Summary
Tetherin is a host defense factor that physically prevents virion release from the plasma membrane. However, the Nef accessory protein of SIV engages the clathrin adaptor AP-2 to downregulate tetherin via its DIWK motif. As human tetherin lacks DIWK, antagonism of tetherin by Nef is a barrier to simian-human transmission of non-human primate lentiviruses. To determine the molecular basis for tetherin counteraction, we reconstituted the AP-2 complex with a simian tetherin and SIV Nef, and determined its structure by cryo-EM. Nef refolds the first α-helix of the β2 subunit of AP-2 to a β hairpin, creating a binding site for the DIWK sequence. The tetherin binding site in Nef is distinct from those of most other Nef substrates, including MHC-I, CD3, and CD4, but overlaps with the site for the restriction factor SERINC5. This structure explains the dependence of SIVs on tetherin DIWK and consequent barrier to human transmission.
Keywords: HIV, SIV, clathrin, adaptor protein, host-factor restriction, trafficking, cryo-EM
eTOC blurb
Simian to human lentiviral transmissions require adaptations in tetherin antagonism by viral Nef proteins. Buffalo et al. present the cryo-EM structure of SIV Nef complexed with tetherin and AP-2, providing insight into the importance of the tetherin DIWK motif and its role as a barrier to SIV spread in humans.
Graphical Abstract
Introduction
Human and simian immunodeficiency viruses (HIV and SIV) depend on factors known as accessory viral proteins for their efficient replication and spread (Collins and Collins, 2014). Lentiviral accessory proteins, including Nef, Vpu, Vif, Vpr, and Vpx, counteract host defenses by redirecting cellular pathways for the benefit of the virus (Collins and Collins, 2014; Malim and Bieniasz, 2012). In some cases, activities of these viral proteins overlap or compensate for one another to ensure viral fitness. The counteraction of the host factor tetherin by lentiviral Nef, Env, and Vpu is a prominent example (Sauter et al., 2009). Tetherin (also known as BST2) is an interferon-induced antiviral restriction factor that physically tethers nascent virions to the plasma membrane of the viral producer cell, preventing release and marking the cell for immune detection (Neil et al., 2008). The sequences of tetherin orthologs vary considerably between even closely related species and tetherin is counteracted by viral accessory proteins in a species-specific manner (Jia et al., 2009; Zhang et al., 2009). Most notably, the lack of an otherwise conserved (G/D)DIWK motif in human tetherin confers resistance to downregulation by the viral protein Nef used by most SIVs to counteract this restriction factor. Thus, antagonism of tetherin is a major barrier to successful simian-human transmission of non-human primate lentiviruses, the origin of HIV (Sauter and Kirchhoff, 2019). Here, we elucidate the structural basis for antagonism of (G/D)DIWK-containing simian tetherin by SIV Nef in near-atomic detail.
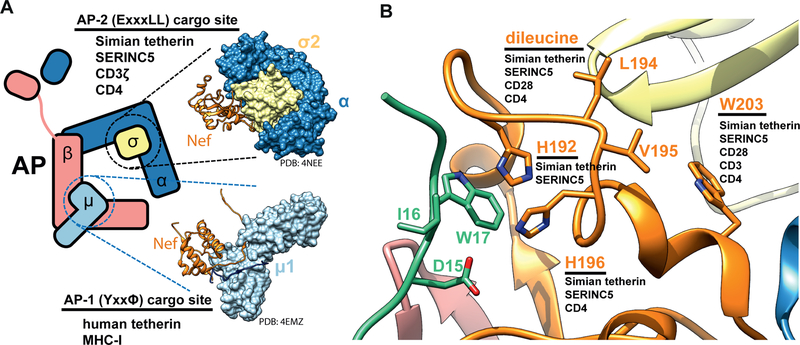
The accessory proteins Nef and Vpu of HIV and SIV reconfigure the membrane protein repertoire of host cells by hijacking clathrin-coated vesicles (CCVs) to sequester and/or degrade target cargoes (Pereira and daSilva, 2016). The heterotetrameric adaptor protein (AP) complexes are primarily responsible for connecting cargo and membranes to clathrin. The trans-Golgi network (TGN) and plasma membrane complexes AP-1 and AP-2, respectively, are major direct targets of Nef and Vpu (Collins and Collins, 2014; Pereira and daSilva, 2016). APs consists of two large solenoidal subunits (α or γ and β), a medium subunit μ, and a small subunit σ (Traub and Bonifacino, 2013). All known host cargoes interact with APs via either tyrosine-based sorting signals (YxxΦ) that bind to the C-terminal domain (CTD) of μ or dileucine signals (ExxxL(L/V)) that bind to a hemicomplex of α/γ and σ (Traub and Bonifacino, 2013). Nef and Vpu both use (ExxxL(L/V)) motifs to hijack AP complexes (Aiken et al., 1994; Bresnahan et al., 1998; Craig et al., 1998; Jia et al., 2014). In solution, the binding sites on the AP complexes for the YxxΦ and ExxxL(L/V) motifs are hidden in the locked conformation of APs (Canagarajah et al., 2013; Jackson et al., 2010). Upon activation by the small GTPase Arf1 at TGN membranes (Ren et al., 2013) or the lipid PI(4,5)P2 at the plasma membrane (PM) (Jackson et al., 2010), the μ CTD undergoes a large conformational change, unlocking and activating the complex.
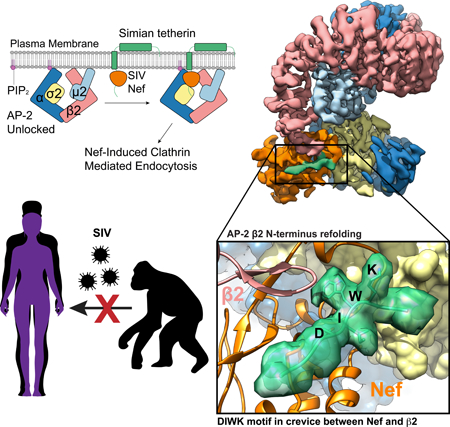
To overcome barriers to cross-species transmission during primate lentiviral evolution, the task of antagonizing tetherin has shifted repeatedly between Nef and Vpu (Sauter and Kirchhoff, 2019). Most SIV including SIVcpz, SIVgor, and SIVsmm, the direct precursors of HIV-1 and HIV-2, respectively, use their Nef protein to target AP-2 at the PM by a mechanism whose details are still unknown (Jia et al., 2009; Sauter et al., 2009; Schmokel et al., 2011; Zhang et al., 2011). Pandemic group M HIV-1 strains switched from Nef to Vpu to hijack AP-1 at the TGN, using its dileucine motif to unlock AP-1 and free the μ CTD to target a variant Tyr motif in the cytosolic tail of tetherin (Jia et al., 2014; Kueck et al., 2015). In contrast, Nef proteins from the epidemic HIV-1 group O evolved to interact with a region in human tetherin sequence adjacent to the (G/D)DIWK deletion bridging the μ and σ subunits of AP-1 in trans and promoting a unique trimeric assembly of AP-1 to sequester tetherin at the TGN (Kluge et al., 2014; Morris et al., 2018). The remaining N and P groups of HIV-1 have gained little if any activity against human tetherin and were only detected in a few individuals. Here, we report on the Nef of SIVsmm infecting sooty mangabeys. SIVsmm uses Nef and its envelope (Env) glycoprotein as tetherin antagonist in is natural simian host and represents the direct ancestor of HIV-2 and of SIVmac causing simian AIDS in rhesus macaques (Sauter and Kirchhoff, 2019). SIVsmm managed to cross the species barrier from monkeys to humans on at least 9 occasions possibly because its Env protein shows some activity against human tetherin (Heusinger et al., 2018). However, the Nef proteins of HIV-2 gained little if any activity against human tetherin (Heusinger et al., 2018; Le Tortorec and Neil, 2009) and HIV-2 shows lower transmission rates and is much less prevalent than HIV-1. We show by cryo-EM and allied methods that simian tetherin is targeted in a unique way by the induced refolding of the β subunit of AP-2 from an α-helix to a β-hairpin, creating a binding site for the (G/D)DIWK motif in a crevice between the new β-hairpin and Nef.
Results
Cooperative interaction between AP-2, simian tetherin and SIVsmm Nef
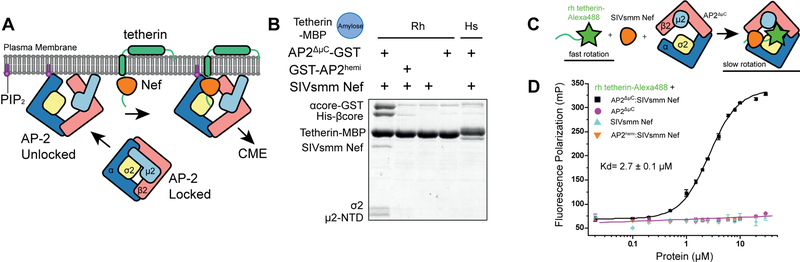
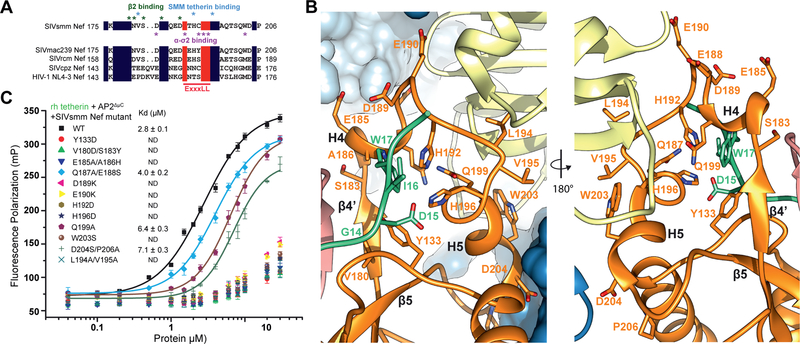
In the absence of the lipid membrane, the AP-2 core adopts a locked conformation that obscures the binding sites for recruiting both physiological cargoes and Nef (Fig. 1A). In order to promote the open conformation of AP-2 in the absence of lipid membranes, we generated the unlocked tetrameric core construct AP2ΔμC, in which the μ2 CTD that is dispensable for Nef binding to AP-2 (Chaudhuri et al., 2007), is deleted. We compared the SIVsmm Nef and simian tetherin binding of this construct to that of the AP-2 α-σ2 hemicomplex (“AP2hemi”). AP2hemi binds tightly to HIV-1 NL4–3 Nef in the absence of cargo (Ren et al., 2014), but binds only very weakly to SIVmac239 Nef (96–237) and rhesus (rh) tetherin (Serra-Moreno et al., 2013). By tetherin tail-MBP pull down assay, we observed a tight cooperative interaction between rh tetherin, SIVsmm Nef and AP2ΔμC in combination. In contrast, the tetherin tail did neither bind to AP2ΔμC or SIVsmm Nef alone nor interact with AP2hemi, even in the presence of SIVsmm Nef (Fig 1B). We used fluorescence polarization (FP) to determine that Alexa488-tetherin tail bound to AP2ΔμC and SIVsmm Nef with Kd = 2.7 μM ± 0.1 μM (Fig 1C, D), while binding to the separate components or to AP2hemi was undetectable. These data indicate that simian tetherin binds to the AP-2 core and SIVsmm Nef in a cooperative and high affinity ternary complex that requires elements of the full AP-2 tetramer.
Figure 1. Reconstitution of the AP2ΔμC:SMM tetherin-SIVsmm Nef complex.
(A) Cartoon representation of AP-2 unlocking at the plasma membrane when it interacts with PI(4,5)P2. Nef recruits tetherin to the unlocked AP-2 complex where clathrin mediated endocytosis (CME) will result in Nef-induced downregulation of tetherin from the plasma membrane. (B) Simian tetherin tail synergistically binds to AP-2 and SIVsmm Nef. Amylose resin was used to pull down tetherin tail-MBP (rhesus or human), SIVsmm Nef and AP2ΔμC or the α-σ2 hemi complex. The pull-down results were visualized by SDS-PAGE and Coomassie staining. (C) Upper panel: Cartoon representation of FP assay. Alexa488 labeled tetherin tail rotates much slower and further causes FP signal increasing while binding to large AP2ΔμC and SIVsmm Nef. Lower panel: Fluorescence polarization result indicates the cooperative interaction between AP2ΔμC, rh tetherin and SIVsmm Nef. Fluorescence polarization (in units of mP) is plotted as a function of AP2ΔμC/SIVsmm Nef concentration (μM) using a logarithmic scale.
