Abstract
Growth cone motility and morphology are based on actin-filament dynamics. Cofilin plays an essential role for the rapid turnover of actin filaments by severing and depolymerizing them. The activity of cofilin is repressed by phosphorylation at Ser3 by LIM kinase (LIMK, in which LIM is an acronym of the three gene products Lin-11, Isl-1, and Mec-3) and is reactivated by dephosphorylation by phosphatases, termed Slingshot (SSH). We investigated the roles of cofilin, LIMK, and SSH in the growth cone motility and morphology and neurite extension by expressing fluorescence protein-labeled cofilin, LIMK1, SSH1, or their mutants in chick dorsal root ganglion (DRG) neurons and then monitoring live images of growth cones by time-lapse video fluorescence microscopy. The expression of LIMK1 remarkably repressed growth cone motility and neurite extension, whereas the expression of SSH1 or a nonphosphorylatable S3A mutant of cofilin enhanced these events. The fan-like shape of growth cones was disorganized by the expression of any of these proteins. The repressive effects on growth cone behavior by LIMK1 expression were significantly rescued by the coexpression of S3A–cofilin or SSH1. These findings suggest that LIMK1 and SSH1 play critical roles in controlling growth cone motility and morphology and neurite extension by regulating the activity of cofilin and may be involved in signaling pathways that regulate stimulus-induced growth cone guidance. Using various mutants of cofilin, we also obtained evidence that the actin-filament-severing activity of cofilin is critical for growth cone motility and neurite extension.
Keywords: LIM kinase, Slingshot, cofilin, actin-depolymerizing factor, growth cone guidance, neurite outgrowth
Introduction
The establishment of a highly ordered neuronal network depends on the precise control of axon guidance during the development of the nervous system. Growth cones at the distal tips of growing axons sense a variety of attractive or repulsive cues from the environment and integrate these signals into changes in their shape and motility, thereby correctly guiding the axons to their targets (Tessier-Lavigne and Goodman, 1996; Mueller, 1999). It has become clear that actin-filament dynamics and reorganization play a central role in the motility, morphology, and directional movement of growth cones (Mitchison and Kirschner, 1988;Tanaka and Sabry, 1995). Various actin-binding proteins and intracellular signaling proteins that regulate the actin cytoskeleton (such as Rho family GTPases) have been found to be involved in growth cone extension/retraction and guidance (Luo, 2000; Song and Poo, 2001). However, molecular mechanisms governing how growth cones translate extracellular guidance signals into the spatially coordinated actin cytoskeletal reorganization for their correct guidance are not well understood.
Cofilin and actin-depolymerizing factor (ADF) are actin-binding proteins that play an essential role in enhancing actin-filament dynamics and reorganization by severing actin filaments and accelerating the depolymerization of actin filaments at their pointed ends (Bamburg, 1999). The activities of cofilin/ADF (hereafter referred to as cofilin) are reversibly regulated by phosphorylation and dephosphorylation at Ser3, with the phosphorylated form being inactive (Agnew et al., 1995). LIM kinases (LIMKs, in which LIM is an acronym of the three gene products Lin-11, Isl-1, and Mec-3), composed of LIMK1 and LIMK2, phosphorylate cofilin specifically at Ser3 and thereby induce actin cytoskeletal reorganization (Arber et al., 1998;Yang et al., 1998). LIMKs are activated by Rho family GTPases via actions of their downstream effectors, such as Rho-associated kinase (ROCK) and p21-activated kinase (PAK) (Arber et al., 1998; Yang et al., 1998; Edwards et al., 1999; Maekawa et al., 1999; Sumi et al., 1999;Ohashi et al., 2000). Thus, LIMKs seem to play a critical role in stimulus-induced actin cytoskeletal remodeling by linking the signal from Rho family GTPases to the change in cofilin activity. Regarding the mechanism related to the dephosphorylation of cofilin, a novel family of protein phosphatases, termed Slingshot (SSH), which specifically dephosphorylate and reactivate cofilin, have been identified (Niwa et al., 2002).
LIMK1 is expressed predominantly in the nervous system of developing mammals (Proschel et al., 1995; Mori et al., 1997). Hemizygotic deletion of the LIMK1 gene is implicated in the impairment of visuospatial cognition in patients with Williams syndrome (Frangiskakis et al., 1996). These findings suggest that LIMK1 and its substrate cofilin play a role in the development of the nervous system. The expression of cofilin, but not its S3E mutant that mimics the phosphorylated form of cofilin, increases the neurite length of primary cultured neurons (Meberg and Bamburg, 2000). Thus, the regulation of cofilin activity by Ser3 phosphorylation seems important for the control of neurite extension. However, there has been no direct evidence for the roles of LIMK and SSH in growth cone motility and extension.
We obtained evidence that LIMK1 and SSH1 play important roles in the control of growth cone motility and morphology and neurite extension principally by regulating the activity of cofilin. We also provide evidence that the actin-filament-severing activity of cofilin is critical for stimulating growth cone motility and neurite extension.
Materials and Methods
cDNA cloning of chick LIMK1.Poly(A)+ RNA from chick whole embryos on embryonic day 7 (E7) was reverse-transcribed into the oligo(dT)-primed cDNA and used as a template for PCR amplification. The 474 bp chick LIMK1 (chLIMK1) cDNA was amplified by PCR using degenerate oligonucleotide primers, 5′-GA(A/G)GTNATGGTNATGAA(A/G)GA-3′ and 5′-GTA(A/G)AGNCCNCGGTAGGT(T/C)AT-3′, corresponding to amino acid residues 363–369 and 514–520 of human LIMK1 (Mizuno et al., 1994). To obtain the 5′- and 3′-terminal sequences of chLIMK1 cDNA, 5′- and 3′-rapid amplification of cDNA ends (RACE) was performed by two-step PCR, using as a template an adaptor-ligated chick embryo E7 cDNA library synthesized using a Marathon cDNA amplification kit (Clontech, Palo Alto, CA). The amplified products were subcloned into pGEM-T vector (Promega, Madison, WI). The chLIMK1 cDNA clones were identified by colony hybridization using a probe of the 474 bp chLIMK1 RT-PCR product, isolated, and subjected to DNA sequence analysis. DNA sequences were determined on both strands of three independent clones using a model 377 DNA sequencer (PE Applied Biosystems, Foster City, CA).
Plasmid construction. To generate the full-length chLIMK1 cDNA, cDNA fragments were combined and inserted into the pUCD2 expression vector. Expression plasmid coding for N-terminally Myc-tagged chLIMK1 was constructed by inserting the full-length chLIMK1 cDNA into the pMYC-C1 mammalian expression vector containing the Myc epitope tag (Amano et al., 2002). Expression plasmid coding for chLIMK1(D467A), with replacement of Asp467 with Ala, was constructed using a site-directed mutagenesis kit (Clontech). Plasmids coding for human SSH-1L (hSSH-1L, hereafter referred to as SSH1) and its phosphatase-inactive mutant, SSH1(CS), with the replacement of Cys393 by Ser, were constructed in a pcDNA3 vector (Invitrogen, Carlsbad, CA), as described previously (Niwa et al., 2002). Plasmids coding for human cofilin and its site-directed mutants (Moriyama and Yahara, 1999, 2002) were constructed in the pUCD2 vector. These plasmids were used to construct the recombinant herpes simplex viruses.
Northern blot analysis. The RNA blot containing 2 μg of poly(A)+ RNA from various chick tissues was hybridized with the 32P-labeled probe of the 0.6 kb EcoNI–EcoRI fragment of chLIMK1 cDNA or the 1.6 kb DraIII fragment of chLIMK2 cDNA under conditions described previously (Ohashi et al., 1994). The filter was also hybridized with the 32P-labeled 2.0 kb cDNA fragment of chick β-actin as a control for the integrity of the RNA. The filter was washed and analyzed, using a BAS1800 Bio-imaging Analyzer (Fujifilm, Tokyo, Japan), as described previously (Ohashi et al., 1994).
Antibodies, immunoprecipitation, and immunoblot analysis. An antibody specific to chLIMK1 (C20) was prepared by immunizing rabbits with keyhole limpet hemocyanin conjugated with the peptide chLK1-C20 (GCGLPPHPELPDTAPHLHPL), corresponding to the C-terminal sequence of chLIMK1. The anti-chLIMK1 antibody was purified on a protein A–Sepharose column (Amersham Biosciences, Arlington Heights, IL) and a HiTrap affinity column (Amersham Biosciences) coupled with the antigenic chLK1-C20 peptide. The antibody specific to the Ser3-phosphorylated form of cofilin (P-cofilin) was prepared as described previously (Toshima et al., 2001a). Anti-Myc epitope monoclonal antibody (9E10) was purchased from Roche Diagnostics (Tokyo, Japan). Immunoprecipitation and immunoblot analysis were performed as described previously (Amano et al., 2002).
In vitro kinase assay. An in vitro kinase assay was performed as described previously (Amano et al., 2002). Briefly, Myc-tagged chLIMK1 or chLIMK1(D467A) expressed in COS-7 cells was immunoprecipitated using a 9E10 anti-Myc antibody and incubated at 30°C for 1 hr in kinase reaction buffer containing 50 μm ATP and 5 μCi of [γ-32P]ATP (3000 Ci/mmol) in the presence of 0.1 mg/ml (His)6-tagged cofilin or its S3A mutant. After centrifugation, supernatants were separated on 15% SDS-PAGE and subjected to autoradiography to visualize32P-labeled cofilin, using a BAS1800 Bio-Image Analyzer (Fujifilm, Tokyo, Japan). The precipitates were separated on 9% SDS-PAGE and analyzed by immunoblotting with 9E10 anti-Myc antibody to visualize Myc-chLIMK1.
Cell staining. HeLa cells were transfected with plasmids coding for Myc-chLIMK1 or Myc-chLIMK1(D467A) by the Lipofectamine (Invitrogen) method, fixed in 4% formaldehyde, and stained with 9E10 anti-Myc antibody followed by FITC-labeled anti-mouse IgG antibody (Chemicon, Temecula, CA) as described previously (Toshima et al., 2001a). To visualize the expression of endogenous chLIMK1, chick E7 DRG explants were cultured for 12 hr, fixed in 4% formaldehyde, and stained with C20 anti-chLIMK1 antibody followed by FITC-labeled anti-rabbit IgG antibody (Chemicon). Cells were also stained for F-actin with rhodamine-labeled phalloidin (Molecular Probes, Eugene, OR). Cells were stained with anti-P-cofilin antibody as described previously (Amano et al., 2002). Cells were photographed on a Leica (Nussloch, Germany) fluorescent microscope (model DMLB).
