Abstract
Recently, prediction of lncRNA-disease associations has attracted more and more attentions. Various computational models have been proposed; however, there is still room to improve the prediction accuracy. In this paper, we propose a kernel fusion method with different types of similarities for the lncRNAs and diseases. The expression similarity and cosine similarity are used for lncRNAs, and the semantic similarity and cosine similarity are used for the diseases. To eliminate the noise effect, a neighbor constraint is enforced to refine all the similarity matrices before fusion. Experimental results show that the proposed similarity kernel fusion (SKF)-LDA method has the superiority performance in terms of AUC values and other measurements. In the schemes of LOOCV and -fold CV, AUC values of SKF-LDA achieve and respectively. In addition, the conducted case studies of three diseases (hepatocellular carcinoma, lung cancer, and prostate cancer) show that SKF-LDA can predict related lncRNAs accurately.
Keywords: lncRNA-disease association, lncRNA similarity, disease similarity, similarity kernel fusion, Laplacian regularized least squares
Introduction
In humans, only about 1.5% of the genome can be encoded into proteins, while the rest are extensively transcribed into non-coding RNAs (ncRNAs).1, 2, 3 Studies have shown that ncRNAs play an important role in human biological mechanisms,4, 5, 6 especially the long non-coding RNAs (lncRNAs), a kind of significant ncRNA with over 200 nt in length.7, 8, 9, 10 In the past years, lncRNAs have been found that influenced transcription, translation, cell cycle, imprinting, splicing, and protein localization;5, 11, 12 for example, intergenic 10 regulates expression of ADAM12- and FANK1-flanking genes through modulation of the chromatin structure in cis.13 Additionally, it has shown that the dysregulations and mutations of some lncRNAs are associated with human diseases,14, 15, 16 such as breast cancer,17, 18 intracranial aneurysm,19, 20 and -thalassemia.21, 22 Though the detail mechanisms of lncRNA are a riddle, we can still use computational models to predict the relationship between lncRNAs and diseases, which can be helpful for disease diagnosis. Besides, the computational methods can be a powerful complementary tool to the biological experiments or can even avoid the time-consuming experiments.23
In the past few years, many effective computational models have been built to calculate potential lncRNA-disease associations.24, 25, 26, 27 We can roughly classify the methods into two categories according to the computational model. The first category of methods predicts the lncRNA-disease associations with designed machine learning models. Chen et al.28 proposed Laplacian regularized least squares for lncRNA-disease association (LRLSLDA) in the semi-supervised learning framework, which is based on the prior that similar diseases are more likely to be associated with functionally similar lncRNAs. Lan et al.29 integrated multiple biological data resources to predict lncRNA-disease associations with a bagging support vector machine (SVM) classifier based on lncRNA similarity and disease similarity. The SIMCLDA30 method uses an inductive matrix to predict lncRNA-disease associations. In this model, the Gaussian interaction profile kernel of lncRNAs is from known lncRNA-disease interactions, and the function similarity of diseases is based on disease-gene and gene-gene ontology associations. Also, the primary feature vectors from constructed feature matrices are used to complete the association matrix based on the inductive matrix completion framework. Another type of method predicts associations based on constructed networks. In an earlier study, Sun et al.31 designed a computational network framework, random walk with restart for lncRNA-disease association (RWRlncD), to predict lncRNA-disease association based on the lncRNA-lncRNA functional similarity network. RWRlncD integrates lncRNA-disease association and disease functional similarity and then uses random walk. Bi-random walks for lncRNA-disease association (BRWLDA)32 applies bi-random walks that take the structural differences into account between lncRNA similarity and disease similarity and build two networks using lncRNA functional similarity and disease semantic similarity. Then multiple the random walks method is used on both networks to predict potential lncRNA-disease associations. Paths with limited lengths for lncRNAs-diseases association (PLLDA) builds lncRNA similarity networks and disease similarity networks based on lncRNA functional similarity and disease semantic similarity.33 PLLDA is based on the method of connecting their pathways and their length in their respective similarity networks. The depth-first search is then used to calculate the probability of lncRNA-disease association. This method is suitable for the prediction of the relation between a new disease and lncRNAs or the relation between a new lncRNA and diseases. However, since the length-based cost function is relatively simple, it is necessary to look for model substitution for machine learning. By combining lncRNA-disease association and gene and disease association, tripartite graph for lncRNAs-diseases association (TPGLDA) can effectively predict the association between lncRNA and disease, but this method relies on the structure among the three parts, and the incomplete data will limit its performance.34
Although the aforementioned methods achieve relatively good results, there is still room for us to improve the accuracy of association prediction. For example, some methods only consider the similarity of lncRNA and disease in one dimension (functional or semantic) and do not fully take the multi-dimensional information into consideration. It is believed that, when more biological knowledge is applied with elaborated fusion method, we can get a more accurate prediction, as proved by previous studies on the prediction of association between microRNA (miRNA) and disease.35, 36 Recently, there are increasingly more state-of-the art computational models37, 38, 39, 40, 41 published in well-known journals that have promoted the study of the association between lncRNA and disease. For example, the integration method we used is inspired by the method of predicting miRNA and disease association.42 Also, the idea of using Laplacian regularization43 to explore the relationship between miRNA and disease inspires us to apply similar ideas to predict the association between lncRNA and disease.
In this study, we present a similarity kernel fusion (SKF) method to predict lncRNA-disease association (SKF-LDA, for short). In the proposed method, two different similarities, the functional similarity and semantic similarity, are utilized with a new fusion approach. The fusion step is built on the refined similarity matrices by a neighbor-based constraint and iterates over the similarity matrices instead of a simply weighted addition. The final lncRNA-disease association is computed by a normal Laplacian regularized least-squares method. To demonstrate the prediction performance of the proposed SKF-LDA, the leave-one-out cross-validation (LOOCV) and 5-fold cross-validation (5-fold CV) frameworks are implemented. The experimental results showed that the proposed SKF-LDA achieves 0.9049 and 0.87430.0050 in terms of AUC value in the scheme of LOOCV and 5-fold CV. Furthermore, in case studies, 9, 10, and 7 out of the top 10 predicted lncRNAs for disease hepatocellular carcinoma, lung cancer, and prostate cancer, respectively, are confirmed by recent research.
