Abstract
The subunits that compose eukaryotic glutamate ion channel receptors have three transmembrane domains (TMs) and terminate with intracellular tails that are important for controlling channel expression and localization. Truncation of NMDA receptor subunits before the final TM showed that this TM and intracellular tail region are necessary to form functional channels. However, it is shown here that these truncated subunits may be partially rescued by coexpressing the final TM and tail as a separate protein. The whole-cell currents so produced are somewhat lower than with full-length subunits, and they do not show the sag characteristic of currents from channels containing NR1 and NR2A subunits in the continued presence of an agonist. In addition, these truncated subunits were joined to full-length subunits to generate tandems. The functional expression of these tandems confirmed the tetrameric structure of NMDA receptors and also suggested that the subunits making up NMDA receptors are arranged as a dimer of dimers in the receptors with a 1-1-2-2 orientation of the subunits in the channel, and not in an alternating pattern of subunits around the pore. These results may redirect future studies into the mechanism of binding and gating in these receptors toward schemes including dimers, and may also be relevant to studies of glutamate receptor ion channels in general.
Keywords: glutamate receptor, stoichiometry, tandem, dimer, NMDA, ion channel, assembly of subunits
Introduction
NMDA receptors are notorious for their long and complex activations, and although the kinetics and gating of NMDA receptors have been well studied (Stern et al., 1992;Behe et al., 1995; Zhang and Auerbach, 1995; Takahashi et al., 1996;Premkumar et al., 1997; Schneggenburger and Ascher, 1997; Wyllie et al., 1998), no complete kinetic mechanism has been made for these channels. A prerequisite for such a mechanism is an understanding of the number and organization of the two types of subunits, NR1 and NR2, required to form functional NMDA channels, which is currently unknown.
Eukaryotic glutamate ion channel receptors (GluRs), including NMDA receptors, are composed of subunits, most likely four in total (Laube et al., 1998; Rosenmund et al., 1998; Chen et al., 1999), each containing three transmembrane domains (TMs) plus a loop region, and ending with an intracellular tail that has been shown to be important for helping the subunits get to the membrane, assemble into channels, and anchor in synapses (Sheng and Pak, 2000). The TMs are usually known as TM1, TM3, and TM4. In addition to homology with each other, the eukaryotic GluRs have some homology with prokaryotic glutamate channels, which contain only two TMs (Chen et al., 1999). The lack of the final TM in bacterial channels, and an intron placed at the junction between the ligand-binding site and the final TM (TM4) in eukaryotes (Wo and Oswald, 1995), suggest that the final TM and cytoplasmic tail are evolutionary additions to the core GluR subunit.
In GluR subunits, deletions of the C tail have profound effects on channel activity and surface expression (Matsuda and Mishina, 2000;Steigerwald et al., 2000). Nuclear localization and endoplasmic reticulum retention sequences in the C tails of NMDA receptor subunits have been discovered recently to be critical for the proper expression of the subunits (Standley et al., 2000; Scott et al., 2001;Xia et al., 2001; Holmes et al., 2002).
In this paper we show that by truncating the subunits and supplying the TM4 tails as separate peptides, the ability of NMDA receptors to function with only two joined TMs in each subunit (as in the prokaryotic channels) can be partially recovered. However, the resulting prokaryotic-style channels have different properties from control channels with all three TMs included in a single peptide.
Normally, the N and C terminals of GluR subunits are on opposite sides of the membrane, so it is not possible to link them together. The ability of truncated channels to form functional channels in the presence of separate TMs provided the opportunity to construct tandems of GluR subunits without requiring the addition of long transmembrane linkers, as was attempted previously (Prybylowski et al., 1999).
In truncated subunits, both terminals are outside the cell membrane, and consequently, the end of a truncated subunit may be linked to the beginning of a full-length subunit to generate a tandem construct. The tandems are used to constrain the number and arrangement of subunits within receptors and to determine which arrangements of subunits are best able to form functional receptors. The results suggest that NMDA receptors are a dimer of dimers, with a pair of glycine-binding NR1 subunits facing a pair of glutamate-binding NR2 subunits.
Materials and Methods
Molecular biology. Standard molecular biology methods were used to generate tandems. The original control DNAs, NR1 (U08261) and NR2A DNA (NM_012573; gifts from R. Schoepfer, University College London, London, UK), were amplified with primers designed to introduce EcoRV and a Kozak sequence (GCCACC) at the 5′ end, and NotI at the 3′ end. EcoRV andNotI were used to subclone the DNAs into pBluescript KS (Stratagene, La Jolla, CA) for PCR-based mutagenesis. Mutagenic primers were used to introduce NheI sites at the beginning of the mature peptide encoding sequences (at nucleotide 289 of U08261for NR1 before R1; at nucleotide 288 of NM_012573 for NR2A before Q1; residue numbering from mature protein), an SpeI site near the 5′ end of the final TM of NR1 (nucleotide 2698 before G804), and anAvrII site 5′ of the final TM of NR2A (nucleotide 2661 before I792). PCR-based mutagenesis was done according to standard protocols, with a round of single-stranded PCR followed byDpnI digests to eliminate nonamplified DNA. The mutations introduced single amino acid changes in the subunits (see Fig.1C). The resulting clones NR1** and NR2A** had normal dose–response curves, and were used to build truncated subunits and tandems. Truncated subunits ended at the SpeI (NR1) or theAvrII (NR2A) sites. TM4-tail clones were constructed by cutting NR1 with NheI and SpeI and NR2A with NheI and AvrII and ligating the two ends together. Tandems were constructed by isolatingNheI–SpeI (NR1) orNheI–AvrII (NR2A) fragments and ligating them into the NheI site of the subunit chosen for the second half of the tandem. All primer and DNA sequences are available from S. Schorge on request.
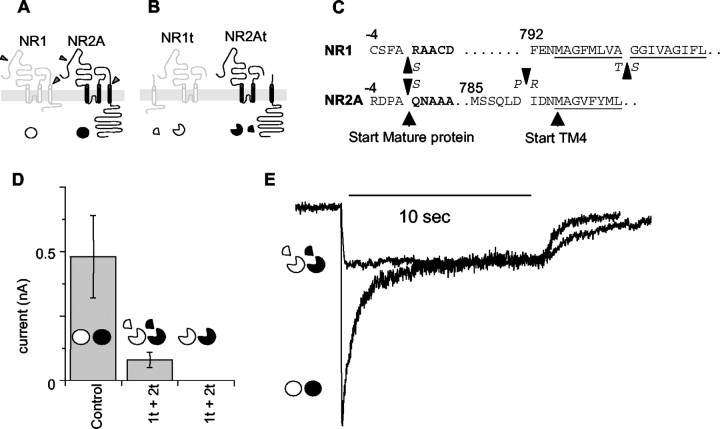
Fig. 1.