Recombinant herpes simplex virus preparation and infection.The recombinant herpes simplex viruses (HSVs) that express yellow fluorescence protein (YFP)- or cyan fluorescence protein (CFP)-fused proteins were constructed as described previously (Sasaki et al., 2002). The cDNA coding for chLIMK1, SSH1, cofilin, actin, or their derivatives was ligated with the cDNA for YFP or CFP (Clontech) and inserted into the pHSV-PrPUC vector containing the immediate early promoter 4/5 and an HSV packaging site (Sasaki et al., 2002). The plasmid was transfected into Vero 2-2 HSV packaging cell lines with Lipofectamine, and the cells were infected with the replicon-defective HSV IE2 deletion mutant 5 dl 1.2 HSV helper virus 1 d later. Recombinant virus was amplified by three rounds of infection and then stored at −80°C. For infection, freshly dissociated DRG explants were allowed to adhere to dishes for 30 min and then incubated with recombinant viral stocks for 12 hr before analysis. The percentage of axons expressing YFP- or CFP-tagged proteins ranged from 30 to 60% in the infected cultures.
Cell culture of DRG neurons. Chick DRG explants were cultured as described previously (Goshima et al., 1995). In brief, DRGs were dissected from E7 chick embryos and placed on a poly-l-lysine- and laminin-coated 35 mm glass-bottom culture dish. The explant was cultured in Ham's F-12 medium containing 10% fetal bovine serum and 10 ng/ml nerve growth factor (NGF) for 12 hr and then treated with serum-free Ham's F12 medium containing 10 ng/ml NGF for 1 hr before time-lapse observation.
Time-lapse-video fluorescence image analysis and quantification. Live growth cones of chick DRG neurons were observed using a Leica inverted fluorescence microscope (model DMIRBE) equipped with PLAN NEO FLUOTAR 40× phase-contrast objective [numerical aperture (NA), 0.75] or PLAN APOCHROMAT 63× oil objective (NA, 1.32) lenses and a YFP- or CFP-optimized filter set (Omega Optical, Brattleboro, VT). Time-lapse fluorescence images were captured every 1–2 min for 16–20 min with 50–100 msec exposures, using a Coolsnap HQ-cooled CCD camera (1300 × 1030 pixels; Roper Scientific, Wetziar, Germany) driven by Q550FW Imaging Software (Leica). Figures were assembled using Adobe Photoshop (Adobe Systems, San Jose, CA). The room temperature was maintained at 37°C. The sequential images were converted to video movies in Audio, Video, still Images (AVI) format at a speed of two frames per second. For quantification of areas of protrusion, a series of images was digitized, and growth cone outlines at 2 min time points were measured by binary segmentation using IPLab image analysis software (Scanalytics, Fairfax, VA), merged, and subtracted. The index of growth cone motility was calculated by dividing the average value of new protrusion areas at 2 min intervals from a total of 10 min of recording of each growth cone (8–15 different growth cones in duplicate or triplicate experiments) by the total growth cone area (calculated as an average of the images at every 2 min). The rate of neurite extension was calculated from the average of the migrating distance of the center of the growth cone during 10 min of recording of each growth cone (8–15 different growth cones). Statistical analyses were performed using Student'st test. Morphologies of growth cones expressing YFP fusion proteins were usually monitored by images from fluorescence microscopy because growth cone outlines observed by fluorescence images were almost equal to those observed by phase-contrast microscopy.
Results
cDNA cloning and characterization of chLIMK1
To examine the role of LIMK1 in chick DRG neurons, we first isolated the cDNA clones coding for chLIMK1, using PCR and 5′- and 3′-RACE. Sequence analysis of the combined 2.8 kb chLIMK1 cDNA revealed that it is composed of a 37 base 5′-noncoding region, a 1986 base open reading frame coding for a protein of 662 aa (Fig.1A), and a 0.8 kb 3′-noncoding region containing a poly(A) tail (GenBank/European Bioinfomatics Institute/DNA Data Bank of Japan accession numberAB073752). The size of the cDNA sequence (∼2.8 kb) coincides with the size of chLIMK1 mRNA (Fig. 1B), thus indicating that the cloned cDNA covers an almost full-length sequence of chLIMK1 mRNA. The deduced amino acid sequence of chLIMK1 has structural features conserved in members of a LIMK family in that it is composed of two LIM domains and a postsynaptic density-95/discs large/zona occludens-1 (PDZ) domain at the N-terminal half and a protein kinase domain at the C-terminal half (Fig. 1A). It is more closely related to the sequence of LIMK1 than to that of LIMK2, with the overall identity to the sequence of human LIMK1 and LIMK2 being 83 and 56%, respectively (Mizuno et al., 1994; Okano et al., 1995). Thus, we referred to the encoded protein as chLIMK1. Compared with human, rat, and mouse LIMK1, chLIMK1 has an extra 8 aa extension (TAPHLHPL) at the C terminal.
Fig. 1.
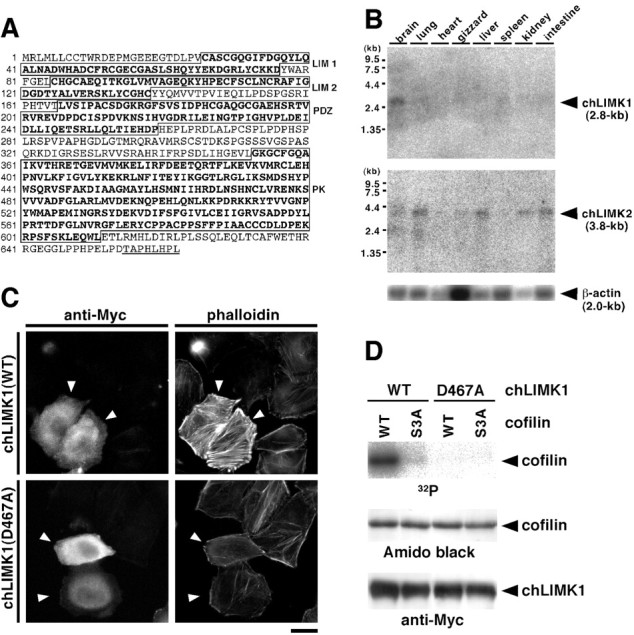
Amino acid sequence, tissue distribution, and actin reorganization and kinase activities of chLIMK1.A, Deduced amino acid sequence of chLIMK1. Amino acid residues are numbered on the left. The regions of two LIM domains (LIM 1, LIM 2), a PDZ domain (PDZ), and a protein kinase domain (PK) are boxed and indicated on the right. A unique C-terminal 8 aa sequence of chLIMK1 is underlined. B, Tissue distribution of chLIMK1 mRNA (top) and chLIMK2 mRNA (middle). Poly(A)+ RNAs (2 μg each) from various chick tissues were subjected to Northern blot analysis, using the chLIMK1 (top) or chLIMK2 (middle) cDNA as a probe. Bottom, Expression of β-actin mRNA, used as a control. Positions of molecular weight markers are indicated on the left.C, Induction of actin reorganization by chLIMK1. HeLa cells were transfected with plasmids coding for Myc-chLIMK1 (top) or Myc-chLIMK1(D467A) (bottom) and costained with anti-Myc antibody (left) and rhodamine-conjugated phalloidin (right).Arrowheads indicate cells expressing Myc-chLIMK1 (top) or Myc-chLIMK1(D467A) (bottom). Scale bar, 20 μm. D, Cofilin-phosphorylating activity of chLIMK1. Myc-tagged WT chLIMK1 or its D467A mutant was expressed in COS-7 cells, immunoprecipitated with anti-Myc antibody, and incubated with [γ-32P]ATP and (His)6-tagged WT cofilin or its S3A mutant. After centrifugation, supernatants were separated on 15% SDS-PAGE and analyzed using autoradiography (top) and Amido Black staining (middle) for cofilin, and precipitates were separated on 9% SDS-PAGE and analyzed by immunoblotting with anti-Myc-antibody (bottom) to detect Myc-chLIMK1 or Myc-chLIMK1(D467A).
Northern blot analysis revealed that a single 2.8 kb chLIMK1 mRNA was expressed predominantly in the brain and faintly in other tissues (Fig.1B, top). In contrast, expression of a 3.8 kb chLIMK2 mRNA was detected in various tissues, including lung, liver, kidney, and intestine (Fig. 1B, middle), as reported previously (Ohashi et al., 1994). Thus, similar to the tissue distribution of LIMK1 and LIMK2 mRNAs in humans and other species (Nunoue et al., 1995; Okano et al., 1995), chLIMK1 mRNA is expressed predominantly in the brain and chLIMK2 mRNA in various tissues.
Previous studies revealed that LIMKs induce actin cytoskeletal reorganization by phosphorylating cofilin at Ser3. To determine whether chLIMK1 has similar activities, Myc epitope-tagged chLIMK1 was expressed in HeLa cells, and actin filaments were visualized by staining with rhodamine-labeled phalloidin. The ectopic expression of chLIMK1 induced significant enhancement of actin stress fibers and accumulation of actin filaments at the cell periphery compared with findings in surrounding chLIMK1-nontransfected cells (Fig.1C, top). In contrast to WT chLIMK1, expression of its kinase-inactive mutant, chLIMK1(D467A), in which a presumptive catalytic residue Asp467 is replaced by alanine, had no apparent effect on actin organization (Fig. 1C,bottom). In vitro kinase reaction revealed that wild-type chLIMK1 phosphorylated wild-type cofilin but not cofilin(S3A), in which Ser3 was replaced by alanine, whereas chLIMK1(D467A) did not phosphorylate either (Fig.1D). Thus, similar to mammalian LIMKs, chLIMK1 has the potential to phosphorylate cofilin specifically at Ser3 and to induce actin reorganization in cultured cells.
