Lactobacilli are dominant members of the healthy female bladder microbiota. Here, we report the complete genome sequences of six Lactobacillus gasseri and three Lactobacillus paragasseri strains isolated from catheterized urine samples. These L. paragasseri genomes are the first publicly available sequences of the species from the bladder.
ABSTRACT
Lactobacilli are dominant members of the healthy female bladder microbiota. Here, we report the complete genome sequences of six Lactobacillus gasseri and three Lactobacillus paragasseri strains isolated from catheterized urine samples. These L. paragasseri genomes are the first publicly available sequences of the species from the bladder.
ANNOUNCEMENT
Lactobacillus is a commensal bacterium in the human body and is a key component of the healthy urinary and vaginal microbiota (1). The family Lactobacillaceae has one of the highest rates of incidence compared to those of other bacterial families in the urinary tract (2). Lactobacillus gasseri is a predominant species in the human microbiota and is able to prevent other bacteria from growing in the same environment, protecting the host from pathogens (3). Lactobacillus paragasseri was classified as a novel species in 2018 (4) and, until now, has not been characterized in the urinary tract.
Catheterized urine samples were collected from women as part of prior institutional review board (IRB)-approved studies (5–9). Bacteria were isolated from these samples using the enhanced quantitative urine culture (EQUC) method (9) and stored at −80°C. We selected nine strains in our collection for whole-genome sequencing; these strains were identified as L. gasseri by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF) mass spectrometry. Freezer stocks for each of the nine strains were first streaked on Columbia colistin-nalidixic acid agar with 5% sheep blood plates (catalog number 221353; BD) and incubated at 35°C in 5% CO2 for 48 hours. A single colony was then selected and grown in MRS liquid medium at 35°C in 5% CO2 for 48 hours. DNA was extracted with the Qiagen DNeasy UltraClean microbial kit, and the DNA was quantified by a Qubit fluorometer. DNA libraries were constructed (Nextera XT library prep kit) and sequenced using the MiSeq reagent kit v2, producing 250-bp paired-end reads (minimum, 266,494 pairs; maximum, 1,342,972 pairs; average, 524,976 pairs). The raw reads were trimmed using Sickle v1.33 (https://github.com/najoshi/sickle) and then assembled with SPAdes v3.13.0 (10) (parameters, “only-assembler” option for k = 55, 77, 99, and 127). The assembled contigs were evaluated for genome completeness and contamination by CheckM v1.0.12 (11), and genome coverage was calculated using BBMap v38.47 (https://sourceforge.net/projects/bbmap/). Genome annotations were performed using PATRIC v3.5.43 (12) and the NCBI Prokaryotic Genome Annotation Pipeline (PGAP) v4.8 (13). The PGAP annotations are published with the deposited genome assemblies. A phylogenetic tree was derived with RAxML v8.2.11 in PATRIC using the PATRIC annotations and the codon tree method (12). Unless otherwise noted, default parameters were used for all software tools.
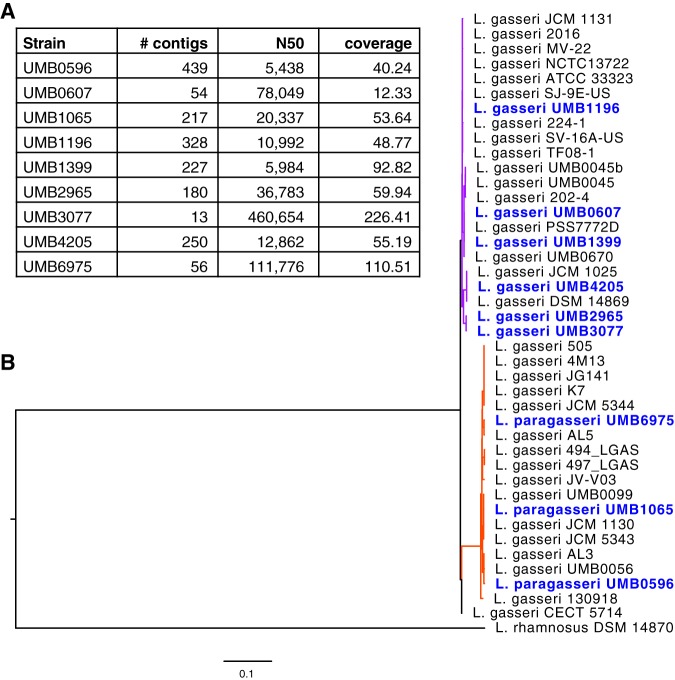
The nine bladder lactobacilli genomes vary in size from 1,041,937 bp (strain UMB1399) to 2,108,391 bp (strain UMB6975) in length, with an average GC content of 35.2%. Assembly statistics are listed in Fig. 1A. Genome assemblies for lactobacilli are particularly challenging given the presence of numerous short repeats throughout the genome (14). As part of NCBI’s quality control process, average nucleotide identity is calculated (15), and three of the genomes (UMB0596, UMB1065, and UMB6975) were reclassified as strains of the species L. paragasseri (98 to 99% identical for over 94% of the genome to the type genome of L. paragasseri (strain JCM 5343 [GenBank accession number AP018549]). The nine bladder lactobacillus genomes were also compared with those of publicly available L. gasseri and L. paragasseri strains in PATRIC (as of July 2019). Figure 1B shows a phylogenetic analysis of these genome comparisons. There is a clear distinction between the six bladder L. gasseri and three bladder L. paragasseri strains. From our phylogenetic analysis, we have identified not only three new strains of L. paragasseri but also other strains presently classified as L. gasseri that are likely members of the L. paragasseri species (Fig. 1B; branches shown in orange).
