Abstract
The emergence of E.coli strains displaying patterns of virulence genes from different pathotypes shows that the current classification of E.coli pathotypes may be not enough, the study aimed to compare the phylogenetic groups and urovirulence genes of uropathogenic Escherichia coli (UPEC) and diarrheagenic E.coli (DEC) strains to extend the knowledge of E.coli classification into different pathotypes. A total of 173 UPEC and DEC strains were examined for phylogenetic typing and urovirulence genes by PCR amplifications. In contrast to most reports, phylogenetic group A was the most prevalent in both UPEC and DEC strains, followed by B2 group. Amplification assays revealed that 89.32% and 94.29% of UPEC and DEC strains, respectively, carried at least one of the urovirulence genes, 49.5% and 31.4% of UPEC and DEC strains, respectively, carried ≥ 2 of the urovirulence genes, fim H gene was the most prevalent (66.9% and 91.4%) in UPEC and DEC strains respectively. Twenty different patterns of virulence genes were identified in UPEC while 5 different patterns in DEC strains. Strains with combined virulence patterns of four or five genes were belonged to phylogenetic group B2. Our finding showed a closer relationship between the DEC and UPEC, so raised the suggestion that some DEC strains might be potential uropathogens. These findings also provide different insights into the phylogenetic classification of E. coli as pathogenic or commensals where group A can be an important pathogenic type as well as into the classification as intestinal or extra- intestinal virulence factors.
Introduction
Escherichia coli are normal inhabitants of gastrointestinal tract of humans and many animals, however some E.coli strains acquired specific virulence genes that enable them to cause intestinal and extra-intestinal infections in humans such as diarrhea and urinary tract infections [1]. Diarrheagenic E.coli (DEC) represent a leading bacterial cause of diarrhea all over the world. DEC pathotypes are characterized by their specific virulence determinants [2]. E.coli strains causing extra-intestinal infections are known as extra-intestinal pathogenic E.coli (ExPEC) [3]. Molecular and epidemiological studies have identified ExPEC as a distinct E. coli type. ExPEC strains usually carry characteristic virulence factors that allow colonization of the host mucosa and conferring their pathogenic potential [4].
Urinary tract infection (UTI) is the most common extra-intestinal infection caused by E.coli [5], which occur mainly due to the spread of Uropathogenic Escherichia coli (UPEC) strains from the intestine to the urinary tract [6]. These strains become pathogenic by acquiring new virulence properties encoded by specific genes, allowing them to colonize host mucosal surfaces and invade the normally sterile urinary tract [5]. Surface virulence factors (adhesins) are very important virulence factor of UPEC as the main attachment factor, P fimbriae is associated with pyelonephritis and is encoded by pap genes. [7]. Other adhesins that act as virulence factors are S fimbrial adhesin, which is coded by sfa genes and Type 1 fimbriae which is encoded by the fim gene cluster[8, 9, 10,11]. A part from adhesins, important virulence factors of UPEC strains are the toxins that cause an inflammatory response. The most important secreted virulence factor is a lipoprotein toxin called α-haemolysin (HlyA) which is encoded by hlyA gene [12]. Regarding phylogenetic typing, E.coli strains are classified into four major phylogenetic groups (phylogroups) named as A, B1, B2, and D [13], moreover these phylogenetic groups are intertwined with virulence patterns [1]. Each E.coli type has characteristic patterns, which allow them to colonize and invade their host [14]. However, exceptions exist where some UPEC strains have been reported to carry DEC markers [15], on the other hand, some DEC strains carry virulence factors associated with UPEC [5, 15]. Interestingly, E.coli strains that carry genetic determinants from different E.coli pathotypes are now termed as “heteropathogenic E.coli” [14, 16, 17], so the current classification of E.coli pathotypes may be not enough [18, 19]. These findings raised the suggestion that some DEC strains might be potential uropathogens. The current study has investigated a collection of DEC and UPEC strains isolated from clinical cases regarding the presence of urovirulence determinants and phylogenetic grouping. The aim was to detect whether DEC strains share virulence properties with the UPEC pathotypes and to recognize their phylogenetic diversity.