Subcellular localization of LIMK1 in chick DRG neurons
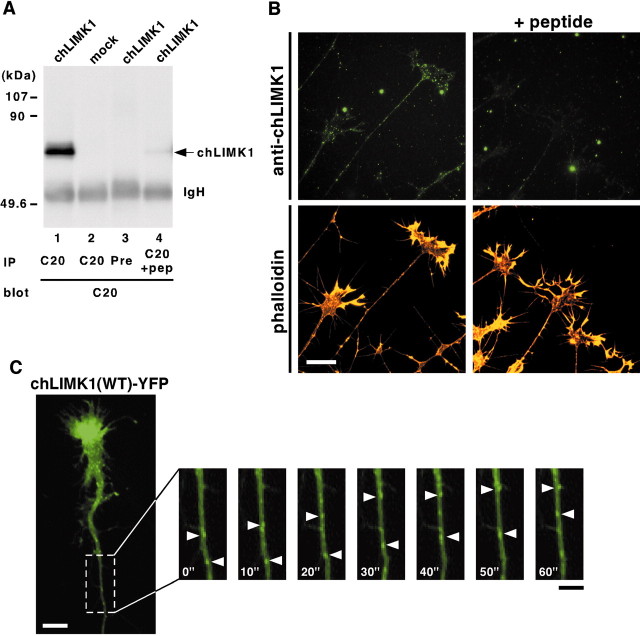
To determine the subcellular localization of chLIMK1 in chick DRG neurons, we prepared an anti-chLIMK1 antibody (C20) raised against the C-terminal 20 aa peptide of chLIMK1. To assess the specificity of the antibody, COS-7 cells were transfected with the plasmid coding for chLIMK1 or the vector plasmid, and then cell lysates were analyzed by immunoprecipitation and immunoblotting with C20 anti-chLIMK1 antibody. One major immunoreactive band with an estimated molecular mass of ∼74 kDa was detected in lysates of COS-7 cells transfected with chLIMK1 cDNA but not in lysates of cells mock-transfected with the vector plasmid (Fig. 2A,lanes 1, 2). This band was not detected and barely detectable when lysates of COS-7 cells transfected with chLIMK1 cDNA were immunoprecipitated with the preimmune serum (Fig.2A, lane 3) and with anti-chLIMK1 antibody preincubated with excess amounts of antigenic peptide (Fig.2A, lane 4), respectively. These results suggest that the C20 anti-chLIMK1 antibody specifically recognizes chLIMK1 protein.
Fig. 2.
Subcellular localization of chLIMK1 in chick DRG neurons. A, Specificity of anti-chLIMK1 antibody. COS-7 cells were transfected with chLIMK1 cDNA expression plasmid (lanes 1, 3, 4) or mock-transfected with pMYC-C1 vector alone (lane 2). Lysates were immunoprecipitated (IP) with C20 anti-chLIMK1 antibody (C20,lanes 1, 2, 4) or preimmune serum (Pre, lane 3), run on SDS-PAGE, and then immunoblotted with C20 anti-chLIMK1 antibody. In lane 4, anti-chLIMK1 antibody was pretreated with excess amounts of antigenic peptide (pep). The positions of molecular weight marker proteins are indicated on the left.IgH, Ig heavy chain. B, Subcellular localization of chLIMK1 in chick DRG neurons. Chick E7 DRG neurons were costained with rhodamine-conjugated phalloidin (bottom) and C20 anti-chLIMK1 antibody (top) in the absence (left) or presence (right) of excess amounts of antigenic peptide. Dot-like staining of endogenous chLIMK1 was specifically detected in growth cones and axonal shafts at thetop left. Scale bar, 20 μm. C, Axonal transport of LIMK1(WT)-YFP. Chick E7 DRG neurons were infected with HSV coding for YFP-fused chLIMK1(WT) and recorded 12 hr later by video fluorescence microscopy. Each frame shows the fluorescence image of YFP at 10 sec intervals. Arrowheads indicate LIMK1(WT)-YFP-containing vesicles moving anterogradely. Also see the supplemental movie (available at www.jneurosci.org), in which retrograde movements can be seen. Scale bars: white, 10 μm; black, 5 μm.
To determine the subcellular localization of chLIMK1, chick E7 DRG explants were cultured for 12 hr and immunostained with C20 anti-chLIMK1 antibody. The antibody stained dot-like signals in growth cones and axon shafts of DRG neurons (Fig. 2B,top left). In contrast, no such signal was detected when DRG neurons were stained with either the anti-chLIMK1 antibody preadsorbed with antigenic peptide (Fig. 2B, top right) or the IgG fraction of the preimmune serum (data not shown), therefore indicating that the dot-like signals observed with anti-chLIMK1 antibody in growth cones and axon shafts represent the localization of endogenous chLIMK1. Phalloidin staining revealed that actin filaments predominantly localize in the lamellipodial and filopodial structures at the periphery of growth cones (Fig.2B, bottom). chLIMK1 only partially colocalized with F-actin at the tips of the growth cones. When chLIMK1–YFP fusion protein was expressed in chick E7 DRG neurons by infection of recombinant HSV encoding chLIMK1-YFP and its localization in neurons was analyzed by time-lapse video fluorescence microscopy, the movements of chLIMK1-YFP particles both toward the axon terminal (anterograde) and back to the cell body (retrograde) were observed within neurites (Fig. 2C; also see a supplemental movie of Fig. 2C). No such particle was detected in DRG neurons expressing control YFP (data not shown), which suggests that LIMK1 dynamically translocates within neurites through anterograde and retrograde axonal flows.
Expression of LIMK1 represses growth cone motility and extension and the growth cone becomes small
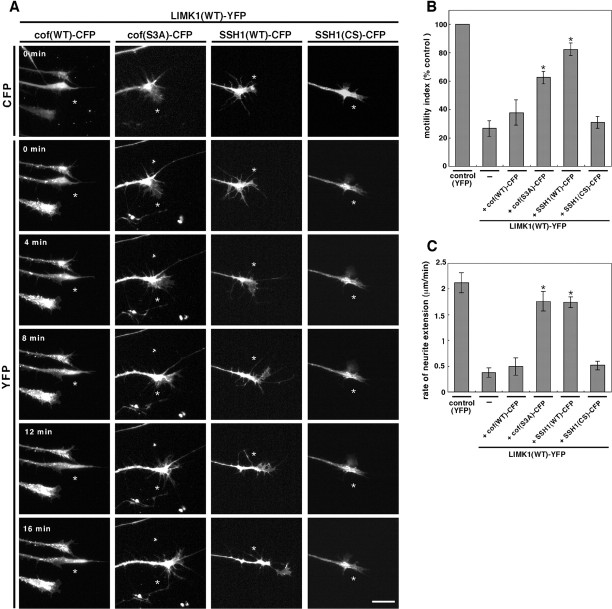
To investigate the role of LIMK1 in the motility, morphology, and extension of growth cones, chLIMK1 or its kinase-inactive D467A mutant was expressed in chick DRG neurons. Chick E7 DRG explants were infected with recombinant HSV coding for CFP-fused LIMK1 or LIMK1(D467A), together with HSV coding for YFP-actin. As a control, DRG neurons were infected with YFP-actin-encoding HSV alone. DRG explants were cultured for 12 hr after infection, and then the growth cone morphology, motility, and extension were monitored by time-lapse video fluorescence microscopy. Representative live images of growth cones are shown in Figure 3 (also see supplemental movies of Fig. 3A–C; available at www.jneurosci.org). Growth cones of control neurons expressing YFP-actin alone had a fan-like extended shape and dynamically changed morphology by incessantly protruding and retracting filopodial and lamellipodial protrusions (Fig.3A), and they went forward very fast (see Fig. 5 for quantitative analyses). In contrast, growth cones of neurons coexpressing wild-type LIMK1-CFP with YFP-actin were sticky and small and had a pestle-like morphology and poorly extended filopodia and lamellipodia (Fig. 3B). Phase-contrast analysis also showed such morphological changes (data not shown). YFP-actin abnormally accumulated in the central region of the growth cones (Fig.3B). Both the motility of growth cones and the rate of neurite extension markedly slowed in LIMK1-expressing neurons compared with the findings in control cells. In contrast, neurons expressing LIMK1(D467A)-CFP had an extended growth cone, but the shape was irregular and the lamellipodial and filopodial extensions were frequently retained on the shaft for a while even after the growth cone had migrated forward (Fig. 3C). The motility of growth cones was slightly reduced in LIMK1(D467A)-expressing neurons compared with findings in control cells (see Fig. 5).
Fig. 3.
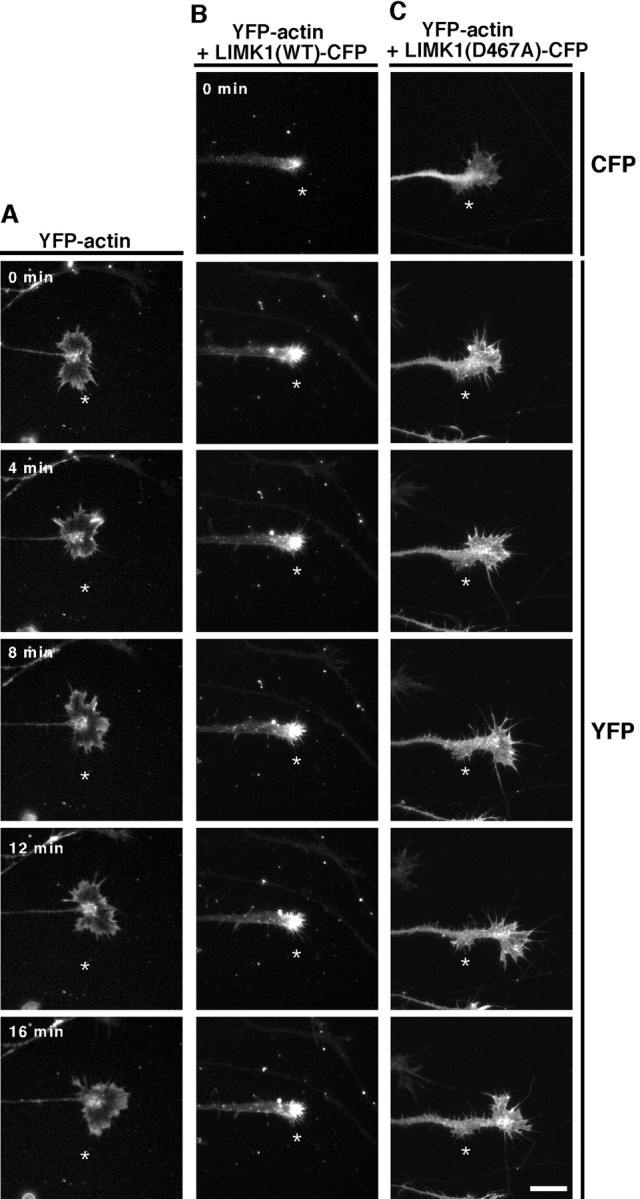
Time-lapse fluorescence images of growth cones expressing YFP-actin alone (A), YFP-actin plus CFP-tagged wild-type chLIMK1 [LIMK1(WT)-CFP] (B), or YFP-actin plus CFP-tagged D467A mutant of chLIMK1 [LIMK1(D467A)-CFP] (C). Chick E7 DRG neurons were infected with HSV encoding YFP-actin (A) or coinfected with HSV encoding YFP-actin plus HSV encoding LIMK1(WT)-CFP (B) or LIMK1(D467A)-CFP (C) and recorded 12 hr later by time-lapse video fluorescence microscopy. Cells expressing LIMK1(WT)-CFP or LIMK1(D467A)-CFP are assigned by CFP fluorescence, as shown in B (top) and C(top). Other frames show the fluorescence images of YFP-actin at 4 min intervals, as indicated in A. Theasterisk in each frame indicates the fixed point. Compared with the control growth cone in A, the growth cone expressing LIMK1(WT) was small and sticky and the growth cone motility and neurite extension were markedly suppressed. The motility of the growth cone expressing LIMK1(D467A) was slightly repressed. Scale bar, 20 μm. Also see the supplemental movies (available atwww.jneurosci.org). Quantitative data of growth cone motility and the rate of neurite extension of DRG neurons expressing YFP, LIMK1(WT)-YFP, or LIMK1(D467A)-YFP are summarized in Figure 5.
