Abstract
Cutaneous syncytial myoepithelioma (CSM) is a rare but distinctive benign variant in the family of myoepithelial neoplasms of skin and soft tissue. CSM has unique morphologic and immunohistochemical features, characterized by intradermal syncytial growth of spindled, ovoid, and histiocytoid cells and consistent staining for S-100 protein and EMA, and differs from other myoepithelial tumors by showing only infrequent keratin staining. Rearrangement of the EWSR1 gene is now known to occur in up to half of all skin and soft tissue myoepithelial tumors, with a wide family of documented fusion partners. In 2013, we reported frequent (80%) EWSR1 rearrangements in CSM, but were unable to identify the fusion partner using available studies at that time. After recent identification of an index case of CSM harboring an EWSR1-PBX3 fusion, we used a combination of targeted RNA sequencing and fluorescence in situ hybridization (FISH) studies to investigate the genetic features of a cohort of CSM. An EWSR1-PBX3 fusion was identified in all 13 cases successfully tested. RNA sequencing was successful in 8/13 cases, all of which were found to have identical breakpoints fusing exon 8 of EWSR1 to exon 5 of PBX3. FISH confirmed both EWSR1 and PBX3 rearrangements in 9/9 cases tested, which included 4 confirmed to have EWSR1-PBX3 fusion by RNA-Seq, 3 cases that failed RNA-Seq, and 2 cases examined by FISH alone. 2 cases failed RNA sequencing but had no additional tissue remaining for FISH studies. Our findings demonstrate that EWSR1-PBX3 fusions occur in most (and possibly all) cases of CSM.
Keywords: myoepithelioma, syncytial, skin, soft tissue, EWSR1, PBX3, RNA-Seq
INTRODUCTION
Cutaneous syncytial myoepithelioma (CSM) is a rare but distinctive variant in the family of myoepithelial neoplasms, having unique morphologic and immunohistochemical features. CSM most commonly arises in the extremities of adult patients (although a wide age range is observed) and occurs more frequently in men than women(1). In contrast to the characteristic morphologically heterogeneous appearance of myoepithelial neoplasms of skin and soft tissue(2–10), CSM is characterized by syncytial intradermal growth of spindled, ovoid, and histiocytoid cells(1, 8, 11). Tumors are benign and show no metastatic risk; local recurrence occurs only rarely, usually after incomplete excision. CSM shows consistent positivity for S-100 protein and EMA. However, unlike most myoepithelial neoplasms, keratin staining is infrequent, being at most focal or multifocal in only up to 10% of cases(1). Notably, CSM is distinct from cutaneous mixed tumor (i.e. chondroid syringoma), which shows ductal differentiation and frequent PLAG1 gene rearrangement (similarly to their salivary counterparts)(12, 13).
The genetic features of myoepithelial neoplasms of skin and soft tissue have increasingly been characterized over the past decade. Rearrangements of the EWSR1 gene are now known to occur in up to half of all benign and malignant myoepithelial tumors of skin, soft tissue, and bone(14, 15). To date, fusion partners have been identified in about 20% of cases, and the varied group of documented partners include POU5F1, PBX1, PBX3, ZNF444, ATF1, and KLF17(14, 16–20), as well as fusion genes with alternate FUS rearrangement(19, 21). In 2013, in a series of 38 CSM, we reported EWSR1 rearrangements in approximately 80% (14/17) of cases tested (1), and subsequent case reports of CSM have also confirmed the presence of EWSR1 rearrangement(11, 22). However, the fusion partner has remained unknown, and in our prior series of CSM, testing was negative for all known fusion partners at that time(1).
Recently, we identified an index case of CSM harboring an EWSR1-PBX3 fusion. Our aim in this current study was to evaluate a cohort of CSM to investigate the frequency of EWSR1-PBX3 fusions and to detect the presence of any alternative fusion genes.