Generation and expression of truncated subunits.A, Schematic of subunit layout showing cut sites in NR1 and NR2a subunits. NR1 is white; NR2A isblack. The symbols underneath, awhite circle for NR1 and a black circlefor NR2A, are used throughout as a guide to the subunits included in each experiment. Full circles indicate full-length subunits. B, The putative membrane arrangement of the truncated subunits NR1t and NR2t and the arrangement of the TM4-tail pieces. Symbols are as in A, withwedges removed from the circles indicating truncation of the main subunits and quarter circle segments indicating TM4-tail peptides expressed as separate peptides linked to their own signal sequence. C, The exact amino acid sequences at the sites at which mutations were introduced to generate the cut sites. The single-letter code also indicates at what point conservative amino acid substitutions were introduced in the truncated subunits. Numbering is as in Anson et al. (1998), with the signal sequence negative from the start site of the mature protein. D, The average relative currents of cells expressing NR1 and NR2A subunits (Control) and truncated subunits with (middle) and without (right) the TM4-tail peptides coexpressed. Currents were elicited with 1 sec applications of 1 mm glutamate and 1 mm glycine 24–48 hr after transfection. Cells used in the chart were all recorded from transfections done at the same time on the same batch of cells. Currents were measured within 1 min of achieving whole-cell configuration. Symbols are as in A. Exact data are summarized in Table 1. 1t + 2t, Truncated NR1 plus truncated NR2 subunits. E, Representative traces from cells expressing control NR1 and NR2A channels and truncated NR1t and NR2t channels. Currents were elicited by a 10 sec pulse of 1 mm glutamate and 1 mm glycine in 2 mm CaCl2. The control, NR1 and NR2A trace (circles) shows the rapid sag characteristic of NR2A-containing NMDA channels. Under identical conditions, currents from cells expressing NR1t and NR2t show almost no sag (circles missing wedges). Currents in the figure are normalized at the 10 sec level.
Cell culture. DNA (1–2 μg of each) was used to transfect 4 × 13 mm plated human embryonic kidney (HEK) cells (clone tsA-201) using a calcium phosphate transfection protocol. Briefly, DNAs were mixed with 55 μl of 340 mmCaCl2, and the mixture was added slowly, with mixing to 75 μl of 2× HBSS. The resulting precipitate was added to cells in glutamine-free medium with 100 μm APV for 12–16 hr. Cells were washed and left for 2 d in glutamine-free medium with APV until recording.
Electrophysiology. Currents were measured at −70 mV (including junction potential) from cells in medium containing (in mm): 150 NaCl, 2.5 KCl, 2 CaCl2, and 10 HEPES, pH adjusted to 7.35 with NaOH. Intracellular solution contained (in mm): 110 K gluconate, 2.5 NaCl, 10 HEPES, and 10 BAPTA-K, pH adjusted to 7.3 with KOH. All electrodes were between 2 and 5 MΩ resistance after fire polishing. Capacitance and series resistances were corrected before recording from each cell, and any cells with series resistances of >20 MΩ were discarded. Recordings were filtered at 3 kHz and recorded to digital audio tape. Currents were then filtered at 100 Hz and sampled at 300 Hz using a CED1401 (Cambridge Electronic Designs, Cambridge, UK) and exported to windows metafiles (programs consam and plotsamp fromhttp://www.ucl.ac.uk/Pharmacology/dc.html).
Results
Expression of truncated subunits
Both the NR1 and the NR2A subunits were truncated at the beginning of the final TM region (TM4 at sites indicated in Fig.1A–C). The same sites used for the truncation of the pore regions (SpeI in NR1 andAvrII in NR2A) were used as the start of the TM4-tail subunits when they were expressed separately. To aid correct processing and assembly, the signal peptides normally found at the beginning of the NR1 and NR2A subunits were preserved at the beginning of the truncated subunits. In addition, identical signal peptide sequences were inserted at the beginning of each of the TM4-tail sequences.
In all of the figures, the NR1 subunit is represented by an open circle, and the NR2A subunit is represented by a filled circle. Truncated subunits are shown by removal of 90° segments, and isolated TM4-tail peptides are shown by a quarter circle segment.
Truncated subunits expressed alone in HEK cells failed to produce currents (Fig. 1D, Table1). However, when these subunits were cotransfected with plasmids containing the TM4-tail portions of the subunits, functional channels were formed (Fig. 1D,E, Table 1). These currents were consistently smaller than those from full-length subunits transfected in simultaneous control experiments (Table 1). It is important to note that currents from cells expressing altered subunits were always compared with control cells from simultaneous experiments, because currents in control cells proved to be variable over time and between batches of HEK cells. To control this variability, currents from cells expressing each combination of tandem or truncated subunits were compared only with currents from cells from the same batch transfected at the same time with control subunits.
Table 1.
TM4-tail region is critical for expression of glutamate currents from truncated channels or tandems made from truncated channels
| DNAs | Mean current | Number | |
|---|---|---|---|
| TM4s | pA ± SEM | ||
| Control | − | 480 ± 160 | 3 |
| NR1t + NR2t | + | 80 ± 30 | 5 |
| NR1t + NR2t | − | 0 ± 0 | 5 |
| Control | − | 180 ± 40 | 3 |
| 1t→1 + 2t→2 | + | 140 ± 60 | 14 |
| 1t→1 + 2t→2 | − | 0 ± 0 | 10 |
| 1t→2 + 2t→1 | + | 180 ± 90 | 13 |
| 1t→2 + 2t→1 | − | 40 ± 40 | 4 |
The first column indicates which subunit DNAs were used in a transfection. The second column shows whether or not the truncated DNAs or tandems were supplemented with the TM4-tail encoding DNAs. When TM4-tail DNAs were included, the TM4 tails of each truncated subunit were included. Controls are from cells transfected with NR1 and NR2A during the same set of experiments. Currents are rounded to 10 pA.
The cells transfected with full-length subunits showed, in the continued presence of agonist, a response that reaches a peak and then declines to a steady-state level as the receptors desensitize (Fig.1E). We refer to this phenomenon as sag, because it is known that the decline in macroscopic current is a poor measure of the fraction of receptors that are in desensitized states (Feltz and Trautmann, 1982). When the truncated NR1 and NR2 subunits, denoted NR1t and NR2t, were coexpressed with the TM4-tail portions of the channels as separate proteins, the currents produced lacked the distinctive sag of NR1–NR2A whole-cell currents (Fig.1E). During a 10 sec application of saturating glutamate and glycine (1 mm each), currents in cells expressing NR1 and NR2A subunits sagged to 34.4 ± 3.1% (n = 5) of their peak. In the same conditions, currents from the truncated subunits coexpressed with the separate TM4 tails remained at 96.2 ± 3.3% (n = 3) of their peak, with virtually no sag at all.
