; data not shown). These dashes are present in all neuronal compartments, indicating that growing MTs are located throughout neurons. Importantly, the distribution of the different GFP+TIP fusion proteins is similar to that observed with the antibodies. Consistent with these data, we found that the MT plus end signal, as detected with EB3-GFP, is brighter than that detected with the other +TIP fusion proteins (data not shown).
EB1-GFP and GFP-CLIP-115 distribute along MTs at higher expression levels (data not shown). At these levels GFP-CLIP-170 aggregates in patches (data not shown), similar to its behavior in transfected non-neuronal cells (Pierre et al., 1994). We therefore concentrated on EB3-GFP for our further analysis, because this fusion protein remains associated with MT distal ends unless expressed at very high levels. Double-labeling studies in the growth cone with antibodies against tyrosinated (unmodified) α-tubulin reveal that EB3-GFP is located at distal segments of a subset of MTs (Fig. 2L–N; one example of MT plus end labeling by EB3-GFP is indicated with anarrow). We also visualized EB3-GFP dashes and actin simultaneously, using anti-actin antibodies. Both EB3-GFP-positive dashes (Fig. 2O) and actin-rich extensions (Fig.2P) are detected clearly. Most, but not all, of the filopodial extensions in growth cones are devoid of EB3-GFP-positive comet-like dashes (Fig. 2Q). These data indicate not only that EB3-GFP is a faithful marker of MT plus ends but that low-level expression of the fusion protein does not influence neuronal growth cone formation to a great extent. Comparison of movements of EB3-GFP dashes in transfected COS-1 cells reveals no aberrant effect of the expressed fusion protein on MT growth rates with respect to other established MT plus end markers (Fig. 3). These data together suggest that EB3-GFP is the best suitable fusion protein for live studies of MT dynamics in transfected neurons.
To investigate growing neuronal MTs, we electroporated hippocampal neurons with EB3-GFP and examined live neurons with the confocal microscope 2–6 d later. Many moving fluorescent dashes are observed in these cells in all neuronal compartments, i.e., cell bodies, neurites, and growth cones (movie 3; available atwww.jneurosci.org). Still images of EB3-GFP movements from movie 3 are depicted in Figure 4. Application of nocodazole and taxol abolishes the EB3-GFP dashes in live neurons (data not shown), again suggesting that EB3-GFP specifically associates with the ends of growing MTs.
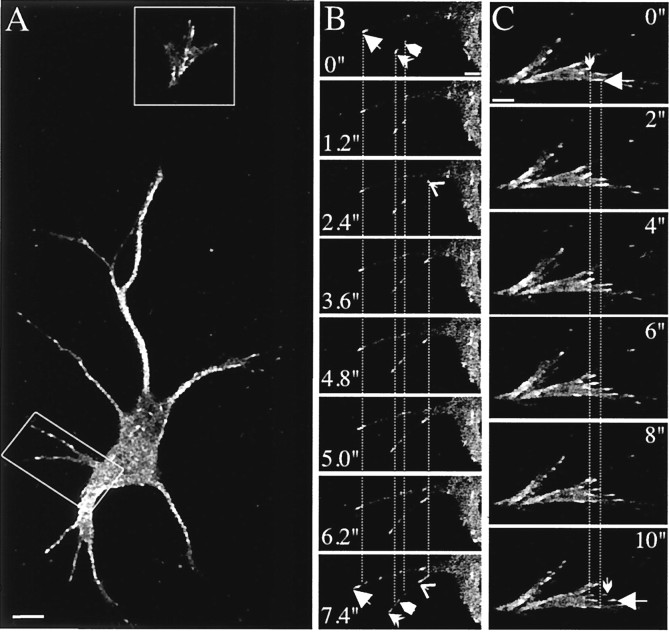
Fig. 4.
EB3-GFP localization in transfected hippocampal neurons. Hippocampal neurons were transfected with EB3-GFP, and live cells were analyzed 3–4 d later by confocal microscopy in a 37°C chamber. EB3-GFP comets are seen to move in all neuronal compartments. This figure contains images of movie 3 (available atwww.jneurosci.org). The rectangles in Arepresent the areas magnified in B (cell body and dendrites) and C (growth cone). The images inB were acquired every 1.2 sec and every 2 sec inC. Arrows and arrowheadsin these panels indicate the beginning and end positions of a selected number of EB3-GFP dashes. Dotted lines help to distinguish the movements of these dashes. The life time of the individual dashes differs, but each moves with an average velocity of ∼0.2 μm/sec. Scale bars: A, 10 μm;B, 5 μm; C, 2 μm.