Results
Overview of Proposed Method
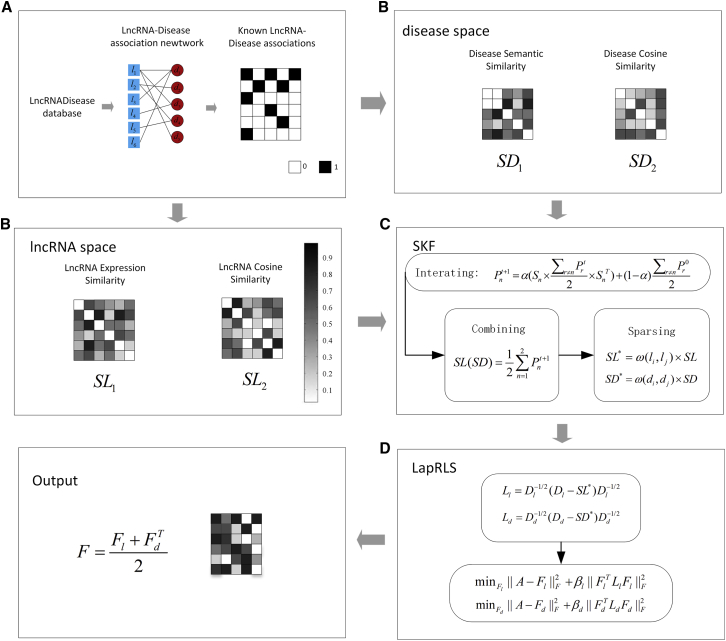
The proposed SKF-LDA method can be summarized in four steps, as shown in Figure 1. First, we construct the lncRNA-disease correlation matrix based on the known lncRNA-disease association. Second, we calculate lncRNA similarity (lncRNA expression similarity, lncRNA cosine similarity) and disease similarity (disease semantic similarity, disease cosine similarity). Third, SKF is used to integrate the two similarities of lncRNAs and diseases. Lastly, we obtain the predicted lncRNA-disease association matrix by the Laplacian regularized least-squares method.
Figure 1.
Flow Chart of SKF-LDA Applied to lncRNA-Disease Association Prediction
(A–D) SKF-LDA consists of four steps: (A) constructing the lncRNA-disease correlation matrix; (B) calculating the two similarities of lncRNA and disease similarity, respectively; (C) using SKF integration similarity; and (D) obtaining the prediction matrix by Laplacian regularized least squares.
We verify the performance of SKF-LDA by LOOCV and 5-fold CV. The idea of LOOCV is to use one of the 540 lncRNA-disease relationships as a test sample and the rest as training set. 5-fold CV randomly divides 540 lncRNA-disease associations into 5 parts; each time, one part is used as a test set and the remaining as training sets. When one dataset is used as the test set, the prior knowledge is removed before calculating the similarity, and the associations in the dataset are regarded as unknown; in other words, the initial 1 is set to be 0. When the final predicted value is higher than the threshold for those who have the associations, the prediction is correct. According to different thresholds, various false-positive rates (FPRs; 1 − specificity) and true-positive rates (TPRs; sensitivity) are obtained. Based on these data, we can plot the receiver operating characteristic (ROC) curve and get the area under the ROC curve (AUC). The prediction ability is perfect if AUC = 1, the prediction ability tends to be random if AUC = 0.5, and AUC = 0 indicates that the forecast result is negative prediction. In addition, we adopt the area under the precision-recall curve (AUPR) as another measurement. In order to verify the accuracy of SKF-LDA, the precision (PRE), sensitivity (SEN), accuracy (ACC), F1-score (F1-score) and Matthews’s correlation coefficient (MCC) are defined as follows:
| (Equation 1) |
| (Equation 2) |
| (Equation 3) |
| (Equation 4) |
and
| (Equation 5) |
where TP represents true positive, TN represents true negative, FP represents false positive, and FN represents false negative.
Parameter Selection
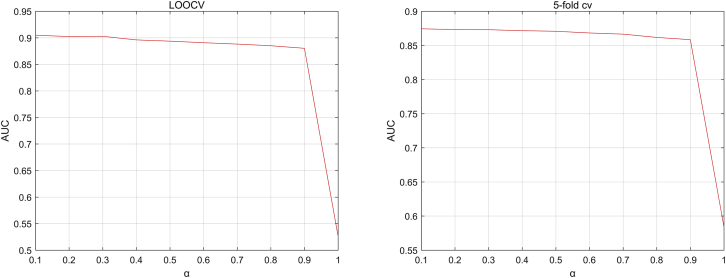
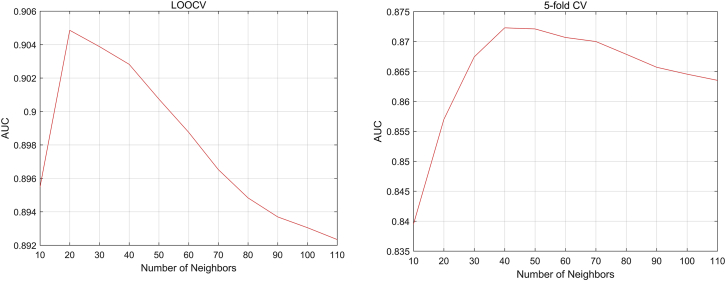
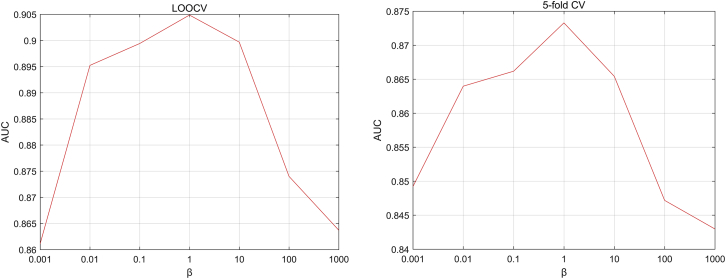
In this paper, there are five parameters: the number of iteration, ; the weight parameter ; the number of neighbors, , in the SKF; and the weight parameters and in Laplacian regularized least squares. In the experiments, the iteration number is set to be , and the value of is set to be 0.1 after parameter tuning. We set the value of as a range from 0.1 to 1 with step 0.1. The AUC values with different s are shown in Figure 2 based on the LOOCV scheme and 5-fold CV scheme. As shown in Figure 2, the AUC values barely change when is in the range of 0.1 to 0.9 and decay fast in the range of 0.9 to 1. Since there are 178 diseases and 115 lncRNAs in our data, the value of ranges from 10 to 110 with step 10. As shown in Figure 3, the highest AUC value is achieved at in the LOOCV scheme and in the 5-fold CV scheme. As for the weighting parameter in Laplacian regularized least squares, previous research has shown that the performance of Laplacian regularization least squares (LapRLS) is robust to the paremeters, so we set to be equal to as , and in the experiments, the value of ranges from to 1,000. As shown in Figure 4, the AUC values in both the LOOCV scheme and 5-fold CV scheme change in a small interval and achieve the highest value when .
Figure 2.
The AUC Values with Different αs
Figure 3.