The successful expression of truncated subunits suggested the possibility that two subunits might be joined to make a single tandem protein. According to the accepted transmembrane arrangement of glutamate receptor subunits, the N terminal of each subunit is outside the cell and the C terminal is inside (Bigge, 1999; Dingledine et al., 1999; Cull-Candy et al., 2001). Therefore, the start of one subunit cannot be linked to the end of another. However, the truncated subunits are predicted to have both ends on the same side of the cell membrane, so they can be linked together to form tandems (Fig.2A). These tandems may be used to investigate the stoichiometry of the receptor.
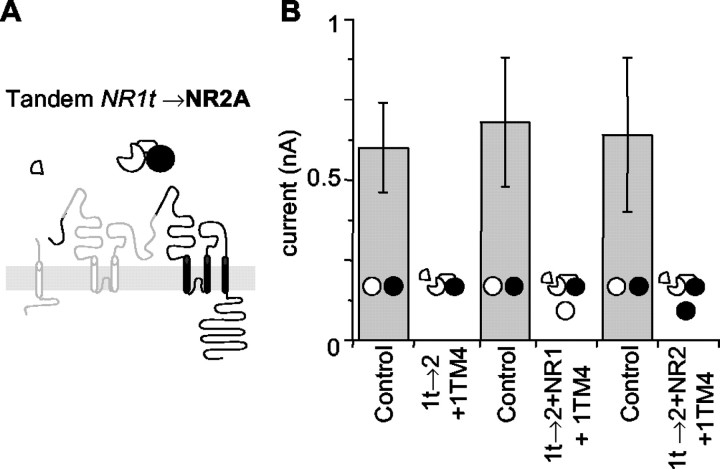
Fig. 2.
1t→2 tandems fail to generate functional channels. A, The presumed membrane orientation of the NR1t→2 tandem. The TM4 tail for NR1, the subunit truncated to form the tandem, is included as a separate subunit. The symbol above indicates the order of subunits in the tandem, with thetruncated subunit missing a 90οsegment forming the start or head of the tandem (NR1, white). NR2, black. B, NR1t→2 tandems do not generate currents. Currents from NR1 and NR2A cells (Control) recorded on the same day as NR1t→2 tandems alone, or with different full-length subunits added, are shown. The NR1t→2 tandem gives no significant currents alone or with any combination of single subunits. Diagrams of tandems are included in the histograms to indicate which DNAs were introduced to the cells for each column. Data are given in Table2.
Expression of individual tandems
Tandems were initially constructed with either the end of NR1t linked to NR2 (1t→2) or NR2t linked to NR1 (2t→1). These tandems were transfected into HEK cells together with the TM4 tail from the corresponding truncated subunit. No currents were observed from the individual tandems with 10 μm glutamate and 10 μm glycine (1 sec application). However, currents were seen when the glycine concentration was increased to levels higher than those needed for control full-length receptors. To minimize the chance that functional channels would be missed during the screening of different constructs simply because agonist concentrations were too low, an arbitrarily high, 1 mm glutamate and 1 mm glycine concentration was used to elicit currents from cells with combinations of tandems. With 1 mm glutamate and 1 mm glycine, the 2t→1 tandem produced currents (in four of five cells) (Fig. 3B). The 1t→2 tandem, in contrast, failed to produce currents in all of the conditions tested (Fig. 2B). Figure 3Dshows repeated 1 sec applications (60 sec apart) of 1 mm glutamate and 1 mmglycine on 2t→1 and control NR1 and NR2A cells; the first application was made within 1 min of entering the whole-cell configuration. Figure3C shows the averages of such currents from multiple cells. The current elicited by the first application in cells expressing the 2t→1 tandem was similar in amplitude to control currents, but during the second application it had dropped to 51 ± 9% (n = 6) of the first application, and after six applications the current in cells expressing the 2t→1 tandem had almost vanished (15 ± 9% of starting level; n = 3) (Fig. 3C). In contrast, this sort of rundown was much slower in the control receptors: the second application produced a current that was 93 ± 4% (n = 5) of the first, and after six applications the current was still 73 ± 3% of the first (n = 3). The channels made by expressing NR1t and NR2t with both TM4 tails had rundown rates comparable with those of control channels (currents at the second application were 95 ± 9% of starting levels; n = 5).
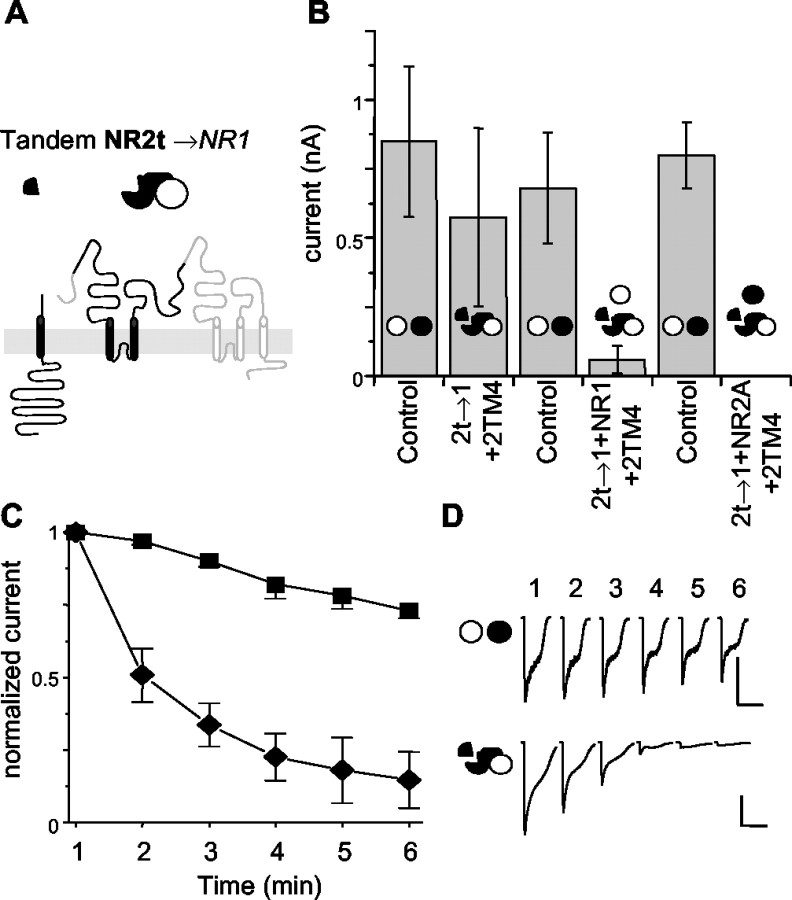
Fig. 3.
