Abstract
Bacteria utilize multiple mechanisms that enable them to gain or acquire resistance to antibiotic therapies during the treatment of infections. In addition, bacteria form biofilms which are surface-attached communities of enriched populations containing persister cells encased within a protective extracellular matrix of biomolecules, leading to chronic and recurring antibiotic-tolerant infections. Antibiotic resistance and tolerance are major global problems that require innovative therapeutic strategies to address the challenges associated with pathogenic bacteria. Historically, natural products have played a critical role in bringing new therapies to the clinic to treat life-threatening bacterial infections. This Perspective provides an overview of antibiotic resistance and tolerance and highlights recent advances (chemistry, biology, drug discovery, and development) from various research programs involved in the discovery of new antibacterial agents inspired by a diverse series of natural product antibiotics.
Graphical Abstract

INTRODUCTION
The discovery and routine administration of antibiotics to treat bacterial infections led to their recognition as “wonder drugs” in the mid-20th century.1–9 To date, antibiotics are still considered one of the most significant interventions in human medicine. The efficacy and selectivity antibiotics exert made many believe that bacterial infections would be a thing of the past. However, instead of the routine eradication of bacterial pathogens in the clinic, we have witnessed an alarming rise in the failure to treat bacterial infections due to the generation of antibiotic-resistant “superbugs”3,5 and antibiotic-tolerant persister cells and biofilms.10–12
Clinically approved antibiotics operate via the selective inhibition of critical targets that impede replication and growth or eradicate bacterial cells. It is interesting to note that relatively few bacterial targets have been exploited for antibiotic treatments in the clinic. The mechanisms of action for our arsenal of antibiotics include the inhibition or disruption of (1) cell wall synthesis (e.g., β-lactams,13 vancomycin14), (2) protein synthesis (e.g., tetracyclines,15 macrolides,16 aminoglycosides,17 linezolid18), (3) RNA poly-merase (e.g., rifamycins19–21), (4) nucleic acid synthesis (e.g., quinolones22–25), (5) folate biosynthesis (e.g., sulfonamides, trimethoprim),26 and (6) cell membrane (e.g., polymyxins;26 Figure 1).4,7
Figure 1.

Antibacterial targets of conventional antibiotics and mechanisms by which bacteria develop resistance to these therapeutic agents.
Upon exposure to antibiotics, bacteria can expediently evade their action using one or more well-defined resistance mechanisms.3–6,8,27,28 Bacterial resistance has led to the catastrophic rise of roughly 2 million annual infections, resulting in 23 000 deaths in the United States.29 In Europe, more than 33 000 deaths were attributed to antibiotic-resistant bacterial infections in 2015.30 Universal acquired resistance mechanisms can prevent antibiotics access to the aforementioned intracellular targets. The cell wall of Gram-negative bacteria consists of inner and outer membranes, which upon mutation can act as a permeability barrier to antibiotics leading to reduced penetration into the cells.31,32 Alternatively, bacteria can actively eliminate, or “pump out”, antibiotics before they bind their corresponding intracellular target via the action of efflux pumps (e.g., efflux of macrolides).31,32 Mutations and modifications of the target’s binding site are, collectively, another resistance mechanism bacteria utilize to acquire resistance as the resulting changes negatively impact drug–target interactions of antibiotic therapies. Specific examples of mutation and modification to impede antibiotic binding sites include (1) mutation and alteration of penicillin binding proteins to negate the effects of penicillin, (2) structural alterations in DNA gyrase to prevent fluoroquinolone action, and (3) methylation of critical residues within the rRNA of bacterial ribosomes to eliminate the binding of aminoglycosides.33–35 Additionally, bacteria can enzymatically modify antibiotics to generate inactive agents, and examples include (1) hydrolysis of β-lactam antibiotics by β-lactamase enzymes to cleave and destroy the critical β-lactam warhead (pharmacophore) required for antibacterial activities and (2) modification, or functionalization (e.g., acylation), of hydroxyl and/or amine groups critical for aminoglycoside antibiotics to hydrogen bond (bind) to bacterial ribosomes by aminoglycoside-inactivating enzymes.29 In addition, bacteria can overproduce targets to bypass the effects of antibiotics (e.g., trimethoprim; Figure 1).29,35
Innate antibiotic tolerance is a distinct microbiological phenomena from acquired resistance and is now recognized as the underlying cause of chronic and recurring bacterial infections.36 Using a communication process known as quorum sensing, bacteria secrete and sense small organic signaling molecules to monitor their population density and coordinate virulent behaviors.37–41 Bacteria use quorum sensing to coordinate surface attachment and subsequent biofilm formation, where dense communities of bacterial cells are encased within matrixes of polysaccharides and extracellular DNA.42–46 Once within a biofilm, bacterial cells divide at a much slower rate compared to their planktonic (free-floating) counterparts or they do not replicate at all. The subpopulation of biofilm cells that enter into a dormant state are known as “persister cells” and are characterized as metabolically inactive.12,47–51 Extracellular polymeric substances (EPSs), or biofilm matrix, provide an additional layer of protection to surface-attached communities, enabling these bacteria to thrive in harsh conditions (e.g., host immune response).52,53
It is important to note that all classes of FDA-approved conventional antibiotics were initially discovered as bacterial growth inhibitors against actively dividing cultures. Therefore, it should be no surprise that clinically used antibiotics are essentially ineffective against nonreplicating biofilm infections. With the 17 million new cases of biofilm infections in addition to >500 000 deaths each year in the United States that result from these infections, there is a critical need to identify compounds that effectively eradicate biofilms through growth-independent mechanisms (Figure 2).10,41
Figure 2.

Presentation of the distinctions between acquired antibiotic resistance and innate antibiotic tolerance.
The current need for new and effective antibiotic therapies has motivated several research groups to initiate drug discovery programs inspired by various antibiotic natural products, which may be known or recently discovered. The Lewis group has created a new technology to facilitate isolation of novel antibiotics from previously inaccessible sources, while the Myers lab has developed novel total synthesis platforms for the discovery of new antibiotics. The Hergenrother group elegantly utilized a collection of diverse small molecules to establish guidelines that will allow the targeting of clinically imposing Gram-negative pathogens. The Miller group has pioneered a Trojan horse strategy and continues to make advances in this area by taking advantage of siderophore systems for bacterial import in unique and elegant ways to impart activity against Gram-negative organisms. Recent work by the Wuest lab has focused on a natural product with narrow-spectrum activity against Pseudomonas aeruginosa. Both broad- and narrow-spectrum-based strategies have been employed, but scientists agree that care must be taken to minimize resistance development against these novel agents. With biofilms being inherently tolerant to current antibiotics, the Lewis group and our lab have identified agents capable of eradicating persister cells and established biofilms. Our group has identified a series of synthetically tunable halogenated phenazine small molecules derived from a marine antibiotic that potently eradicates bacterial biofilms through a rapid iron starvation mechanism. Fortunately, recent advances in chemistry, molecular biology, and chemical biology have yielded a bevy of powerful techniques that can be employed to better understand problems associated with pathogenic bacteria and enable the discovery of novel natural-product-inspired therapies. This Perspective will detail each of these innovative and exciting antibiotic discovery strategies (Figure 3).
Figure 3.

Strategies to address the problems associated with antibiotic resistance and tolerance presented in this Perspective.
TEIXOBACTIN, A PROMISING NEW ANTIBIOTIC: iChip DISCOVERY, TOTAL SYNTHESIS, AND NOVEL ANALOGUES
Bacteria compete for finite resources in their environment and often produce antibiotics to compete with each other. Many antimicrobials used in the clinic today have been derived from natural products produced by bacteria. Large numbers of organisms cannot be grown successfully through traditional culturing methods and are thus named “unculturable”.1 It is believed that approximately 99% of all species in external environments are uncultured.54 However, recent development of iChip technology by the Lewis group has enabled the growth of previously uncultured bacteria (Figure 4).54,55 This new technology has enhanced the potential for many more types of bacteria to be investigated and screened for the identification of new antibiotics.
Figure 4.

iChip (multichannel device) technology developed by Lewis and co-workers to culture previously “unculturable” bacteria, which led to the discovery of the new antibiotic teixobactin 1.
By use of the iChip, extracts from 10 000 bacterial isolates were grown and screened, which led to one isolate (β-proteobacteria, named Eleftheria terrae) that demonstrated antibacterial activity against S. aureus. From the Eleftheria terrae extract, teixobactin 1 was isolated and structurally elucidated using mass spectrometry, NMR, and Marfey’s analysis.55 Teixobactin 1 proved to be a unique antibiotic containing a cyclic depsipeptide moiety composed of four amino acid residues, including an enduracididine residue, that are appended to a seven amino acid linear chain bearing a methylphenylalanine. It is also interesting to note that 1 has four d-amino acid residues incorporated into its impressive structure. The biosynthetic gene cluster consists of two nonribosomal peptide synthetase (NRPS) genes named txo1 and txo2 where 11 modules are encoded. The depsipeptide 1 showed excellent antibacterial activities in minimum inhibitory concentration (MIC) assays against significant, Gram-positive pathogens, including S. aureus (MIC = 0.25 μg/mL against methicillin-sensitive and methicillin-resistant strains; 1 also demonstrated antibacterial activities against intermediate vancomycin-resistant S. aureus, VISA), enterococci (MIC = 0.5 μg/mL, vancomycin-resistant enterococci strains, VRE), M. tuberculosis (MIC = 0.125 μg/mL), Clostridium dif f icile (MIC = 0.005 μg/mL), and Streptococcus pneumoniae (MIC ≤ 0.03 μg/mL, penicillin-resistant strain) as reported by Lewis and co-workers.54
A common technique for determining the mechanism of action for antibiotics is to generate resistant mutants. Lewis and colleagues were not able to develop resistant mutants of S. aureus or M. tuberculosis, indicating that teixobactin’s target may not be a protein. This hypothesis was supported by the inhibition of peptidoglycan synthesis by 1, while no effect was observed on biosynthesis of DNA, RNA, or proteins. Because true vancomycin resistance in S. aureus is rare and vancomycin is known to bind lipid II, Lewis and co-workers hypothesized that teixobactin could also exert its activity through interaction with this target. When whole cells (S. aureus) were treated with 1, a significant amount of the peptidoglycan precursor undecaprenyl-N-acetylmuramic acid-pentapeptide (UDP-Mur-NAc-pentapeptide) accumulated in a similar fashion to cells treated with vancomycin. Competition enzyme binding assays indicated that 1 does not inhibit MurG, FemX, or PBP2 enzymes directly, while the addition of lipid II prevented 1 from inhibiting growth against S. aureus. These combined results demonstrate that 1 specifically interacts with the peptidoglycan precursor rather than interfering with the activity of the enzymes. Teixobactin retained activity against VRE which has a modified lipid II, suggesting that it binds at a different site than vancomycin.54
The therapeutic potential of 1 was also reported by Lewis and co-workers.54 Teixobactin retained its antibacterial potency in the presence of serum and demonstrated good chemical and microsomal stability and low toxicity. The pharmacokinetic parameters showed that after a single 20 mg per kg dose (iv injection) of 1 in mice, serum concentrations were maintained above MIC values for 4 h. Animal efficacy studies were then performed with 1 in a mouse septicemia model against MRSA at a dose that leads to 90% death; however, treatment with 1 (iv at 20 mg/kg, 1 h after infection) resulted in the survival of all mice, and a later experiment determined the PD50 (protective dose at which half of the animals survive) of 1 to be 0.2 mg/kg. 1 was also shown to be efficacious in a thigh infection of S. aureus and a lung infection model of S. pneumoniae, causing a 6 log10 reduction of colony forming units (CFU) per mL.54
Following the initial report of teixobactin’s discovery, two total syntheses of 1 have been published utilizing solid-phase technology in separate reports by Payne and co-workers56 and Li and co-workers.57 In both total syntheses of 1, macro-lactamization at the less hindered Thr8-Ala9 connection has proved to be the fruitful cyclization approach (see conversion of 5 to 6, Figure 5A, by Li and co-workers) as opposed to a macrolactonization strategy. Following the macrolactamization step, Li and co-workers coupled linear peptide 3 to cyclic peptide 6 using a key ligation step in pyridine/acetic acid before final HPLC conversion to 1. Yuan and co-workers developed a stereoselective and scalable synthetic route to the rare l-allo-enduracididine (L-allo-End)58 moiety of 1, which played a critical role in the total synthesis reported by Li and co-workers.
Figure 5.
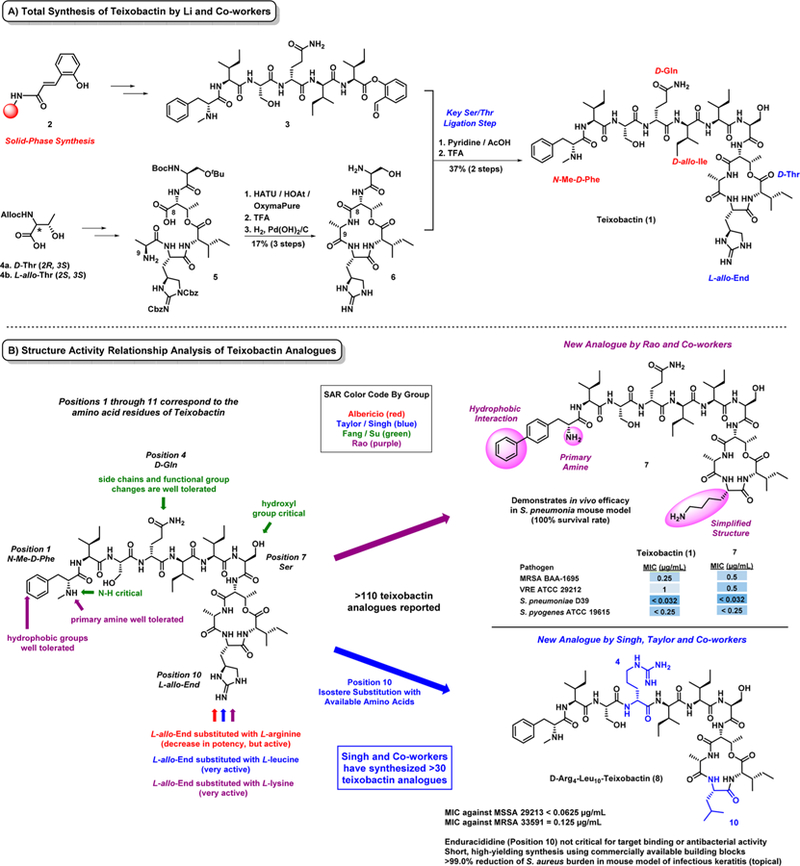
(A) Total synthesis of the new antibiotic teixobactin 1 and (B) structure–activity relationship (SAR) insights.
