Abstract
Hormones and nuclear receptors (NRs) play important roles in brain development and function. The recently identified steroid receptor coactivator (SRC) family contains three homologous members that can enhance transcriptional activities of NRs and certain non-NR transcription factors. To study the role of SRC-1 in brain development and function, we examined the spatial and temporal expression patterns of SRC-1 and characterized the phenotypes of brain development and function in SRC-1 knock-out (SRC-1−/−) mice. In the adult mouse brain, SRC-1 is highly expressed in the olfactory bulb, hippocampus, piriform cortex, amygdala, hypothalamus, cerebellum, and brainstem. Multiple behavioral tests revealed that SRC-1−/−mice exhibit normal hippocampal function but moderate motor dysfunction. The behavior phenotypes correlate with the spatial distribution of the SRC family members. In most brain structures where SRC-1 is expressed, SRC-2 is expressed at lower levels; however, SRC-3 mRNA is detectable only in the hippocampus. In the adult cerebellum, Purkinje cells (PCs) preferentially express SRC-1 over SRC-2, but SRC-2 mRNA is slightly elevated in the SRC-1−/−PCs. During embryonic development, SRC-1 is expressed in the cerebellar primordium. SRC-2 is expressed in PCs after postnatal day (P) 10. Time course analysis revealed that the precursors of SRC-1−/− PCs were generated ∼2 d later than wild-type precursor cells. A further delay in SRC-1−/− PC maturation was detected at the neonatal stage. The morphology and number of SRC-1−/− PCs were equivalent to wild type by P10; this timing correlated with the early expression of SRC-2 in the SRC-1−/−PCs. These results demonstrate that the relative levels of SRC expression are region specific, and the degree of overlapping expression may influence their functional redundancy. Disruption of SRC-1 specifically delays the PC development and maturation in early stages and results in moderate motor dysfunction in adulthood.
Keywords: coactivator, nuclear receptor, gene expression, animal model, motor function, Purkinje cell
Introduction
Steroid, thyroid, and retinoid hormones play important roles in brain development and function (McEwen, 1994; Lopes da Silva and Burbach, 1995). The gonadal steroids affect sexual differentiation of the brain and regulate reproductive behaviors by influencing neuroendocrine functions (Tetel, 2000; McEwen, 2001). The adrenal steroids are required for survival of the pyramidal neurons in the dentate gyrus, but the stress-induced increase of glucocorticoids is believed to produce atrophy of dendrites of CA3 pyramidal neurons in the hippocampus (McEwen, 1994). In addition, hypothyroidism is accompanied by a significant reduction in Purkinje cell dendritic arborization and synaptogenesis (Koibuchi and Chin, 2000). In the brain, most hormonal actions are mediated by their cognate nuclear receptors (NRs), which are ligand-regulated transcription factors (Tsai and O'Malley, 1994). Therefore, disruption of either estrogen or progesterone receptors in female mice completely diminishes their sexual receptivity (Mani et al., 1997; Rissman et al., 1997). Furthermore, the glucocorticoid receptor is essential for the regulation of hypothalamus–pituitary–adrenal axis and stress response (Reichardt et al., 2000). In addition, severe defects in brain development and function have been observed in mice lacking individual orphan NRs, including retinoid-related orphan receptor α (RORα), chicken ovalbumin upstream promoter transcription factor 1, nuclear-related receptor 1, and rev-erbAα (Zetterstrom et al., 1997;Steinmayr et al., 1998; Zhou et al., 1999; Chomez et al., 2000).
The transcriptional activities of NRs are mediated by interaction with several classes of coactivators in a ligand-dependent manner (Glass and Rosenfeld, 2000; McKenna and O'Malley, 2002; Xu and O'Malley, 2002). The first and best characterized steroid receptor coactivator (SRC) family contains three homologous members including SRC-1 (Onate et al., 1995), SRC-2 (also known as GRIP1 and TIF2) (Hong et al., 1996; Voegel et al., 1996), and SRC-3 (also known as p/CIP, RAC3, AIB1, ACTR, and TRAM-1) (Anzick et al., 1997; Chen et al., 1997; Li et al., 1997; Takeshita et al., 1997; Torchia et al., 1997;Suen et al., 1998). Biochemical experiments have revealed that SRC coactivators function in several ways. These include recruitment of histone acetyltransferases such as CREB-binding protein (CBP), adenovirus E1A-binding protein p300, and p300/CBP-associated factor (p/CAF) (Torchia et al., 1997; Li et al., 2000), histone methyltransferases such as coactivator-associated arginine methyltransferase-1 and protein arginineN-methyltransferase-1 for chromatin remodeling (Koh et al., 2001), interaction with other coactivators such as activating signal cointegrator 2 (ASC-2), steroid receptor RNA activator (SRA), and RNA-binding DEAD-box proteins (p72/p68) (Lanz et al., 1999; Lee et al., 1999; Watanabe et al., 2001), and contact with certain general transcription factors such as transcription factor IIB and TATA-binding protein (Takeshita et al., 1996).
To study the in vivo function of SRC-1, we generated SRC-1 null (SRC-1−/−) mice by gene targeting. We found that although SRC-1−/−mice had normal body size, their hormone target tissues such as the uterus, prostate, testis, and mammary gland exhibited decreased growth and development in response to steroid hormones (Xu et al., 1998). A partial resistance to thyroid hormone (TH) was also observed in SRC-1−/−mice (Weiss et al., 1999). These results validated SRC-1 as an in vivo NR coactivator. However, the role of SRC-1 in brain development and function remained to be characterized.
In this study, we compared the spatiotemporal expression patterns of all three SRC coactivators in brains of wild-type (WT) and SRC-1−/−mice and characterized the role of SRC-1 in brain development and function. We show that SRC coactivators are preferentially expressed in particular brain regions and disruption of SRC-1 causes delayed development of Purkinje cells at the neonatal stage and moderate motor dysfunction in adulthood.
Materials and Methods
Animals. SRC-1−/−mice were initially generated from embryonic stem (ES) cells with a targeted deletion of ∼9 kb of SRC-1 genomic sequence encoding 446 amino acids from Met-381 to Thr-826. The targeting event in ES cells eliminated all functional domains of SRC-1 for transcriptional activation, histone acetyltransferase activity, and interactions with NRs and other coactivators such as CBP/p300 and p/CAF (Xu et al., 1998). To achieve a homogenous genetic background, the initial SRC-1 mouse line with hybrid genetic background of C57BL/6J and 129/SVJ stains was backcrossed to C57BL/6J for six generations. Subsequently, WT and age-matched SRC-1−/−mice for this study were produced from heterozygous breeding pairs. Mice were weaned at 3 weeks after birth and allowed free access to water and lab chow. They were housed in an environment with a controlled temperature of 19°C and 12 hr alternating darkness and artificial light cycles. All mice used for behavioral tests were 15–20 weeks old. All parts of apparatus for behavioral tests were cleaned with deionized water and 70% ethanol after each trial. Behavioral tests were performed between 1 and 6 P.M. in a soundproof room in which external noise was greatly reduced (∼45 dB at 500 Hz). The test room was equipped with an air cleaning and air conditioning system. All animal experiments were performed according to approved protocols at Baylor College of Medicine.
Hanging wire test. Eight WT and 11 SRC-1−/−mice were tested on their ability to hang from a screen with wire bars (10 × 18 cm area, 1 mm in diameter spaced 1 cm apart). After mice were placed on the bars, the screen was waved gently in the air three times to force the mice to grip the wires. The screen was turned upside down and latency to fall (maximum 60 sec) was measured. Mice that fell in <10 sec were provided a second trial.
Rotarod test. Mice were placed on a rod (3 cm in diameter), and the rod was accelerated from 4 to 40 rpm in 5 min. The time periods that individual mice spent on the rod without falling were recorded. Three trials with a 30 min intertrial interval were performed every day for 3 d. Twenty WT and 20 age-matched SRC-1−/−mice were tested.
Morris water task and swim speed. A circular polypropylene pool (120 cm in diameter) was filled with opaque water. In the hidden platform version, a 10 cm-diameter platform was submerged 1 cm below the water surface. Ten WT and 9 SRC-1−/−mice were initially guided to the platform and allowed to remain for 1 min. Subsequently, they were placed in the pool and allowed to search for the platform for 1 min using visible external cues. Each mouse was allowed to stay on the platform for 20 sec before the mouse was carried back to its home cage. Escape latency, the time to find the platform, was recorded in each trial. Each trial was initiated from a different quadrant of the pool, and four consecutive trials with 60 min intervals were performed per day for 7 d. The latencies of the four trials in the same day were averaged and used for statistical analysis. Probe trial was performed after 7 d training with a computerized video tracking device (HVS Image) to track mouse movement. In the probe trial, the platform was removed, and the trained mice were allowed to search for the platform for 1 min. The latencies to find the platform position, the swim distances, and the swim speeds were measured in four consecutive trials with 15 min intervals.
Real-time RT-PCR. Real-time RT-PCR was performed with the One Step Master Mix reagent (Applied Biosystems, Foster City, CA) and total RNA samples by using the ABI 7700 Sequence Detection System (Applied Biosystems). In the measurement of SRC-1 mRNA, the sequences of the forward and reverse primers and the TaqMan probe were tatctctccagcccatggtgt, caaagttcccttggttgttgc, and tcatccacgttgccaccatcca. In the measurement of SRC-2 mRNA, the forward and reverse primers and the TaqMan probe were gcagcacaggaaatagccatagt, catggccctcgctgagg, and accaacagttccctcaatgcactgcaa. In the measurement of SRC-3 mRNA, the forward and reverse primers and the TaqMan probe were agcaaaggccacaagaaactg, ggtcaaggaggaatggcctc, and cagttactcacgtgctcctccgacgac. Parallel measurements of the 18S RNA were performed for each sample to serve as endogenous controls. Relative expression levels of SRC coactivators were determined by the ratios of SRC mRNA levels to 18S RNA levels.