The AUC Values with Different Values of the Number of Neighbors (K)
Figure 4.
The AUC Values with Different βs
Comparison with Other Fusion Methods
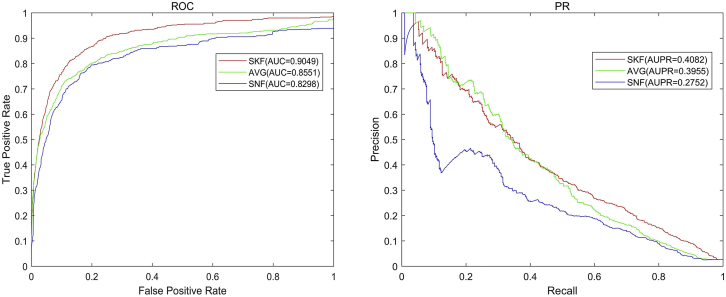
To verify the superiority of the SKF, we compared SKF with two common fusion methods: average kernel fusion (AVG) and similarity network fusion (SNF).44 We plotted the ROC curve and the precision recall (PR) curve of three methods based on LOOCV. As shown in Figure 5, the AUC values of SKF, AVG, and SNF are 0.9049, 0.8511, and 0.8298, respectively. The AUPR values of SKF, AVG, and SNF are 0.4082, 0.3955, and 0.2752, respectively. In summary, SKF performs better than other fusion methods in terms of the prediction association accuracy between lncRNA and disease.
Figure 5.
The ROC Curve and the PR Curve of Three Integration Methods
Comparison with Single Similarity
In this paper, we combined different types of similarity for both lncRNAs and diseases. To demonstrate the benefit of the combination, we performed a series of comparison experiments, including all combinations of one single similarity for the lncRNA and the disease. The AUC values of different combinations in LOOCV and the 5-fold CV scheme are shown in Table 1, from which we can see that the proposed SKF-LDA achieves the highest AUC values.
Table 1.
The AUC Values of SKF-LDA and Other Single Similarity in LOOCV and 5-fold CV Scheme
| lncRNA Similarity | Disease Similarity | LOOCV | 5-fold CV |
|---|---|---|---|
| Expression | semantic | 0.8512 | 0.8476 ± 0.0034 |
| Expression | cosine | 0.8630 | 0.8375 ± 0.0046 |
| Cosine | semantic | 0.8835 | 0.8502 ± 0.0073 |
| Expression | cosine | 0.8754 | 0.8519 ± 0.0070 |
| Expression + cosine | semantic + cosine | 0.9049 | 0.8743 ± 0.0050 |
Comparison with and without Neighbor Constraint
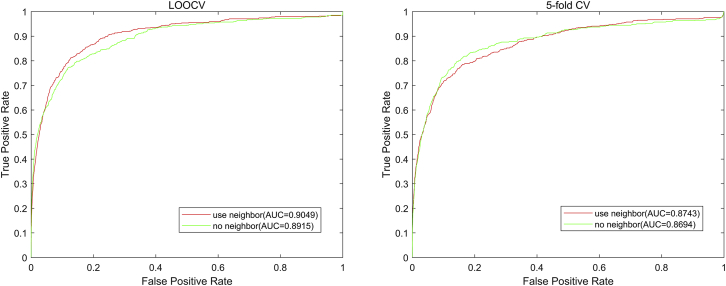
In this paper, we add a neighbor constraint to eliminate the noise effect. To demonstrate the benefit of the neighbor constraint, we tested the case without the neighbor constraint in comparison, as shown in Figure 6, which achieved 0.8915 and 0.86940.0035 in LOOCV and 5-fold CV, respectively; and the one with the neighbor constraint achieved 0.9049 and 0.87430.0050, respectively, which validates the effect of the neighbor constraint.
Figure 6.
The ROC Curve and the PR Curve When Using Neighbor and without Neighbor Constraint
Comparison with Other Methods
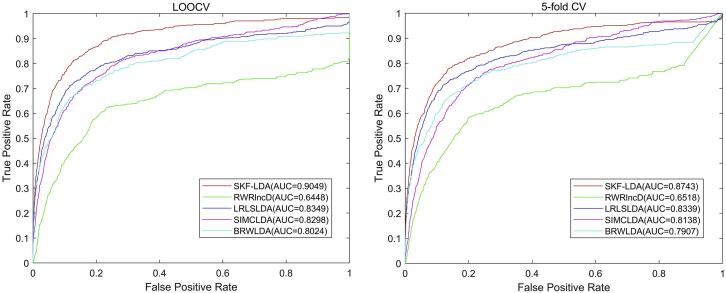
To further validate the advantage of SKF-LDA, we compared our method with four state-of-the-art methods, namely, RWRlncD,31 LRLSLDA,28 SIMCLDA30 and BRWLDA.32 As shown in Figure 7, based on the LOOCV scheme, the AUC values of RWRlncD, LRLSLDA, SIMCLDA, and BRWLDA are 0.6448, 0.8349, 0.8298, and 0.8024, respectively, while the proposed SKF-LDA method achieves 0.9049, which is much better than the others. In the 5-fold CV scheme, the AUC values of RWRlncD, LRLSLDA, SIMCLDA, and BRWLDA are 0.6518, 0.8339, 0.8138, and 0.7907, respectively, while ours is 0.8743. The AUPR measurement is also used to evaluate different methods. The AUPRs of SKF-LDA, RWRlncD, LRLSLDA, SIMCLDA, and BRWLDA are 0.4081, 0.0808, 0.3343, 0.2555, and 0.3068, respectively. Meanwhile, we set two stringency levels to evaluate predictive performance as shown in Table 2. When the stringency level of specificity is set as , the PRE, sensitivity, accuracy, F1-score, and MCC of SKF-LDA are 0.4884, 0.3519, 0.97318, 0.5206, and 0.4013. When , the PRE, sensitivity, accuracy, F1-score, and MCC of SKF-LDA are 0.2407, 0.5852, 0.9404, 0.7383, and 0.3501, which are higher than those of RWRlncD, LRLSLDA, SIMCLDA, and BRWLDA.
Figure 7.
The ROC Curve and AUC Values of Different Methods in LOOCV and 5-fold CV Scheme: SKF-LDA, RWRlncD, LRLSLDA, SIMCLDA, and BRWLDA
Table 2.