MATERIALS AND METHODS
Index Case and Selection of CSM Cohort
The index case (case 1) was a 59-year-old woman who underwent biopsy of a skin lesion on the left shin. Immunohistochemical workup showed EMA positivity and focal weak S-100 staining (Table 1). Targeted RNA sequencing was performed on formalin-fixed paraffin embedded (FFPE) tissue sections, as part of the diagnostic work-up. Detection of EWSR1-PBX3 fusion prompted a retrospective search for a cohort of CSM. Fourteen additional cases of CSM diagnosed between 2013–2018 were selected from the consultation archives of one author (C.D.M.F.; Brigham and Women’s Hospital, Boston, MA), for which FFPE sections were available for RNA sequencing (n=12) and/or fluorescence in situ hybridization (FISH) (n=9). Immunohistochemical studies performed during diagnostic workup were reviewed. None of these cases were included in the prior study (1).
Table 1:
Clinicopathologic Features of this Cutaneous Syncytial Myoepithelioma Cohort
| Case | Clinicopathologic Features | Immunohistochemistry | Molecular Results | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Sex | Age (yrs) | Site | Tumor size (cm) | EMA | S-100 | Keratin | RNA-Seq | FISH | |
| 1 | F | 59 | Leg | 0.7 | Pos | Focal | Neg | EWSR1-PBX3 | - |
| 2 | F | 19 | Back | 0.6 | Pos | Pos | Neg | Failed | EWSR1, PBX3 |
| 3 | M | 47 | Buttock | 0.6 | Pos | Pos | Focal | Failed | EWSR1, PBX3 |
| 4 | F | 30 | Thigh | 0.4 | Pos | Pos | - | EWSR1-PBX3 | EWSR1, PBX3 |
| 5 | M | 17 | Arm | 0.4 | Pos | Pos | Neg | Failed* | - |
| 6 | F | 38 | Thigh | 0.3 | Pos | Pos | Neg | Failed* | - |
| 7 | M | 70 | Shoulder | 0.4 | Pos | Pos | Neg | Failed | EWSR1, PBX3 |
| 8 | M | 36 | Back | 0.5 | Pos | Pos | Neg | EWSR1-PBX3 | EWSR1, PBX3 |
| 9 | M | 56 | Thigh | 0.5 | Pos | Focal | Neg | EWSR1-PBX3 | - |
| 10 | F | 47 | Back | 0.3 | Pos | Pos | Focal | EWSR1-PBX3 | EWSR1, PBX3 |
| 11 | M | 30 | Thigh | 0.9 | Pos | Focal | - | EWSR1-PBX3 | - |
| 12 | F | 35 | Arm | 0.5 | Pos | Focal | Neg | EWSR1-PBX3 | EWSR1, PBX3 |
| 13 | F | 46 | Toe | 0.3 | Pos | MF | Neg | EWSR1-PBX3 | - |
| 14 | M | 22 | Arm | 0.5 | Pos | Pos | - | - | EWSR1, PBX3 |
| 15 | F | 24 | Thigh | 0.6 | Pos | MF | - | - | EWSR1, PBX3 |
F, female; M, male; Pos, positive; Neg, negative; MF, multifocal.
Insufficient material left for FISH analysis
RNA Sequencing
For case 1, RNA was extracted from 4 10 µm FFPE tissue scrolls. For the remaining 12 cases, 4 µm FFPE sections were cut onto positively charged glass slides and 3–5 slides scraped to provide tissue for RNA extraction. RNA was prepared using the ExpressArt FFPE Clear RNA Ready kit (Amsbio, Cambridge, MA), and total RNA quantified using the Qubit RNA HS Assay kit (Thermofisher Scientific, Mississauga, ON). RNA-Seq libraries were prepared with the TruSight RNA Fusion Panel (Illumina, San Diego, CA), and an input of 20–100 ng RNA. Each sample was sequenced with 76 base-pair paired-end reads on an Illumina MiSeq at 8 samples per flow cell (~3 million reads per sample). Results were analyzed using two pipelines; STAR aligner with Manta fusion caller, and BOWTIE2 alignment with the JAFFA fusion caller(23, 24).