2t→1 tandems produce currents that run down quickly. A, The presumed membrane organization of the NR2t→1 tandem with its symbol above. The TM4 tail for NR2A, the subunit truncated to form this tandem, is included as a separate subunit. The symbol above is as in Figure2A, with black for NR2-derived portions and white for NR1. B, NR2t→1 tandems are able to produce unstable currents. As in Figure2B, cells expressing the NR2t→1 tandem were compared with cells transfected with NR1 and NR2A subunits in the same experiment. The 2t→1 alone was able to generate currents similar in amplitude to the control cells transfected on the same day. The addition of either NR1 or NR2A alone to the NR2t→1 tandem caused a dramatic drop in the currents produced by these cells.C, Currents derived from the NR2t→1 tandem rundown rapidly compared with control NR1 and NR2A cells. One second applications of 1 mm glutamate and 1 mm glycine were given at 1 min intervals starting within 1 min of obtaining the whole-cell configuration. All levels are normalized to the first application. The NR2t→1 tandem was supplemented with the NR2 TM4-tail segment in all cells. Squares, Controls;diamonds, 2t→1 only. D, Traces from representative cells showing rundown of NR2t→1 relative to control. The top traces are from a control cell, and thebottom traces are from a cell expressing NR2t→1. Calibration: 500 pA, 5 sec.
The remarkable instability of the 2t→1 channels argues that although these tandems could produce currents in some cells, they are not easily incorporated into functional channels. A common problem of tandem constructs is residual flexibility or aberrant ordering of subunits theoretically constrained by the tandems to a certain order (Liman et al., 1992; McCormack et al., 1992). The 1t→2 tandem used here seems genuinely restricted (Fig. 2B, Table2), given its inability to produce currents alone or when mixed with either NR1 or NR2A. If either subunit within this tandem was able to function independently, the coexpression of the appropriate control subunit should permit that subunit to form channels. However, in the 1t→2 tandem neither control subunit was able to help form functional channels, suggesting that neither of the subunits in the tandem was able to pair with the coexpressed control subunits. The 2t→1 tandem, however, does produce a current when expressed alone (Fig. 3, Table 2). This unstable current may be attributable to some flexibility between the subunits of this tandem. It is possible that two subunits connected in this tandem contribute to two separate channels (Fig.4C) or are cleaved by proteases (Fig. 4B). In either case, the 2t→1 tandem, although able to form channels, seems unable to produce the type of stable currents seen in control cells.
Table 2.
Single tandems and monomers do not give sustained currents
| DNAs | Mean current | Number |
|---|---|---|
| pA ± SEM | ||
| Control | 600 ± 140 | 5 |
| 1t→2 | 0 ± 0 | 5 |
| Control | 680 ± 200 | 6 |
| 1t→2 + NR1 | 0 ± 0 | 5 |
| Control | 640 ± 240 | 5 |
| 1t→2 + NR2 | 0 ± 0 | 6 |
| Control | 630 ± 300 | 7 |
| 2t→1 | 360 ± 250* | 6 |
| Control | 680 ± 200 | 5 |
| 2t→1 + NR1 | 60 ± 50 | 5 |
| Control | 800 ± 120 | 5 |
| 2t→1 + NR2A | 0 ± 0 | 5 |
Currents from 2t→1 had rapid rundown.
The first column shows which subunit DNAs were included in transfections. The control is NR1 and NR2A, from the same set of experiments in which tandem recordings were done. In each case the TM4-tail DNAs were included in each mix for each subunit truncated in tandems. Currents are rounded to 10 pA.
Fig. 4.

Predicted and aberrant arrangements of subunits included in tandem constructs. A, The theoretical arrangements of expressed single subunits and tandems and their ability to generate functional currents. Diagrams are as used in all figures;open areas represent NR1-derived elements; filled areas represent NR2A-derived elements. In all cases thesubunit missing a 90° segment is the start (the N terminal or head of a tandem), and the full circle is the end of the tandem. Arrangements shown in A are the organizations expected to be permitted by tandems without stretching or distorting the linker. B, Some of the possible ways that subunits in tandems may be able to violate the arrangements shown inA. The letters N and C are used to show the N and C terminals of the tandems forming a channel. Diagrams are as in A, except that gray-scale subunits indicate that the normally intracellular region of the tandem has flipped to the outside of the cell (flip in membrane). The side of the tandem used to form the pore has been chosen arbitrarily.C, Subunits within a single tandem may participate in the formation of two different channels. A chain of channels may be formed by subunits within a tandem participating in separate channels. The arrangement shown would allow a dimer of dimers organization of subunits within channels, although only the 2t→1 tandem is used.
In addition to testing the ability of individual subunits within a tandem to function independently, the addition of full-length control subunits to each tandem would allow a range of possible orientations of subunits to be tested. Each of the tandems was supplemented with either full-length NR1 or NR2A subunits as well as the TM4s. The addition of these single subunits would allow the receptors to form as pentamers (one tandem and three control subunits) or as tetramers (one tandem and two control subunits) that contained a single copy of one sort of subunit plus three copies of the other sort (tandems alone would allow two of each sort, as shown in Fig. 4). However, none of these combinations of tandems and single control subunits were able to produce significant currents (Figs. 2B,3B): it seems that 3:1 combinations either do not form or do not function.
Tandems may be combined to form stable channels
In an attempt to restore sustained currents of the sort seen in control cells, both the 2t→1 and 1t→2 tandems were expressed simultaneously in cells, together with both TM4 tails. In these cells the tandems were able to produce robust currents (Fig.5). The second response (at 1 min) (Fig.5C, bottom two traces) showed some rundown (to 80 ± 5% of starting levels; n = 9) (Fig.5B). After 6 min the response had dropped to 58 ± 10% (n = 9), somewhat lower than the control currents but much larger than the currents remaining (15% after 6 min) in cells expressing 2t→1 alone. It would be expected that these cells, expressing a mixture of 1t→2 and 2t→1 tandems, could produce three sorts of channels: (1) 1t→2 alone (not functional), (2) 2t→1 alone (which run down rapidly), and (3) channels with one 1t→2 and one 2t→1. As this range of possibilities suggests, the currents seen in cells expressing both 1t→2 and 2t→1 had a more variable rate of rundown than either control or cells transfected with 2t→1 alone (Fig. 5B). Examples from two cells are shown in Figure5C, which shows one (middle) with relatively little rundown and one (bottom) with more pronounced rundown. The overall intermediate amount of rundown seen in these cells could result primarily from channels with 2t→1 alone, so it is likely that channels made of both 1t→2 and 2t→1 tandems run down little, if at all.
Fig. 5.
