Abstract
In order to maintain a state of self-renewal, yet retain the ability to rapidly differentiate in response to external signals, pluripotent cells exert tight control over gene expression at many levels. Recent studies have suggested that N6-methyladenosine (m6A) RNA methylation, one of the most abundant post-transcriptional modifications, is important for both pluripotency and differentiation. In this review, we summarize the current state of the m6A field, with emphasis on the impact of writers, erasers and readers of m6A on RNA metabolism and stem cell biology.
Keywords: Stem cells, pluripotency, development, m6A methylation, RNA, post-transcriptional modification
1. Introduction
Transitions from one cell type to another require complex, carefully executed maneuvers to coordinate global changes in gene expression and ensure the desired outcome is achieved efficiently and appropriately. Although transcriptional changes are integral to most cell fate transitions, transcription takes time to induce or repress and must be coordinated with post-transcriptional regulation to facilitate rapid remodeling of the transcriptome in response to extracellular cues[1–6].
Cells utilize several levels of post-transcriptional regulation to coordinate gene expression changes. For example, RNA-binding proteins (RBPs) and microRNAs (miRNAs) exploit sequence elements shared between target mRNAs to enable co-regulation of several functionally related transcripts (~10-500 transcripts may be impacted). At the same time, ubiquitous and crucial modifications such as the 5’ cap and poly(A) tail have a vast influence on gene expression through recruitment of proteins such as eIF4E, and poly(A) binding protein to virtually all RNA polymerase II transcripts. Internal m6A methylation resides between these two levels of regulation allowing regulation of thousands of transcripts in response to developmental and other cues while leaving unmethylated transcripts unaffected.
Importantly, while methylation can dramatically influence RNA metabolism, it is not required for any aspect of the RNA life cycle. This means that otherwise identical transcripts may experience different levels of modification and, as a result, may behave as distinct populations in response to stimuli (Figure 1).
Figure 1: The impact of m6A on mRNA abundance on mRNA stability.
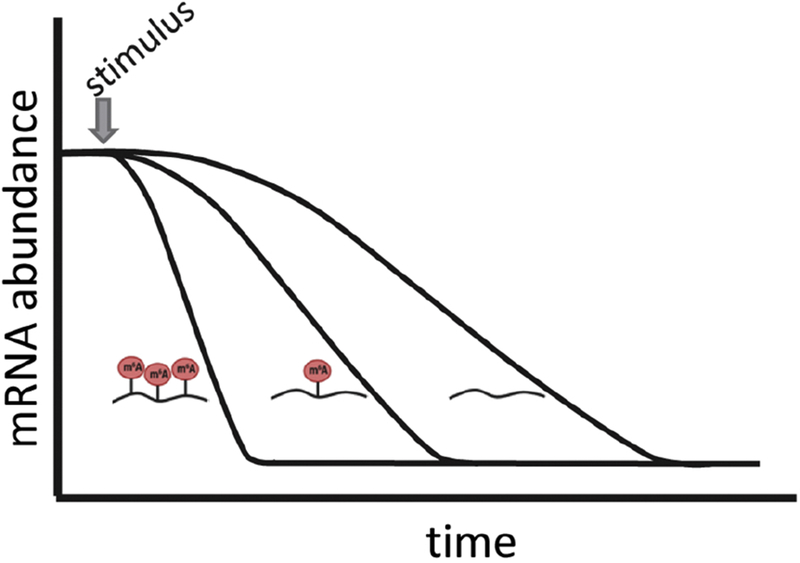
Different subpopulations of transcripts from the same gene can exhibit different responses based on their level of modification. Following stimulation, the sharpest change in mRNA abundance is seen for transcripts with clusters of m6A. Transcripts with single m6A modification may experience a more gradual decrease, while transcripts with no m6A are the slowest to change.
Virtually all RNA species including messenger RNAs (mRNAs), transfer RNAs (tRNAs), ribosomal RNAs (rRNA), miRNAs and long non-coding RNAs (lncRNAs) can experience methylation at various positions (m1A, m5C, 2’-O-methyl etc). Of these modifications, N6-methyladenosine (m6A) has the best characterized effects on gene expression and was the first shown to modulate mRNA abundance[7,8], m6A modification can modulate the fate of an mRNA at the level of splicing[9], cleavage/polyadenylation[10], subcellular localization[11], decay[12] and translation[13](Figure 2). Moreover, m6A is the most prevalent internal mRNA modification in many eukaryotic species [7,8,14–16]. Although m6A was discovered over 40 years ago[17–19], its cellular function was only characterized through recent advancements in antibody-based precipitation and high-throughput sequencing. Since 2012, ~10,000 m6A sites have been documented in over a quarter of human transcripts[7,8,20]. The precise impact m6A marks have on RNA metabolism and gene expression is dictated by the actions of methyltransferases (‘writers’), demethylases (‘erasers’) and m6A binding-proteins (‘readers’), many of which are now known to play roles in pluripotency and development.
Figure 2: m6A influences mRNA function from cradle to grave.
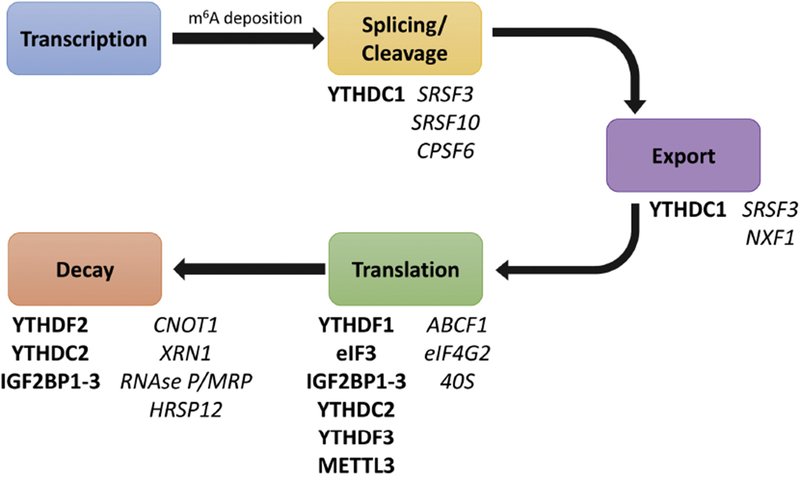
Recognition of m6A by methyl-reader proteins, bolded, can influence various RNA processes including splicing/cleavage, nuclear export, translation and decay. Often, reader proteins interact with or recruit effector proteins, italicized, to carry out a given function. They can also compete with miRNAs and RBPs.
The goal of this review is to provide an up-to-date overview of the factors and mechanisms within the m6A methylation pathway and then to focus on the influence m6A methylation has on stem cell maintenance/differentiation and cell state transitions.
2. The How, Where, When and Why of m6A Writing
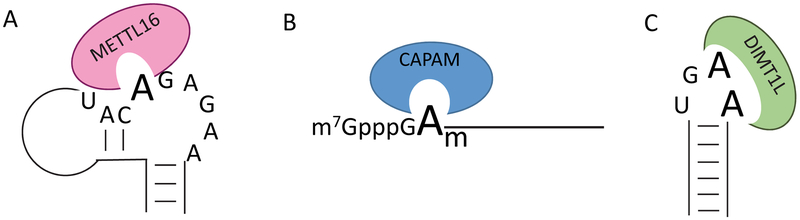
The N6-position of adenosine in RNAs can be methylated by four separate methyltransferase activities: CAPAM (also known as PCIF1) targets 2’-O-methyladenosine found adjacent to the 5’ cap to produce m6Am[21,22], DIMT1L catalyzes dimethylation two adjacent adenosines in18S rRNA[23,24], whereas METTL16[25–30] (both in Box 1) and the METTL3/METTL14[31] heterodimer are responsible for internal m6A modifications. For brevity, we will be focusing on the METTL3/METTL14 methyltransferase which installs the vast majority of internal m6A marks on mRNAs and lncRNAs. The remaining methyltransferase activities are described in more detail in Box 1. METTL3/METTL14 is part of the surprisingly large m6A writer complex which can be divided into two sub-complexes: the m6A-METTL complex (MAC), which comprises METTL3 and METTL14[31], and the m6A-METTL-associated complex (MACOM), comprising WTAP, ZC3H13, RBM15/15B, VIRMA and HAKAI[32–36](Figure 3).
Box:
Other ways to deposit m6A
While we have focused primarily on METTL3-mediated m6A deposition, it is important to note that there are other enzymes capable of installing methyl groups on the N6 position of adenosine. In each case the substrate/site of modification is distinct from that recognized by the MAC.
METTL16 METTL16 is in the same family as METTL3 and METTL14 but targets a smaller set of transcripts at specific sites containing a UACAGAGAA motif and unique secondary structure where the adenosine is bulged out[27,29,192](Panel A). To date, targets of METTL16 include non-coding RNAs, lncRNAs and pre-mRNAs[25]. METTL16-mediated methylation of U6 snRNA facilitates snRNA-pre-mRNA interactions during splicing[25], while methylation in the 3’ UTR of the SAM synthetase, MAT2A, regulates alternative splicing to control SAM homeostasis[27]. In the latter example, the methylated adenosine is recognized by YTHDC1 under high SAM conditions to down-regulate MAT2A expression[28], while prolonged association of METTL16 under low SAM conditions causes intron retention and eventually decay of the MAT2A mRNA. Loss of METTL16 leads to massive effects on the transcriptome and embryonic lethality in mice[193], primarily because of dysregulation of SAM homeostasis. It is not known whether m6A deposited by METTL16 can be removed by ALKBH5 or FTO.
CAPAM/PCIF1 Recently, CAPAM was identified as the enzyme responsible for modifying 2’O-methyladenosine adjacent to the 5’ cap of RNAP II transcripts, including mRNAs and snRNAs, to generate m6Am[21,194](Panel B). Being adjacent to the cap allows m6Am to promote translation of capped mRNAs independent of eIF4E[21]. m6Am is also crucial for proper snRNA biogenesis[82], which in turn affects mRNA splicing. Notably, m6Am is removed by FTO[82] and therefore effects of FTO on splicing may be largely due to hyper-methylation of snRNAs. As some immunoprecipitation-based approaches fail to distinguish internal 5’ UTR m6A from m6Am, existing research on the effects of m6A at the 5’ end of mRNAs must be carefully interpreted.
DIM1/DIMT1L N6-dimethylation of 18S rRNA is performed by Dimlp in yeast[24] and its homolog DIMT1L in mammals[23] at two adjacent positions in a highly conserved region near the decoding site. Interestingly, association of the methyltransferase with the 18S rRNA is required for rRNA processing and catalysis is delayed until after cleavage occurs. This requirement ensures that all cleaved rRNAs are committed to modification.
Box Figure: Alternative methyltransferases recognize distinct properties of their target RNAs.
A: METTL16 targets a unique structure found in U6 snRNA, MALAT lncRNA, and the 3’ UTR of the MAT2A mRNA as well as other RNAs. B: CAPAM targets the 2’O-methyladenozyne found adjacent to the 5’ cap in many RNA polymerase II transcripts. C: DIMT1L targets two adjacent adenosine residues in a stem-loop structure of 18S rRNA.
Figure 3: The m6A writer complex.
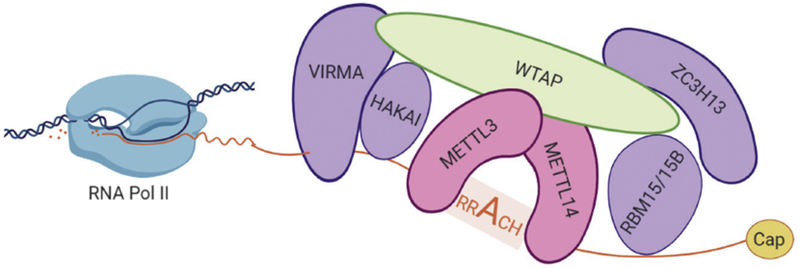
The m6A writer complex targets RNAs as they are transcribed. The m6A-METTL complex (MAC; shown in pink), consisting of the METTL3/14 heterodimer, is the catalytically active subcomplex which methylates adenosine at sites matching the RRACH consensus. WTAP is the major scaffolding protein that connects the MAC and m6A-METTL-associated complex (MACOM). The composition and/or activity of MACOM subunits can influence the distribution of m6A across a transcript. VIRMA and HAKAI favor 3’ methylation, while RBM15/15B facilitates 5’ methylation.
In order to perform catalysis, the m6A writer must be localized appropriately within the cell and directed to appropriate sites on each RNA. Furthermore, activity must be regulated such that methylation is restricted to the appropriate developmental stage and/or RNA region. We discuss how these functions are achieved below.
2.1. Methyltransferase Activity (How is m6A added?)
The MAC is responsible for the methyltransferase activity of the writer complex[31,37]. Briefly, this reaction entails the transfer of a methyl group, from an S-Adenosyl Methionine (SAM) molecule, onto the sixth nitrogen of the adenosine base. METTL3 is the sole catalytic subunit and the only one able to bind the SAM methyl donor[38–41]. While METTL3 possesses weak methyltransferase activity in vitro, the METTL3/METTL14 heterodimer exhibits substantially higher catalytic activity[31]. Although METTL14 does not perform catalysis, it is essential for methyltransferase activity, providing structural stability, and allosterically enhancing the methyltransferase activity of METTL3[37–39]. Interactions between METTL3 and METTL14 form a cavity where the RNA substrate is bound[38–40]. The methyltransferase domains of METTL3 and METTL14 are highly conserved among eukaryotes, with functional homologs in Drosophila (Ime4 and Cg7818, respectively)[16,42], Arabidopsis (Mta and Mtb, respectively)[36,43] and S.cerevisiae (IME4 and KAR4; although the role of KAR4 remains uncharacterized)[44,45]. Consistent with their high level of conservation, these proteins are essential for viability in mammals and plants, and play vital roles in embryonic development, sex determination and gametogenesis[46–54]. Depletion of either protein in human cell lines results in a marked global decrease in m6A levels[31,32].
2.2. Methyltransferase Specificity (Where on the mRNA?)
When it comes to methylation, not all adenosines are created equal. The context of the adenosine is critical in determining where and whether a transcript is modified. MAC target sites are enriched for the short and redundant consensus m6A motif, RRACH[31,32,55]. However, the expected occurrence of the RRACH motif in RNAs (~1 modification per 85 nucleotides) is much higher than the observed one m6A modification per ~1000 nucleotides[7,8,20,56]. Only a fraction of adenosines found within this motif are actually modified, supporting the existence of additional layers of regulation. Such regulation likely includes selective recruitment of the MAC to specific RNA substrates and is influenced by the surrounding sequence context (such as structure, and availability of binding sites for competing factors).
Recent studies are starting to reveal how MAC is recruited to specific RNAs. There is evidence for interactions between MAC and chromatin/transcription factors including CEBPZ[57], SMAD2/3[6] and ZFP217[49]. These interactions facilitate (CEBPZ, SMAD2/3) or repress (ZFP217) m6A modification of transcripts derived from promoters dependent on these transcription factors. For example, in stem cells SMAD2/3 is activated by TGFβ signaling and binds promoters of genes involved in early cell fate, including NANOG. It then recruits WTAP and MAC to allow RNA methylation. Marking these transcripts with m6A helps prime stem cells for a timely exit from pluripotency[6]. More broadly, an interaction between METTL14 and trimethylated histone H3 (H3K36me3) may help recruit MAC to nascent RNAs/RNA polymerase II (RNAP II)[58] and thereby promote m6A deposition. These observations suggest that transcripts can be selected for modification as they are transcribed and also supports that the epigenome can directly influence the epitranscriptome.