In situ hybridization. Mice were decapitated and their brains were frozen immediately with O.C.T. compound in an ethanol–dry-ice bath. Sections were cut at 12 μm on a cryostat and thaw mounted on Superfrost Plus slides (VWR Scientific, West Chester, PA). The sections were fixed in 4% paraformaldehyde as described previously (Nishihara et al., 1998). To prepare DNA templates for transcribing riboprobes, individual mouse cDNA fragments corresponding to a 527 bp sequence of SRC-1 (nucleotides 3910–4436, GenBank U56920), a 446 bp sequence of SRC-2 (nucleotides 4017–4462, GenBank AF000582), and a 498 bp sequence of SRC-3 (nucleotides 3377–3874, GenBankAF000581) were subcloned into EcoRI/BamHI sites of the Bluescript II SK plasmid, respectively. These plasmids were linearized with either BamHI or EcoRI digestion. The 35S-UTP-labeled antisense or sense riboprobes were transcribed using T7 or T3 RNA polymerase, respectively. Hybridization solution was made of 1 × 107 cpm/ml of labeled probe, 50% formamide, 0.3 m NaCl, 10% dextran sulfate, 20 mm Tris-HCl, pH 8.0, 10 mmsodium phosphate, pH 8.0, 5 mm EDTA, 1× Denhardt's solution, and 500 μg/ml yeast tRNA. Each slide was applied with 100 μl of hybridization solution. All slides were coverslipped and incubated at 55°C for 16 hr in a humidified chamber. The slides with sections were washed twice at 65°C for 30 min in 2× SSC containing 50% formamide and 100 mm β-mercaptoethanol, and three times at 37°C in 10 mm Tris-HCl, pH 7.5, containing 0.5m NaCl and 5 mm EDTA. Sections were digested with ribonuclease A (20 μg/ml) at 37°C for 30 min. Slides were dehydrated in an ethanol series and dried at room temperature. Then, slides were dipped in the photographic emulsion (Kodak NTB2), exposed at 4°C for 6 d, developed in Kodak D-19, and fixed with Kodak Fix. These sections were counterstained with hematoxylin, dehydrated through a graded series of ethanol, and coverslipped.
Paraffin sections were used when immunohistochemical staining was required after in situ hybridization. Sections were dewaxed, rehydrated, and treated with proteinase K (20 μg/ml) in 50 mm Tris-HCl, pH 7.5, and 5 mm EDTA for 7.5 min before hybridization was performed as described above. After hybridization, sections were digested with ribonuclease A and kept in PBS until immunohistochemical staining. Autoradiography of the immunostained sections was performed as described above.
Immunohistochemistry. Mice were anesthetized with Avertin and perfused through the heart with 4% paraformaldehyde in PBS. The brain was removed and kept in the same fixative for 16 hr at 4°C. Samples were dehydrated in ethanol series, cleared in xylene, and embedded in paraffin. Sections were cut at 7 μm. For SRC-1 immunostaining, sections were dewaxed, rehydrated, and boiled for 10 min in 10 mm citrate buffer, pH 6.0 (Nishihara et al., 2000). After washing in PBS, endogenous peroxidase was blocked using 0.3% H2O2 in methanol for 15 min. To reduce nonspecific binding of antibodies, sections were washed in PBS again and preincubated with 500 μg/ml goat IgG and 5% bovine serum albumin (BSA) in PBS for 60 min at room temperature. Sections were incubated in a 1:400 dilution of the polyclonal SRC-1 antibodies (Santa Cruz Biotechnology, Santa Cruz CA) overnight at 4°C. After being washed in PBS with 0.075% Brij 35 (Sigma, St. Louis, MO), sections were further incubated with the horseradish peroxidase (HRP)–goat anti-mouse IgG (1:500 dilution) for 2 hr at room temperature. Sections were washed again in PBS with 0.075% Brij 35. The sites of HRP were visualized by 3,3′-diaminobenzidine (DAB, Sigma). Finally, sections were counterstained with methyl green.
Immunostaining of the calbindin and synaptophysin was performed using the Histomouse kit (Zymed Laboratories, South San Francisco, CA). Monoclonal antibodies against calbindin-D28k (Sigma) and synaptophysin (Sigma) were diluted at 1:500 and 1:50, respectively. Sections were treated with DAB and counterstained with methyl green.
BrdU labeling of the Purkinje cell precursors during embryonic development was performed as described previously with slight modifications (Hayashi et al., 1988; Vogel et al., 2000). Briefly, successful mating between SRC-1 heterozygous males and females was identified by checking coitus plugs in the morning. The day when the coitus plug was found was defined as 0.5 d postcoitum (d.p.c.). On 10.5, 12.5, and 13.5 d.p.c., a single dose of BrdU (50 mg/kg body weight) was injected intraperitoneally into the pregnant females. BrdU labeling of Purkinje cells was examined on postnatal day (P) 5. All pups were genotyped by PCR as described previously (Xu et al., 1998). Pups were decapitated, and their brains were dissected and fixed in 10% formalin overnight at room temperature. Samples were embedded in paraffin and sectioned as described above. Deparaffinized and rehydrated sections were immersed in methanol containing 3% H2O2 to inactivate endogenous peroxidase. The sections were treated with 0.02% pepsin and 0.01N HCl in PBS for 60 min at 37°C and denatured with 2N HCl for 45 min at room temperature. These sections were neutralized in 0.1m sodium borate for 45 min at room temperature and washed with PBS. BrdU incorporation was detected using a monoclonal antibody specific to BrdU (Sigma) at 1:50 dilution.
Cerebellar neuronal culture and immunocytochemistry. Dissociated cerebellar neuronal cultures were prepared from the cerebellum of mouse embryo on 18 d.p.c. according to the protocol for Purkinje cell culture (Furuya et al., 1998). In brief, 5.0 × 106 cells/ml were plated onto glass coverslips (12 mm in diameter; Becton Dickinson Labware, Mountain View, CA) coated with poly-l-lysine and cultured in DMEM/Ham's F-12 medium containing bovine insulin (10 μg/ml), BSA (100 μg/ml), gentamicin (10 μg/ml), glutamycine (200 μg/ml), human apotransferrin (100 μg/ml), putrescine (100 nm), sodium selenite (30 nm), progesterone (20 nm), tri-iodothyronine (T3) (0.5 ng/ml), and fetal bovine serum (10%). One day after plating, the medium was changed to a serum-free medium. The cultures were maintained at 37°C in a humidified incubator supplied with 5% CO2. Half of the culture medium was changed weekly with the same fresh medium.
For immunostaining, cells were fixed with 4% paraformaldehyde in PBS for 30 min at 4°C and treated with PBS containing 0.05% Triton X-100, 500 μg/ml goat IgG, and 1% BSA for 30 min at room temperature. Cells were incubated with a monoclonal antibody against calbindin-D28k (Sigma) at 1:400 dilution for 60 min at room temperature and then with a biotinylated anti-mouse IgG. The biotinylated secondary antibody was visualized by a streptavidin peroxidase conjugate (Vectastain, Vector Laboratories, Burlingame, CA) and subsequent addition of DAB.
Data analysis. Statistical comparisons between two groups were made using unpaired Student's t tests. ANOVA with repeated measure was used in the analysis of behavioral experiments. Experimental observers were blinded to mouse genotypes in all behavioral studies. All values are expressed as mean ± SEM.
Results
Expression of SRC-1 in the adult mouse brain
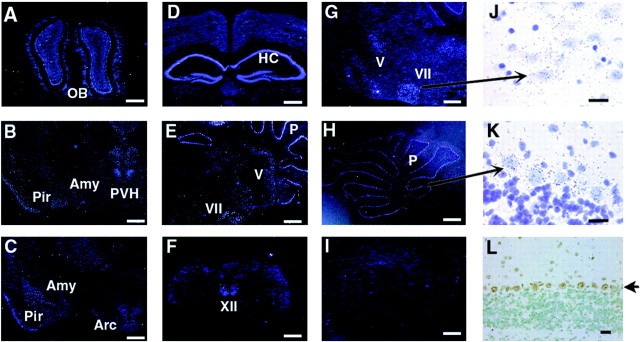
To determine potential regions of SRC-1 function in the brain, the expression pattern of SRC-1 mRNA in the adult mouse brain was examined by in situ hybridization. SRC-1 was expressed at high levels in the olfactory bulb, piriform cortex, hippocampus (CA1, CA2, and CA3 regions and dentate gyrus), amygdala complex, hypothalamus (relatively high in the paraventricular nucleus), cerebellum, and brainstem (Fig.1A–H). Areas with strong hybridization signals in the brainstem include the facial nucleus (Fig.1E,G,J), trigeminal nucleus (both motor and mesencephalic nuclei) (Fig.1E,G), and hypogrossal nucleus (Fig. 1F). In the cerebellum, SRC-1 was selectively expressed in the Purkinje cell layer but was undetectable in the granule and molecule layers (Fig.1E,H,K). The SRC-1 protein was specifically detected in nuclei of Purkinje cells by immunohistochemical analysis (Fig. 1L). In addition, moderate levels of SRC-1 mRNA were observed in the thalamus and isocortex (Fig. 1B–D). These data were substantiated by the absence of SRC-1 mRNA signal in SRC-1−/−mice when the same antisense probe was used in SRC-1−/−mice (Fig. 1I) and when a sense control probe was used in WT mice (data not shown). These results indicate that the relative expression levels of SRC-1 are dependent on specific brain regions, suggesting that SRC-1 may have a functional preference in those brain regions with higher levels of SRC-1.