Results of Different Methods
| Measurement | SKF-LDA | RWRlncD31 | LRLSLDA28 | SIMCLDA30 | BRWLDA32 |
|---|---|---|---|---|---|
| AUC | 0.9049 | 0.6448 | 0.8349 | 0.8298 | 0.8024 |
| AUPR | 0.4082 | 0.0808 | 0.3343 | 0.2555 | 0.3068 |
| Sp = 99% | |||||
| PRE | 0.4884 | 0.1076 | 0.4472 | 0.3539 | 0.4413 |
| Sensitivity | 0.3519 | 0.0444 | 0.2982 | 0.2019 | 0.2926 |
| Accuracy | 0.9732 | 0.9651 | 0.9718 | 0.9692 | 0.9716 |
| F1-score | 0.5206 | 0.0851 | 0.4593 | 0.3359 | 0.4527 |
| MCC | 0.4013 | 0.0532 | 0.3513 | 0.2526 | 0.3455 |
| Sp = 95% | |||||
| PRE | 0.2407 | 0.1293 | 0.2283 | 0.2037 | 0.2100 |
| Sensitivity | 0.5852 | 0.2741 | 0.5463 | 0.4722 | 0.4907 |
| Accuracy | 0.9404 | 0.9321 | 0.9393 | 0.9374 | 0.9379 |
| F1-score | 0.7383 | 0.4302 | 0.7066 | 0.6415 | 0.6584 |
| MCC | 0.3501 | 0.1563 | 0.3271 | 0.2823 | 0.2937 |
Case Studies
To validate the ability of SKF-LDA to predict lncRNA-disease associations, case studies were conducted for three human diseases: lung cancer, hepatocellular carcinoma, and prostate cancer. The top 10 predicted lncRNAs of each disease are confirmed by two other databases: Lnc2Cancer45 and MNDR.46
Lung cancer is one of the leading cancers that cause death. The death rate for lung cancer is nearly , as its malignancy has the highest numbers among all cancers.47 Therefore, it is necessary to study the biological mechanism and the cause of lung cancer. Here, in our experiments, 9 of the top 10 lncRNA-lung cancer forecast results by SKF-LDA are confirmed by known databases, as shown in Table 3. GAS5 is a novel lung cancer biomarker that is related to the diagnosis and prognosis of lung cancer patients.48, 49 CCAT2 not only promotes the non-small-cell lung cancer production but also is one specific lncRNA of lung adenocarcinoma.50 UCA1 is overexpressed in lung cancer cells, because it induces resistance to T790M in the AKT/mTOR pathway of non-small-cell lung cancer.51
Table 3.
The Top 10 lncRNA Candidates Predicted for Lung Cancer
| Rank | lncRNA | Disease | Evidence |
|---|---|---|---|
| 1 | GAS5 | lung cancer | lnc2Cancer2, MNDR |
| 2 | CCAT2 | lung cancer | MNDR |
| 3 | UCA1 | lung cancer | lnc2Cancer2, MNDR |
| 4 | HULC | lung cancer | unconfirmed |
| 5 | SPRY4-IT1 | lung cancer | MNDR |
| 6 | CCAT1 | lung cancer | MNDR |
| 7 | PVT1 | lung cancer | lnc2Cancer2, MNDR |
| 8 | NEAT1 | lung cancer | lnc2Cancer2, MNDR |
| 9 | XIST | lung cancer | MNDR |
| 10 | HNF1A-AS1 | lung cancer | MNDR |
Hepatocellular carcinoma (HCC) is one of the most often seen types of cancer in the world. Since many HCC patients are already in advanced stages of cancer when they are diagnosed, it is urgent to understand the principle of HCC and improve early diagnosis ability.52 Studies have shown that lncRNAs have a vital effect on human HCC.53 In this study, the top 10 lncRNAs of lncRNA-HCC results based on SKF-LDA are confirmed by known databases and related literature as shown in Table 4. Studies have shown that GAS5 is downregulated in most cell cancer patients and can be regarded as an important prognostic factor for HCC.54, 55 In addition, UCA1 promotes the development of HCC by inhibiting miR-216b and activating the FGFR1/ERK signaling pathway.56 PVT1 is upregulated during the liver development and contributes to HCC by affecting the lncRNA-hPVT1/NOP2 pathway.57
Table 4.
The Top 10 lncRNA Candidates Predicted for Hepatocelluar Carcinoma
| Rank | lncRNA | Disease | Evidence |
|---|---|---|---|
| 1 | GAS5 | hepatocelluar carcinoma | lnc2Cancer2, MNDR |
| 2 | UCA1 | hepatocelluar carcinoma | lnc2Cancer2, MNDR |
| 3 | PVT1 | hepatocelluar carcinoma | lnc2Cancer2, MNDR |
| 4 | CCAT2 | hepatocelluar carcinoma | lnc2Cancer2, MNDR |
| 5 | CDKN2B-AS1 | hepatocelluar carcinoma | MNDR |
| 6 | CCAT1 | hepatocelluar carcinoma | lnc2Cancer2, MNDR |
| 7 | BANCR | hepatocelluar carcinoma | lnc2Cancer2, MNDR |
| 8 | PTENP1 | hepatocelluar carcinoma | lnc2Cancer2, MNDR |
| 9 | SPRY4-IT1 | hepatocelluar carcinoma | lnc2Cancer2, MNDR |
| 10 | NEAT1 | hepatocelluar carcinoma | lnc2Cancer2, MNDR |
Prostate cancer is also a common form of malignancy among males and accounts for the second leading cause of cancer fatality.58 The ability to explain the principles of prostate cancer from a genetic perspective will help us to diagnose and prevent prostate cancer. 4 of the top 5 lncRNAs are successfully found in the databases, while 7 of the top 10 lncRNAs are found in the databases based on SKF-LDA, as shown in Table 5. Different variants of CDKN2B-AS1 are associated with prostate cancer.59 CCAT2 is upregulated in prostate cancer patients and affects the development of prostate cancer by changing the epithelial-mesenchymal transition.50 Among prostate cancer patients, the XIST gene locus is hypomethylated. This phenomenon may contribute to a further realization of the biological mechanism of prostate cancer.60
Table 5.
The Top 10 lncRNA Candidates Predicted for Prostate Cancer
| Rank | lncRNA | Disease | Evidence |
|---|---|---|---|
| 1 | CDKN2B-AS1 | prostate cancer | MNDR |
| 2 | CCAT2 | prostate cancer | lnc2Cancer2, MNDR |
| 3 | XIST | prostate cancer | lnc2Cancer2, MNDR |
| 4 | PTENP1 | prostate cancer | lnc2Cancer2, MNDR |
| 5 | LSINCT5 | prostate cancer | unconfirmed |
| 6 | IGF2-AS | prostate cancer | unconfirmed |
| 7 | SPRY4-IT1 | prostate cancer | lnc2Cancer2, MNDR |
| 8 | MINA | prostate cancer | unconfirmed |
| 9 | CCAT1 | prostate cancer | lnc2Cancer2 |
| 10 | BANCR | prostate cancer | MNDR |
Discussion
Numerous literatures have shown that lncRNA is of great importance in disease. Studying the relationship between lncRNA and disease not only helps us to realize the fundamentals behind disease but also contributes to the prognosis and prevention of disease. Since the current biological experimental methods are time consuming, many lncRNA-disease predictive models have emerged. In this paper, the proposed SKF-LDA method combines both the expression similarity with cosine similarity for lncRNAs and the semantic similarity with cosine similarity for diseases with an effective fusion method. Compared with the other four methods, SKF-LDA performs better in terms of AUC and AUPR in the LOOCV and 5-fold CV schemes.