Once MAC is recruited to an mRNA, there is further regulation to determine which sites within that transcript will be modified. Specificity is conferred by METTL3 via its two CCCH-type zinc finger domains [41] and METTL14 through a C-terminal domain containing RGG repeats[37]. Recent observations imply that the sequence context, and its match to the preferred consensus may be a driving factor determining the distribution and extent of m6A modification (i.e. the m6A signal is ‘hard-coded’ into the mRNA sequence). In further support of this hypothesis, a remarkable number of m6A sites, and the flanking sequences, are conserved across vertebrates[7,59], and in some cases even in organisms as distant as yeast[60].
It is important to note that m6A is not uniformly distributed across the length of the transcript (Figure 4). There are distinct preferences for the 5’ and 3’ ends[7,8,61,62]. Interestingly, the consensus RRACH motif is also enriched in these areas, particularly near the stop codon[59], and H3K36me3 chromatin marks also show a skewed enrichment around stop codons[58], which may contribute to enhanced MAC recruitment. In addition, the FTO demethylase may prune the methyl group from sites at the 5’ end[63– 65] resulting in an overall enrichment of m6A at the 3’ end of RNAs. In exons, the vast majority of m6A marks are located internally, residing at least 50 nucleotides away from splice junctions[66]. Moreover, there is an enrichment of m6A in long (>200 nucleotides) internal exons[7,59,66,67]. The purpose and mechanism by which this is achieved is unclear. Interestingly, introns are rarely methylated when compared to exons, despite containing nearly three times the amount of RNA[20,66]. One explanation for this observation could be that the splicing machinery outcompetes or has priority over the m6A writer complex for access to nascent RNA.
Figure 4: Factors influencing the distribution of m6A and m6Am on mammalian mRNAs.
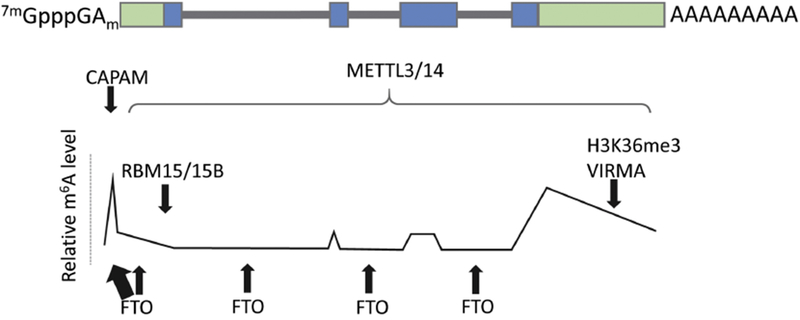
The overall m6A landscape in physiological conditions is determined primarily by the distribution of consensus motifs recognized by the methyltransferases (CAPAM, METTL3/14). However, it can be influenced by demethylases such as FTO, which prunes m6A from introns and across the transcript. Moreover, certain components of the MACOM, namely RBM15/15B and VIRMA, can favor methylation at the 5’ and 3’ ends respectively. Finally, there is evidence that histone modifications can influence m6A deposition.
As well as being concentrated in specific regions of the RNA, m6A sites are often found in clusters, similar to DNA methylation sites in CpG islands[8,20,56,59,66]. Clustering may be achieved in part through an association between the METTL3 and RNAP II that facilitates methylation when transcription slows[68], but also inherently requires that the available consensus sites be clustered.
Finally, the MAC can be directed to a specific location on an RNA through interaction with components of the MACOM, some of which are RNA-binding proteins. Specifically, VIRMA recruits the m6A writer complex to sites near the stop codon and within the 3’ UTR[34], while RBM15 and RBM15B binding sites are enriched in the 5’ UTR of transcripts[61]. RBM15/15B both bind U-rich sequences through canonical RRMs, whereas VIRMA lacks a characterized RNA binding domain, but may be recruited to mRNAs through interaction with the cleavage and polyadenylation machinery[34]. Interestingly, depletion of VIRMA in human cells resulted not only in a decrease in m6A marks near the stop codon and within the 3’ UTR, but also a significant increase in methylation at 5’ UTRs[34]. This suggests that RBM15/15B and VIRMA may compete for recruitment of the writer complex to different regions of a transcript. HAKAI, an E3 ubiquitin ligase, may also guide MACOM to specific sites or modulate activity of the complex, but more work is needed to uncover its exact role[36]. Changes in expression or activity of proteins like these may facilitate developmental changes in the distribution of m6A, such as the switch from 3’ UTR to 5’ UTR m6A seen during cerebellum development in mice[69].
To sum up, the MAC is recruited to specific RNAs/RNAP II through interactions with histones and transcription factors. m6A is then deposited on RNAP II transcripts in positions that are selected in large part on the basis of how close they match the preferred motif for METTL3/METTL14. However, the final distribution of m6A is also influenced through recruitment and repression of the methyltransferase by RNA-binding proteins, and histone marks, as well as through removal of the modification at the 5’ end by the demethylase, FTO[63,70]. As a result, m6A is concentrated towards the 3’ end of RNAs.
2.3. Methyltransferase Localization
It is clear that m6A modification occurs early in the life of an mRNA[66,71] and this is consistent with the observation that the writer complex accumulates in nuclear speckles. These membraneless organelles congregate near sites of active transcription and serve as a reservoir for factors that participate in transcription and pre-mRNA processing[72]. WTAP, the third subunit of the core methyltransferase, plays no characterized role in catalysis or substrate selection but is essential for proper MAC localization to nuclear speckles[31], as is ZC3H13[73]. Importantly, if the MAC is not appropriately localized, it does not function: depletion of WTAP or ZC3H13 results in a profound global reduction in m6A methylation[31,32,73].
2.4. Regulating the Writer
Cells modulate m6A profiles in response to differentiation cues and external signals such as heat shock, DNA damage or stress[65,74–77]. The complexity of the m6A writer allows for many levels of regulation; changes in abundance of individual subunits, and post-translational modifications likely influence activity at different stages of development and differentiation. However, to date we have only scratched the surface in identifying how such regulation is achieved. METTL3, being the catalytically active subunit, is an obvious target for regulation, and is targeted by miRNAs[78] and SUMOylation[79]. Other subunits appear to be dependent on METTL3 for their stability or expression[80] and can also be impacted by post-translational modifications (PTMs)[37]. However, there are still many knowledge gaps with respect to how these events regulate methyltransferase activity and the m6A landscape overall.
3. m6A erasure
Modifications of nucleic acids, including the 5’ cap and poly(A) tail on RNA and methylation of cytosine in DNA, are generally reversible; their removal is catalyzed by dedicated enzymes (decapping enzymes, deadenylases, and T5-methylcytosine hydroxylases, respectively) with well characterized consequences such as decay/inactivation of RNAs and de-repression of gene expression. The m6A modification is also reversible, through the action of two enzymes in the α-ketoglutarate-dependent dioxygenase (AlkB) family (ALKBH5 and FTO/ALKBH9). Both ALKBH5 and FTO remove internal m6A modifications, and thus can directly oppose the action of METTL3. However, as discussed below, ALKBH5 activity is restricted both temporally and spatially, suggesting that it is not a global regulator of all methylated mRNAs. A major impact of FTO may arise from its ability to target m6Am adjacent to the 5’ cap structure, especially in snRNAs[81,82](see Box 1).
Like other AlkB proteins, ALKBH5 and FTO utilize an α-ketoglutarate (αKG) domain to recognize the m6A-modified RNA, and also bind Fe(II) and αKG cofactors[70,83,84]. In addition, they possess a nucleotide recognition lid that interacts with the RNA substrate. Upon substrate recognition the demethylases catalyze oxidative dealkylation of m6Ato regenerate an unmodified adenosine[84–88]. The ALKBH5 protein is localized to nuclear speckles and is thought to act upon nascent mRNAs[84], whereas FTO shuttles between nucleus and cytoplasm[65,89] and can affect both nascent and mature transcripts[63,66,70,81,89,90]. Importantly, although the AlkB protein family is conserved across eukaryotic and bacterial domains, ALKBH5 and FTO are found only in vertebrates[84,91], although Arabidopsis have two alternative demethylases, ALKBH9B[92] and ALKBH10B[93]. The fact that eraser proteins are absent from lower eukaryotic organisms, which have both writers and readers of m6A, suggests that demethylation is unlikely to play a general role in reversing all m6A modifications but rather may have evolved to act on specific substrates and/or in response to external signals.
In support of this idea, knockout of ALKBH5 has no discernable effect on overall health in mice but results in profound defects in spermatogenesis, leading to male infertility[84]. Although ALKBH5 has the potential to broadly counteract the action of METTL3, it appears this is not its primary role in most cell types. Outside of the testes, the impact of ALKBH5-mediated demethylation on overall levels of m6A is modest[84] and m6A status of mRNAs does not vary greatly as they mature and enter the cytoplasm[66]. These two observations again support that only a subset of m6A modifications are targeted for removal by ALKBH5. However, it is not clear how selectivity may be achieved as the evidence for any sequence preference is weak[84,94]. Although the overall influence of demethylation by ALKBH5 in most normal cells is modest based on the robust health of knockout mice[84,95], the protein is upregulated in hypoxia[96] and in certain cancer cells[97] and modulates expression of number of transcripts such as TFEB, FOXM1, and NANOG[97–99]. Thus, for some transcripts and responses ALKBH5 may be important.
In contrast to ALKBH5, FTO knockout has quite dramatic effects in mice with links to obesity and neurogenesis[69,91,100,101] although some phenotypes may be due to indirect effects on neighboring genes[102,103]. Since the recent discovery that FTO specifically targets m6Am to reverse methylation adjacent to the cap[81], it is not so clear what proportion of FTO’s impact on gene expression is due to removal of m6Am (see Box) versus m6A or even m1A[89]. It should also be noted that MeRIP-seq experiments are, at least in part, at the mercy of m6A antibody specificity which currently have difficulties distinguishing between m6A and m6Am[81,104]. There is evidence to suggest FTO may prune m6A from the 5’ UTR of specific mRNAs in the nucleus and that this activity is blocked during stress[63–65,75](see below). This is consistent with m6A being more abundant at the 3’ end and around the stop codon in mammals, while being prevalent around both the start and stop codons in Arabidopsis[15], which lack FTO[15,91]. Conversely, there is also evidence that FTO acts on m6A sites outside of the 5’ UTR [105,106], suggesting that the demethylase activity of FTO is not confined to any one region of transcripts. In addition, FTO associates with intronic sequences which may partially explain why introns exhibit significantly lower levels of modification than exons[107](Figure 4). However, it is hard to distinguish whether effects of FTO on splicing are due to altered function of snRNAs (which have m6Am cap structures and are essential components of the splicing machinery) versus changes in binding of RBPs that are recruited or repressed by m6A (see below for more details).
Overall, demethylation may have a significant effect on expression and/or processing of specific transcripts, or during certain cellular responses, but more research is required to tease out when in the RNA life cycle demethylation occurs and how widespread its influence really is. This topic has been debated recently[64,66,81,108,109].
4. How m6A affects RNA fate: Functions of the m6A readers
m6A alone is of little consequence to an RNA transcript; deposition of m6A does not immediately trigger degradation or activate translation. Rather, m6A influences RNA secondary structure and association with proteins and other RNAs to direct altered processing[9,10], export[11], translation[13,110,111] or decay[12,110,112,113](See Figure 2). The presence of m6A favors a single stranded conformation and can therefore disrupt stem-loop structures[114] in a process termed ‘m6A switching’[115]. In some cases, this can allow enhanced binding of proteins like HNRNPC, which, in turn, alters abundance and alternative splicing of target mRNAs[116,117]. Alternatively, m6A can block proteins from binding[62,118,119]. For example, inhibition of Human antigen R (HuR or ELAVL1) binding, due to the presence of m6A, leaves mRNAs susceptible to miRNA-mediated degradation[120]. Although indirect influences on protein and miRNA binding have a significant impact, for brevity, this section will focus on the recruitment of proteins that directly recognize m6A (a.k.a. m6A readers) and their effects on RNA metabolism.
4.1. Reader Recognition of m6A
The best characterized m6A readers are members of the YTH domain-containing family of proteins, namely YTHDF1-3, YTHDC1 and 2 in mammals. The YTH domain was originally identified in the splicing factor YT521-B/YTHDC1[121,122], and is now known to confer m6A-dependent RNA-binding activity[123,124]. In all cases, the preference for m6A-modified RNA is specified by an aromatic cage that forms a hydrophobic pocket around the modified nucleotide[125–128]. Beyond this, it is not clear whether each YTH protein recognizes a distinct motif that allows it to act on a unique population of mRNAs, or if binding is dictated primarily by relative availability and affinity of each family member. Furthermore, whether YTH domains can accommodate m6Am adjacent to the 5’ cap has not been explored. Although there is some evidence for sequence preference outside of the clear requirement for m6A, this must be evaluated critically, as experiments involving capture and sequencing of RNAs bound by YTH proteins inevitably give a motif that overlaps with that of the MAC (i.e. RRACH)[9,12,13,111,129]. For YTHDC1, in vitro and structural analyses support that there is a specific interaction with the G at the −1 position that does not occur in other YTH proteins[125,126]. Other interactions between the RNA and YTH domain[113,127,128], and possibly regions outside of the YTH domain[12,126,130], could confer additional sequence specificity. Such interactions must be important at some level, as RNAs less than 5 nucleotides long bind poorly to YTH proteins compared to longer transcripts[126]. Additional specificity may also be derived from interactions of the YTH proteins with factors associating elsewhere on the RNA. For example, YTHDC1 interacts with 3’ end formation factors which could favor binding towards the 3’ end of mRNAs[10]. The subcellular location of the RNA also influences its accessibility to different YTH proteins. In the nucleus, YTHDC1 has a much higher concentration[9] whereas the other YTH proteins are more abundant in the cytoplasm[12,13,110,129]. At this point, the extent to which the sequence context of an m6A site dictates reader binding potential and the subsequent fate of a modified RNA is an area in need of further investigation. Moreover, while the YTH domain accommodates m6A, there is evidence that these proteins also associate with N1-methyladenosine modified substrates[131] and that binding can occur independent of METTL3-mediated methylation[132]. It is also worth considering that multiple YTH proteins are programmed to recognize the same or adjacent methylated residues, as YTHDF1-3 have numerous shared targets[61,110,111]. These proteins appear to cooperate to modulate translation and decay, but it is not clear whether multiple YTH proteins bind simultaneously to the RNA substrate at adjacent m6A residues, or if the first YTH protein to bind engages the others through protein-protein interactions (Figure 5).
Figure 5: Competition and Collaboration between Reader Proteins.
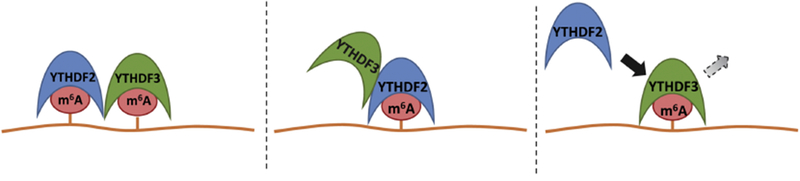
A: Reader proteins bind adjacent m6A and recruit effectors to simultaneously influence multiple aspects of metabolism. B: Reader protein binds m6A and recruits other readers along with effectors, again allowing simultaneous impact on multiple pathways. C: Reader proteins compete for single m6A and shuttle mRNA down different pathways.