Materials and methods
A total of 173 E.coli strains were included in the study were recovered from patients of both sex and different ages presenting symptomatic UTIs and diarrhea. A total of 103 isolates were recovered from urine samples of patients with UTIs (UPEC) during outpatient treatment (patients who visited an acute day ward were considered to be outpatients). E.coli UTI was diagnosed by clinical symptoms such frequency, urgency, dysuria, small-volume voids or lower abdominal pain in addition to urine culture with a colony count >105 CFU E.coli/ml in midstream urine sample. Seventy strains were recovered from the stool samples of patients with diarrhea (DEC) during outpatient treatment. Diarrhea was diagnosed by, passage of loose stools for three times or more daily, in addition to one or more of characteristic clinical symptoms (nausea, vomiting, abdominal pain or cramps, fecal urgency, or dysentery). The samples of the study were collected from outpatients' clinics, Minia University Hospitals, Egypt during the period from January to March 2018.
Ethics statement
The study protocol was approved by Minia Faculty of Medicine Review Board (code: 46 A at 2/1/2018). Written informed consents were obtained from all patients for the use of their samples.
Bacterial isolates
Urine samples were cultured on chromogenic media (CHROMagar™ Orientation, Paris, France), while diarrhea samples were cultured on MacConkey and EMB agar. The isolates were confirmed as E.coli by standard bacteriological and biochemical tests including indole, urease, citrate and sugar fermentation tests. E. coli as the sole urine and stool Cultures microorganism were only included. Strains confirmed as E. coli were kept in trypticase soy broth with sterilized 15% glycerol at—20°C. The DEC strains were identified as enteroaggregative E. coli (EAEC) by PCR technique [20].
DNA extraction
DNA was extracted by using the QIAamp DNA extraction Mini kit (Qiagen, Hilden, Germany) according to manufacturer’s instructions. DNA was used immediately or stored at—20°C until used.
Phylogenetic analysis
E. coli isolates were classified into phylogenetic groups by targeting two marker genes (chuA and yjaA) and a DNA fragment TSPE4.C2 (Table 1) by triplex PCR as described previously [21]. Additional sub-grouping scheme proposed by Branger et al, 2005 was used [22].
Table 1. Primers sequences used for PCR assays.
| Genes | Primer sequence | Size fragment (bp) | Reference |
|---|---|---|---|
| CVD432 |
CTGGCGAAAGACTGTATCAT CAATGTATAGAAATCCGCTGTT |
630 | [20] |
| ChuA | F-GACGAACCAACGGTCAGGAT R-TGCCGCCAGTACCAAAGACA |
279 | [21] |
| yjaA | F-TGAAGTGTCAGGAGACGCTG R-ATGGAGAATGCGTTCCTCAAC |
211 | [21] |
| TspE4C2 | F-GAGTAATGTCGGGGCATTCA R-CGCGCCAACAAAGTATTACG |
154 | [21] |
| fimH | F: TGCAGAACGGATAAGCCGTGG R: GCAGTCACCTGCCCTCCGGTA |
506 | [23] |
| Sfa (sfa/foc) | F: CTCCGGAGAACTGGGTGCATCTTAC R: CGGAGGAGTAATTACAAACCTGGCA |
410 | [24] |
| pap A | F: ATGGCAGTGGTGTTTTGGTG R:CGTCCCACCATACGTGCTCTTC |
720 | [23] |
| pap E/F | F: GCAACAGCAACGCTGGTTGCATCAT R: AGAGAGAGCCACTCTTATACGGACA |
336 | [25] |
| hly A | F: AACAAGGATAAGCACTGTTCTGGCT R: ACCATATAAGCGGTCATTCCCGTCA |
1170 | [25] |
| iroN | F AAGTCAAAGCAGGGGTTGCCCG R GACGCCGACATTAAGACGCAG |
665 | [26] |
Detection of virulence genes
Specific primers were used to amplify sequences of 6 different virulence genes. Primer sequences and predicted sizes of the PCR products are shown in (Table 1). The amplification reactions were carried out using Biometra, UNO II thermal cycler (Goettingen, Germany) under the following conditions: initial denaturation at 95°C for 5 min, followed by 35 cycles of: 30 s at 94°C for, 30s at 63°C, then 30 s at 72°C, with a final extension step at 72°C for 5 min. PCR was performed in a 25 ml reaction mixture containing1 ul of template DNA (*100 ng/ml), 12.5 ml of PCR mastermix (Maxima Hot Start Green PCR Master Mix, USA), and 1 ul (10 pmol) of each primer and 9.5 ml of nuclease free water. PCR products were resolved on 2% agarose gel and visualized under a UV transilluminator (Biometra).