FIG 1.
Genome assembly statistics (A) and phylogenetic tree (B) of six L. gasseri and three L. paragasseri strains. Genomes sequenced in this study are shown in blue. Branches belonging to the L. gasseri clade are shown in purple, and the L. paragasseri clade’s branches are shown in orange.
Data availability.
This whole-genome shotgun project has been deposited in GenBank under the accession numbers VNFS00000000 (UMB4205), VNFT00000000 (UMB2965), VNFU00000000 (UMB1196), VNFY00000000 (UMB0607), VNGC00000000 (UMB3077), and VNGD00000000 (UMB1399) for the six L. gasseri strains and VNFQ00000000 (UMB0596), VNFV00000000 (UMB1065), and VNFX00000000 (UMB6975) for the three L. paragasseri strains. The versions described in this paper are the first versions. Raw sequence data are publicly available for the six L. gasseri strains (SRA accession numbers SRR9695707, SRR9695712, SRR9695713, SRR9695714, SRR9695719, and SRR9695724) and the three L. paragasseri strains (accession numbers SRR9695720, SRR9695721, and SRR9695723).
ACKNOWLEDGMENTS
This work was conducted as part of Loyola University of Chicago’s Department of Biology Bacterial Genomics course.
For prior patient recruitment, we acknowledge the Loyola Urinary Education and Research Collaborative (LUEREC) and the patients who provided the samples for this study. We also thank Roberto Limeira at Loyola’s Genomics Facility for his assistance in sequencing these isolates.
T.M.-E. is funded through Loyola’s Carbon Research Fellowship.
REFERENCES
- 1.Hammes WP, Vogel RF. 1995. The genus Lactobacillus, p 19–54. In Wood BJB, Holzapfel WH (ed), The genera of lactic acid bacteria. Springer, Boston, MA. [Google Scholar]
- 2.Thomas-White K, Forster SC, Kumar N, Van Kuiken M, Putonti C, Stares MD, Hilt EE, Price TK, Wolfe AJ, Lawley TD. 2018. Culturing of female bladder bacteria reveals an interconnected urogenital microbiota. Nat Commun 9:1557. doi: 10.1038/s41467-018-03968-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boris S, Suárez JE, Barbés C. 1997. Characterization of the aggregation promoting factor from Lactobacillus gasseri, a vaginal isolate. J Appl Microbiol 83:413–420. doi: 10.1046/j.1365-2672.1997.00250.x. [DOI] [PubMed] [Google Scholar]
- 4.Tanizawa Y, Tada I, Kobayashi H, Endo A, Maeno S, Toyoda A, Arita M, Nakamura Y, Sakamoto M, Ohkuma M, Tohno M. 2018. Lactobacillus paragasseri sp. nov., a sister taxon of Lactobacillus gasseri, based on whole-genome sequence analyses. Int J Syst Evol Microbiol 68:3512–3517. doi: 10.1099/ijsem.0.003020. [DOI] [PubMed] [Google Scholar]
- 5.Thomas-White KJ, Hilt EE, Fok C, Pearce MM, Mueller ER, Kliethermes S, Jacobs K, Zilliox MJ, Brincat C, Price TK, Kuffel G, Schreckenberger P, Gai X, Brubaker L, Wolfe AJ. 2016. Incontinence medication response relates to the female urinary microbiota. Int Urogynecol J 27:723–733. doi: 10.1007/s00192-015-2847-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pearce MM, Hilt EE, Rosenfeld AB, Zilliox MJ, Thomas-White K, Fok C, Kliethermes S, Schreckenberger PC, Brubaker L, Gai X, Wolfe AJ. 2014. The female urinary microbiome: a comparison of women with and without urgency urinary incontinence. mBio 5:e01283. doi: 10.1128/mBio.01283-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pearce MM, Zilliox MJ, Rosenfeld AB, Thomas-White KJ, Richter HE, Nager CW, Visco AG, Nygaard IE, Barber MD, Schaffer J, Moalli P, Sung VW, Smith AL, Rogers R, Nolen TL, Wallace D, Meikle SF, Gai X, Wolfe AJ, Brubaker L, Pelvic Floor Disorders Network . 