Although the majority of research on m6A readers and their biological functions focuses on YTH proteins, other proteins with different RNA binding domains have been implicated as m6A readers. The best supported of these are the insulin-like growth factor 2 mRNA-binding proteins (IGF2BP1-3)[133,134], the eIF3 complex[135] and PRRC2A[136]. In these cases, there is compelling evidence for preferential binding of each factor to m6A modified RNAs, but the structural basis of these interactions is unknown as all available crystal structures were derived using unmodified substrates. FMR1, or FMRP, has also been implicated as a potential m6A reader[119,137], but binding of FMR1 to m6A sites may be influenced more by the sequence context rather than the modification itself [138].
4.2. m6A effects on RNA splicing
The timing of m6A deposition and how it interfaces with other nuclear RNA processing events like splicing and polyadenylation is the subject of intense investigation and also has fueled debate[64,66,108]. There is an apparent disconnect between the distribution of m6A (primarily in exons, and away from exon junctions) and its ability to influence splicing. One reason for the discrepancy may be that conclusions about the distribution of m6A are drawn from analysis of m6A across the whole transcriptome, whereas effects on splicing are uncovered by scrutinizing splicing on a gene by gene basis. It is also possible that our maps of m6A are not accurate due to bias and limitations of the approaches used to identify m6A sites as well as variation between different cell types[109]. And finally, it is important to remember that most experiments only assess the abundance of different mRNA isoforms at steady state, and cannot distinguish whether variation is caused by differential synthesis (i.e. changes in splice site choice) or differential stability. Although deposition patterns are not consistent with splicing regulation being the primary function for m6A, and METTL3 depletion has only modest effects on splicing, for certain transcripts methylation occurs near sequence elements important for splicing and has a dramatic influence on splice site selection. A significant portion of this regulation is achieved through recruitment of a methyl-reader that was first identified as a splicing factor, YTHDC1/YT521B[9]. Splicing may also be influenced by m6A-mediated changes in RNA structure that can favor or inhibit association of other RNA binding proteins like hnRNPA2B1[139].
Among the YTH domain-containing family of proteins, YTHDC1 is the only one that is constitutively enriched in the nucleus under physiological conditions[9]; it accumulates in foci termed YT bodies that are found near active transcription sites and adjacent to splicing speckles[121,140]. YTHDC1 directly interacts with splicing factors SRSF3 and SRFS10 to modulate exon inclusion events. One model asserts binding of SRSF3 to YTHDC1 facilitates its recruitment to splicing substrates and simultaneously prevents association of SRSF10 via steric hindrance[9]. SRSF3 is more abundant and favors exon inclusion, thus SRSF10 is only able to promote exon skipping when methylation or YTHDC1 function are disrupted. Indeed, YTHDC1 deficiency in mouse oocytes affected splicing of almost 2000 genes[10]. Overall, YTHDC1 may not directly influence splice site choice, but by binding to m6A-modified RNAs and interacting with splicing regulators it can have a profound effect on processing events.
4.3. Cleavage and Polyadenylation
Given that m6A modification is concentrated in the 3’ UTR and around the stop codon, it is well positioned to influence 3’ end processing. YTHDC1 interacts with CPSF6, a subunit of the Cleavage Factor Im complex that is responsible for proper polyadenylation[141]. Furthermore, loss of YTHDC1 results in extensive alteration of polyadenylation sites in mouse oocytes[10]. Depletion of METTL3 and VIRMA also induces changes in polyadenylation site[34], but further study is required to determine whether the effects of reduced methylation are mediated entirely through loss of YTHDC1 binding or if other factors are involved.
4.4. Nuclear Export
One of the most dramatic effects of m6A is on RNA export. Inhibition of methylation delays export of mature mRNAs[142] and knockdown of the ALKBFI5 demethylase accelerates export[84]. Recent studies have discovered that in HeLa cells YTHDC1 mediates this facet of m6A function by recruiting SRSF3, which in turn interacts with the nuclear RNA export factor 1 (NXF1) to facilitate export of methylated mRNAs[11]. Interestingly, SRSF3 has been identified as a key regulator of pluripotency in murine iPSCs, where it binds to NANOG mRNA and facilitates its export, as well as modulating splicing and expression of numerous pluripotency genes[143]. In the future, it will be interesting to assess how much of this regulation relies on recruitment by YTHDC1.
4.5. Regulation of YTHDC1
As outlined above, YTHDC1 is crucial for the efficient and accurate processing and export of methylated mRNAs and is the major direct reader of m6A in the nucleus. It is therefore worth highlighting that YTHDC1 is a substrate for a number of tyrosine kinases including FYN, c-Src and c-Abl and phosphorylation of YTHDC1 influences both its localization and function. The phosphorylated form of YTHDC1 is dispersed throughout the nucleoplasm, insoluble, and unable to influence splice site selection[144]. At this time it is not known when or where phosphorylation naturally occurs but, based on the observation that YT bodies disperse during mitosis, it seems likely that activity of YTHDC1 is modulated in a cell-cycle dependent manner[144].
4.6. m6A effects on translation
Once a methylated mRNA reaches the cytoplasm, it becomes available for binding by other YTH proteins although whether, and how, exchange of readers occurs is unknown. In the cytoplasmic compartment, m6A modulates translation and decay of modified transcripts. YTHDF1[13,145–147], YTHDF3[110,111], YTHDC2[129,148,149] and even the methyl-writer, METTL3[150], have all been implicated as translation regulators, but are recruited to different regions of the mRNA and interact with different components of the translation machinery.
4.6.1. 5’ UTR-mediated regulation:
m6A methylation in the 5’ UTR promotes cap-independent translation[135] and influences selection of the translation start site[106]. Cap-independent/m6A-dependent translation is achieved by recruiting the translation initiation factor, eIF3 which in turn engages the 40S ribosomal subunit[135]. There is evidence that the ATP binding protein, ABCF1, and YTHDF3 also play a role[151]. During stress, cap-dependent translation is often blocked, thus this m6A-dependent mechanism facilitates recovery by directing translation of chaperones and other required factors. As m6A is deposited on capped RNAs, it also has the capacity to compete or collaborate with canonical cap-dependent translation under physiological/non-stress conditions[151]. Importantly, 5’ UTR m6A is specifically increased during heat shock with no effect observed in the body or 3’ UTR of mRNAs[135]. This is achieved through relocalization of the normally cytoplasmic YTHDF2 to the nucleus where it specifically protects 5’ m6A sites from demethylation by FTO[65,75]. Increased m6A methylation in the 5’ UTR is also observed during oxidative stress which induces accumulation of many translation-arrested transcripts in cytoplasmic bodies known as stress granules (SGs). Under these conditions, 5’ UTR m6A facilitates translation silencing and SG localization through binding of YTHDF3[152]. There is obviously still more to be learned about these mechanisms: How are 5’ UTR m6A sites are selectively protected or modified? How do oxidative and heat stress both induce 5’ UTR methylation but have opposing effects on translation of m6A modified mRNAs? It will also be interesting to investigate whether these novel translation mechanisms are important during development and differentiation.
4.6.2. 3’ UTR/Coding Region methylation:
As noted above, the majority of m6A marks are clustered around the stop codon and in the 3’ UTR, where they are ideally situated to influence both translation and decay[7,8,56]. Both METTL3 and YTHDF1 have been independently implicated as readers of m6A located at the 3’ end of mRNAs, and both were reported to act by recruiting eIF3[13,150]. Tethering experiments suggest that YTHDF1 and METTL3 proteins function in separate pathways rather than one enlisting the other. However, it is not clear why eIF3 requires participation of other factors to recognize m6A near the stop codon when it appears to bind m6A in the 5’ UTR directly[135]. In addition, the precise mechanism by which METTL3 interacts with an mRNA to promote translation requires further investigation. There is evidence from tethering assays that the translation-promoting activity of METTL3 does not require methyltransferase activity or METTL14/WTAP[150], but it is not clear whether METTL14/WTAP are required for association of METTL3 with RNAs in the cytoplasm. It is also not known whether the MAC is removed from the mRNA after m6A deposition, with METTL3 reuniting with the transcript at a later time to promote translation. Alternatively, METTL3/MAC may be retained on some sites after it catalyzes the methylation reaction, while being ejected from other sites to allow binding of other factors.
YTHDF1 and YTHDF3 share many substrates and appear to cooperate to promote translation. However YTHDF3 does not influence translation on its own, although it does exhibit interactions with ribosomal proteins[110,111]. In contrast, when bound to circular RNAs, YTHDF3 promotes translation by acting in concert with elF4G2[153].
A third member of the YTH family, YTHDC2, also directly interacts with m6A-modified transcripts to facilitate translation. YTHDC2 is extremely important for germline development and specifically for the switch from mitosis to meiosis in male germ cells[113,149,154]. Tethered YTHDC2 enhances translation and loss of YTHDC2 results in a marked decrease in translational efficiency of its targets[129]. Moreover, YTHDC2 interacts directly with the small ribosomal subunit[148]. It is hypothesized that the RNA helicase activity of YTHDC2 might facilitate access for translation initiation factors and/or the ribosome[113,148,155].
Beyond the YTH family of proteins, IGF2BPs also co-localize with SG markers[134] and sequester target transcripts to stress granules[156], suggesting they could perform a similar triaging role to that of YTHDF3. IGF2BP1 specifically regulates the translation of MYC mRNA in an m6A-dependent manner through a coding region element that contains m6A sites[134]. The IGF2BPs have previously been linked to several mechanisms of translation regulation[156], including of viral transcripts[157]. However, the precise mechanism for m6A dependent regulation by IGF2BPs remains elusive.
4.7. m6A effects on decay
The final effect of m6A on the RNA life cycle is its impact on RNA decay rates. Multiple studies have shown that m6A methylated transcripts are less stable than their unmodified counterparts[12,20,50,74,110,112], and YTHDF2 is the primary culprit causing this destabilization[12]. Under physiological conditions, a majority of YTHDF2 binding sites are found near the stop codon and within the 3’ UTR[12], which likely reflects the natural distribution of m6A[7,8]. Upon substrate binding YTHDF2 accelerates the degradation of target transcripts through direct recruitment of the CCR4-NOT deadenylase complex[112] via an interaction between YTHDF2 and the CNOT1 subunit of the deadenylase. As deadenylation is the first and rate-limiting step in decay of most mRNAs, modulating this step has a significant effect on overall mRNA decay rates[158]. At the same time, YTHDF2 can also work with the HRSP12 RBP to recruit RNAse P/MRP endonuclease[159]. YTHDF2-mediated decay represses transcripts encoding cell cycle regulators such as Cyclin A2 (CCNA2) and CDK2[101], and also clears populations of unwanted mRNAs such as maternal transcripts during oocyte maturation[2] and transcripts favoring self-renewal in differentiating hematopoietic stem cells (HSCs)[160]. YTHDF2 plays an important role in clearing RNAs during cell state transitions such as the maternal-to-zygotic[1] and the epithelial-mesenchymal transitions[161].
While YTHDF2 makes the most obvious contribution to m6A dependent mRNA decay, there is evidence that other YTH proteins also interface with the mRNA decay machinery. YTHDF2 and YTHDF3 both enhance deadenylation somewhat when tethered to a reporter mRNA[112], but this might perhaps be explained by observations that YTH proteins, and particularly YTHDF3, can augment the binding of other family members (such as YTHDF2) to their substrates[110]. YTHDC2 is also able to down-regulate its targets, presumably by destabilizing them[129], and has been implicated as an RNA stability factor in the germline, where it interacts with a meiosis-specific protein known as MEIOC[162,163]. The interaction of MEIOC and YTHDC2 with mitosis-promoting transcripts, including CCNA2 enhances their decay and facilitates a sharp transition into meiosis[149,163]. These experiments indicate a primitive role for m6A in meiosis that may be conserved from yeast to humans, as m6A deposition only occurs during meiosis in S. cerevisiae[164–166]. The exact mechanism by which YTHDC2 influences decay in meiotic cells needs more study. While there is strong evidence for direct interaction between YTHDC2 and the 5’−3’ exoribonuclease XRN1[148], XRN1 can only act on mRNAs that have already been committed to destruction by endonucleolytic cleavage or decapping[167]. Furthermore, XRN1 is required for meiosis in S. cerevisiae, but its catalytic activity is dispensable[168], suggesting that the interaction with XRN1 may not be directly responsible for the ability of YTHDC2 to enhance mRNA turnover.
The IGF2BP family of proteins also modulate the decay of m6A-modified transcripts but unlike YTHDF2 and YTHDC2, IGF2BPs stabilize methylated transcripts[134]. In some cases, this is achieved through sequestration of methylated mRNAs into stress granules[156], but stabilization can also be caused by impeding binding of destabilizing factors such as miRNAs[133]. It is also worth noting that m6A on its own can also modulate mRNA stability by perturbing binding of factors with overlapping target sites. For example, the presence of m6A impedes HuR binding and thereby favors association miRNAs at neighboring sites to enhance decay of developmental transcripts such as IGF2BP3 and FGF5[120].
Regulation of reader activity can be achieved through standard mechanisms including gene expression, subcellular localization and PTMs. In terms of gene expression, YTHDF2 is repressed by two miRNAs[169,170], while YTHDC2 is inducible by TNFα[171]. Previously mentioned observations of YTHDF2 re-localizing to the nucleus during heat shock[65,75] and PTMs on YTHDC1[144] infer there may be other instances of regulation awaiting discovery. Future studies regarding the mechanisms and situations by which reader activity is regulated will provide useful insight about the impact m6A has on global gene expression.
5. How does m6A methylation of mRNAs influence pluripotency and development?
To recap, m6A is added to a large proportion of mRNAs and non-coding RNAs in many different tissues and cell types and can influence most, if not all, aspects of RNA metabolism. It is not surprising then that m6A methylation has a great influence on both cell and organism function. However, despite the almost ubiquitous nature of m6A modification, its power is observed most clearly in stem cells and during development[50,51,74,120,172], when large-scale coordinated changes in gene expression occur to determine cell fate. The effects of reducing m6A methylation are highly cell-type dependent, suggesting that methyltransferase activity, the RNA targets it modifies, and/or the activity/abundance of methyl-readers are regulated in a cell-specific manner. Ultimately though, a common theme is that cell fate decisions are skewed following loss of methylation[50,52,53,172–176], suggesting that m6A helps maintain stem cells in a balanced state where they are able to self-renew but also primed to rapidly and efficiently differentiate down one or more pathways. When either methylation itself, or the activity of methyl-readers, is perturbed, the balance tips and cells are committed to a specific fate or permanently detained in the self-renewal program. The effects of m6A on developmental decisions appear to occur through at least three mechanisms, which are discussed below.
5.1. Tipping the balance of dosage-sensitive master regulators
In many cases, methylation of mRNAs encoding one or two dosage sensitive master regulators (generally signaling or transcription factors) is pivotal. If expression of these key targets rises above, or falls below a threshold it pushes the cell to differentiate, or alternatively into a permanent state of self-renewal or quiescence.
Some recently characterized examples are shown in Figure 6. In bone marrow mesenchymal stem cells (BMSCs) knockout of METTL3 strongly favors differentiation down the adipocyte lineage over osteoblast production, leading to osteoporosis. The Parathyroid Hormone Receptor (PTH1R) mRNA is a key target of METTL3-mediated methylation in BMSCs; its translation is compromised following METTL3 knockdown and this disrupts the PTH signaling axis[175](Figure 6A). The importance of m6A modification of this single mRNA for osteogenesis is highlighted by the fact that restoration of PTH1R expression partially rescues osteogenic gene expression in METTL3-deficient cells[175].