Fig. 1.
Expression of SRC-1 mRNA in the adult mouse brain.In situ hybridization was performed with35S-labeled SRC-1 antisense RNA probe. Coronal (A–D, F,L) and sagittal (E,G–K) brain sections were prepared from 10-week-old WT mice (A–H,J, K). As a negative control,in situ hybridization was performed under identical conditions on the sagittal section (I) prepared from the cerebellum region of SRC-1−/−mice of the same age. A–I andJ–L were taken under dark-field and bright-field lighting conditions, respectively. J andK were stained with hematoxylin after in situ hybridization, and their images were taken at higher magnifications from the corresponding areas indicated inG and H. Arrows indicate the facial nucleus in J and Purkinje cells inK and L. In L, SRC-1 protein (brown color) was specifically detected in the nuclei of Purkinje cells by immunostaining. Scale bars:A–I, 600 μm;J–L, 25 μm. OB, Olfactory bulb; Amy, amygdala complex;Pir, piriform cortex; PVH, paraventricular nucleus of the hypothalamus; Arc, arcuate nucleus; HC, hippocampus; P, Purkinje cell layer; V, trigeminal nucleus;VII, facial nucleus; XII, hypoglossal nucleus.
SRC-1−/−mice exhibited a moderate motor dysfunction
To explore the possible role of SRC-1 in brain development, the general brain structure and histology of the adult SRC-1−/−mice were compared with their sex- and age-matched WT mice by staining serial sections of the entire brain with hematoxylin and eosin (H&E) or Nissl. However, no gross morphological defect was observed in the adult brains of SRC-1−/−mice (data not shown). To assess the role of SRC-1 in brain function, we subsequently performed a battery of behavioral tests to compare specific neural functions between WT and SRC-1−/−mice. Motor coordination, strength, and balance were assessed by hanging wire and rotarod tests. In the hanging wire test, SRC-1−/−mice fell 29% earlier than WT mice (Fig.2A). Statistical analysis showed significant differences (p = 0.03) in latencies to fall between WT and SRC-1−/−mice. The average body weight of SRC-1−/−mice is similar to WT mice (Xu et al., 1998), so differential weights cannot account for the difference.
Fig. 2.
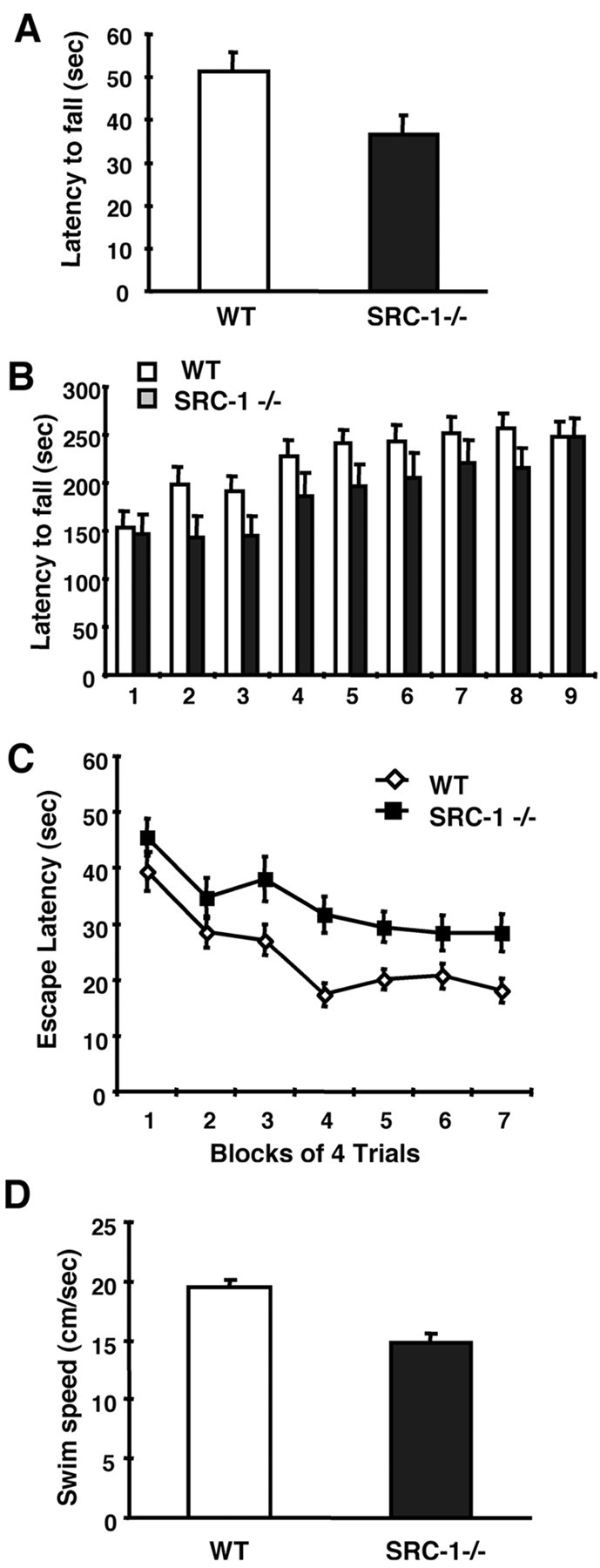
Behavioral analyses. A, Hanging wire test. WT (n = 8) and SRC-1−/−(n = 11) mice were placed on bars and turned upside down. The latency to fall was measured. B, Rotarod test. WT (n = 20) and SRC-1−/−(n = 20) mice were trained three times per day for a period of 3 d. Latency to fall from the accelerating rotarod was measured. C, Morris water task. WT (n = 10) and SRC-1−/−(n = 9) mice were trained with four trials per day for 7 d. Mean escape latency was measured in the hidden platform version of the Morris water task. D, Swim speed. WT (n = 9) and SRC-1−/−(n = 9) mice were allowed to swim in the pool for 60 sec. Mean swim speed was measured from four consecutive trials.
In the rotarod test, a total of nine trials were recorded (Fig.2B). SRC-1−/−mice exhibited identical latencies to fall in the first and last trials of this experiment. However, a shorter latency to fall was detected consistently in SRC-1−/−mice from trials 2 thru 9. For trials 2 and 3, WT mice significantly improved their performance and stayed on the rotarod for ∼200 sec (compared with 150 sec in trial 1). In contrast, SRC-1−/−mice did not improve and remained on the rotarod for the same amount of time as in trial 1 (150 sec). In addition, WT mice rapidly reached the maximum value of their latency to fall after four trials. Although SRC-1−/−mice gradually improved their performance in the rotarod test, they did not reach a maximum value of latency until the ninth trial (Fig.2B). These results suggest that SRC-1 is required for normal motor learning.
Because SRC-1 was highly expressed in regions of the hippocampus and amygdala, we assayed the animals in several different learning and memory tests. During training in the Morris water maze (Morris et al., 1982), the SRC-1−/−mice had significantly delayed escape latencies compared with WT mice (F(1,17) = 8.3; p = 0.01) (Fig. 2C). The increased escape latency could result from an impairment of learning and memory, or from a dysfunction of motor skills required by the test. To assess spatial memory after training, the platform was removed from the pool after the 28th trial. Both WT and SRC-1−/−mice searched for the missing platform selectively during this probe trial in the quadrant where the platform was located previously (data not shown). Tests of cued and contextual fear conditioning and passive avoidance also failed to reveal any impairment of learning and memory (data not shown).
We next analyzed swim speed for WT and SRC-1−/−mice in the absence of the platform. SRC-1−/−mice showed a significantly slower swim speed compared with WT mice (14.9 ± 0.8 vs 19.5 ± 0.6 cm/sec; p = 0.0002) (Fig. 2D). Therefore, the relatively poor performance of the null mice during training in the Morris water task can be attributed to a slower swim speed from a motor impairment.
Because of the limitations of the above tests, we have recognized the difficulties in distinguishing a motor deficit from muscle weakness. In an attempt to search for answers, we extracted RNA samples from the femoral muscle and quantitatively measured the expression levels of the SRC family members by performing TaqMan Real-Time RT-PCR. The relative amounts of SRC-1, SRC-2, and SRC-3 mRNAs in the skeletal muscle were 4.6:1.0:267. Therefore, the total number of SRC-1 mRNA molecules in the skeletal muscle (∼2.4 molecules per nanogram total RNA) was ∼60-fold lower than the number of SRC-3 mRNA molecules (∼144 molecules per nanogram total RNA). Real-Time RT-PCR is an extremely sensitive technique providing very accurate measurements. When in situ hybridization was used, SRC-1 mRNA could not be detected in the skeletal muscle (data not shown). Furthermore, although the SRC-3 mRNA level was much higher than SRC-1 and SRC-2 in the skeletal muscle when measured by real-time RT-PCR, the level of SRC-3 expression in the muscle was still lower than the detectable threshold of X-gal staining in the reporter mice with a LacZ knock-in (Xu et al., 2000). Thus, it can be concluded that SRC-1 is expressed at extremely low levels in the skeletal muscle, and therefore it is very unlikely that disruption of SRC-1 will directly affect the muscle strength. Collectively, our results from the hanging wire test, rotarod test, Morris water task, and gene expression analyses suggest that SRC-1-deficient mice exhibit a moderate motor dysfunction.