Many of the EB3-GFP-positive fluorescent stretches in the neuronal cell body appear to move randomly (movie 3; available atwww.jneurosci.org), consistent with results that use fixed neurons (Fig. 2). After entry into neurites the MT growth becomes restricted to the long axis and is therefore directional. Within neurites some comets touch the plasma membrane and then disappear, whereas other dashes move alongside the membrane for a number of frames. Most of the dashes move from the cell body toward the distal neurites (forward or anterograde growth); however, backward or retrograde movement (i.e., back to the cell body) is observed also. Retrograde MT growth constitutes ∼35% of the total moving dashes or frames in proximal neurites of transfected hippocampal neurons (Table1). Retrograde movements generally decline in more distal parts of the neurons (Table 1).
Table 1.
Forward and backward movement of EB3-GFP in neurons
| Compartment | Dashes | EB3-GFP displacements | Displacements per dash1-d | |||||
|---|---|---|---|---|---|---|---|---|
| na | % f1-b | % b1-c | n | % f | % b | f | b | |
| Hippocampal cultures | ||||||||
| Proximal neurite | 26 | 65 | 35 | 119 | 71 | 29 | 4.9 | 3.9 |
| Distal neurite | 15 | 87 | 13 | 55 | 87 | 12 | 3.7 | 3.5 |
| Distal axon | 9 | 100 | 0 | 37 | 100 | 0 | 4.1 | |
| Distal axon1-e | 47 | 83 | 17 | 137 | 83 | 17 | 2.9 | 2.9 |
| Growth cone | 7 | 100 | 0 | 28 | 100 | 0 | 4 | |
| PC cultures | ||||||||
| Proximal dendrites | 54 | 65 | 35 | 145 | 64 | 36 | 2.6 | 2.7 |
| Distal dendrites | 51 | 75 | 25 | 151 | 74 | 26 | 2.9 | 3 |
| Axon 1 | 20 | 100 | 0 | 65 | 100 | 0 | 3.25 | |
| Axon 2 | 13 | 85 | 15 | 40 | 83 | 17 | 3 | 3.5 |
Number (n) of dashes observed.
Percentage (%) of forward (f) moving dashes.
Percentage (%) of backward (b) moving dashes.
EB3-GFP displacements per dash were calculated by dividing the number of displacements by the number of dashes. With an average acquisition time of 2.5 sec/frame and average velocity of 0.22 μm/sec, the average displacement per dash is ∼1.65 μm.
Distal axon near growth cone.
Behavior of EB3-GFP in differentiated Purkinje cell neurons
As an alternative to the hippocampal system we studied cerebellar PCs, because these neurons have a morphologically distinct dendritic compartment as compared with hippocampal neurons. Using immunostaining against calbindin-D28K, a marker specific for PCs in cerebellar cultures, we confirmed that healthy, viable cells with good dendritic arborizations and distinguishable axons are detectable as early as after 10 DIV (data not shown). Staining with a mixture of the three anti-CLIP antibodies indicates that these proteins are present in the cell bodies and dendrites of PCs (Fig.5A,B). In peripheral dendrites the labeling pattern resembles the decoration of MT ends that we observed in other cell cultures (Fig. 5B; seearrowheads). MT plus end staining also is detected with EB antibodies (data not shown).
Fig. 5.
CLIP and EB3-GFP localizations in Purkinje cells.A, B, PC neurons of 10–17 d in culture were fixed and incubated with a mixture of antibodies recognizing both CLIP-115 and CLIP-170. A, The distinct initial dendritic arborization of the PCs is highly visible, indicating CLIP accumulation in this region of PCs. B, A comet-like labeling is visible in the more peripheral parts of the dendrite (examples of comet-like dashes are indicated by arrowheads).C, D, PC neurons of 10–17 DIV were infected with SFV-EB3-GFP, and the cells were analyzed 5–8 hr after infection by confocal microscopy in a 37°C chamber. C, Movements of EB3-GFP dashes in an axon are shown (each dash is indicated by a distinct arrow). Images were acquired every 2 sec. Anterograde movement of EB3-GFP dashes toward the growth cone is toward the right. D, Images (derived from movie 4; available atwww.eur.nl/fgg/ch1/galjart/jon.html) demonstrate bidirectional movement of EB3-GFP dashes in dendrites (each dash is indicated by a distinctarrow).
We used the SFV vehicle (Lundstrom et al., 2001b) to deliver EB3-GFP to highly differentiated PC neurons in culture. With this infection system live imaging studies of EB3-GFP movements are optimal ∼5–8 hr after infection, because at later time points expression of the fusion protein becomes too high and MTs are bundled completely. At 5–8 hr after infection we observed bidirectional EB3-GFP movements in PC dendrites [Fig. 5D, Table 1; see also movies 4 (www.eur.nl/fgg/ch1/galjart/jon.html) and 5 (available atwww.jneurosci.org)], whereas in axons most, but not all, movements are unidirectional (Fig. 5C; Table 1). A similar movement is detected in axons of infected hippocampal neurons of 10 DIV (movie 6; available at www.jneurosci.org). These results indicate that the directionality of EB3-GFP movements actually reflects the organization of the MT cytoskeleton in the different neuronal compartments (Baas et al., 1988, 1989).