CryoEM structure of the AP2ΔμC, SMM tetherin and SIVsmm Nef complex
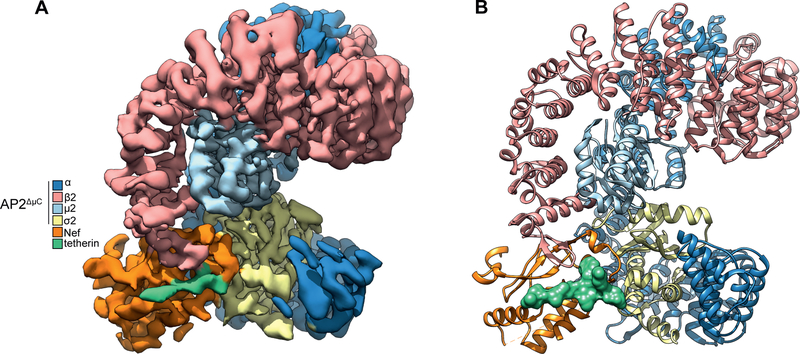
The structure of the complex of AP2ΔμC cross-linked to the tetherin-SIVsmm Nef complex was determined by cryo-EM at a resolution of 3.8 Å (Fig. 2A, B, S1–3, Movie S1). Side-chain density was visible throughout most of the density map, including most of all four subunits of AP-2, SIVsmm Nef, and SMM tetherin. An atomic model was built on the basis of the crystal structures of unlocked AP-2 (Jackson et al., 2010) and NL4–3 HIV-1 Nef bound to AP2hemi (Ren et al., 2014). The tetherin tail and unique regions of SIVsmm Nef were built ab initio (Fig. 3A–C). The conformation of the dileucine loop in the cross-linked SIVsmm Nef complex is essentially identical to the conformation previously seen in the uncrosslinked NL4–3 complex (Ren et al., 2014) (Fig. 3D), showing that the presence of the crosslink did not perturb the structure. The overall conformation of AP2ΔμC is similar to that of the unlocked AP-2 core (Jackson et al., 2010), with the pivotal exception of the β2 N-terminus.
Figure 2. Atomic model of the AP2ΔμC:SMM tetherin-SIVsmm Nef complex.
(D) Cryo-EM density map of AP2ΔμC bound to SIVsmm Nef and the SMM tetherin cytoplasmic tail (E) Ribbon model of AP2ΔμC bound to SIVsmm Nef and the SMM tetherin cytoplasmic tail build from the AP2ΔμC:SMM tetherin-SIVsmm Nef cryo-EM density map. See also Figure S1–3, Table S1
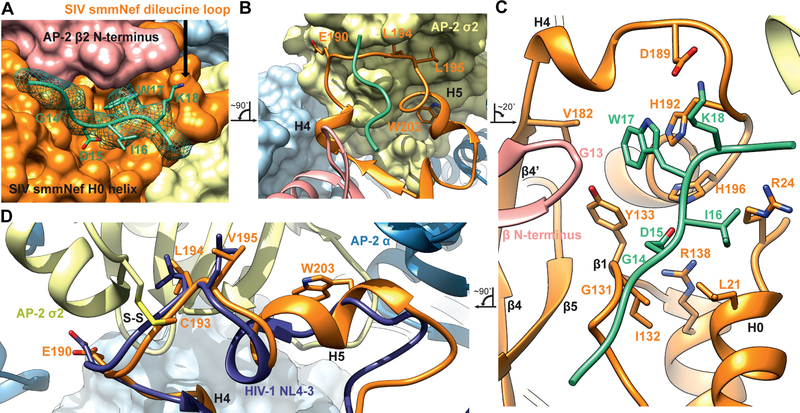
Figure 3. Structural model for cooperative binding of SMM tetherin and to SIVsmm Nef to AP2ΔμC.
(A) Tetherin 14GDIWK18 motif rests in a pocket created by the SIVsmm Nef dileucine loop. The GDIWK motif is further sandwiched by the AP2ΔμC β2 N-terminus and SIVsmm Nef H0 helix. The cryo-EM map density is depicted as a mesh around the SMM tetherin peptide while the Nef and AP-2 are depicted as space filling surface representations. (B) Ribbon representation of the SIVsmm Nef dileucine loop (190ExxxLV195) bound to the dileucine binding site of the AP2ΔμC α-σ2. SMM tetherin peptide is shown to highlight its position relative to the SIVsmm Nef dileucine loop. Fig 3B is rotated ~90° counterclockwise to Fig 3A. (C) Hydrophobic pocket or SIV Nef created by both the SIVsmm Nef dileucine loop and Nef core with the SMM tetherin GDIWK peptide bound and sandwiched by the AP2ΔμC β2 N-terminus and SIVsmm Nef H0 helix. Fig 3C is rotated ~20° clockwise to Fig 3B. Interacting residues are highlighted. (D) Dileucine loop alignment between SIVsmm Nef of the AP2ΔμC:SMM tetherin: SIVsmm Nef complex and HIV NL4–3 Nef bound to the AP-2 α-σ2 hemi complex (PDB: 4NEE). Alignment was performed between AP2ΔμC α-σ2 and AP-2 α-σ2 hemi. Fig 3D is rotated ~90° counterclockwise to Fig 3C. Disulfide bond is labeled S-S.
SIVsmm Nef refolds the N-terminus of β2-adaptin
An interface was observed between the β2 subunit and SMM tetherin (Fig 3A), that was not found in previous structures of NL4–3 Nef complexes with APs (Jia et al., 2012; Morris et al., 2018; Ren et al., 2014; Shen et al., 2015). Despite a large conformational change upon unlocking, the N-terminus of β2 (residues 13–24) comprises helix α1, preceded by random coil. When AP2ΔμC binds to SMM tetherin and SIVsmm Nef, the flexible loop (residues 6–12) along with the first turn of α1 (residues 14–17) refold into a β hairpin structure not previously described in AP structures (Fig. 4A, Movie S2). This hairpin conformation appears to be stabilized by direct interactions with both SIVsmm Nef and the SMM tetherin tail (Fig. 4A, B).
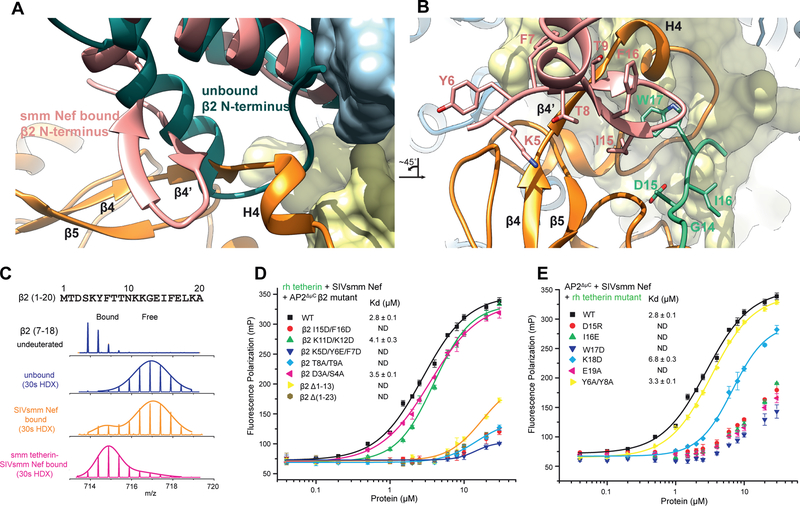
Figure 4. Structural comparison and analysis of the β2-adaptin refolding at the SIVsmm Nef interface.
(A) Alignment of unlocked AP-2 β2 from PDB:2XA7 onto the AP2ΔμC β2 of the AP2ΔμC:SMM tetherin: SIVsmm Nef structure. (B) β2 mutants in β/Nef interface disrupt cooperative tetherin binding, measured by Fluorescence polarization assay. Figure 4B is rotated ~45° counterclockwise to Fig 4A. (C) Mass spectra of the peptide from β2 (7–18) in AP2ΔμC complex upon SIVsmm Nef (orange) or SMM tetherin-SIVsmm Nef (magenta) binding, compared to unbound state (blue). Gaussian fit on one or two peaks is used to represent the distribution of peak heights of the ion peaks across the m/z values and overlay with mass spectra (straight line). (D) FP assay screens the key residues in tetherin tail. See also Figure S4
To corroborate the refolding of the β2 N-terminus, we used hydrogen-deuterium exchange coupled to mass spectrometry (HDX-MS) to monitor changes in the dynamics of AP-2 protein upon binding tetherin and SIVsmm Nef. When bound to SIVsmm Nef only, the β2 N-terminus underwent rapid exchange, similar to the unbound state (Fig. 4C). However, when bound to both tetherin and Nef, β2 (peptide 7–18) underwent 40% less HD exchange (30 s) (Fig. 4C, S4A). Slower deuteration was also seen in α and σ2 residues in SIVsmm Nef binding regions (Fig. S4B, C), as expected. The finding that protection from amide HD exchange in the β2 N-terminus depends on the presence of both SIVsmm Nef and tetherin strongly supports the conclusions drawn from the cryo-EM structure that the presence of these two molecules together is required for refolding of this region.
Side-chains of the induced β2 hairpin present a wall of exposed hydrophobic surface for interactions with tetherin and Nef. These include Tyr6, Phe7, Thr8, Thr9, and Ile15 (Fig. 4B). We generated double and triple mutations to test the role of β2 N-terminal residues in the interface (Fig. 4D). Mutation of residues in both the first (residue 5–7 and 8–9) and second β-strands (residue 15–16) disrupted complex formation. Mutation of Lys11 and Lys12 at the turn connecting the two strands had no effect, however, consistent with their high solvent exposure.
SIVsmm Nef residues 179–189 are the ones most intimately involved in interacting with the β2 hairpin structure (Fig. 4A, B, Fig. 5A, B). This key section is well-conserved in HIV-1 Nef (Fig. 5A). The SIVsmm Nef main chain from residues 179–182 folds into a β strand that complements the β2 hairpin, completing a three-stranded β-sheet spanning SIVsmm Nef and β2. SIVsmm Nef side-chains of Val180, Ser183, Glu185, and Ala186 all interact with β2 (Fig. 5B). The mutations made to disrupt the function of this region, V180D/S183Y and E185A/A186H, severely reduced binding as judged by FP (Fig. 5C). Others immediately following these, including Gln187, Glu188, Asp189, and Glu190 (the E of the EXXXL(L/V) motif) contact SMM tetherin and σ2, and lead into the heart of the dileucine highlighting how this short stretch of Nef sequence forms a central nexus between all of the other elements of the complex (Fig. 5B). The relatively solvent exposed residues 187–188 seem to be non-essential, while mutation of Asp189 or Glu190 have strong reductions in binding (Fig. 5C). Collectively, the cryo-EM, HDX-MS, binding, and mutational data lead to a clear and consistent picture of how tetherin and Nef-induced refolding of the β2 N-terminus creates a binding site for the (G/D)DIWK motif in the cytoplasmic tail of simian tetherin.
Figure 5. Structural analysis of the SIVsmm Nef tetherin binding pocket.