Expression of other members of the SRC family in the brain
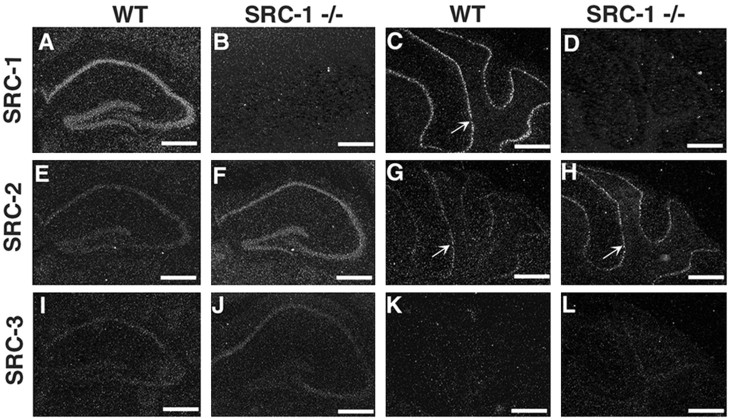
Because SRC-1−/−mice exhibit only a moderate motor dysfunction, we reasoned that other members of the SRC family may provide an overlapping function in the brain. To address this possibility, the spatial expression patterns of all SRC members in the brain were compared by in situhybridization. We found that SRC-2 mRNA was coexpressed with SRC-1 at lower levels in multiple brain structures, including the olfactory bulb, piriform cortex, hippocampus (CA1, CA2, and CA3 regions and dentate gyrus), amygdala complex, hypothalamus, and cerebellum (Fig.3) (data not shown). In contrast, SRC-3 mRNA was detected at low levels only in the CA1, CA2, and CA3 regions of the hippocampus (Fig. 3) (data not shown). These results demonstrate that all three SRC family members are coexpressed in the hippocampus, although their relative expression levels are different. SRC-1 and SRC-2 have overlapping expression patterns in the adult mouse brain, although in general, SRC-1 is expressed at higher levels.
Fig. 3.
Expression of SRC-2 and SRC-3 transcripts in the hippocampus and cerebellum of SRC-1−/−mice. Expression of all SRC family members was analyzed by in situ hybridization in the hippocampus (A, B, E, F, I, J) and cerebellum (C, D, G, H, K, L). Serial coronal sections were prepared from 10-week-old WT (A, C, E, G, I, K) and SRC-1−/−(B, D, F, H, J, L) mice. Note that hybridization signals in F, J, and H(SRC-1−/−) are stronger than those signals in E, I, and G (WT), respectively. SRC-3 mRNA was undetectable in the cerebellum (K,L). Arrows in C,G, and H indicate the Purkinje cell layer. Scale bar, 250 μm.
Next, the question regarding whether the absence of the SRC-1 gene product might alter the expression levels of SRC-2 and SRC-3 was addressed. Interestingly, although SRC-2 was expressed at low levels in the hippocampus and cerebellar Purkinje cells of WT mice, its expression was observed at higher levels in those equivalent brain regions of SRC-1−/−mice (Fig. 3, compare E, F and G,H). In the hippocampus of SRC-1−/−mice, SRC-3 was also expressed at slightly higher levels than in WT mice (Fig. 3, compare I, J); however, the SRC-3 signal remained undetectable in the cerebellum of SRC-1−/−mice (Fig. 3K,L). Therefore, the disruption of the SRC-1 gene results in an apparent upregulation of other SRC family members in specific regions of the brain.
Delayed development of Purkinje cells in SRC-1−/−mice
Because the coexpression and upregulation of other SRC members in the SRC-1−/−mouse brain may compensate for the loss of SRC-1 function during neural development and correct early developmental defects at a later stage, brain morphologies at earlier stages after birth including P0, P5, and P10 were examined. Although no other abnormal structures in the brain could be identified, the Purkinje cell layer in the cerebellum was apparently missing at P0 on the SRC-1−/−cerebellar sections stained with H&E (data not shown). To confirm this observation, immunostaining of calbindin, a Purkinje cell-specific marker, was performed (Anderson et al., 1998). In WT pups, calbindin-positive Purkinje cells were detected at P0 and at all later stages examined (Fig.4A,B). In contrast, the calbindin-positive cells were nearly undetectable at P0 in SRC-1−/−mice (Fig. 4C). At P3, the calbindin-positive Purkinje cells in some SRC-1−/−pups became detectable on the midsagittal sections (data not shown). At P5, the density and total number of Purkinje cells in SRC-1−/−pups were still significantly lower than those observed in WT pups. At P10, the number of calbindin-positive Purkinje cells in SRC-1−/−mice finally reached comparable levels with their age-matched WT pups (Fig. 4B,D). These observations were further confirmed by the quantitative counting of calbindin-positive Purkinje cells in the cerebellum of WT and SRC-1−/−mice at P0, P5, and P10 (Table 1). These results indicate that Purkinje cell development is significantly delayed at the neonatal stages in mice lacking SRC-1.
Fig. 4.

Delayed neonatal development of Purkinje cells in SRC-1−/−mice. Immunostaining with anti-calbindin antibody (A–D) and with anti-synaptophysin antibody (E–H) was performed on the midsagittal cerebellar sections. Mouse ages and genotypes are as indicated. Note that the calbindin-positive Purkinje cells in C and the synaptophysin signal in G were undetectable. Scale bars, 50 μm.
Table 1.
Number of Purkinje cells in wild-type and SRC-1−/− mice1-a
| Age | Genotype | n1-b | PC/section1-c |
|---|---|---|---|
| P0 | WT | 5 | 375 ± 20 |
| SRC-1−/− | 5 | 15 ± 4.61-d | |
| P5 | WT | 5 | 639 ± 16 |
| SRC-1−/− | 5 | 508 ± 201-e | |
| P10 | WT | 6 | 682 ± 24 |
| SRC-1−/− | 6 | 637 ± 32 |
Midsagittal sections of the cerebella were prepared from WT and SRC-1−/− mice with the ages indicated. The calbindin-positive Purkinje cells were identified by immunostaining.
n, Number of animals in each genotype group.
PC/section, Number of Purkinje cells per section was presented as mean ± SEM. The average number of Purkinje cells per section for each mouse was calculated according to the counts of Purkinje cell on three adjacent midsagittal sections. The mean value of Purkinje cells per section for each genotype was the average of all mice in the same genotype group.
p < 0.001 when compared with WT in the t test.
p < 0.005 when compared with WT in the t test.
The development of synaptic connections of the Purkinje cells was further examined by immunostaining of synaptophysin, an abundant glycoprotein localized in developing presynaptic termini (Leclerc et al., 1989). At P0, the immunostaining of synaptophysin was undetectable in the cerebellum of SRC-1−/−mice but was clearly detected in Purkinje cells of wild-type mice (Fig.4E,G). At P10, synaptophysin immunostaining of the puncta around Purkinje cells and of the connections with granule cells showed no gross differences between WT and SRC-1−/−mice (Fig. 4F,H). This observation is consistent with the results obtained from calbindin staining described above. Collectively, these results prove that disruption of the SRC-1 gene in the mouse brain delays Purkinje cell development.
Temporal expression of SRC coactivators during Purkinje cell development
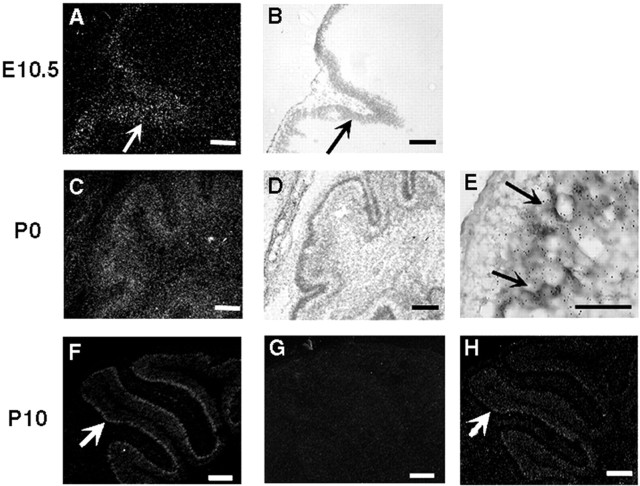
To define the temporal involvement of SRC coactivators in origination and differentiation of Purkinje cells, the temporal expression patterns of the SRC family members during Purkinje cell development were examined. SRC-1 mRNA was expressed in the cerebellar primordium since 10.5 d.p.c. (Fig.5A,B). At P0, although the Purkinje cells had not lined into a clear cell layer at this stage, high levels of SRC-1 mRNA were detected only in the calbindin-positive Purkinje cells (Fig.5C–E). At P10, SRC-1 expression was clearly detected in the Purkinje cell layer (Fig. 5F). In contrast, SRC-2 mRNA was undetectable in Purkinje cells of WT mice at P10 (Fig. 5G), whereas low levels of SRC-2 expression were observed in the Purkinje cells of the adult brain in WT mice (Fig.3G). Interestingly, in the Purkinje cells of SRC-1−/−mice, SRC-2 mRNA clearly became detectable at P10 (Fig.5H), suggesting that SRC-2 was expressed earlier in the Purkinje cells of SRC-1−/−mice. In addition, SRC-3 mRNA was undetectable in the Purkinje cells byin situ hybridization. These results suggest that SRC-1 is the major member of the SRC family to be involved in the Purkinje cell development at prenatal and neonatal stages. The earlier expression of SRC-2 in the Purkinje cells of SRC-1−/−mice at neonatal stage may replace SRC-1 to support Purkinje cell differentiation.