Atypical behavior of EB3-GFP in neurons
Although most of the EB3-GFP comets move either anterogradely or retrogradely and in growth cones some dashes appear to extend into filopodia-like extensions, on rare occasions we observed pausing and/or oscillating GFP-positive dashes (see, for example, some of the EB3-GFP dashes in the growth cone in movie 3; available at www.jneurosci.org). In these cases we cannot determine whether EB3-GFP is on a pausing, shrinking, or growing MT end (which is pulled backward) because both anterograde and retrograde movements of MTs have been observed in growth cones (Dent et al., 1999).
Studies with SFV-mediated EB3-GFP delivery revealed movement of the fusion protein in highly dynamic hippocampal axons (movie 7; available at www.jneurosci.org), with a morphology strikingly similar to that of the recently described retracting axon (He et al., 2002). Some of the EB3-GFP dashes at the end of the axonal shaft of these typical axons move on curved tracks, which might represent curved and bent MTs. In addition, rapid EB3-GFP excursions into filopodia-like protrusions can be observed. In most instances the EB3-GFP signal disappears with collapse of the filopodium. We conclude that even under conditions that resemble axonal retraction EB3-GFP dashes (and, by implication, MT growth) persist.
Analysis of EB3-GFP movements
EB3-GFP dashes in neurons are observed on average for two to five frames or displacements (see average number of displacements per EB3-GFP dash in Table 1). The average number of displacements per dash is similar for forward and backward movements. For quantitation of the speed of EB3-GFP movements (and thus MT growth rates), we included only dashes that could be followed clearly for three frames or more. In electroporated hippocampal neurons the distance that these EB3-GFP dashes travel is comparable, irrespective of the compartment analyzed (Fig. 6A). In these neurons ∼30–50% of the dashes can be followed for 2 μm (Fig.6A). Taken together, the data in Table 1 and Figure6A indicate that under normal culture conditions EB3-GFP movements are similar in the different neuronal compartments.
Fig. 6.
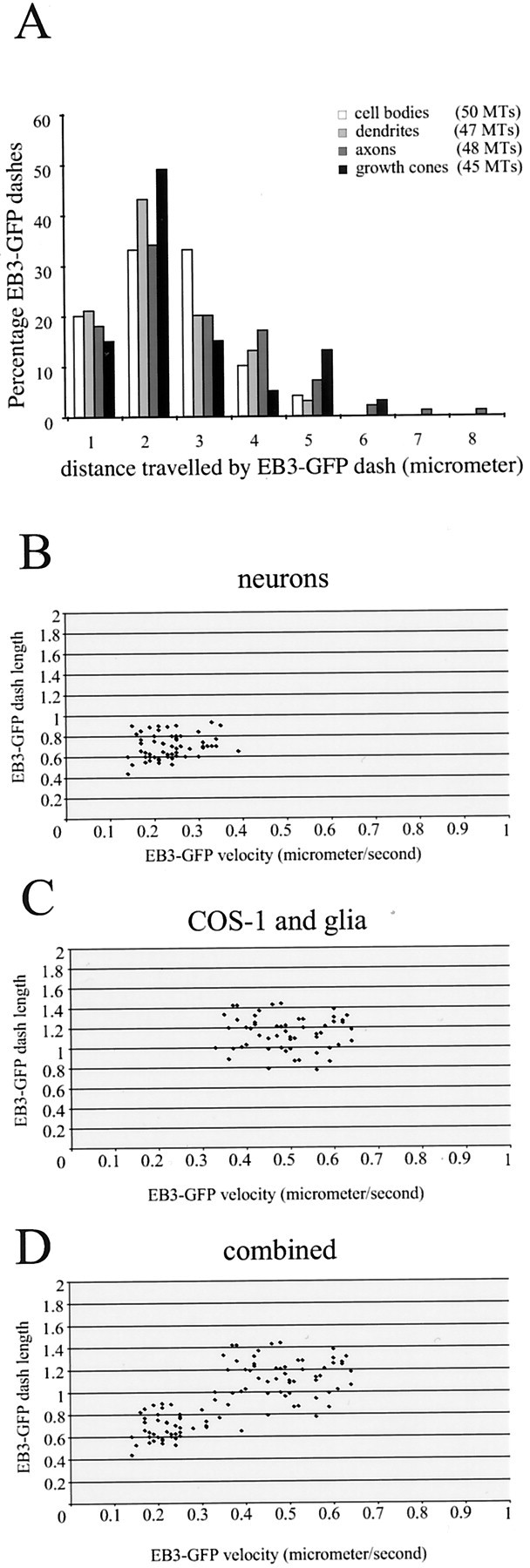
Distances traveled by EB3-GFP dashes in transfected neurons. A, Hippocampal neurons were transfected with EB3-GFP, and the cells were analyzed 3–4 d later by confocal microscopy in a 37°C chamber. The distance that individual EB3-GFP dashes could be followed was measured in different neuronal compartments (cell bodies, dendrites, axons, and growth cones). The total number of individual MTs that were counted is indicated. In all compartments the distance traveled by EB3-GFP dashes is comparable.B–D, The average speed and length of a selected number of EB3-GFP dashes derived from different measurements in neurons (B), COS-1 cells and glia (C), and the combined data (D) are plotted.