Fluorescence in Situ Hybridization
FISH was performed on 9 cases from FFPE 4-µm sections by applying custom probes using bacterial artificial chromosomes (BAC), covering and flanking EWSR1 and PBX3, using previously detailed methods(18). Briefly, BAC clones were chosen according to the USCS genome browser (http://genome.uscs.edu) and obtained from BACPAC sources of Children’s Hospital of Oakland Research Institute (CHORI) (Oakland, CA) (http://bacpac.chori.org). DNA from individual BACs was isolated according to the manufacturer’s instructions, and labeled with different fluorochromes in a nick translation reaction, denatured, and hybridized to pretreated slides. The slides were then incubated, washed, and mounted with DAPI in an antifade solution. The genomic location of each BAC set was verified by hybridizing them to normal metaphase chromosomes. 200 successive tumor cell nuclei were examined using a Zeiss fluorescence microscope (Zeiss Axioplan, Oberkochen, Germany), controlled by Isis 5 software (Metasystems). A positive score was interpreted when at least 20% of interphase nuclei showed the break-apart signal. Nuclei with incomplete signal sets were not scored.
RESULTS
Clinicopathologic Features
The clinicopathologic features of the cohort are summarized in Table 1. The cohort consisted of 7 men and 8 women, with an age range of 17 to 70 years (median, 36 years). Tumors arose on the lower extremity (n=7, including 1 on toe), arm (n=4), and trunk (n=4). Lesions ranged from 0.3 cm to 0.9 cm (median, 0.5 cm). Tumors were well-circumscribed, but unencapsulated, dermal lesions comprised of solid and sheet-like syncytial growth of ovoid-to-spindled and histiocytoid cells with palely eosinophilic cytoplasm (Fig 1A). Tumor cells had uniform appearing ovoid or round nuclei with fine chromatin, smooth nuclear membranes, and small or inconspicuous nucleoli (Fig 1B–C). No significant atypia was seen, and necrosis was absent. Mitotic activity was generally low (range, 0–2 per 10 high-power fields (median count, 0). Five cases showed adipocytic metaplasia (Fig 1D). Entrapment of adnexal structures was common, as were lymphocytic infiltrates within and around the tumor, often surrounding vessels.
Figure 1.
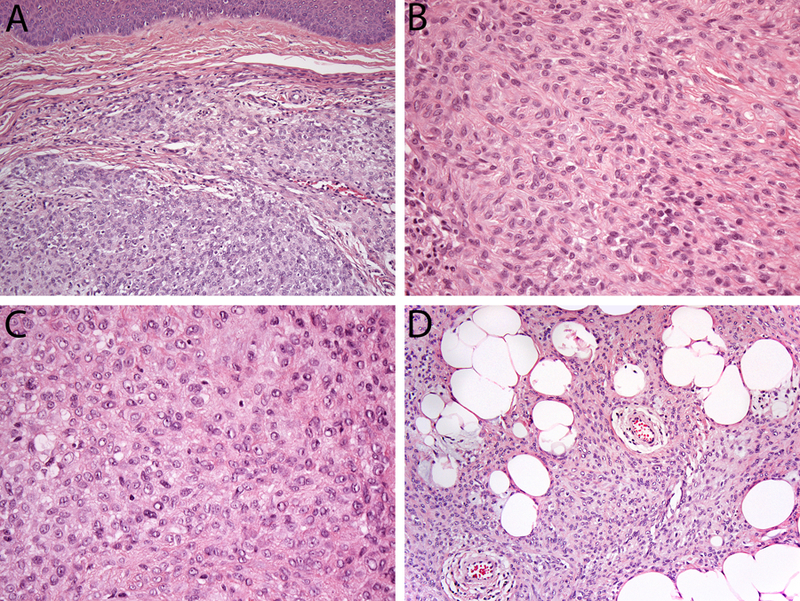
Features of cutaneous syncytial myoepithelioma. (A) Tumors are well-circumscribed and show syncytial growth of uniform ovoid, spindled, and histiocytoid cells (Case 8). Tumors cells have palely eosinophilic cytoplasm, and uniform nuclei that are ovoid (B, case 14) or round (C, case 7), with small or inconspicuous nuclei. Heterologous adipocytic metaplasia was present in a subset of cases (D, Case 11).