Fig. 5.
Temporal expression of SRC-1 and SRC-2 mRNAs at early cerebellar developmental stages. Sagittal sections through the cerebellar primordium at embryonic day (E) 10.5 (A, B) and the cerebellum at P0 (C–E) and P10 (F–H) were prepared from WT (A–G) and SRC-1−/−pups (H). The expression of SRC-1 (A–F) and SRC-2 (G,H) was examined by in situhybridization using 35S-labeled antisense RNA probes. Images in A, C, andF–H were photographed under dark-field lighting. The bright-field images in B andD were photographed from the same areas shown inA and C, respectively.Arrows in A and B indicate the expression of SRC-1 in the cerebellar primordium. Eis a bright-field image of the premature cerebellum at P0 in higher magnification, showing the colocalization of the immunostaining signals of calbindin (brown color) and the in situ hybridization signals of SRC-1 (small black particles) in the Purkinje cells (arrows).Arrows in F and H indicate the expression of SRC-1 (F) and SRC-2 (H) in the Purkinje cell layer. Note that SRC-2 expression was negative in WT pups at P10 (G) but positive in SRC-1−/−pups at the same stage (H). Scale bars:A–D, 100 μm; E, 50 μm;F–H, 300 μm.
Birth of Purkinje cell precursors in SRC-1−/−mice
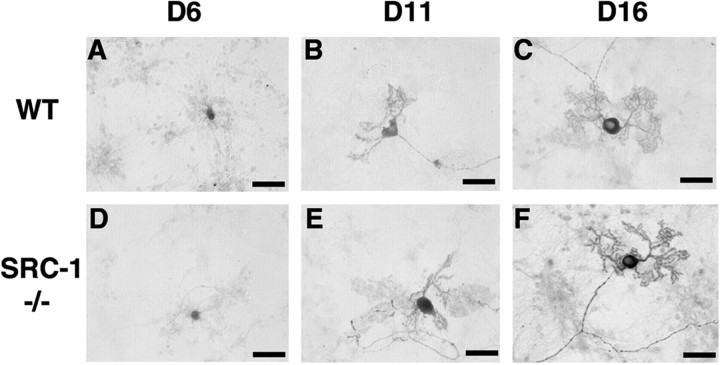
To search for the cellular mechanisms responsible for the delayed development of Purkinje cells in SRC-1-deficient mice, several experiments were performed. First, programmed cell death was analyzed by performing terminal deoxynucleotidyl transferase-mediated biotinylated UTP nick end labeling assays, and no increase of apoptotic cells was detected in the cerebellum of SRC-1−/−mice at any developmental stage examined (data not shown). Second, the differentiation potential of SRC-1-deficient Purkinje cells was analyzed in an in vitro culture system (Furuya et al., 1998). Cells were isolated from the cerebellum of 18-d-old embryos and treated with T3 and progesterone to induce differentiation in culture. Successful differentiation of Purkinje cells in culture was verified by calbindin immunostaining. After 6 d, large globular cell bodies of Purkinje cells were observed in cultures derived from both WT and SRC-1−/− mice (Fig.6A,D). During 11–16 d in culture, both WT and SRC-1−/− Purkinje cells developed their typical mature morphology, exhibited as a large globular cell body with thick apical dendrites and numerous branches (Fig.6B,C,E,F). These results suggest that immature Purkinje cells from SRC-1−/−embryos can differentiate normally into mature Purkinje cells in culture.
Fig. 6.
Purkinje cell differentiation in culture. Cerebellar cells were isolated from 18-d-old WT (A–C) and SRC-1−/−(D–F) mouse embryos. Purkinje cell bodies and their dendritic branches at 6, 11, and 16 d in culture were identified by immunocytochemical staining for calbindin. Scale bars, 50 μm.
Finally, the time course for the generation of Purkinje cell precursors during early embryonic development was studied. During mouse cerebellar development, the precursors of Purkinje cells and neurons in deep cerebellar nuclei originate in the ventricular zone during 11–13 d.p.c. These precursors start to migrate from the ventricular zone to the location just beneath the proliferating granule cells during 14–17 d.p.c. (Hatten and Heintz, 1995). Because SRC-1 is expressed in the cerebellar primordium (Fig. 5A) at early stages of cerebellar development, the absence of the SRC-1 protein in SRC-1−/−mice may affect the generation of Purkinje cell precursors. To define the developmental time course of Purkinje cell precursors in WT and SRC-1−/−mice, the birth of Purkinje cells was followed by BrdU labeling (Vogel et al., 2000). Cerebellar precursor cells were labeled at 10.5, 12.5, or 13.5 d.p.c. by injecting a single dose of BrdU into SRC-1 heterozygous pregnant females. The incorporated BrdU in the nuclear DNA was examined by immunostaining at P5 when calbindin-positive Purkinje cells became reliably detectable in both WT and SRC-1−/−mice (Fig. 7A). A large number of BrdU-positive Purkinje cells were clearly detected at P5 in WT pups that received BrdU at 10.5 d.p.c. In contrast, no BrdU-positive Purkinje cells could be detected at P5 in SRC-1−/−littermates that received identical amounts of BrdU at 10.5 d.p.c. (Fig.7Ba,Bb,C). When BrdU was injected at 12.5 d.p.c., the number of BrdU-labeled Purkinje cells at P5 in SRC-1−/−-deficient mice was threefold higher than in their WT littermates (Fig.7Bc,Bd,C). However, when BrdU was given at 13.5 d.p.c., only a small number of BrdU-labeled Purkinje cells were observed at P5 in both WT and SRC-1−/−mice (Fig. 7Be,Bf,C). These results demonstrate that the birth of Purkinje cell precursors in SRC-1−/−mice is delayed by ∼2 d at early embryonic developmental stages, suggesting that the delayed Purkinje cell development at the neonatal stage is caused partially by the delayed birth of Purkinje cell precursors at earlier embryonic developmental stages.
Fig. 7.
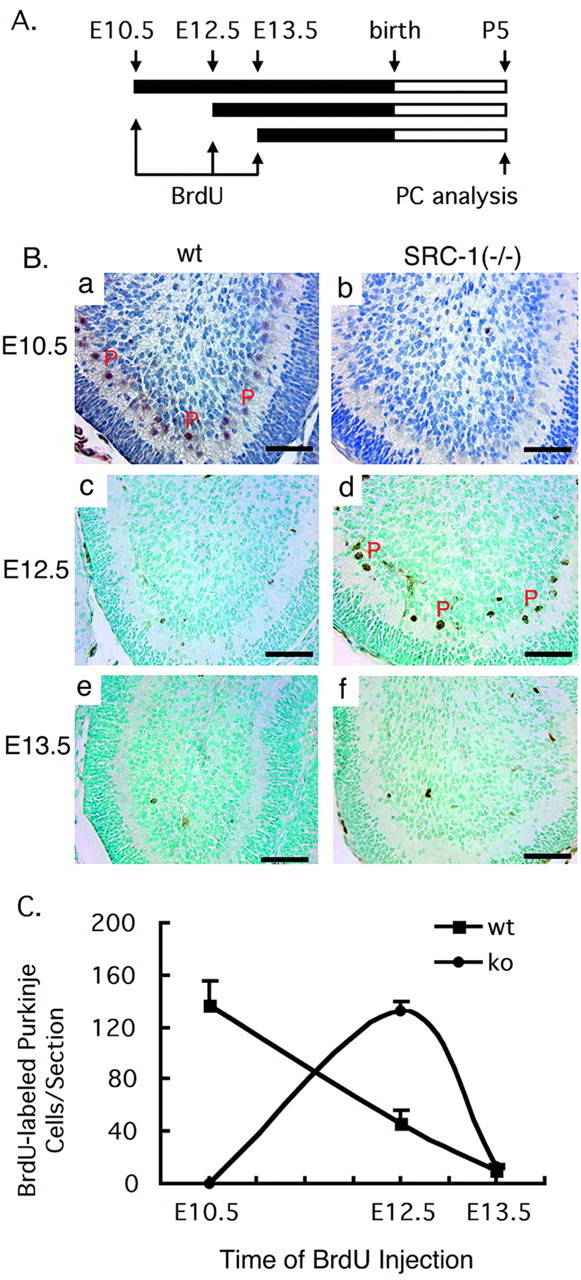
Delayed generation of Purkinje cell precursors.A, Time course for treatment and analysis. Precursor cells were labeled by injecting a single dose of BrdU into multiple pregnant SRC-1+/− females at 10.5, 12.5, and 13.5 d.p.c. Cerebellum samples were obtained at P5 for analysis of BrdU-labeled Purkinje cells (PCs). B, The nuclei of BrdU-labeled PCs on the midsagittal sections of cerebella were identified by immunostaining with an antibody specific to BrdU. After antibody reactions, the slides were counterstained either with hematoxylin (a, b) or with methyl green (c–f). Note that BrdU-positive PCs, indicated byP, were observed primarily at E10.5 for WT mice (a) and at E12.5 for SRC-1−/− mice (d). Scale bars, 50 μm. C, Quantitative analysis of BrdU-labeled PCs. BrdU-labeled PCs on three adjacent midsagittal sections prepared from each cerebellum at P5 were counted. Each average number of BrdU-labeled PCs per section for each genotype and each time point was calculated from a total of 12 sections, which were prepared from four mice. When BrdU was given at E10.5 (10.5 d.p.c.), the number of BrdU-labeled PCs at P5 in WT mice was significantly higher than in knock-out (ko) (SRC-1−/−) mice (p < 0.001; t test). However, when BrdU was given at E12.5, the number of BrdU-labeled PCs at P5 in WT mice was significantly lower than in knock-out (SRC-1−/−) mice (p < 0.001; t test). There were no statistical differences between two groups when BrdU was given at E13.5.