Velocities of EB3-GFP movements were calculated in electroporated hippocampal neurons and glial cells of 2–6 DIV, in infected differentiated PCs and neighboring glial cells of >10 DIV, and in the three examples of retracting hippocampal axons (10 DIV) that we observed (Table 2). As a control for the values of MT growth dynamics in neurons, obtained with EB3-GFP, we electroporated neurons with yellow fluorescent protein-tubulin but could not get transfected neurons with a recognizable fluorescent MT array (data not shown). We therefore electroporated neurons with EB1-GFP (movie 8; www.eur.nl/fgg/ch1/galjart/jon.html) and compared its behavior to that of EB3-GFP. Because EB1 and EB3 are highly similar proteins and might have similar effects on the MT network, we also wanted to transfect neurons with a reliable plus end marker that is structurally unrelated to EB3. For this we chose a mutant form of GFP-CLIP-170 (GFP-CLIP-170ΔHind), which lacks the C-terminal metal-binding motif, because studies in non-neuronal cells have shown that this domain is responsible for the formation of patches of overexpressed protein at the cell periphery (Pierre et al., 1994). When expressed in neurons, GFP-CLIP-170ΔHind does not aggregate in patches as does full-length GFP-CLIP-170 but, instead, moves in comet-like dashes (movie 9; www.eur.nl/fgg/ch1/galjart/jon.html).
The measurements show that the average velocity of EB3-GFP movement is very similar in electroporated hippocampal neurons, infected PCs, and in the single Golgi neuron taken along for evaluation (Table 2). In addition, the velocities of EB1-GFP (0.22 ± 0.07 μm/sec; 39 dashes measured) and GFP-CLIP-170ΔHind (0.25 ± 0.05 μm/sec; 35 dashes measured) in electroporated hippocampal neurons are also comparable. EB3-GFP speeds do not vary remarkably within different neuronal compartments, and there is no significant difference in average speed between comets moving in opposite directions (Table 2). The velocities measured in neurons are approximately one-half the speed of movement of EB3-GFP-labeled plus ends in COS-1 cells (compare Fig.3, Table 1). One concern is that slight differences in the system set up and/or culture conditions might cause variations in EB3-GFP velocities in different cell types. We therefore measured velocities in neurons and glia cultured in the same chamber. EB3-GFP speeds in glia are comparable to the values derived from COS-1 cells (Table 1). These data indicate that MT growth rates are generally lower in neurons as compared with glia and COS-1 cells. However, in the three retracting axons the average velocity of EB3-GFP dashes is increased significantly as compared with normal values (Table 1; p < 0.001), yet it is still significantly lower than the values measured in glia (Table 1; p < 0.001).
In the original studies on GFP-CLIP-170 (Perez et al., 1999) and EB1-GFP (Mimori-Kiyosue et al., 2000b), the authors plotted the length of an individual comet versus the speed of movement of that particular dash. A stochastic association and dissociation behavior was detected for both GFP fusion proteins, leading to variation in the length of the individual dashes over time. Still, an increased speed of the dash generally was associated with an increased length of the comet tail. We measured average lengths and corresponding speeds of a large number of the dashes used for the calculations in Table 1 (Fig.6B–D). These data suggest that an increased speed of movement, as observed in the population of EB3-GFP dashes derived from COS-1 cells and glia, correlates with an increased length of EB3-GFP staining of a MT. The average speed of the selected population of COS-1 and glial dashes is 0.49 ± 0.09 μm/sec, which corresponds to an average length of 1.14 ± 0.16 μm. For the neuronal dashes these values are 0.23 ± 0.06 μm/sec and 0.70 ± 0.11 μm, respectively.