All cases of CSM showed expression of EMA and S-100. EMA staining was diffuse in all cases (15/15), while S-100 protein was strong in most cases but with some cases showing focal (n=4) or multifocal (n=2) staining. Keratin staining was focal in 2/11 cases tested, all others being negative. GFAP expression was present in 1/8 cases examined. SOX10 was negative in two cases tested (0/2).
RNA Sequencing
13 cases of CSM underwent RNA sequencing (including the index case). EWSR1-PBX3 fusions were identified in 7 additional cases (cases 4, 8, 9, 10, 11, 12 and 13), for a total of 8/13 cases confirmed by RNA-Seq. Five cases failed due to low sequencing quality metrics; of these, 3 cases had material available for FISH (see below) and 2 had insufficient material for further molecular testing. All 8 cases with EWSR1-PBX3 fusions had identical breakpoints involving exon 8 of EWSR1 fused to exon 5 of PBX3 (Fig 2).
Figure 2.
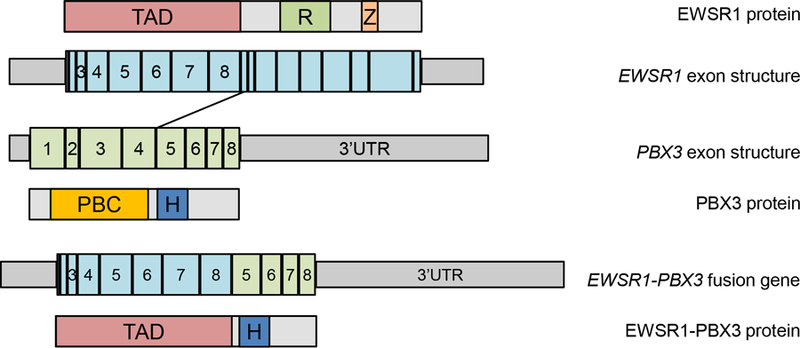
Schematic illustrating exon and protein structure of EWSR1 (top), PBX3 (middle), and the EWSR1-PBX3 fusion product found in cutaneous syncytial myoepithelioma (bottom). Grey boxes in the exon structure indicate 5’ and 3’ untranslated regions (UTRs). TAD – Transactivation domain, R – RRM RNA recognition motif, Z- Zinc Finger, PBC – PBC domain, H- Homeodomain DNA binding domain. The EWSR1-PBX3 gene fusion between EWSR1 exon 8 and PBX3 exon 5 results in a chimeric protein containing the transactivation domain of EWSR1 fused to the DNA binding homeodomain of PBX3.
EWSR1 and PBX3 FISH Findings
FISH testing for EWSR1 and PBX3 were performed to independently verify the presence of the fusion gene in 9 cases, which included 3 cases that failed RNA-Seq (cases 2, 3, 7), 4 confirmed to have EWSR1-PBX3 fusion (cases 4, 8, 10, 12), and 2 cases examined by FISH alone (cases 14, 15). All 9 cases showed rearrangement of both EWSR1 and PBX3 (Fig 3).
Figure 3.