Other important reference indices show a perfect performance of SKF-LDA as well. To further validate the accuracy of the SKF-LDA, we predicted three diseases (lung cancer, HCC, prostate cancer) based on the forecast result by SKF-LDA. We found that the prediction success rates reached 90%, 100%, and 70%, respectively. The reason for the excellent performance of the SKF-LDA method is mainly due to several reasons as follows. First, SKF-LDA integrates two lncRNA similarities and two disease similarities, which provide us with rich biological information. Second, we integrate different similarities with the neighbor constraint, which will eliminate the noise data in the known dataset. Finally, the final lncRNA-disease correlation prediction matrix is obtained by solving an optimization model based on the Laplacian operator normalization, which has shown its successful application in many other related problems.
Still, the proposed method has some shortcomings. The original lncRNA-disease association matrix is a sparse matrix. There were only 540 associations for 115 lncRNAs and 178 diseases; that is to say, there are only three associations per disease, which is not enough and unstable for the forecast result. Meanwhile, there are only two similarities in the current integration, and more biological knowledge can be applied in the future.
Materials and Methods
Human lncRNA-Disease Association Dataset
The lncRNADisease database61 is used as the known lncRNA-disease association dataset, which contains 687 confirmed lncRNA-disease associations between 369 lncRNAs and 247 diseases. After eliminating lncRNAs without an expression profile62 and diseases without disease ontology,63 540 known lncRNA-disease associations including 115 lncRNAs and 178 diseases were obtained. From the aforementioned known associations, we can get the lncRNA-disease adjacency matrix , where and are the number of lncRNAs and diseases, respectively, and each row of matrix represents one lncRNA, while each column denotes one disease. 0 in indicates that the relationship between lncRNA and disease is still unknown, and 1 in indicates that lncRNA has some relationship to disease . The definition of matrix is as follows:
| (Equation 6) |
Similarity Kernels for lncRNAs and Diseases
The proposed method is based on the currently accepted hypothesis that lncRNAs with similar functionality tend to be associated with diseases with semantic or phenotypic similarities, and vice versa. Therefore, it is very important to get the similarity kernels for both the lncRNAs and the diseases, which can provide lncRNA-disease associations with more accuracy. In this paper, first, we will compute the expression similarity and cosine similarity for the lncRNAs. Second, we will get the semantic similarity and cosine similarity for the diseases. Then, a kernel fusion method is applied to all similarity kernels. At last, based on the integrated lncRNA similarity kernel matrix and the disease similarity matrix, the Laplacian regularized least-squares method is used to get the final lncRNA-disease associations.
lncRNA Expression Similarity
The expression profiles of the lncRNAs are downloaded from ArrayExpression: E-MEXP-3783,64 in which more than 1.5 million expression profiles are collected by high-throughput sequencing. The Spearman correlation is used to calculate the expression similarity between different lncRNAs.28, 65 The matrix denotes the similarity of lncRNA expression, where element represents the similarity degree between lncRNA and lncRNA ; values range from 0 to 1.
Disease Semantic Similarity
The disease semantic is very important information in characterizing a disease. The directed acyclic graph (DAG) has been studied to calculate the semantic similarity of diseases and shows great performance.66, 67 In this paper, the semantic similarity is also used as one dimension of disease similarity. The raw data of semantic similarity are downloaded from the U.S. National Library of Medicine. Based on medical subject heading (MeSH) description information, a DAG, , can be constructed, where denotes that the ancestor node of disease including itself, is the corresponding connection of .68 The disease semantic similarity between disease and its ancestor disease is calculated as follows:
| (Equation 7) |
where the disease , is the weight parameter of the semantic similarity of diseases, and by default.
Also, we can define the semantic value for each disease as follows:
| (Equation 8) |
With the similarity and semantic value defined, the semantic similarity matrix can be calculated. The similarity between arbitrary disease and disease is computed as follows:
| (Equation 9) |
Cosine Similarity for lncRNAs and Diseases
The expression profile similarity for lncRNAs and the semantic similarity for diseases are two commonly used similarity kernels.30, 68, 69 To better improve the similarity kernels, one more dimensional similarity is used in the proposed method. Previous studies have showed that cosine similarity is successfully applied to collaborative filtering recommendation algorithms,70, 71 which inspired us to combine such similarity into the lncRNA-disease association prediction.
The principle of lncRNA cosine similarity is based on the assumption that if lncRNA and lncRNA are similar to each other, then, in the lncRNA-disease association matrix, pattern and pattern should be similar to each other. The same assumption should also be true for diseases, and the cosine similarity between lncRNA and lncRNA is calculated as follows:
| (Equation 10) |
where represents the ith row of the lncRNA-disease association matrix and contains the relationship of all the diseases to lncRNAs .
Similarly, the cosine similarity between disease and disease can be calculated as follows:
| (Equation 11) |
where is the cosine similarity matrix for diseases, and represents the ith column of the lncRNA-disease association matrix .
SKF for lncRNAs and Diseases
Now we have two lncRNA similarity kernels (lncRNA expression similarity and lncRNA cosine similarity) and two disease similarity kernels (disease semantic similarity and disease cosine similarity). Next, we use SKF to integrate the two lncRNA similarity kernels . In the first step, we normalize each lncRNA similarity kernels as follows:
| (Equation 12) |
where represents the set of lncRNAs, and represents the normalized kernel and satisfies .
In the second step, we create a neighbor-constraint kernel for two lncRNA similarity kernels as follows:
| (Equation 13) |
where denotes a neighbor-constraint kernel and satisfies . Here, the neighbor of lncRNA is defined by the most similar lncRNAs to .
In the third step, we mix up the normalized similarity kernel and the neighbor-constraint kernel literally, as follows:
| (Equation 14) |
where denotes the nth kernel obtained after th iterations, denotes the initial value of , and denotes the weight parameter. After th iterations, the final kernel is obtained as follows:
| (Equation 15) |
In the fourth step, one more weighted matrix is added to the embedding of more neighbor information. The weighted matrix is as follows:
| (Equation 16) |
Finally, we get the integrated lncRNA similarity kernel matrix as follows:
| (Equation 17) |
Similarly, we can get the integrated disease similarity kernel matrix .