2015. The female urinary microbiome in urgency urinary incontinence. Am J Obstet Gynecol 213:347.e1–347.e11. doi: 10.1016/j.ajog.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Price TK, Dune T, Hilt EE, Thomas-White KJ, Kliethermes S, Brincat C, Brubaker L, Wolfe AJ, Mueller ER, Schreckenberger PC. 2016. The clinical urine culture: enhanced techniques improve detection of clinically relevant microorganisms. J Clin Microbiol 54:1216–1222. doi: 10.1128/JCM.00044-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hilt EE, McKinley K, Pearce MM, Rosenfeld AB, Zilliox MJ, Mueller ER, Brubaker L, Gai X, Wolfe AJ, Schreckenberger PC. 2014. Urine is not sterile: use of enhanced urine culture techniques to detect resident bacterial flora in the adult female bladder. J Clin Microbiol 52:871–876. doi: 10.1128/JCM.02876-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. 2015. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25:1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wattam AR, Brettin T, Davis JJ, Gerdes S, Kenyon R, Machi D, Mao C, Olson R, Overbeek R, Pusch GD, Shukla MP, Stevens R, Vonstein V, Warren A, Xia F, Yoo H. 2018. Assembly, annotation, and comparative genomics in PATRIC, the all bacterial bioinformatics resource center. Methods Mol Biol 1704:79–101. doi: 10.1007/978-1-4939-7463-4_4. [DOI] [PubMed] [Google Scholar]
- 13.Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky L, Lomsadze A, Pruitt KD, Borodovsky M, Ostell J. 2016. NCBI Prokaryotic Genome Annotation Pipeline. Nucleic Acids Res 44:6614–6624. doi: 10.1093/nar/gkw569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmid M, Muri J, Melidis D, Varadarajan AR, Somerville V, Wicki A, Moser A, Bourqui M, Wenzel C, Eugster-Meier E, Frey JE, Irmler S, Ahrens CH. 2018. Comparative genomics of completely sequenced Lactobacillus helveticus genomes provides insights into strain-specific genes and resolves metagenomics data down to the strain level. Front Microbiol 9:63. doi: 10.3389/fmicb.2018.00063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ciufo S, Kannan S, Sharma S, Badretdin A, Clark K, Turner S, Brover S, Schoch CL, Kimchi A, DiCuccio M. 2018. Using average nucleotide identity to improve taxonomic assignments in prokaryotic genomes at the NCBI. Int J Syst Evol Microbiol 68:2386–2392. doi: 10.1099/ijsem.0.002809. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This whole-genome shotgun project has been deposited in GenBank under the accession numbers VNFS00000000 (UMB4205), VNFT00000000 (UMB2965), VNFU00000000 (UMB1196), VNFY00000000 (UMB0607), VNGC00000000 (UMB3077), and VNGD00000000 (UMB1399) for the six L. gasseri strains and VNFQ00000000 (UMB0596), VNFV00000000 (UMB1065), and VNFX00000000 (UMB6975) for the three L. paragasseri strains. The versions described in this paper are the first versions. Raw sequence data are publicly available for the six L. gasseri strains (SRA accession numbers SRR9695707, SRR9695712, SRR9695713, SRR9695714, SRR9695719, and SRR9695724) and the three L. paragasseri strains (accession numbers SRR9695720, SRR9695721, and SRR9695723).