Discussion
The importance of hormones and their cognate NRs for the development and function of the CNS caused us to investigate the potential roles and molecular mechanisms that the newly identified NR coactivators have in the brain. In this study, a number of experiments were designed to characterize the role of SRC-1 in brain development and function. We first identified the potential sites of SRC-1 function in the adult mouse brain by examining the expression of the SRC-1 transcript. We found that SRC-1 is preferentially expressed in specific brain regions. Although the expression pattern of SRC-1 in the mouse brain is similar to that observed in the rat brain (Meijer et al., 2000), the identification of Purkinje cell-specific expression of SRC-1 mRNA and protein in the mouse cerebellum is a novel finding. These SRC-1-positive brain regions contain high levels of multiple NRs (Lopes da Silva and Burbach, 1995; Morimoto et al., 1996; Shughrue et al., 1997). The overlapping of SRC-1 and NRs in these specific brain regions highlights their potential functional partnership in mediating hormone actions. In addition, in the brain of mouse embryos, SRC-1 is expressed much more broadly, with its highest expression localized in the olfactory epithelium (Misiti et al., 1999). These results suggest that SRC-1 function in the brain may be both region specific and developmental stage specific.
Next, we addressed whether SRC-1 is required for generation and maintenance of mature brain structure by analyzing the brain histology of adult SRC-1−/−mice. Although serial brain sections were examined carefully, no structural defects were observed in the brains of both male and female adult SRC-1−/−mice. This observation indicates that SRC-1 is not essential for the formation and maintenance of mature mouse brain morphology.
Subsequently, we compared the brain function of SRC-1−/−mice with age-matched WT mice using a battery of well established behavior tests. SRC-1−/−mice performed poorly in the hanging wire tests, suggesting that SRC-1−/−mice may have a dysfunction of motor strength. In the rotarod tests, although SRC-1−/−mice initially showed coordination comparable with that of WT mice, serial trials revealed that SRC-1−/−mice made significantly slower improvement in their performance on the rotarod, suggesting that the motor learning and the motor performance of SRC-1−/−mice are partially impaired. In the Morris water maze, SRC-1−/−mice exhibited significantly longer escape latencies when compared with WT mice, which was primarily attributable to a slower swim speed, as demonstrated by further analysis. Because SRC-1 is expressed at extremely low levels in the skeletal muscle, the sum of these results suggests that SRC-1−/−mice have a moderate motor dysfunction.
No other abnormalities in brain function, such as dysfunction of spatial learning, could be detected in SRC-1−/−mice, although SRC-1 was highly expressed in functional regions including hippocampus and amygdala. Because in vitro studies have indicated that SRC coactivators have overlapping function in coactivation of multiple transcription factors when expressed in the same type of cells (Xu and O'Malley, 2002), and we have reported that SRC-2 mRNA is more abundant in total RNA samples from SRC-1−/−mouse brains (Xu et al., 1998), we have hypothesized that other members of the SRC family may compensate for the loss of SRC-1 during brain development and function.
To test our hypothesis, spatial distributions and relative expression levels for all three SRC coactivators in brain regions of both WT and SRC-1−/−mice were compared in parallel experiments. Our results demonstrated the following. (1) In WT mice, SRC-1 is more abundantly expressed than other SRC family members in multiple brain regions described above. SRC-2 is expressed at lower levels in all brain regions where SRC-1 is expressed, except in the brainstem. SRC-3 mRNA is detectable at a very low level only in the hippocampus by in situ hybridization. By using a much more sensitive method (X-gal staining), SRC-3 expression can also be detected in the olfactory bulb in addition to the hippocampus (Xu et al., 2000). (2) In SRC-1−/−mice, SRC-2 expression is slightly elevated, at least in the hippocampus and cerebellum. SRC-3 expression is also slightly elevated in the hippocampus. When one or more of the family members are disrupted, the partially overlapping expression of SRCs and upregulation of other SRC members may provide compensatory mechanisms to support brain development and function. The coexpression patterns of all SRCs in the hippocampus of WT mice may explain, at least in part, why SRC-1−/−mice can still maintain their normal hippocampal function in the learning and memory tests. The notion of functional redundancy among SRC family members is further supported by the lethal phenotype of the double SRC-1 and SRC-2 knock-out mice, although each individual knock-out mouse line shows nearly normal life span (data not shown).
Purkinje cells serve as the sole output neurons for the cerebellar cortex and affect motor neuron activity. Motor learning tasks are mediated by Purkinje cells in the cerebellum and are frequently implicated in altered function of Purkinje cells (Linden, 1994; Raymond et al., 1996). Because SRC-1 is highly and specifically expressed in Purkinje cells of the cerebellum, the lack of SRC-1 may affect Purkinje cell function. Therefore, the moderate dysfunction of motor learning observed in adult SRC-1−/−mice can be attributed to the absence of SRC-1 in Purkinje cells. However, it is also possible that motor dysfunction in adulthood is a result of developmental problems at early stages. To test this possibility, we examined brain development at late embryonic and neonatal stages, with our focus on the brain regions expressing high levels of SRC-1. Indeed, a delay in Purkinje cell development was identified at the neonatal stage in SRC-1−/−mice. Both calbindin and synaptophysin molecular markers for Purkinje cells were undetectable in the cerebella of SRC-1−/−pups at P0 although these two molecules were detected in their WT littermates as expected (Leclerc et al., 1989; Anderson et al., 1998). Analysis of the time course for the origination of Purkinje cell precursors by BrdU labeling made it clear that the delay in Purkinje cell development at the neonatal stage is related to a deficit in the generation of precursor cells during 10.5–12.5 d.p.c. in SRC-1−/−embryos. During this developmental stage, SRC-1 is expressed in the cerebellar primordium in WT embryos, suggesting that removal of SRC-1 from this region may directly affect the generation of Purkinje cell precursors. However, once the precursors of Purkinje cells in SRC-1−/−embryos were generated, they were able to differentiate into Purkinje cells as WT precursors did when analyzed in a culture system (Maeda et al., 1989; Nakanishi et al., 1991).
The time course analysis revealed that the generation of SRC-1−/−Purkinje cell precursors was delayed ∼2 d when compared with WT precursors. In SRC-1−/−pups at neonatal stages, the calbindin-positive Purkinje cells were nearly undetectable at P0 and became detectable at P3. Even at P5, the number of calbindin-stained Purkinje cells in SRC-1−/−pups was still significantly lower than that in WT pups. The number of Purkinje cells in SRC-1−/−mice finally reached that of their WT littermates during P5–P10. These observations indicate that the time differences between WT and SRC-1−/−pups with respect to the final morphological maturation of Purkinje cells after birth are longer than the delay in the generation of Purkinje precursors. To uncover the underlying mechanisms, we further compared the temporal expression patterns of SRC coactivators in Purkinje cells. Our analysis indicates that SRC-1 transcripts exist in cerebellar primordium in embryos and in Purkinje cells after birth, but SRC-2 mRNA was undetectable in Purkinje cells even at P10. Similar to adult mice, SRC-3 was undetectable in the cerebellum at these stages byin situ hybridization. The different temporal expression patterns of SRCs suggest that SRC-1 is the major player in the SRC family for Purkinje cell development at embryonic and neonatal stages, and SRC-2 may participate at a later stage. More interestingly, SRC-2 mRNA became detectable at P10 in SRC-1−/−Purkinje cells, suggesting that this earlier expression of SRC-2 in SRC-1−/−Purkinje cells might play a compensatory role to support the maturation of Purkinje cells in SRC-1−/−mice.
The critical roles of RORα and TH in Purkinje cell development have been well established. RORα heterozygous mutant mice show accelerated dendritic atrophy and cell loss, suggesting that RORα has a role in mature Purkinje cells (Zanjani et al., 1992). RORα homozygous mutants exhibit severe tremors and body imbalance caused by a cell-autonomous defect in the development of Purkinje cells (Hamilton et al., 1996;Dussault et al., 1998). These RORα-deficient Purkinje cells show immature morphology and synaptic arrangement and a reduction in numbers. Hypothyroidism causes reduced dendritic arborization of Purkinje cells and less synaptogenesis at neonatal stage similar to that seen in RORα mutant mice (Koibuchi and Chin, 2000). Interestingly, TH treatment alters the timing of expression of the RORα gene in Purkinje cells, and RORα mutation blocks Purkinje cell response to TH (Messer, 1988; Koibuchi and Chin, 1998). Disruption of SRC-1 may affect Purkinje cell development and maturation caused by a partial impairment of the RORα and TR functions on the basis of the following findings: (1) the members in the SRC family serve as transcriptional coactivators for RORα and TR (Takeshita et al., 1996,1997; Atkins et al., 1999); (2) SRC-1 is coexpressed with RORα and TR in the Purkinje cells; (3) only SRC-1 in the SRC family is expressed in the cerebellar primordium and Purkinje cells during embryonic and neonatal stages; and (4) the SRC-1-deficient mice exhibit thyroid hormone resistance (Weiss et al., 1999). In addition to NRs, SRC-1 deficiency may also affect the function of some other transcription factors during Purkinje cell development because SRC-1 also interacts with and augments a number of non-NR transcription factors such as AP-1, serum response factor, and NF-κB (Xu and O'Malley, 2002). Characterization of the specific molecular targets of SRC-1 and the degree of the functional impairments of related transcription factors in SRC-1−/−mice during Purkinje cell development deserve additional studies.