Figure 6: m6A methylation influences key events during development and differentiation.
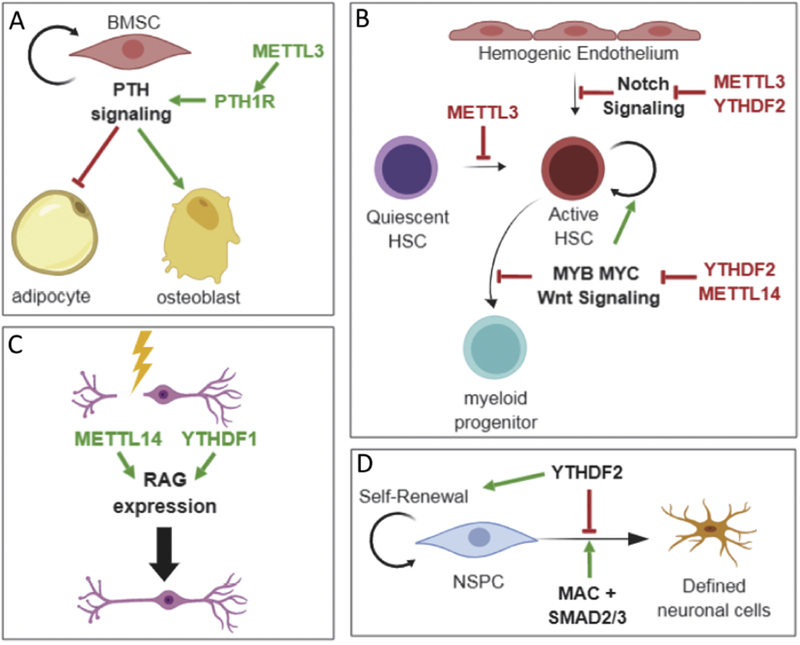
A: A: In bone marrow stem cells (BMSC), translation of the parathyroid hormone receptor (PTH1R) mRNA is regulated by METTL3-mediated methylation to favor production of osteoblasts. B: B: m6A influences hematopoiesis at multiple steps. C: Following nerve injury, regeneration associated genes are subject to m6A-mediated regulation to promote recovery. D: Neural Progenitor Stem Cells (NPSCs) rely on m6A methylation for self-renewal and for a sharp transition upon receipt of a cue to differentiate. See text for more details.
Methylation is also important in myeloid differentiation (Figure 6B). Loss of METTL3 results in activation of quiescent HSCs to a proliferative state[177], while METTL14[178] and YTHDF2[160,179] are required for self-renewal of active HSCs - silencing either gene favors myeloid differentiation. Two key targets of METTL14 in this process are the MYB and MYC transcription factor mRNAs whose expression is down-regulated following METTL14 depletion[178]. Moreover, upstream of this, m6A deposition on the NOTCH1 mRNA can promote its destabilization via YTHDF2 and repress Notch signaling. This favors the endothelial-hematopoietic transition[172,180].
The above examples highlight that some genes are uniquely sensitive to methylation of their transcripts. Further study is required to determine what makes these genes so responsive. It could be simply that the proteins encoded by these genes are very sensitive to small changes in expression. Alternatively, the extent of methylation and/or context of the methylation site(s) could directly influence recruitment of readers and effector proteins to these transcripts.
5.2. Clearing unwanted mRNAs from the transcriptome
During differentiation and development, massive changes in gene expression occur which require that existing transcripts are cleared to make way for those being induced to specify the new cell type. Recent studies have found that m6A methylation, through recruitment of the decay promoting factor YTHDF2, plays vital roles in clearing unwanted transcripts. During the maternal-zygotic transition in zebrafish, YTHDF2 associates with over one third of maternally loaded mRNAs and induces their decay[1]. Similarly, maternal mRNA clearance during mouse oocyte maturation is disrupted following loss of YTHDF2[2]. Interestingly, newly transcribed zygotic mRNAs are m6A methylated, but their abundance is not generally affected by removal of YTHDF2[1]. Further investigation will be needed to determine what distinguishes the maternally supplied methylated mRNAs from those derived from the zygotic genome. It is possible that both populations are targeted for decay but that zygotic transcription is down-regulated to compensate for loss of YTHDF2. Alternatively, the proteins deposited on mRNAs during zygotic transcription are likely to be different than those found on maternal transcripts and could influence m6A recognition and impact. It is important to note that although m6A clearly plays a role in clearance of some maternal mRNAs, it is not an essential one, as Ythdf2−/− embryos are generally viable, although they do exhibit delayed development[1,2]. Evidence suggests that there are multiple redundant mechanisms for maternal mRNA clearance which ensures robustness and reduces the influence of random variation[181].
In the examples above, m6A induces clearance of a population of mRNAs at a specific developmental transition. In other scenarios, m6A facilitates a phenomenon known as transcriptional pre-patterning, where mRNAs specifying an alternative cell fate are actively transcribed but constantly targeted for m6A-mediated decay by YTHDF2 before being translated[182]. This can create a balanced state where stem cells are poised for rapid differentiation while still maintaining a stem cell phenotype. Strong evidence for this role of m6A comes from studies of neural development, where destabilization of neuron-specific mRNAs by m6A tagging and recruitment of YTHDF2 may both clear these transcripts in neural stem cells and facilitate their coordinated upregulation following induction of neuronal differentiation[174]. Although differentiation can still occur in the absence of methylation, it is less robust, at least in part due to prolonged cell cycle progression[174,182].
5.3. Potentiating transcriptional changes for sharp transitions
It is clear that m6A is deposited on many mRNAs, and thus has a widespread impact on overall gene expression. In this respect, m6A modification is reminiscent of miRNA-mediated regulation. miRNAs fine tune gene expression, buffer against noise and confer robustness on biological systems[183]; all roles that can also be attributed to methylation. Specifically, while transcriptional changes are central to initiating cell fate transitions during development, methylation and its downstream effects are key to ensuring these transitions are carried through in a sharp and coordinated fashion.
An excellent example is seen in a recent study evaluating the impact of m6A during nerve regeneration following injury. Robust axon regeneration requires transcription and translation of regeneration-associated genes (RAGs), many of which are enriched among mRNAs tagged with m6A following nerve injury. Both the site and level of methylation on RAG mRNAs is altered following injury. Importantly, upregulation of RAG proteins is delayed following deletion of METTL14. This delay can be attributed, in part, to reduced recruitment of the translation regulator, YTHDF1, to the RAG mRNAs, but reduced expression of translation factors may also be an influence[145]. Overall, axon regeneration is more robust in the presence of a functional m6A pathway; recovery of damaged axons is impaired following loss of METTL14 or YTHDF1 (figure 6C).
Methylation is also used to facilitate neuroectoderm differentiation through a mechanism activated by TGFβ signaling[6]. TGFβ signaling is required to maintain a pluripotent state and functions by activating SMAD2/3 transcription factors. The MAC is recruited by activated SMAD2/3 to its target genes, resulting in enhanced methylation and destabilization of several mRNAs whose transcription is induced by TGFβ signaling, including that encoding the pluripotency factor NANOG[6]. It is well known that unstable transcripts respond more rapidly to external cues[184,185], thus by destabilizing its target mRNAs, methylation facilitates sharper down-regulation of pluripotency factors when TGFβ signaling is inhibited. This in turn allows timely exit from pluripotency and efficient entry into the neuroectoderm differentiation program (Figure 6D).
6. Where does the m6A field go from here?
The ability to study m6A and its impact on biological processes became readily available relatively recently, through advances in next generation sequencing technologies that allow isolation of methylated RNAs (MeRIP-Seq)[7,8]. Since then, adaptations have been implemented to increase the resolution[20,56,186,187], but the method remains at the mercy of antibody specificity, leaving it vulnerable to nonspecific signals or sequencing artifacts[104]. The reliance on antibodies also prevents quantitative assessment of m6A levels on a global scale. Other methods, like SCARLET[188], are able to accurately quantify m6A at a specific site, but the method is laborious, works only on relatively abundant transcripts and can only characterize one site at a time. The very recent introduction of sequencing approaches relying on m6A-sensitive endonucleases[59,60] and nanopore-based direct RNA sequencing[189] will undoubtedly reveal more information regarding the stoichiometry of m6A modification at individual sites. As these techniques are adopted, new insights into how m6A stoichiometry changes during development and in response to stimuli will be revealed, as well as a better understanding of relative roles of the writers and erasers in defining the m6A landscape under different conditions.
There are many avenues for future research connected to the m6A epitranscriptome and its impacts on development and pluripotency. We have much to learn about how m6A deposition integrates with other events that are dependent on the same SAM methyl-donor such as histone and DNA methylation. Through the SAM donor and methyltransferase activities, methylation is used to connect stem cell metabolism to gene expression at multiple levels. The effects of SAM depletion on pluripotent cells include impaired self-renewal, with cells entering a state poised for differentiation and, if deprivation is prolonged, eventually apoptosis[190]. The naïve-to-primed hESC transition is also dependent on SAM levels[191]. Methionine deprivation (which leads to SAM depletion) is known to influence the modification of DNA and histones, but the effects of reducing this metabolite on METTL3/14 and the major pathway of m6A deposition are virtually unstudied. Given the compelling evidence that RNA methylation is integral to self-renewal and differentiation of stem cells, it seems likely that it contributes to the effects of SAM depletion on these processes and may be tightly coordinated with similar modifications occurring at the chromatin level.
While there has been significant progress in identifying components of the m6A writing machinery, and numerous readers have been characterized, the interactions between these proteins need to be better defined. For example, the MACOM subunits may compete for binding to the MAC which could result in an array of methyltransferase complexes with different preferences and expression patterns. This in turn could account for changes in the m6A landscape that have been observed in response to stimuli. At the level of m6A reading, there is evidence that readers cooperate, but we have not yet determined whether each must bind m6A directly for recruitment, whether different readers have unique site preferences, or whether reader proteins are exchanged as the mRNP matures and progresses to translation and eventually decay.
Finally, while connections between m6A and development are clear, the exact role of m6A in facilitating massive reorganization of the transcriptome must be further dissected to understand how one set of methylated transcripts are targeted for destruction (e.g. maternal RNAs) while others (e.g. zygotic transcripts) are not. It could be that the m6A distribution changes and/or that different readers/effectors are recruited as transcription changes. Sifting through the links between chromatin/transcription and m6A deposition will likely give important insights in this area.
At this point it is clear that m6A methylation is an essential means of coordinating and regulating gene expression during cell state transitions. With advances in technology, and increased attention, it seems inevitable that this avenue of investigation will lead to a much clearer understanding of pluripotency and differentiation and also lead to novel therapeutic approaches for cancer and other diseases.
Supplementary Material
Highlights.
m6A is an abundant post-transcriptional modification that enables rapid responses to external cues
The m6A methylation pathway consists of readers, writers and erasers
m6A modifications impact multiple facets of RNA metabolism
m6A regulation is crucial for stem cell differentiation and development
Acknowledgements
This work was supported by the National Institutes of Health, National Institute for General Medical Sciences Award #GM114247 to CJW and JW. AMH was supported in part through NSF-NRT Award # 1450032 (Principal Investigator: Tom Chen) and an NSF Graduate Research Fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing Interests
The authors declare no competing interests.