In addition to total synthesis campaigns, solid-phase chemistry has enabled initial medicinal chemistry efforts of 1 by multiple groups, including Albericio,59 Singh,60–63 Su and Fang,64 Rao,65 Li,66 and Chan.67 Several of these groups have probed the structural requirements of the l-allo-enduracididine moiety at position 10 of 1. If this unique residue is not required for antibacterial activities, then more simplified analogues of 1 can be prepared more readily. Albericio and co-workers were the first to report the replacement of the unusual l-allo-enduracididine residue of 1 by substituting with L-arginine; however, this new analogue lost significant antibacterial activities against S. aureus (MIC = 1.6 μg/mL) and B. subtilis (MIC = 0.40 μg/mL) based on MIC value comparison (1 reported MIC values with ≥8-fold more potent activities when compared to this analogue).59 Fang and Su and co-workers synthesized nine teixobactin analogues, which establish that (1) a guanidine or amine group at position 10, (2) the hydroxyl group of serine (position 7), (3) the N–H proton of the terminal phenylalanine (position 1) were all critical for antibacterial activity. During these investigations, Fang and Su also reported that position 4 can tolerate various side chain size and functional group content.64
The initial reports of teixobactin analogues gave insightful information into the SAR of 1; however, a “first wave” of synthetic analogues reported by Albericio, Su, Fang, and co-workers59,64 lost a substantial amount of antibacterial activity and appeared to be less than exciting lead molecules. Rao and co-workers65 recently reported the synthesis of 21 new teixobactin analogues, including simplified and equipotent analogue 7 (Figure 5B). This work demonstrated that the N-Me-d-Phe residue at position 1 of teixobactin (1) tolerates additional hydrophobic groups on the phenyl ring and (2) does not require the methyl group on the amine, as the primary amine was well tolerated. These structural modifications combined with a l-lysine substitution of the l-allo-enduracididine residue at position 10 resulted in equipotent analogue 7, which also demonstrated outstanding in vivo efficacy in a S. pneumonia mouse model of infection (100% survival rate of mice infected with bacterial loads that lead to 90% death in controls).
DEFINING RULES TO TARGET GRAM-NEGATIVE BACTERIA AND CONVERTING A DEOXYNYBOMYCIN ANALOGUE TO A BROAD-SPECTRUM ANTIBIOTIC
Gram-negative pathogens are a major clinical problem due to their lack of sensitivity to many commonly used anti-biotics.68,69 Quinolones were the last new class of antibacterial agents active against Gram-negative bacteria introduced into the clinic in 1968.1 Despite extensive high-throughput screening efforts against Escherichia coli with large compound libraries of synthetic compounds, no lead compounds were translated into clinical agents.70 With two cellular membranes and a lipopolysaccharide-coated outer membrane, Gram-negative bacteria pose significant challenges for compound entry.68,71–73 Compounds capable of penetrating the outer membrane do so through porins, which are narrow trans-membrane proteins lined with charged amino acids. Once inside Gram-negative bacterial cells, small molecules are susceptible to removal through the function of efflux pumps; therefore, in order for a compound to accumulate within Gram-negative bacteria and engage its intracellular target, compounds are required to enter through porins faster than being removed via efflux.68
Hergenrother and co-workers noted that despite initial efforts by others, we have a limited understanding of the physicochemical properties enabling compound accumulation in Gram-negative bacteria.68,72,73 A structurally diverse set of 100 compounds with unique scaffolds rapidly accessed from available natural products were used to systematically analyze small molecule accumulation in E. coli (whole cells) using a liquid chromatography with tandem mass spectroscopy (LC–MS/MS) method. This approach using whole cells allowed multiple variables affecting compound accumulation to be accounted for, such as multiple porins, efflux pumps, and lipopolysaccharide composition. Conventional antibiotics with known high (i.e., tetracycline) and low (i.e., novobiocin) accumulation properties were evaluated as controls and reported to validate the method.68
Natural-product-derived compounds were selected for this study as most clinically used antibacterial drugs are either natural products or derivatives. The compound collection utilized by the Hergenrother lab was synthesized by this team, so these compounds are synthetically accessible with tunable properties to enable structure–activity relationships geared toward accumulation in Gram-negative bacteria. The library of 100 compounds utilized for this work, which was generated utilizing the “complexity-to-diversity” approach,74–78 included positively charged, negatively charged, and neutral compounds.68 The chemical structure, molecular weight, ClogD7.4, and charge of all compounds were tracked with accumulation results in E. coli.
From the initial accumulation studies, the primary factor that governed accumulation in E. coli was charge, with 12 of 41 positively charged compounds being the most likely to accumulate. Interestingly, 8 of the 12 positively charged compounds were primary amines.68 In follow-up SAR investigations, replacement of the primary amine with different functional groups (e.g., carboxylic acid, amide, alcohol) or amines bearing more substitutions significantly reduced accumulation in E. coli. Although the presence of a primary amine was important for accumulation, it alone was insufficient; therefore, chemoinformatics was utilized to understand which factors contribute to amine accumulation. This work revealed that the flexibility, as measured by the number of rotatable bonds, and shape of a compound are important factors that govern accumulation. Compound shape was described by “globularity” to inform the three-dimensionality. Shape was scored on a scale from 0 globularity for a “flat” compound (e.g., benzene) to 1 globularity for a “spherical” compound (e.g., adamantane).
On the basis of the extensive data set generated by Hergenrother and co-workers, the following guiding principles for accumulation in E. coli were developed: (1) compounds are most likely to accumulate if they contain a nonsterically encumbered amine, (2) contain some nonpolar functionality (typically not a problem for organic compounds), (3) contain a rigid structure and (4) have low globularity.68 These guidelines are referred to as the “eNTRy rules” (N, ionizable nitrogen, primary amine is best; T, low three-dimensionality, globularity score of ≤0.25; R, relatively rigid, ≤5 rotatable bonds).73 These guidelines were validated retrospectively with antibacterial agents presented by O’Shea and Moser.79 In addition, the role of porins was examined by testing the high-accumulation antibiotic controls and test compounds in a strain bearing a diminished number of porins.68 As predicted, a decrease in accumulation was observed for all compounds tested in the strain with fewer porins. Molecular modeling was performed with select test compounds and antibacterial agents that transverse the major porin of E. coli, OmpF, revealing a key interaction between the necessary primary, or non-hindered, amine and acidic residues (most often Asp113) that allow high accumulating compounds.
These well-defined guidelines were instructive in converting an analogue of the Gram-positive antibiotic deoxynybomycin (DNM, 9) into the broad-spectrum agent, 6DNM-NH3 (11, Figure 6).68 DNM is an inhibitor of DNA gyrase,80 however, only active against Gram-positive pathogens, presumably due to minimal accumulation in Gram-negative bacteria.68 DNM possesses shape and rigidity features that align with the eNTRy rules; however, no amine group is present. Analogue 6DNM 10 possesses an aliphatic bridge expanded by one methylene unit to give a six-membered ring, compared to DNM’s (9) five-membered ring.
Figure 6.

eNTRy rules defined by Hergenrother and co-workers that enabled the successful conversion of the Gram-positive antibacterial agent 6DNM 10 into 6DNM-NH3 11, a new broad-spectrum antibacterial agent.
As one would expect based on the small structural differences between analogues, DNM 9 and 6DNM 10 possess the same activity profile (S. aureus MIC = 0.06–1 μg/mL; E. coli MIC > 32 μg/mL). However, 6DNM-NH3 11 has incorporated a primary amine appended to the aliphatic region of the six-membered ring, and as predicted based on the eNTRy rules, 11 demonstrates significantly higher levels of accumulation compared to 6DNM 10 (11, accumulation = 1114 nmol per 1012 CFU; 10, accumulation = 298 nmol per 1012 CFU). In line with eNTRy rule design and observed increases regarding E. coli accumulation, 6DNM-NH3 demonstrates broad-spectrum antibacterial activities (11, S. aureus MIC = 0.03–0.5 μg/mL; E. coli MIC = 0.5–16 μg/mL), including activities against several Gram-negative pathogens (11, A. baumannii MIC = 2–16 μg/mL; K. pneumoniae MIC = 1–8 μg/mL; E. cloacae MIC = 0.5–4 μg/mL; P. aeruginosa MIC = 16 μg/mL).68 Steered molecular dynamics simulations indicated that 6DNM-NH3 11 was able to pass through the porin OmpF with the assistance of the key interaction between the primary amine of 11 and Asp113 within the porin channel; however, 6DNM 10 was incapable of this interaction and required distortion in Asp113 and neighboring residues to allow passage. This ground-breaking work will undoubtedly pave the way for the discovery and development of antibiotics against Gram-negative pathogens.72
DEVELOPING TOTAL SYNTHESIS PLATFORMS FOR NEW, FULLY SYNTHETIC ANTIBIOTICS
One of the primary methods to discover and develop new antibacterial agents over the past 60 years has been semisynthesis, which is chemical synthesis using natural products (e.g., antibiotics) as starting points.81 Early examples of semisynthesis in the antibacterial arena include the conversion of (1) streptomycin to dihydrostreptomycin (via a chemoselective reduction of the aldehyde in streptomycin to a primary alcohol to yield a new, more stable antibacterial agent) and (2) chlorotetracycline to tetracycline (via hydro-genation; tetracycline was later isolated). With these early success stories, medicinal chemists realized the value of semisynthesis for antibacterial discovery and utilized this approach to advance other antibiotic classes (e.g., macrolides, synthetic transformation of erythromycin to azithromycin in four steps); however, this approach has significant limitations as deep-seated modifications are not always accessible via semisynthesis. Myers and co-workers are working to address these limitations by developing modular total synthesis platforms, and they have made significant progress generating novel tetracycline81–88 and macrolide antibiotics89–93 bearing structural modifications not possible through current semisynthetic approaches.
In the mid-1990s, the Myers lab began work on fully synthetic tetracycline antibiotics.82 This program led to the implementation of a high-yielding, diastereoselective Michael–Claisen annulation between benzoate esters related to 13 and chiral “AB enone” 14, resulting in the constructive C-ring “stitching” of new tetracycline antibiotics.81–86 The “diversity element” of this route can be clearly seen in structure 15 (Figure 7), which bears four positions poised for structural modification. On the basis of structure–activity relationship knowledge, the diversity element tracks to positions that do not negatively impact bacterial ribosome inhibition by tetracyclines. This incredible chemistry platform has enabled >3000 fully synthetic tetracyclines to date,81,82,86 several of which have proven to be promising clinical candidates (e.g., 24, 25) including ervacycline 26 (XERAVA), which recently received FDA approval for the treatment of complicated intra-abdominal infections (cIAI).94 As presented in Figure 7D, ervacycline 26 has very potent, broad-spectrum activity against several pathogenic bacteria, including methicillin-resistant Staphylococcus aureus.
Figure 7.

(A) Myers’ retrosynthetic analysis of tetracycline. (B) Key Michael–Claisen annulation step for fully synthetic tetracycline antibiotics. (C) Mechanism of Michael–Claisen annulation reaction. (D) Antibacterial profiles of fully synthetic tetracycline antibiotics, including FDA-approved eravacycline 26.
The Myers lab has also recently established a total synthesis platform for the discovery of new macrolide antibiotics.89–93 Myers and co-workers reported the generation of >300 fully synthetic macrolides (FSMs) that incorporate structural design features that combine structure–activity relationship knowledge (macrolactone, C9 ketone/amine replacement, 14- or 15-membered macrocycle, incorporation of desosamine sugar at C5) with resistance liabilities (replacement of C3-clasinose sugar with ketone to give ketolides; Figure 8A) to inform the synthesis of new macrolides.92 Eight simple building blocks (30–33, 38–40, 47; Figure 8B) were utilized to assemble key left-handed fragment 44 (amine) and right-handed fragment 43 (ketone), which were then joined, or converged, through a stereoselective reductive amination reaction followed by sequential macrolactonization and click reactions to yield 48 (a 14-membered azaketolide). The Myers team noted that many past macrolide synthesis efforts have failed at the critical macrolactonization step; therefore, special attention was made to rigidify the substrate via the cyclic carbamate group, which led to an impressive 78% yield for the macrolactonization step (on gram scale). Several clinically effective and promising macrolides have cyclic carbamate moieties fused to the C12/ C13 position of the macrolide, including telithromycin, cethromycin, and solithromycin 50. An additional benefit of the cyclic carbamate moiety is that the aniline residue of solithromycin 50 is known to bind a unique pocket in the bacterial ribosome,92 making such structures better inhibitors of this target and more effective antibacterial agents.
Figure 8.
(A) Overview of the structure–activity relationships related to macrolide antibiotics with Myers’ design elements regarding new, fully synthetic macrolides. (B) Overview of the convergent chemical synthesis approach to access fully synthetic macrolide small molecules.
A series of 15-membered azaketolides, including FSM 49, was synthesized using a similar route to the 14-membered azaketolides (Figure 8B).92 This new route required modification to only three of the building blocks (34, 35, and 41, the original glycosyl donor reported by Woodward et al.95) used to synthesize the 14-membered azaketolides. The key left-handed fragment 44 was also utilized in this route and underwent an analogous (1) reductive amination with right-hand aldehyde 45, (2) macrolactonization (94% yield, 1.9 g scale), (3) click reaction sequence to afford 15-membered azaketolide 49. Myers and co-workers also utilized the key right-hand aldehyde 45 to access a series of 14-membered ketolides, including the clinical candidate solithromycin 50 and approved drug telithromycin (not shown). Intermediate 42, which was accessed through a stereoselective addition of ketone 36 with alkynyl lithium 37, was transformed into a Grignard reagent via the hydromagnesiation of the acetylenic moiety and subsequently reacted with aldehyde 45 to join the key left- and right-hand fragments of the ketolide series. Following a subsequent oxidation/deprotection sequence, the key macrolactionization step was successfully carried out in 66% yield (1.7 g scale) to produce the desired 14-membered macrolactone scaffold. A final reaction sequence involving C2 fluorination and installation of the cyclic carbamate (bearing the important aniline residue) led to the synthesis of solithromycin 50 in 14 steps and 16% yield from 34 and 35.
This total synthesis platform was utilized to generate >300 FSM antibiotic candidates by varying building blocks in addition to modifying readily diversifiable structural features at multiple positions around the macrolactone architecture.92 This proved to be a powerful synthetic medicinal chemistry tactic as deep-seated variations at select positions within FSM scaffolds were accessed, which are not possible using semisynthetic approaches. Following chemical synthesis, 305 FSMs were evaluated for antibacterial activities against a diverse panel of Gram-positive and Gram-negative pathogens. Potent antibacterial agents were identified among each of the FSM subclasses (14-membered azaketolides, 15-membered azaketolides, 14-membered ketolides).
Following the initial screen, the most promising FSMs were evaluated against an expanded panel of bacteria with genetically characterized resistance mechanisms to erythromycin (ermB, ribosome-modifying methyltransferase; mefA, efflux; Figure 9) and other antibiotics (MRSA and VRE strains).92 Several analogues (e.g., 49, 51, 52; Figure 9) demonstrated significant improvements in activity against this macrolide-resistant panel of pathogenic bacteria. FSM 51 (FSM-100573) is a 14-membered ketolide that demonstrated remarkable activities during these investigations. In fact, 51 outperformed every current clinically used macrolide antibiotic against extremely challenging strains, such as S. pneumoniae with both ermB and mefA genes. FSM 51 also demonstrated significant improvements against Gram-negative pathogens, such as A. baumannii and P. aeruginosa.
Figure 9.

Antibacterial profiles of select FSM analogues identified from microbiological studies. Several analogues demonstrate high antibacterial potencies and overcome specific macrolide resistance mechanisms. Resistant strain details are the following: iErmA, inducible erythromycin ribosome methyltransferase A; MsrA, macrolide streptogramin resistance efflux pump A; ErmB, erythromycin ribosome methyltransferase B; MefA, macrolide efflux protein A.