Sustained currents are produced by mixtures of tandems allowing a 1-1-2-2 organization of subunits in receptors.A, Mixtures of pairs of tandems were able to generate sustained currents. Currents from cells expressing a mix of 1t→2 and 2t→1 (columns 2 and 3) or 1t→1 and 2t→2 (columns 4 and 5) are shown compared with currents from cells transfected with full-length NR1 and NR2A (Control). For clarity, the diagrams indicate the DNAs included as well as the axis labels. All currents resulted from 1 sec applications of 1 mm glutamate and 1 mm glycine applied within 1 min of obtaining a whole-cell configuration. In the presence of both of the TM4 tails (columns 2 and 4), both mixtures of tandems generated currents that were not significantly different from control cells. In the absence of the TM4 tails, the 1t→2 and 2t→1 mixture did elicit small currents in two of four cells. The 1t→1 and 2t→2 cells did not have currents when the TM4 tails were not coexpressed (0 of 10 cells; full data in Table 1). B, Currents generated from mixtures of tandems are more stable than those from 2t→1 alone. The rundown was measured as in Figure 3C.Squares, Control; diamonds, 2t→1 only;down triangles, 2t→1 with 1t→2 and both TM4 tails;up triangles, 1t→1 with 2t→2 with both TM4 tails. All currents are normalized to the first application. C, Representative traces from cells expressing mixtures of tandems. Thebottom two traces are from cells expressing both 2t→1 with 1t→2, showing different levels of rundown in this population. The top traces are from a representative cell expressing 1t→1 with 2t→2. Calibration: 250 pA, 5 sec for all traces.
The production of a component of large (similar to control) and sustained currents from a mixture of 1t→2 with 2t→1 raises the possibility that the stable channels can be formed by the two types of tandems coming together head to tail in a 1t→2/2t→1 orientation around the pore (Figs. 4B,5A). This new orientation could be formed only if both tandems were included. The production of functional channels by a single tandem would favor an alternating pattern of subunits (e.g., 1→2, 1→2 around the pore), unless the flexibility in the tandem backbone allowed a subunit within a tandem to fold around (or to flip out) so that the tandems could align in a head-to-head orientation (e.g., 1→2, 2←1). This orientation would require the backbone of the tandem to rotate 180° (Fig. 4B). It is possible that such a rotation underlies the transient currents that were seen in cells containing 2t→1 only (Fig. 3). Cells with 1t→2 alone produced no currents (Fig. 2).
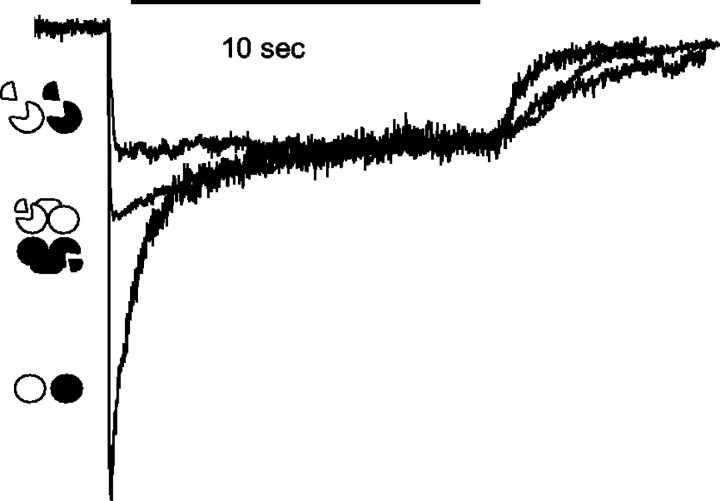
Finally, to eliminate the possibility of having two sorts of channels (e.g., currents arising from 2t→1 alone, as well as from both 1t→2 plus 2t→1), a new set of tandems was constructed that linked NR1t to NR1 (1t→1) and NR2t to NR2 (2t→2). These tandems, when coexpressed with both TM4 tails, produced sustained currents in HEK cells that were almost as big as the controls (Fig. 5A) and showed rundown that differed little from that seen in controls (Fig. 5B,C,top trace). The second response (at 1 min) was 94 ± 4% of the first (n = 3), very similar to that seen in control (93 ± 4%; n = 5). Even after 6 min the 1t→1 plus 2t→2 currents remained similar to control (69 ± 13%; n = 3). Unlike the currents from 1t→2 plus 2t→1, which had some small currents in the absence of the TM4 tails, the currents from 1t→1 and 2t→2 absolutely required the TM4 tails to produce currents. In the 1t→2 plus 2t→1 (expressed without the addition of TM4 tails) cells, two of five cells had currents, only one of which was >50 pA, whereas in the 1t→1 plus 2t→2 cells, none of nine had currents of >10 pA when the TM4 tails were not included. The complete dependence on coexpression of the TM4s is similar to that seen in the case of the separate truncated subunits not joined together as tandems (Fig. 1D, Table 1). The fact that the 1t→1 plus 2t→2 combination also required the TM4 tails suggests that the truncated subunits within the tandems contributed to the channels. The sag observed during long (10 sec) applications of glutamate also was intermediate between the truncated subunits and control subunits (Fig.6). During a 10 sec application of glycine and glutamate (1 mm each), the current in 1t→1 and 2t→2 cells sagged to 64 ± 3.6% of peak (n = 4). In comparison, currents from control cells dropped to 34% and currents from truncated channels remained at 96% of starting levels. The fact that the sag was different in tandem-expressing cells from that seen in cells expressing only truncated channels suggests that the former were not simply using only the first, truncated subunits of each tandem to form channels, a problem suggested to occur in some tandem studies (Liman et al., 1992;McCormack et al., 1992). The presence of an intermediate amount of sag indicates that the full-length subunits in the 1t→1 plus 2t→2 tandems also contributed to the whole-cell currents in these cells. However, notice that the sag was measured at a fixed agonist concentration (rather than, say, a fixed response), so it cannot yet be interpreted fully.
Fig. 6.
Normalized currents from cells expressing 1t→1 with 2t→2 and both TM4 tails show an intermediate level of sag. Representative traces, as in Figure 1E, from 10 sec applications of 1 mm glutamate and 1 mmglycine show an intermediate level of sag in cells containing 1t→1 and 2t→2 with TM4 tails. Circles, Full-length NR1 and NR2A; circles missing wedges, NR1t and NR2t;connected symbols, 1t→1 with 2t→2. All currents are normalized to the 10 sec level.
Together, these data suggest that the 1t→1 and 2t→2 tandems with TM4 tails assemble together to form channels arranged as two dimers with the subunits arranged in a 1-1-2-2 orientation around the pore.
Not surprisingly, the currents resulting from transfecting DNA tandems were different from those resulting from full-length control DNAs or the truncated DNAs. The potencies of glutamate and glycine were dramatically shifted in the tandems relative to controls. In cells expressing 1t→1 plus 2t→2 with both TM4 tails, currents were sustained and the potency of glycine decreased to an EC50 of 153 ± 16 μm(n = 3), compared with 2.05 ± 0.60 μm (n = 4) for control (NR1 plus NR2A). In contrast, the potency of glutamate was greatly increased; the EC50 was probably <10 nm (this is comparable with contaminant glutamate concentrations, making it difficult to determine an EC50 value; data not shown). Given the close proximity of the putative S2 ligand-binding regions in the subunits to the sites used to truncate and link the subunits into a tandem, some disruption of the binding sites is expected. Therefore, it is not surprising that the EC50 values for glutamate and glycine are altered. However, the channels formed by the tandem mixture, 1t→1 and 2t→2, do behave, qualitatively, like control channels, in that currents were sustained and were blocked by 100 μm APV, 1 mmMg2+, and 100 μmkynurenic acid (data not shown).