Statistical analysis
The chi -square test or the Fisher’s exact test was used. P <0.05 was considered statistically significant (two-tailed).
Results
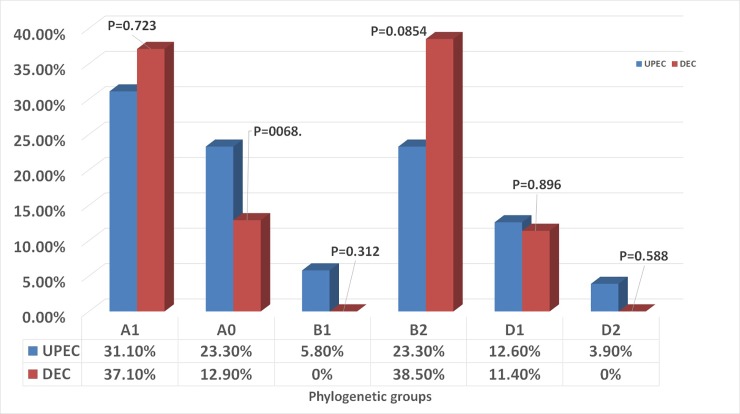
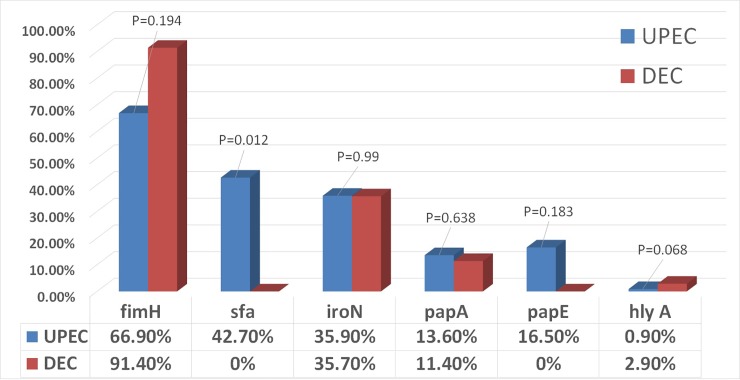
Phylogenetic grouping and virulence genes were characterized in103 UPEC isolates and 70 DEC isolates (EAEC) using PCR assay. Regarding the phylogenetic analysis, the predominant groups were A1 (31.1% UPEC, 37.1% DEC) with no significant difference (p value = 0.723); A0 (23.3%UPEC, 12.9%DEC; p value = 0.068) followed by group B2 then group D1. Phylogenetic groups D2 and B1 were detected only in UPEC isolates and were not detected in DEC isolates. There were no significant difference between phylogenetic distribution in UPE and DEC isolates (Fig 1 and S1 Fig). Six virulence genes were examined in103 UPEC isolates and 70 DEC isolates (EAEC) to compare between the virulence repertoires of them. The six studied virulence genes were detected in UPEC isolates while only 4 genes were detected in DEC isolates. The most frequently detected virulence gene in UPEC and DEC isolates was fimH (UPEC isolates: n = 69/103, 66.9%; DEC isolates: n = 64/70, 91.4%). The frequencies of iroN gene were detected in a similar percentage for both types of isolates; (UPEC: 37/103, 35.9%; DEC: 25/70, 35.7%). papA gene was detected in a percentage of (UPEC isolates: 14/103, 13.5%; DEC isolates: 8/70, 11.4%) and hlyA gene was found in only one isolate of UPEC (0.9%) and 2 isolates of DEC (2.9%). The sfa and papE genes were detected in UPEC isolates in (44/103, 42.7%) and (17/103, 17.1%) respectively but were not detected in DEC isolates at all. There were no significant differences between virulence genes in UPEC and DEC isolates, except for sfa gene (p value = 0.012) (Fig 2 and S2–S6 Figs).
Fig 1. Phylogenetic distribution of UPEC and DEC isolates.