This incredible synthesis platform has created unprecedented opportunities to explore and develop novel macrolide antibiotics. The synthesis approach to FSMs is highly convergent from multiple building blocks; however, key advanced left-handed and right-handed intermediates were strategically utilized in divergent pathways to access multiple FSM subclasses (e.g., left-handed fragment 44 was used to synthesize 14- and 15-membered azaketolide scaffolds; right-handed fragment 45 was used to synthesize 15-membered azaketolide and 14-membered ketolide scaffolds). Similar to Myers’ tetracycline synthesis platform, one can envision accessing thousands of FSM from the chemistry described here. Myers and co-workers have truly inspired scientists and clinicians with this ground-breaking work, which has great potential to lead to new clinical agents that overcome bacterial resistance in human patients.
PROMYSALIN: A SPECIES-SPECIFIC ANTIBIOTIC THAT TARGETS SUCCINATE DEHYDROGENASE IN Pseudomonas aeruginosa
Broad-spectrum antibiotics target essential pathways in bacteria, including cell wall and protein synthesis, and are capable of treating a wide range of infections.96 However, misuse and prolonged exposure to broad-spectrum antibiotics can result in undesirable side effects (e.g., inflammatory diseases, obesity) as commensal population dynamics can be dramatically altered by these agents.96–98 “Narrow-spectrum” antibiotics (e.g., amoxicillin) target large subsets of bacteria, such as Gram-positive versus Gram-negative bacteria, or anaerobes versus aerobes; however, species-specific agents against significant human pathogens would be extremely useful and could avoid the undesired side effects that result from the misuse of our current antibiotic arsenal.96
Over the past several years, Wuest and co-workers have developed an exciting medicinal chemistry and chemical biology program around the species-selective agent promysalin 53 (Figure 10A), which targets the major Gram-negative pathogen Pseudomonas aeruginosa.96,99–103 The multispecies community found in the root systems of various plants is known as the rhizosphere,104,105 and in this environment, bacteria are known to utilize chemical warfare strategies for colonization and to defend themselves. Pseudomonads are highly prevalent in the rhizosphere, and these bacteria produce an array of secondary metabolites with biological activities that promote survival, including antibiotics, virulence factors, biosurfactants, siderophores (to obtain iron from their environment).99 Promysalin is a secondary metabolite isolated from Pseudomonas putida found in rice plants.106 This natural product induces swarming and biofilm formation in P. putida; however, it demonstrates unique species-specific antibacterial activity against P. aeruginosa (growth inhibition, IC50 = 0.83 μg/mL; note, MIC values have not been reported for the promysalin work described here).
Figure 10.

(A) Retrosynthesis and (B) synthetic scheme to the Pseudomonas aeruginosa selective natural product promysalin by Wuest and co-workers. (C) Representative promysalin analogues synthesized using diverted total synthesis to probe structure–activity relationships against P. aeruginosa (strain PA14).
Wuest and co-workers reported a convergent and concise total synthesis of (−)-promysalin 53 (Figure 10).99 In the original isolation paper by De Mot and co-workers,106 the three stereogenic centers of promysalin were not defined. Therefore, Wuest’s total synthesis of promysalin involved a unique combination of bioinformatics (reannotation of the biosynthetic gene cluster via AntiSMASH computation) and stereoselective approaches (reagent control, substrate selection; Figure 10B) to yield four diastereomers of promysalin for structural elucidation and biological assessment.
All four diastereomers of the myristic acid fragment 60 (diastereomers 60a–d) of promysalin were synthesized using a stereoselective, five-step synthetic route.99 First, Evans’ oxazolidinone 54 underwent a diastereoselective hydroxylation (oxidation) upon treatment with sodium hexamethyldisilazane followed by addition Davis oxaziridine 58, then a TBS-protection afforded 59. This was followed by a cross-metathesis reaction with homoallylic alcohol 55, and then hydrogenation of the resulting olefin (not shown) and final ammonolysis of the oxazolidinone moiety with ammonium hydroxide afforded amide 60. EDC-coupling of the alcohol in 60 with acid (−)-62 and then silyl-group removal with tetrabutylammonium fluoride afforded promysalin 53. All four diastereomers of 60 (60a–d) were synthesized and coupled to (−)-62 to afford all possible diastereomers at C2 and C8 of the myristic acid moiety of promysalin (53a–d).
With the four diastereomers of 53 in hand, NMR comparison and antibacterial assays were utilized in concert to unequivocally identify diastereomer 53a as the natural product. Unfortunately, no optical rotation or authentic sample was available to compare; however, Wuest and co-workers rationalized that the correct enantiomer would exhibit the most potent biological activity.99 Promysalin 53a, which had identical spectral data to those reported in De Mot’s isolation paper, proved to have the most potent antibacterial activities against P. aeruginosa strain PA14, which demonstrated an IC50 value of 125 nM (IC50 = 1.8 μM, initial report) and an IC50 value of 1 μM against PAO1. The other diastereomers 53b–d demonstrated >10-to >60-fold less potency against P. aeruginosa strain PA14 than 53a. On the basis of these findings, the absolute stereochemistry of natural promysalin (−)-53 was assigned as (2R,8R,16S).
By use of the total synthesis of promysalin 53 as a platform, a series of 16 related analogues was generated via diverted total synthesis.96 This approach allowed several structural features to be probed (i.e., ester linker, proline ring size and functionality, hydroxyl group on the myristic acid fragment) to gain structure–activity relationship information regarding 53. From these investigations, several interesting structural features were identified that led to a complete abolishment of antibacterial activity against PA14, including (1) proline ring size and saturation (analogue 63) and (2) substitution of the ester group with an amide (analogue 64). In contrast, removal of the C2 hydroxyl group (analogue 65) and introduction of a vinyl fluoride on the proline moiety (analogue 66) resulted in promysalin analogues with improved antibacterial activities against PA14.
The SAR insights gained from the diverted synthesis of promysalin analogues enabled the design, synthesis, and utilization of diazirine photoprobe 67, which retained activity, in affinity-based protein profiling (AfBPP) studies.101 Active promysalin photoprobe 67, inactive photoprobe 68 (to identify nonspecific protein binding partners), and promysalin (for competition purposes) were utilized in an elaborate series of experiments (involving irradiation of photoactive probes in cell lysate, subsequent click reaction with biotin azide, pull-down, dimethyl isotope labeling, and LC–MS/MS measurement; Figure 11) that led to the identification of succinate dehydrogenase (SdhC) as the target of promysalin. Further validation that SdhC was the target of 53 was carried out in vitro and via whole genome sequencing of resistant mutants. These findings demonstrate that targeting the tricarboxylic acid (TCA) cycle could be useful in developing narrow, species-selective antibiotic therapies and warrant additional studies regarding SdhC as an antibiotic target.101–103
Figure 11.

Experimental workflow for pull-down experiments using photoactive diazirines 67 and 68 to identify succinate dehydrogenase (SdhC) as the biological target of species-specific antibiotic promysalin 53.
RECENT ADVANCES OF SIDEROPHORE CONJUGATES FOR “TROJAN HORSE” TARGETED DELIVERY OF ANTIBIOTICS TO PATHOGENIC BACTERIA
Bacteria are able to successfully establish an infection when they experience unlimited growth and proliferation within a host.107 In order for bacteria to proliferate, adequate supply of key nutrients is required. Iron is a crucial nutrient that bacteria require to thrive and is known to play critical roles in energy metabolism, stabilization of protein structures, and oxygen transport.107–109 Unfortunately, iron is not membrane soluble and, therefore, is unable to diffuse through bacterial membranes, so acquisition of this key nutrient has its challenges; however, bacteria synthesize and utilize specialized organic molecules known as siderophores to acquire iron from their surroundings.107,108 Bacteria secrete specific siderophores into their environment where they tightly bind iron(III). The resulting iron(III)-siderophore complex is then recognized by specific cell-surface receptors on bacteria, which shuttles the complex inside the bacterial cell. Once within the reductive cytoplasm of the bacterium, iron(III) is reduced to iron(II), resulting in a loss of affinity by the siderophore and subsequent release of iron(II) for unitization by the bacterial cell.107
Bacteria utilize highly specific siderophore systems that can be exploited for bacterial targeting and drug delivery. For instance, the Gram-positive pathogen S. aureus utilizes two siderophores, staphyloferrin A and staphyloferrin B (69, Figure 12A).110 Enterobactin (71, Figure 12B) is a siderophore produced by multiple Gram-negative pathogens, including E. coli and S. typhimurium.111 P. aeruginosa and other pathogens have effectively developed receptors to recognize and transport iron(III)-siderophore complexes from other bacterial species (xenosiderophores) providing a competitive growth advantage. Although enterobactin is not produced by P. aeruginosa, this siderophore can promote iron uptake in this pathogen.111
Figure 12.

(A) Structure and iron(III) complex with the S. aureus siderophore staphyloferrin B. (B) Enterobactin and iron(III) bound complex utilized by multiple Gram-negative pathogens for iron acquisition. (C) Utilization of iron uptake systems as a platform for antibiotic drug delivery by Miller and co-workers (“Trojan horse” conjugate 74 selectively and potently targets P. aeruginosa via iron uptake systems).
These iron-specific uptake systems have provided a promising platform for targeted antibiotic delivery to select pathogens, especially for Gram-negative pathogens that present challenges regarding compound penetration as a result of two cellular membranes. Miller and co-workers have pioneered this area with much success in recent years with several inspiring examples of potent and targeted antibiotic delivery using a “Trojan horse” strategy that links antibiotics to various synthetic siderophores (sideromycins).111–118 One example is presented in Figure 12C, where Miller and colleagues designed triscatecholate sideromycin 73, inspired by catechol-based siderophore enterobactin 71, which is utilized to target P. aeruginosa.111 Sideromycin 73 was designed to have a linker moiety for attaching an antibiotic agent, in this case amoxicillin (ampicillin was also attached, not shown) to afford “Trojan horse” conjugate 74 (note: acetate groups were installed on catechol moieties to prevent pharmacological side effects and serve as prodrugs while circumventing potential methylation via a catechol O-methyl-transferase that would result in the loss of iron binding capabilities of the siderophore). Trojan horse conjugate 74 demonstrated potent antibacterial activities against P. aeruginosa strains in iron-deficient media, which resulted from the hijacking of energy-dependent active bacterial iron uptake systems required for bacterial growth under these conditions.111 During these studies, MIC values for 74 ranged from 0.05 to 0.39 μM under iron-deficient media compared to MIC values that ranged from 12.5 to 50 μM in iron-rich media (note: amoxicillin was inactive against these P. aeruginosa strains, MIC > 100 μM).
A more detailed schematic of the Trojan horse strategy to target Gram-negative pathogens can be found in Figure 13A. Recently, Miller’s group has published two outstanding examples of Trojan horse conjugates hijacking iron uptake systems that enable Gram-positive antibiotics (an oxazolidinone117 and daptomycin;116 Figure 13B and Figure 13C) to effectively target Gram-negative pathogens, including Acinetobacter baumannii. These impressive examples of Trojan horse antibiotic delivery also showcase “releasable linkers” versus “nonreleasable linkers” that are an important design component in these systems.
Figure 13.
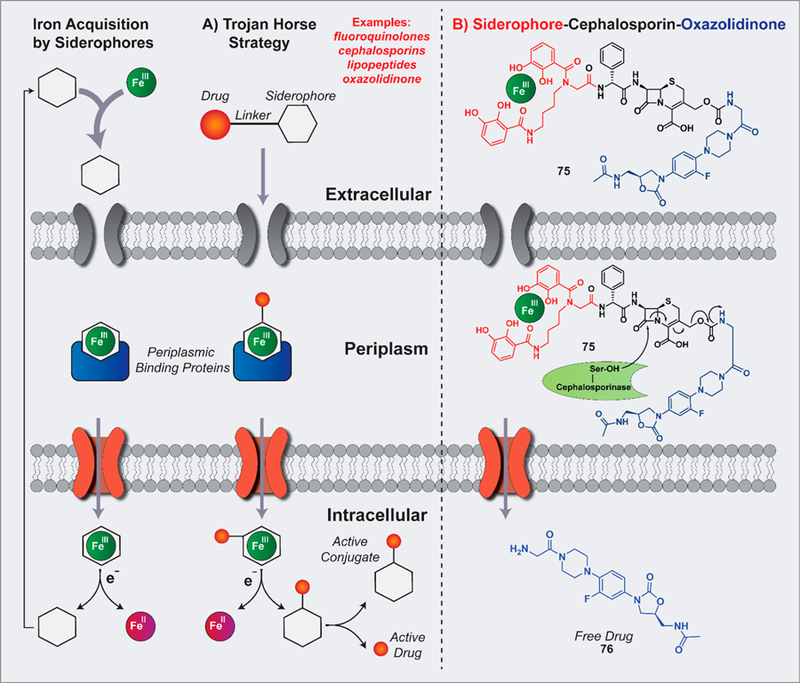
(A) Illustration of the Trojan horse strategy for targeted antibiotic delivery. (B) Example of Trojan horse strategy with a suicide siderophore–cephalosporin–oxazolidinone 75 (via releasable linker) recently reported by Miller and co-workers.
Oxazolidinones target bacterial ribosomes and are effective at treating Gram-positive infections; however, these agents are inactive against most Gram-negative pathogens as they are unable to penetrate their outer membrane, or if they do permeate the Gram-negative bacteria, they are rapidly effluxed.117 This spectrum of activity along with these resistance mechanisms (lack of penetration, efflux) makes oxazolidinones ideal candidates for Trojan horse conjugation. With that, sideromycin 75 was designed as a dual active agent containing siderophore–cephalosporin–oxazolidinone components, which combine the Trojan horse entry of Gram-negative pathogens with β-lactamase triggered release upon destructive ring-opening of the cephalosporin moiety to liberate an active oxazolidinone (76; Figure 13B).117
In the arsenal of Trojan horse conjugate linkers, it is important to have releasable linker options to avoid conjugate structures that perturb critical drug–target interactions, ultimately preventing antibiotic activities. Early attempts to develop releasable linkers included functional groups that were hydrolyzable (i.e., ester cleavage by esterases112) or reductively cyclized (i.e., trimethyl lock bioreductive activation,115 not shown) and led to problems with premature antibiotic release before iron-uptake dependent penetration of Gram-negative bacteria.117 The cephalosporin linker in 75 requires the enzymatic activity of intracellular β-lactamases for “suicide” antibiotic release, bypassing premature release issues presented by other releasable linker systems. In initial proof-of-concept experiments, 75 underwent rapid and complete hydrolysis by purified ADC-1 β-lactamases, which are ADC enzymes found in A. baumannii that are capable of hydrolyzing cephalosporin antibiotics.117
The siderophore–cephalosporin–oxazolidinone 75 demonstrated impressive antibacterial activities against multiple Gram-negative pathogens, including A. baumannii (multiple, drug-resistant isolates; MIC = 0.4–6.25 μM), E. coli (MIC < 0.025 μM), and P. aeruginosa (MIC = 0.2–0.4 μM).117 The corresponding oxazolidinone 76, alone or when linked directly to the siderophore moiety (without cephalosporin to release 76), was inactive against these pathogens (MIC from >50 to >500 μM). Interestingly, the siderophore–cephalosporin moiety alone (without oxazolidinone 76; structure not shown) has moderate to good activities against some pathogens but is dramatically enhanced with all three moieties (75) intact against A. baumannii. For instance, against A. baumannii ATCC BAA 1800, the siderophore-cephalosporin components report MIC = 25 μM versus MIC = 0.8 μM for siderophore–cephalosporin–oxazolidinone 75, demonstrating a clear structure–activity relationship for the necessity of all three structural components to display potent antibacterial activities at a level that is clinically relevant.