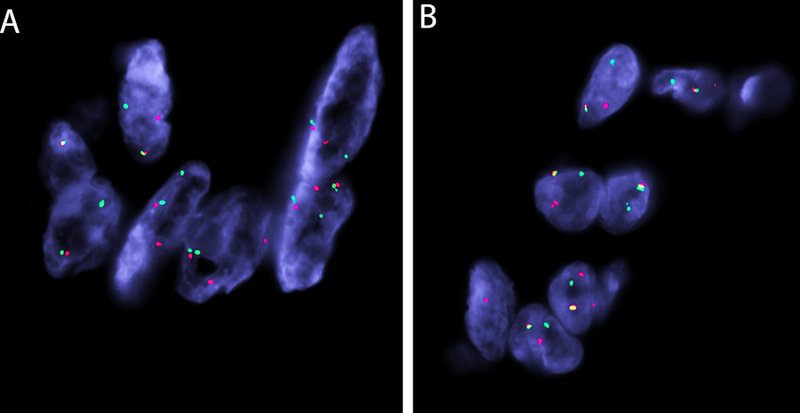
FISH break-apart assays demonstrating split signals for EWSR1 gene (A) and PBX3 (B) genes (B) (red, centromeric; green telomeric, for both genes).
DISCUSSION
Herein we report that EWSR1-PBX3 fusions characterize the vast majority of CSMs. In 2013, we first reported the presence of EWSR1 rearrangement in 82% (14/17) cases of CSM(1). We had excluded all known fusion partners in myoepithelial neoplasms at the time, including PBX1, ZNF444, POU5F1, DUX4, ATF1, CREB1, NR4A3, DDIT3, and NFATc2, and concluded that CSMs were likely characterized by a novel fusion gene. Subsequently in 2015, EWSR1-PBX3 fusions were identified in 3 myoepithelial tumors (2 in bone and 1 in soft tissue)(18), however, no PBX3 rearrangements were detected in the 3 CSM cases with EWSR1-rearrangements included in that study(18). While it is possible that these cases may harbor separate, unidentified novel fusions, it is also likely that tissue degradation of older archival material was a factor.
Myoepithelial neoplasms of skin and soft tissue are characterized by histologic heterogeneity, both intratumorally, and between different tumors, with patterns including spindled, ovoid, epithelioid, and clear cell morphology and a combination of reticular, trabecular, and nested growth, as well as variable expression of keratin, EMA, S-100 protein, and GFAP (2, 3, 5, 6, 8, 9, 15). A variably chondromyxoid or hyalinized stroma is common and often a useful diagnostic feature. Heterologous differentiation occurs in up to 15% of myoepithelial neoplasms and is most commonly chondro-osseous(2, 6, 9). In contrast, CSM show a distinctive appearance that is relatively constant between tumors. While showing variably spindled, ovoid, and histiocytoid morphology, CSM tumor cells overall appear uniform with palely eosinophilic cytoplasm and consistent syncytial growth. CSM lacks ductal differentiation, a feature of cutaneous mixed tumors. Moreover, unlike other myoepithelial tumors, adipocytic metaplasia is the most commonly observed type of heterologous differentiation (1, 8, 11), and the immunophenotype is reliably positive for EMA and S-100 protein, and usually negative for keratin.
Given the consistent morphology and immunophenotype of CSM, their genetic uniformity, with nearly all cases showing EWSR1-PBX3 gene fusions, is not entirely surprising. Studies have suggested that certain fusion genes are associated with specific morphologic features, such as tumors with EWSR1-POU5F1 showing epithelioid cells with clear cytoplasm in a solid and nested growth pattern surrounded by thin fibrous septa(14). Among myoepithelial neoplasms with EWSR1-PBX1 fusions, a subset have either a predominant appearance (or foci) of a deceptively bland spindle cell proliferation(14), as was also seen in 2008 in the first ever reported myoepithelial tumor of soft tissue with a fusion gene (EWSR1-PBX1)(16). To date, EWSR1-PBX3 fusions are rare in neoplasia, having been reported in 3 intraosseous myoepitheliomas, arising in the tibia, fibula(18), and fourth metatarsal(25), and one soft tissue myoepithelial tumor of the finger(18), as well as a single case of retroperitoneal leiomyoma(26), thus any typical morphologic features characterizing these tumors requires a larger sample size to ascertain with any certainty.