Fig 2. Distribution of the virulence genes among UPEC & DEC isolates.
Based on the distribution of the various genes, all the studied strains exhibited 21 virulence gene patterns, referred to as Ec (Table 2). UPEC isolates showed 20 Ec patterns while DEC isolates showed 5 Ec patterns only. Ec1 was characterized by the presence of the fimH gene only which was the most noted pattern and found in 24 UPEC isolates (23.3%) and 44 DEC isolates (62.9%). Ec3 (fimH & iroN genes) was the most frequent combined virulence pattern (11, 10.67% of UPEC and 14, 20%) of DEC isolates) followed by Ec5 (fimH &sfa genes; 9, 8.7% of UPEC isolates) and Ec10 (fimH, papE &iroN), (3, 2.91% of UPEC; 6, 8.57% of DEC isolates. The distribution of virulence patterns in phylogenic groups are summarized in Table 2.
Table 2. Virulence patterns (EC) distribution among Phylogenetic groups of UPEC and DEC E.coli isolates.
| Virulence pattern | Phylogenetic group | Isolates N (%) |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| A0 | A1 | B1 | B2 2 | B2 3 | D1 | D2 | UPEC | DEC | |
| Ec1(UPEC/DEC) | 9 /5 | 8/16 | 1/- | 2/6 | 1/11 | 3/6 | - | 24 (23.3%) | 44 (62.9%) |
| Ec2 | 2 | 2 | - | 1 | 6 | 1 | - | 12(11.65%) | 0 |
| Ec3(UPEC/DEC) | 2/2 | 5/8 | 1 | -/2 | 1/2 | 2 | - | 11(10.67%) | 14 (20%) |
| Ec4(UPEC/DEC) | 5/2 | 3/2 | 1 | - | - | 1 | 1 | 11(10.67%) | 4 (5.71%) |
| Ec5 | 3 | 5 | - | - | - | 1 | - | 9 (8.7%) | 0 |
| Ec6 | - | 2 | - | 2 | 2 | - | - | 6 (5.8%) | 0 |
| Ec7 | - | 1 | 2 | 2 | - | - | - | 5 (4.85%) | 0 |
| Ec8 | - | 1 | - | - | - | 2 | - | 3 (2.91%) | 0 |
| Ec9 | - | - | 1 | - | - | 2 | - | 3 (2.91%) | 0 |
| Ec10(UPEC/DEC) | - | - | - | 1/2 | -/2 | -/2 | 2 | 3 (2.91%) | 6 (8.57%) |
| Ec11 | - | - | - | - | - | 1 | 1 | 2 (1.94%) | 0 |
| Ec12 | - | 1 | - | 1 | - | - | - | 2 (1.94%) | 0 |
| Ec13 | 2 | - | - | - | - | - | - | 2 (1.94%) | 0 |
| Ec14 | - | - | - | - | 2 | - | - | 2 (1.94%) | 0 |
| Ec15 | - | - | - | 2 | - | - | - | 2 (1.94%) | 0 |
| Ec16 | - | 2 | - | - | - | - | - | 2 (1.94%) | 0 |
| Ec17 | - | 1 | - | - | - | - | - | 1 (0.97%) | 0 |
| Ec18 | - | 1 | - | - | - | - | - | 1 (0.97%) | 0 |
| Ec19 | 1 | - | - | - | - | - | - | 1 (0.97%) | 0 |
| Ec20 | - | - | - | - | 1 | - | - | 11 (0.97%) | 0 |
| Ec21(UPEC/DEC) | - | - | - | -/2 | - | - | - | 0 | 2 (2.86%) |
| total(UPEC/DEC) | 24/9 | 32/26 | 6/- | 11/12 | 13/15 | 13/8 | 4/- | 103(100%) | 70(100%) |
UPEC, uropathogenic E.coli; DEC, Diarrheagenic E.coli; Ec1include (fimH gene); Ec2 (sfa gene); Ec3 (fimH and iroN genes); Ec4 (no genes detected); Ec5 (fimH and sfa); Ec6 (fimH, sfa, papE and iroN genes); Ec7 ((fimH, sfa and iroN genes); Ec8 (fimH and papA); Ec9 (papE); Ec10 (fimH, papA and iroN genes); Ec11 (sfa and iroN genes); Ec12 (fimH, sfa, papE); Ec13 (sfa, papA and iroN genes); Ec14 (sfa, papA, papE and iroN genes); Ec15 (fimH, sfa, papA, papE and iroN genes); Ec16 ((fimH, papE and iroN genes), Ec17 (papA); Ec18 (papE); Ec19 ((fimH, sfa, papA); Ec20 (fimH, sfa, iroN and hly); Ec21 (fimH, iroN and hly genes).