(A) Sequence alignment of Nef dileucine loops between SIVsmm Nef and other Nef species. Key residues in SIVsmm Nef for tetherin and AP-2 binding are highlighted. (B) Ribbon representation of SIVsmm Nef mutations investigated with SMM tetherin GDIW motif shown for reference. (C) FP assay indicates that Nef mutations in multi interfaces impair tetherin binding. See also Figure S5
The (G/D)DIWK binding pocket
EM density was resolved for residues 11–19 of SMM tetherin (Fig. 3A), with Ile16 and Trp17 serving as landmarks for the register and direction of the tetherin tail. Trp17 is the linchpin of the binding site, and its mutation leads to a severe loss in affinity for the complex (Fig. 4E). Its side-chain is wedged into a pocket between the turn of the AP-2 β2 subunit N-terminal hairpin on the one hand, and SIVsmm Nef strands β1 and β4 and the dileucine loop on the other. The Cα of β2 subunit Gly13 is in direct contact with the tetherin Trp side-chain (Fig. 3C), the closest direct contact between tetherin and AP-2. Tyr133 from Nef strand β1 forms one side of the Trp indole-binding pocket. The remainder of the pocket is formed by residues from Nef strand β4′ and H4, and most critically, the back side of the dileucine loop. Nef strand β4′ contributes Val182 and H4 contributes Ala186. While Nef residues Leu194 and Val195 are the critical plug the connects Nef to the “socket” of the dileucine binding site in σ2, His192 and His196, which point in the opposite direction, are key parts of the Trp binding pocket. The His192 side-chain imidazole stacks against the Trp indole, forming the most extensive interaction of any residue in the pocket.
Of the remaining tetherin side-chains observed, Ile16 forms van der Waals contacts with the Leu21 and Arg24 side chains of the Nef N-terminal H0 (Fig. 3C). Density for Asp15 was missing, as often seen in EM density and as seen for most of the Asp and Glu in this reconstruction. On the basis of the surrounding stereochemical constraints, the side-chain was placed such that salt bridges are formed both with His196 of the Nef dileucine loop, and with Arg138 of Nef H2 (Fig. 3C). Mutation of either Asp15 or Ile16 disrupts complex formation (Fig. 4E), consistent with the multiple structural interactions. Single mutations in other residues had little impact on binding, with the exception of Lys18 and Glu19 (Fig. 4E). Lys18 forms a salt bridge with Nef Asp189 (Fig. 3C), which likely accounts for the 2.5-fold reduction in affinity in the K18D mutant. Density for the surface-exposed Glu19 was not visualized, however, so the structural basis for the effect of E19A is unclear. These data are gratifying consistent with biological observations of the critical role of the (G/D)DIWK motif for counteraction by SIV Nef.
α-σ2 engagement by SIVsmm Nef
The dileucine loop of SIVsmm Nef interacts with the AP-2 α-σ2 hemicomplex in a similar manner to HIV-1 NL4–3 Nef (21), but the SIVsmm Nef helix H5 is more regular than its NL4–3 counterpart and has additional interactions. SIVsmm Nef Trp203 anchors the C-terminus of H5 to a pocket formed by residues 60–62 of σ2, which are protected from HDX on complex formation (Fig. S4C). The mutation W203S severely impacts complex formation (Fig. 5C). SIVsmm Nef Asp204 and Pro206 also make SIV-specific interactions with σ2 (Fig. 5B). Consistent with the structural observations, HDX-MS showed a very high level of protection for the Nef dileucine loop when the complete ternary complex was formed (Fig. S5). These findings show that SIVsmm Nef is capable in forming a tight and highly ordered complex with α-σ2 via the dileucine loop.
Residues specifically disrupting anti-tetherin activity of SIVsmm Nef
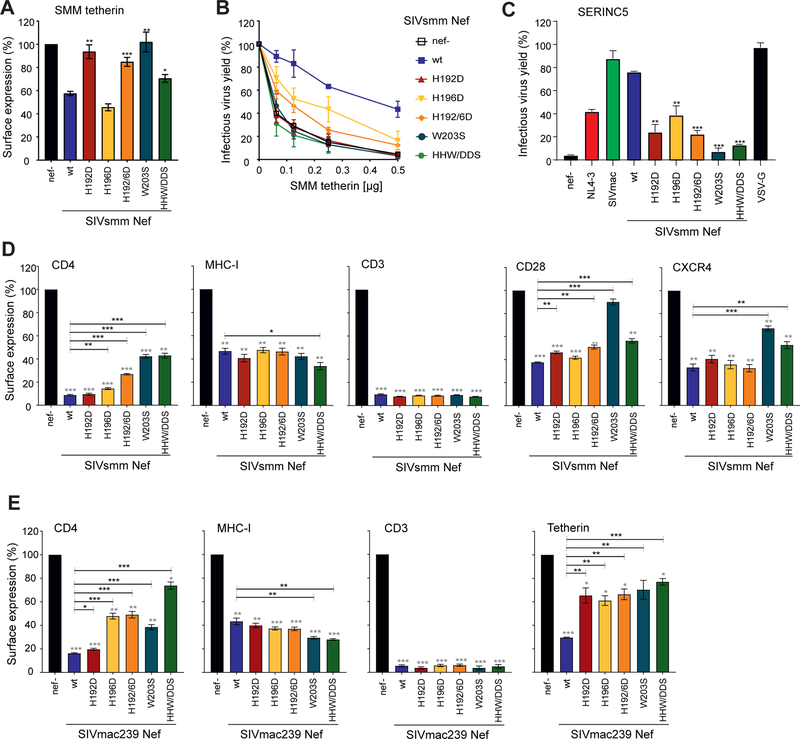
To examine the relevance of the specific residues for Nef function, we introduced single or combined mutations of H192D, H196D, W203S, and their combinations, into the bicistronic pCG expression vector and in proviral HIV-1 NL4–3 constructs coexpressing the SIVsmm Fwr1 Nef protein and eGFP via an IRES element. H192D and H196D were designed to disrupt interactions with tetherin Trp17 and Asp15, respectively. W203S was designed to disengage Nef from the α-σ2 subcomplex. Western blot analysis showed that the mutant Nef proteins were efficiently expressed (Fig. S6A). One exception was that the Y133D substitution, another mutant made to probe the Trp17 binding site, showed lower levels of expression than the parental SIVsmm Nef and was thus excluded from functional analyses. To test anti-tetherin activity, we cotransfected HEK293T cells with env- and vpu-deficient HIV-1 NL4–3 proviral constructs containing wild type or mutant SIVsmm nef alleles, as well as SMM tetherin and Env expression or control plasmid. Subsequently, we measured tetherin surface levels, infectious virus yield and p24 antigen release two days later. Mutation of H192D or W203S disrupted the ability of SIVsmm Nef to downregulate cell surface expression of SMM tetherin (Fig. 6A) and to enhance infectious virus yield (Fig. 6B) and p24 antigen release (Fig. S6B). In comparison, H196D did not affect the ability of SIVsmm Nef to downregulate SMM tetherin but resulted in a phenotype intermediate between wt and nef-defective SIVsmm in infectious virus yield assays. From these phenotypes we conclude that Nef engagement with the AP-2 α-σ2 subcomplex and with tetherin Trp17 is essential for SMM tetherin downregulation, while tetherin Asp15 engagement is less so.
Figure 6. Residue specificity in tetherin antagonism by SIVsmm Nef.
(A, B) Cell surface expression of tetherin (A) and infectious virus yield (B) from HEK293T cells following cotransfection with an env- and vpu-deficient HIV-1 NL4–3 proviral construct encoding the indicated nef alleles and eGFP via an IRES and varying amounts of plasmids expressing SMM tetherin. (A) To measure tetherin modulation, cells were stained with an APC-conjugated antibody against tetherin (Biolegend, 348410) or an isotype control (mouse IgG1-κ, Biolegend, 400121). Tetherin surface expression was analyzed by flow cytometry and the APC mean fluorescence intensity (MFI) of cells coexpressing Nef and eGFP was normalized to the negative control (nef-; 100%). Mean of three to six independent experiments ± SEM. (B) Infectious virus yield was determined by infection of TZM-bl indicator cells and normalized to that detected in the absence of tetherin (100%). Mean of three to six independent experiments measured in triplicates ± SEM. (C) HEK293T cells were co-transfected with vpu- and env-defective HIV-1 NL4–3 constructs encoding the indicated Nef proteins, a construct expressing the HIV-1 Env glycoprotein, and SERINC5 expression or empty control plasmids. Shown are the mean levels of infectious virus production in the presence of transient SERINC5 expression relative to those obtained in cells transfected with the control vector (100%). Mean of three to six independent experiments measured in triplicates ± SEM. (D) To measure receptor modulation, human PBMCs were transduced with VSV-G pseudoytped env- and vpu-deficient HIV-1 NL4–3 IRES-eGFP constructs containing the indicated nef alleles. Cell surface expression of infected (i.e. eGFP positive) cells was normalized to uninfected (i.e. eGFP negative) cells within the same sample before normalization to the negative control (nef-, 100%). Mean of three independent experiments ± SEM. P-values represent differences from the nef-defective (grey stars) or wt SIVsmm (black stars) controls. (E) Rhesus PBMCs were transduced with VSV-G pseudoytped SIVmac239 IRES-eGFP constructs containing the indicated nef alleles. Modulation of CD4, MHC-I, CD3 and tetherin was quantified as described in the legend to figure 6D. Shown are mean values (± SEM) obtained for PBMCs derived from three different rhesus macaques. P-values represent differences from the nef-defective (grey stars) or wt SIVsmm (black stars) controls. *, p < 0.05; **, p < 0.01; ***, p < 0.001. See also Figure S6
To further analyze the impact of the mutations on SIVsmm Nef function, we examined their effect on viral infectivity in the presence of SERINC5, a recently described restriction factor that impairs virion infectivity (Rosa et al., 2015; Usami et al., 2015). In contrast to tetherin antagonism, Nef counteracts SERINC5 and modulates various immune receptors in a species-independent manner (Rosa et al., 2015; Usami et al., 2015). Coexpression of SERINC5 reduced infectivity of the nef-defective control HIV-1 construct by >95% but did not affect the infectiousness of VSV-G pseudotyped viral particles (Fig. 6C). The H192D mutation impaired and W203S almost fully disrupted the anti-SERINC5 activity of SIVsmm Nef, while H196D resulted in an intermediate phenotype (Fig. 6C). As seen for SMM tetherin downregulation, the AP-2 α-σ2 subcomplex and Trp17 binding pocket are both essential, while the Asp15 binding site also contributes. These data suggest that SERINC5 and SMM tetherin downregulation involve a common mode of AP-2 α-σ2 engagement (Fig. 7). Moreover, it suggests that residues of SERINC5 bind to Nef through the backside of a similar conformation of the dileucine loop as SMM tetherin (Fig. 7).
Figure 7. Comparative structural analysis of Nef-mediated downregulation events.
(A) A structural model highlighting the two major cargo binding sites targeted by Nef, the dileucine binding site (ExxxLL) and the tyrosine binding site (YxxΦ), and the host factors downregulated by interactions at these sites. (B) A structural model highlighting the Nef mutations that had effects on Nef-dependent downregulation of host tetherin, SERINC5 and immune receptors.
To determine the impact of the above-mentioned amino acid changes on the ability of SIVsmm Nef to modulate various cellular immune receptors, we transduced human peripheral blood mononuclear cells (PBMCs) with VSV-G pseudotyped env- and vpu-deficient HIV-1 NL4–3 based IRES-eGFP constructs containing different nef alleles and performed flow cytometric analysis (Fig. S6C). In agreement with published data (Schmokel et al., 2009), the parental SIVsmm Nef efficiently downregulated CD4, CD3, and (to a lesser extent) MHC-I, CD28, and CXCR4 in primary human cells (Fig. 6D). Although mutation of H192D fully disrupted the anti-tetherin and anti-SERINC5 activities of SIVsmm Nef, it did not reduce its ability to downregulate any of these immune receptors from the cell surface (Figs. 6D, S6C). Similarly, mutation of H196D had little if any effect on the efficiency of Nef-mediated modulation of CD3, MHC-I, CD28, and CXCR4, although it moderately attenuated down-modulation of CD4. In comparison, W203S strongly reduced downregulation of CD4 and CXCR4 and fully disrupted the effect on CD28, while having no effect on modulation of MHC-I or CD3 (Fig. 6D). Similar observations on the role of SIVmac Nef residues His196 and Trp203 were recently reported in the context of MHC escape mutants (Schouest et al., 2018). The W203S data show that AP-2 α-σ2 engagement is a relatively general theme in the downregulation of many Nef substrates, including SMM tetherin, SERINC5, CD4, CD28, and CXCR4. On the other hand, the usage of the backside of the dileucine loop as probed by H192D appears to be limited to a much smaller set of substrates comprising, so far just SMM tetherin and SERINC5 (Fig. 7).