Discussion
The two main conclusions from this work are (1) that TM4 regions of the NMDA receptor can function when detached from the rest of the subunit and (2) the implication from tandem studies that the receptor consists of an NR1 dimer and an NR2 dimer.
The finding that the presence of detached TM4 proteins allows functional receptors to be inserted into the cell membrane is novel, but cannot at present be interpreted in structural terms. The most obvious change in the receptor made from truncated subunits and separate TM4 proteins is the near-complete abolition of the decline in response seen normally in the continuous presence of agonist. Therefore, it seems that the C-terminal end of the molecule affects “desensitization” in some way, but little more can be said at the moment. The importance of this region in receptor gating is also supported by recent studies of mutations in the TM4 domain that also show profound effects on channel desensitization (Ren et al., 2003). However, desensitization takes many forms in the NMDA receptor (for review, see Dingledine et al., 1999) and is affected by changes in many parts of the molecule (Villarroel et al., 1998; Kohda et al., 2000;Krupp et al., 2002).
Although the role of the TM4 in controlling sag is completely disrupted when the link between the channel pore and TM4 is broken, the role of the TM4 tail in controlling channel assembly and localization is partially able to compensate for the separation. The autonomous function of the TM4 tails is circumstantial evidence for the separate evolutionary origins of the first two TMs and the final TM and tail. Possibly, as postulated by Wo and Oswald (1995), these regions originated as separate proteins that chaperoned the GluR subunits to the membranes.
The data from the tandem subunits are strongly suggestive of a 1-1-2-2 pair of dimers orientation of the subunits within the receptor. In the past, tandems have been used to study the stoichiometry of several types of channels, including GABA receptors (Im et al., 1995; Baumann et al., 2001), cyclic nucleotide-gated channels (Gordon and Zagotta, 1995; Liu et al., 1998; Morrill and MacKinnon, 1999; He et al., 2000), voltage-gated chloride channels (Fahlke et al., 1998), and K channels (Isacoff et al., 1990; Zheng and Sigworth, 1998). However, subunits supposedly constrained by tandems have occasionally been found to behave independently (Liman et al., 1992; McCormack et al., 1992). To surmount this problem, the NMDA tandems were tested in many combinations to allow any independence to be revealed. One tandem did show behavior that, in the context of the rest of the results, indicated that subunits within it could behave somewhat independently.
The 2t→1 tandem was able to form functional channels alone. This would imply the expression of channels with an alternating 1-2-1-2 orientation of subunits around the pore. However, the instability of the channels formed from these receptors makes it unlikely that they are occurring in as favorable a configuration as the 1-1-2-2 receptors. It is possible that the 2t→1 receptors are able to form functional channels by assembling into a dimer of dimers orientation by allowing outlying subunits within individual tandems to participate in forming separate channels (Fig. 4C). This would allow the 2t→1 tandems alone to form channels composed of a dimer of dimers, but these channels would be forced to align in a chain or raft of linked channels, which might explain their instability on opening after agonist applications. Alternatively, these tandems simply were not restrictive enough for the subunits, allowing them to twist and stretch into different orientations, as shown in Figure4B. The remaining tandems did seem to be restrictive, so the evidence of the 2t→1 tandem alone is not enough to disprove the evidence of the remaining tandems, and it is unlikely that the NMDA receptor subunits are not constrained to a specific stoichiometry within the receptor; this is particularly so given the inability of the 1t→2 tandem to form functional receptors unless paired with the 2t→1 tandem.
The advantage of using tandems to determine receptor subunit stoichiometry is that not only can they reveal the number of subunits in the receptor, already convincingly shown to be four by other investigators (Laube et al., 1998; Rosenmund et al., 1998; Chen et al., 1999), but they may identify the number of each type of subunits (two NR1 and two NR2), as opposed to 3:1 or 1:3; finally, they also reveal the organization of the two types of subunits into two pairs, or 1-1-2-2, as opposed to alternating around the pore in a 1-2-1-2 manner. The two-pair orientation may have dramatic effects on the mechanism of channel gating and binding.
The evidence from the tandems, with robust expression occurring only when the tandems would favor a dimer of dimers orientation, supports the idea that these channels are indeed arranged as a dimer of dimers in the membrane. These data from tandems are supported by previous studies with other methods, which have already suggested such a possibility. Crystal studies of isolated GluR-binding domains suggest that these domains may be configured as dimers rather than monomers when purified. The pair-forming nature of the glutamate binding motif appears to have been conserved from bacterial glutamate-binding domains (Hsiao et al., 1996), through G-protein-coupled glutamate receptors (Kunishima et al., 2000), to the ion-channel receptors (Kuusinen et al., 1999; Armstrong and Gouaux, 2000). The importance of the pairing of the binding domains for the binding and subsequent gating of the channels is unknown. However, it is worth noting that mechanisms with two independent dimers generate many discrete states with closely spaced time constants, which could make the fitting of a few exponentials to macroscopic currents or to open time distributions quite misleading.
In addition, it has already been suggested that the subunits in AMPA GluRs may behave as independent dimers during gating (Robert et al., 2001; Bowie and Lange, 2002; Sun et al., 2002). It would not be surprising now to find that the subunits in NMDA receptors also behave functionally as dimers. However, the tandem data show that the NMDA receptors are most likely organized into two different dimers. They are either nonsymmetrical 1-2 and 2-1 dimers or a dimer of two NR1 subunits and a second dimer of two NR2 subunits. This opens the possibility that the roles of glutamate and glycine are different in the receptor.
The organization of receptors into dimers may be derived from outside of the GluR family, from their relatives (the K channels), because it has also been shown recently that K channels are formed by the dimerization of two dimers (Tu and Deutsch, 1999); thus, the formation of dimers might be a general theme in receptor assembly as well as function. It has already been shown that NR1 subunits are able to form homodimers before binding NR2 subunits and reaching the surface of the cell (Meddows et al., 2001).
In conclusion, the proposed number and organization of the subunits within the channels provide an essential starting point for future efforts to describe quantitatively the behavior of these channels. In particular, it will be important to discern whether the pairs of subunits act independently or are somehow cooperative.
Footnotes
This work was supported by the Medical Research Council and the Wellcome Trust. We thank Paul Groot-Kormelink and Lucia Sivilotti for generous assistance with laboratory space and materials. The original NR1 and NR2A clones were gifts from R. Schoepfer.