Discussion
Each E.coli type has characteristic phylogenetic and virulence patterns, which allow them to colonize and invade their host [14]. The phylogenetic analysis classify E.coli pathotypes into 4 major phylogenetic groups A, B1, B2 and D [13]. Several previous studies have reported that B2 and D phylogroups are the most common among UPEC isolates [27, 28], while isolates in phylogenetic groups A and B1 were mostly identified as commensal E.coli isolates [29]. Our study has reported different findings, where the predominant phylogenetic group in UPEC isolates was A1 (31.1%), followed by B2 (23.3%), A0 (23.3%), and lastly D1 (12.6%), that agree with some previous studies [30, 31]. The predominance of phylogenetic group A in UPEC isolates which is usually associated with commensal strains suggesting that the gastrointestinal tract is the main source of strains that colonize the urinary tract [32, 33]. The distribution of virulence genes and phylogenetic types varies among different countries, for example, group A was the most prevalent in Russia [34], and also in China [35] in UPEC, so our findings can be explained by geographical variation.
Phylogenetic group A was also the predominant in DEC isolates of the current study, where the frequency of A1 type was (37.1%) and A0 was (12.9%). The predominance of phylogenetic group A in DEC isolates suggesting that the source of gastrointestinal tract infection can be endogenous even in immunocompetent persons. Although several previous reports showed that group B2 E.coli strains are rare in fecal samples of healthy persons [36, 37], group B2 was recorded in a high percent (38.5%) in our DEC isolates so our data reveals that acquiring the group B2 strain is important for developing of infection. On the other hand, previous studies, which investigated DEC isolates in Costa Rica [38] and Peru [39], showed that most of the isolates belonged to B1 and D groups respectively that disagrees with our study and reflects the diversity of DEC isolates in different countries. In order to find possible link between strain phylogeny and virulence genes, we analyzed an overall virulence profile (fimH, sfa, iroN, pap A, papE and hly A genes) in UPEC and DEC strains. The six studied virulence genes were detected in UPEC isolates while only 4 genes were detected in DEC isolates. The most frequent gene was fimH gene (66.9% of UPEC isolates and 91.4% of DEC). A similar prevalence (65.9%) was detected in a previous study in Egypt [40] and also in other countries [41, 42]. For the other virulence genes sfa, iroN, papA, papE and hly A; the prevalence in our study was 42.7%, 35.9%, 13.6%, 16.5%, and 0.9% respectively in UPEC isolates. Our results were near to some studies in percent of sfa and iroN but the percent of pap and hlyl genes was lower [42]. Our results showed that the UPEC strains have different virulence profiles compared with other studies, suggesting that the virulence profile depends on the regional geography and climate or may be other factors.
The frequencies of urovirulence genes, iroN and papA detection in DEC were similar to that of UPEC isolates. There were no significant differences between virulence genes in UPEC and DEC isolates, except for sfa gene (p value = 0.012). About 66/70, 94.3% of DEC isolates carry at least one urovirulence gene. These findings showed a closer relationship between DEC specially EAEC and UPEC that may be explained by the remarkable genome plasticity or gene transfer in E.coli strains which leads to appearance of virulent strains displaying virulence genes from different pathotypes in a single isolate [18]. This interesting finding has shown that the classical classification of strains into pathotypes of E.coli is limited and inaccurate [19]. Our results showed that virulence genes were more prevalent in phylogroups A, so group A can be pathogenic and the other phylogoups as well. These findings also provide different insights into classification of E.coli pathotypes. A close relationship between the DEC and UPEC strains was reported, showing that DEC particularly the EAEC pathotype is an emerging enteropathogen which can cause intestinal and extra -intestinal infections, particularly in developing countries.