References
- [1].Zhao BS, Wang X, Beadell AV, Lu Z, Shi H, Kuuspalu A, Ho RK, He C, m6A-dependent maternal mRNA clearance facilitates zebrafish maternal-to-zygotic transition, Nature. 542 (2017) 475–478. doi: 10.1038/nature21355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Ivanova I, Much C, Di Giacomo M, Azzi C, Morgan M, Moreira PN, Monahan J, Carrieri C, Enright AJ, O’Carroll D, The RNA m 6 A Reader YTHDF2 Is Essential for the Post-transcriptional Regulation of the Maternal Transcriptome and Oocyte Competence, Mol. Cell (2017). doi: 10.1016/j.molcel.2017.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Oh Y, Park J, Kim J-I, Chang M-Y, Lee S-H, Cho Y-H, Hwang J, Lin28B and miR-142-3p regulate neuronal differentiation by modulating Staufen1 expression, Cell Death Differ. 25 (2018) 432–443. doi: 10.1038/cdd.2017.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Kami D, Kitani T, Nakamura A, Wakui N, Mizutani R, Ohue M, Kametani F, Akimitsu N, Gojo S, The DEAD-box RNA-binding protein DDX6 regulates parental RNA decay for cellular reprogramming to pluripotency, PLoS One. 13 (2018) e0203708. doi: 10.1371/journal.pone.0203708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Lloret-Llinares M, Karadoulama E, Chen Y, Wojenski LA, Villafano GJ, Bornholdt J, Andersson R, Core L, Sandelin A, Jensen TH, The RNA exosome contributes to gene expression regulation during stem cell differentiation, Nucleic Acids Res. 46 (2018) 11502–11513. doi: 10.1093/nar/gky817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Bertero A, Brown S, Madrigal P, Osnato A, Ortmann D, Yiangou L, Kadiwala J, Hubner NC, de los Mozos IR, Sadée C, Lenaerts A-S, Nakanoh S, Grandy R, Farnell E, Ule J, Stunnenberg HG, Mendjan S, Vallier L, The SMAD2/3 interactome reveals that TGFβ controls m6A mRNA methylation in pluripotency, Nature. 555 (2018) 256–259. doi: 10.1038/nature25784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, Sorek R, Rechavi G, Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq., Nature. 485 (2012) 201–6. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- [8].Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR, Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons., Cell. 149 (2012) 1635–46. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Xiao W, Adhikari S, Dahal U, Chen Y-S, Hao Y-J, Sun B-F, Sun H-Y, Li A, Ping X-L, Lai W-Y, Wang X, Ma H-L, Huang C-M, Yang Y, Huang N, Jiang G-B, Wang H-L, Zhou Q, Wang X-J, Zhao Y-L, Yang Y-G, Nuclear m(6)A Reader YTHDC1 Regulates mRNA Splicing., Mol. Cell 61 (2016) 507–19. doi: 10.1016/j.molcel.2016.01.012. [DOI] [PubMed] [Google Scholar]
- [10].Kasowitz SD, Ma J, Anderson SJ, Leu NA, Xu Y, Gregory BD, Schultz RM, Wang PJ, Nuclear m6A reader YTHDC1 regulates alternative polyadenylation and splicing during mouse oocyte development, PLOS Genet. 14 (2018) e1007412. doi: 10.1371/journal.pgen.1007412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Roundtree IA, Luo G-Z, Zhang Z, Wang X, Zhou T, Cui Y, Sha J, Huang X, Guerrero L, Xie P, He E, Shen B, He C, YTHDC1 Mediates Nuclear Export of N6-methyladenosine Methylated mRNAs, Elife. 6 (2017). doi: 10.7554/eLife.31311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, Ren B, Pan T, He C, N6-methyladenosine-dependent regulation of messenger RNA stability., Nature. 505 (2014) 117–20. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, Weng X, Chen K, Shi H, He C, N6-methyladenosine Modulates Messenger RNA Translation Efficiency, Cell. 161 (2015) 1388–1399. doi: 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Bodi Z, Button JD, Grierson D, Fray RG, Yeast targets for mRNA methylation, Nucleic Acids Res. 38 (2010) 5327–5335. doi: 10.1093/nar/gkq266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Luo G-Z, MacQueen A, Zheng G, Duan H, Dore LC, Lu Z, Liu J, Chen K, Jia G, Bergelson J, He C, Unique features of the m6A methylome in Arabidopsis thaliana, Nat. Commun. 5 (2014) 5630. doi: 10.1038/ncomms6630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Lence T, Soller M, Roignant J-Y, A fly view on the roles and mechanisms of the m6A mRNA modification and its players, RNA Biol. 14 (2017) 1232. doi: 10.1080/15476286.2017.1307484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Perry RP, Kelley DE, Existence of methylated messenger RNA in mouse L cells, Cell. 1 (1974) 37–42. doi: 10.1016/0092-8674(74)90153-6. [DOI] [Google Scholar]
- [18].Wei CM, Gershowitz A, Moss B, 5’-Terminal and internal methylated nucleotide sequences in HeLa cell mRNA., Biochemistry. 15 (1976) 397–401. [DOI] [PubMed] [Google Scholar]
- [19].Desrosiers R, Friderici K, Rottman F, Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells., Proc. Natl. Acad. Sci. U. S. A 71 (1974) 3971–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Chen K, Lu Z, Wang X, Fu Y, Luo G-Z, Liu N, Han D, Dominissini D, Dai Q, Pan T, He C, High-resolution N(6) -methyladenosine (m(6) A) map using photo-crosslinking-assisted m(6) A sequencing., Angew. Chem. Int. Ed. Engl 54 (2015) 1587–90. doi: 10.1002/anie.201410647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Akichika S, Hirano S, Shichino Y, Suzuki T, Nishimasu H, Ishitani R, Sugita A, Hirose Y, Iwasaki S, Nureki O, Suzuki T, Cap-specific terminal N 6 -methylation of RNA by an RNA polymerase II-associated methyltransferase, Science. 363 (2019). doi: 10.1126/science.aav0080. [DOI] [PubMed] [Google Scholar]
- [22].Cowling VH, CAPAM: The mRNA Cap Adenosine N6-Methyltransferase, Trends Biochem. Sci 44 (2019) 183–185. doi: 10.1016/j.tibs.2019.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Zorbas C, Nicolas E, Wacheul L, Huvelle E, Heurgué-Hamard V, Lafontaine DLJ, The human 18S rRNA base methyltransferases DIMT1L and WBSCR22-TRMT112 but not rRNA modification are required for ribosome biogenesis., Mol. Biol. Cell 26 (2015) 2080–95. doi: 10.1091/mbc.E15-02-0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Lafontaine D, Vandenhaute J, Tollervey D, The 18S rRNA dimethylase Dim1p is required for pre-ribosomal RNA processing in yeast., Genes Dev. 9 (1995) 2470–81. [DOI] [PubMed] [Google Scholar]
- [25].Warda AS, Kretschmer J, Hackert P, Lenz C, Urlaub H, Höbartner C, Sloan KE, Bohnsack MT, Human METTL16 is a N6-methyladenosine (m6A) methyltransferase that targets pre-mRNAs and various non-coding RNAs., EMBO Rep. 18 (2017) 2004–2014. doi: 10.15252/embr.201744940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Brown JA, Kinzig CG, DeGregorio SJ, Steitz JA, Methyltransferase-like protein 16 binds the 3’-terminal triple helix of MALAT1 long noncoding RNA, Proc. Natl. Acad. Sci 113 (2016) 14013–14018. doi: 10.1073/pnas.1614759113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Pendleton KE, Chen B, Liu K, Hunter ΟV, Xie Y, Tu BP, Conrad NK, The U6 snRNA m6A Methyltransferase METTL16 Regulates SAM Synthetase Intron Retention, Cell. 169 (2017) 824–835. e14. doi: 10.1016/J.CELL.2017.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Shima H, Matsumoto M, Ishigami Y, Ebina M, Muto A, Sato Y, Kumagai S, Ochiai K, Suzuki T, Igarashi K, S-Adenosylmethionine Synthesis Is Regulated by Selective N6-Adenosine Methylation and mRNA Degradation Involving METTL16 and YTHDC1, Cell Rep. 21 (2017) 3354–3363. doi: 10.1016/J.CELREP.2017.11.092. [DOI] [PubMed] [Google Scholar]
- [29].Doxtader KA, Wang P, Scarborough AM, Seo D, Conrad NK, Nam Y, Structural Basis for Regulation of METTL16, an S-Adenosylmethionine Homeostasis Factor, Mol. Cell. 71 (2018) 1001–1011. e4. doi: 10.1016/J.MOLCEL.2018.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Ruszkowska A, Ruszkowski M, Dauter Z, Brown JA, Structural insights into the RNA methyltransferase domain of METTL16, Sci. Rep. 8 (2018) 5311. doi: 10.1038/s41598-018-23608-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, Jia G, Yu M, Lu Z, Deng X, Dai Q, Chen W, He C, A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation., Nat. Chem. Biol 10 (2014) 93–5. doi: 10.1038/nchembio.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Ping X-L, Sun B-F, Wang L, Xiao W, Yang X, Wang W-J, Adhikari S, Shi Y, Lv Y, Chen Y-S, Zhao X, Li A, Yang Y-GY, Dahal U, Lou X-M, Liu X, Huang J, Yuan W-P, Zhu X-F, Cheng T, Zhao Y-L, Wang X, Danielsen JMR, Liu F, Yang Y-GY, Rendtlew Danielsen JM, Liu F, Yang Y-GY, Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase., Cell Res. 24 (2014) 177–89. doi: 10.1038/cr.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Guo J, Tang H-W, Li J, Perrimon N, Yan D, Xio is a component of the Drosophila sex determination pathway and RNA N6-methyladenosine methyltransferase complex., Proc. Natl. Acad. Sci. U. S. A 115 (2018) 3674–3679. doi: 10.1073/pnas.1720945115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Yue Y, Liu J, Cui X, Cao J, Luo G, Zhang Z, Cheng T, Gao M, Shu X, Ma H, Wang F, Wang X, Shen B, Wang Y, Feng X, He C, Liu J, VIRMA mediates preferential m6A mRNA methylation in 3’UTR and near stop codon and associates with alternative polyadenylation, Cell Discov. 4 (2018) 10. doi: 10.1038/s41421-018-0019-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Knuckles P, Lence T, Haussmann IU, Jacob D, Kreim N, Carl SH, Masiello I, Hares T, Villaseñor R, Hess D, Andrade-Navarro MA, Biggiogera M, Helm M, Soller M, Bühler M, Roignant J-Y, Zc3h13/Flacc is required for adenosine methylation by bridging the mRNA-binding factor Rbm15/Spenito to the m6 A machinery component Wtap/Fl(2)d, Genes Dev. (2018). doi: 10.1101/gad.309146.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Růžička K, Zhang M, Campilho A, Bodi Z, Kashif M, Saleh M, Eeckhout D, El-Showk S, Li H, Zhong S, De Jaeger G, Mongan NP, Hejátko J, Helariutta Y, Fray RG, Identification of factors required for m6 A mRNA methylation in Arabidopsis reveals a role for the conserved E3 ubiquitin ligase HAKAI, New Phytol. 215 (2017) 157–172. doi: 10.1111/nph.14586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Schöller E, Weichmann F, Treiber T, Ringle S, Treiber N, Hatley A, Feederle R, Bruckmann A, Meister G, Interactions, localization, and phosphorylation of the m6A generating METTL3-METTL14-WTAP complex, RNA. 24 (2018) 499–512. doi: 10.1261/rna.064063.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Wang X, Feng J, Xue Y, Guan Z, Zhang D, Liu Z, Gong Z, Wang Q, Huang J, Tang C, Zou T, Yin P, Structural basis of N6-adenosine methylation by the METTL3-METTL14 complex, Nature. 534 (2016) 575–8. doi: 10.1038/nature18298. [DOI] [PubMed] [Google Scholar]
- [39].Wang P, Doxtader KA, Nam Y, Structural Basis for Cooperative Function of Mettl3 and Mettl14 Methyltransferases, Mol. Cell 63 (2016) 306–317. doi: 10.1016/j.molcel.2016.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Śledź P, Jinek M, Structural insights into the molecular mechanism of the m6A writer complex, Elife. 5 (2016). doi: 10.7554/eLife.18434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Huang J, Dong X, Gong Z, Qin L-Y, Yang S, Zhu Y-L, Wang X, Zhang D, Zou T, Yin P, Tang C, Solution structure of the RNA recognition domain of METTL3-METTL14 N6-methyladenosine methyltransferase, Protein Cell. (2018) 1–13. doi: 10.1007/s13238-018-0518-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Hongay CF, Orr-Weaver TL, Drosophila Inducer of MEiosis 4 (IME4) is required for Notch signaling during oogenesis, Proc. Natl. Acad. Sci 108 (2011) 14855–14860. doi: 10.1073/pnas.1111577108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Zhong S, Li H, Bodi Z, Button J, Vespa L, Herzog M, Fray RG, MTA Is an Arabidopsis Messenger RNA Adenosine Methylase and Interacts with a Homolog of a Sex-specific Splicing Factor, PLANT CELL ONLINE. 20 (2008) 1278–1288. doi: 10.1105/tpc.108.058883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Clancy MJ, Shambaugh ME, Timpte CS, Bokar JA, Induction of sporulation in Saccharomyces cerevisiae leads to the formation of N6-methyladenosine in mRNA: a potential mechanism for the activity of the IME4 gene., Nucleic Acids Res. 30 (2002) 4509–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Lence T, Paolantoni C, Worpenberg L, Roignant J-Y, Mechanistic insights into m6A RNA enzymes, Biochim. Biophys. Acta - Gene Regul. Mech (2018). doi: 10.1016/J.BBAGRM.2018.10.014. [DOI] [PubMed] [Google Scholar]
- [46].Bodi Z, Zhong S, Mehra S, Song J, Graham N, Li H, May S, Fray RG, Adenosine Methylation in Arabidopsis mRNA is Associated with the 3’ End and Reduced Levels Cause Developmental Defects, Front. Plant Sci 3 (2012) 48. doi: 10.3389/fpls.2012.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Kan L, Grozhik AV, Vedanayagam J, Patil DP, Pang N, Lim K-S, Huang Y-C, Joseph B, Lin C-J, Despic V, Guo J, Yan D, Kondo S, Deng W-M, Dedon PC, Jaffrey SR, Lai EC, The m6A pathway facilitates sex determination in Drosophila, Nat. Commun 8 (2017) 15737. doi: 10.1038/ncomms15737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Schwartz S, Agarwala SD, Mumbach MR, Jovanovic M, Mertins P, Shishkin A, Tabach Y, Mikkelsen TS, Satija R, Ruvkun G, Carr SA, Lander ES, Hnk GR, Regev A, High-Resolution Mapping Reveals a Conserved, Widespread, Dynamic mRNA Methylation Program in Yeast Meiosis, Cell. 155 (2013) 1409–1421. doi: 10.1016/j.cell.2013.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Aguilo F, Zhang F, Sancho A, Hdalgo M, Di Cecilia S, Vashisht A, Lee D-F, Chen C-H, Rengasamy M, Andino B, Jahouh F, Roman A, Krig SR, Wang R, Zhang W, Wohlschlegel JA, Wang J, Walsh MJ, Coordination of m(6)A mRNA Methylation and Gene Transcription by ZFP217 Regulates Pluripotency and Reprogramming., Cell Stem Cell. 17 (2015) 689–704. doi: 10.1016/j.stem.2015.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Geula S, Moshitch-Moshkovitz S, Dominissini D, Mansour AA, Kol N, Salmon-Divon M, Hershkovitz V, Peer E, Mor N, Manor YS, Ben-Haim MS, Eyal E, Yunger S, Pinto Y, Jaitin DA, Viukov S, Rais Y, Krupalnik V, Chomsky E, Zerbib M, Maza I, Rechavi Y, Massarwa R, Hanna S, Amit I, Levanon EY, Amariglio N, Stern-Ginossar N, Novershtern N, Rechavi G, Hanna JH, m6A mRNA methylation facilitates resolution of naive pluripotency toward differentiation., Science. 347 (2015) 1002–6. doi: 10.1126/science.1261417. [DOI] [PubMed] [Google Scholar]