The PBX transcription factors (PBX1, PBX2, PBX2, PBX4) are members of the TALE (three amino acid loop extension) homeobox gene family and are involved in regulating gene expression during development through their ability to form DNA-binding heterodimers that interact with a wide range of transcription factors, including the HOX family(27). The PBX genes show a high degree of homology(28) and are involved in the differentiation of urogenital system and steroidogenesis in the adrenal cortex(27). PBX1, the most extensively characterized PBX transcription factor, was first discovered as part of the E2A-PBX1 fusion characteristic of a subset of pre-B-cell acute lymphoblastic leukemia(29). The oncogenic role of PBX3 is currently less well understood, but this gene has been shown to be overexpressed in cell lines of colorectal cancer(30), prostate cancer(31), and cervical cancer(32), and upregulated in gastric cancer resections(33). The EWSR1-PBX3 gene fusion in CSM is predicted to result in a chimeric protein containing the transactivation domain of EWSR1 fused to the DNA binding homeodomain of PBX3 (Figure 2). This is presumed to lead to dysregulation of PBX3 target genes.
Despite the distinctive morphologic and immunohistochemical features of CSM, the diagnosis may be challenging due to the rarity of these tumors, and, on rare occasion, detection of EWSR1-PBX3 fusion may potentially be diagnostically helpful, particularly in distinguishing CSM from more clinically significant lesions. The differential diagnosis includes epithelioid benign fibrous histiocytoma, juvenile xanthogranuloma, epithelioid sarcoma, and melanocytic lesions. Immunohistochemistry is useful in most cases in resolving the differential diagnosis. Epithelioid benign fibrous histiocytoma shows frequent EMA positivity, but is negative for S-100 and harbors ALK rearrangements that can be identified by immunohistochemistry, FISH, and/or RNA-Seq(34, 35). Juvenile xanthogranuloma (especially “early stage” lesions) are positive for histiocytic markers and do not express conventional myoepithelial markers. Epithelioid sarcoma is positive for EMA, but typically shows more keratin expression as well as CD34 positivity (in 50% of cases) and S-100-negativitiy. Most cases of epithelioid sarcoma show loss of INI1/SMARCB1 expression(36), frequently correlating with homozygous deletions of SMARCB1(37). Among melanocytic tumors, Spitz nevus and spitzoid melanoma may arise in the differential; both express S-100 and melanocytic differentiation can be confirmed by HMB-45 and Mart1.
In summary, we identified recurrent EWSR1-PBX3 fusions in CSM. In contrast to the broad spectrum of myoepithelial neoplasms, CSM shows more uniform immunophenotypic and genetic features. Whether other fusion variants exist in CSM remains to be determined, and the molecular spectrum, if any, may be expanded in the future.
Footnotes
Financial disclosure and conflict of interest: The authors declare no relevant conflicts of interest. This project was supported in part by: P50 CA140146–01 (CRA); P50 CA217694 (CRA); P30-CA008748 (CRA); Kristen Ann Carr Foundation (CRA); Cycle for Survival (CRA).