Correspondence should be addressed to Dr. David Colquhoun, Pharmacology Department, University College London, Gower Street, London WC1E 6BT, UK. E-mail: d.colquhoun@ucl.ac.uk.
References
- 1.Anson LC, Chen PE, Wyllie DJ, Colquhoun D, Schoepfer R. Identification of amino acid residues of the NR2A subunit that control glutamate potency in recombinant NR1/NR2A NMDA receptors. J Neurosci. 1998;18:581–589. doi: 10.1523/JNEUROSCI.18-02-00581.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armstrong N, Gouaux E. Mechanisms for activation and antagonism of an AMPA-sensitive glutamate receptor: crystal structures of the GluR2 ligand binding core. Neuron. 2000;28:165–181. doi: 10.1016/s0896-6273(00)00094-5. [DOI] [PubMed] [Google Scholar]
- 3.Baumann SW, Baur R, Sigel E. Subunit arrangement of γ-aminobutyric acid type A receptors. J Biol Chem. 2001;276:36275–36280. doi: 10.1074/jbc.M105240200. [DOI] [PubMed] [Google Scholar]
- 4.Behe P, Stern P, Wyllie DJ, Nassar M, Schoepfer R, Colquhoun D. Determination of NMDA NR1 subunit copy number in recombinant NMDA receptors. Proc R Soc Lond B Biol Sci. 1995;262:205–213. doi: 10.1098/rspb.1995.0197. [DOI] [PubMed] [Google Scholar]
- 5.Bigge CF. Ionotropic glutamate receptors. Curr Opin Chem Biol. 1999;3:441–447. doi: 10.1016/S1367-5931(99)80065-9. [DOI] [PubMed] [Google Scholar]
- 6.Bowie D, Lange GD. Functional stoichiometry of glutamate receptor desensitization. J Neurosci. 2002;22:3392–3403. doi: 10.1523/JNEUROSCI.22-09-03392.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen GQ, Cui C, Mayer ML, Gouaux E. Functional characterization of a potassium-selective prokaryotic glutamate receptor. Nature. 1999;402:817–821. doi: 10.1038/45568. [DOI] [PubMed] [Google Scholar]
- 8.Cull-Candy S, Brickley S, Farrant M. NMDA receptor subunits: diversity, development and disease. Curr Opin Neurobiol. 2001;11:327–335. doi: 10.1016/s0959-4388(00)00215-4. [DOI] [PubMed] [Google Scholar]
- 9.Dingledine R, Borges K, Bowie D, Traynelis SF. The glutamate receptor ion channels. Pharmacol Rev. 1999;51:7–61. [PubMed] [Google Scholar]
- 10.Fahlke C, Rhodes TH, Desai RR, George AL., Jr Pore stoichiometry of a voltage-gated chloride channel. Nature. 1998;394:687–690. doi: 10.1038/29319. [DOI] [PubMed] [Google Scholar]
- 11.Feltz A, Trautmann A. Desensitization at the frog neuromuscular junction: a biphasic process. J Physiol (Lond) 1982;322:257–272. doi: 10.1113/jphysiol.1982.sp014036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gordon SE, Zagotta WN. Subunit interactions in coordination of Ni2+ in cyclic nucleotide-gated channels. Proc Natl Acad Sci USA. 1995;92:10222–10226. doi: 10.1073/pnas.92.22.10222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He Y, Ruiz M, Karpen JW. Constraining the subunit order of rod cyclic nucleotide-gated channels reveals a diagonal arrangement of like subunits. Proc Natl Acad Sci USA. 2000;97:895–900. doi: 10.1073/pnas.97.2.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holmes KD, Mattar P, Marsh DR, Jordan V, Weaver LC, Dekaban GA. The C-terminal C1 cassette of the N-methyl-d-aspartate receptor 1 subunit contains a bi-partite nuclear localization sequence. J Neurochem. 2002;81:1152–1165. doi: 10.1046/j.1471-4159.2002.00865.x. [DOI] [PubMed] [Google Scholar]
- 15.Hsiao CD, Sun YJ, Rose J, Wang BC. The crystal structure of glutamine-binding protein from Escherichia coli. J Mol Biol. 1996;262:225–242. doi: 10.1006/jmbi.1996.0509. [DOI] [PubMed] [Google Scholar]
- 16.Im WB, Pregenzer JF, Binder JA, Dillon GH, Alberts GL. Chloride channel expression with the tandem construct of α 6-β 2 GABAA receptor subunit requires a monomeric subunit of α6 or γ2. J Biol Chem. 1995;270:26063–26066. doi: 10.1074/jbc.270.44.26063. [DOI] [PubMed] [Google Scholar]
- 17.Isacoff EY, Jan YN, Jan LY. Evidence for the formation of heteromultimeric potassium channels in Xenopus oocytes. Nature. 1990;345:530–534. doi: 10.1038/345530a0. [DOI] [PubMed] [Google Scholar]
- 18.Kohda K, Wang Y, Yuzaki M. Mutation of a glutamate receptor motif reveals its role in gating and delta2 receptor channel properties. Nat Neurosci. 2000;3:315–322. doi: 10.1038/73877. [DOI] [PubMed] [Google Scholar]
- 19.Krupp JJ, Vissel B, Thomas CG, Heinemann SF, Westbrook GL. Calcineurin acts via the C-terminus of NR2A to modulate desensitization of NMDA receptors. Neuropharmacology. 2002;42:593–602. doi: 10.1016/s0028-3908(02)00031-x. [DOI] [PubMed] [Google Scholar]
- 20.Kunishima N, Shimada Y, Tsuji Y, Sato T, Yamamoto M, Kumasaka T, Nakanishi S, Jingami H, Morikawa K. Structural basis of glutamate recognition by a dimeric metabotropic glutamate receptor. Nature. 2000;407:971–977. doi: 10.1038/35039564. [DOI] [PubMed] [Google Scholar]
- 21.Kuusinen A, Abele R, Madden DR, Keinanen K. Oligomerization and ligand-binding properties of the ectodomain of the α-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid receptor subunit GluRD. J Biol Chem. 1999;274:28937–28943. doi: 10.1074/jbc.274.41.28937. [DOI] [PubMed] [Google Scholar]
- 22.Laube B, Kuhse J, Betz H. Evidence for a tetrameric structure of recombinant NMDA receptors. J Neurosci. 1998;18:2954–2961. doi: 10.1523/JNEUROSCI.18-08-02954.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liman ER, Tytgat J, Hess P. Subunit stoichiometry of a mammalian K+ channel determined by construction of multimeric cDNAs. Neuron. 1992;9:861–871. doi: 10.1016/0896-6273(92)90239-a. [DOI] [PubMed] [Google Scholar]
- 24.Liu DT, Tibbs GR, Paoletti P, Siegelbaum SA. Constraining ligand-binding site stoichiometry suggests that a cyclic nucleotide-gated channel is composed of two functional dimers. Neuron. 1998;21:235–248. doi: 10.1016/s0896-6273(00)80530-9. [DOI] [PubMed] [Google Scholar]