Fig. 5.
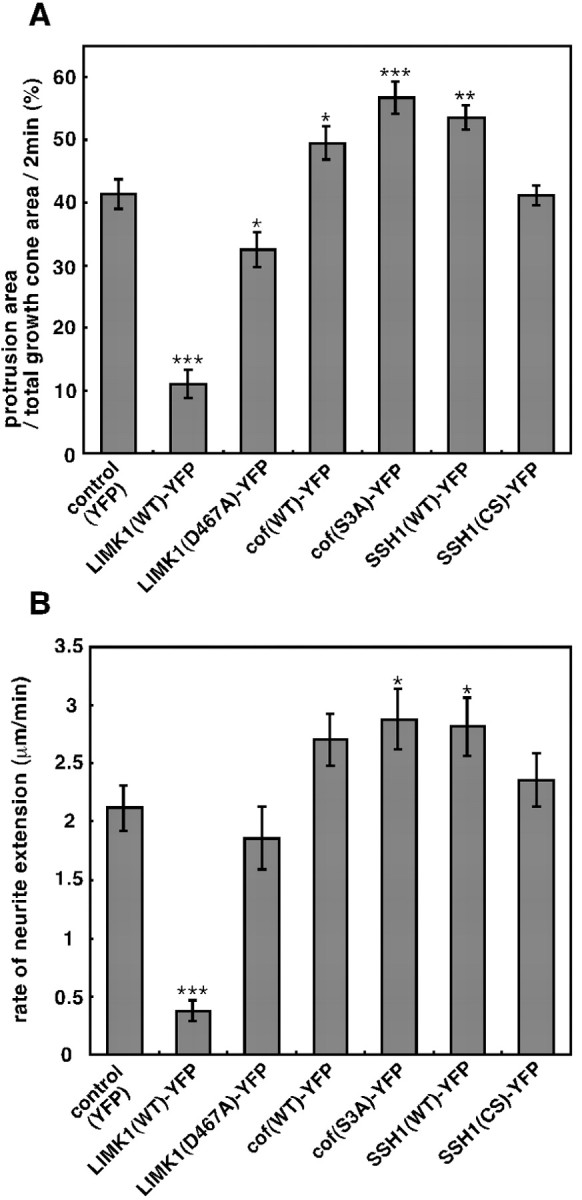
Quantitative analyses of the effects of overexpression of LIMK1, cofilin, SSH1, or their mutants on the growth cone motility and the rate of neurite extension of chick DRG neurons.A, Motility of growth cones. Data represent the mean percentage values of the newly protruded areas of growth cones every 2 min, divided by the mean value of total areas of growth cones during 10 min of observation. B, Average rate of neurite extension, calculated by measuring the migration distance of the growth cone center for 10 min. Data represent means ± SEM from 8 to 15 different growth cones in duplicate or triplicate experiments. *p < 0.05, **p < 0.005, and ***p < 0.001 compared with the control YFP-expressing cells.
To quantify the motility of the growth cone, DRG neurons were infected with HSV coding for LIMK1-YFP, LIMK1(D467A)-YFP, or YFP as a control, and then the growth cone motility was monitored by time-lapse video fluorescence microscopy. Individual images were digitized, and growth cone outlines between at 2 min time points were compared to calculate areas of new protrusion of the growth cone perimeter. We calculated the average areas of new protrusion at 2 min intervals from a total of 10 min of recording of each growth cone (8–15 different growth cones from two to three experiments), divided them by the total growth cone area (calculated as an average of the images of the 2 min time points), and used the average percentages of protrusion area per total growth cone area per 2 min as the index of growth cone motility. As shown in Figure 5A, the motility of growth cones expressing LIMK1-YFP (11.1 ± 2.3%) was significantly lower than that for control growth cones expressing YFP (41.3 ± 2.4%), which clearly shows that overexpression of LIMK1 remarkably reduced the motility of the growth cones. In contrast, the motility of the growth cones expressing LIMK1(D467A)-YFP (32.5 ± 2.8%) was slightly lower than that of control cells (see Fig.5A). We also quantified the rate of neurite extension by calculating the average migrating distance of the center of growth cone during 10 min of recording of each growth cone (8–15 different growth cones from two to three experiments). As shown in Figure 5B, the average rate of neurite extension of LIMK1-expressing neurons was 0.38 μm/min (calculated from eight growth cones), which corresponds to only 18% of the average neurite extension rate of control YFP-expressing growth cones (2.1 μm/min, calculated from 10 growth cones). No statistically significant change was observed in the rate of neurite extension for LIMK1(D467A)-expressing neurons compared with that of control neurons (see Fig. 5B). These findings suggest that overexpression of LIMK1 suppresses growth cone motility and neurite extension and makes the growth cone small and sticky, in a manner dependent on its kinase activity.
Expression of S3A–cofilin increases growth cone motility and extension, and the growth cone becomes slender and branchy
The overexpression of cofilin increases neurite length in rat cortical neurons and chick spinal cord neurons (Meberg and Bamburg, 2000). We examined the effects of cofilin overexpression on the motility and morphology of growth cones of chick DRG neurons. YFP-fused cofilin or its S3A mutant was expressed in DRG neurons and growth cone behavior was analyzed using time-lapse video fluorescence microscopy (Fig. 4A,B; also see supplemental movies of Fig. 4A,B; available at www.jneurosci.org). Cofilin(S3A) was used as a constitutively active form because its phosphorylation site Ser3 is mutated to the nonphosphorylatable alanine. The growth cones expressing wild-type or S3A-cofilin were slender and too labile to form the fan-like extended shape; they rapidly protruded and retracted filopodial extensions. Protrusions from these growth cones were less sticky on the substratum, and the growth cones had the tendency to form branches and migrate faster than control growth cones, although most branches were retracted and finally taken up into the shaft during or after the growth cones had migrated forward. As summarized in Figure5, quantitative analyses revealed that both the index of growth cone motility (56.7 ± 2.5%) and the rate of neurite extension (2.9 μm/min) of neurons expressing cofilin(S3A) were significantly higher than in control cells. The expression of wild-type cofilin had similar effects but was less effective (49.5 ± 2.7%; 2.7 μm/min). In contrast, expression of an S3D mutant of cofilin, which mimics the phosphorylated form of cofilin by the replacement of Ser3 by Asp, had no apparent effect on motility, morphology, and extension speed of growth cones (data not shown). These results suggest that cofilin plays an important role in promoting growth cone motility and the rate of neurite extension, and these activities of cofilin are strictly regulated by phosphorylation at Ser3.
Fig. 4.
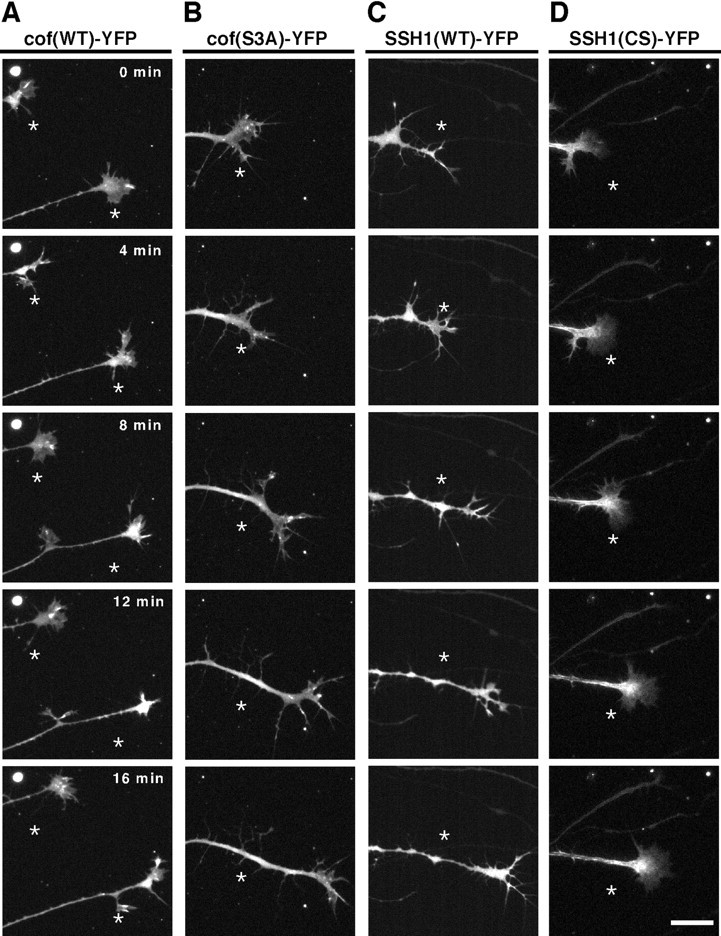
Time-lapse fluorescence images of growth cones expressing YFP-tagged wild-type cofilin [cof(WT)] (A), its S3A mutant [cof(S3A)] (B), wild-type SSH1 [SSH1(WT)] (C), or its C393S mutant [SSH1(CS)] (D). Chick E7 DRG neurons were infected with HSV coding for the respective YFP-fused proteins, cultured for 12 hr, and recorded by time-lapse video fluorescence microscopy. Each frame shows the fluorescence image of YFP at 4 min intervals, as indicated. Theasterisk in each frame indicates the fixed point. Compared with the control growth cone in Figure 3A, the growth cones expressing cof(WT), cof(S3A), or SSH1(WT) were slender and branchy, and their growth cone motility and neurite extension rate increased. No apparent effect was observed in the motility and morphology of the growth cone expressing SSH1(CS). Scale bar, 20 μm. Also see the supplemental movies (available at www.jneurosci.org). Quantitative data of growth cone motility and the rate of neurite extension are summarized in Figure 5.