Nef counteracts tetherin in a species-specific manner, precluding meaningful analysis of tetherin antagonism by SIVsmm Nef in human-derived PBMCs. Via studies in primary rhesus-derived T cells, we thus introduced the above-mentioned mutations in Nef in the infectious molecular SIVmac239 clone that is commonly used for studies on AIDS pathogenesis and vaccine development in the SIV/macaque model. Similar to the results obtained for the SIVsmm Nef (Fig. 6A–D), mutation of H192D impaired the ability of SIVmac239 Nef to counteract rhesus tetherin but had little if any disruptive effect on downmodulation of CD4, MHC-I and CD3 in rhesus PBMCs (Fig. 6E, S6D). In comparison, mutation of H196D or W203S significantly reduced anti-tetherin activity as well as the efficiency of CD4 downmodulation (Fig. 6E). Altogether, our results confirmed the key role of H192, H196 and W203 for tetherin counteraction by SIV Nef in primary virally infected monkey cells. The structural findings thus reflect general features of Nef function in SIV from two different monkey species in cultured and primary cells.
Discussion
It was discovered a decade ago that the sensitivity of simian tetherin to antagonism by SIV Nefs maps to a (G/D)DIWK motif in the N-terminal tail of tetherin that is missing in its human ortholog (Jia et al., 2009; Sauter et al., 2009; Zhang et al., 2009). The structure explains how each of the four residues in the most conserved central DIWK part of this motif interact with AP-2 and SIVsmm Nef, and is beautifully congruent with the biological data. The structural and binding data explain how the absence of this motif in human tetherin renders it resistant to downregulation from the cell surface by SIV Nefs. The inability of SIV to antagonize human tetherin is now considered to be one of the major hurdles to successful simian-human zoonotic transmission (Sauter and Kirchhoff, 2019). Our data thus provide the structural basis for a transmission barrier that helps to protect humans from efficient primate lentiviral transmission.
The involvement of the AP-2 β2 subunit in recognizing both Nef and cargo sets the SIVsmm Nef and tetherin interaction apart from all other physiological or Nef-hijacked sorting events reported to date. All known physiological cargoes that bind to AP cores do so through either the Tyr or dileucine motif binding sites in the μ and the σ subunits, respectively (Fig 7A) (Traub and Bonifacino, 2013). Nef downregulation of MHC-I (Le Gall et al., 1998) occurs principally through the Tyr binding site on the CTD of μ1. The difference is consistent with the null effect of all of the SIVsmm Nef mutants tested on MHC-I downregulation. CD4 downregulation by Nef- appears to be mediated by a site on Nef wedged between strand β1 and helix H2 (Manrique et al., 2017), which is not in direct contact with AP-2. SIV tetherin downregulation shares with CD4, CD28, and CXCR4 a dependence on the engagement of the dileucine loop (Aiken et al., 1994; Bresnahan et al., 1998; Craig et al., 1998; Leonard et al., 2011) with the α-σ2 hemicomplex, consistent with the W203S phenotype in tetherin and CD4 downregulation. It is remarkable that such a small protein as Nef can bind and hijack the AP complexes in so many different ways. This multifunctionality confers robustness to viral mutation and host variation.
Our results suggest that many downregulatory functions of Nef are structurally and genetically separable. Perhaps most notably, mutation of H192D impairs the ability of SIVsmm and SIVmac Nef to antagonize tetherin and SERINC5 during the late stage of the viral replication cycle but had no disruptive effect on the modulation of various immune receptors (CD4, CD3, CD28, MHC-I and CXCR4) required for viral immune evasion throughout the viral life cycle (Fig 7B). Thus, this mutation may help probe the relevance of early vs. late Nef functions for viral replication and pathogenicity in vivo in non-human primate models for AIDS.
His192 is conserved in most monkey SIVs but not the ape and human viruses. On the other hand His196 is conserved across all Nefs, and H196D is defective in SERINC5 downregulation in human cells. The proximity of His196 to the β2 hairpin interface, and the conservation in HIV-1 of the Nef strand β4 involved in hairpin refolding suggests that the β2 hairpin participates in HIV-1 Nef downregulation of SERINC5. The apparent lack of a normal physiological role for the β2 hairpin in AP-2-dependent cargo sorting suggests that this interface could be targeted therapeutically by Nef antagonists to restore SERINC5 expression and block viral infectivity.
Different HIV strains have adapted to their human hosts with different degrees of success through both Nef-dependent and independent mechanisms. HIV-1 O-Nefs adapted to target a tetherin sequence adjacent to the DIWK deletion (Kluge et al., 2014) in conjunction with the μ1 subunit of AP-1. When bound to human tetherin and μ1, HIV-1 O-Nef bridges the β1- μ1 and γ1-σ1 hemicomplexes of two different AP-1 tetramers in trans. This trans bridging mode, in contrast to the cis bridge described here for SIVsmm Nef, leads to AP-1 trimerization and formation of a structure capable of sequestering tetherin at the TGN (Morris et al., 2018). HIV-2 Nefs never recovered the ability to target tetherin, and HIV-2 is dependent on Env for tetherin counteraction. The pandemic M-group HIV evolved a potent Vpu counteraction of a tetherin, which however, uses some of the same principles as Nef-dependent counteraction. HIV-1 M-Vpu has a dileucine that can engage the γ1-σ1 hemicomplex of AP-1 and targets the same DIWK deletion-proximal sequence in tetherin and the same site on μ1 as HIV-1 O-Nefs (Jia et al., 2014). All of the adaptions whereby HIVs counteract human tetherin involved major changes in mechanism, either using Nef to hijack a different AP complex through different sites, or using other proteins entirely. This illustrates that the barrier caused by the absence of DIWK in human tetherin cannot be overcome simply by modifying and repurposing the pre-existing DIWK binding site in SIV Nefs. On the other hand, the fact that the barrier ultimately was overcome repeatedly and in different ways highlights the multiplicity of entry points to the host trafficking machinery and the versatility of viral proteins through evolution.
STAR METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
All plasmids generated in this study are outlined in the Key Resources Table and are available upon request. Further information and requests for resources should be directed to and will be fulfilled by the Lead Contact, Xuefeng Ren (snowren@berkeley.edu).
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| APC anti-human CD317 (BST2, Tetherin) | Biolegend | Cat#348410: RRID: AB_2067121 |
| PerCP-Cy™5.5 Mouse Anti-Human CD4 | BD bioscience | Cat#552838; RRID: AB_394488 |
| V450 Mouse Anti-Human CD3 | BD bioscience | Cat#560351; RRID: AB_1645168 |
| PE Mouse Anti-Human HLA-ABC | BD bioscience | Cat#555553; RRID: AB_395936 |
| CD4 Monoclonal Antibody (S3.5), APC | Thermo Fisher | Cat#MHCD0405; RRID: AB_1473570 |
| PE Mouse Anti-Human CD3 | BD bioscience | Cat#555333; RRID: AB_395740 |
| Monoclonal Mouse Anti-Human HLA-ABC Antigen/RPE |
Dako | Cat#R7000; RRID: AB_11192092 |
| PE Mouse Anti-Human CD28 | BD bioscience | Cat#348047; RRID: AB_400368 |
| APC Mouse Anti-Human CD184 | BD bioscience | Cat#555976; RRID: AB_398616 |
| APC Mouse IgG1, κ Isotype Ctrl (FC) Antibody | Biolegend | Cat#400121; RRID: AB_326443 |
| PerCP-Cy™5.5 Mouse IgG1 κ Isotype Control | BD bioscience | Cat#552834; RRID: AB_394484 |
| V450 Mouse IgG1, κ Isotype Control | BD bioscience | Cat#560373; RRID: AB_1645606 |
| PE Mouse IgG1, κ Isotype Control | BD bioscience | Cat#555749; RRID: AB_396091 |
| Bacterial and Virus Strains | ||
| pCG-SIVsmm FWR1 nef H192D C-AU1 IRES EGFP | This Paper | N/A |
| pCG-SIVsmm FWR1 nef H196D C-AU1 IRES EGFP | This Paper | N/A |
| pCG-SIVsmm FWR1 nef H192D/H196D C-AU1 IRES EGFP | This Paper | N/A |
| pCG-SIVsmm FWR1 nef W203S C-AU1 IRES EGFP | This Paper | N/A |
| pCG-SIVsmm FWR1 nef H192D/H196D/W203S C-AU1 IRES EGFP | This Paper | N/A |
| pBR-NL43 Δvpu env* nef* IRES-eGFP | Wildum et al. 2006 | N/A |
| pBR-NL43 Δvpu env* IRES-eGFP | Wildum et al. 2006 | N/A |
| pBR-NL43 Δvpu env* IRES-eGFP_SIVsmmFwr1 nef | Wildum et al. 2006 | N/A |
| pBR-NL43 Δvpu env* IRES-eGFP_SIVsmmFwr1 nef H192D | This Paper | N/A |
| pBR-NL43 Δvpu env* IRES-eGFP_SIVsmmFwr1 nef H196D | This Paper | N/A |
| pBR-NL43 Δvpu env* IRES-eGFP_SIVsmmFwr1 nef H192D/H196D | This Paper | N/A |
| pBR-NL43 Δvpu env* IRES-eGFP_SIVsmmFwr1 nef W203S | This Paper | N/A |
| pBR-NL43 Δvpu env* IRES-eGFP_SIVsmmFwr1 nef H192D/H196D/W203S | This Paper | N/A |
| pBR-NL43 Δvpu env* IRES-eGFP SIVmac239 nef | Wildum et al. 2006 | N/A |
| pBR-SIVmac239 IRES-eGFP | Schindler et al. 2006 | N/A |
| pBR-SIVmac239 nef* IRES-eGFP | Schindler et al. 2006 | N/A |
| pBR-SIVmac239_IRES-eGFP nef H192D | This Paper | N/A |
| pBR-SIVmac239_IRES-eGFP nef H196D | This Paper | N/A |
| pBR-SIVmac239_IRES-eGFP nef H192D/H196D | This Paper | N/A |
| pBR-SIVmac239_IRES-eGFP nef W203S | This Paper | N/A |
| pBR-SIVmac239_IRES-eGFP nef H192D/H196D/W203S | This Paper | N/A |
| pHIT-G-VSV-G | Fouchier et al. 1997 | N/A |
| pBJ6 (empty) | Rosa et al. 2015 | N/A |
| pBJ6 SERINC5-HA | Rosa et al. 2015 | N/A |
| pCAGGS_HIV-1 M NL4–3 Env (137 FLAG-tag) | Braun et al. 2019 | N/A |
| pCG-HIV-1 NL4–3 nef 3* IRES-DsRed | Mack et al. 2017 | N/A |
| pCG-rhesus G tetherin DsRed | Mack et al. 2017 | N/A |
| pCG-SIVsmm1 tetherin Fit2 | This Paper | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| 2,2’-Dipyridyldisulfide (2-PDS) | Alfa Aesar | A11118, CAS: 2127-03-9 |
| AP-2 β2 1–591 rat | This Paper | P62944 |
| AP-2 σ2 Rat | This Paper | P62744 |
| AP-2 μ2 1–141 mouse | This Paper | Q3TWV4 |
| AP-2 α 1–621 rat | This Paper | Q66HM2 |