- 25.Matsuda I, Mishina M. Identification of a juxtamembrane segment of the glutamate receptor delta2 subunit required for the plasma membrane localization. Biochem Biophys Res Commun. 2000;275:565–571. doi: 10.1006/bbrc.2000.3351. [DOI] [PubMed] [Google Scholar]
- 26.McCormack K, Lin L, Iverson LE, Tanouye MA, Sigworth FJ. Tandem linkage of Shaker K+ channel subunits does not ensure the stoichiometry of expressed channels. Biophys J. 1992;63:1406–1411. doi: 10.1016/S0006-3495(92)81703-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meddows E, Le Bourdelles B, Grimwood S, Wafford K, Sandhu S, Whiting P, McIlhinney RA. Identification of molecular determinants that are important in the assembly of N-methyl-d-aspartate receptors. J Biol Chem. 2001;276:18795–18803. doi: 10.1074/jbc.M101382200. [DOI] [PubMed] [Google Scholar]
- 28.Morrill JA, MacKinnon R. Isolation of a single carboxyl-carboxylate proton binding site in the pore of a cyclic nucleotide-gated channel. J Gen Physiol. 1999;114:71–83. doi: 10.1085/jgp.114.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Premkumar LS, Qin F, Auerbach A. Subconductance states of a mutant NMDA receptor channel kinetics, calcium, and voltage dependence. J Gen Physiol. 1997;109:181–189. doi: 10.1085/jgp.109.2.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prybylowski K, Vicini S, Wang J, Kirkness E, Wolfe BB. Tandem proteins to study NMDA receptor stoichiometry. Soc Neurosci Abstr. 1999;25:1487. [Google Scholar]
- 31.Ren H, Honse Y, Karp BJ, Lipsky RH, Peoples RW. A site in the fourth membrane-associated domain of the N-methyl-d-aspartate receptor regulates desensitization and ion channel gating. J Biol Chem. 2003;278:276–283. doi: 10.1074/jbc.M209486200. [DOI] [PubMed] [Google Scholar]
- 32.Robert A, Irizarry SN, Hughes TE, Howe JR. Subunit interactions and AMPA receptor desensitization. J Neurosci. 2001;21:5574–5586. doi: 10.1523/JNEUROSCI.21-15-05574.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosenmund C, Stern-Bach Y, Stevens CF. The tetrameric structure of a glutamate receptor channel. Science. 1998;280:1596–1599. doi: 10.1126/science.280.5369.1596. [DOI] [PubMed] [Google Scholar]
- 34.Schneggenburger R, Ascher P. Coupling of permeation and gating in an NMDA-channel pore mutant. Neuron. 1997;18:167–177. doi: 10.1016/s0896-6273(01)80055-6. [DOI] [PubMed] [Google Scholar]
- 35.Scott DB, Blanpied TA, Swanson GT, Zhang C, Ehlers MD. An NMDA receptor ER retention signal regulated by phosphorylation and alternative splicing. J Neurosci. 2001;21:3063–3072. doi: 10.1523/JNEUROSCI.21-09-03063.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sheng M, Pak DT. Ligand-gated ion channel interactions with cytoskeletal and signaling proteins. Annu Rev Physiol. 2000;62:755–778. doi: 10.1146/annurev.physiol.62.1.755. [DOI] [PubMed] [Google Scholar]
- 37.Standley S, Roche KW, McCallum J, Sans N, Wenthold RJ. PDZ domain suppression of an ER retention signal in NMDA receptor NR1 splice variants. Neuron. 2000;28:887–898. doi: 10.1016/s0896-6273(00)00161-6. [DOI] [PubMed] [Google Scholar]
- 38.Steigerwald F, Schulz TW, Schenker LT, Kennedy MB, Seeburg PH, Kohr G. C-terminal truncation of NR2A subunits impairs synaptic but not extrasynaptic localization of NMDA receptors. J Neurosci. 2000;20:4573–4581. doi: 10.1523/JNEUROSCI.20-12-04573.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stern P, Behe P, Schoepfer R, Colquhoun D. Single-channel conductances of NMDA receptors expressed from cloned cDNAs: comparison with native receptors. Proc R Soc Lond B Biol Sci. 1992;250:271–277. doi: 10.1098/rspb.1992.0159. [DOI] [PubMed] [Google Scholar]
- 40.Sun Y, Olson R, Horning M, Armstrong N, Mayer M, Gouaux E. Mechanism of glutamate receptor desensitization. Nature. 2002;417:245–253. doi: 10.1038/417245a. [DOI] [PubMed] [Google Scholar]
- 41.Takahashi T, Feldmeyer D, Suzuki N, Onodera K, Cull-Candy SG, Sakimura K, Mishina M. Functional correlation of NMDA receptor ε subunits expression with the properties of single-channel and synaptic currents in the developing cerebellum. J Neurosci. 1996;16:4376–4382. doi: 10.1523/JNEUROSCI.16-14-04376.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tu L, Deutsch C. Evidence for dimerization of dimers in K+ channel assembly. Biophys J. 1999;76:2004–2017. doi: 10.1016/S0006-3495(99)77358-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Villarroel A, Regalado MP, Lerma J. Glycine-independent NMDA receptor desensitization: localization of structural determinants. Neuron. 1998;20:329–339. doi: 10.1016/s0896-6273(00)80460-2. [DOI] [PubMed] [Google Scholar]
- 44.Wo ZG, Oswald RE. Unraveling the modular design of glutamate-gated ion channels. Trends Neurosci. 1995;18:161–168. doi: 10.1016/0166-2236(95)93895-5. [DOI] [PubMed] [Google Scholar]
- 45.Wyllie DJ, Behe P, Colquhoun D. Single-channel activations and concentration jumps: comparison of recombinant NR1a/NR2A and NR1a/NR2D NMDA receptors. J Physiol (Lond) 1998;510:1–18. doi: 10.1111/j.1469-7793.1998.001bz.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xia H, Hornby ZD, Malenka RC. An ER retention signal explains differences in surface expression of NMDA and AMPA receptor subunits. Neuropharmacology. 2001;41:714–723. doi: 10.1016/s0028-3908(01)00103-4. [DOI] [PubMed] [Google Scholar]
- 47.Zhang Y, Auerbach A. Kinetic properties of NMDA receptors in embryonic Xenopus spinal neurons. J Neurophysiol. 1995;74:153–161. doi: 10.1152/jn.1995.74.1.153. [DOI] [PubMed] [Google Scholar]
- 48.Zheng J, Sigworth FJ. Intermediate conductances during deactivation of heteromultimeric Shaker potassium channels. J Gen Physiol. 1998;112:457–474. doi: 10.1085/jgp.112.4.457. [DOI] [PMC free article] [PubMed] [Google Scholar]