Expression of SSH1 increases growth cone motility and extension, and the growth cone becomes slender and branchy
We recently identified SSHs, a family of protein phosphatases that specifically dephosphorylate and reactivate cofilin (Niwa et al., 2002). To examine the role of cofilin dephosphorylation in growth cone motility and morphology, we constructed recombinant HSV encoding YFP-fused human SSH1 (SSH1-YFP) and infected it into chick DRG neurons. Growth cones expressing SSH1-YFP had a slender and branchy morphology, similar to cones expressing cofilin(S3A), and significantly increased the motility and the speed of neurite extension (Fig. 4C; also see supplemental movie of Fig. 4C). The growth cone motility index (53.6 ± 2.0%) and the rate of neurite extension (2.8 μm/min) of neurons expressing SSH1 were similar to those of neurons expressing cofilin(S3A) (Fig. 5). The expression of SSH1 probably altered the behavior of growth cones by dephosphorylation and activation of endogenous cofilin. On the contrary, the expression of SSH1(CS), a phosphatase-inactive mutant of SSH1, in which the presumptive catalytic residue Cys393 was replaced by serine, had no apparent effect on the morphology and motility of growth cones (Fig.4D; also see supplemental movie of Fig.4D). Growth cones expressing SSH1(CS) had the fan-like extended shape, similar to findings in control cells. No statistically significant difference in growth cone motility and the rate of neurite extension was observed between SSH1(CS)-expressing cells and control cells (Fig. 5). Thus, SSH1 enhanced the motility and extension of growth cones in a manner dependent on the phosphatase activity.
Expression of cofilin(S3A) or SSH1 rescues the inhibitory effects of LIMK1 expression on growth cone motility and extension
To determine whether the inhibitory effects of LIMK1 expression on growth cone motility and extension are mediated by the phosphorylation and inactivation of cofilin, we expressed LIMK1-YFP together with CFP-tagged wild-type or S3A-cofilin in DRG neurons and analyzed the behavior of growth cones using time-lapse video fluorescence microscopy. In contrast to LIMK1-expressing growth cones that have a pestle-like shape and migrate slowly (Fig. 3B), growth cones expressing both LIMK1 and cofilin(S3A) had a well extended morphology, with filopodia and lamellipodia that dynamically protrude and retract (Fig. 6A), similar to findings in control cells expressing YFP-actin alone (Fig.3A). Quantitative analyses revealed that the coexpression of S3A-cofilin significantly rescued the inhibitory effects of LIMK1 expression on growth cone motility and the rate of neurite extension, whereas the coexpression of wild-type cofilin had only a minor effect (Fig. 6B,C). These results strongly suggest that the effects of LIMK1 overexpression were primarily caused by the excessive phosphorylation of cofilin.
Fig. 6.
Overexpression of cofilin(S3A) or wild-type SSH1 partially rescues the inhibitory effects of LIMK1 on the motility and extension speed of growth cones. A, Chick E7 DRG neurons were coinfected with HSV encoding chLIMK1(WT)-YFP plus HSV encoding CFP-tagged cofilin(WT), cofilin(S3A), SSH1(WT), or SSH1(CS), cultured for 12 hr, and recorded using time-lapse video fluorescence microscopy. The expression of CFP-tagged proteins was assigned by CFP fluorescence, as shown at the top. Other frames show the fluorescence images of LIMK1(WT)-YFP at 4 min intervals, as indicated. Theasterisk in each frame indicates the fixed point. Scale bar, 20 μm. B, C, Quantitative analysis of the effects of coexpression of LIMK1 with cofilin, SSH1, or their mutants on the motility and extension speed of growth cones. B, The motility index was determined as in Figure 5A and is expressed in percentages, with the value of the control growth cone expressing YFP alone taken as 100%. C, Rate of neurite extension was determined as in Figure 5B. Data are means ± SEM from 8 to 15 different growth cones in duplicate or triplicate experiments. *p < 0.001 compared with the cells expressing LIMK1(WT)-YFP alone.
We next examined whether the expression of SSH1 could rescue the inhibitory effects of LIMK1 expression on growth cone motility and morphology. As shown in Figure 6, coexpression of wild-type SSH1 significantly reverted the effects of LIMK1. Growth cones expressing both LIMK1 and SSH1 had an extended morphology with lamellipodial and filopodial protrusions (Fig. 6A), and the growth cone motility and rate of neurite extension were almost comparable with those of control cells expressing YFP alone (Fig.6B,C). In contrast, the coexpression of a phosphatase-inactive mutant, SSH1(CS), did not revert the LIMK1-induced suppressive effects on growth cone motility and neurite extension. These results suggest that SSH1 has the potential to dephosphorylate and reactivate cofilin that is phosphorylated by LIMK1 overexpression, and that the precise control of cofilin phosphorylation and dephosphorylation by LIMK and SSH plays an important role in regulating growth cone motility and morphology and neurite extension.
Expression of LIMK1 increases the level of cofilin phosphorylation in DRG neurons, and the coexpression of SSH1 neutralizes it
To further assess the role of LIMK1 expression on growth cone motility and extension, we analyzed the level of the ectopically expressed LIMK1 in DRG neurons in comparison with the amount of endogenous LIMK1. Lysates from DRG neurons infected with HSV coding for YFP-tagged chLIMK1 or control YFP were immunoprecipitated with C20 anti-chLIMK1 antibody, and the amounts of endogenous and exogenous LIMK1 in immunoprecipitates were estimated by immunoblotting with C20 anti-chLIMK1 antibody and measuring in vitro kinase activities (Fig. 7A). Densitometric analysis of immunoblots revealed that the amount of YFP-chLIMK1 of ∼100 kDa was ∼15-fold higher than that of endogenous LIMK1 of 74 kDa. Comparison of the relative kinase activities also indicated that DRG neurons expressing YFP-chLIMK1 exhibited ∼11-fold higher LIMK1 kinase activity compared with the neurons expressing control YFP. The high level of expression of exogenous LIMK1 may explain why the coexpression of wild-type cofilin had only a minor effect on LIMK1-induced suppression of growth cone motility.
Fig. 7.
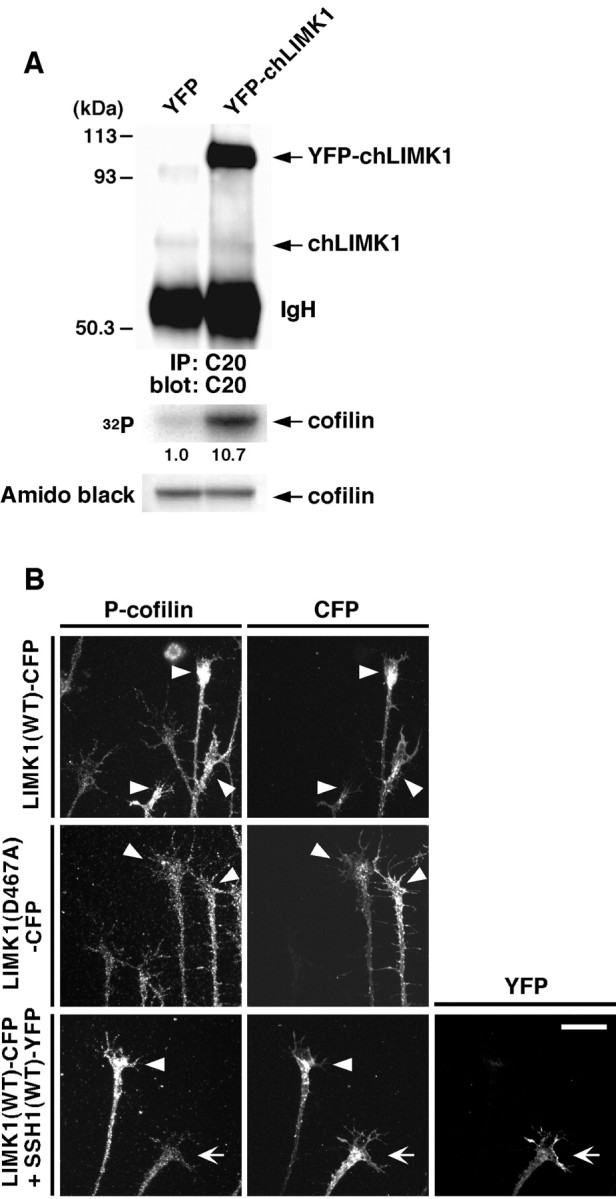
The level of expression of endogenous and exogenous LIMK1 and the level of cofilin phosphorylation in LIMK1- and SSH1-expressing DRG neurons. A, The level of LIMK1 expression. Chick E7 DRG neurons were infected with HSV encoding YFP or YFP-chLIMK1. After 12 hr, lysates were immunoprecipitated (IP) and immunoblotted with C20 anti-chLIMK1 antibody. Densitometric analysis revealed that the amount of YFP-chLIMK1 was ∼15-fold higher than that of endogenous chLIMK1. The level of LIMK1 was also estimated by in vitro kinase assay. Immunoprecipitates were subjected to in vitro kinase reaction using (His)6-cofilin as a substrate, as described in Figure 1D. The kinase activity of endogenous chLIMK1 in control YFP-expressing cells was taken as 1.0.IgH, Ig heavy chain. B, The level of cofilin phosphorylation. Chick E7 DRG neurons were infected with HSV encoding chLIMK1(WT)-CFP or chLIMK1(D467A)-CFP or coinfected with HSV encoding chLIMK1(WT)-CFP and HSV encoding SSH1(WT)-YFP. After 12 hr of culture, cells were fixed and immunostained with anti-P-cofilin antibody. The expression of LIMK1 and SSH1 was assigned by CFP and YFP fluorescence. Arrowheads indicate cells expressing chLIMK1(WT)-CFP or chLIMK1(D467A)-CFP. An arrowindicates the cell coexpressing chLIMK1(WT)-CFP and SSH1(WT)-YFP. Scale bar, 20 μm.