In separate work, Miller and co-workers coupled a mixed synthetic ligand analogue of A. baumannii siderophore fimsbactin with daptomycin to give conjugate 79.116 Daptomycin is a large, negatively charged lipopeptide anti-biotic that has significant value as a therapeutic agent against Gram-positive pathogens only. Although daptomycin’s mechanism is not fully understood, it has been found to bind and disrupt bacterial cell membranes, resulting in rapid depolarization, concomitant ion efflux and dysregulation of nucleic acid and protein synthesis, and subsequent bactericidal death.119 Prior structure–activity relationship investigations revealed that acylation of the primary amine of the ornithine residue in daptomycin was well-tolerated, and thus the Miller group used this information in the design of daptomycin-siderophore conjugate 79.116 Regarding chemical synthesis, the trichloroethyl chloroformate (Troc) protecting group of 77 was removed using zinc in the presence of glutaric anhydride to afford 78 as a benzyl protected siderophore bearing a (nonreleasable) glutarate linker for subsequent conjugation (Figure 14). A mixed anhydride of 78 was generated in situ before subjecting the siderophore-linker moiety to daptomycin under aqueous reaction conditions for conjugation via amide bond formation. Final hydrogenolytic removal of the benzyl groups provided target daptomycin-siderophore conjugate 79.116
Figure 14.
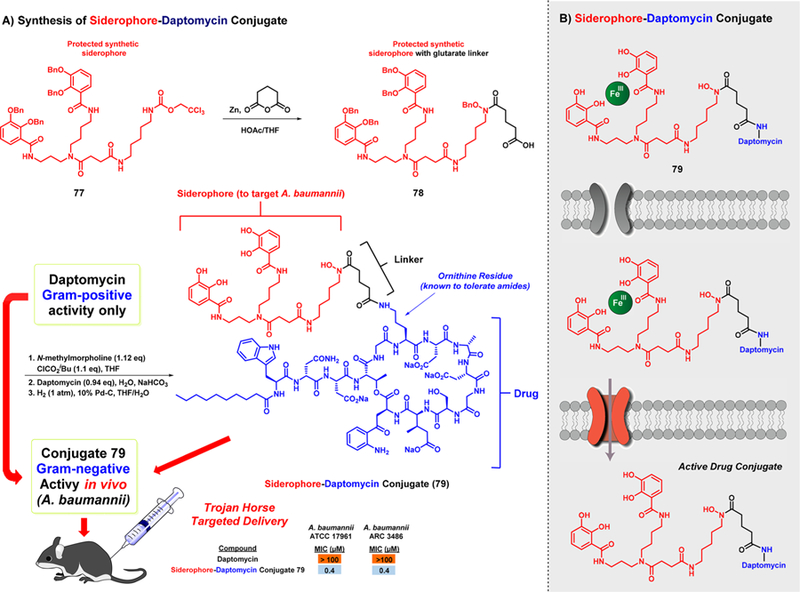
Chemical synthesis and targeted A. baumannii activities of siderophore–daptomycin conjugate 79.
Siderophore-daptomycin conjugate 79 was shown to bind iron(III) stoichiometrically and demonstrated profound effectiveness against A. baumannii strains (MIC = 0.4–0.8 μM).116 As predicted, daptomycin itself was inactive against A. baumannii strains (MIC > 100 μM) when tested alongside conjugate 79. To demonstrate the importance of iron-binding and uptake regarding conjugate 79, its synthetic precursor bearing bis benzyl group protected catechol moieties (structure not shown) proved to be completely inactive against A. baumannii (MIC > 50 μM). When tested against other Gram-negative pathogens, such as P. aeruginosa, Burkholderia multivorans, and Escherichia coli, conjugate 79 proved to be inactive (MIC > 100 μM), demonstrating the selective targeting of this Trojan horse conjugate toward A. baumannii.
Following in vitro assessment, Trojan horse conjugate 79 was advanced to testing in mice.116 Intravenous administration of 79 at 250 mg/kg in ICR mice resulted in no observed adverse effects. Following favorable toxicity studies in mice, conjugate 79 was evaluated for in vivo efficacy in a sepsis model of infection in female ICR mice using A. baumannii 17961. Mice were infected with A. baumannii via intraperitoneal (ip) administration, and treatments with conjugate 79 (5, 10, 25 mg/kg) were provided intravenously (iv) 30 min and 24.5 h after infection. In this study, all six mice that did not receive treatment with 79 died after 1 day; however, mice treated with 79 at 10 and 25 mg/kg resulted in survival in 4 of 5 mice (at each test concentration) while only 1 of 5 mice survived with 79 at 5 mg/kg, demonstrating a dose-dependent in vivo efficacy against A. baumannii. This is an outstanding demonstration of the clinical applications that the Trojan horse strategy can be utilized to target Gram-negative pathogens in human patients.
Cefiderocol (S-649266; structure not shown) is a catechol-bearing siderophore–cephalosporin conjugate that demonstrates potent antibacterial activities against a broad range of Gram-negative pathogens (e.g., carbepenem-resistant Enterobacteriaceae, P. aeruginosa, A. baumannii).120 Cefiderocol has been advanced to clinical studies in human patients, demonstrating the translational potential of the Trojan horse strategy. In a clinical trial for the treatment of complicated urinary tract infection with antibiotic-resistant Gram-negative pathogens, cefiderocol was shown to be noninferior when compared to imipenem–cilastatin treated patients.121 Currently, cefiderocol is being investigated in clinical trials for hospital-acquired pneumonia and carbapenem-resistant infections. Collectively, this body of work aimed at targeting iron uptake mechanisms in bacteria to deliver antibiotics in a Trojan horse fashion is a promising approach to overcome antibiotic-resistant infections.
ClpP PROTEASE-ACTIVATING AGENT ADEP4 FOR PERSISTER CELL AND BIOFILM ERADICATION
Isolated from Strepomyces hawaiiensis, the acyldepsipeptides (ADEPs) are a class of natural product antibiotics that have been shown to exhibit potent activities against Gram-positive bacteria including methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant Enterococci.122,123 The antibacterial activities for the natural product ADEP1 (80, Figure 15) and related synthetic analogues were evaluated by Brötz-Oesterhelt and co-workers, wherein ADEP1 reported a 8.76 μM IC50 value against MRSA. In addition, synthetic analogues ADEP2 (81) and ADEP4 (82) also reported therein demonstrated markedly improved activity (500 nM and 64.9 nM, respectively).124 Furthermore, ADEP1 and ADEP4 were efficacious in vivo against both S. aureus and E. faecalis infection models. ADEP4 demonstrated the ability to rescue 80% of mice from mortal S. aureus infections following a single dose of 12.5 mg/kg. The group used genomic analyses to identify the target for ADEP analogues as ClpP (caseinolytic protease), a highly conserved, cylindrical bacterial peptidase.124
Figure 15.

Structures of the natural product, ADEP1 (80), its synthetic counterparts, and the key intramolecular hydrogen bonding required for activity. Structural modifications have been highlighted with different colors on each synthetic analogue.
The ClpP serine peptidase employs an energy-dependent process implicated in biological functions such as protein quality control, degradation of transient regulatory proteins, and clearance of cellular debris following conditions of bacterial stress.125,126 Under normal homeostatic conditions, ClpP binds to ATP-dependent cochaperones (e.g., ATPases) which help regulate the unfolding and translocation of proteins into the proteolytic sites of ClpP. Crystal structures of ADEP1 (80) bound to Escherichia coli ClpP showed that ADEP analogues occupy the site of ATPase binding.127 The ClpP–ADEP interaction promotes the conversion of the ClpP entrance pore from a closed-to an open-gate form (Figure 16). This allosteric control of the ClpP barrel conformation bypasses the need for an associated cochaperone (an energy dependent process) and permits the unregulated entrance of proteins into the proteolytic active site.128 The resultant increase of ClpP activity diverts proteolysis from native physiological proteins to nascent peptides, leading to inhibition of bacterial cell division.
Figure 16.
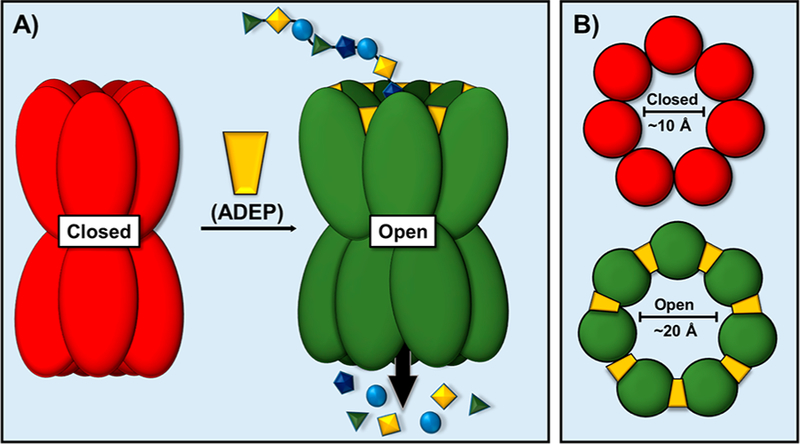
(A) ADEPs bind to Clp-ATPase binding sites, promoting the entry of unfolded proteins into proteolytic sites, causing indiscriminate degradation of nascent polypeptides, ultimately leading to self-digestion and bacterial cell death. (B) The binding of ADEP analogues engenders the open conformation of the ClpP protease.
In a departure from prior studies (evaluating ADEP-promoted protein degradation against rapidly dividing cells), Lewis and co-workers utilized proteomic analysis to demonstrate that stationary phase MRSA exposed to ADEP4 (82) resulted in a reduced abundance of 24% of cellular proteins compared to the untreated control.129 Since stationary phase populations of S. aureus are typically nondividing (and thus notoriously difficult to treat), this finding unveiled a potential therapeutic intervention against dormant persister cells. To evaluate the effect of ADEP4 against dormant bacteria, a population of persister cells was treated with the agent and it was found that ADEP4 was capable of eradicating persister cells to the limit of detection. Conversely, rifampicin reported no effect on persister cell viability.
ADEP4 also reported remarkable killing of stationary phase S. aureus following 2 days of treatment, demonstrating a 4 log reduction of viable bacterial cell counts.129 Since ClpP is a nonessential protein for S. aureus, the null clpP (the gene encoding the ClpP protease) mutation frequency is high (∼10−6), and thus, rebounding populations were observed after day 3 of the experiments. It was surmised that formation of resistant mutants could be suppressed via coadministration of ADEP4 with conventional antibiotics. This co-treatment strategy eradicated persister cells to the limit of detection with no concomitant population rebound. It was found that although treatment of a clpP mutant strain with rifampicin demonstrated similar MIC values as that seen against of the wild-type strain, the mutant strains were less capable of producing persister cells (by an order of 10-to 100-fold). The conclusion was that clpP mutations (e.g., those resulting from ADEP treatment) may render the bacteria less fit and thus more susceptible to other antibiotics.
By use of 96-well plate, non-Calgary Biofilm Device assays, the ability of the ADEP4–rifampicin combination to eradicate bacterial biofilms was also evaluated.129 Following a 24-h S. aureus UAMS-1 biofilm establishment, wells were treated with the ADEP4–rifampicin combination (6.49 μM ADEP4, 10 × MIC; 0.49 μM rifampicin, 10 × MIC), resulting in eradicated biofilms (>4 log-fold reduction) following 3 days of treatment. The ADEP4–rifampicin combination was also shown to be highly efficacious in deep-seated in vivo infection models. Following a 24 h infection period, histopathology was used to confirm the adherence of bacterial biofilms to mouse thigh muscle tissue. Treatment of the infection with the ADEP4–rifampicin combination resulted in an eradication of the biofilm-associated infection within 24 h (representing a >4 log-fold reduction). In contrast, rifampicin, vancomycin, or a combination of both could not eradicate the infection although decreased viable cell counts were observed.
The LaFleur group was also able to show that the ADEP–antibiotic combination strategy could be expanded to include additional clinically used antibiotics.130 LeFleur and co-workers reported ADEP4 activity against stationary phase multidrug-resistant E. faecalis V583 when treated in combination with ampicillin, ciprofloxacin, daptomycin, oritavancin, or tigecycline. In these cases, the ADEP4–antibiotic combination yielded a 5 log reduction in bacterial cell survival. This was especially interesting as the activity was observed regardless of the mechanistic class of partnered antibiotic. Expression and purification of E. faecium ClpP permitted the use of a fluorometric monitoring assay used to evaluate casein degradation wherein it was reported that ADEP4 activated ClpP with an EC50 of 0.53 μM. A crystal structure was also disclosed, highlighting the binding of ADEP4 to the hydrophobic pockets located between subunits at the apical and distal ends of ClpP. In a murine model of peritoneal septicemia, treatment with ADEP4 (50 mg/kg) and ampicillin (50 mg/kg) yielded a 2 log10 greater reduction in average bacterial burden when compared to antibiotic monotherapy and a 4 log10 reduction of viable bacterial cells relative to the vehicle control.
Encouraged by the in vivo antibacterial efficacy of ADEP analogues, several groups have put forth medicinal chemistry efforts to improve on this therapeutic strategy. In an attempt to reduce entropic binding penalties, Sello and co-workers disclosed an enhancement of ADEP analogues via introduction of conformational restriction.131 This strategy was realized via replacement of key macrocyclic residues. The result of the rigidified ADEP analogues was a 20-fold improvement of in vitro S. aureus activity from MIC = 649 nM 129 for ADEP4 compared to 30.6 nM for ADEP1g (83). Building upon the library of known antibacterial ADEP analogues, the Duerfeldt group described an attempt to remedy a potential metabolic liability of the ADEP series (i.e., the readily hydrolyzed ester within the lactone core).132 On the basis of the reported crystal structures of ADEPs bound to ClpP, it seemed evident that replacement of the ester with an amide may have no influence on the interaction of the ADEPs with ClpP. Alas, Duerfeldt and co-workers were surprised to learn that amide-containing ADEP analogues 85 and 86 were markedly less active (by up to 100-fold) than the corresponding ester ADEP analogue 84. It was surmised that although the ester oxygen may not participate in molecular interactions with the target, its replacement with an amide nitrogen disrupts intramolecular hydrogen bonds (87, Figure 15) and thus compromises the compact conformation of the active agent (which presumably assists in membrane permeability). Although no biofilm eradication or in vivo assays were conducted as part of these recent studies, the reported novel agents demonstrate that the ADEP-based therapeutic approaches are fruitful pursuits and initial efforts to advance these agents toward clinical use have been very promising.