Laplacian Regularized Least Squares for lncRNA-Disease Association
With the lncRNA similarity matrix and the disease similarity matrix obtained by the SKF method, LapRLS is used to predict the potential lncRNA-disease association. From the view of lncRNAs, we can build the minimization model as follows:
| (Equation 18) |
where is the Frobenius norm; is the initial known lncRNA-disease association matrix; is the weighting parameter of LapRLS; is the correlation matrix in the lncRNA space; and is the normalized similarity matrix, where is the diagonal matrix obtained by summing the elements of each row of the lncRNA similarity matrix . The first objective function in Equation 18 is to make sure that the obtained new correlation matrix is similar to the known one. The second objective function is to make sure that the obtained correlation matrix is smooth over the lncRNA space. We can solve Equation 18 by calculating the derivative of the objective function as follows:44
| (Equation 19) |
Similarly, we can obtain the optimal correlation matrix in the disease space as follows:
| (Equation 20) |
Finally, we integrate the prediction matrix from the lncRNA and disease space and obtain the final prediction association matrix as follows:
| (Equation 21) |
Author Contributions
Conceptualization, G.X. and T.M.; Formal Analysis, T.M.; Investigation and Methodology, T.M. and Y.L.; Resources, T.M.; Project Administration, G.X. and Y.L.; Supervision, G.X. and Z.L.; Visualization, T.M.; Writing – Original Draft, T.M.; Writing – Review & Editing, G.X., T.M., Y.L., and Z.L.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (618002072 and 61702112); the Natural Science Foundation of Guangdong Province (2018A030313389); the Science and Technology Plan Project of Guangdong Province (2017A040405050, 2016B030306004, and 2016B030301008); the Science and Technology Project of Guangzhou City (201902020012, 201902010034, and 201907010021); and the Science and Technology Project of Guangdong Province (2018B030323026).
Contributor Information
Yu Luo, Email: yuluo@gdut.edu.cn.
Zhenguo Liu, Email: liuzg1340@126.com.
References
- 1.Claverie J.M. Fewer genes, more noncoding RNA. Science. 2005;309:1529–1530. doi: 10.1126/science.1116800. [DOI] [PubMed] [Google Scholar]
- 2.Birney E., Stamatoyannopoulos J.A., Dutta A., Guigó R., Gingeras T.R., Margulies E.H., Weng Z., Snyder M., Dermitzakis E.T., Thurman R.E., ENCODE Project Consortium. NISC Comparative Sequencing Program. Baylor College of Medicine Human Genome Sequencing Center. Washington University Genome Sequencing Center. Broad Institute. Children’s Hospital Oakland Research Institute Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., International Human Genome Sequencing Consortium Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 4.Kapranov P., Cheng J., Dike S., Nix D.A., Duttagupta R., Willingham A.T., Stadler P.F., Hertel J., Hackermüller J., Hofacker I.L. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 5.Esteller M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 6.Zhao Q., Zhang Y., Hu H., Ren G., Zhang W., Liu H. IRWNRLPI: integrating random walk and neighborhood regularized logistic matrix factorization for lncRNA-protein interaction prediction. Front. Genet. 2018;9:239. doi: 10.3389/fgene.2018.00239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu H., Zhu C., Ai H., Zhang L., Zhao J., Zhao Q., Liu H. LPI-ETSLP: lncRNA-protein interaction prediction using eigenvalue transformation-based semi-supervised link prediction. Mol. Biosyst. 2017;13:1781–1787. doi: 10.1039/c7mb00290d. [DOI] [PubMed] [Google Scholar]
- 8.Hu H., Zhang L., Ai H., Zhang H., Fan Y., Zhao Q., Liu H. HLPI-Ensemble: prediction of human lncRNA-protein interactions based on ensemble strategy. RNA Biol. 2018;15:797–806. doi: 10.1080/15476286.2018.1457935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao Q., Liang D., Hu H., Ren G., Liu H. RWLPAP: random walk for lncRNA-protein associations prediction. Protein Pept. Lett. 2018;25:830–837. doi: 10.2174/0929866525666180905104904. [DOI] [PubMed] [Google Scholar]
- 10.Zhao Q., Yu H., Ming Z., Hu H., Ren G., Liu H. The bipartite network projection-recommended algorithm for predicting long non-coding RNA-protein interactions. Mol. Ther. Nucleic Acids. 2018;13:464–471. doi: 10.1016/j.omtn.2018.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma L., Li A., Zou D., Xu X., Xia L., Yu J., Bajic V.B., Zhang Z. LncRNAWiki: harnessing community knowledge in collaborative curation of human long non-coding RNAs. Nucleic Acids Res. 2015;43(Database issue, D1):D187–D192. doi: 10.1093/nar/gku1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gibb E.A., Brown C.J., Lam W.L. The functional role of long non-coding RNA in human carcinomas. Mol. Cancer. 2011;10:38. doi: 10.1186/1476-4598-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clark M.B., Mattick J.S. Long noncoding RNAs in cell biology. Semin. Cell Dev. Biol. 2011;22:366–376. doi: 10.1016/j.semcdb.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 14.Huppi K., Pitt J.J., Wahlberg B.M., Caplen N.J. The 8q24 gene desert: an oasis of non-coding transcriptional activity. Front. Genet. 2012;3:69. doi: 10.3389/fgene.2012.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li L., Liu B., Wapinski O.L., Tsai M.C., Qu K., Zhang J., Carlson J.C., Lin M., Fang F., Gupta R.A. Targeted disruption of Hotair leads to homeotic transformation and gene derepression. Cell Rep. 2013;5:3–12. doi: 10.1016/j.celrep.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wapinski O., Chang H.Y. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 17.Tseng Y.Y., Moriarity B.S., Gong W., Akiyama R., Tiwari A., Kawakami H., Ronning P., Reuland B., Guenther K., Beadnell T.C. PVT1 dependence in cancer with MYC copy-number increase. Nature. 2014;512:82–86. doi: 10.1038/nature13311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi S.J., Wang L.J., Yu B., Li Y.H., Jin Y., Bai X.Z. LncRNA-ATB promotes trastuzumab resistance and invasion-metastasis cascade in breast cancer. Oncotarget. 2015;6:11652–11663. doi: 10.18632/oncotarget.3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pasmant E., Sabbagh A., Vidaud M., Bièche I. ANRIL, a long, noncoding RNA, is an unexpected major hotspot in GWAS. FASEB J. 2011;25:444–448. doi: 10.1096/fj.10-172452. [DOI] [PubMed] [Google Scholar]