References
- 1.Jo VY, Antonescu CR, Zhang L, et al. Cutaneous syncytial myoepithelioma: clinicopathologic characterization in a series of 38 cases. Am J Surg Pathol 2013;37:710–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kilpatrick SE, Hitchcock MG, Kraus MD, et al. Mixed tumors and myoepitheliomas of soft tissue: a clinicopathologic study of 19 cases with a unifying concept. Am J Surg Pathol 1997;21:13–22. [DOI] [PubMed] [Google Scholar]
- 3.Michal M, Miettinen M. Myoepitheliomas of the skin and soft tissues. Report of 12 cases. Virchows Arch 1999;434:393–400. [DOI] [PubMed] [Google Scholar]
- 4.Kutzner H, Mentzel T, Kaddu S, et al. Cutaneous myoepithelioma: an under-recognized cutaneous neoplasm composed of myoepithelial cells. Am J Surg Pathol 2001;25:348–355. [DOI] [PubMed] [Google Scholar]
- 5.Mentzel T, Requena L, Kaddu S, et al. Cutaneous myoepithelial neoplasms: clinicopathologic and immunohistochemical study of 20 cases suggesting a continuous spectrum ranging from benign mixed tumor of the skin to cutaneous myoepithelioma and myoepithelial carcinoma. J Cutan Pathol 2003;30:294–302. [DOI] [PubMed] [Google Scholar]
- 6.Hornick JL, Fletcher CD. Myoepithelial tumors of soft tissue: a clinicopathologic and immunohistochemical study of 101 cases with evaluation of prognostic parameters. Am J Surg Pathol 2003;27:1183–1196. [DOI] [PubMed] [Google Scholar]
- 7.Neto AG, Pineda-Daboin K, Luna MA. Myoepithelioma of the soft tissue of the head and neck: a case report and review of the literature. Head Neck 2004;26:470–473. [DOI] [PubMed] [Google Scholar]
- 8.Hornick JL, Fletcher CD. Cutaneous myoepithelioma: a clinicopathologic and immunohistochemical study of 14 cases. Hum Pathol 2004;35:14–24. [DOI] [PubMed] [Google Scholar]
- 9.Gleason BC, Fletcher CD. Myoepithelial carcinoma of soft tissue in children: an aggressive neoplasm analyzed in a series of 29 cases. Am J Surg Pathol 2007;31:1813–1824. [DOI] [PubMed] [Google Scholar]
- 10.Tanahashi J, Kashima K, Daa T, et al. A case of cutaneous myoepithelial carcinoma. J Cutan Pathol 2007;34:648–653. [DOI] [PubMed] [Google Scholar]
- 11.Alomari AK, Brown N, Andea AA, et al. Cutaneous syncytial myoepithelioma: A recently described neoplasm which may mimic nevoid melanoma and epithelioid sarcoma. J Cutan Pathol 2017;44:892–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bahrami A, Dalton JD, Krane JF, et al. A subset of cutaneous and soft tissue mixed tumors are genetically linked to their salivary gland counterpart. Genes, chromosomes & cancer 2012;51:140–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Antonescu CR, Zhang L, Shao SY, et al. Frequent PLAG1 gene rearrangements in skin and soft tissue myoepithelioma with ductal differentiation. Genes Chromosomes Cancer 2013;52:675–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Antonescu CR, Zhang L, Chang NE, et al. EWSR1-POU5F1 fusion in soft tissue myoepithelial tumors. A molecular analysis of sixty-six cases, including soft tissue, bone, and visceral lesions, showing common involvement of the EWSR1 gene. Genes Chromosomes Cancer 2010;49:1114–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flucke U, Palmedo G, Blankenhorn N, et al. EWSR1 gene rearrangement occurs in a subset of cutaneous myoepithelial tumors: a study of 18 cases. Mod Pathol 2011;24:1444–1450. [DOI] [PubMed] [Google Scholar]
- 16.Brandal P, Panagopoulos I, Bjerkehagen B, et al. Detection of a t(1;22)(q23;q12) translocation leading to an EWSR1-PBX1 fusion gene in a myoepithelioma. Genes Chromosomes Cancer 2008;47:558–564. [DOI] [PubMed] [Google Scholar]
- 17.Brandal P, Panagopoulos I, Bjerkehagen B, et al. t(19;22)(q13;q12) Translocation leading to the novel fusion gene EWSR1-ZNF444 in soft tissue myoepithelial carcinoma. Genes, chromosomes & cancer 2009;48:1051–1056. [DOI] [PubMed] [Google Scholar]