HALOGENATED PHENAZINE BIOFILM-ERADICATING AGENTS
Cystic fibrosis (CF) patients are plagued with chronic lung infections, which became a source of chemical inspiration for our lab to identify new biofilm-eradicating agents. Many young CF patients are known to experience initial S. aureus lung infections.133 As these patients age, P. aeruginosa subsequently co-infects the lung and is believed to eradicate S. aureus using a series of phenazine antibiotics, including pyocyanin.133–137 We believe that the initial S. aureus infections establish surface-attached biofilms on the inside of lung tissues as CF patients have lung infections that span multiple years.
In our initial work, we synthesized a diverse series of 13 phenazine compounds, including five naturally occurring phenazines.138 With literature reports of pyocyanin as the main antibiotic of this class responsible for activities against S. aureus, we were sure to include this compound in our initial screening collection. Interestingly, in our initial antibacterial screens against S. aureus and S. epidermidis, we found that 2-bromo-1-hydroxyphenazine, a marine Streptomyces derived phenazine, demonstrated good antibacterial activities with MIC values of 6.25 μM against these pathogens (pyocyanin reported MICs of 50 μM).138 In addition, we found that related synthetic analogue 2,4-dibromo-1-hydroxyphenazine (not shown) demonstrated improved antibacterial activities against S. aureus and S. epidermidis with MICs of 1.56 μM. In later work, we found that 2,4-dibromo-1-hydroxyphenazine was able to eradicate methicillin-resistant Staphylococcus aureus biofilms and reported a minimum biofilm eradication concentration, or MBEC, of ∼100 μM (note: when tested alongside 2,4-dibromo-1-hydroxyphenazine, vancomycin reported an MBEC of >2000 μM in Calgary Biofilm Device assays despite an MBEC of 4 μM against planktonic cells in the same experiment, demonstrating a significant level of biofilm tolerance toward vancomycin).139
After demonstrating that 2,4-dibromo-1-hydroxyphenazine eradicates MRSA biofilms, we worked to develop modular, convergent synthetic routes to new “halogenated phenazine” (HP; Figure 17A, see structure 95) small molecules.140–143 Chemical synthesis pathways involving phenylenediamine (88) condensation with quinone 89,140,141 a Wohl–Aue reaction142 (aniline 90 condensed with 2-nitroanisole 91), and a Buchwald–Hartwig cross-coupling/reductive cyclization path-way143 (fusing aniline 92 with 2-bromo-3-nitroanisole 93) enabled rapid access to a diversity of 1-methoxyphenazines (94) with various substitutions at the 6–9 positions of the HP scaffold (95). A final boron tribromide (BBr3) demethylation and N-bromosuccinimide (NBS) bromination sequence afforded target HPs 95. These synthetic efforts and subsequent antibacterial studies have led to the identification of several highly potent biofilm-eradicating agents, including HPs 96–98, which demonstrate up to 50-fold more potent biofilm killing activities in Calgary Biofilm Device assays compared to parent 2,4-dibromo-1-hydroxyphenazine (e.g., 98 reported an MBEC of 2.35 μM against MRSA biofilms143).
Figure 17.
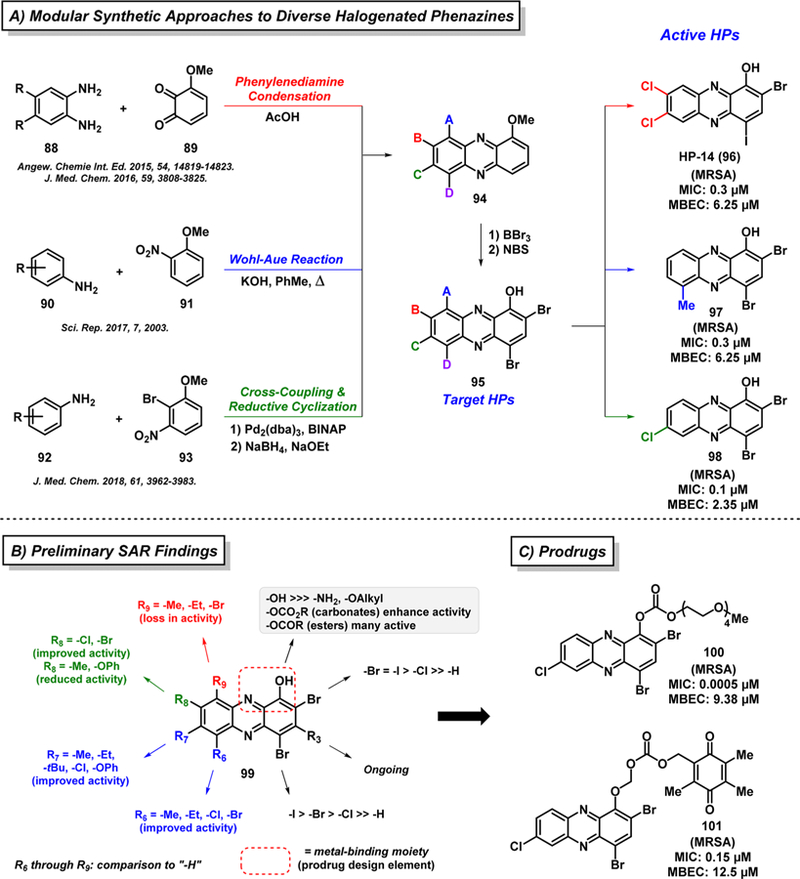
Chemical synthesis of halogenated phenazines and preliminary SAR findings.
These efforts have enabled a detailed structure–activity relationship understanding regarding halogenated phenazine antibacterial agents (see 99, Figure 17B).138–143 In general, substitution at the 6- and 7-positions of the HP scaffold is typically well-tolerated and enhances antibacterial activities compared to the corresponding unsubstituted HP; however, the 8-position of the HP scaffold can influence antibacterial potency in a positive (e.g., halogens improve activity) or negative (e.g., methyl reduces activity) fashion. Interestingly, we found that substitution at the 9-position of the HP scaffold completely abolishes all antibacterial activities. We later discovered the reason for a loss in activity regarding 9- substituted HPs after learning that active HP compounds bind metal(II) cations between the hydroxyl oxygen and adjacent nitrogen (forming a five-membered chelate upon metal binding), which plays a critical role in the antibacterial mode of action. Substituents at the 9-position of the HP scaffold, including a relatively small methyl group, impede the critical metal-chelation event for antibacterial activities.140 Interestingly, general metal-chelating agents (EDTA and TPEN) have been tested alongside HPs and are unable to eradicate bacterial biofilms, highlighting the unique metal-dependent mechanism behind HP small molecules.141–143
In ongoing efforts to translate HP biofilm-eradicating agents for clinical applications, we are using the SAR knowledge of the HP scaffold to pursue multiple prodrug strategies to functionalize the phenolic hydroxyl group to abolish metal-chelation until selective release within bacterial cells.142,143 This approach provides a platform for extensive developments, including (1) tuning of physicochemical properties (e.g., installation of PEG group to increase water solubility142,143) and (2) diverse functional triggers for HP release (e.g., bioreduction of the quinone in 101 is required to liberate the corresponding HP agent143). Despite initial success with HPs 100 and 101, continued prodrug efforts are ongoing with the goal of developing an effective clinical agent for biofilm infections.
In separate work, we used HP-14 (96, MBEC = 6.25 μM) as a probe molecule for transcriptomic analysis of treated and untreated MRSA biofilms using RNA-seq technology.144 This platform allowed us to define new cellular targets and pathways critical to biofilm survival, which is challenging to study as these surface-attached bacterial communities are composed of slow-growing or nonreplicating cells. Treating an established MRSA biofilm with HP-14 at low concentration (0.625 μM; 1/ 10 MBEC) for 20 h enabled the identification of >200 gene transcripts that were either up- or down-regulated (≥2.0-fold change in gene expression) compared to vehicle control.
Using a WoPPER gene cluster analysis tool, we identified 37 gene clusters with alterations in MRSA biofilm gene expression profiles following 20 h treatment with HP-14.144 Upon exposure of HP-14 to MRSA biofilms, six gene clusters involved in iron acquisition were found to be dramatically up-regulated (“activated”) via WoPPER analysis, including: hts/sfa (staphyloferrin A; siderophore), sir/sbn (staphyloferrin B), isd (iron-regulated surface determinant; heme iron acquisition), MW0695 (hypothetical protein, similar to ferrichrome ABC transporters), and f hu (ferric hydroxamate uptake; two gene clusters). Following validation of these results using RT-qPCR experiments, a time-course assessment of MRSA biofilms treated with HP-14 (0.625 μM; 1/10 MBEC) revealed that four iron acquisition gene clusters (isd, sbn, sfa, MW0695) were activated in 1 h. The rapid activation of multiple iron uptake gene clusters by low concentrations of HP-14 is profound, as bacterial biofilms are notorious for their dormant phenotypes. Following 4 and 8 h of HP-14 exposure at 0.625 μM (1/10 MBEC), MRSA biofilms demonstrated more significant activation levels of these iron uptake genes (e.g., isdB, 22.3-fold activation after 4 h). Interestingly, EDTA and TPEN (5 μM; general metal-chelating agents) were unable to activate iron acquisition genes in MRSA biofilms,144 which aligns with our previous observations that these metal-chelating agents are unable to eradicate MRSA biofilms (MBEC of >2000 μM) in Calgary Biofilm Device assays.141–143
On the basis of RNA-seq findings, we believe HP-14 induces rapid iron starvation in MRSA biofilms, which leads to eradication of these surface-attached bacteria.144 With the lipophilic properties of HP-14 (cLogP = 6.25) combined with potent MRSA biofilm eradication activity (MBEC = 6.25 μM), an iron chelating moiety, and rapid activation of multiple iron uptake gene clusters, we believe that this HP rapidly diffuses into biofilm cells and binds iron(II) following the intracellular release of iron(II) from a siderophore or heme. A proposed scheme of HP-14 inducing iron starvation and MRSA biofilm eradication is presented in Figure 18 and aligns with our WoPPER gene cluster analysis and subsequent RT-qPCR results in time-course studies. In addition, we believe that EDTA and TPEN are not able to efficiently penetrate biofilm cells, which is why they are unable to activate iron uptake genes or eradicate biofilms when tested alongside active HP biofilm-eradicating agents. This work not only demonstrates HP-14’s ability to rapidly induce iron starvation in MRSA biofilms but highlights an impressive level of sensitivity along with rapid response rates of bacterial biofilms to small molecule threats. In addition, these findings suggest that iron starvation of surface-attached bacterial communities could serve as the Achilles heel to persistent and recurring biofilm infections in the clinic.
Figure 18.

Proposed mechanism of MRSA biofilm eradication by HP-14, based on RNA-seq findings.
CONCLUSIONS
Despite increasing concerns regarding antibiotic resistance, antibiotic drug discovery is entering a dynamic and exciting new era. The multidisciplinary approaches of the research programs presented here have, in part, contributed to the reinvigoration of natural-product-based research for antibiotic discovery. Other impactful programs inspired by natural products, yet not included in this Perspectives article, include Genentech’s optimized arylomycin analogues for Gram-negative pathogens145 and Boger et al.’s vancomycin work.146–150 The classic tools of synthetic chemistry, microbiology, and chemical biology have been applied in novel ways to answer questions about bacterial pathogenesis and to design innovative therapeutic approaches for combatting infections. Significant challenges lie ahead, but these research efforts along with others have helped pave the way for critical and desperately needed discoveries to address problems presented by antibiotic resistance and tolerance.
ACKNOWLEDGMENTS
We acknowledge the National Institute of General Medical Sciences of the National Institutes of Health for providing financial support (Grant R35GM128621 to R.W.H.). A.T.G. and G.M.B. were each awarded University of Florida Graduate School Fellowships. A.C.-R. is supported by a postdoctoral fellowship from CONACYT (CVU Grant 346860).
ABBREVIATIONS USED
- Accum
accumulation
- AcOH
acetic acid
- ADEP
acyldepsipeptide (antibacterial agent)
- AfBPP
affinity-based protein profiling
- Asp
aspartic acid
- aq
aqueous
- BBr3
boron tribromide
- BINAP
2,2′-bis(diphenylphosphino)-1,1′-binaphthyl
- Bn
benzyl
- Boc
tert-butyloxycarbonyl
- C
carbon
- ClpP
caseinolytic protease
- CF
cystic fibrosis
- CFU
colony forming unit (viable bacterial count)
- DHF
dihydrofolate
- DIPEA
N,N-diisopropylethylamine
- DMAP
4-dimethylaminopyridine
- DMF
dimethylformamide
- DMP
Dess-Martin periodinane
- DMPU
N,N′-dimethylpropyleneurea
- DNA
deoxyribonucleic acid
- DNM
deoxynybomycin
- EDC
1-ethyl-3-(3-dimethylaminopropyl)carbodiimide
- EDTA
ethylenediaminetetraacetic acid
- ETC
electron transport chain
- EtOAc
ethyl acetate
- FDA
Food and Drug Administration
- Fe
iron
- HATU
hexafluorophosphate azabenzotriazole tetramethyluranium
- Glob
globularity
- HF
hydrogen fluoride
- HOAt
1-hydroxy-7-azabenzotriazole
- HP
halogenated phenazine
- Isd
heme-iron complex
- KOH
potassium hydroxide
- LCMS/MS
liquid chromatography-mass spectrometry
- LDA
lithium diisopropylamide
- LiOH
lithium hydroxide
- Lys
lysine
- MBEC
minimum biofilm eradication concentration
- mg/kg
milligrams per kilogram
- MIC
minimum inhibitory concentration (lowest concentration that demonstrates complete bacterial growth inhibition)
- MRSA
methicillin-resistant Staphylococcus aureus
- NaBH4
sodium borohydride
- NaHMDS
sodium bis(trimethylsilyl)amide
- NaOEt
sodium ethoxide
- NBS
N-bromosuccinimide
- nmol
nanomole
- NMR
nuclear magnetic resonance
- NRPS
non-ribosomal peptide synthetase
- PABA
p-aminobenzoate
- Pd
palladium
- Phe
phenylalanine
- PhMe
toluene
- rRNA
ribosomal ribonucleic acid
- RB
rotatable bond (rigidity)
- Ru
ruthenium
- SAR
structure-activity relationship
- sbn
staphyloferrin B
- SdhC
succinate dehydrogenase C subunit
- SEM
2-(trimethylsilyl)ethoxymethyl
- Ser
serine
- sfa
staphyloferrin A
- TBAF
tetrabutylammonium fluoride
- TBDPS
tert-butyldiphenylsilyl
- TBS
tert-butyldimethylsilyl
- TBSCl
tert-butyldimethylsilyl chloride
- TCA
tricarboxylic acid
- Tf
triflate
- TFA
trifluoroacetic acid
- THF
tetrahydrofolate
- THF
tetrahydrofuran
- Thr
threonine
- TMEDA
tetramethylethylenediamine
- TMS
trimethylsilyl
- TPEN
N,N,N′,N′-tetrakis(2-pyridinylmethyl)-1,2-ethanediamine
- Troc
trichloroethyl chloroformate
- μg/mL
micrograms per milliliter
- μM
micromolar
- VRE
vancomycin-resistant enterococci
- WGS
whole genome sequencing
- Zn
zinc
Biographies
Yasmeen Abouelhassan received a Ph.D. in Medicinal Chemistry with Prof. Robert W. Huigens at the University of Florida in 2018. She received her B.S. in Pharmaceutical Sciences from Cairo University, Egypt, in 2012. Her Ph.D. research focused on the identification of novel biofilm-eradicating agents against Gram-positive bacteria. This research led to the discovery of phenazine antibiotic derivatives as potent methicillin-resistant S. aureus biofilm eradicators operating through a novel iron starvation mechanism.