| AP-2 α 1–396 rat | This Paper | Q66HM2 |
| sooty mangabey tetherin 4–26 C2S SIVsmm Nef 1–251 C55A chimera | This Paper | C3VHQ4/DQ092758 |
| Nef C55A SIVsmm | This Paper | DQ092758 |
| tetherin 2–26 rhesus macaque | This Paper | G0WVJ8 |
| Deposited Data | ||
| AP2ΔμC:SMM tetherin-SIVsmm Nef EM map | This Paper | EMD-20217 |
| AP2ΔμC:SMM tetherin-SIVsmm Nef EM PDB | This Paper | 6OWT |
| Experimental Models: Cell Lines | ||
| Human: HEK293T cells | ATCC | Cat# CRL-3216; RRID: CVCL_0063 |
| Human: TZM-bl cells | NIH | Cat# 8129; RRID: CVCL_B478 |
| Oligonucleotides | ||
| Primers used to generate pCG IRES eGFP expression plasmids and HIV-1 IRES eGFP proviral constructs harboring SIVsmmFwr1 nef | See table S2 of this paper | N/A |
| Primers used to generate SIVmac239 IRES eGFP proviral constructs harboring mutations in nef | See table S3 of this paper | N/A |
| Recombinant DNA | ||
| Rat AP-2S1/Mm AP2M1/Rat AP2A1-TEVsite-GST/His-TEVsite-Rat AP2B1 in pST39 vector | This Paper | AP2ΔμC |
| AP2S1: N97C/C99S in AP2ΔμC in pST39 vector | This Paper | AP2ΔμCss |
| AP2S1: N97C/C99S in AP2ΔμC in pST39 vector | This Paper | AP2ΔμC-β D3A/S5A |
| AP2B1: K5D/K6E/F7D/pST39 in pST39 vector | This Paper | AP2ΔμC-β K5D/K6E/F7D |
| AP2B1: T8A/T9A in pST39 vector | This Paper | AP2ΔμC-β T8A/T9A |
| AP2B1: K11D/K12D in pST39 vector | This Paper | AP2ΔμC-β K11D/K12D |
| AP2B1: I15D/F16D in pST39 construct | This Paper | AP2ΔμC -β I15D/F16D |
| AP2B1: residue (1–13) truncation in pST39 construct | This Paper | AP2ΔμC-β Δ(1–13) |
| AP2B1: residue (1–23) truncation in pST39 construct | This Paper | AP2ΔμC-β Δ(1–23) |
| Rat AP2S1 (1–143)/GST-TEVsite-Rat AP2A1 (1–396) in pST39 construct | This Paper | GST-α-σ2 hemi |
| Rh Tetherin (2–26)-MBP in LIC 2BT vector | This Paper | His-rh Tetherin-MBP |
| Rh Tetherin (2–26) wild type in pGST2 vector (BamHI/XhoI) | This Paper | GST-rh Tetherin-WT |
| Rh Tetherin: Y6A/Y8A in pGST2 vector | This Paper | GST-rh Tetherin-Y6A/Y8A |
| Rh Tetherin: D15R/pGST2 in pGST2 vector | This Paper | GST-rh Tetherin-D15R |
| Rh Tetherin: I16E in pGST2 vector | This Paper | GST-rh Tetherin-I16E |
| Rh Tetherin: W17D in pGST2 vector | This Paper | GST-rh Tetherin-W17D |
| Rh Tetherin: K18D in pGST2 vector | This Paper | GST-rh Tetherin-K18D |
| Rh Tetherin: E19A in pGST2 vector | This Paper | GST-rh Tetherin-E19A |
| SIVsmm Nef-C55A in LIC 2BT vector | This Paper | His-SIVsmm Nef-WT |
| SIVsmm Nef: Y133D in LIC 2BT vector | This Paper | SIVsmm Nef-Y133D |
| SIVsmm Nef: V180D/S183Y in LIC 2BT vector | This Paper | SIVsmm Nef-V180D/S183Y |
| SIVsmm Nef: E185A/A186H in LIC 2BT vector | This Paper | SIVsmm Nef-E185A/A186H |
| SIVsmm Nef: Q187A/E188S in LIC 2BT vector | This Paper | SIVsmm Nef-Q187A/E188S |
| SIVsmm Nef: D189K in LIC 2BT vector | This Paper | SIVsmm Nef-D189K |
| SIVsmm Nef: E190K in LIC 2BT vector | This Paper | SIVsmm Nef-E190K |
| SIVsmm Nef: H192D in LIC 2BT vector | This Paper | SIVsmm Nef-H192D |
| SIVsmm Nef: L194A/V195A in LIC 2BT vector | This Paper | SIVsmm Nef-L194A/V195A |
| SIVsmm Nef: H196D in LIC 2BT vector | This Paper | SIVsmm Nef-H196D |
| SIVsmm Nef: Q199A in LIC 2BT vector | This Paper | SIVsmm Nef-Q199A |
| SIVsmm Nef: W203S in LIC 2BT vector | This Paper | SIVsmm Nef-W203S |
| SIVsmm Nef: D204S/P206A in LIC 2BT vector | This Paper | SIVsmm Nef-D204S/P206A |
| Smm tetherin (4–26)-C9S/C25S-10aa linker (GSDEASEGSG)-SIVsmm Nef (1–251)-C55A-6xHis in pMBP2 vector (BamHI/XhoI) | This Paper | smm tetherin-SIVsmm Nef-6His |
| smm tetherin-C2S-10aa linker-SIVsmm Nef-C55A-Δ(28–36)-6xHis in pMBP2 vector (BamHI/XhoI) | This Paper | smm tetherin-SIVsmm Nef-ΔL1 |
| 10aa linker (GSDEASEGSG)-SIVsmm Nef (1–251)-C55A-Δ(28–36)-6xHis in pMBP2 vector (BamHI/XhoI) | This Paper | 10link-SIVsmm Nef-ΔL1 |
| Software and Algorithms | ||
| MotionCor2–1.0.0 | Zheng et al., 2017 | http://msg.ucsf.edu/em/software/motioncor2.html |
| Gct-v1.06 | Zhang, 2016 | https://www.mrc-lmb.cam.ac.uk/kzhang/Gctf/ |
| CTFIND4.1.12 | Rohou and Grigorieff, 2015 | http://grigoriefflab.janelia.org/ctffind4 |
| Relion-3.06 | Zivanov et al., 2018 | https://github.com/3dem/relion |
| cryoSPARC v2.8.0 | Punjani et al., 2017 | https://cryosparc.com |
| UCSF Chimera1.13.1 | Pettersen et al., 2004 | https://www.cgl.ucsf.edu/chimera/download.html |
| PyEM | Daniel Asarnow, UCSF; Github | https://github.com/asarnow/pyem |
| PyEM support scripts | This Paper | https://github.com/simonfromm/miscEM |
| Coot 0.9 | Emsley et al., 2010 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| Phenix 1.12 | Adams et al., 2010 | https://www.phenix-online.org/download/ |
| emRinger | Barad et al., 2015 | https://github.com/fraser-lab/EMRinger |
| MolProbity | Davis et al., 2007 | Phenix package |
| HDExaminer | Sierra Analytics, Modesto, CA | http://massspec.com |
| Proteome Discoverer 2.1 | Thermo Scientific, Waltham, MA | http://www.thermofisher.com/ |
| StatView version 4.0 | Abacus Concepts, Berkeley, CA | http://www.macintoshrepository.org/10824-statview-4-0 |
| Origin | OriginLab, Northampton, MA | http://www.originlab.com |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell lines
Human Embryonic Kidney (HEK) 293T cells (DuBridge et al., 1987) were obtained from the American Type Culture Collection (ATCC) and TZM-bl reporter cells (Platt et al., 1998) were kindly provided by Drs. Kappes and Wu and Tranzyme Inc. through the NIH AIDS Reagent Program. Both cell lines were cultured in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% heat-inactivated fetal calf serum (FCS), 2 mM L-glutamine, 100 units/ml penicillin and 100 μg/ml streptomycin. TZM-bl cells express CD4, CCR5 and CXCR4 and contain the β-galactosidase gene under the control of the HIV-1 promoter.
Primary human cell cultures
Experiments involving human blood were reviewed and approved by the Institutional Review Board (i.e. the Ethics Committee of Ulm University). Individuals and/or their legal guardians provided written informed consent prior to donating blood and all human-derived samples were anonymized before use, therefore the age and gender of the donors is not known to the experimenters. Peripheral blood mononuclear cells (PBMCs) from healthy human donors were isolated using lymphocyte separation medium (Biocoll separating solution; Biochrom), stimulated for 3 days with phytohemagglutinin (PHA; 2 μg/ml), and cultured in RPMI 1640 medium with 10% fetal calf serum and 10 ng/ml interleukin-2 (IL-2).
METHOD DETAILS
Plasmid construction
The four subunits of AP2ΔμC were subcloned into pST39 vector by restriction cloning (Selleck and Tan, 2008). DNAs coding for the four subunits of AP2ΔμC were as follows: rat σ2 full-length, mouse μ2 (1–141), rat α (1–621) as a C-terminal GST fusion, and rat β2 (1–591) fused to an N-terminal 6xHis tag. TEV cleavage sites were introduced between the affinity tags and the protein. The above protein sequences have been compared with that of sooty mangabey (SMM). The following are 100% identical: Rat β2 (uniprot: P62944) vs SMM β2 (A0A2K5LRZ0); Rat σ2 (P62744) vs SMM σ2 (A0A2K5LIR6); Mouse μ2 (Q3TWV4) and SMM μ2 (A0A2K5NQG4). There are four differences between the Rat α (Q66HM2) and SMM α (A0A2K5NR50) core, which are at residues 8, 137, 403 and 557. None of these residues are in the σ2 or Nef binding interface. GST tagged AP2hemi consists of rat α (1–396) with an N-terminal GST tag and rat σ2 full-length, the minimal construct binding to HIV-1 NL4–3 Nef (Ren et al., 2014). For all structural and in vitro experiments, the SIVsmm Fwr1 Nef C55A construct bearing the single Cys193 was used, which is referred to as wild-type of SIVsmm Nef. All bacterial expression constructs used in this study are listed in the Key Resources Table.
For MBP pull down or binding assay, SIVsmm Nef wild-type and mutants were subcloned into LIC 2BT vector (Macrolab, UC Berkeley), and were expressed as TEV-cleavable N-terminal 6xHis fusions. DNAs coding for the tetherin tail (2–26) from rhesus macaque or sooty mangabey were codon-optimized using DNAworks server (Hoover and Lubkowski, 2002) for E. coli expression. Rhesus tetherin was first fused to a C-terminal MBP tag and then subcloned into the LIC 2BT vector, which was used to generate proteins for MBP pull down assays. Rhesus tetherin was also subcloned into pGST2, yielding a fusion with an N-terminal GST and a TEV cleavage site for FP assays (Sheffield et al., 1999). For cryo-EM, a PCR fragment encoding sooty mangabey tetherin (4–26)-C2S-10aa linker-SIVsmm Nef (1–251)-C55A was subcloned into pMBP2, and the protein was expressed with an N-terminal TEV cleavable MBP tag and C-terminal uncleavable 6xHis tag.
Protein expression and purification
Wild-type and mutant AP2ΔμC complexes were expressed in BL21 (DE3) pLysS strain (Promega, Madison, WI) by induction at 23 °C overnight. The cells were lysed by sonication in 50 mM Tris pH 8.0, 300 mM NaCl, 10% glycerol, 3 mM β-mercaptoethanol (βME), 20 mM imidazole and 2 mM PMSF. The clarified lysate was first purified on a Ni-NTA column (Qiagen, Valencia, CA). The eluate was further purified on glutathione-Sepharose 4B resin (GE healthcare, Piscataway, NJ). After TEV cleavage at 4 °C overnight, the sample was concentrated and then loaded onto a HiLoad 16/60 Superdex 200 column (GE healthcare) in the sample buffer of 20 mM Tris pH 8.0, 150 mM NaCl, 0.1 mM TCEP. The AP2ΔμC fractions were passed through 1 ml of glutathione-Sepharose 4B resin to capture the GST tag, concentrated, and flash-frozen in liquid N2.
His-tagged SIVsmm Nef constructs were expressed in BL21 (DE3) star cells (Life technologies, Grand Island, NY), and induced with 0.3 mM IPTG at 25 °C overnight. Purification was carried out using Ni-NTA resin, and samples were eluted with 200 mM imidazole/500 mM NaCl Tris buffer, pH 8. The eluate was subjected to a HiLoad 16/60 Superdex 75 column in high salt buffer of 20 mM Tris pH 8, 500 mM NaCl, 0.1 mM TCEP.