Supporting information
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
Acknowledgments
We would like to thank the staff members of Minia University hospital for helping with the collection of samples
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The authors received no specific funding for this work.
References
- 1.Kaper JB, Nataro JP, Mobley HL. Pathogenic Escherichia coli. Nat Rev Microbiol. 2004; 2(2):123–40. 10.1038/nrmicro818 [DOI] [PubMed] [Google Scholar]
- 2.Abba K, Sinfield R, Hart CA, Garner P. Pathogens associated with persistent diarrhea in children in low and middle income countries: systematic review. BMC Infect Dis. 2009; 9: 1–15. 10.1186/1471-2334-9-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campos ACC, Andrade NL, Ferdous M, Chlebowicz MA, Santos CC, Correal JCD, et al. Comprehensive Molecular Characterization of Escherichia coli Isolates from Urine Samples of Hospitalized Patients in Rio de Janeiro, Brazil. Front Microbiol. 2018; 16: 9 243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cyoia PS, Rodrigues GR, Nishio EK, Medeiros LP, Koga VL, Pereira AP, et al. Presence of virulence genes and pathogenicity islands in extraintestinal pathogenic Escherichia coli isolates from Brazil. J. Infect. Dev. Ctries. 2015; 9: 1068–1075. 10.3855/jidc.6683 [DOI] [PubMed] [Google Scholar]
- 5.Lara FB, Nery DR, de Oliveira PM, Araujo ML, Carvalho FR, Messias-Silva LC, et al. Virulence Markers and Phylogenetic Analysis of Escherichia coli Strains with Hybrid EAEC/UPEC Genotypes Recovered from Sporadic Cases of Extraintestinal Infections. Front Microbiol. 2017; (3)8:146 10.3389/fmicb.2017.00146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xie J, Foxman B, Zhang L, Marrs CF. Molecular epidemiologic identification of Escherichia coli genes that are potentially involved in movement of the organism from the intestinal tract to the vagina and bladder. J Clin Microbiol. 2006; 44: 2434–2441. 10.1128/JCM.00397-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Snyder JA, Lloyd AL, Lockatell CV, Johnson DE, Mobley HL. Role of phase variation of type 1 fimbriae in a uropathogenic Escherichia coli cystitis isolate during urinary tract infection. Infect Immun. 2006; 74(2):1387–93. 10.1128/IAI.74.2.1387-1393.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alizade H, Ghanbarpour R, and Aflatoonian MR. Virulence genotyping of Escherichia coli isolates from diarrheic and urinary tract infections in relation to phylogeny in southeast of Iran. Trop. Biomed. 2014; 31: 174–182. [PubMed] [Google Scholar]
- 9.Stordeur P, Bree A, Mainil J, Moulin-Schouleur M. Pathogenicity of pap-negative avian Escherichia coli isolated from septicemic lesions. Microbes Infect. 2004; 6(7): 637–45. 10.1016/j.micinf.2004.03.006 [DOI] [PubMed] [Google Scholar]
- 10.Simms AN, Mobley HL. Multiple genes repress motility in uropathogenic Escherichia coli constitutively expressing type 1 fimbriae. J Bacteriol. 2008; 190(10): 3747–56. 10.1128/JB.01870-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tarchouna M, Ferjani A, Ben-Selma W, Boukadida J. Distribution of uropathogenic virulence genes in Escherichia coli isolated from patients with urinary tract infection. Int J Infect Dis. 2013; 17(6):e450–3. 10.1016/j.ijid.2013.01.025 [DOI] [PubMed] [Google Scholar]