We next examined the level of cofilin phosphorylation in DRG neurons expressing LIMK1 and SSH1 by immunostaining using the antibody that specifically recognizes the Ser3-phosphorylated form of cofilin (P-cofilin) (Toshima et al., 2001a). As shown in Figure 7B, the expression of LIMK1(WT) significantly increased the level of P-cofilin in the growth cones of DRG neurons compared with the findings of surrounding nonexpressing neurons. The expression of kinase-inactive LIMK1(D467A) had no apparent effect on the level of P-cofilin. When SSH1(WT) was coexpressed with LIMK1(WT) in DRG neurons, the level of P-cofilin was reduced to the level in control cells. These observations also provide evidence that LIMK1 and SSH1 regulate growth cone motility and neurite extension by phosphorylating and dephosphorylating cofilin, respectively.
Actin-filament-severing activity of cofilin is critical for growth cone motility and extension
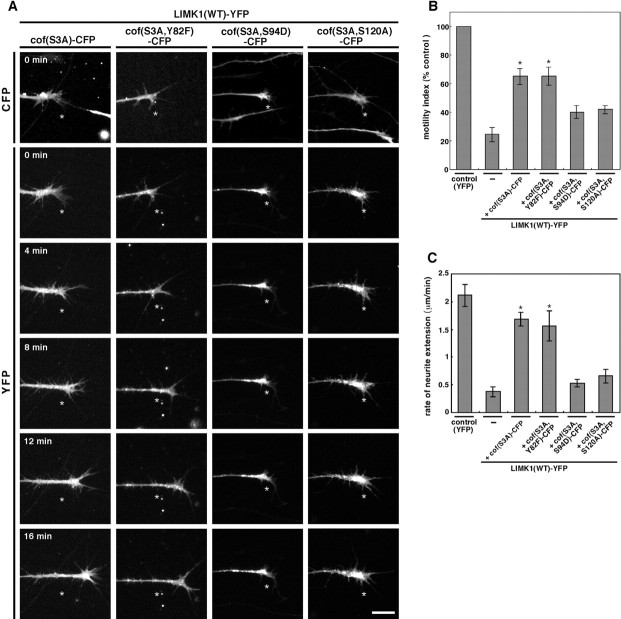
Cofilin can sever actin filaments and accelerate the release of actin monomers from the pointed ends of actin filaments (Bamburg, 1999). To examine which of these two activities plays a dominant role in growth cone motility and extension, we expressed site-directed cofilin mutants, which harbor defects of severing and depolymerization activities differentially, together with LIMK1 and analyzed the extent to which these cofilin mutants rescue the inhibitory effects of LIMK1 on growth cone motility and extension. We used (S3A, Y82F)-, (S3A, S94D)-, and (S3A, S120A)-cofilin, in which amino acids Tyr82, Ser94, and Ser120 were replaced by Phe, Asp, and Ala, respectively, in addition to the replacement of Ser3 by Ala. Biochemical studies revealed that Y82F-cofilin has decreased depolymerizing activity but retains the severing activity, S94D-cofilin retains the effective depolymerizing activity but is defective in severing activity, and S120A-cofilin has very reduced depolymerizing and severing activities (Moriyama and Yahara, 1999, 2002). As shown in Figure8, (S3A, Y82F)-cofilin significantly rescued the inhibitory effects of LIMK1 on growth cone motility and extension to levels similar to those rescued by S3A-cofilin. However, either (S3A, S94D)-cofilin or (S3A, S120A)-cofilin much less efficiently rescued the effects of LIMK1 expression, resulting in lower levels of growth cone motility and speed of neurite extension. These results suggest that the actin-filament-severing activity rather than the actin-depolymerizing activity plays a dominant role for cofilin to promote growth cone motility and extension.
Fig. 8.
The potential for cofilin mutants to rescue the inhibitory effect of LIMK1 on the motility and extension speed of growth cones. A, Chick E7 DRG neurons were coinfected with HSV encoding LIMK1-YFP and HSV encoding CFP-tagged cofilin mutants and recorded 12 hr later using time-lapse video fluorescence microscopy. The expression of CFP-tagged cofilin mutant proteins was assigned by CFP fluorescence, as shown at the top. Other frames show the fluorescence images of LIMK1(WT)-YFP at 4 min intervals, as indicated. The asterisk in each frame indicates the fixed point. Scale bar, 20 μm. B, C, Quantitative analysis of the effects of coexpression of LIMK1 with cofilin mutants on the motility and extension speed of growth cones. The motility index of growth cones (B) and the rate of neurite extension (C) were determined as in Figure 6. Data are means ± SEM from 8 to 15 different growth cones in duplicate or triplicate experiments. *p < 0.001 compared with the control cells expressing LIMK1(WT)-YFP alone.
Discussion
Actin-filament dynamics and reorganization are central for growth cone motility, morphology, and extension (Mitchison and Kirschner, 1988; Tanaka and Sabry, 1995; Luo, 2000; Song and Poo, 2001). Because cofilin is an essential regulator of actin-filament dynamics and is highly expressed in growth cones, it seems to play a critical role in growth cone motility and extension (Bamburg and Bray, 1987; Kuhn et al., 2000; Meberg, 2000). Meberg and Bamburg (2000) reported thatXenopus ADF/cofilin increased the length of neurite outgrowth when expressed in rat cortical neurons or chick spinal cord neurons in primary culture. In accordance with their results, we found in the present study that the expression of cofilin or its nonphosphorylatable (constitutively active) S3A-mutant in chick DRG neurons increases growth cone motility and the rate of neurite extension, based on the analysis of live images of growth cones expressing fluorescence-labeled proteins. We also show that the expression of LIMK1 increased the level of cofilin phosphorylation and drastically suppressed growth cone motility and extension in a manner dependent on the kinase activity. The suppressive effect of LIMK1 was noticeably recovered by the coexpression of cofilin(S3A), thus indicating that ectopically expressed LIMK1 inhibits growth cone motility and extension primarily by phosphorylating and inactivating cellular cofilin. Together, these observations suggest that cofilin plays an essential role in maintaining and promoting growth cone motility and neurite extension.
SSHs were found recently to be a novel family of protein phosphatases that dephosphorylate and reactivate cofilin in whole animals, in cultured cells, and in cell-free assays (Niwa et al., 2002). InDrosophila, the loss of ssh function increased the levels of phospho-cofilin and F-actin and disorganized epidermal cell morphogenesis, including the bifurcation phenotypes of the bristles and wing hairs, after whichslingshot was named. In mammalian cells, the expression of human SSH1, a member of three human SSH homologs, decreased the level of phospho-cofilin and suppressed LIMK1-induced actin assembly (Niwa et al., 2002). In the present study, we found that the expression of SSH1 in DRG neurons increases growth cone motility and extension and alters the shape of the growth cone, which becomes slender and branchy, phenotypes similar to those induced by cofilin(S3A) expression. The coexpression of SSH1 with LIMK1 significantly reduced the level of P-cofilin and notably recovered the inhibitory effects of LIMK1 on growth cone motility and extension. These findings also support the notion that SSH1 functions as a cofilin phosphatase in vivoand suggest that SSH1 plays an important role in regulating growth cone dynamics by dephosphorylating and activating cellular cofilin. It is likely that the modulation of cofilin activity by LIMK and SSH alters growth cone morphology and dynamics, thereby controlling the speed and direction of growth cone movement.
However, it is noted that the suppressive effects of LIMK1 on growth cone motility and extension were not fully recovered by the coexpression of cofilin(S3A) or SSH1, although the expression of cofilin(S3A) or SSH1 alone elevated these parameters. These observations may suggest the possibility that LIMK1 has targets other than cofilin for the suppression of growth cone motility.
Cofilin is thought to play a critical role in actin-filament dynamics and remodeling by severing actin filaments and accelerating the depolymerization rate of actin monomers from the pointed ends of actin filaments (Bamburg, 1999; Chen et al., 2000; Pollard et al., 2000). Severance of actin filaments by cofilin results in an increase in the number of free barbed ends from which actin filaments can elongate to protrude lamellipodial and filopodial extensions (Chan et al., 2000;Zebda et al., 2000). Acceleration of the off-rate of actin monomers from the pointed ends by cofilin results in an increase in the concentration of actin monomers, which can support the rapid turnover of actin filaments by coupling with the barbed end polymerization of actin monomers at the tip of the protrusions (Carlier et al., 1997). The relative importance of actin-filament severing and depolymerizing activities of cofilin in executing various cell activities is a much debated point (Carlier et al., 1999; McGrath et al., 2000; Condeelis, 2001). To address the question of which of these two activities plays a dominant role in growth cone motility and extension, we examined the potential of various cofilin mutants, which exhibit severing and depolymerizing activities differentially, to rescue the inhibitory effect of LIMK1 on growth cone motility and extension. Cofilin(S3A, Y82F), which is defective only in depolymerizing activity, significantly recovered the inhibitory effect of LIMK1 to a level similar to that of cofilin(S3A), but cofilin(S3A, S94D), which is defective only in severing activity, exhibited a much lesser effect. These results suggest that the actin-filament-severing activity of cofilin is primarily important to maintain and promote growth cone motility and extension, and that the depolymerizing activity alone is not sufficient to do so. In studies using non-neuronal cells, the inhibition of cofilin activity by the microinjection of function-blocking antibodies against cofilin or by the overexpression of LIMK1 suppressed the appearance of free barbed ends at the leading edge and inhibited the lamellipodium extension after epidermal growth factor stimulation, even in the presence of abundant free actin monomers, which indicates that cofilin is involved in lamellipodium protrusion by increasing the number of free barbed ends through its severing activity (Chan et al., 2000; Zebda et al., 2000). Mutational analyses in yeast also indicated that the severing but not the depolymerizing activity of cofilin is indispensable for yeast cell viability (Moriyama and Yahara, 1999, 2002). These findings suggest that the severing activity of cofilin is more critical for stimulating the motility and extension of growth cones, as well as for supporting lamellipodium protrusions of non-neuronal cells and yeast cell growth. However, depending on the cell type and the extent of motility, it is still possible that cofilin plays a primary role as a depolymerizing factor to accelerate the rate of the actin filament treadmill and in supplying free actin monomers for barbed end polymerization, if the concentration of free actin monomers is limited at the leading edges of motile cells (Condeelis, 2001).