All MBP tagged proteins were expressed in BL21 (DE3) star cells and grown at 30 °C overnight. Cells were induced at 35 °C for 2 hours. The purification of His-tagged tetherin-MBP proteins was carried out by Ni-NTA column, size-exclusion in a HiLoad 16/60 Superdex 200 column, and then a second Ni-NTA column. SMM tetherin-10aa linker-SIVsmm Nef proteins were first purified using a Ni-NTA column, the eluate was diluted 5 times in Q buffer A (30 mM Tris pH 8), and then loaded onto a HiTrap Q HP 5ml column (GE healthcare, Piscataway, NJ). The elution on Q column was performed with a 10 CV linear gradient from 0–1 M NaCl in Q buffer A. The sample fractions were pooled together for TEV cleavage at 4 °C overnight. The TEV cleaved samples was adjusted to final 20 mM imidazole and 500 mM NaCl, and then passed through the second Ni column to remove free MBP tag. The proteins were eluted in 500 mM NaCl/200 mM imidazole buffer and subjected to a 16/60 Superdex 75 column in the same high salt buffer.
GST-tagged tetherin constructs were expressed in BL21 (DE3) star cells, and induced with 0.3mM IPTG at 20 °C overnight. The cleared lysate was purified using glutathione-Sepharose 4B resin. The eluate was concentrated to 1–1.5 ml, and 150 μl of TEV was added to cleave samples at room temperature for 4–5 hours. The cut proteins were then loaded onto a 16/60 Superdex 75 column in the same sample buffer to separate tetherin peptides from MBP and TEV proteins.
MBP pull down assay
35 μg of recombinant His-tetherin-MBP proteins were incubated with His- and GST-tagged AP2ΔμC (1 μM) or/and SIVsmm Nef proteins (6 μM) at 4 °C overnight in 20 mM Tris pH 8, 150 mM NaCl, 0.1 mM TCEP. Then 30 μl Amylose resin (New England Biolabs, Ipswich, MA) was to the mixture and rocked at 4 °C for 2 hours. The beads were washed 4 times, mixed with 50 μl of 2x lithium dodecylsulfate (LDS)/BME buffer and heated at 90°C for 3 min. 19 μl of each sample was subjected to SDS/PAGE gel.
Tetherin peptide fluorescent labeling
The wild-type of rh tetherin peptide used in the FP assay consists of residue (2–26): APILYDYRKMPMGDIWKEDGDKRCK. The single cysteine at the C-terminus was used for labeling. The rh tetherin tail differs by only two residues from the SMM tetherin tail, which are both outside the binding region. Alexa Fluor 488 C5 Maleimide (Thermo Fisher Scientific, Waltham, MA) was dissolved in dimethyl sulfoxide (DMSO). 100 μM of diluted peptide in 1x PBS pH 7.4 was reacted with dye at 1:1.2 molar ratio for 40 min at room temperature. The conjugation reaction was quenched with 100 mM βME at 15 min at room temperature. The samples were then subjected to a HiTrap desalting column in the sample buffer. The Alexa488 labeling efficiency of each peptide was between 96% and 100%.
Fluorescence polarization binding assay
Assays were conducted in Greiner 384-well black microplates (784076, Greiner, Monroe, NC). SIVsmm Nef proteins were exchanged from high salt buffer to the sample buffer using a Zeba spin desalting column (7K MWCO, Thermo Fisher Scientific). After desalting, the stock concentration of SIVsmm Nef was adjusted to 80 μM. The Alexa488 labeled tetherin tail peptide was incubated with SIVsmm Nef and AP2ΔμC in a total volume of 20 μl for 30 min at room temperature. The final concentration of labeled peptide was fixed at 50 nM, while SIVsmm Nef and AP2ΔμC were kept at a 1:1 molar ratio and titrated from 0.04 to 30 μM in this assay. FP values (mP) were measured using a Synergy H4 microplate reader (excitation at 485 nm and emission at 528 nm, BioTek, Winooski, VT), and plotted as a function of protein concentration using a logarithmic scale (Du, 2015; Moerke, 2009). Measurements were repeated at three times and the data were processed using Origin (OriginLab, Northampton, MA). The binding constant (Kd) was fitted using a one-site model.
SMM tetherin-SIVsmm Nef: AP2ΔμC cross-linking
In order to prevent dissociation of the AP-2:Nef complex, intermolecular disulfide cross linking activated by 2,2’-Dipyridyldisulfide (2-PDS) was used to stabilize the SIVsmm Nef-AP-2 interaction. Ser163 in the Nef dileucine loop is within 3Å of Asn97 in σ2 (PDB: 4NEE) (Ren et al., 2014). The equivalent residue in SIVsmm Nef is Cys193, thus we generated the σ2 N97C/C99S mutation (denoted AP2ΔμCSS following cross-linking) to induce this intermolecular disulfide bond formation. AP2ΔμCSS proteins were treated with final 20 mM DTT at 30 °C for 30min, followed by desalting into cross-linking buffer (20 mM Tris pH 8, 150 mM NaCl) using Zeba spin desalting column. SMM tetherin-SIVsmm Nef fusion proteins were also reduced by 20 mM DTT at 30 °C for 30min, followed by desalting into the buffer of 20 mM Tris pH 8, 300 mM NaCl using Zeba spin column. After elution from the desalting column, tetherin-Nef fusion proteins were treated with final 3 mM 2,2’-Dipyridyldisulfide (2-PDS), and incubated at room temperature for 30min (Olsen and Lima, 2013). The activated tetherin-Nef samples were desalted into 20mM Tris PH 8, 150mM NaCl to remove excess 2,2’-Dipyridyldisulfide. Then the activated samples (final 1.5 μM) were immediately mixed with desalted AP2ΔμCss (final 1 μM) and incubated at RT for 30min. The cross-linking mixture was further purified over a HiTrap Q HP 1ml column, and then subjected to a Superdex 200 10/300 GL column in cross-linking buffer. The AP2ΔμCSS:SMM tetherin (4–26)-C2S-10aa-SIVsmm Nef (1–251)-C55A complex eluted as a single homogenous peak in gel filtration, and no additional products were observed on SDS-PAGE.
Cryo-EM Data Collection
AP2ΔμCSS:SMM tetherin (4–26)-C2S-10aa-SIVsmm Nef (1–251)-C55A samples for data collection were prepared on 2/1-thick C-flat holey carbon copper 300-mesh grids that were prepared by plasma cleaning for 30 s at 25 mA using a Pelco easiGlow glow discharger (Ted Pella, Inc.). Complex at 0.6 mgmL-1 was applied in a 4 μL drop to one side of the grid, followed by a 30 second wait time, then plunge frozen into liquid ethane using a Vitrobot mark VI. The humidity was controlled at 100%, 22°C and the grids blotted with Whatman #1 paper using a blot force of 6 for 5 s. Two rounds of data collected (dataset 1 and 2) were performed on a Titan Krios (FEI; BACEM UCB) at a nominal magnification of 105,000 and a magnified pixel size of 1.149 Å pix-1. The dose rate was 6.08 e-/Å2/sec at the sample. Data collection was carried out using SerialEM. Dataset 1, consisting of 1647 movies, used stage shift navigation with 3 exposures per hole and focusing for each hole. Dataset 2 consisted of 10,248 micrographs using image shift navigation. 3 shots per hole were collected in a 3×3 grid of holes, focusing once per 9 holes. The defocus range was 1.00 to 2.5 m. Movies were acquired on a K2 direct electron detector (Gatan) in super-resolution counting mode with a dose fractionated frame rate of 250 ms and total collection time of 8250 ms.
Cryo-EM Data Processing
Figure S2 shows the cryo-EM data processing workflow. In brief, gain-corrected movies from two data sets (dataset 1 and 2) were motion corrected using the Relion-3 (Zivanov et al., 2018) MotionCor2 (Zheng et al., 2017) wrapper. Corrected micrographs were inspected visually and micrographs with apparent defects, such as ice, debris contamination, or exposures of the carbon support film, were excluded from further processing. The contrast transfer function (CTF) of non-dose weighted micrographs was estimated using Gctf (Zhang, 2016) for dataset 1 and CTFfind4 (Rohou and Grigorieff, 2015) for the dataset 2. After an additional visual cleanup, 622,487 and 5,696,434 particles were picked from 1618 and 9333 micrographs using Gautomatch and Relion Laplacian particle picking for datasets 1 and 2, respectively. Particles were extracted from datasets 1 and 2 with a 256 pixel box size from dose-weighted micrographs and subjected to a single round of 2D classification at 4-fold binning in Relion-3.0 (Kimanius et al., 2016; Scheres, 2012) and cryoSPARC v2 (Punjani et al., 2017), respectively. Obvious “Junk” was removed resulting in 4,107,380 refined particles. A 4 class Ab-initio 3D reconstruction was performed in cryoSPARC v2 on dataset 1, resulting in 2 heterogeneous 3D classes (class 1 and 2), where class 1 was the object of refinement, and two “junk” classes. Using 3D class 1 and class 2, multiple rounds of heterogeneous 3D refinement were performed on dataset 1 resulting in a set of 215,685 particles in class 1 which refined to 4.25 Å resolution. These 215,685 particles were then transferred to Relion-3.0. When needed, cryoSPARC v2 database files were converted using UCSF PyEM (https://github.com/asarnow/pyem) and in-house written scripts (https://github.com/simonfromm/miscEM) into star files for the use as inputs in Relion3.0. A single round of 3D classification was run against two low resolution maps simulated from a refined coordinate model built into the original 4.25 Å structure, one containing Nef and the N-terminus of the β2 subunit of AP2ΔμCSS (class 1) and one lacking both (class 2). This focused 3D classification was run with a low pass filter of 20 Å, τ = 20, and no angular refinement. 90,285 particles were classified to generate the map containing Nef and the N-terminus of the β2 subunit of AP2ΔμC. After further refinement in cryoSPARC v2, final map contained 84,834 particles and refined to 3.7 Å.
The refined particles of dataset 2 were processed using an iterative 2D classification and heterogeneous refinement workflow similarly to dataset 1 (Fig S2). Eventually, 300,243 particles were subject to a round of focused 3D classification in Relion-3.0 identical to the one performed on dataset 1. The resulting 129,701 particles in class 1 were retained and used for a 3D auto-refine run in Relion-3.0 which served as the basis for per-particle CTF refinement and Bayesian polishing in Relion-3.0. After further refinement of the “shiny” particles in cryoSPARC v2, the remaining 128,842 particles of were combined with the 84,834 particles remaining from dataset 1 to give a final dataset of 212,541 particles. Two further rounds of 2D classification and selection yielded 159,406 particles. A final round of homogeneous as well as non-uniform refinement was performed separately in cryoSPARC v2 (Fig S2, S3B). The Non-uniform reconstructions reached a resolution of 3.8 Å and the homogeneous reconstruction reached a resolution of 3.9 Å, both according to the FSC 0.143 cutoff. Both maps showed improved density in the regions of primary interest compared to the prior 3.7 Å map reconstruction. After visual inspection of both reconstructions, the map resulting from non-uniform refinement was chosen for atomic model building and coordinate refinement, and density thereof is shown throughout the manuscript.
Model Building and Refinement
Rigid body fitting was performed in UCSF Chimera (Pettersen et al., 2004) using as starting models PDB entries 4NEE and 2XA7, followed by manual adjustment in Coot (Emsley et al., 2010). To help alleviate local differences in map quality, two differently filtered and sharpened maps were used during the model building process. Model building in Coot and real-space refinement in phenix (Afonine et al., 2018) was used to improve the model geometry against both a locally filtered and b-factor sharpened map generated through the cryoSPARC v2 filtering/sharpening routine (applied b-factor: −145.0 Å2) and a map generated from the Relion3.0 PostProcess routine (applied b-factor: −77.4 Å2; low-pass filter: 3.8 Å). Real-space refinement as implemented in phenix with a target resolution of 3.8 Å, secondary structure restraints and a weight of 1 was used iteratively with manual adjustments in coot to refine the atomic coordinate model. EMRinger scores were calculated (Barad et al., 2015) to identify regions requiring further remodeling in Coot. Where clear residue side-chain density was lacking, side chains were not modeled.