- 12.Wiles TJ, and Mulvey MA. The RTX pore-forming toxin α-hemolysin of uropathogenic Escherichia coli: progress and perspectives. Future Microbiology. 2013; 8(1): 73–84. 10.2217/fmb.12.131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doumith M, Day MJ, Hope, Wain J, and Woodford N. Improved multiplex PCR strategy for rapid assignment of the four major Escherichia coli phylogenetic groups. J. Clin. Microbiol. 2012; 50: 3108–3110. 10.1128/JCM.01468-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bielaszewska M, Schiller R, Lammers L, Bauwens A, Fruth A, Middendorf B, et al. Heteropathogenic virulence and phylogeny reveal phased pathogenic metamorphosis in Escherichia coli O2:H6. EMBO Mol. Med. 2014; 6: 347–357. 10.1002/emmm.201303133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abe CM, Salvador FA, Falsetti IN, Vieira MA, Blanco J, Blanco JE, et al. Uropathogenic Escherichia coli (UPEC) strains may carry virulence properties of diarrhoeagenic E. coli. FEMS Immunol. Med. Microbiol. 2008; 52: 397–406. 10.1111/j.1574-695X.2008.00388.x [DOI] [PubMed] [Google Scholar]
- 16.Toval F, Köhler C-D, Vogel U, Wagenlehner F, Mellmann A, Fruth A, et al. Characterization of Escherichia coli isolates from hospital inpatients or outpatients with urinary tract infection. J.Clin.Microbiol. 2014; 52: 407–418. 10.1128/JCM.02069-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ang CW, Bouts H, Rossen JW, VanderKuip M, VanHeerde M, and Bökenkamp A. Diarrhea, urosepsis and hemolytic uremic syndrome caused by the same heteropathogenic Escherichia coli strain. Pediatr. Infect.Dis. J. 2016; 35, 1045–1047. 10.1097/INF.0000000000001226 [DOI] [PubMed] [Google Scholar]
- 18.Gomes TAT, Elias WP, Scaletsky ICA, Guth BEC, Rodrigues JF, Piazza RMF, et al. Diarrheagenic Escherichiacoli. BrazilianJ. Microbiol. 2016; 47: 3–30. [Google Scholar]
- 19.Robins-Browne RM, Holt KE, Ingle DJ, Hocking DM, Yang J, and Tauschek M. Are Escherichia coli pathotypes still relevant in the Era of Whole-genome sequencing? Front. Cell.Infect. Microbiol. 2016; 6:141 10.3389/fcimb.2016.00141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmidt H, Knop C, Franke S, Aleksic S, Heesemann J, Karch H. Development of PCR for screening of enteroaggregative Escherichia coli. J Clin Microbiol. 1995; 33(3): 701–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clermont O, Bonacorsi S, Bingen E. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl Environ Microbiol. 2000; 66(10):4555–8. 10.1128/aem.66.10.4555-4558.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Branger C, Zamfir O, Geoffroy S, Laurans G, Arlet G, Thien HV, et al. Genetic background of Escherichia coli and extended-spectrum beta-lactamase type. Emerg Infect Dis. 2005; 11: 54–61. 10.3201/eid1101.040257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Le Bouguenec CM, Archambaud and A. Labigne. Rapid and specific detection of the pap, afa, and sfa adhesin-encoding operons in uropathogenic Escherichia coli strains by polymerase chain reaction. J. Clin. Microbiol. 1992; 30:1189–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamamoto S, Terai A, Yuri K, Kurazono H, Takeda Y, Yoshida O. Detection of urovirulence factors in Escherichia coli by multiplex polymerase chain reaction. FEMS Immunol Med Microbiol. 1995; 12(2): 85–90. 10.1111/j.1574-695X.1995.tb00179.x [DOI] [PubMed] [Google Scholar]
- 25.Johnson JR, Stell AL. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J Infect Dis. 20000; 181: 261–72. 10.1086/315217 [DOI] [PubMed] [Google Scholar]
- 26.Johnson JR, Russo TA, Tarr PI, Carlino U, Bilge SS, Vary JC, et al. Molecular epidemiological and phylogenetic associations of two novel putative virulence genes, iha and iroN among Escherichia coli isolates from patients with urosepsis. Infect Immun. vol. 2000; 68: 3040–7. 10.1128/iai.68.5.3040-3047.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee JH, Subhadra B, Son YJ, Kim DH, Park HS, Kim JM, et al. Phylogenetic group distributions, virulence factors and antimicrobial resistance properties of uropathogenic Escherichia coli strains isolated from patients with urinary tract infections in South Korea. Lett Appl Microbiol. 2016; 62(1):84–90. 10.1111/lam.12517 [DOI] [PubMed] [Google Scholar]