Discussion
Here we have investigated the behavior of the distal ends of growing MTs in cultured hippocampal and Purkinje cell neurons mainly by following the movement of GFP-tagged EB3. Our conclusions are based on the assumption that EB3-GFP reliably marks the tips of growing MTs without changing MT growth parameters. Several observations support these statements. First, we show that endogenous EB3 colocalizes with the verified plus end marker EB1 at MT distal ends in cultured hippocampal neurons and glia. The MT plus end association of EB3 is confirmed further by using anti-tubulin antibodies. In contrast to the other +TIPs that have been analyzed, EB3 accumulates preferentially at neuronal plus ends with respect to glial cells, and no other intracellular structure is detected in neurons by the novel anti-EB3 antibodies. The MT plus end association of EB3 is sensitive to nocodazole and taxol, drugs that perturb MT dynamics and that have been shown to abolish MT plus end localization of several +TIPs (Perez et al., 1999; Mimori-Kiyosue et al., 2000b; Akhmanova et al., 2001). Costaining of EB3-GFP and tubulin in fixed neurons and COS-1 cells suggests that the comet-like GFP signals are located at the ends of MTs, in line with the endogenous EB3 localization. The addition of taxol and nocodazole abolishes EB3-GFP localization to MT distal ends, similar to the effect on endogenous EB3 and EB1. Furthermore, EB3-GFP, EB1-GFP, and mutant GFP-CLIP-170 show very similar behavior in transfected hippocampal neurons, suggesting that MT growth rates are not affected by low-level expression of these proteins. In a recent study MT growth rates were measured in the same cell (or cytoplast) that had been injected with fluorescent tubulin and that expressed GFP-CLIP-170 (Komarova et al., 2002a). MT growth rates (∼0.30 μm/sec) were similar with the use of both fluorescent proteins, validating the use of +TIPS as markers for MT growth. Finally, a short report has appeared recently, describing the behavior of EB1-GFP in a neuroblastoma cell line (Morrison et al., 2002). Although this study is less detailed than our report, the properties and behavior of EB1-GFP in cells with neuronal features are consistent with our results in primary cultured hippocampal and PC neurons.
The average distance traveled by an EB3-GFP dash is not directly comparable to the average length of growth of an individual MT. For example, two consecutive and nearby EB3-GFP growth events that we record as separate in fact may represent a growth–pause (or slow growth)–growth event of one individual MT. Moreover, because we used a confocal imaging system for our analysis, in the neuronal cell body some of the plus ends may move in and out of focus because the thickness of the optical slice is ∼1 μm. Thus we may not capture a complete growth spurt of an individual MT in the cell body. Note that in both examples that have been mentioned the EB3-GFP-derived results underestimate the true value of the average length of an MT growth event. Thus it is likely that the latter are higher than the average distance traveled by EB3-GFP, which, on the basis of the data in Table1 and Figure 6A, we estimate to be ∼1–2 μm. The average length of MTs in young hippocampal neurons is ∼4 μm both in minor processes and in an early axon (Yu and Baas, 1994). Although these studies were performed in rat embryonic neurons, they do correlate with our data and suggest that the movement of EB3-GFP reflects the growth of MTs in differentiating neurons.
Except in the case of retracting axons, we find that the speed of growing MTs and the average duration of growth events, as measured by the movement of EB3-GFP, are similar in all neuronal compartments, i.e., cell bodies, dendrites, axons, and growth cones. The graphs in Figure 6B–D indicate a correlation between length of distal MT end staining and speed of EB3-GFP movement and do not suggest the existence of more than one type of EB3-GFP movement. For example, if a significant proportion of EB3-GFP would associate with shrinking MTs, one would expect two populations of dashes to come forward in the graph in Figure 6B, because MT shrinking rates are higher than MT polymerization rates (Komarova et al., 2002a). Similarly, if a significant population of EB3-GFP molecules was transported by molecular motors, we also would expect to see this as a separate population in the graph, but we do not. We therefore propose that similar mechanisms underlie the dynamic association of EB3-GFP with MTs in neurons and non-neuronal cells.
Both in hippocampal neurons of 2–6 DIV and differentiated Purkinje cells we observed anterograde and retrograde movements of EB3-GFP. Most bidirectional EB3-GFP movements were detected in proximal dendrites, whereas in axons the vast majority of dashes moved toward the growth cone (with the noted exceptions). The directionality of EB3-GFP dash movement agrees with observations in rodent hippocampal neurons, in which the differentiation of the axonal and dendritic compartments is accompanied by changes in the polarity of MTs such that in (proximal) dendrites MT polarity is mixed, with approximately one-half of the plus ends distal to the cell body, whereas in axons all MTs have a plus end distal orientation (Baas et al., 1988, 1989). This is further evidence for a preferential association of EB3-GFP with the ends of growing MTs. Taken together, our data suggest that local MT polymerization occurs throughout neurons, irrespective of MT organization. These polymerization events may contribute to the formation of the MT network. However, because EB3-GFP is only a marker for growing MT plus ends, our analysis does not address the issue of movement of MT polymer in axons and dendrites or what percentage it constitutes of the MT assembly process (Baas, 1997; Chang et al., 1999; Terada et al., 2000).
In young hippocampal axons there is a shift toward shorter and longer MTs, and the total content of MTs in the early axon increases ∼10-fold as compared with minor processes (Yu and Baas, 1994). As the axon grows further, MT mass increases even more. We do not find an obvious increase in the amount of EB3-GFP dashes in axons as compared with minor processes in electroporated hippocampal neurons of 2–6 DIV. This suggests that preferential MT stabilization in axons as compared with dendrites, and not increased MT polymerization frequencies, accounts for the increase in total MT polymer. Indeed it has been found that the proportion of stable, nocodazole-resistant MTs in axons is more than twice as high as that in dendrites (Baas et al., 1991) and that preferential stabilization of MTs occurs in the proximal part of axons (Baas et al., 1993). In addition, the labile MT proportion in axons is found as distinct domains at the plus ends of stable MTs (Baas and Black, 1990), and it has been shown that the plus ends of stable MTs serve as the sole nucleating structures for MTs in the axon (Baas and Ahmad, 1992). These data suggest that MT polymerization can occur on existing MTs in axons.