The final model-map cross correlation was 0.75, with a map-versus-model FSC0.5 of 4.5 Å (Fig S3D). Model geometry was validated using MolProbity (Davis et al., 2007). All map and model statistics are detailed in Table S1. A cross-validation test was performed to assess overfitting. In brief, atoms of the final coordinate model were perturbed by an average of 0.5 Å and one round of automated real-space-refinement in phenix against one half map of the gold-standard refinement was performed. The resulting map-vs-model FSC curves of the refined model against the same half map (FSCwork) and the second half map (FSCtest) do not diverge significantly indicating the absence of overfitting (Fig. S3E). Table S1 and Figure S3F summarize the completely assembled AP2ΔμCSS:SMM tetherin-SIVsmm Nef coordinate model, with S3F showing representative density for all AP-2 subunits, SIV smmNef and SMM tetherin.
HDX-MS
Uncrosslinked SIVsmm Nef-ΔL1–6His was used in all HDX studies. The construct used for HDX contained a deletion of residues 28–36, which are non-conserved in SIVcpz Nef or HIV-1 NL4–3 Nef, and whose presence reduced the stability of the complex. AP2ΔμC was incubated with SMM tetherin-SIVsmm Nef-ΔL1 or 10aa-SIVsmm Nef-ΔL1 at 4 °C overnight. The mixture was purified using Ni-NTA resin, and loaded on a Suerpdex 200 10/300 GL column. Fractions were pooled and concentrated to 20 μM. Amide HDX-MS was initiated by a 20-fold dilution of 20 μM sample stock into a D2O buffer containing 20 mM HEPES (pD 7.2), 200 mM NaCl and 0.5 mM TCEP at room temperature. After intervals of 30 s-120 s, exchange was quenched at 0 °C with the addition of ice-cold quench buffer (400 mM KH2PO4/H3PO4, pH 2.2). The samples were then injected onto an HPLC (Agilent 1100) with in-line peptic digestion and desalting. Desalted peptides were eluted and directly analyzed by an Orbitrap Discovery mass spectrometer (Thermo Scientific). Peptides were identified with tandem MS/MS experiments and a Proteome Discoverer 2.1 (Thermo Scientific) search. Mass analysis of peptide centroids was performed using HDExaminer (Sierra Analytics, Modesto, CA), followed by manual verification of each peptide. The relative deuteron content of the peptic peptides was determined from the centroid of the molecular ion isotope envelope.
Proviral constructs
Overlap-extension PCR was used to replace the NL4–3 nef in the IRES-eGFP HIV-1 construct by SIVsmm nef genes as described (Schindler et al., 2006). The integrity of all PCR-derived inserts was confirmed by sequencing. The control HIV-1 NL4–3 env* Δvpu IRES-eGFP constructs expressing the NL4–3 and SIVmac239 Nefs or containing a disrupted nef gene have been previously described (Wildum et al., 2006). All proviral constructs used in this study are listed in the Key Resources Table.
Expression vectors
Nef genes were amplified by PCR with flanking primers introducing XbaI and MluI restriction sites for cloning into the bi-cistronic CMV promoter-based pCG IRES eGFP vector as described (Schindler et al., 2006). pCG SMM tetherin and pBJ6 human SERINC5-HA protein expression vectors have been previously described (Kmiec et al., 2018; Munch et al., 2007).
Western blots
To examine the expression of SIVsmm Nef proteins, HEK293T cells were transfected in 12-well dishes with 2.5 μg DNA of pCG IRES eGFP vectors expressing AU1-tagged Nefs. Two days post-transfection, cells were lysed and proteins separated, blotted and detected as previously described (Kmiec et al., 2018).
Flow cytometry
CD4, MHC-I, CD28, CD3, CXCR4 and tetherin surface levels and GFP reporter molecules in PBMCs were measured as described previously (Joas et al., 2018). To analyze cell surface receptor modulation in primary cells, PBMCs were infected with VSV-G-pseudotyped HIV-1 NL4–3 IRES eGFP expressing different nef alleles. Three days post infection, cells were stained for CD4 (Life Technologies, MHCD0405), CD3 (BD, 555333), MHCI (Dako, R7000), CD28 (BD, 348047), or CXCR4 (BD, 555976) and analyzed by fluorescence-activated cell sorting (FACS). To calculate the specific effect of Nef on receptor cell surface expression, the mean fluorescence intensities obtained for cells coexpressing Nef and GFP were normalized to those obtained with the nef* construct expressing GFP only (100%).
Virus stocks
To generate virus stocks, HEK293T cells were co-transfected with the proviral HIV-1 NL4–3 constructs encoding containing various nef alleles and a plasmid (pHIT-G) expressing VSV-G to achieve comparably high infection levels for flow cytometric analysis and replication kinetics. Two days post-transfection, supernatants containing infectious virus were harvested. The amount of HIV-1 capsid protein was quantified by p24 antigen ELISA for normalization of the virus dose.
Viral infectivity
Virus infectivity was determined using TZM-bl cells. Briefly, the cells were sown out in 96-well dishes in a volume of 100 μl and infected with virus stocks produced by transfected HEK293T cells. Three days post-infection, viral infectivity was detected using the Gal-Screen kit from Applied Biosystems as recommended by the manufacturer. β-galactosidase activity was quantified as relative light units per second using the Orion microplate luminometer.
SERINC5 antagonism
To measure Nef-mediated SERINC5 counteraction, HEK293T cells were co-transfected using calcium phosphate with 1.5 μg of HIV-1 NL4–3 env* Δvpu IRES GFP reporter proviral constructs containing various nef alleles, 0.05μg pCAGGS_NL4–3 FLAG-tag env or 0.25μg pHIT VSV-g expression plasmid, and 1.25 μg pBJ6-SERINC5-HA expression plasmid or pBJ6-empty vector (12-well format). The HIV-1 NL4–3 env* Δvpu nef- construct was used as a negative control, whereas HIV-1 NL4–3 env* Δvpu nef- pseudotyped with VSV-G as well as HIV-1 NL43 env* Δvpu nef+ IRES eGFP and HIV-1 NL43 env* Δvpu SIVmac239 nef IRES eGFP served as positive controls. Two days post-transfection cell supernatants were harvested and infectious HIV-1 yield was quantified by TZM-bl infection assay. Yields obtained in the presence of pBJ6 SERINC5-HA were normalized to the corresponding pBJ6 empty vector controls (100%) for each virus.
Tetherin antagonism
To measure Nef-mediated SMM tetherin counteraction, HEK293T cells were co-transfected using calcium phosphate method with 4 μg HIV-1 NL4–3 provirus lacking Vpu expression (Δvpu), 1 μg Nef expression vector or empty vector as well as increasing amounts of SMM tetherin DNA. Two days post-transfection, supernatants and cells were harvested. Infectious HIV-1 yield was quantified by a 96-well TZM-bl infection assay whereas p24 concentration was determined by p24 ELISA quantification of cell free (CF) and cell associated (CA) capsid p24 antigen. Release was calculated as the percentage of CF p24 out of total (CF + CA) produced p24 antigen.
SIVmac239 Nef analysis
HEK293T cells were co-transfected with 5 μg of SIVmac239 IRES-eGFP proviral constructs encoding wild-type or mutant Nef proteins and 1 μg VSV-G expression vector. Two days post-transfection, virus stocks were harvested and concentrated six times using Amicon Ultra-15 centrifugal filter units (100 kDa). Rhesus PBMCs were isolated from rhesus monkey whole blood (provided by Simian lab Europe, Strasbourg) by Ficoll gradient centrifugation and stimulated with IL-2 and PHA for 3 days prior to infection. Three million cells were infected with a total of 300 μl virus stock using spinoculation at 1.200 rpm and 37°C for 2 hours. After spinoculation, 300 μl supernatant were removed before the cells were washed in 3 ml DPBS to remove input virus. Three days post infection, cell surface expression of CD4, CD3, MHC-I and tetherin was determined by flow cytometry. Antibodies used were: CD4 PerCP-Cy5.5-coupled, BD, 552838; CD3 V450-coupled, BD, 560351; MHC-I PE-coupled, BD, 555553; tetherin APC-coupled, Biolegend, 348410 or the respective isotype controls (IgG1κ PerCP-Cy5.5-coupled, BD, 552834; IgG1κ V450-coupled, BD, 560373; IgG1κ PE-coupled, BD, 555749; IgG1κ APC-coupled, Biolegend, 400122).
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis
For assays performed in Figures 1,3–5, mean measurements were processed using Origin (OriginLab, Northampton, MA). The mean activities in Figure 6 and Figure S6 were compared using Student’s t-test. Similar results were obtained with the Mann Whitney U test. The software package StatView version 4.0 (Abacus Concepts, Berkeley, CA) was used for these calculations. All data in these figures are shown as the mean of at least three independent experiments ± SEM. Significance differences are indicated as: *, p < 0.05; **, p < 0.01; ***, p < 0.001. Statistical parameters are specified in the figure legends.
DATA AND CODE AVAILABILITY
Image processing and modeling software
All software and code is available as detailed in the key resources table.
Cryo-EM map, raw data and model accession numbers
EM density has been deposited in the EMDB with accession code EMD-20217. Atomic coordinates have been deposited in the PDB with the accession code 6OWT. See also Table S1
Supplementary Material
Highlights.
Cryo-EM structure of AP-2, simian tetherin, and SIVsmm Nef complex at 3.8 Å resolution ·
Nef refolds the β2 subunit of AP-2 for binding of the simian tetherin DIWK sequence
Tetherin binding site is distinct from those of Nef substrates MHC-I, CD3, and CD4
Tetherin binding site on Nef overlaps with the site for SERINC5 binding
Acknowledgments
We thank D. Toso, S. Fromm, and A. Yokom for cryo-EM support and advice and D. Sauter for comments on the manuscript. Access to the FEI Titan Krios was provided through the BACEM UCB facility. This research was supported by NIH grants R01 AI120691 (X. R.), P50 GM082250 (J. H. H.), F32 GM125209 (C. Z. B.) and an Advanced ERC grant ‘Anti-Virome’ and DFG-funded SFB 1279 and SPP 1923 (F. K.). Additional support was provided by a generous gift from the Andrew Dougherty Vision Foundation (C.Z.B.)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors declare no competing interests.