Aaron T. Garrison received his Ph.D. in Medicinal Chemistry under Professor Robert W. Huigens III at the University of Florida in 2017 where he synthesized potent halogenated phenazine and quinoline small molecules, including diverse prodrug analogues of lead compounds. Thereafter, he accepted a postdoctoral position with Professor Craig W. Lindsley at the Vanderbilt Center for Neuro-science Drug Discovery. His research is presently focused on the design and synthesis of negative allosteric modulators of the muscarinic acetylcholine receptor subtype 5 (M5) for the treatment of opioid and ethanol dependence. His additional research projects include the development of synthetic methodology to facilitate the discovery of novel therapeutics.
Hongfen Yang received a B.S. in Chemistry from Hubei University in Wuhan, China. She then moved to the University of Chinese Academy of Sciences in Beijing under the guidance of Professor Wu, where she obtained a M.S. in Bioinorganic Chemistry. Currently, Hongfen is a fourth year graduate student in the Medicinal Chemistry Department at the University of Florida under the supervision of Professor Huigens. Her doctoral research involves the design, chemical synthesis, and biological evaluation of new halogenated phenazine biofilm-eradicating agents.
Alejandra Chávez-Riveros obtained her M.S. and Ph.D. in Chemical Sciences from the National Autonomous University of Mexico (Universidad Nacional Autónoma de México, UNAM). Her doctoral work focused on the design and synthesis of steroidal 5α-reductase inhibitors under the mentorship of Professor Eugene Bratoeff. She went to Evestra Inc. for a predoctoral internship, where she synthesized antiprogestins. Upon graduation with a Ph.D. in 2016, she joined Professor Luis D. Miranda’s group at the Institute of Chemistry UNAM for postdoctoral studies and developed the synthesis of novel macrocycles, which demonstrate anti-inflammatory activities. She is currently a Postdoctoral Research Associate in Professor Huigens’ lab at the University of Florida in the Department of Medicinal Chemistry, where she is developing new synthetic methodologies for ring distortion of indole alkaloids.
Gena M. Burch is the Kenneth F. Finger Memorial Fellow in the Infectious Disease and Pharmacokinetics Laboratory at the University of Florida. She received a PharmD in 2016, followed by a M.S. in Medicinal Chemistry in 2018 from the University of Florida under the supervision of Dr. Robert W. Huigens III working on the discovery and development of novel antibacterial and biofilm-eradicating agents. Her current research includes liquid chromatography–tandem mass spectrometry method development for the determination of serum concentrations for antimicrobial drugs. Her clinical research efforts are focused on therapeutic drug monitoring and clinical pharmacokinetics for patients with bacterial and mycobacterial infections.
Robert W. Huigens III received a Ph.D. in Chemistry with Professor Christian Melander at NC State University in 2009. He then moved to the University of Illinois of Urbana–Champaign as an American Cancer Society Postdoctoral Fellow under the guidance of Professor Paul Hergenrother. In 2013, Huigens began his independent career as an Assistant Professor of Medicinal Chemistry at the University of Florida. The Huigens lab has interdisciplinary drug discovery programs inspired by natural products that aim to (1) discover novel antibacterial agents that eradicate bacterial biofilms and (2) rapidly generate architecturally complex and diverse compound libraries from readily available indole alkaloids.
Footnotes
The authors declare no competing financial interest.
REFERENCES
- (1).Lewis K Platforms for Antibiotic Discovery. Nat. Rev. Drug Discovery 2013, 12, 371–387. [DOI] [PubMed] [Google Scholar]
- (2).Brown ED; Wright GD Antibacterial Drug Discovery in the Resistance Era. Nature 2016, 529, 336–343. [DOI] [PubMed] [Google Scholar]
- (3).Blair JMA; Webber MA; Baylay AJ; Ogbolu DO; Piddock LJV Molecular Mechanisms of Antibiotic Resistance. Nat. Rev. Microbiol 2015, 13, 42–51. [DOI] [PubMed] [Google Scholar]
- (4).Rossiter SE; Fletcher MH; Wuest WM Natural Products as Platforms To Overcome Antibiotic Resistance. Chem. Rev 2017, 117, 12415–12474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Ali J; Rafiq QA; Ratcliffe E Antimicrobial Resistance Mechanisms and Potential Synthetic Treatments. Futur. Sci. OA 2018, 4, FSO290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Zaman SB; Hussain MA; Nye R; Mehta V; Mamun KT; Hossain N A Review on Antibiotic Resistance: Alarm Bells Are Ringing. Cureus 2017, 9, e1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Kohanski MA; Dwyer DJ; Collins JJ How Antibiotics Kill Bacteria: From Targets to Networks. Nat. Rev. Microbiol 2010, 8, 423–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Martinez JL General Principles of Antibiotic Resistance in Bacteria. Drug Discovery Today: Technol 2014, 11, 33–39. [DOI] [PubMed] [Google Scholar]
- (9).Wright GD Molecular Mechanisms of Antibiotic Resistance. Chem. Commun 2011, 47, 4055–4061. [DOI] [PubMed] [Google Scholar]
- (10).Garrison AT; Huigens RW Eradicating Bacterial Biofilms with Natural Products and Their Inspired Analogues That Operate Through Unique Mechanisms. Curr. Top. Med. Chem 2017, 17, 1954–1964. [PubMed] [Google Scholar]
- (11).Lewis K Persister Cells, Dormancy and Infectious Disease. Nat. Rev. Microbiol 2007, 5, 48–56. [DOI] [PubMed] [Google Scholar]
- (12).Lewis K Persister Cells. Annu. Rev. Microbiol 2010, 64, 357–372. [DOI] [PubMed] [Google Scholar]
- (13).Marín M; Gudiol F Beta-Lactam Antibiotics. Enferm. Infecc. Microbiol. Clin 2003, 21, 42–55. [DOI] [PubMed] [Google Scholar]
- (14).Hammes WP; Neuhaus FC On the Mechanism of Action of Vancomycin: Inhibition of Peptidoglycan Synthesis in Gaffkya Homari. Antimicrob. Agents Chemother 1974, 6, 722–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Chopra I; Roberts M Tetracycline Antibiotics: Mode of Action, Applications, Molecular Biology, and Epidemiology of Bacterial Resistance. Microbiol. Mol. Biol. Rev 2001, 65, 232–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Leclercq R Mechanisms of Resistance to Macrolides and Lincosamides: Nature of the Resistance Elements and Their Clinical Implications. Clin. Infect. Dis 2002, 34, 482–492. [DOI] [PubMed] [Google Scholar]
- (17).Mingeot-Leclercq MP; Glupczynski Y; Tulkens PM Aminoglycosides: Activity and Resistance. Antimicrob. Agents Chemother 1999, 43, 727–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Weigelt J; Itani K; Stevens D; Lau W; Dryden M; Knirsch C; Linezolid CSSTI Study Group. Linezolid versus Vancomycin in Treatment of Complicated Skin and Soft Tissue Infections. Antimicrob. Agents Chemother 2005, 49, 2260–2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Maggi N; Pasqualucci CR; Ballotta R; Sensi P Rifampicin: A New Orally Active Rifamycin. Chemotherapy 2004, 11, 285–292. [DOI] [PubMed] [Google Scholar]
- (20).Wehrli W; Knüsel F; Schmid K; Staehelin M Interaction of Rifamycin with Bacterial RNA Polymerase. Proc. Natl. Acad. Sci. U. S. A 1968, 61, 667–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Nagaoka H; Kishi Y Further Synthetic Studies on Rifamycin S. Tetrahedron 1981, 37, 3873–3888. [Google Scholar]
- (22).Palumbo M; Gatto B; Zagotto G; Palù G On the Mechanism of Action of Quinolone Drugs. Trends Microbiol 1993, 1, 232–235. [DOI] [PubMed] [Google Scholar]
- (23).Blondeau JM Fluoroquinolones: Mechanism of Action, Classification, and Development of Resistance. Surv. Ophthalmol 2004, 49, S73–S78. [DOI] [PubMed] [Google Scholar]
- (24).Bearden DT; Danziger LH Mechanism of Action of and Resistance to Quinolones. Pharmacotherapy 2001, 21, 224S–232S. [DOI] [PubMed] [Google Scholar]
- (25).Fàbrega A; Madurga S; Giralt E; Vila J Mechanism of Action of and Resistance to Quinolones. Microb. Biotechnol 2009, 2, 40–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Bushby SRM; Hitchings GH Trimethoprim, A Sulphonamide Potentiator. Br. J. Pharmacol. Chemother 1968, 33, 72–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Sinha S; Shen X; Gallazzi F; Li Q; Zmijewski JW; Lancaster JR; Gates KS Generation of Reactive Oxygen Species Mediated by 1-Hydroxyphenazine, a Virulence Factor of Pseudomonas aeruginosa. Chem. Res. Toxicol 2015, 28, 175–181. [DOI] [PubMed] [Google Scholar]
- (28).Davies J; Davies D Origins and Evolution of Antibiotic Resistance. Microbiol. Mol. Biol. Rev 2010, 74, 417–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Munita JM; Arias CA Mechanisms of Antibiotic Resistance. Microbiol. Spectrum 2016, 4, VMBF-0016–2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Cassini A; Högberg LD; Plachouras D; Quattrocchi A; Hoxha A; Simonsen GS; Colomb-Cotinat M; Kretzschmar ME; Devleesschauwer B; Cecchini M; Ouakrim DA; Oliveira TC; Struelens MJ; Svetens C; Monnet DL; Burden of AMR Collaborative Group. Attributable Deaths and Disability-Adjusted Life-Years Caused by Infections with Antibiotic-Resistant Bacteria in the EU and the European Economic Area in 2015: A Population-Level Modelling Analysis. Lancet. Infect. Dis 2019, 19, 56–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Du D; Wang-Kan X; Neuberger A; van Veen HW; Pos KM; Piddock LJV; Luisi BF Multidrug Efflux Pumps: Structure, Function and Regulation. Nat. Rev. Microbiol 2018, 16, 523–539. [DOI] [PubMed] [Google Scholar]
- (32).Delcour AH Outer Membrane Permeability and Antibiotic Resistance. Biochim. Biophys. Acta, Proteins Proteomics 2009, 1794, 808–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Redgrave LS; Sutton SB; Webber MA; Piddock LJV Fluoroquinolone Resistance: Mechanisms, Impact on Bacteria, and Role in Evolutionary Success. Trends Microbiol 2014, 22, 438–445. [DOI] [PubMed] [Google Scholar]
- (34).Pechère JC Macrolide Resistance Mechanisms in Gram-Positive Cocci. Int. J. Antimicrob. Agents 2001, 18 (Suppl. 1), 25–28. [DOI] [PubMed] [Google Scholar]
- (35).Wilson DN Ribosome-Targeting Antibiotics and Mechanisms of Bacterial Resistance. Nat. Rev. Microbiol 2014, 12, 35–48. [DOI] [PubMed] [Google Scholar]
- (36).Brauner A; Fridman O; Gefen O; Balaban NQ Distinguishing Between Resistance, Tolerance and Persistence to Antibiotic Treatment. Nat. Rev. Microbiol 2016, 14, 320–330. [DOI] [PubMed] [Google Scholar]
- (37).Miller MB; Bassler BL Quorum Sensing in Bacteria. Annu. Rev. Microbiol 2001, 55, 165–199. [DOI] [PubMed] [Google Scholar]
- (38).Solano C; Echeverz M; Lasa I Biofilm Dispersion and Quorum Sensing. Curr. Opin. Microbiol 2014, 18, 96–104. [DOI] [PubMed] [Google Scholar]
- (39).Defoirdt T; Brackman G; Coenye T Quorum Sensing Inhibitors: How Strong Is the Evidence? Trends Microbiol 2013, 21, 619–624. [DOI] [PubMed] [Google Scholar]
- (40).Waters CM; Bassler BL Quorum Sensing: Cell-to-Cell Communication in Bacteria. Annu. Rev. Cell Dev. Biol 2005, 21, 319–346. [DOI] [PubMed] [Google Scholar]
- (41).Worthington RJ; Richards JJ; Melander C Small Molecule Control of Bacterial Biofilms. Org. Biomol. Chem 2012, 10, 7457–7474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Flemming H-C; Wingender J; Szewzyk U; Steinberg P; Rice SA; Kjelleberg S Biofilms: An Emergent Form of Bacterial Life. Nat. Rev. Microbiol 2016, 14, 563–575. [DOI] [PubMed] [Google Scholar]
- (43).Bjarnsholt T The Role of Bacterial Biofilms in Chronic Infections. APMIS Suppl 2013, 121, 1–51. [DOI] [PubMed] [Google Scholar]