- [51].Wang Y, Li Y, Yue M, Wang J, Kumar S, Wechsler-Reya RJ, Zhang Z, Ogawa Y, Kellis M, Duester G, Zhao JC, N6-methyladenosine RNA modification regulates embryonic neural stem cell self-renewal through histone modifications, Nat. Neurosci. (2018). doi: 10.1038/s41593-017-0057-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Lin Z, Hsu PJ, Xing X, Fang J, Lu Z, Zou Q, Zhang K-J, Zhang X, Zhou Y, Zhang T, Zhang Y, Song W, Jia G, Yang X, He C, Tong M-H, Mettl3-/Mettll4-mediated mRNA N6-methyladenosine modulates murine spermatogenesis, Cell Res. 27 (2017) 1216–1230. doi: 10.1038/cr.2017.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Xu K, Yang Y, Feng G-H, Sun B-F, Chen J-Q, Li Y-F, Chen Y-S, Zhang X-X, Wang C-X, Jiang L-Y, Liu C, Zhang Z-Y, Wang X-J, Zhou Q, Yang Y-G, Li W, Mettl3-mediated m6A regulates spermatogonial differentiation and meiosis initiation, Cell Res. 27 (2017) 1100–1114. doi: 10.1038/cr.2017.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Haussmann IU, Bodi Z, Sanchez-Moran E, Mongan NP, Archer N, Fray RG, Soller M, m6A potentiates Sxl alternative pre-mRNA splicing for robust Drosophila sex determination, Nature. 540 (2016) 301–304. doi: 10.1038/nature20577. [DOI] [PubMed] [Google Scholar]
- [55].Rottman FM, Bokar JA, Narayan P, Shambaugh ME, Ludwiczak R, N6-adenosine methylation in mRNA: substrate specificity and enzyme complexity., Biochimie. 76 (1994) 1109–14. [DOI] [PubMed] [Google Scholar]
- [56].Linder B, Grozhik AV, Olarerin-George AO, Meydan C, Mason CE, Jaffrey SR, Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome., Nat. Methods. 12 (2015) 767–772. doi: 10.1038/nmeth.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Barbieri I, Tzelepis K, Pandolfini L, Shi J, Millan-Zambrano G, Robson SC, Aspris D, Migliori V, Bannister AJ, Han N, De Braekeleer E, Ponstingl H, Hendrick A, Vakoc CR, Vassiliou GS, Kouzarides T, Promoter-bound METTL3 maintains myeloid leukaemia by m6A-dependent translation control, Nature. 552 (2017) 126–131. doi: 10.1038/nature24678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Huang H, Weng H, Zhou K, Wu T, Zhao BS, Sun M, Chen Z, Deng X, Xiao G, Auer F, Klemm L, Wu H, Zuo Z, Qin X, Dong Y, Zhou Y, Qin H, Tao S, Du J, Liu J, Lu Z, Yin H, Mesquita A, Yuan CL, Hu Y-C, Sun W, Su R, Dong L, Shen C, Li C, Qing Y, Jiang X, Wu X, Sun M, Guan J-L, Qu L, Wei M, Miischen M, Huang G, He C, Yang J, Chen J, Histone H3 trimethylation at lysine 36 guides m6A RNA modification co-transcriptionally, Nature. (2019) 1. doi: 10.1038/s41586-019-1016-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Zhang Z, Chen L-Q, Zhao Y-L, Yang C-G, Roundtree IA, Zhang Z, Ren J, Xie W, He C, Luo G-Z, Single-base mapping of m6A by an antibody-independent method, BioRxiv. (2019) 575555. doi: 10.1101/575555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Garcia-Campos MA, Edelheit S, Toth U, Shachar R, Nir R, Lasman L, Brandis A, Hanna JH, Rossmanith W, Schwartz S, Deciphering the ‘m6A code’ via quantitative profiling of m6A at single-nucleotide resolution, BioRxiv. (2019) 571679. doi: 10.1101/571679. [DOI] [Google Scholar]
- [61].Patil DP, Chen C-K, Pickering BF, Chow A, Jackson C, Guttman M, Jaffrey SR, m6A RNA methylation promotes XIST-mediated transcriptional repression, Nature. 537 (2016) 369–373. doi: 10.1038/nature19342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Liu N, Zhou K.l., Parisien M, Dai Q, Diatchenko L, Pan T, N 6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein, Nucleic Acids Res. 45 (2017) 6051–6063. doi: 10.1093/nar/gkx141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Zhao X, Yang Y, Sun B-F, Shi Y, Yang X, Xiao W, Hao Y-J, Ping X-L, Chen Y-S, Wang W-J, Jin KX, Wang X, Huang C-M, Fu Y, Ge X-M, Song S-H, Jeong HS, Yanagisawa H, Niu Y, Jia G-F, Wu W, Tong W-M, Okamoto A, He C, Danielsen JMR, Wang X-J, Yang Y-G, FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis, Cell Res. 24 (2014) 1403–1419. doi: 10.1038/cr.2014.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Zhao BS, Nachtergaele S, Roundtree IA, He C, Our views of dynamic N6-methyladenosine RNA methylation., RNA. 24 (2018) 268–272. doi: 10.1261/rna.064295.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Zhou J, Wan J, Gao X, Zhang X, Jaffrey SR, Qian S-B, Dynamic m(6)A mRNA methylation directs translational control of heat shock response, Nature. 526 (2015) 591–594. doi:doi: 10.1016/j.chom.2016.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Ke S, Pandya-Jones A, Saito Y, Fak JJ, Vagbp CB, Geula S, Hanna JH, Black DL, Darnell JE, Darnell RB, m(6)A mRNA modifications are deposited in nascent pre-mRNA and are not required for splicing but do specify cytoplasmic turnover., Genes Dev. 31 (2017) 990–1006. doi: 10.1101/gad.301036.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Schwartz S, Mumbach MR, Jovanovic M, Wang T, Maciag K, Bushkin GG, Mertins P, Ter-Ovanesyan D, Habib N, Cacchiarelli D, Sanjana NE, Freinkman E, Pacold ME, Satija R, Mikkelsen TS, Hacohen N, Zhang F, Carr SA, Lander ES, Regev A, Perturbation of m6A Writers Reveals Two Distinct Classes of mRNA Methylation at Internal and 5’ Sites, Cell Rep. 8 (2014) 284–296. doi: 10.1016/j.celrep.2014.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Slobodin B, Han R, Calderone V, Vrielink JAFOFO, Loayza-Puch F, Elkon R, Agami R, Transcription Impacts the Efficiency of mRNA Translation via Co-transcriptional N6-adenosine Methylation, Cell. 169 (2017) 326–337. e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Ma C, Chang M, Lv H, Zhang Z-W, Zhang W, He X, Wu G, Zhao S, Zhang Y, Wang D, Teng X, Liu C, Li Q, Klungland A, Niu Y, Song S, Tong W-M, RNA m6A methylation participates in regulation of postnatal development of the mouse cerebellum, Genome Biol. 19 (2018) 68. doi: 10.1186/s13059-018-1435-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang YY-G, Yi C, Lindahl T, Pan T, Yang YY-G, He C, N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO., Nat. Chem. Biol 7 (2011) 885–7. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Knuckles P, Carl SH, Musheev M, Niehrs C, Wenger A, Biihler M, RNA fate determination through cotranscriptional adenosine methylation and microprocessor binding, Nat. Struct. Mol. Biol (2017). doi: 10.1038/nsmb.3419. [DOI] [PubMed] [Google Scholar]
- [72].Galganski L, Urbanek MO, Krzyzosiak WJ, Nuclear speckles: molecular organization, biological function and role in disease, Nucleic Acids Res. 45 (2017) 10350–10368. doi: 10.1093/nar/gkx759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Wen J, Lv R, Ma H, Shen H, He C, Wang J, Jiao F, Liu H, Yang P, Tan L, Lan F, Shi YG, He C, Shi Y, Diao J, Zc3hl3 Regulates Nuclear RNA m 6 A Methylation and Mouse Embryonic Stem Cell Self-Renewal, Mol. Cell 69 (2018) 1028–1038. e6. doi: 10.1016/j.molcel.2018.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Batista PJ, Molinie B, Wang J, Qu K, Zhang J, Li L, Bouley DM, Lujan E, Haddad B, Daneshvar K, Carter AC, Hynn RA, Zhou C, Lim K-S, Dedon P, Wernig M, Mullen AC, Xing Y, Giallourakis CC, Chang HY, m(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells., Cell Stem Cell. 15 (2014) 707–19. doi: 10.1016/j.stem.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Yu J, Li Y, Wang T, Zhong X, modification of N6-methyladenosine RNA methylation on heat shock protein expression, PLoS One. 13 (2018) e0198604. doi: 10.1371/journal.pone.0198604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Engel M, Eggert C, Kaplick PM, Eder M, Roh S, Tietze L, Namendorf C, Arloth J, Weber P, Rex-Haffner M, Geula S, Jakovcevski M, Hanna JH, Leshkowitz D, Uhr M, Wotjak CT, Schmidt ΜV, Deussing JM, Binder EB, Chen A, The Role of m6A/m-RNA Methylation in Stress Response Regulation, Neuron. 99 (2018) 389–403. e9. doi: 10.1016/j.neuron.2018.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Xiang Y, Laurent B, Hsu C-H, Nachtergaele S, Lu Z, Sheng W, Xu C, Chen H, Ouyang J, Wang S, Ling D, Hsu P-H, Zou L, Jambhekar A, He C, Shi Y, RNA m6A methylation regulates the ultraviolet-induced DNA damage response, Nature. 543 (2017) 573–576. doi: 10.1038/nature21671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Du M, Zhang Y, Mao Y, Mou J, Zhao J, Xue Q, Wang D, Huang J, Gao S, Gao Y, MiR-33a suppresses proliferation of NSCLC cells via targeting METTL3 mRNA, Biochem. Biophys. Res. Commun 482 (2017) 582–589. doi: 10.1016/J.BBRC.2016.11.077. [DOI] [PubMed] [Google Scholar]
- [79].Du Y, Hou G, Zhang H, Dou J, He J, Guo Y, Li L, Chen R, Wang Y, Deng R, Huang J, Jiang B, Xu M, Cheng J, Chen G-Q, Zhao X, Yu J, SUMOylation of the m6A-RNA methyltransferase METTL3 modulates its function, Nucleic Acids Res. (2018). doi: 10.1093/nar/gky156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Sorci M, lanniello Z, Cruciani S, Larivera S, Ginistrelli LC, Capuano E, Marchioni M, Fazi F, Fatica A, METTL3 regulates WTAP protein homeostasis, Cell Death Dis. 9 (2018) 796. doi: 10.1038/s41419-018-0843-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Mauer J, Luo X, Blanjoie A, Jiao X, Grozhik AV, Patil DP, Linder B, Pickering BF, Vasseur J-J, Chen Q, Gross SS, Elemento O, Debart F, Kiledjian M, Jaffrey SR, Reversible methylation of m6Am in the 5’ cap controls mRNA stability, Nature. 541 (2017) 371–375. doi: 10.1038/nature21022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Mauer J, Sindelar M, Despic V, Guez T, Hawley BR, Vasseur J-J, Rentmeister A, Gross SS, Pellizzoni L, Debart F, Goodarzi H, Jaffrey SR, FTO controls reversible m6Am RNA methylation during snRNA biogenesis, Nat. Chem. Biol 15 (2019). doi: 10.1038/s41589-019-0231-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Fedeles B.l., Singh V, Delaney JC, Li D, Essigmann JM, The AlkB Family of Fe(II)/α-Ketoglutarate-dependent Dioxygenases: Repairing Nucleic Acid Alkylation Damage and Beyond, J Biol. Chem 290 (2015) 20734–20742. doi: 10.1074/jbc.R115.656462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang C-M, Li CJ, Vågbø CB, Shi Y, Wang W-L, Song S-H, Lu Z, Bosmans RPG, Dai Q, Hao Y-J, Yang X, Zhao W-M, Tong W-M, Wang X-J, Bogdan F, Furu K, Fu Y, Jia G, Zhao X, Liu J, Krokan HE, Klungland A, Yang Y-G, He C, ALKBH5 Is a Mammalian RNA Demethylase that Impacts RNA Metabolism and Mouse Fertility, Mol. Cell 49 (2013) 18–29. doi: 10.1016/J.MOLCEL.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Baltz AG, Munschauer M, Schwanhäusser B, Vasile A, Murakawa Y, Schueler M, Youngs N, Penfold-Brown D, Drew K, Milek M, Wyler E, Bonneau R, Selbach M, Dieterich C, Landthaler M, The mRNA-Bound Proteome and Its Global Occupancy Profile on Protein-Coding Transcripts, Mol Cell. 46 (2012) 674–690. doi: 10.1016/j.molcel.2012.05.021. [DOI] [PubMed] [Google Scholar]
- [86].Feng C, Liu Y, Wang G, Deng Z, Zhang Q, Wu W, Tong Y, Cheng C, Chen Z, Crystal structures of the human RNA demethylase Alkbh5 reveal basis for substrate recognition., J. Biol. Chem 289 (2014) 11571–83. doi: 10.1074/jbc.M113.546168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Xu C, Liu K, Tempel W, Demetriades M, Aik W, Schofield CJ, Min J, Structures of human ALKBH5 demethylase reveal a unique binding mode for specific single-stranded N6-methyladenosine RNA demethylation., J. Biol. Chem 289 (2014) 17299–311. doi: 10.1074/jbc.M114.550350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Shen L, Song C-X, He C, Zhang Y, Mechanism and Function of Oxidative Reversal of DNA and RNA Methylation, Annu. Rev. Biochem 83 (2014) 585–614. doi: 10.1146/annurev-biochem-060713-035513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Wei J, Liu F, Lu Z, Fei Q, Ai Y, He PC, Shi H, Cui X, Su R, Klungland A, Jia G, Chen J, He C, Differential m6A, m6Am, and m1A Demethylation Mediated by FTO in the Cell Nucleus and Cytoplasm, Mol. Cell. 71 (2018) 973–985. e5. doi: 10.1016/J.MOLCEL.2018.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Su R, Dong L, Li C, Nachtergaele S, Wunderlich M, Qing Y, Deng X, Wang Y, Weng X, Hu C, Yu M, Skibbe J, Dai Q, Zou D, Wu T, Yu K, Weng H, Huang H, Ferchen K, Qin X, Zhang B, Qi J, Sasaki AT, Plas DR, Bradner JE, Wei M, Marcucci G, Jiang X, Mulloy JC, Jin J, He C, Chen J, R-2HG Exhibits Anti-tumor Activity by Targeting FTO/m 6 A/MYC/CEBPA Signaling, Cell. 172 (2018) 90–105. e23. doi: 10.1016/j.cell.2017.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Robbens S, Rouzé P, Cock JM, Spring J, Worden AZ, Van de Peer Y, The FTO Gene, Implicated in Human Obesity, Is Found Only in Vertebrates and Marine Algae, J. Mol. Evol 66 (2008) 80–84. doi: 10.1007/s00239-007-9059-z. [DOI] [PubMed] [Google Scholar]