Using SFV-mediated EB3-GFP expression, we were able to document MT growth events in highly differentiated PC (and hippocampal) neurons, and we also recorded EB3-GFP movements in three retracting hippocampal axons. Consistent with the result that MT polymer mass does not diminish significantly in these axons (He et al., 2002), we detected EB3-GFP excursions into the growth cones of these axons, one of which was in the process of collapsing during the recording session (movie 7; available at www.jneurosci.org). Interestingly, MT growth rates are significantly higher in the three retracting axons that we observed as compared with MT growth rates under more static conditions. These increased MT growth rates must be balanced to maintain MT mass. We therefore hypothesize that MT dynamics are affected under conditions of axonal retraction, which may contribute to the curving and bending of MTs observed in these axons.
MT growth rates are determined in part by the concentration of free tubulin present in the cytoplasm of a cell. In neurons the balance of MTs versus free tubulin may be tilted in favor of the MTs such that less cytoplasmic tubulin is present than in non-neuronal cells. This may explain the lower neuronal MT growth rates we have observed. Another remarkable feat is the altered distribution of +TIPs at MT distal ends in cultured neurons versus non-neuronal cells. We have shown recently that a reduction in the levels of one of these +TIPS (i.e., CLIP-115) leads to neuronal dysfunction (Hoogenraad et al., 2002). The development of EB3-GFP as a tool for measuring MT growth rates will allow us to determine in more detail the consequences of a deletion of CLIP-115, as well as that of other +TIPs, on MT growth dynamics in mammalian neurons. This will be particularly interesting in view of the recent report that low-dose application of taxol and nocodazole abolishes the sensitivity to extracellular guidance cues of growth cones from embryonic Xenopus spinal neurons, whereas focal applications of these drugs cause growth cone turning without addition of the cues (Buck and Zheng, 2002).
Footnotes
This research was supported by the Netherlands Organization for Scientific Research (NWO; Grants NWO-MW 903.47.067, NWO-ALW 810.67.012, and NWO-SLW 805–33.311-P), The Dutch Royal Academy of Sciences, and Erasmus University. We thank Dr. Carlos Dotti for his help with setting up the neuronal hippocampal cultures and Michael van der Reijden and Dr. Gerard Borst for their assistance with the generation of the EB3-GFP virus.
Correspondence should be addressed to Dr. Niels Galjart, MGC Department of Cell Biology and Genetics, Erasmus University, P.O. Box 1738, 3000 DR Rotterdam, The Netherlands. E-mail: galjart@ch1.fgg.eur.nl.
References
- 1.Akhmanova A, Hoogenraad CC, Drabek K, Stepanova T, Dortland B, Verkerk T, Vermeulen W, Burgering BM, De Zeeuw CI, Grosveld F, Galjart N. CLASPs are CLIP-115 and -170 associating proteins involved in the regional regulation of microtubule dynamics in motile fibroblasts. Cell. 2001;104:923–935. doi: 10.1016/s0092-8674(01)00288-4. [DOI] [PubMed] [Google Scholar]
- 2.Baas PW. Microtubules and axonal growth. Curr Opin Cell Biol. 1997;9:29–36. doi: 10.1016/s0955-0674(97)80148-2. [DOI] [PubMed] [Google Scholar]
- 3.Baas PW, Ahmad FJ. The plus ends of stable microtubules are the exclusive nucleating structures for microtubules in the axon. J Cell Biol. 1992;116:1231–1241. doi: 10.1083/jcb.116.5.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baas PW, Black MM. Individual microtubules in the axon consist of domains that differ in both composition and stability. J Cell Biol. 1990;111:495–509. doi: 10.1083/jcb.111.2.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baas PW, Deitch JS, Black MM, Banker GA. Polarity orientation of microtubules in hippocampal neurons: uniformity in the axon and nonuniformity in the dendrite. Proc Natl Acad Sci USA. 1988;85:8335–8339. doi: 10.1073/pnas.85.21.8335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baas PW, Black MM, Banker GA. Changes in microtubule polarity orientation during the development of hippocampal neurons in culture. J Cell Biol. 1989;109:3085–3094. doi: 10.1083/jcb.109.6.3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baas PW, Slaughter T, Brown A, Black MM. Microtubule dynamics in axons and dendrites. J Neurosci Res. 1991;30:134–153. doi: 10.1002/jnr.490300115. [DOI] [PubMed] [Google Scholar]