In summary, the expression of individual SRC members in the mouse brain is spatially and temporally regulated. Their partial overlapping expression patterns may determine their functional redundancy during brain development and function. In the cerebellum, SRC-1 is selectively expressed in Purkinje cells. Disruption of the SRC-1 gene in mice delays the generation of Purkinje cell precursors at an early embryonic stage and further delays the maturation of Purkinje cells after birth. Although the morphology of SRC-1−/−Purkinje cells develops to the same extent as WT Purkinje cells by P10, adult SRC-1−/−mice still exhibit moderate motor dysfunction, suggesting that the abnormal development of Purkinje cells at early stage may have a negative impact on the cerebellar function in adulthood. For the first time, these findings demonstrate that SRC coactivators play an important role in cerebellar Purkinje cell development and motor function.
Footnotes
This work was supported by National Institutes of Health Grants U54-HD-07495 (B.W.O.) and DK58242 (J.X.), by National Institute of Mental Health Grant IMH 60420 (R.L.D.), and by a Research Award (E.N.) from the Uehara Memorial Foundation. We thank Drs. M.-J. Tsai, F. DeMayo, H. Y. Zoghbi, F. A. Pereira, and N. J. McKenna for valuable discussions.
Correspondence should be addressed to Dr. Jianming Xu, Department of Molecular and Cellular Biology, Baylor College of Medicine. One Baylor Plaza, Houston, TX 77030. E-mail:jxu@bcm.tmc.edu.
References
- 1.Anderson GW, Larson RJ, Oas DR, Sandhofer CR, Schwartz HL, Mariash CN, Oppenheimer JH. Chicken ovalbumin upstream promoter-transcription factor (COUP-TF) modulates expression of the Purkinje cell protein-2 gene. A potential role for COUP-TF in repressing premature thyroid hormone action in the developing brain. J Biol Chem. 1998;273:16391–16399. doi: 10.1074/jbc.273.26.16391. [DOI] [PubMed] [Google Scholar]
- 2.Anzick SL, Kononen J, Walker RL, Azorsa DO, Tanner MM, Guan XY, Sauter G, Kallioniemi OP, Trent JM, Meltzer PS. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science. 1997;277:965–968. doi: 10.1126/science.277.5328.965. [DOI] [PubMed] [Google Scholar]
- 3.Atkins GB, Hu X, Guenther MG, Rachez C, Freedman LP, Lazar MA. Coactivators for the orphan nuclear receptor RORalpha. Mol Endocrinol. 1999;13:1550–1557. doi: 10.1210/mend.13.9.0343. [DOI] [PubMed] [Google Scholar]
- 4.Chen H, Lin RJ, Schiltz RL, Chakravarti D, Nash A, Nagy L, Privalsky ML, Nakatani Y, Evans RM. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- 5.Chomez P, Neveu I, Mansen A, Kiesler E, Larsson L, Vennstrom B, Arenas E. Increased cell death and delayed development in the cerebellum of mice lacking the rev-erbA(alpha) orphan receptor. Development. 2000;127:1489–1498. doi: 10.1242/dev.127.7.1489. [DOI] [PubMed] [Google Scholar]
- 6.Dussault I, Fawcett D, Matthyssen A, Bader JA, Giguere V. Orphan nuclear receptor ROR alpha-deficient mice display the cerebellar defects of staggerer. Mech Dev. 1998;70:147–153. doi: 10.1016/s0925-4773(97)00187-1. [DOI] [PubMed] [Google Scholar]
- 7.Furuya S, Makino A, Hirabayashi Y. An improved method for culturing cerebellar Purkinje cells with differentiated dendrites under a mixed monolayer setting. Brain Res Brain Res Protoc. 1998;3:192–198. doi: 10.1016/s1385-299x(98)00040-3. [DOI] [PubMed] [Google Scholar]
- 8.Glass CK, Rosenfeld MG. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 2000;14:121–141. [PubMed] [Google Scholar]
- 9.Hamilton BA, Frankel WN, Kerrebrock AW, Hawkins TL, FitzHugh W, Kusumi K, Russell LB, Mueller KL, van Berkel V, Birren BW, Kruglyak L, Lander ES. Disruption of the nuclear hormone receptor RORalpha in staggerer mice. Nature. 1996;379:736–739. doi: 10.1038/379736a0. [DOI] [PubMed] [Google Scholar]
- 10.Hatten ME, Heintz N. Mechanisms of neural patterning and specification in the developing cerebellum. Annu Rev Neurosci. 1995;18:385–408. doi: 10.1146/annurev.ne.18.030195.002125. [DOI] [PubMed] [Google Scholar]
- 11.Hayashi Y, Koike M, Matsutani M, Hoshino T. Effects of fixation time and enzymatic digestion on immunohistochemical demonstration of bromodeoxyuridine in formalin-fixed, paraffin-embedded tissue. J Histochem Cytochem. 1988;36:511–514. doi: 10.1177/36.5.3282006. [DOI] [PubMed] [Google Scholar]
- 12.Hong H, Kohli K, Trivedi A, Johnson DL, Stallcup MR. GRIP1, a novel mouse protein that serves as a transcriptional coactivator in yeast for the hormone binding domains of steroid receptors. Proc Natl Acad Sci USA. 1996;93:4948–4952. doi: 10.1073/pnas.93.10.4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koh SS, Chen D, Lee YH, Stallcup MR. Synergistic enhancement of nuclear receptor function by p160 coactivators and two coactivators with protein methyltransferase activities. J Biol Chem. 2001;276:1089–1098. doi: 10.1074/jbc.M004228200. [DOI] [PubMed] [Google Scholar]
- 14.Koibuchi N, Chin WW. ROR alpha gene expression in the perinatal rat cerebellum: ontogeny and thyroid hormone regulation. Endocrinology. 1998;139:2335–2341. doi: 10.1210/endo.139.5.6013. [DOI] [PubMed] [Google Scholar]
- 15.Koibuchi N, Chin WW. Thyroid hormone action and brain development. Trends Endocrinol Metab. 2000;11:123–128. doi: 10.1016/s1043-2760(00)00238-1. [DOI] [PubMed] [Google Scholar]
- 16.Lanz RB, McKenna NJ, Onate SA, Albrecht U, Wong J, Tsai SY, Tsai MJ, O'Malley BW. A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell. 1999;97:17–27. doi: 10.1016/s0092-8674(00)80711-4. [DOI] [PubMed] [Google Scholar]
- 17.Leclerc N, Beesley PW, Brown I, Colonnier M, Gurd JW, Paladino T, Hawkes R. Synaptophysin expression during synaptogenesis in the rat cerebellar cortex. J Comp Neurol. 1989;280:197–212. doi: 10.1002/cne.902800204. [DOI] [PubMed] [Google Scholar]
- 18.Lee SK, Anzick SL, Choi JE, Bubendorf L, Guan XY, Jung YK, Kallioniemi OP, Kononen J, Trent JM, Azorsa D, Jhun BH, Cheong JH, Lee YC, Meltzer PS, Lee JW. A nuclear factor, ASC-2, as a cancer-amplified transcriptional coactivator essential for ligand-dependent transactivation by nuclear receptors in vivo. J Biol Chem. 1999;274:34283–34293. doi: 10.1074/jbc.274.48.34283. [DOI] [PubMed] [Google Scholar]
- 19.Li H, Gomes PJ, Chen JD. RAC3, a steroid/nuclear receptor-associated coactivator that is related to SRC-1 and TIF2. Proc Natl Acad Sci USA. 1997;94:8479–8484. doi: 10.1073/pnas.94.16.8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li J, O'Malley BW, Wong J. p300 requires its histone acetyltransferase activity and SRC-1 interaction domain to facilitate thyroid hormone receptor activation in chromatin. Mol Cell Biol. 2000;20:2031–2042. doi: 10.1128/mcb.20.6.2031-2042.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Linden DJ. Long-term synaptic depression in the mammalian brain. Neuron. 1994;12:457–472. doi: 10.1016/0896-6273(94)90205-4. [DOI] [PubMed] [Google Scholar]
- 22.Lopes da Silva S, Burbach JP. The nuclear hormone-receptor family in the brain: classics and orphans. Trends Neurosci. 1995;18:542–548. doi: 10.1016/0166-2236(95)98376-a. [DOI] [PubMed] [Google Scholar]
- 23.Maeda N, Niinobe M, Inoue Y, Mikoshiba K. Developmental expression and intracellular location of P400 protein characteristic of Purkinje cells in the mouse cerebellum. Dev Biol. 1989;133:67–76. doi: 10.1016/0012-1606(89)90297-2. [DOI] [PubMed] [Google Scholar]
- 24.Mani SK, Blaustein JD, O'Malley BW. Progesterone receptor function from a behavioral perspective. Horm Behav. 1997;31:244–255. doi: 10.1006/hbeh.1997.1393. [DOI] [PubMed] [Google Scholar]
- 25.McEwen BS. Steroid hormone actions on the brain: when is the genome involved? Horm Behav. 1994;28:396–405. doi: 10.1006/hbeh.1994.1036. [DOI] [PubMed] [Google Scholar]
- 26.McEwen BS. Invited review: estrogens effects on the brain: multiple sites and molecular mechanisms. J Appl Physiol. 2001;91:2785–2801. doi: 10.1152/jappl.2001.91.6.2785. [DOI] [PubMed] [Google Scholar]