- 28.Munkhdelger Y, Gunregjav N, Dorjpurev A, Juniichiro N. and Sarantuya J. Detection of virulence genes, phylogenetic group and antibiotic resistance of uropathogenic Escherichia coli in Mongolia. J. Infect. Dev. Ctries. 2017; 11(1): 51–57 10.3855/jidc.7903 [DOI] [PubMed] [Google Scholar]
- 29.Katouli M. Population structure of gut Escherichia coli and its role in development of extra-intestinal infections. Iran. J. Microbiol. 2010; 2: 59–72. [PMC free article] [PubMed] [Google Scholar]
- 30.Piatti G, Mannini A, Balistreri M, & Schito M. Virulence factors in urinary Escherichia coli strains: phylogenetic background and quinolone and fluoroquinolone resistance. Journal of Clinical Microbiology. 2008; 46(2): 480–487. 10.1128/JCM.01488-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adib N, Ghanbarpour R, Solatzadeh H, Alizade H. Antibiotic resistance profile and virulence genes of uropathogenic Escherichia coli isolates in relation to phylogeny. Tropical Biomedicine. 2014; 31(1): 17–25. [PubMed] [Google Scholar]
- 32.Moreno E, Andreu A, Perez T, Sabate M, Johnson JR, Prats G. Relationship between Escherichia coli strains causing urinary tract infection in women and the dominant faecal flora of the same hosts. Epidemiology and Infection. 2006; 134(5): 1015–1023. 10.1017/S0950268806005917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.López-Banda DA, Carrillo-Casas EM, Leyva-Leyva M, Orozco-Hoyuela G, Manjarrez-Hernández AH, Arroyo-Escalante S, et al. Identification of Virulence Factors Genes in Escherichia coli Isolates from Women with Urinary Tract Infection in Mexico. Biomed Res Int. 2014; Article ID 959206, 10 P. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grude N, Potaturkina-Nesterova NI, Jenkins A, Strand L, Nowrouzian FL, Nyhus J, Kristiansen BEA. Comparison of phylogenetic group, virulence factors and antibiotic resistance in Russian and Norwegian isolates of Escherichia coli from urinary tract infection. Clin. Microbiol. Infect. 2007; 13: 208–211. 10.1111/j.1469-0691.2006.01584.x [DOI] [PubMed] [Google Scholar]
- 35.Tong Y, Sun S, Chi Y. Virulence genotype and phylogenetic groups in relation to chinese herb resistance among Escherichia coli from patients with acute pyelonephritis. Afr. J. Tradit. Complement Altern. Med. 2014; 11: 234–238 10.4314/ajtcam.v11i3.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Duriez P, Clermont O, Bonacorsi S, Bingen E, Chaventre A, Elion J, et al. Commensal Escherichia coli isolates are phylogenetically distributed among geographically distinct human populations. Microbiology. 2001; 147(6):1671–6. [DOI] [PubMed] [Google Scholar]
- 37.Blyton MD, Cornall SJ, Kennedy K, Colligon P, Gordon DM. Sex-dependent competitive dominance of phylogenetic group B2 Escherichia coli strains within human hosts. Environ Microbiol Rep. 2014; 6(6): 605–10. [DOI] [PubMed] [Google Scholar]
- 38.Pérez C, Gómez-Duarte OG, Arias ML. Diarrheagenic Escherichia coli in children from Costa Rica. Am J Trop Med Hyg. 2010; 83: 292–297. 10.4269/ajtmh.2010.09-0412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mosquito S, Pons MJ, Riveros M, Ruiz J, Ochoa TJ. Diarrheagenic Escherichia coli phylogroups are associated with antibiotic resistance and duration of diarrheal episode. Sci World J. 2015; 610403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zaki ME and Elewa A. Evaluation of Uropathogenic Virulence Genes in Escherichia coli Isolated from Children with Urinary Tract Infection. International Journal of Advanced Research. 2015; 3(3): 165–173. [Google Scholar]
- 41.Karimian A, Momtaz H, Madani M. Detection of uropathogenic Escherichia coli virulence factors in patients with urinary tract infections in Iran. African Journal of Microbiology. 2012; 6(39): 6811–6816 [Google Scholar]
- 42.Jalali HR, Eslami G, Fallah F, Pourbakhsh A. Genotyping of Virulence Factors of Uropathogenic Escherichia coli by PCR. Novel Biomed. 2015; 3(4):177–81. [Google Scholar]