- 8.Baas PW, Ahmad FJ, Pienkowski TP, Brown A, Black MM. Sites of microtubule stabilization for the axon. J Neurosci. 1993;13:2177–2185. doi: 10.1523/JNEUROSCI.13-05-02177.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buck KB, Zheng JQ. Growth cone turning induced by direct local modification of microtubule dynamics. J Neurosci. 2002;22:9358–9367. doi: 10.1523/JNEUROSCI.22-21-09358.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang S, Svitkina TM, Borisy GG, Popov SV. Speckle microscopic evaluation of microtubule transport in growing nerve processes. Nat Cell Biol. 1999;1:399–403. doi: 10.1038/15629. [DOI] [PubMed] [Google Scholar]
- 11.Coquelle FM, Caspi M, Cordelieres FP, Dompierre JP, Dujardin DL, Koifman C, Martin P, Hoogenraad CC, Akhmanova A, Galjart N, De Mey JR, Reiner O. LIS1, CLIP-170's key to the dynein/dynactin pathway. Mol Cell Biol. 2002;22:3089–3102. doi: 10.1128/MCB.22.9.3089-3102.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Hoop MJ, Meyn L, Dotti CG. Culturing hippocampal neurons and astrocytes from fetal rat brain. In: Celis JE, editor. Cell biology: a laboratory handbook, 2nd Ed. Academic; San Diego: 1998. pp. 154–163. [Google Scholar]
- 13.Dent EW, Callaway JL, Szebenyi G, Baas PW, Kalil K. Reorganization and movement of microtubules in axonal growth cones and developing interstitial branches. J Neurosci. 1999;19:8894–8908. doi: 10.1523/JNEUROSCI.19-20-08894.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Zeeuw CI, Hoogenraad CC, Goedknegt E, Hertzberg E, Neubauer A, Grosveld F, Galjart N. CLIP-115, a novel brain-specific cytoplasmic linker protein, mediates the localization of dendritic lamellar bodies. Neuron. 1997;19:1187–1199. doi: 10.1016/s0896-6273(00)80411-0. [DOI] [PubMed] [Google Scholar]
- 15.Dotti CG, Sullivan CA, Banker GA. The establishment of polarity by hippocampal neurons in culture. J Neurosci. 1988;8:1454–1468. doi: 10.1523/JNEUROSCI.08-04-01454.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Downing KH. Structural basis for the interaction of tubulin with proteins and drugs that affect microtubule dynamics. Annu Rev Cell Dev Biol. 2000;16:89–111. doi: 10.1146/annurev.cellbio.16.1.89. [DOI] [PubMed] [Google Scholar]
- 17.Ehrengruber MU, Lundstrom K, Schweitzer C, Heuss C, Schlesinger S, Gahwiler BH. Recombinant Semliki Forest virus and Sindbis virus efficiently infect neurons in hippocampal slice cultures. Proc Natl Acad Sci USA. 1999;96:7041–7046. doi: 10.1073/pnas.96.12.7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han G, Liu B, Zhang J, Zuo W, Morris NR, Xiang X. The Aspergillus cytoplasmic dynein heavy chain and NUDF localize to microtubule ends and affect microtubule dynamics. Curr Biol. 2001;11:719–724. doi: 10.1016/s0960-9822(01)00200-7. [DOI] [PubMed] [Google Scholar]
- 19.He Y, Yu W, Baas PW. Microtubule reconfiguration during axonal retraction induced by nitric oxide. J Neurosci. 2002;22:5982–5991. doi: 10.1523/JNEUROSCI.22-14-05982.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoogenraad CC, Akhmanova A, Grosveld F, De Zeeuw CI, Galjart N. Functional analysis of CLIP-115 and its binding to microtubules. J Cell Sci. 2000;113:2285–2297. doi: 10.1242/jcs.113.12.2285. [DOI] [PubMed] [Google Scholar]
- 21.Hoogenraad CC, Koekkoek B, Akhmanova A, Krugers H, Dortland B, Miedema M, van Alphen A, Kistler WM, Jaegle M, Koutsourakis M, Van Camp N, Verhoye M, van der Linden A, Kaverina I, Grosveld F, De Zeeuw CI, Galjart N. Targeted mutation of Cyln2 in the Williams syndrome critical region links CLIP-115 haploinsufficiency to neurodevelopmental abnormalities in mice. Nat Genet. 2002;32:116–127. doi: 10.1038/ng954. [DOI] [PubMed] [Google Scholar]
- 22.Komarova YA, Vorobjev IA, Borisy GG. Life cycle of MTs: persistent growth in the cell interior, asymmetric transition frequencies and effects of the cell boundary. J Cell Sci. 2002a;115:3527–3539. doi: 10.1242/jcs.115.17.3527. [DOI] [PubMed] [Google Scholar]