- 18.Agaram NP, Chen HW, Zhang L, et al. EWSR1-PBX3: a novel gene fusion in myoepithelial tumors. Genes Chromosomes Cancer 2015;54:63–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang SC, Chen HW, Zhang L, et al. Novel FUS-KLF17 and EWSR1-KLF17 fusions in myoepithelial tumors. Genes Chromosomes Cancer 2015;54:267–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flucke U, Mentzel T, Verdijk MA, et al. EWSR1-ATF1 chimeric transcript in a myoepithelial tumor of soft tissue: a case report. Hum Pathol 2012;43:764–768. [DOI] [PubMed] [Google Scholar]
- 21.Puls F, Arbajian E, Magnusson L, et al. Myoepithelioma of bone with a novel FUS-POU5F1 fusion gene. Histopathology 2014;65:917–922. [DOI] [PubMed] [Google Scholar]
- 22.Lee JH, Huang HY, Lan J, et al. Cutaneous syncytial myoepithelioma: A case report with emphasis on the differential diagnosis of problematic dermal tumors. Oncol Lett 2015;9:2275–2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu S, Tsai WH, Ding Y, et al. Comprehensive evaluation of fusion transcript detection algorithms and a meta-caller to combine top performing methods in paired-end RNA-seq data. Nucleic Acids Res 2016;44:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen X, Schulz-Trieglaff O, Shaw R, et al. Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics 2016;32:1220–1222. [DOI] [PubMed] [Google Scholar]
- 25.Yun S, Kim SH, Cho HS, et al. EWSR1-PBX3 fused myoepithelioma arising in metatarsal bone: Case report and review of the literature. Pathol Int 2019;69:42–47. [DOI] [PubMed] [Google Scholar]
- 26.Panagopoulos I, Gorunova L, Bjerkehagen B, et al. Fusion of the genes EWSR1 and PBX3 in retroperitoneal leiomyoma with t(9;22)(q33;q12). PLoS One 2015;10:e0124288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laurent A, Bihan R, Omilli F, et al. PBX proteins: much more than Hox cofactors. Int J Dev Biol 2008;52:9–20. [DOI] [PubMed] [Google Scholar]
- 28.Monica K, Galili N, Nourse J, et al. PBX2 and PBX3, new homeobox genes with extensive homology to the human proto-oncogene PBX1. Mol Cell Biol 1991;11:6149–6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kamps MP, Look AT, Baltimore D. The human t(1;19) translocation in pre-B ALL produces multiple nuclear E2A-Pbx1 fusion proteins with differing transforming potentials. Genes Dev 1991;5:358–368. [DOI] [PubMed] [Google Scholar]
- 30.Han HB, Gu J, Ji DB, et al. PBX3 promotes migration and invasion of colorectal cancer cells via activation of MAPK/ERK signaling pathway. World J Gastroenterol 2014;20:18260–18270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ramberg H, Alshbib A, Berge V, et al. Regulation of PBX3 expression by androgen and Let-7d in prostate cancer. Mol Cancer 2011;10:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H, Sun G, Liu C, et al. PBX3 is associated with proliferation and poor prognosis in patients with cervical cancer. Onco Targets Ther 2017;10:5685–5694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Y, Sun Z, Zhu Z, et al. PBX3 is overexpressed in gastric cancer and regulates cell proliferation. Tumour Biol 2014;35:4363–4368. [DOI] [PubMed] [Google Scholar]
- 34.Doyle LA, Marino-Enriquez A, Fletcher CD, et al. ALK rearrangement and overexpression in epithelioid fibrous histiocytoma. Mod Pathol 2015;28:904–912. [DOI] [PubMed] [Google Scholar]
- 35.Dickson BC, Swanson D, Charames GS, et al. Epithelioid fibrous histiocytoma: molecular characterization of ALK fusion partners in 23 cases. Mod Pathol 2018;31:753–762. [DOI] [PubMed] [Google Scholar]
- 36.Hornick JL, Dal Cin P, Fletcher CD. Loss of INI1 expression is characteristic of both conventional and proximal-type epithelioid sarcoma. Am J Surg Pathol 2009;33:542–550. [DOI] [PubMed] [Google Scholar]
- 37.Le Loarer F, Zhang L, Fletcher CD, et al. Consistent SMARCB1 homozygous deletions in epithelioid sarcoma and in a subset of myoepithelial carcinomas can be reliably detected by FISH in archival material. Genes Chromosomes Cancer 2014;53:475–486. [DOI] [PMC free article] [PubMed] [Google Scholar]