- (44).Donlan RM Biofilms: Microbial Life on Surfaces. Emerging Infect. Dis 2002, 8, 881–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Donlan RM; Costerton JW Biofilms: Survival Mechanisms of Clinically Relevant Microorganisms. Clin. Microbiol. Rev 2002, 15, 167–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Hall-Stoodley L; Costerton JW; Stoodley P Bacterial Biofilms: From the Natural Environment to Infectious Diseases. Nat. Rev. Microbiol 2004, 2, 95–108. [DOI] [PubMed] [Google Scholar]
- (47).Lewis K; Shan Y Persister Awakening. Mol. Cell 2016, 63, 3–4. [DOI] [PubMed] [Google Scholar]
- (48).Lewis K Persister Cells: Molecular Mechanism Related to Antibiotic Tolerance. Handb. Exp. Pharmacol 2012, 211, 121–133. [DOI] [PubMed] [Google Scholar]
- (49).Keren I; Kaldalu N; Spoering A; Wang Y; Lewis K Persister Cells and Tolerance to Antimicrobials. FEMS Microbiol. Lett 2004, 230, 13–18. [DOI] [PubMed] [Google Scholar]
- (50).Wood TK Combatting Bacterial Persister Cells. Biotechnol. Bioeng 2016, 113, 476–483. [DOI] [PubMed] [Google Scholar]
- (51).Defraine V; Fauvart M; Michiels J Fighting Bacterial Persistence: Current and Emerging Anti-Persister Strategies and Therapeutics. Drug Resist. Updates 2018, 38, 12–26. [DOI] [PubMed] [Google Scholar]
- (52).Stewart PS Mechanisms of Antibiotic Resistance in Bacterial Biofilms. Int. J. Med. Microbiol 2002, 292, 107–113. [DOI] [PubMed] [Google Scholar]
- (53).Fux CA; Costerton JW; Stewart PS; Stoodley P Survival Strategies of Infectious Biofilms. Trends Microbiol 2005, 13, 34–40. [DOI] [PubMed] [Google Scholar]
- (54).Ling LL; Schneider T; Peoples AJ; Spoering AL; Engels I; Conlon BP; Mueller A; Schäberle TF; Hughes DE; Epstein S; Jones M; Lazarides L; Steadman VA; Cohen DR; Felix CR; Fetterman KA; Millett WP; Nitti AG; Zullo AM; Chen C; Lewis K A New Antibiotic Kills Pathogens without Detectable Resistance. Nature 2015, 517, 455–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Nichols D; Cahoon N; Trakhtenberg EM; Pham L; Mehta A; Belanger A; Kanigan T; Lewis K; Epstein SS Use of Ichip for High-Throughput In Situ Cultivation of “Uncultivable” Microbial Species. Appl. Environ. Microbiol 2010, 76, 2445–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Giltrap AM; Dowman LJ; Nagalingam G; Ochoa JL; Linington RG; Britton WJ; Payne RJ Total Synthesis of Teixobactin. Org. Lett 2016, 18, 2788–2791. [DOI] [PubMed] [Google Scholar]
- (57).Jin K; Sam IH; Po KHL; Lin D; Ghazvini Zadeh EH; Chen S; Yuan Y; Li X Total Synthesis of Teixobactin. Nat. Commun 2016, 7, 12394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Craig W; Chen J; Richardson D; Thorpe R; Yuan Y A Highly Stereoselective and Scalable Synthesis of L-allo-Enduracididine. Org. Lett 2015, 17, 4620–4623. [DOI] [PubMed] [Google Scholar]
- (59).Jad YE; Acosta GA; Naicker T; Ramtahal M; El-Faham A; Govender T; Kruger HG; de la Torre BG; Albericio F Synthesis and Biological Evaluation of a Teixobactin Analogue. Org. Lett 2015, 17, 6182–6185. [DOI] [PubMed] [Google Scholar]
- (60).Parmar A; Iyer A; Vincent CS; Van Lysebetten D; Prior SH; Madder A; Taylor EJ; Singh I Efficient Total Syntheses and Biological Activities of Two Teixobactin Analogues. Chem. Commun 2016, 52, 6060–6063. [DOI] [PubMed] [Google Scholar]
- (61).Parmar A; Iyer A; Lloyd DG; Vincent CS; Prior SH; Madder A; Taylor EJ; Singh I Synthesis of Potent Teixobactin Analogues against Methicillin-Resistant Staphylococcus aureus (MRSA) through the Replacement of L-allo-Enduracididine with Its Isosteres. Chem. Commun 2017, 53, 7788–7791. [DOI] [PubMed] [Google Scholar]
- (62).Parmar A; Iyer A; Prior SH; Lloyd DG; Goh ETL; Vincent CS; Palmai-Pallag T; Bachrati CZ; Breukink E; Madder A; Lakshminarayanan R; Taylor EJ; Singh I Teixobactin Analogues Reveal Enduracididine to be Non-essential for Highly Potent Antibacterial Activity and Lipid II Binding. Chem. Sci 2017, 8, 8183–8192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Parmar A; Lakshminarayanan R; Iyer A; Mayandi V; Goh ETL; Lloyd DG; Chalasani MLS; Verma NK; Prior SH; Beurman RW; Madder A; Taylor EJ; Singh I Design and Synthesis of Highly Potent Teixobactin Analogues against Staphylococcus aureus, Methicillin-Resistant Staphylococus aureus (MRSA), and Vancomycin-Resistant Enterococci (VRE) In Vitro and In Vivo. J. Med. Chem 2018, 61, 2009–2017. [DOI] [PubMed] [Google Scholar]
- (64).Wu C; Pan Z; Yao G; Wang W; Fang L; Su W Synthesis and Structure–Activity Relationship Studies of Teixobactin Analogues. RSC Adv 2017, 7, 1923–1926. [Google Scholar]
- (65).Zong Y; Sun X; Gao H; Meyer KJ; Lewis K; Rao Y Developing Equipotent Teixobactin Analogues against Drug-Resistant Bacteria and Discovering a Hydrophobic Interaction Between Lipid II and Teixobactin. J. Med. Chem 2018, 61, 3409–3421. [DOI] [PubMed] [Google Scholar]
- (66).Jin K; Po KHL; Kong WY; Lo CH; Lo CW; Lam HY; Sirinimal A; Reuven JA; Chen S; Li X Synthesis and Antibacterial Studies of Teixtobactin Analogues with Non-Isostere Substitution of Enduracididine. Bioorg. Med. Chem 2018, 26, 1062–1068. [DOI] [PubMed] [Google Scholar]
- (67).Ng V; Kuehne SA; Chan WC Rational Design and Synthesis of Modified Teixobactin Analogues: In Vitro Antibacterial Activity against Staphylococcus aurues, Propionibacterium acnes and Pseudomonas aeruginosa. Chem. - Eur. J 2018, 24, 9136–9147. [DOI] [PubMed] [Google Scholar]
- (68).Richter MF; Drown BS; Riley AP; Garcia A; Shirai T; Svec RL; Hergenrother PJ Predictive Compound Accumulation Rules Yield a Broad-Spectrum Antibiotic. Nature 2017, 545, 299–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Fischbach MA; Walsh CT Antibiotics for Emerging Pathogens. Science 2009, 325, 1089–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Payne DJ; Gwynn MN; Holmes DJ; Pompliano DL Drugs for Bad Bugs: Confronting the Challenges of Antibacterial Discovery. Nat. Rev. Drug Discovery 2007, 6, 29–40. [DOI] [PubMed] [Google Scholar]
- (71).Nikaido H Molecular Basis of Bacterial Outer Membrane Permeability Revisited. Microbiol. Mol. Biol. Rev 2003, 67, 593–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Richter MF; Hergenrother PJ Broad-Spectrum Antibiotics, A Call for Chemists. Chem 2017, 3, 10–13. [Google Scholar]
- (73).Richter MF; Hergenrother PJ The Challenge of Converting Gram-Positive-Only Compounds into Broad-Spectrum Antibiotics. Ann. N. Y. Acad. Sci 2019, 1435, 18–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Huigens RW; Morrison KC; Hicklin RW; Flood TA; Richter MF; Hergenrother PJ A Ring-Distortion Strategy to Construct Stereochemically Complex and Structurally Diverse Compounds from Natural Products. Nat. Chem 2013, 5, 195–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).Hicklin RW; López Silva TL; Hergenrother PJ Synthesis of Bridged Oxafenestranes from Pleuromutilin. Angew. Chem., Int. Ed 2014, 53, 9880–9883. [DOI] [PubMed] [Google Scholar]
- (76).Rafferty RJ; Hicklin RW; Maloof KA; Hergenrother PJ Synthesis of Complex and Diverse Compounds through Ring Distortion of Abietic Acid. Angew. Chem., Int. Ed 2014, 53, 220–224. [DOI] [PubMed] [Google Scholar]
- (77).Garcia A; Drown BS; Hergenrother PJ Access to a Structurally Complex Compound Collection via Ring Distortion of the Alkaloid Sinomenine. Org. Lett 2016, 18, 4852–4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78).Tasker SZ; Cowfer AE; Hergenrother PJ Preparation of Structurally Diverse Compounds from the Natural Product Lycorine. Org. Lett 2018, 20, 5894–5898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).O’Shea R; Moser HE Physicochemical Properties of Antibacterial Compounds: Implications for Drug Discovery. J. Med. Chem 2008, 51, 2871–2878. [DOI] [PubMed] [Google Scholar]
- (80).Parkinson EI; Bair JS; Nakamura B; Lee HY; Kuttab HI; Southgate EH; Lezmi S; Lau GW; Hergenrother PJ Deoxynybomycins Inhibit Mutant DNA Gyrase and Rescue Mice Infected with Fluoroquinolone-Resistant Bacteria. Nat. Commun 2015, 6, 6947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (81).Wright PM; Seiple IB; Myers AG The Evolving Role of Chemical Synthesis in Antibacterial Drug Discovery. Angew. Chem., Int. Ed 2014, 53, 8840–8869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (82).Liu F; Myers AG Development of a Platform for the Discovery and Practical Synthesis of New Tetracycline Antibiotics. Curr. Opin. Chem. Biol 2016, 32, 48–57. [DOI] [PubMed] [Google Scholar]
- (83).Charest MG; Lerner CD; Brubaker JD; Siegel DR; Myers AG A Convergent Enantioselective Route to Structurally Diverse 6-Deoxytetracycline Antibiotics. Science 2005, 308, 395–398. [DOI] [PubMed] [Google Scholar]
- (84).Brubaker JD; Myers AG A Practical, Enantioselective Synthetic Route to a Key Precursor to the Tetracycline Antibiotics. Org. Lett 2007, 9, 3523–3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (85).Sun C; Wang Q; Brubaker JD; Wright PM; Lerner CD; Noson K; Charest M; Siegel DR; Wang Y-M; Myers AG A Robust Platform for the Synthesis of New Tetracycline Antibiotics. J. Am. Chem. Soc 2008, 130, 17913–17927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (86).Wright PM; Myers AG Methodological Advances Permit the Stereocontrolled Construction of Diverse Fully Synthetic Tetracyclines Containing an All-Carbon Quaternary Center at Position C5a. Tetrahedron 2011, 67, 9853–9869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (87).Sun C; Hunt DK; Clark RB; Lofland D; O’Brien WJ; Plamondon L; Xiao X-Y Synthesis and Antibacterial Activity of Pentacyclines: A Novel Class of Tetracycline Analogs. J. Med. Chem 2011, 54, 3704–3731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (88).Clark RB; He M; Fyfe C; Lofland D; O’Brien WJ; Plamondon L; Sutcliffe JA; Xiao X-Y 8-Azatetracyclines: Synthesis and Evaluation of a Novel Class of Tetracycline Antibacterial Agents. J. Med. Chem 2011, 54, 1511–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (89).Zhang Z; Kitamura Y; Myers A An Efficient Directed Claisen Reaction Allows for Rapid Construction of 5,6-Disubstituted 1,3-Dioxin-4-Ones. Synthesis 2015, 47, 2709–2712. [Google Scholar]
- (90).Seiple I; Hog D; Myers A Practical Protocols for the Preparation of Highly Enantioenriched Silyl Ethers of (R)-3-Hydroxypentan-2-one, Building Blocks for the Synthesis of Macrolide Antibiotics. Synlett 2015, 27, 57–60. [Google Scholar]
- (91).Zhang Z; Fukuzaki T; Myers AG Synthesis of D-Desosamine and Analogs by Rapid Assembly of 3-Amino Sugars. Angew. Chem., Int. Ed 2016, 55, 523–527. [DOI] [PubMed] [Google Scholar]
- (92).Seiple IB; Zhang Z; Jakubec P; Langlois-Mercier A; Wright PM; Hog DT; Yabu K; Allu SR; Fukuzaki T; Carlsen PN; Kitamura Y; Zhou X; Condakes ML; SzczypiĔski FT; Green WD; Myers AG A Platform for the Discovery of New Macrolide Antibiotics. Nature 2016, 533, 338–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (93).Hogan PC; Chen C-L; Mulvihill KM; Lawrence JF; Moorhead E; Rickmeier J; Myers AG Large-Scale Preparation of Key Building Blocks for the Manufacture of Fully Synthetic Macrolide Antibiotics. J. Antibiot 2018, 71, 318–325. [DOI] [PubMed] [Google Scholar]
- (94).Tetraphase Pharmaceuticals Announces FDA Approval of XERAVA (Eravacycline) for Complicated Intra-Abdominal Infections (cIAI). Tetraphase Pharmaceuticals https://ir.tphase.com/news-releases/news-release-details/tetraphase-pharmaceuticals-announces-fda-approval-xeravatm (accessed Jan 21, 2019).