- 23.Komarova YA, Akhmanova AS, Kojima S, Galjart N, Borisy GG. Cytoplasmic linker proteins promote microtubule rescue in vivo. J Cell Biol. 2002b;159:589–599. doi: 10.1083/jcb.200208058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lundstrom K, Rotmann D, Hermann D, Schneider EM, Ehrengruber MU. Novel mutant Semliki Forest virus vectors: gene expression and localization studies in neuronal cells. Histochem Cell Biol. 2001a;115:83–91. doi: 10.1007/s004180000223. [DOI] [PubMed] [Google Scholar]
- 25.Lundstrom K, Schweitzer C, Rotmann D, Hermann D, Schneider EM, Ehrengruber MU. Semliki Forest virus vectors: efficient vehicles for in vitro and in vivo gene delivery. FEBS Lett. 2001b;504:99–103. doi: 10.1016/s0014-5793(01)02707-7. [DOI] [PubMed] [Google Scholar]
- 26.Mimori-Kiyosue Y, Shiina N, Tsukita S. Adenomatous polyposis coli (APC) protein moves along microtubules and concentrates at their growing ends in epithelial cells. J Cell Biol. 2000a;148:505–518. doi: 10.1083/jcb.148.3.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mimori-Kiyosue Y, Shiina N, Tsukita S. The dynamic behavior of the APC-binding protein EB1 on the distal ends of microtubules. Curr Biol. 2000b;10:865–868. doi: 10.1016/s0960-9822(00)00600-x. [DOI] [PubMed] [Google Scholar]
- 28.Morrison EE, Moncur PM, Askham JM. EB1 identifies sites of microtubule polymerisation during neurite development. Brain Res Mol Brain Res. 2002;98:145–152. doi: 10.1016/s0169-328x(01)00290-x. [DOI] [PubMed] [Google Scholar]
- 29.Nakagawa H, Koyama K, Murata Y, Morito M, Akiyama T, Nakamura Y. EB3, a novel member of the EB1 family preferentially expressed in the central nervous system, binds to a CNS-specific APC homologue. Oncogene. 2000;19:210–216. doi: 10.1038/sj.onc.1203308. [DOI] [PubMed] [Google Scholar]
- 30.Perez F, Diamantopoulos GS, Stalder R, Kreis TE. CLIP-170 highlights growing microtubule ends in vivo. Cell. 1999;96:517–527. doi: 10.1016/s0092-8674(00)80656-x. [DOI] [PubMed] [Google Scholar]
- 31.Pierre P, Pepperkok R, Kreis TE. Molecular characterization of two functional domains of CLIP-170 in vivo. J Cell Sci. 1994;107:1909–1920. doi: 10.1242/jcs.107.7.1909. [DOI] [PubMed] [Google Scholar]
- 32.Rickard JE, Kreis TE. Binding of pp170 to microtubules is regulated by phosphorylation. J Biol Chem. 1991;266:17597–17605. [PubMed] [Google Scholar]
- 33.Rogers SL, Rogers GC, Sharp DJ, Vale RD. Drosophila EB1 is important for proper assembly, dynamics, and positioning of the mitotic spindle. J Cell Biol. 2002;158:873–884. doi: 10.1083/jcb.200202032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schuyler SC, Pellman D. Microtubule “plus-end-tracking proteins”: the end is just the beginning. Cell. 2001;105:421–424. doi: 10.1016/s0092-8674(01)00364-6. [DOI] [PubMed] [Google Scholar]
- 35.Su LK, Qi Y. Characterization of human MAPRE genes and their proteins. Genomics. 2001;71:142–149. doi: 10.1006/geno.2000.6428. [DOI] [PubMed] [Google Scholar]
- 36.Terada S, Kinjo M, Hirokawa N. Oligomeric tubulin in large transporting complex is transported via kinesin in squid giant axons. Cell. 2000;103:141–155. doi: 10.1016/s0092-8674(00)00094-5. [DOI] [PubMed] [Google Scholar]
- 37.Tirnauer JS, Grego S, Salmon ED, Mitchison TJ. EB1–microtubule interactions in Xenopus egg extracts: role of EB1 in microtubule stabilization and mechanisms of targeting to microtubules. Mol Biol Cell. 2002;13:3614–3626. doi: 10.1091/mbc.E02-04-0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Valetti C, Wetzel DM, Schrader M, Hasbani MJ, Gill SR, Kreis TE, Schroer TA. Role of dynactin in endocytic traffic: effects of dynamitin overexpression and colocalization with CLIP-170. Mol Biol Cell. 1999;10:4107–4120. doi: 10.1091/mbc.10.12.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vaughan KT, Tynan SH, Faulkner NE, Echeverri CJ, Vallee RB. Colocalization of cytoplasmic dynein with dynactin and CLIP-170 at microtubule distal ends. J Cell Sci. 1999;112:1437–1447. doi: 10.1242/jcs.112.10.1437. [DOI] [PubMed] [Google Scholar]
- 40.Vaughan PS, Miura P, Henderson M, Byrne B, Vaughan KT. A role for regulated binding of p150Glued to microtubule plus ends in organelle transport. J Cell Biol. 2002;158:305–319. doi: 10.1083/jcb.200201029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu W, Baas PW. Changes in microtubule number and length during axon differentiation. J Neurosci. 1994;14:2818–2829. doi: 10.1523/JNEUROSCI.14-05-02818.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]