- 20.Li H., Wang W., Zhang L., Lan Q., Wang J., Cao Y., Zhao J. Identification of a long noncoding RNA-associated competing endogenous RNA network in intracranial aneurysm. World Neurosurg. 2017;97:684–692.e4. doi: 10.1016/j.wneu.2016.10.016. [DOI] [PubMed] [Google Scholar]
- 21.Halvorsen M., Martin J.S., Broadaway S., Laederach A. Disease-associated mutations that alter the RNA structural ensemble. PLoS Genet. 2010;6:e1001074. doi: 10.1371/journal.pgen.1001074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bhan A., Hussain I., Ansari K.I., Kasiri S., Bashyal A., Mandal S.S. Antisense transcript long noncoding RNA (lncRNA) HOTAIR is transcriptionally induced by estradiol. J. Mol. Biol. 2013;425:3707–3722. doi: 10.1016/j.jmb.2013.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yan C., Xie H., Liu S., Yin J., Zhang Y., Dai Q. Effective Uyghur language text detection in complex background images for traffic prompt identification. IEEE Trans. Intell. Transp. Syst. 2017;19:220–229. [Google Scholar]
- 24.Chen X. Predicting lncRNA-disease associations and constructing lncRNA functional similarity network based on the information of miRNA. Sci. Rep. 2015;5:13186. doi: 10.1038/srep13186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen X., Yan C.C., Zhang X., You Z.H. Long non-coding RNAs and complex diseases: from experimental results to computational models. Brief. Bioinform. 2017;18:558–576. doi: 10.1093/bib/bbw060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang J., Zhang Z., Chen Z., Deng L. Integrating multiple heterogeneous networks for novel lncRNA-disease association inference. IEEE/ACM Trans. Comput. Biol. Bioinform. 2019;16:396–406. doi: 10.1109/TCBB.2017.2701379. [DOI] [PubMed] [Google Scholar]
- 27.Chen X., Sun Y.Z., Guan N.N., Qu J., Huang Z.A., Zhu Z.X., Li J.Q. Computational models for lncRNA function prediction and functional similarity calculation. Brief. Funct. Genomics. 2019;18:58–82. doi: 10.1093/bfgp/ely031. [DOI] [PubMed] [Google Scholar]
- 28.Chen X., Yan G.Y. Novel human lncRNA-disease association inference based on lncRNA expression profiles. Bioinformatics. 2013;29:2617–2624. doi: 10.1093/bioinformatics/btt426. [DOI] [PubMed] [Google Scholar]
- 29.Lan W., Li M., Zhao K., Liu J., Wu F.X., Pan Y., Wang J. LDAP: a web server for lncRNA-disease association prediction. Bioinformatics. 2017;33:458–460. doi: 10.1093/bioinformatics/btw639. [DOI] [PubMed] [Google Scholar]
- 30.Lu C., Yang M., Luo F., Wu F.X., Li M., Pan Y., Li Y., Wang J. Prediction of lncRNA-disease associations based on inductive matrix completion. Bioinformatics. 2018;34:3357–3364. doi: 10.1093/bioinformatics/bty327. [DOI] [PubMed] [Google Scholar]
- 31.Sun J., Shi H., Wang Z., Zhang C., Liu L., Wang L., He W., Hao D., Liu S., Zhou M. Inferring novel lncRNA-disease associations based on a random walk model of a lncRNA functional similarity network. Mol. Biosyst. 2014;10:2074–2081. doi: 10.1039/c3mb70608g. [DOI] [PubMed] [Google Scholar]
- 32.Yu G., Fu G., Lu C., Ren Y., Wang J. BRWLDA: bi-random walks for predicting lncRNA-disease associations. Oncotarget. 2017;8:60429–60446. doi: 10.18632/oncotarget.19588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiao X., Zhu W., Liao B., Xu J., Gu C., Ji B., Yao Y., Peng L., Yang J. BPLLDA: predicting lncRNA-disease associations based on simple paths with limited lengths on a heterogeneous network. Front. Genet. 2018;9:411. doi: 10.3389/fgene.2018.00411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ding L., Wang M., Sun D., Li A. TPGLDA: Novel prediction of associations between lncRNAs and diseases via lncRNA-disease-gene tripartite graph. Sci. Rep. 2018;8:1065. doi: 10.1038/s41598-018-19357-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen X., Yan G.Y. Semi-supervised learning for potential human microRNA-disease associations inference. Sci. Rep. 2014;4:5501. doi: 10.1038/srep05501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen X., Xie D., Zhao Q., You Z.H. MicroRNAs and complex diseases: from experimental results to computational models. Brief. Bioinform. 2019;20:515–539. doi: 10.1093/bib/bbx130. [DOI] [PubMed] [Google Scholar]
- 37.You Z.H., Huang Z.A., Zhu Z., Yan G.Y., Li Z.W., Wen Z., Chen X. PBMDA: A novel and effective path-based computational model for miRNA-disease association prediction. PLoS Comput. Biol. 2017;13:e1005455. doi: 10.1371/journal.pcbi.1005455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen X., Wang C.C., Yin J., You Z.H. Novel human miRNA-disease association inference based on random forest. Mol. Ther. Nucleic Acids. 2018;13:568–579. doi: 10.1016/j.omtn.2018.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen X., Wang L., Qu J., Guan N.N., Li J.Q. Predicting miRNA-disease association based on inductive matrix completion. Bioinformatics. 2018;34:4256–4265. doi: 10.1093/bioinformatics/bty503. [DOI] [PubMed] [Google Scholar]
- 40.Chen X., Yin J., Qu J., Huang L. MDHGI: matrix decomposition and heterogeneous graph inference for miRNA-disease association prediction. PLoS Comput. Biol. 2018;14:e1006418. doi: 10.1371/journal.pcbi.1006418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen X., Xie D., Wang L., Zhao Q., You Z.H., Liu H. BNPMDA: bipartite network projection for MiRNA–disease association prediction. Bioinformatics. 2018;34:3178–3186. doi: 10.1093/bioinformatics/bty333. [DOI] [PubMed] [Google Scholar]