- [92].Martínez-Pérez M, Aparicio F, López-Gresa MP, Bellés JM, Sánchez-Navarro JA, Pallás V, Arabidopsis m6 A demethylase activity modulates viral infection of a plant virus and the m6 A abundance in its genomic RNAs, Proc. Natl. Acad. Sci 114 (2017) 10755–10760. doi: 10.1073/pnas.1703139114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Duan H-C, Wei L-H, Zhang C, Wang Y, Chen L, Lu Z, Chen PR, He C, Jia G, ALKBH10B Is an RNA N6 -Methyladenosine Demethylase Affecting Arabidopsis Floral Transition, Plant Cell. 29 (2017) 2995–3011. doi: 10.1105/tpc.16.00912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Zou S, Toh JDW, Wong KHQ, Gao Y-G, Hong W, Woon ECY, N6-Methyladenosine: a conformational marker that regulates the substrate specificity of human demethylases FTO and ALKBH5, Sci. Rep 6 (2016) 25677. doi: 10.1038/srep25677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Tang C, Klukovich R, Peng H, Wang Z, Yu T, Zhang Y, Zheng H, Klungland A, Yan W, ALKBH5-dependent m6A demethylation controls splicing and stability of long 3’-UTR mRNAs in male germ cells, Proc. Natl. Acad. Sci 115 (2018) E325–E333. doi: 10.1073/pnas.1717794115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Thalhammer A, Bencokova Z, Poole R, Loenarz C, Adam J, O’Flaherty L, Schödel J, Mole D, Giaslakiotis K, Schofield CJ, Hammond EM, Ratcliffe PJ, Pollard PJ, Human AlkB Homologue 5 Is a Nuclear 2-Oxoglutarate Dependent Oxygenase and a Direct Target of Hypoxia-Inducible Factor 1α (HIF-1α), PLoS One. 6 (2011) e16210. doi: 10.1371/journal.pone.0016210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Zhang S, Zhao BS, Zhou A, Lin K, Zheng S, Lu Z, Chen Y, Sulman EP, Xie K, Bögler O, Majumder S, He C, Huang S, m 6 A Demethylase ALKBH5 Maintains Tumorigenicity of Glioblastoma Stem-like Cells by Sustaining FOXM1 Expression and Cell Proliferation Program, Cancer Cell. 31 (2017) 591–606. e6. doi: 10.1016/j.ccell.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Song H, Feng X, Zhang H, Luo Y, Huang J, Lin M, Jin J, Ding X, Wu S, Huang H, Yu T, Zhang M, Hong H, Yao S, Zhao Y, Zhang Z, METTL3 and ALKBH5 oppositely regulate m6 A modification of TFEB mRNA, which dictates the fate of hypoxia/reoxygenation-treated cardiomyocytes, Autophagy. (2019) 1–19. doi: 10.1080/15548627.2019.1586246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Zhang C, Samanta D, Lu H, Bullen JW, Zhang H, Chen I, He X, Semenza GL, Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m6A-demethylation of NANOG mRNA., Proc. Natl. Acad. Sci. U. S. A 113 (2016) E2047–56. doi: 10.1073/pnas.1602883113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Church C, Moir L, McMurray F, Girard C, Banks GT, Teboul L, Wells S, Brüning JC, Nolan PM, Ashcroft FM, Cox RD, Overexpression of Fto leads to increased food intake and results in obesity, Nat. Genet 42 (2010) 1086–1092. doi: 10.1038/ng.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Wu R, Liu Y, Yao Y, Zhao Y, Bi Z, Jiang Q, Liu Q, Cai M, Wang F, Wang Y, Wang X, FTO regulates adipogenesis by controlling cell cycle progression via m6A-YTHDF2 dependent mechanism, Biochim. Biophys. Acta - Mol. Cell Biol. Lipids 1863 (2018) 1323–1330. doi: 10.1016/J.BBALIP.2018.08.008. [DOI] [PubMed] [Google Scholar]
- [102].Fawcett KA, Barroso I, The genetics of obesity: FTO leads the way, Trends Genet. 26 (2010) 266–274. doi: 10.1016/J.TIG.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Tung YCL, Yeo GSH, O’Rahilly S, Coll AP, Obesity and FTO: Changing Focus at a Complex Locus, Cell Metab. 20 (2014) 710–718. doi: 10.1016/J.CMET.2014.09.010. [DOI] [PubMed] [Google Scholar]
- [104].McIntyre ABR, Gokhale NS, Cerchietti L, Jaffrey SR, Horner SM, Mason CE, Limits in the detection of m6A changes using MeRIP/m6A-seq, BioRxiv. (2019) 657130. doi: 10.1101/657130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Hess ME, Hess S, Meyer KD, Verhagen LAW, Koch L, Brönneke HS, Dietrich MO, Jordan SD, Saletore Y, Elemento O, Belgardt BF, Franz T, Horvath TL, Rüther U, Jaffrey SR, Kloppenburg P, Brüning JC, The fat mass and obesity associated gene (Fto) regulates activity of the dopaminergic midbrain circuitry, Nat. Neurosci. 16 (2013) 1042–1048. doi: 10.1038/nn.3449. [DOI] [PubMed] [Google Scholar]
- [106].Zhou J, Wan J, Shu XE, Mao Y, Liu X-M, Yuan X, Zhang X, Hess ME, Briining JC, Qian S-B, N6-Methyladenosine Guides mRNA Alternative Translation during Integrated Stress Response, Mol. Cell. 69 (2018) 636–647. e7. doi: 10.1016/j.molcel.2018.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Bartosovic M, Molares HC, Gregorova P, Hrossova D, Kudla G, Vanacova S, N6-methyladenosine demethylase FTO targets pre-mRNAs and regulates alternative splicing and 3’-end processing, Nucleic Acids Res. 45 (2017) 11356–11370. doi: 10.1093/nar/gkx778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Darnell RB, Ke S, Darnell JE, Pre-mRNA processing includes N6 methylation of adenosine residues that are retained in mRNA exons and the fallacy of “RNA epigenetics”., RNA. 24 (2018) 262–267. doi: 10.1261/rna.065219.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Mauer J, Jaffrey SR, FTO, m 6 A m , and the hypothesis of reversible epitranscriptomic mRNA modifications, FEBS Lett. 592 (2018) 2012–2022. doi: 10.1002/1873-3468.13092. [DOI] [PubMed] [Google Scholar]
- [110].Shi H, Wang X, Lu Z, Zhao BS, Ma H, Hsu PJ, Liu C, He C, YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA, Cell Res. 27 (2017) 315–328. doi: 10.1038/cr.2017.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Li A, Chen Y-S, Ping X-L, Yang X, Xiao W, Yang Y, Sun H-Y, Zhu Q, Baidya P, Wang X, Bhattarai DP, Zhao Y-L, Sun B-F, Yang Y-G, Cytoplasmic m6A reader YTHDF3 promotes mRNA translation, Cell Res. 27 (2017) 444–447. doi: 10.1038/cr.2017.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Du H, Zhao Y, He J, Zhang Y, Xi H, Liu M, Ma J, Wu L, YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex, Nat. Commun. 7 (2016) 12626. doi: 10.1038/ncommsl2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Wojtas MN, Pandey RR, Mendel M, Homolka D, Sachidanandam R, Pillai RS, Regulation of m 6 A Transcripts by the 3’→5’ RNA Helicase YTHDC2 Is Essential for a Successful Meiotic Program in the Mammalian Germline, Mol. Cell. 68 (2017) 374–387. e12. doi: 10.1016/j.molcel.2017.09.021. [DOI] [PubMed] [Google Scholar]
- [114].Roost C, Lynch SR, Batista PJ, Qu K, Chang HY, Kool ET, Structure and thermodynamics of N6-methyladenosine in RNA: a spring-loaded base modification., J. Am. Chem. Soc 137 (2015) 2107–15. doi: 10.1021/ja513080v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Liu N, Dai Q, Zheng G, He C, Parisien M, Pan T, N6-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions, Nature. 518 (2015) 560–564. doi: 10.1038/nature14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Liu N, Dai Q, Zheng G, He C, Parisien M, Pan T, N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions., Nature. 518 (2015) 560–4. doi: 10.1038/nature14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Zhou K.l., Parisien M, Dai Q, Liu N, Diatchenko L, Sachleben JR, Pan T, N6-methyladenosine modification in a long non-coding RNA hairpin predisposes its conformation to protein binding., J. Mol. Biol (2015). doi: 10.1016/j.jmb.2015.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Wu B, Su S, Patil DP, Liu H, Gan J, Jaffrey SR, Ma J, Molecular basis for the specific and multivariant recognitions of RNA substrates by human hnRNP A2/B1, Nat. Commun. 9 (2018) 420. doi: 10.1038/s41467-017-02770-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Arguello AE, DeLiberto AN, Kleiner RE, RNA Chemical Proteomics Reveals the N 6 - Methyladenosine (m 6 A)-Regulated Protein-RNA Interactome, J. Am. Chem. Soc 139 (2017) 17249–17252. doi: 10.1021/jacs.7b09213. [DOI] [PubMed] [Google Scholar]
- [120].Wang Y, Li Y, Toth J.l., Petroski MD, Zhang Z, Zhao JC, N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells., Nat. Cell Biol 16 (2014) 191–8. doi: 10.1038/ncb2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Zhang B, zur Hausen A, Orlowska-Volk M, Jäger M, Bettendorf H, Stamm S, Hirschfeld M, Yiqin O, Tong X, Gitsch G, Stickeler E, Alternative Splicing-Related Factor YT521, Int. J. Gynecol. Cancer 20 (2010) 492–499. doi: 10.1111/IGC.0b013e3181d66ffe. [DOI] [PubMed] [Google Scholar]
- [122].Imai Y, Matsuo N, Ogawa S, Tohyama M, Takagi T, Cloning of a gene, YT521, for a novel RNA splicing-related protein induced by hypoxia/reoxygenation, Mol. Brain Res. 53 (1998) 33–40. doi: 10.1016/S0169-328X(97)00262-3. [DOI] [PubMed] [Google Scholar]
- [123].Stoilov P, Rafalska I, Stamm S, YTH: a new domain in nuclear proteins., Trends Biochem. Sci. 27 (2002) 495–7. http://www.ncbi.nlm.nih.gov/pubmed/12368078 (accessed March 4, 2019). [DOI] [PubMed] [Google Scholar]
- [124].Zhang Z, Theler D, Kaminska KH, Hiller M, de la Grange P, Pudimat R, Rafalska I, Heinrich B, Bujnicki JM, Allain FH-T, Stamm S, The YTH domain is a novel RNA binding domain., J. Biol. Chem 285 (2010) 14701–10. doi: 10.1074/jbc.M110.104711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Xu C, Wang X, Liu K, Roundtree IA, Tempel W, Li Y, Lu Z, He C, Min J, Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain, Nat. Chem. Biol 10 (2014) 927–929. doi: 10.1038/nchembio.1654. [DOI] [PubMed] [Google Scholar]
- [126].Xu C, Liu K, Ahmed H, Loppnau P, Schapira M, Min J, Structural Basis for the Discriminative Recognition of N6-Methyladenosine RNA by the Human YT521-B Homology Domain Family of Proteins., J. Biol. Chem 290 (2015) 24902–13. doi: 10.1074/jbc.M115.680389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Li F, Zhao D, Wu J, Shi Y, Structure of the YTH domain of human YTHDF2 in complex with an m6A mononucleotide reveals an aromatic cage for m6A recognition, Cell Res. 24 (2014) 1490–1492. doi: 10.1038/cr.2014.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Zhu T, Roundtree IA, Wang P, Wang X, Wang L, Sun C, Tian Y, Li J, He C, Xu Y, Crystal structure of the YTH domain of YTHDF2 reveals mechanism for recognition of N6-methyladenosine, Cell Res. 24 (2014) 1493–1496. doi: 10.1038/cr.2014.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Hsu PJ, Zhu Y, Ma H, Guo Y, Shi X, Liu Y, Qi M, Lu Z, Shi H, Wang J, Cheng Y, Luo G, Dai Q, Liu M, Guo X, Sha J, Shen B, He C, Ythdc2 is an N6-methyladenosine binding protein that regulates mammalian spermatogenesis, Cell Res. (2017). doi: 10.1038/cr.2017.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Lao N, Barron N, Cross-talk between m6A and m1A regulators, YTHDF2 and ALKBH3 fine-tunes mRNA expression, BioRxiv. (2019) 589747. doi: 10.1101/589747. [DOI] [Google Scholar]
- [131].Dai X, Wang T, Gonzalez G, Wang Y, Identification of YTH Domain-Containing Proteins as the Readers for N 1-Methyladenosine in RNA, Anal. Chem. 90 (2018) 6380–6384. doi: 10.1021/acs.analchem.8b01703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Zhang Y, Wang X, Zhang X, Wang J, Ma Y, Zhang L, Cao X, RNA-binding protein YTHDF3 suppresses interferon-dependent antiviral responses by promoting FOX03 translation., Proc. Natl. Acad. Sci. U. S. A 116 (2019) 976–981. doi: 10.1073/pnas.1812536116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Miiller S, GlaR M, Singh AK, Haase J, Bley N, Fuchs T, Lederer M, Dahl A, Huang H, Chen J, Posern G, Hiittelmaier S, IGF2BP1 promotes SRF-dependent transcription in cancer in a m6A-and miRNA-dependent manner, Nucleic Acids Res. (2018). doi: 10.1093/nar/gkyl012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Huang H, Weng H, Sun W, Qin X, Shi H, Wu H, Zhao BS, Mesquita A, Liu C, Yuan CL, Hu Y-C, Hüttelmaier S, Skibbe JR, Su R, Deng X, Dong L, Sun M, Li C, Nachtergaele S, Wang Y, Hu C, Ferchen K, Greis KD, Jiang X, Wei M, Qu L, Guan J-L, He C, Yang J, Chen J, Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation, Nat. Cell Biol. 20 (2018) 285–295. doi: 10.1038/s41556-018-0045-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Meyer KD, Patil DP, Zhou J, Zinoviev A, Skabkin MA, Elemento O, Pestova TV, Qian S-B, Jaffrey SR, 5’ UTR m6A Promotes Cap-Independent Translation, Cell. 163 (2015) 999–1010. doi: 10.1016/j.cell.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Wu R, Li A, Sun B, Sun J-G, Zhang J, Zhang T, Chen Y, Xiao Y, Gao Y, Zhang Q, Ma J, Yang X, Liao Y, Lai W-Y, Qi X, Wang S, Shu Y, Wang H-L, Wang F, Yang Y-G, Yuan Z, A novel m6A reader Prrc2a controls oligodendroglial specification and myelination, Cell Res. 29 (2019) 23–41. doi: 10.1038/s41422-018-0113-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Edupuganti RR, Geiger S, Lindeboom RGH, Shi H, Hsu PJ, Lu Z, Wang S-Y, Baltissen MPA, Jansen PWTC, Rossa M, Miiller M, Stunnenberg HG, He C, Carell T, Vermeulen M, N6-methyladenosine (m6A) recruits and repels proteins to regulate mRNA homeostasis, Nat. Struct. Mol. Biol 24 (2017) 870–878. doi: 10.1038/nsmb.3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Zhang F, Kang Y, Wang M, Li Y, Xu T, Yang W, Song H, Wu H, Shu Q, Jin P, Fragile X mental retardation protein modulates the stability of its m6A-marked messenger RNA targets, Hum. Mol. Genet 27 (2018) 3936–3950. doi: 10.1093/hmg/ddy292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Alarcón CR, Goodarzi H, Lee H, Liu X, Tavazoie S, Tavazoie SF, HNRNPA2B1 Is a Mediator of m6A-Dependent Nuclear RNA Processing Events, Cell. 162 (2015) 1299–1308. doi: 10.1016/j.cell.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Hirschfeld M, Zhang B, Jaeger M, Stamm S, Erbes T, Mayer S, Tong X, Stickeler E, Hypoxia-dependent mRNA expression pattern of splicing factor YT521 and its impact on oncological important target gene expression, Mol. Carcinog. 53 (2014) 883–892. doi: 10.1002/mc.22045. [DOI] [PubMed] [Google Scholar]
- [141].Gruber AR, Martin G, Keller W, Zavolan M, Cleavage factor I m is a key regulator of 3’ UTR length, RNA Biol. 9 (2012) 1405–1412. doi: 10.4161/rna.22570. [DOI] [PubMed] [Google Scholar]
- [142].Camper SA, Albers RJ, Coward JK, Rottman FM, Effect of undermethylation on mRNA cytoplasmic appearance and half-life., Mol. Cell. Biol 4 (1984) 538–43. doi: 10.1128/MCB.4.3.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Ratnadiwakara M, Archer SK, Dent C.l., Ruiz De Los Mozos I, Beilharz TH, Knaupp AS, Nefzger CM, Polo JM, Anko M-L, SRSF3 promotes pluripotency through Nanog mRNA export and coordination of the pluripotency gene expression program, Elife. 7 (2018). doi: 10.7554/eLife.37419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Rafalska I, Zhang Z, Benderska N, Wolff H, Hartmann AM, Brack-Werner R, Stamm S, The intranuclear localization and function of YT521-B is regulated by tyrosine phosphorylation, Hum. Mol. Genet 13 (2004) 1535–1549. doi: 10.1093/hmg/ddhl67. [DOI] [PubMed] [Google Scholar]