LIMK1 is predominantly expressed in the nervous system of developing mammals (Proschel et al., 1995; Mori et al., 1997). We also found the expression of LIMK1 protein in chick embryo DRG neurons. The hemizygotic deletion of the LIMK1 gene locus on human chromosome 7q11.23 is implicated in the impairment of visuospatial constructive cognition in Williams syndrome (Frangiskakis et al., 1996). LIMK1 knock-out mice exhibited significant abnormalities in the morphology of spines and growth cones and brain functions, including enhanced hippocampal long-term potentiation and altered fear response and space learning (Meng et al., 2002). Growth cones of hippocampal neurons cultured from LIMK1-deficient mice were greatly reduced in size or were absent compared with findings in neurons from control animals. These data suggest that LIMK1 and its substrate cofilin play a significant role in the development and physiological functions of the nervous system. In contrast, the development of the nervous system in LIMK1 knock-out mice is grossly normal, which might be attributable to the existence of the other cofilin kinases, such as LIMK2, testicular protein kinase 1 (TESK1), and TESK2 (Amano et al., 2001;Toshima et al., 2001a,c). In fact, the content of the phosphorylated cofilin was significantly reduced but not nil in the brains of LIMK1 knock-out mice (Meng et al., 2002). The expression of LIMK2 and TESK1 in the nervous system (Mori et al., 1997; Toshima et al., 2001b) also supports this notion. Thus, LIMK1 seems to function cooperatively with other members of the LIMK/TESK family to regulate cofilin activity and actin-filament dynamics in the development and function of the nervous system.
Rho family small GTPases, such as Rho, Rac, and Cdc42, are key regulators that mediate signals from extracellular stimuli to the actin cytoskeletal reorganization (Hall, 1998). Several lines of evidence implicate Rho family GTPases and their downstream effectors in growth cone extension/retraction and guidance (Luo, 2000; Dickson, 2001; Song and Poo, 2001). For example, Rac and Rho may mediate the growth cone collapse induced by semaphorins and ephrins, respectively (Jin and Strittmatter, 1997; Wahl et al., 2000). ROCK mediates the effects of Rho on axon retraction and the inhibition of axonogenesis (Bito et al., 2000; Wahl et al., 2000). Because LIMKs are activated by PAK and ROCK, downstream effectors of Rac and Rho, they are probably involved in such processes by linking signals of Rho family GTPases to changes in cofilin activity, which in turn regulate actin-filament dynamics. Indeed, cofilin phosphorylation by LIMK1 has been shown to play a critical role in semaphorin 3A-induced growth cone collapse (Aizawa et al., 2001). In a similar manner, LIMK1-induced cofilin phosphorylation may be more generally involved in growth cone collapse and retraction, including those induced by ephrin and Nogo (Fournier and Strittmatter, 2001). In contrast to LIMK activation, mechanisms governing SSH activity remain unknown. Although previous studies have revealed that cofilin dephosphorylation is induced in response to several extracellular stimuli (Moon and Drubin, 1995), it has not been determined whether SSH activation is involved in these processes. Future studies on upstream signaling pathways regulating the activity of SSH will provide additional insight into mechanisms related to the stimulus-induced regulation of cofilin activity and also lead to a better understanding of how extracellular signals regulate the actin cytoskeleton for growth cone outgrowth and guidance.
Extracellular attractive or repulsive guidance cues induce directional movement of growth cones, which is probably supported by the spatial and temporal regulation of actin-filament dynamics and stability in growth cones. Our results suggest that cofilin is a positive regulator for growth cone motility and neurite extension, and that LIMK and SSH oppositely regulate the activity of cellular cofilin. Thus, local and temporal regulation of LIMK and SSH activities seems to play a critical role in growth cone guidance through their action on cofilin. In this respect, it will be interesting to determine the localization of active forms of LIMK and SSH in growth cones and whether or not extracellular guidance cues induce the spatially graded activation or specific localization of SSH and LIMK.
Footnotes
This work was supported by a grant-in-aid for Creative Scientific Research from the Japan Society of the Promotion of Science Research and a grant-in-aid for Scientific Research from the Ministry of Education, Science, Technology, Sports, and Culture of Japan. We thank Dr. Rachael L. Neve (Harvard Medical School) for providing the herpes simplex virus; Kazumichi Goto, Masahiro Yamamoto, and Dai Tadokoro (Tohoku University) for assistance; and Mariko Ohara for comments.
Correspondence should be addressed to Dr. Kensaku Mizuno, Department of Biomolecular Sciences, Graduate School of Life Sciences, Tohoku University, Sendai 980-8578, Japan. E-mail:kmizuno@biology.tohoku.ac.jp.
References
- 1.Agnew BJ, Minamide LS, Bamburg JR. Reactivation of phosphorylated actin depolymerizing factor and identification of the regulatory site. J Biol Chem. 1995;270:17582–17587. doi: 10.1074/jbc.270.29.17582. [DOI] [PubMed] [Google Scholar]
- 2.Aizawa H, Wakatsuki S, Ishii A, Moriyama K, Sasaki Y, Ohashi K, Sekine-Aizawa Y, Sehara-Fujisawa A, Mizuno K, Goshima Y, Yahara I. Phosphorylation of cofilin by LIM-kinase is necessary for semaphorin 3A-induced growth cone collapse. Nat Neurosci. 2001;4:367–373. doi: 10.1038/86011. [DOI] [PubMed] [Google Scholar]
- 3.Amano T, Tanabe K, Eto T, Narumiya S, Mizuno K. LIM-kinase 2 induces formation of stress fibres, focal adhesions and membrane blebs, dependent on its activation by Rho-associated kinase-catalysed phosphorylation at threonine-505. Biochem J. 2001;354:149–159. doi: 10.1042/0264-6021:3540149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amano T, Kaji N, Ohashi K, Mizuno K. Mitosis-specific activation of LIM motif-containing protein kinase and roles of cofilin phosphorylation and dephosphorylation in mitosis. J Biol Chem. 2002;277:22093–22102. doi: 10.1074/jbc.M201444200. [DOI] [PubMed] [Google Scholar]
- 5.Arber S, Barbayannis FA, Hanser H, Schneider C, Stanyon CA, Bernard O, Caroni P. Regulation of actin dynamics through phosphorylation of cofilin by LIM-kinase. Nature. 1998;393:805–809. doi: 10.1038/31729. [DOI] [PubMed] [Google Scholar]
- 6.Bamburg JR. Proteins of the ADF/cofilin family: essential regulators of actin dynamics. Annu Rev Cell Dev Biol. 1999;15:185–230. doi: 10.1146/annurev.cellbio.15.1.185. [DOI] [PubMed] [Google Scholar]
- 7.Bamburg JR, Bray D. Distribution and cellular localization of actin depolymerizing factor. J Cell Biol. 1987;105:2817–2825. doi: 10.1083/jcb.105.6.2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bito H, Furuyashiki T, Ishihara H, Shibasaki Y, Ohashi K, Mizuno K, Maekawa M, Ishizaki T, Narumiya S. A critical role for a Rho-associated kinase, p160ROCK, in determining axon outgrowth in mammalian CNS neurons. Neuron. 2000;26:431–441. doi: 10.1016/s0896-6273(00)81175-7. [DOI] [PubMed] [Google Scholar]
- 9.Carlier MF, Laurent V, Santolini J, Melki R, Didry D, Xia GX, Hong Y, Chua NH, Pantaloni D. Actin depolymerizing factor (ADF/cofilin) enhances the rate of filament turnover: implication in actin-based motility. J Cell Biol. 1997;136:1307–1322. doi: 10.1083/jcb.136.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carlier MF, Ressad F, Pantaloni D. Control of actin dynamics in cell motility: role of ADF/cofilin. J Biol Chem. 1999;274:33827–33830. doi: 10.1074/jbc.274.48.33827. [DOI] [PubMed] [Google Scholar]
- 11.Chan AY, Bailly M, Zebda N, Segall JE, Condeelis JS. Role of cofilin in epidermal growth factor-stimulated actin polymerization and lamellipod protrusion. J Cell Biol. 2000;148:531–542. doi: 10.1083/jcb.148.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen H, Bernstein BW, Bamburg JR. Regulating actin-filament dynamics in vivo. Trends Biochem Sci. 2000;25:19–23. doi: 10.1016/s0968-0004(99)01511-x. [DOI] [PubMed] [Google Scholar]
- 13.Condeelis J. How is actin polymerization nucleated in vivo? Trends Cell Biol. 2001;11:288–293. doi: 10.1016/s0962-8924(01)02008-6. [DOI] [PubMed] [Google Scholar]
- 14.Dickson BJ. Rho GTPases in growth cone guidance. Curr Opin Neurobiol. 2001;11:103–110. doi: 10.1016/s0959-4388(00)00180-x. [DOI] [PubMed] [Google Scholar]
- 15.Edwards DC, Sanders LC, Bokoch GM, Gill GN. Activation of LIM-kinase by Pak1 couples Rac/Cdc42 GTPases signaling to actin cytoskeletal dynamics. Nat Cell Biol. 1999;1:253–259. doi: 10.1038/12963. [DOI] [PubMed] [Google Scholar]