- (95).Woodward RB; Logusch E; Nambiar KP; Sakan K; Ward DE; Au-Yeung BW; Balaram P; Browne LJ; Card PJ; Chen CH Asymmetric Total Synthesis of Erythromycin. 3. Total Synthesis of Erythromycin. J. Am. Chem. Soc 1981, 103, 3215–3217. [Google Scholar]
- (96).Steele AD; Keohane CE; Knouse KW; Rossiter SE; Williams SJ; Wuest WM Diverted Total Synthesis of Promysalin Analogs Demonstrates That an Iron-Binding Motif Is Responsible for Its Narrow-Spectrum Antibacterial Activity. J. Am. Chem. Soc 2016, 138, 5833–5836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (97).Cox LM; Yamanishi S; Sohn J; Alekseyenko AV; Leung JM; Cho I; Kim SG; Li H; Gao Z; Mahana D; Zárate Rodriguez JG; Rogers AB; Robine N; Loke P; Blaser MJ Altering the Intestinal Microbiota During a Critical Developmental Window Has Lasting Metabolic Consequences. Cell 2014, 158, 705–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (98).Gerber JS; Ross RK; Bryan M; Localio AR; Szymczak JE; Wasserman R; Barkman D; Odeniyi F; Conaboy K; Bell L; Zaoutis TE; Fiks AG Association of Broad- vs Narrow-Spectrum Antibiotics With Treatment Failure, Adverse Events, and Quality of Life in Children With Acute Respiratory Tract Infections. JAMA 2017, 318, 2325–2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (99).Steele AD; Knouse KW; Keohane CE; Wuest WM Total Synthesis and Biological Investigation of (−)-Promysalin. J. Am. Chem. Soc 2015, 137, 7314–7317. [DOI] [PubMed] [Google Scholar]
- (100).Knouse KW; Wuest WM The Enantioselective Synthesis and Biological Evaluation of Chimeric Promysalin Analogs Facilitated by Diverted Total Synthesis. J. Antibiot 2016, 69, 337–339. [DOI] [PubMed] [Google Scholar]
- (101).Keohane CE; Steele AD; Fetzer C; Khowsathit J; Van Tyne D; Moynie L; Gilmore MS; Karanicolas J; Sieber SA; Wuest WM Promysalin Elicits Species-Selective Inhibition of Pseudomonas aeruginosa by Targeting Succinate Dehydrogenase. J. Am. Chem. Soc 2018, 140, 1774–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (102).Giglio KM; Keohane CE; Stodghill PV; Steele AD; Fetzer C; Sieber SA; Filiatrault MJ; Wuest WM Transcriptomic Profiling Suggests That Promysalin Alters the Metabolic Flux, Motility, and Iron Regulation in Pseudomonas putida KT2440. ACS Infect. Dis 2018, 4, 1179–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (103).Shapiro JA; Kaplan AR; Wuest WM From General to Specific: Can Pseudomonas Primary Metabolism Be Exploited for Narrow-Spectrum Antibiotics? ChemBioChem 2019, 20, 34–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (104).Philippot L; Raaijmakers JM; Lemanceau P; van der Putten WH Going Back to the Roots: The Microbial Ecology of the Rhizosphere. Nat. Rev. Microbiol 2013, 11, 789–799. [DOI] [PubMed] [Google Scholar]
- (105).Mendes R; Garbeva P; Raaijmakers JM The Rhizosphere Microbiome: Significance of Plant Beneficial, Plant Pathogenic, and Human Pathogenic Microorganisms. FEMS Microbiol. Rev 2013, 37, 634–663. [DOI] [PubMed] [Google Scholar]
- (106).Li W; Estrada-de los Santos P; Matthijs S; Xie G-L; Busson R; Cornelis P; Rozenski J; De Mot R Promysalin, a Salicylate-Containing Pseudomonas putida Antibiotic, Promotes Surface Colonization and Selectively Targets Other Pseudomonas. Chem. Biol 2011, 18, 1320–1330. [DOI] [PubMed] [Google Scholar]
- (107).Bilitewski U; Blodgett JAV; Duhme-Klair A-K; Dallavalle S; Laschat S; Routledge A; Schobert R Chemical and Biological Aspects of Nutritional Immunity-Perspectives for New Anti-Infectives that Target Iron Uptake Systems. Angew. Chem., Int. Ed 2017, 56, 14360–14382. [DOI] [PubMed] [Google Scholar]
- (108).Wilson BR; Bogdan AR; Miyazawa M; Hashimoto K; Tsuji Y Siderophores in Iron Metabolism: From Mechanism to Therapy Potential. Trends Mol. Med 2016, 22, 1077–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (109).Lyles KV; Eichenbaum Z From Host Heme To Iron: The Expanding Spectrum of Heme Degrading Enzymes Used by Pathogenic Bacteria. Front. Cell. Infect. Microbiol 2018, 8, 198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (110).Madsen JLH; Johnstone TC; Nolan EM Chemical Synthesis of Staphyloferrin B Afforts Insight into the Molecular Structure, Iron Chelation, and Biological Activity of a Polycarboxylate Siderophore Deployed by the Human Pathogen Staphylococcus aureus. J. Am. Chem. Soc 2015, 137, 9117–9127. [DOI] [PubMed] [Google Scholar]
- (111).Ji C; Miller PA; Miller MJ Iron Transport-Mediated Drug Delivery: Practical Syntheses and In Vitro Antibacterial Studies of Tris-Catecholate Siderophore-Aminopenicillin Conjugates Reveals Selectivity Potent Antipseudomonal Activity. J. Am. Chem. Soc 2012, 134, 9898–9901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (112).Ji C; Miller MJ Chemical Syntheses and In Vitro Antibacterial Activity of Two Desferrioxamine B-Ciprofloxacin Conjugates with Potential Esterase and Phosphatase Triggered Drug Release Linkers. Bioorg. Med. Chem 2012, 20, 3828–3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (113).Wencewicz TA; Miller MJ Biscatecholate-Monohydrox-amate Mixed Ligand Siderophore-Carbacephalosporin Conjugates are Selective Sideromycin Antibiotics that Target Acinetobacter baumannii. J. Med. Chem 2013, 56, 4044–4052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (114).Wencewicz TA; Long TE; Möllmann U; Miller MJ Trihydroxamate Siderophore-Fluoroquinolone Conjugates Are Selective Sideromycin Antibiotics that Target Staphylococcus aureus. Bioconjugate Chem 2013, 24, 473–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (115).Ji C; Miller MJ Siderophore-Fluoroquinolone Conjugates Containing Potential Reduction-Triggered Linkers for Drug Release: Synthesis and Antibacterial Activity. BioMetals 2015, 28, 541–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (116).Ghosh M; Miller PA; Möllmann U; Claypool WD; Schroeder VA; Wolter WR; Suckow M; Yu H; Li S; Huang W; Zajicek J; Miller MJ Targeted Antibiotic Delivery: Selective Siderophore Conjugation with Daptomycin Confers Potent Activity against Multidrug Resistant Acinetobacter Baumannii Both in Vitro and in Vivo. J. Med. Chem 2017, 60, 4577–4583. [DOI] [PubMed] [Google Scholar]
- (117).Liu R; Miller PA; Vakulenko SB; Stewart NK; Boggess WC; Miller MJ A Synthetic Dual Drug Sideromycin Induces Gram-Negative Bacteria To Commit Suicide with a Gram-Positive Antibiotic. J. Med. Chem 2018, 61, 3845–3854. [DOI] [PubMed] [Google Scholar]
- (118).Ghosh M; Lin Y-M; Miller PA; Möllmann U; Boggess WC; Miller MJ Siderophore Conjugates of Daptomycin are Potent Inhibitors of Carbapenem Resistant Strains of Acinetobacter baumannii. ACS Infect. Dis 2018, 4, 1529–1535. [DOI] [PubMed] [Google Scholar]
- (119).Taylor SD; Palmer M The Action Mechanism of Daptomycin. Bioorg. Med. Chem 2016, 24, 6253–6268. [DOI] [PubMed] [Google Scholar]
- (120).Ito A; Sato T; Ota M; Takemura M; Nishikawa T; Toba S; Kohira N; Miyagawa S; Ishibashi N; Matsumoto S; Nakamura R; Tsuji M; Yamano Y In Vitro Antibacterial Properties of Cefiderocol, a Novel Siderophore Cephalosporin, against Gram-Negative Bacteria. Antimicrob. Agents Chemother 2018, 62, e01454–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (121).Portsmouth S; van Veenhuyzen D; Echols R; Machida M; Ferreira JCA; Ariyasu M; Tenke P; Nagata TD Cefiderocol Versus Imipenem-Cilastatin for the Treatment of Complicated Urinary Tract Infections Caused by Gram-Negative Uropathogens: A Phase 2, Randomised, Double-Blind, Non-Inferiority Trial. Lancet Infect. Dis 2018, 18, 1319–1328. [DOI] [PubMed] [Google Scholar]
- (122).Hinzen B; Raddatz S; Paulsen H; Lampe T; Schumacher A; Häbich D; Hellwig V; Benet-Buchholz J; Endermann R; Labischinski H; Brötz-Oesterhelt H Medicinal Chemistry Optimization of Acyldepsipeptides of the Enopeptin Class Antibiotics. ChemMedChem 2006, 1, 689–693. [DOI] [PubMed] [Google Scholar]
- (123).Malik IT; Brötz-Oesterhelt H Conformational Control of the Bacterial Clp Protease by Natural Product Antibiotics. Nat. Prod. Rep 2017, 34, 815–831. [DOI] [PubMed] [Google Scholar]
- (124).Brötz-Oesterhelt H; Beyer D; Kroll H-P; Endermann R; Ladel C; Schroeder W; Hinzen B; Raddatz S; Paulsen H; Henninger K; Bandow JE; Sahl H-G; Labischinski H Dysregulation of Bacterial Proteolytic Machinery by a New Class of Antibiotics. Nat. Med 2005, 11, 1082–1087. [DOI] [PubMed] [Google Scholar]
- (125).Lee B-G; Park EY; Lee K-E; Jeon H; Sung KH; Paulsen H; Rübsamen-Schaeff H; Brötz-Oesterhelt H; Song HK Structures of ClpP in Complex with Acyldepsipeptide Antibiotics Reveal Its Activation Mechanism. Nat. Struct. Mol. Biol 2010, 17, 471–478. [DOI] [PubMed] [Google Scholar]
- (126).Brötz-Oesterhelt H; Sass P Bacterial Caseinolytic Proteases as Novel Targets for Antibacterial Treatment. Int. J. Med. Microbiol 2014, 304, 23–30. [DOI] [PubMed] [Google Scholar]
- (127).Li DHS; Chung YS; Gloyd M; Joseph E; Ghirlando R; Wright GD; Cheng YQ; Maurizi MR; Guarné A; Ortega J Acyldepsipeptide Antibiotics Induce the Formation of a Structured Axial Channel in ClpP: A Model for the ClpX/ClpA-Bound State of ClpP. Chem. Biol 2010, 17, 959–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (128).Gersch M; Famulla K; Dahmen M; Göbl C; Malik I; Richter K; Korotkov VS; Sass P; Rübsamen-Schaeff H; Madl T; Brötz-Oesterhelt H; Sieber SA AAA+ Chaperones and Acyldepsipeptides Activate the ClpP Protease via Conformational Control. Nat. Commun 2015, 6, 6320. [DOI] [PubMed] [Google Scholar]
- (129).Conlon BP; Nakayasu ES; Fleck LE; LaFleur MD; Isabella VM; Coleman K; Leonard SN; Smith RD; Adkins JN; Lewis K Activated ClpP Kills Persisters and Eradicates a Chronic Biofilm Infection. Nature 2013, 503, 365–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (130).Brown Gandt A; Griffith EC; Lister IM; Billings LL; Han A; Tangallapally R; Zhao Y; Singh AP; Lee RE; LaFleur MD In Vivo and In Vitro Effects of a ClpP-Activating Antibiotic against Vancomycin-Resistant Enterococci. Antimicrob. Agents Chemother 2018, 62, e00424–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (131).Carney DW; Schmitz KR; Truong JV; Sauer RT; Sello JK Restriction of the Conformational Dynamics of the Cyclic Acyldepsipeptide Antibiotics Improves Their Antibacterial Activity. J. Am. Chem. Soc 2014, 136, 1922–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (132).Li Y; Lavey NP; Coker JA; Knobbe JE; Truong DC; Yu H; Lin Y-S; Nimmo SL; Duerfeldt AS Consequences of Depsipeptide Substitution on the ClpP Activation Activity of Antibacterial Acyldepsipeptides. ACS Med. Chem. Lett 2017, 8, 1171–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (133).Machan ZA; Pitt TL; White W; Watson D; Taylor GW; Cole PJ; Wilson R Interaction Between Pseudomonas aeruginosa and Staphylococcus aureus: Description of an Antistaphylococcal Substance. J. Med. Microbiol 1991, 34, 213–217. [DOI] [PubMed] [Google Scholar]
- (134).Laursen JB; Nielsen J Phenazine Natural Products: Biosynthesis, Synthetic Analogues, and Biological Activity. Chem. Rev 2004, 104, 1663–1685. [DOI] [PubMed] [Google Scholar]
- (135).Price-Whelan A; Dietrich LEP; Newman DK Rethinking “Secondary” Metabolism: Physiological Roles for Phenazine Antibiotics. Nat. Chem. Biol 2006, 2, 71–78. [DOI] [PubMed] [Google Scholar]
- (136).Dietrich LEP; Okegbe C; Price-Whelan A; Sakhtah H; Hunter RC; Newman DK Bacterial Community Morphogenesis Is Intimately Linked to the Intracellular Redox State. J. Bacteriol 2013, 195, 1371–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (137).Guttenberger N; Blankenfeldt W; Breinbauer R Recent Developments in the Isolation, Biological Function, Biosynthesis, and Synthesis of Phenazine Natural Products. Bioorg. Med. Chem 2017, 25, 6149–6166. [DOI] [PubMed] [Google Scholar]
- (138).Borrero NV; Bai F; Perez C; Duong BQ; Rocca JR; Jin S; Huigens RW III. Phenazine Antibiotic Inspired Discovery of Potent Bromophenazine Antibacterial Agents against Staphylococcus aureus and Staphylococcus epidermidis. Org. Biomol. Chem 2014, 12, 881–886. [DOI] [PubMed] [Google Scholar]
- (139).Garrison AT; Bai F; Abouelhassan Y; Paciaroni NG; Jin S; Huigens RW III. Bromophenazine Derivatives with Potent Inhibition, Dispersion and Eradication Activities against Staphylococcus aureus Biofilms. RSC Adv 2015, 5, 1120–1124. [Google Scholar]
- (140).Garrison AT; Abouelhassan Y; Kallifidas D; Bai F; Ukhanova M; Mai V; Jin S; Luesch H; Huigens RW III. Halogenated Phenazines that Potently Eradicate Biofilms, MRSA Persister Cells in Non-Biofilm Cultures and Mycobacterium tuber-culosis. Angew. Chem., Int. Ed 2015, 54, 14819–14823. [DOI] [PubMed] [Google Scholar]
- (141).Garrison AT; Abouelhassan Y; Norwood VM IV; Kallifidas D; Bai F; Nguyen M; Rolfe M; Burch GM; Jin S; Luesch H; Huigens RW III. Structure-Activity Relationships of a Diverse Class of Halogenated Phenazines that Targets Persistent, Antibiotic-Tolerant Bacterial Biofilms and Mycobacterium tuberculosis. J. Med. Chem 2016, 59, 3808–3825. [DOI] [PubMed] [Google Scholar]
- (142).Yang H; Abouelhassan Y; Burch GM; Kallifidas D; Huang G; Yousaf H; Jin S; Luesch H; Huigens RW III. A Highly Potent Class of Halogenated Phenazine Antibacterial and Biofilm-Eradicating Agents Accessed through a Modular Wohl-Aue Synthesis. Sci. Rep 2017, 7, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (143).Garrison AT; Abouelhassan Y; Kallifidas D; Tan H; Kim YS; Jin S; Luesch H; Huigens RW III. An Efficient Buchwald-Hartwig/Reductive Cyclization for the Scaffold Diversification of Halogenated Phenazines: Potent Antibacterial Targeting, Biofilm Eradication and Prodrug Exploration. J. Med. Chem 2018, 61, 3962–3983. [DOI] [PubMed] [Google Scholar]
- (144).Abouelhassan Y; Zhang Y; Jin S; Huigens RW III. Transcript Profiling of MRSA Biofilms Treated with a Halogenated Phenazine Eradicating Agent: A Platform for Defining Cellular Targets and Pathways Critical to Biofilm Survival. Angew. Chem., Int. Ed 2018, 57, 15523–15528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (145).Smith PA; Koehler MFT; Girgis HS; Yan D; Chen Y; Chen Y; Crawford JJ; Durk MR; Higuchi RI; Kang J; Murray J; Paraselli P; Park S; Phung W; Quinn JG; Roberts TC; Rougé L; Schwarz JB; Skippington E; Wai J; Xu M; Yu Z; Zhang H; Tan MW; Heise CE Optimized Arylomycins are a New Class of Gram-Negative Antibiotics. Nature 2018, 561, 189–194. [DOI] [PubMed] [Google Scholar]
- (146).Pinchman JR; Boger DL Probing the Role of the Vancomycin E-ring Aryl Chloride: Selective Divergent Synthesis and Evaluation of Alternatively Substituted E-ring Analogues. J. Med. Chem 2013, 56, 4116–4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (147).Okano A; Nakayama A; Schammel AW; Boger DL Total Synthesis of [Ψ[C(═NH)NH]Tpg4]Vancomycin and its (4-Chlorobiphenyl)methyl Derivative: Impact of Peripheral Modifications on Vancomycin Analogues Redesigned for Dual D-Ala-D-Ala and D-Ala-D-Lac Binding. J. Am. Chem. Soc 2014, 136, 13522–13525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (148).Okano A; Nakayama A; Wu K; Lindsey EA; Schammel AW; Feng Y; Collins KC; Boger DL Total Synthesis and Initial Evaluation of [Ψ[C(═S)NH]Tpg4]Vancomycin, [Ψ[C(═NH)NH]Tpg4]Vancomycin, [Ψ[CH2NH]Tpg4]Vancomycin, and Their (4-Chlorobiphenyl)methyl Derivatives: Synergistic Binding Pocket and Peripheral Modifications for the Glycopeptide Antibiotics. J. Am. Chem. Soc 2015, 137, 3693–3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (149).Okano A; Isley NA; Boger DL Peripheral Modifications of [Ψ[CH2NH]Tpg4]Vancomycin with Added Synergistic Mechanisms of Action Provide Durable and Potent Antibiotics. Proc. Natl. Acad. Sci. U. S. A 2017, 114, E5052–E5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (150).Okano A; Isley NA; Boger DL Total Synthesis of Vancomycin-Related Glycopeptide Antibiotics and Key Analogues. Chem. Rev 2017, 117, 11952–11993. [DOI] [PMC free article] [PubMed] [Google Scholar]