- 27.McKenna NJ, O'Malley BW. Combinatorial control of gene expression by nuclear receptors and coregulators. Cell. 2002;108:465–474. doi: 10.1016/s0092-8674(02)00641-4. [DOI] [PubMed] [Google Scholar]
- 28.Meijer OC, Steenbergen PJ, De Kloet ER. Differential expression and regional distribution of steroid receptor coactivators SRC-1 and SRC-2 in brain and pituitary. Endocrinology. 2000;141:2192–2199. doi: 10.1210/endo.141.6.7489. [DOI] [PubMed] [Google Scholar]
- 29.Messer A. Thyroxine injections do not cause premature induction of thymidine kinase in sg/sg mice. J Neurochem. 1988;51:888–891. doi: 10.1111/j.1471-4159.1988.tb01825.x. [DOI] [PubMed] [Google Scholar]
- 30.Misiti S, Koibuchi N, Bei M, Farsetti A, Chin WW. Expression of steroid receptor coactivator-1 mRNA in the developing mouse embryo: a possible role in olfactory epithelium development. Endocrinology. 1999;140:1957–1960. doi: 10.1210/endo.140.4.6782. [DOI] [PubMed] [Google Scholar]
- 31.Morimoto M, Morita N, Ozawa H, Yokoyama K, Kawata M. Distribution of glucocorticoid receptor immunoreactivity and mRNA in the rat brain: an immunohistochemical and in situ hybridization study. Neurosci Res. 1996;26:235–269. doi: 10.1016/s0168-0102(96)01105-4. [DOI] [PubMed] [Google Scholar]
- 32.Morris RG, Garrud P, Rawlins JN, O'Keefe J. Place navigation impaired in rats with hippocampal lesions. Nature. 1982;297:681–683. doi: 10.1038/297681a0. [DOI] [PubMed] [Google Scholar]
- 33.Nakanishi S, Maeda N, Mikoshiba K. Immunohistochemical localization of an inositol 1,4,5-trisphosphate receptor, P400, in neural tissue: studies in developing and adult mouse brain. J Neurosci. 1991;11:2075–2086. doi: 10.1523/JNEUROSCI.11-07-02075.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nishihara E, Furuyama T, Yamashita S, Mori N. Expression of copper trafficking genes in the mouse brain. NeuroReport. 1998;9:3259–3263. doi: 10.1097/00001756-199810050-00023. [DOI] [PubMed] [Google Scholar]
- 35.Nishihara E, Nagayama Y, Inoue S, Hiroi H, Muramatsu M, Yamashita S, Koji T. Ontogenetic changes in the expression of estrogen receptor alpha and beta in rat pituitary gland detected by immunohistochemistry. Endocrinology. 2000;141:615–620. doi: 10.1210/endo.141.2.7330. [DOI] [PubMed] [Google Scholar]
- 36.Onate SA, Tsai SY, Tsai MJ, O'Malley BW. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science. 1995;270:1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- 37.Raymond JL, Lisberger SG, Mauk MD. The cerebellum: a neuronal learning machine? Science. 1996;272:1126–1131. doi: 10.1126/science.272.5265.1126. [DOI] [PubMed] [Google Scholar]
- 38.Reichardt HM, Tronche F, Bauer A, Schutz G. Molecular genetic analysis of glucocorticoid signaling using the Cre/loxP system. Biol Chem. 2000;381:961–964. doi: 10.1515/BC.2000.118. [DOI] [PubMed] [Google Scholar]
- 39.Rissman EF, Early AH, Taylor JA, Korach KS, Lubahn DB. Estrogen receptors are essential for female sexual receptivity. Endocrinology. 1997;138:507–510. doi: 10.1210/endo.138.1.4985. [DOI] [PubMed] [Google Scholar]
- 40.Shughrue PJ, Lane MV, Merchenthaler I. Comparative distribution of estrogen receptor-alpha and -beta mRNA in the rat central nervous system. J Comp Neurol. 1997;388:507–525. doi: 10.1002/(sici)1096-9861(19971201)388:4<507::aid-cne1>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 41.Steinmayr M, Andre E, Conquet F, Rondi-Reig L, Delhaye-Bouchaud N, Auclair N, Daniel H, Crepel F, Mariani J, Sotelo C, Becker-Andre M. staggerer phenotype in retinoid-related orphan receptor alpha-deficient mice. Proc Natl Acad Sci USA. 1998;95:3960–3965. doi: 10.1073/pnas.95.7.3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suen CS, Berrodin TJ, Mastroeni R, Cheskis BJ, Lyttle CR, Frail DE. A transcriptional coactivator, steroid receptor coactivator-3, selectively augments steroid receptor transcriptional activity. J Biol Chem. 1998;273:27645–27653. doi: 10.1074/jbc.273.42.27645. [DOI] [PubMed] [Google Scholar]
- 43.Takeshita A, Yen PM, Misiti S, Cardona GR, Liu Y, Chin WW. Molecular cloning and properties of a full-length putative thyroid hormone receptor coactivator. Endocrinology. 1996;137:3594–3597. doi: 10.1210/endo.137.8.8754792. [DOI] [PubMed] [Google Scholar]
- 44.Takeshita A, Cardona GR, Koibuchi N, Suen CS, Chin WW. TRAM-1, A novel 160-kDa thyroid hormone receptor activator molecule, exhibits distinct properties from steroid receptor coactivator-1. J Biol Chem. 1997;272:27629–27634. doi: 10.1074/jbc.272.44.27629. [DOI] [PubMed] [Google Scholar]
- 45.Tetel MJ. Nuclear receptor coactivators in neuroendocrine function. J Neuroendocrinol. 2000;12:927–932. doi: 10.1046/j.1365-2826.2000.00557.x. [DOI] [PubMed] [Google Scholar]
- 46.Torchia J, Rose DW, Inostroza J, Kamei Y, Westin S, Glass CK, Rosenfeld MG. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature. 1997;387:677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- 47.Tsai MJ, O'Malley BW. Molecular mechanisms of action of steroid/thyroid receptor superfamily members. Annu Rev Biochem. 1994;63:451–486. doi: 10.1146/annurev.bi.63.070194.002315. [DOI] [PubMed] [Google Scholar]
- 48.Voegel JJ, Heine MJ, Zechel C, Chambon P, Gronemeyer H. TIF2, a 160 kDa transcriptional mediator for the ligand-dependent activation function AF-2 of nuclear receptors. EMBO J. 1996;15:3667–3675. [PMC free article] [PubMed] [Google Scholar]
- 49.Vogel MW, Sinclair M, Qiu D, Fan H. Purkinje cell fate in staggerer mutants: agenesis versus cell death. J Neurobiol. 2000;42:323–337. doi: 10.1002/(sici)1097-4695(20000215)42:3<323::aid-neu4>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 50.Watanabe M, Yanagisawa J, Kitagawa H, Takeyama K, Ogawa S, Arao Y, Suzawa M, Kobayashi Y, Yano T, Yoshikawa H, Masuhiro Y, Kato S. A subfamily of RNA-binding DEAD-box proteins acts as an estrogen receptor alpha coactivator through the N-terminal activation domain (AF-1) with an RNA coactivator, SRA. EMBO J. 2001;20:1341–1352. doi: 10.1093/emboj/20.6.1341. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 51.Weiss RE, Xu J, Ning G, Pohlenz J, O'Malley BW, Refetoff S. Mice deficient in the steroid receptor co-activator 1 (SRC-1) are resistant to thyroid hormone. EMBO J. 1999;18:1900–1904. doi: 10.1093/emboj/18.7.1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu J, O'Malley BW. Molecular mechanisms and cellular biology of the steroid receptor coactivator (SRC) family in steroid receptor function. Rev Endocr Metab Disord. 2002;3:183–190. doi: 10.1023/a:1020016208071. [DOI] [PubMed] [Google Scholar]
- 53.Xu J, Qiu Y, DeMayo FJ, Tsai SY, Tsai MJ, O'Malley BW. Partial hormone resistance in mice with disruption of the steroid receptor coactivator-1 (SRC-1) gene. Science. 1998;279:1922–1925. doi: 10.1126/science.279.5358.1922. [DOI] [PubMed] [Google Scholar]
- 54.Xu J, Liao L, Ning G, Yoshida-Komiya H, Deng C, O'Malley BW. The steroid receptor coactivator SRC-3 (p/CIP/RAC3/AIB1/ACTR/TRAM-1) is required for normal growth, puberty, female reproductive function, and mammary gland development. Proc Natl Acad Sci USA. 2000;97:6379–6384. doi: 10.1073/pnas.120166297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zanjani HS, Mariani J, Delhaye-Bouchaud N, Herrup K. Neuronal cell loss in heterozygous staggerer mutant mice: a model for genetic contributions to the aging process. Brain Res Dev Brain Res. 1992;67:153–160. doi: 10.1016/0165-3806(92)90216-j. [DOI] [PubMed] [Google Scholar]
- 56.Zetterstrom RH, Solomin L, Jansson L, Hoffer BJ, Olson L, Perlmann T. Dopamine neuron agenesis in Nurr1-deficient mice. Science. 1997;276:248–250. doi: 10.1126/science.276.5310.248. [DOI] [PubMed] [Google Scholar]
- 57.Zhou C, Qiu Y, Pereira FA, Crair MC, Tsai SY, Tsai MJ. The nuclear orphan receptor COUP-TFI is required for differentiation of subplate neurons and guidance of thalamocortical axons. Neuron. 1999;24:847–859. doi: 10.1016/s0896-6273(00)81032-6. [DOI] [PubMed] [Google Scholar]