- 16.Fournier AE, Strittmatter SM. Repulsive factors and axon regeneration in the CNS. Curr Opin Neurobiol. 2001;11:89–94. doi: 10.1016/s0959-4388(00)00178-1. [DOI] [PubMed] [Google Scholar]
- 17.Frangiskakis JM, Ewart AK, Morris CA, Mervis CB, Bertrand J, Robinson BF, Klein BP, Ensing GJ, Everett LA, Green ED, Proschel C, Gutowski NJ, Noble M, Atkinson DL, Odelberg SJ, Keating MT. LIM-kinase 1 hemizygosity implicated in impaired visuospatial constructive cognition. Cell. 1996;86:59–69. doi: 10.1016/s0092-8674(00)80077-x. [DOI] [PubMed] [Google Scholar]
- 18.Goshima Y, Nakamura F, Strittmatter P, Strittmatter SM. Collapsin-induced growth cone collapse mediated by an intracellular protein related to UNC-33. Nature. 1995;376:509–514. doi: 10.1038/376509a0. [DOI] [PubMed] [Google Scholar]
- 19.Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- 20.Jin Z, Strittmatter SM. Rac1 mediates collapsin-1-induced growth cone collapse. J Neurosci. 1997;17:6256–6263. doi: 10.1523/JNEUROSCI.17-16-06256.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuhn TB, Meberg PJ, Brown MD, Bernstein BW, Minamide LS, Jensen JR, Okada K, Soda EA, Bamburg JR. Regulating actin dynamics in neuronal growth cones by ADF/cofilin and rho family GTPases. J Neurobiol. 2000;44:126–144. [PubMed] [Google Scholar]
- 22.Luo L. Rho GTPases in neuronal morphogenesis. Nat Rev Neurosci. 2000;1:173–180. doi: 10.1038/35044547. [DOI] [PubMed] [Google Scholar]
- 23.Maekawa M, Ishizaki T, Boku S, Watanabe N, Fujita A, Iwamatsu A, Obinata T, Ohashi K, Mizuno K, Narumiya S. Signaling from Rho to the actin cytoskeleton through protein kinases ROCK and LIM-kinase. Science. 1999;285:895–898. doi: 10.1126/science.285.5429.895. [DOI] [PubMed] [Google Scholar]
- 24.McGrath JL, Osborn EA, Tardy YS, Dewey CF, Jr, Hartwig JH. Regulation of the actin cycle in vivo by actin filament severing. Proc Natl Acad Sci USA. 2000;97:6532–6537. doi: 10.1073/pnas.100023397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meberg PJ. Signal-regulated ADF/cofilin activity and growth cone motility. Mol Neurobiol. 2000;21:97–107. doi: 10.1385/MN:21:1-2:097. [DOI] [PubMed] [Google Scholar]
- 26.Meberg PJ, Bamburg JR. Increase in neurite outgrowth mediated by overexpression of actin depolymerizing factor. J Neurosci. 2000;20:2459–2469. doi: 10.1523/JNEUROSCI.20-07-02459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meng Y, Zhang Y, Tregoubov V, Janus C, Cruz L, Jackson M, Lu WY, MacDonald JF, Wang JY, Falls DL, Jia Z. Abnormal spine morphology and enhanced LTP in LIMK-1 knockout mice. Neuron. 2002;35:121–133. doi: 10.1016/s0896-6273(02)00758-4. [DOI] [PubMed] [Google Scholar]
- 28.Mitchison T, Kirschner M. Cytoskeletal dynamics and nerve growth. Neuron. 1988;1:761–772. doi: 10.1016/0896-6273(88)90124-9. [DOI] [PubMed] [Google Scholar]
- 29.Mizuno K, Okano I, Ohashi K, Nunoue K, Kuma K, Miyata T, Nakamura T. Identification of a human cDNA encoding a novel protein kinase with two repeats of the LIM/double zinc finger motif. Oncogene. 1994;9:1605–1612. [PubMed] [Google Scholar]
- 30.Moon A, Drubin DG. The ADF/cofilin proteins: stimulus-responsive modulators of actin dynamics. Mol Biol Cell. 1995;6:1423–1431. doi: 10.1091/mbc.6.11.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mori T, Okano I, Mizuno K, Tohyama M, Wanaka A. Comparison of tissue distribution of two novel serine/threonine kinase genes containing the LIM motif (LIMK-1 and LIMK-2) in the developing rat. Mol Brain Res. 1997;45:247–254. doi: 10.1016/s0169-328x(96)00257-4. [DOI] [PubMed] [Google Scholar]
- 32.Moriyama K, Yahara I. Two activities of cofilin, severing and accelerating directional depolymerization of actin filaments, are affected differentially by mutations around the actin-binding helix. EMBO J. 1999;18:6752–6761. doi: 10.1093/emboj/18.23.6752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moriyama K, Yahara I. The actin-severing activity of cofilin is exerted by the interplay of three distinct sites on cofilin and essential for cell viability. Biochem J. 2002;365:147–155. doi: 10.1042/bj20020231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mueller BK. Growth cone guidance: first steps towards a deeper understanding. Annu Rev Neurosci. 1999;22:351–388. doi: 10.1146/annurev.neuro.22.1.351. [DOI] [PubMed] [Google Scholar]
- 35.Niwa R, Nagata-Ohashi K, Takeichi M, Mizuno K, Uemura T. Control of actin reorganization by Slingshot, a family of phosphatases that dephosphorylate ADF/cofilin. Cell. 2002;108:233–246. doi: 10.1016/s0092-8674(01)00638-9. [DOI] [PubMed] [Google Scholar]
- 36.Nunoue K, Ohashi K, Okano I, Mizuno K. LIMK-1 and LIMK-2, two members of a LIM motif-containing protein kinase family. Oncogene. 1995;11:701–710. [PubMed] [Google Scholar]
- 37.Ohashi K, Toshima J, Tajinda K, Nakamura T, Mizuno K. Molecular cloning of a chick lung cDNA encoding a novel protein kinase with N-terminal two LIM/double zinc finger motifs. J Biochem. 1994;111:636–642. doi: 10.1093/oxfordjournals.jbchem.a124573. [DOI] [PubMed] [Google Scholar]
- 38.Ohashi K, Nagata K, Maekawa M, Ishizaki T, Narumiya S, Mizuno K. Rho-associated kinase ROCK activates LIM-kinase 1 by phosphorylation at threonine 508 within the activation loop. J Biol Chem. 2000;275:3577–3582. doi: 10.1074/jbc.275.5.3577. [DOI] [PubMed] [Google Scholar]
- 39.Okano I, Hiraoka J, Otera H, Nunoue K, Ohashi K, Iwashita S, Hirai M, Mizuno K. Identification and characterization of a novel family of serine/threonine kinases containing two N-terminal LIM motifs. J Biol Chem. 1995;270:31321–31330. doi: 10.1074/jbc.270.52.31321. [DOI] [PubMed] [Google Scholar]
- 40.Pollard T, Blanchoin L, Mullins RD. Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu Rev Biophys Biomol Struct. 2000;29:545–576. doi: 10.1146/annurev.biophys.29.1.545. [DOI] [PubMed] [Google Scholar]
- 41.Proschel C, Blouin MJ, Gutowski NJ, Ludwig R, Noble M. Limk1 is predominantly expressed in neural tissues and phosphorylates serine, threonine and tyrosine residues in vitro. Oncogene. 1995;11:1271–1281. [PubMed] [Google Scholar]
- 42.Sasaki Y, Cheng C, Uchida Y, Nakajima O, Ohshima T, Yagi T, Taniguchi M, Nakayama T, Kishida R, Kudo Y, Ohno S, Nakamura F, Goshima Y. Fyn and Cdk5 mediate semaphorin-3A signaling, which is involved in regulation of dendrite orientation in cerebral cortex. Neuron. 2002;35:907–920. doi: 10.1016/s0896-6273(02)00857-7. [DOI] [PubMed] [Google Scholar]
- 43.Song HJ, Poo MM. The cell biology of neuronal navigation. Nat Cell Biol. 2001;3:E81–E88. doi: 10.1038/35060164. [DOI] [PubMed] [Google Scholar]
- 44.Sumi T, Matsumoto K, Takai Y, Nakamura T. Cofilin phosphorylation and actin cytoskeletal dynamics regulated by rho- and Cdc42-activated LIM-kinase-2. J Cell Biol. 1999;147:1519–1532. doi: 10.1083/jcb.147.7.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tanaka E, Sabry J. Making the connection: cytoskeletal rearrangements during growth cone guidance. Cell. 1995;83:171–176. doi: 10.1016/0092-8674(95)90158-2. [DOI] [PubMed] [Google Scholar]
- 46.Tessier-Lavigne M, Goodman CS. The molecular biology of axon guidance. Science. 1996;274:1123–1133. doi: 10.1126/science.274.5290.1123. [DOI] [PubMed] [Google Scholar]
- 47.Toshima J, Toshima JY, Amano T, Yang N, Narumiya S, Mizuno K. Cofilin phosphorylation by protein kinase testicular protein kinase 1 and its role in integrin-mediated actin reorganization and focal adhesion formation. Mol Biol Cell. 2001a;12:1131–1145. doi: 10.1091/mbc.12.4.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Toshima J, Toshima JY, Suzuki M, Noda T, Mizuno K. Cell-type-specific expression of a TESK1 promoter-linked lacZ gene in transgenic mice. Biochem Biophys Res Commun. 2001b;286:566–573. doi: 10.1006/bbrc.2001.5404. [DOI] [PubMed] [Google Scholar]
- 49.Toshima J, Toshima JY, Takeuchi K, Mori R, Mizuno K. Cofilin phosphorylation and actin reorganization activities of testicular protein kinase 2 and its predominant expression in testicular Sertoli cells. J Biol Chem. 2001c;276:31449–31458. doi: 10.1074/jbc.M102988200. [DOI] [PubMed] [Google Scholar]
- 50.Wahl S, Barth H, Ciossek T, Aktories K, Mueller BK. Ephrin-A5 induces collapse of growth cones by activating Rho and Rho kinase. J Cell Biol. 2000;149:263–270. doi: 10.1083/jcb.149.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang N, Higuchi O, Ohashi K, Nagata K, Wada A, Kangawa K, Nishida E, Mizuno K. Cofilin phosphorylation by LIM-kinase 1 and its role in Rac-mediated actin reorganization. Nature. 1998;393:809–812. doi: 10.1038/31735. [DOI] [PubMed] [Google Scholar]
- 52.Zebda N, Bernard O, Bailly M, Welti S, Lawrence DS, Condeelis JS. Phosphorylation of ADF/cofilin abolishes EGF-induced actin nucleation at the leading edge and subsequent lamellipod extension. J Cell Biol. 2000;151:1119–1128. doi: 10.1083/jcb.151.5.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]