- [145].Weng Y-L, Wang X, An R, Cassin J, Vissers C, Liu Y, Liu Y, Xu T, Wang X, Wong SZH, Joseph J, Dore LC, Dong Q, Zheng W, Jin P, Wu H, Shen B, Zhuang X, He C, Liu K, Song H, Ming G, Epitranscriptomic m 6 A Regulation of Axon Regeneration in the Adult Mammalian Nervous System, Neuron. 97 (2018) 313–325. e6. doi: 10.1016/j.neuron.2017.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Zhuang M, Li X, Zhu J, Zhang J, Niu F, Liang F, Chen M, Li D, Han P, Ji S-J, The m6A reader YTHDF1 regulates axon guidance through translational control of Robo3.1 expression, Nucleic Acids Res. 47 (2019) 4765–4777. doi: 10.1093/nar/gkzl57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Shi H, Zhang X, Weng Y-L, Lu Z, Liu Y, Lu Z, Li J, Hao P, Zhang Y, Zhang F, Wu Y, Delgado JY, Su Y, Patel MJ, Cao X, Shen B, Huang X, Ming G, Zhuang X, Song H, He C, Zhou T, m6A facilitates hippocampus-dependent learning and memory through YTHDF1, Nature. 563 (2018) 249–253. doi: 10.1038/s41586-018-0666-l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Kretschmer J, Rao H, Hackert P, Sloan KE, Hobartner C, Bohnsack MT, The m6A reader protein YTHDC2 interacts with the small ribosomal subunit and the 5’−3’ exoribonuclease XRN1., RNA. (2018) rna.064238.117. doi: 10.1261/rna.064238.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Bailey AS, Batista PJ, Gold RS, Chen YG, de Rooij DG, Chang HY, Fuller MT, The conserved RNA helicase YTHDC2 regulates the transition from proliferation to differentiation in the germline, Elife. 6 (2017). doi: 10.7554/eLife.26116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Lin S, Choe J, Du P, Triboulet R, Gregory R.l., The m6A Methyltransferase METTL3 Promotes Translation in Human Cancer Cells, Mol. Cell. 62 (2016) 335–345. doi: 10.1016/j.molcel.2016.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Coots RA, Liu X-M, Mao Y, Dong L, Zhou J, Wan J, Zhang X, Qian S-B, m6A Facilitates elF4F-Independent mRNA Translation., Mol. Cell. 68 (2017) 504–514. e7. doi: 10.1016/j.molcel.2017.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Anders M, Chelysheva I, Goebel I, Trenkner T, Zhou J, Mao Y, Verzini S, Qian S-B, Ignatova Z, Dynamic m 6 A methylation facilitates mRNA triaging to stress granules, Life Sci. Alliance. 1 (2018) e201800113. doi: 10.26508/lsa.201800113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, Jin Y, Yang Y, Chen L, Wang Y, Wong CC, Xiao X, Wang Z, Extensive translation of circular RNAs driven by N6-methyladenosine, Cell Res. (2017). doi: 10.1038/cr.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Jain D, Puno MR, Meydan C, Lailler N, Mason CE, Lima CD, Anderson KV, Keeney S, ketu mutant mice uncover an essential meiotic function for the ancient RNA helicase YTHDC2, Elife. 7 (2018). doi: 10.7554/eLife.30919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Tanabe A, Tanikawa K, Tsunetomi M, Takai K, Ikeda H, Konno J, Torigoe T, Maeda H, Kutomi G, Okita K, Mori M, Sahara H, RNA helicase YTHDC2 promotes cancer metastasis via the enhancement of the Efficiency by which HIF-1α mRNA is translated, Cancer Lett. 376 (2016) 34–42. doi: 10.1016/j.canlet.2016.02.022. [DOI] [PubMed] [Google Scholar]
- [156].Degrauwe N, Suva M-L, Janiszewska M, Riggi N, Stamenkovic I, IMPs: an RNA-binding protein family that provides a link between stem cell maintenance in normal development and cancer., Genes Dev. 30 (2016) 2459–2474. doi: 10.1101/gad.287540.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Weinlich S, Hüttelmaier S, Schierhorn A, Behrens S-E, Ostareck-Lederer A, Ostareck DH, IGF2BP1 enhances HCV IRES-mediated translation initiation via the 3’UTR., RNA. 15 (2009) 1528–42. doi: 10.1261/rna.1578409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Łabno A, Tomecki R, Dziembowski A, Cytoplasmic RNA decay pathways - Enzymes and mechanisms, Biochim. Biophys. Acta - Mol. Cell Res 1863 (2016) 3125–3147. doi: 10.1016/j.bbamcr.2016.09.023. [DOI] [PubMed] [Google Scholar]
- [159].Park OH, Ha H, Lee Y, Boo SH, Kwon DH, Song HK, Kim YK, Endoribonucleolytic Cleavage of m6A-Containing RNAs by RNase P/MRP Complex, Mol. Cell (2019). doi: 10.1016/J.MOLCEL.2019.02.034. [DOI] [PubMed] [Google Scholar]
- [160].Li Z, Qian P, Shao W, Shi H, He XC, Gogol M, Yu Z, Wang Y, Qi M, Zhu Y, Perry JM, Zhang K, Tao F, Zhou K, Hu D, Han Y, Zhao C, Alexander R, Xu H, Chen S, Peak A, Hall K, Peterson M, Perera A, Haug JS, Parmely T, Li H, Shen B, Zeitlinger J, He C, Li L, Suppression of m6A reader Ythdf2 promotes hematopoietic stem cell expansion, Cell Res. (2018). doi: 10.1038/s41422-018-0072-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Chen J, Sun Y, Xu X, Wang D, He J, Zhou H, Lu Y, Zeng J, Du F, Gong A, Xu M, YTH domain family 2 orchestrates epithelial-mesenchymal transition/proliferation dichotomy in pancreatic cancer cells, Cell Cycle. 16 (2017) 2259–2271. doi: 10.1080/15384101.2017.1380125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Abby E, Tourpin S, Ribeiro J, Daniel K, Messiaen S, Moison D, Guerquin J, Gaillard J-C, Armengaud J, Langa F, Toth A, Martini E, Livera G, Implementation of meiosis prophase I programme requires a conserved retinoid-independent stabilizer of meiotic transcripts, Nat. Commun. 7 (2016) 10324. doi: 10.1038/ncommsl0324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Soh YQS, Mikedis MM, Kojima M, Godfrey AK, de Rooij DG, Page DC, Meioc maintains an extended meiotic prophase I in mice, PLOS Genet. 13 (2017) e1006704. doi: 10.1371/journal.pgen.1006704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Bodi Z, Bottley A, Archer N, May ST, Fray RG, Yeast m6A Methylated mRNAs Are Enriched on Translating Ribosomes during Meiosis, and under Rapamycin Treatment, PLoS One. 10 (2015) e0132090. doi: 10.1371/journal.pone.0132090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Chia M, van Werven FJ, Temporal Expression of a Master Regulator Drives Synchronous Sporulation in Budding Yeast., G3 (Bethesda). 6 (2016) 3553–3560. doi: 10.1534/g3.116.034983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Agarwala SD, Blitzblau HG, Hochwagen A, Hnk GR, RNA Methylation by the MIS Complex Regulates a Cell Fate Decision in Yeast, PLoS Genet. 8 (2012) e1002732. doi: 10.1371/journal.pgen.1002732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Braun JE, Truffault V, Boland A, Huntzinger E, Chang C-T, Haas G, Weichenrieder O, Coles M, Izaurralde E, A direct interaction between DCP1 and XRN1 couples mRNA decapping to 5’ exonucleolytic degradation, Nat. Struct. Mol. Biol 19 (2012) 1324–1331. doi: 10.1038/nsmb.2413. [DOI] [PubMed] [Google Scholar]
- [168].Solinger JA, Pascolini D, Heyer WD, Active-site mutations in the Xrnlp exoribonuclease of Saccharomyces cerevisiae reveal a specific role in meiosis., Mol. Cell. Biol 19 (1999) 5930–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Yang Z, Li J, Feng G, Gao S, Wang Y, Zhang S, Liu Y, Ye L, Li Y, Zhang X, MicroRNA-145 Modulates N6 methyladenosine Levels by Targeting the 3’ Untranslated mRNA Region of the N6-methyladenosine binding YTH Domain Family 2 Protein, J. Biol. Chem (2017) jbc.M116.749689. doi: 10.1074/jbc.M116.749689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Li J, Meng S, Xu M, Wang S, He L, Xu X, Wang X, Xie L, Downregulation of N6-methyladenosine binding YTHDF2 protein mediated by miR-493-3p suppresses prostate cancer by elevating N6-methyladenosine levels., Oncotarget. 9 (2018) 3752–3764. doi: 10.18632/oncotarget.23365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Tanabe A, Konno J, Tanikawa K, Sahara H, Transcriptional machinery of TNF-α-inducible YTH domain containing 2 (YTHDC2) gene, Gene. 535 (2014) 24–32. doi: 10.1016/j.gene.2013.11.005. [DOI] [PubMed] [Google Scholar]
- [172].Zhang C, Chen Y, Sun B, Wang L, Yang Y, Ma D, Lv J, Heng J, Ding Y, Xue Y, Lu X, Xiao W, Yang YG, Liu F, m6A modulates haematopoietic stem and progenitor cell specification, Nature. (2017). doi: 10.1038/nature23883. [DOI] [PubMed] [Google Scholar]
- [173].Chen J, Zhang Y-C, Huang C, Shen H, Sun B, Cheng X, Zhang Y-J, Yang Y-G, Shu Q, Yang Y, Li X, m6A Regulates Neurogenesis and Neuronal Development by Modulating Histone Methyltransferase Ezh2, Genomics. Proteomics Bioinformatics. (2019). doi: 10.1016/J.GPB.2018.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Li M, Zhao X, Wang W, Shi H, Pan Q, Lu Z, Perez SP, Suganthan R, He C, Bjpras M, Klungland A, Ythdf2-mediated m6A mRNA clearance modulates neural development in mice, Genome Biol. 19 (2018) 69. doi: 10.1186/sl3059-018-1436-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Wu Y, Xie L, Wang M, Xiong Q, Guo Y, Liang Y, Li J, Sheng R, Deng P, Wang Y, Zheng R, Jiang Y, Ye L, Chen Q, Zhou X, Lin S, Yuan Q, Mettl3-mediated m6A RNA methylation regulates the fate of bone marrow mesenchymal stem cells and osteoporosis, Nat. Commun. 9 (2018) 4772. doi: 10.1038/s41467-018-06898-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Wang C-X, Cui G-S, Liu X, Xu K, Wang M, Zhang X-X, Jiang L-Y, Li A, Yang Y, Lai W-Y, Sun B-F, Jiang G-B, Wang H-L, Tong W-M, Li W, Wang X-J, Yang Y-G, Zhou Q, METTL3-mediated m6A modification is required for cerebellar development, PLOS Biol. 16 (2018) e2004880. doi: 10.1371/journal.pbio.2004880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Yao QJ, Sang L, Lin M, Yin X, Dong W, Gong Y, Zhou BO, Mettl3-Mettll4 methyltransferase complex regulates the quiescence of adult hematopoietic stem cells, Cell Res. 28 (2018) 952–954. doi: 10.1038/s41422-018-0062-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Weng H, Huang H, Wu H, Qin X, Zhao BS, Dong L, Shi H, Skibbe J, Shen C, Hu C, Sheng Y, Wang Y, Wunderlich M, Zhang B, Dore LC, Su R, Deng X, Ferchen K, Li C, Sun M, Lu Z, Jiang X, Marcucci G, Mulloy JC, Yang J, Qian Z, Wei M, He C, Chen J, METTL14 Inhibits Hematopoietic Stem/Progenitor Differentiation and Promotes Leukemogenesis via mRNA m6A modification, Cell Stem Cell. 22 (2018) 191–205. e9. doi: 10.1016/J.STEM.2017.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Wang H, Zuo H, Liu J, Wen F, Gao Y, Zhu X, Liu B, Xiao F, Wang W, Huang G, Shen B, Ju Z, Loss of YTHDF2-mediated m6A-dependent mRNA clearance facilitates hematopoietic stem cell regeneration, Cell Res. 28 (2018) 1035–1038. doi: 10.1038/s41422-018-0082-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Lv J, Zhang Y, Gao S, Zhang C, Chen Y, Li W, Yang Y-G, Zhou Q, Liu F, Endothelial-specific m6A modulates mouse hematopoietic stem and progenitor cell development via Notch signaling, Cell Res. 28 (2018) 249–252. doi: 10.1038/cr.2017.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Sha Q-Q, Zhang J, Fan H-Y, A story of birth and death: mRNA translation and clearance at the onset of maternal-to-zygotic transition in mammalst, Biol. Reprod. (2019). doi: 10.1093/biolre/ioz012. [DOI] [PubMed] [Google Scholar]
- [182].Yoon K-J, Ringeling FR, Vissers C, Jacob F, Pokrass M, Jimenez-Cyrus D, Su Y, Kim N-S, Zhu Y, Zheng L, Kim S, Wang X, Dore LC, Jin P, Regot S, Zhuang X, Canzar S, He C, Ming G, Song H, Temporal Control of Mammalian Cortical Neurogenesis by m6A Methylation, Cell. 171 (2017) 877–889. e17. doi: 10.1016/J.CELL.2017.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].Ebert MS, Sharp PA, Roles for MicroRNAs in Conferring Robustness to Biological Processes, Cell. 149 (2012) 515–524. doi: 10.1016/j.cell.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Crisp PA, Ganguly DR, Smith AB, Murray KD, Estavillo GM, Searle I, Ford E, Bogdanović O, Lister R, Borevitz JO, Eichten SR, Pogson BJ, Rapid Recovery Gene Downregulation during Excess-Light Stress and Recovery in Arabidopsis., Plant Cell. 29 (2017) 1836–1863. doi: 10.1105/tpc.16.00828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].Eichelbaum K, Krijgsveld J, Rapid temporal dynamics of transcription, protein synthesis, and secretion during macrophage activation., Mol. Cell. Proteomics 13 (2014) 792–810. doi: 10.1074/mcp.M113.030916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [186].Ke S, Alemu EA, Mertens C, Gantman EC, Fak JJ, Mele A, Haripal B, Zucker-Scharff I, Moore MJ, Park CY, Vågbø CB, Kusśnierczyk A, Klungland A, Darnell JE, Darnell RB, A majority of m 6 A residues are in the last exons, allowing the potential for 3’ UTR regulation, Genes Dev. 29 (2015) 2037–2053. doi: 10.1101/gad.269415.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [187].Molinie B, Wang J, Lim KS, Hillebrand R, Lu Z-X, Van Wittenberghe N, Howard BD, Daneshvar K, Mullen AC, Dedon P, Xing Y, Giallourakis CC, m(6)A-LAIC-seq reveals the census and complexity of the m(6)A epitranscriptome., Nat. Methods. (2016). doi: 10.1038/nmeth.3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [188].Liu N, Parisien M, Dai Q, Zheng G, He C, Pan T, Probing N6-methyladenosine RNA modification status at single nucleotide resolution in mRNA and long noncoding RNA., RNA. 19 (2013) 1848–56. doi: 10.1261/rna.041178.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [189].Liu H, Begik O, Lucas MC, Mason CE, Schwartz S, Mattick JS, Smith MA, Novoa EM, Accurate detection of m6A RNA modifications in native RNA sequences, BioRxiv. (2019) 525741. doi: 10.1101/525741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [190].Shiraki N, Shiraki Y, Tsuyama T, Obata F, Miura M, Nagae G, Aburatani H, Kume K, Endo F, Kume S, Methionine Metabolism Regulates Maintenance and Differentiation of Human Pluripotent Stem Cells, Cell Metab. 19 (2014) 780–794. doi: 10.1016/j.cmet.2014.03.017. [DOI] [PubMed] [Google Scholar]
- [191].Sperber H, Mathieu J, Wang Y, Ferreccio A, Hesson J, Xu Z, Hscher KA, Devi A, Detraux D, Gu H, Battle SL, Showalter M, Valensisi C, Bielas JH, Ericson NG, Margaretha L, Robitaille AM, Margineantu D, Hehn O, Hockenbery D, Blau CA, Raftery D, Margolin AA, Hawkins RD, Moon RT, Ware CB, Ruohola-Baker H, The metabolome regulates the epigenetic landscape during naive-to-primed human embryonic stem cell transition, Nat. Cell Biol. 17 (2015) 1523–1535. doi: 10.1038/ncb3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [192].Pendleton KE, Park S-K, Hunter OV, Bresson SM, Conrad NK, Balance between MAT2A intron detention and splicing is determined cotranscriptionally., RNA. 24 (2018) 778–786. doi: 10.1261/rna.064899.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [193].Mendel M, Chen K-M, Homolka D, Gos P, Pandey RR, McCarthy AA, Pillai RS, Methylation of Structured RNA by the m6A Writer METTL16 Is Essential for Mouse Embryonic Development, Mol. Cell. 71 (2018) 986–1000. e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [194].Sun H, Zhang M, Li K, Bai D, Yi C, Cap-specific, terminal N6-methylation by a mammalian m6Am methyltransferase, Cell Res. 29 (2019) 80–82. doi: 10.1038/s41422-018-0117-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.