References
- Afonine PV, Poon BK, Read RJ, Sobolev OV, Terwilliger TC, Urzhumtsev A, and Adams PD (2018). Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr D Struct Biol 74, 531–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiken C, Konner J, Landau NR, Lenburg ME, and Trono D (1994). Nef induces CD4 endocytosis: requirement for a critical dileucine motif in the membrane-proximal CD4 cytoplasmic domain. Cell 76, 853–864. [DOI] [PubMed] [Google Scholar]
- Barad BA, Echols N, Wang RY, Cheng Y, DiMaio F, Adams PD, and Fraser JS (2015). EMRinger: side chain-directed model and map validation for 3D cryo-electron microscopy. Nat Methods 12, 943–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bresnahan PA, Yonemoto W, Ferrell S, Williams-Herman D, Geleziunas R, and Greene WC (1998). A dileucine motif in HIV-1 Nef acts as an internalization signal for CD4 downregulation and binds the AP-1 clathrin adaptor. Curr Biol 8, 1235–1238. [DOI] [PubMed] [Google Scholar]
- Canagarajah BJ, Ren X, Bonifacino JS, and Hurley JH (2013). The clathrin adaptor complexes as a paradigm for membrane-associated allostery. Protein Sci 22, 517–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri R, Lindwasser OW, Smith WJ, Hurley JH, and Bonifacino JS (2007). Downregulation of CD4 by human immunodeficiency virus type 1 Nef is dependent on clathrin and involves direct interaction of Nef with the AP2 clathrin adaptor. J Virol 81, 3877–3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins DR, and Collins KL (2014). HIV-1 accessory proteins adapt cellular adaptors to facilitate immune evasion. PLoS Pathog 10, e1003851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig HM, Pandori MW, and Guatelli JC (1998). Interaction of HIV-1 Nef with the cellular dileucine-based sorting pathway is required for CD4 down-regulation and optimal viral infectivity. Proc Natl Acad Sci U S A 95, 11229–11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB 3rd, Snoeyink J, Richardson JS, et al. (2007). MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res 35, W375–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y (2015). Fluorescence polarization assay to quantify protein-protein interactions in an HTS format. Methods Mol Biol 1278, 529–544. [DOI] [PubMed] [Google Scholar]
- DuBridge RB, Tang P, Hsia HC, Leong PM, Miller JH, and Calos MP (1987). Analysis of mutation in human cells by using an Epstein-Barr virus shuttle system. Molecular and cellular biology 7, 379–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, and Cowtan K (2010). Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouchier RA, Meyer BE, Simon JH, Fischer U, and Malim MH (1997). HIV-1 infection of non-dividing cells: evidence that the amino-terminal basic region of the viral matrix protein is important for Gag processing but not for post-entry nuclear import. EMBO J 16, 4531–4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heusinger E, Deppe K, Sette P, Krapp C, Kmiec D, Kluge SF, Marx PA, Apetrei C, Kirchhoff F, and Sauter D (2018). Preadaptation of Simian Immunodeficiency Virus SIVsmm Facilitated Env-Mediated Counteraction of Human Tetherin by Human Immunodeficiency Virus Type 2. J Virol 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoover DM, and Lubkowski J (2002). DNAWorks: an automated method for designing oligonucleotides for PCR-based gene synthesis. Nucleic Acids Res 30, e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson LP, Kelly BT, McCoy AJ, Gaffry T, James LC, Collins BM, Honing S, Evans PR, and Owen DJ (2010). A large-scale conformational change couples membrane recruitment to cargo binding in the AP2 clathrin adaptor complex. Cell 141, 1220–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia B, Serra-Moreno R, Neidermyer W, Rahmberg A, Mackey J, Fofana IB, Johnson WE, Westmoreland S, and Evans DT (2009). Species-specific activity of SIV Nef and HIV-1 Vpu in overcoming restriction by tetherin/BST2. PLoS Pathog 5, e1000429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia X, Singh R, Homann S, Yang H, Guatelli J, and Xiong Y (2012). Structural basis of evasion of cellular adaptive immunity by HIV-1 Nef. Nat Struct Mol Biol 19, 701–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia X, Weber E, Tokarev A, Lewinski M, Rizk M, Suarez M, Guatelli J, and Xiong Y (2014). Structural basis of HIV-1 Vpu-mediated BST2 antagonism via hijacking of the clathrin adaptor protein complex 1. Elife 3, e02362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joas S, Parrish EH, Gnanadurai CW, Lump E, Sturzel CM, Parrish NF, Learn GH, Sauermann U, Neumann B, Rensing KM, et al. (2018). Species-specific host factors rather than virus-intrinsic virulence determine primate lentiviral pathogenicity. Nat Commun 9, 1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimanius D, Forsberg BO, Scheres SH, and Lindahl E (2016). Accelerated cryo-EM structure determination with parallelisation using GPUs in RELION-2. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluge SF, Mack K, Iyer SS, Pujol FM, Heigele A, Learn GH, Usmani SM, Sauter D, Joas S, Hotter D, et al. (2014). Nef proteins of epidemic HIV-1 group O strains antagonize human tetherin. Cell Host Microbe 16, 639–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kmiec D, Akbil B, Ananth S, Hotter D, Sparrer KMJ, Sturzel CM, Trautz B, Ayouba A, Peeters M, Yao Z, et al. (2018). SIVcol Nef counteracts SERINC5 by promoting its proteasomal degradation but does not efficiently enhance HIV-1 replication in human CD4+ T cells and lymphoid tissue. PLoS Pathog 14, e1007269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kueck T, Foster TL, Weinelt J, Sumner JC, Pickering S, and Neil SJ (2015). Serine Phosphorylation of HIV-1 Vpu and Its Binding to Tetherin Regulates Interaction with Clathrin Adaptors. PLoS Pathog 11, e1005141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Gall S, Erdtmann L, Benichou S, Berlioz-Torrent C, Liu L, Benarous R, Heard JM, and Schwartz O (1998). Nef interacts with the mu subunit of clathrin adaptor complexes and reveals a cryptic sorting signal in MHC I molecules. Immunity 8, 483–495. [DOI] [PubMed] [Google Scholar]
- Le Tortorec A, and Neil SJ (2009). Antagonism to and intracellular sequestration of human tetherin by the human immunodeficiency virus type 2 envelope glycoprotein. J Virol 83, 11966–11978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard JA, Filzen T, Carter CC, Schaefer M, and Collins KL (2011). HIV-1 Nef disrupts intracellular trafficking of major histocompatibility complex class I, CD4, CD8, and CD28 by distinct pathways that share common elements. J Virol 85, 6867–6881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack K, Starz K, Sauter D, Langer S, Bibollet-Ruche F, Learn GH, Sturzel CM, Leoz M, Plantier JC, Geyer M, et al. (2017). Efficient Vpu-Mediated Tetherin Antagonism by an HIV-1 Group O Strain. J Virol 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malim MH, and Bieniasz PD (2012). HIV Restriction Factors and Mechanisms of Evasion. Cold Spring Harbor perspectives in medicine 2, a006940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manrique S, Sauter D, Horenkamp FA, Lulf S, Yu H, Hotter D, Anand K, Kirchhoff F, and Geyer M (2017). Endocytic sorting motif interactions involved in Nef-mediated downmodulation of CD4 and CD3. Nat Commun 8, 442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moerke NJ (2009). Fluorescence Polarization (FP) Assays for Monitoring Peptide-Protein or Nucleic Acid-Protein Binding. Curr Protoc Chem Biol 1, 1–15. [DOI] [PubMed] [Google Scholar]
- Morris KL, Buffalo CZ, Sturzel CM, Heusinger E, Kirchhoff F, Ren X, and Hurley JH (2018). HIV-1 Nefs Are Cargo-Sensitive AP-1 Trimerization Switches in Tetherin Downregulation. Cell 174, 659–671 e614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munch J, Rajan D, Schindler M, Specht A, Rucker E, Novembre FJ, Nerrienet E, Muller-Trutwin MC, Peeters M, Hahn BH, et al. (2007). Nef-mediated enhancement of virion infectivity and stimulation of viral replication are fundamental properties of primate lentiviruses. J Virol 81, 13852–13864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neil SJ, Zang T, and Bieniasz PD (2008). Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 451, 425–430. [DOI] [PubMed] [Google Scholar]
- Olsen SK, and Lima CD (2013). Structure of a ubiquitin E1-E2 complex: insights to E1-E2 thioester transfer. Mol Cell 49, 884–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira EA, and daSilva LL (2016). HIV-1 Nef: Taking Control of Protein Trafficking. Traffic 17, 976–996. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004). UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- Platt EJ, Wehrly K, Kuhmann SE, Chesebro B, and Kabat D (1998). Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J Virol 72, 2855–2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punjani A, Rubinstein JL, Fleet DJ, and Brubaker MA (2017). cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods 14, 290–296. [DOI] [PubMed] [Google Scholar]
- Ren X, Farias GG, Canagarajah BJ, Bonifacino JS, and Hurley JH (2013). Structural basis for recruitment and activation of the AP-1 clathrin adaptor complex by Arf1. Cell 152, 755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X, Park SY, Bonifacino JS, and Hurley JH (2014). How HIV-1 Nef hijacks the AP-2 clathrin adaptor to downregulate CD4. Elife 3, e01754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohou A, and Grigorieff N (2015). CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol 192, 216–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa A, Chande A, Ziglio S, De Sanctis V, Bertorelli R, Goh SL, McCauley SM, Nowosielska A, Antonarakis SE, Luban J, et al. (2015). HIV-1 Nef promotes infection by excluding SERINC5 from virion incorporation. Nature 526, 212–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauter D, and Kirchhoff F (2019). Key Viral Adaptations Preceding the AIDS Pandemic. Cell Host Microbe 25, 27–38. [DOI] [PubMed] [Google Scholar]
- Sauter D, Schindler M, Specht A, Landford WN, Munch J, Kim KA, Votteler J, Schubert U, Bibollet-Ruche F, Keele BF, et al. (2009). Tetherin-driven adaptation of Vpu and Nef function and the evolution of pandemic and nonpandemic HIV-1 strains. Cell Host Microbe 6, 409–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SH (2012). RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol 180, 519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler M, Munch J, Kutsch O, Li H, Santiago ML, Bibollet-Ruche F, Muller-Trutwin MC, Novembre FJ, Peeters M, Courgnaud V, et al. (2006). Nef-mediated suppression of T cell activation was lost in a lentiviral lineage that gave rise to HIV-1. Cell 125, 1055–1067. [DOI] [PubMed] [Google Scholar]
- Schmokel J, Li H, Bailes E, Schindler M, Silvestri G, Hahn BH, Apetrei C, and Kirchhoff F (2009). Conservation of Nef function across highly diverse lineages of SIVsmm. Retrovirology 6, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmokel J, Sauter D, Schindler M, Leendertz FH, Bailes E, Dazza MC, Saragosti S, Bibollet-Ruche F, Peeters M, Hahn BH, et al. (2011). The presence of a vpu gene and the lack of Nef-mediated downmodulation of T cell receptor-CD3 are not always linked in primate lentiviruses. J Virol 85, 742–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schouest B, Weiler AM, Janaka SK, Myers TA, Das A, Wilder SC, Furlott J, Baddoo M, Flemington EK, Rakasz EG, et al. (2018). Maintenance of AP-2-Dependent Functional Activities of Nef Restricts Pathways of Immune Escape from CD8 T Lymphocyte Responses. J Virol 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selleck W, and Tan S (2008). Recombinant protein complex expression in E. coli. Curr Protoc Protein Sci Chapter 5, Unit 5 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra-Moreno R, Zimmermann K, Stern LJ, and Evans DT (2013). Tetherin/BST-2 antagonism by Nef depends on a direct physical interaction between Nef and tetherin, and on clathrin-mediated endocytosis. PLoS Pathog 9, e1003487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheffield P, Garrard S, and Derewenda Z (1999). Overcoming expression and purification problems of RhoGDI using a family of “parallel” expression vectors. Protein Expr. Purif 15, 34–39. [DOI] [PubMed] [Google Scholar]
- Shen QT, Ren X, Zhang R, Lee IH, and Hurley JH (2015). HIV-1 Nef hijacks clathrin coats by stabilizing AP-1:Arf1 polygons. Science 350, aac5137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traub LM, and Bonifacino JS (2013). Cargo Recognition in Clathrin-Mediated Endocytosis. Cold Spring Harbor Perspectives in Biology 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usami Y, Wu Y, and Gottlinger HG (2015). SERINC3 and SERINC5 restrict HIV-1 infectivity and are counteracted by Nef. Nature 526, 218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildum S, Schindler M, Munch J, and Kirchhoff F (2006). Contribution of Vpu, Env, and Nef to CD4 down-modulation and resistance of human immunodeficiency virus type 1-infected T cells to superinfection. J Virol 80, 8047–8059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Landford WN, Ng M, McNatt MW, Bieniasz PD, and Hatziioannou T (2011). SIV Nef proteins recruit the AP-2 complex to antagonize Tetherin and facilitate virion release. PLoS Pathog 7, e1002039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Wilson SJ, Landford WC, Virgen B, Gregory D, Johnson MC, Munch J, Kirchhoff F, Bieniasz PD, and Hatziioannou T (2009). Nef proteins from simian immunodeficiency viruses are tetherin antagonists. Cell Host Microbe 6, 54–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K (2016). Gctf: Real-time CTF determination and correction. J Struct Biol 193, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, and Agard DA (2017). MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14, 331–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivanov J, Nakane T, Forsberg BO, Kimanius D, Hagen WJ, Lindahl E, and Scheres SH (2018). New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Image processing and modeling software
All software and code is available as detailed in the key resources table.
Cryo-EM map, raw data and model accession numbers
EM density has been deposited in the EMDB with accession code EMD-20217. Atomic coordinates have been deposited in the PDB with the accession code 6OWT. See also Table S1