- 42.Jiang L., Ding Y., Tang J., Guo F. MDA-SKF: similarity kernel fusion for accurately discovering miRNA-disease association. Front. Genet. 2018;9:618. doi: 10.3389/fgene.2018.00618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen X., Huang L. LRSSLMDA: Laplacian regularized sparse subspace learning for MiRNA-disease association prediction. PLoS Comput. Biol. 2017;13:e1005912. doi: 10.1371/journal.pcbi.1005912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xia Z., Wu L.Y., Zhou X., Wong S.T. Semi-supervised drug-protein interaction prediction from heterogeneous biological spaces. BMC Syst. Biol. 2010;4(Suppl 2):S6. doi: 10.1186/1752-0509-4-S2-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ning S., Zhang J., Wang P., Zhi H., Wang J., Liu Y., Gao Y., Guo M., Yue M., Wang L., Li X. Lnc2Cancer: a manually curated database of experimentally supported lncRNAs associated with various human cancers. Nucleic Acids Res. 2016;44(D1):D980–D985. doi: 10.1093/nar/gkv1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cui T., Zhang L., Huang Y., Yi Y., Tan P., Zhao Y., Hu Y., Xu L., Li E., Wang D. MNDR v2.0: an updated resource of ncRNA-disease associations in mammals. Nucleic Acids Res. 2018;46(D1):D371–D374. doi: 10.1093/nar/gkx1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parkin D.M., Bray F., Ferlay J., Pisani P. Global cancer statistics, 2002. CA Cancer J. Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 48.Liang W., Lv T., Shi X., Liu H., Zhu Q., Zeng J., Yang W., Yin J., Song Y. Circulating long noncoding RNA GAS5 is a novel biomarker for the diagnosis of nonsmall cell lung cancer. Medicine (Baltimore) 2016;95:e4608. doi: 10.1097/MD.0000000000004608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ma C., Shi X., Zhu Q., Li Q., Liu Y., Yao Y., Song Y. The growth arrest-specific transcript 5 (GAS5): a pivotal tumor suppressor long noncoding RNA in human cancers. Tumour Biol. 2016;37:1437–1444. doi: 10.1007/s13277-015-4521-9. [DOI] [PubMed] [Google Scholar]
- 50.Zheng J., Zhao S., He X., Zheng Z., Bai W., Duan Y., Cheng S., Wang J., Liu X., Zhang G. The up-regulation of long non-coding RNA CCAT2 indicates a poor prognosis for prostate cancer and promotes metastasis by affecting epithelial-mesenchymal transition. Biochem. Biophys. Res. Commun. 2016;480:508–514. doi: 10.1016/j.bbrc.2016.08.120. [DOI] [PubMed] [Google Scholar]
- 51.Cheng N., Cai W., Ren S., Li X., Wang Q., Pan H., Zhao M., Li J., Zhang Y., Zhao C. Long non-coding RNA UCA1 induces non-T790M acquired resistance to EGFR-TKIs by activating the AKT/mTOR pathway in EGFR-mutant non-small cell lung cancer. Oncotarget. 2015;6:23582–23593. doi: 10.18632/oncotarget.4361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Llovet J.M. Updated treatment approach to hepatocellular carcinoma. J. Gastroenterol. 2005;40:225–235. doi: 10.1007/s00535-005-1566-3. [DOI] [PubMed] [Google Scholar]
- 53.Li C., Chen J., Zhang K., Feng B., Wang R., Chen L. Progress and prospects of long noncoding RNAs (lncRNAs) in hepatocellular carcinoma. Cell. Physiol. Biochem. 2015;36:423–434. doi: 10.1159/000430109. [DOI] [PubMed] [Google Scholar]
- 54.Tu Z.Q., Li R.J., Mei J.Z., Li X.H. Down-regulation of long non-coding RNA GAS5 is associated with the prognosis of hepatocellular carcinoma. Int. J. Clin. Exp. Pathol. 2014;7:4303–4309. [PMC free article] [PubMed] [Google Scholar]
- 55.Hu L., Ye H., Huang G., Luo F., Liu Y., Liu Y., Yang X., Shen J., Liu Q., Zhang J. Long noncoding RNA GAS5 suppresses the migration and invasion of hepatocellular carcinoma cells via miR-21. Tumour Biol. 2016;37:2691–2702. doi: 10.1007/s13277-015-4111-x. [DOI] [PubMed] [Google Scholar]
- 56.Wang F., Ying H.Q., He B.S., Pan Y.Q., Deng Q.W., Sun H.L., Chen J., Liu X., Wang S.K. Upregulated lncRNA-UCA1 contributes to progression of hepatocellular carcinoma through inhibition of miR-216b and activation of FGFR1/ERK signaling pathway. Oncotarget. 2015;6:7899–7917. doi: 10.18632/oncotarget.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang F., Yuan J.H., Wang S.B., Yang F., Yuan S.X., Ye C., Yang N., Zhou W.P., Li W.L., Li W., Sun S.H. Oncofetal long noncoding RNA PVT1 promotes proliferation and stem cell-like property of hepatocellular carcinoma cells by stabilizing NOP2. Hepatology. 2014;60:1278–1290. doi: 10.1002/hep.27239. [DOI] [PubMed] [Google Scholar]
- 58.Dall’Era M.A., Cooperberg M.R., Chan J.M., Davies B.J., Albertsen P.C., Klotz L.H., Warlick C.A., Holmberg L., Bailey D.E., Jr., Wallace M.E. Active surveillance for early-stage prostate cancer: review of the current literature. Cancer. 2008;112:1650–1659. doi: 10.1002/cncr.23373. [DOI] [PubMed] [Google Scholar]
- 59.Fehringer G., Kraft P., Pharoah P.D., Eeles R.A., Chatterjee N., Schumacher F.R., Schildkraut J.M., Lindström S., Brennan P., Bickeböller H., Ovarian Cancer Association Consortium (OCAC) PRACTICAL Consortium. Hereditary Breast and Ovarian Cancer Research Group Netherlands (HEBON) Colorectal Transdisciplinary (CORECT) Study. African American Breast Cancer Consortium (AABC); and African Ancestry Prostate Cancer Consortium (AAPC) Cross-cancer genome-wide analysis of lung, ovary, breast, prostate, and colorectal cancer reveals novel pleiotropic associations. Cancer Res. 2016;76:5103–5114. doi: 10.1158/0008-5472.CAN-15-2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Laner T., Schulz W.A., Engers R., Müller M., Florl A.R. Hypomethylation of the XIST gene promoter in prostate cancer. Oncol. Res. 2005;15:257–264. doi: 10.3727/096504005776404607. [DOI] [PubMed] [Google Scholar]
- 61.Chen G., Wang Z., Wang D., Qiu C., Liu M., Chen X., Zhang Q., Yan G., Cui Q. LncRNADisease: a database for long-non-coding RNA-associated diseases. Nucleic Acids Res. 2013;41(Database issue, D1):D983–D986. doi: 10.1093/nar/gks1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang X., Sun S., Pu J.K.S., Tsang A.C., Lee D., Man V.O., Lui W.M., Wong S.T., Leung G.K. Long non-coding RNA expression profiles predict clinical phenotypes in glioma. Neurobiol. Dis. 2012;48:1–8. doi: 10.1016/j.nbd.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 63.Schriml L.M., Arze C., Nadendla S., Chang Y.W., Mazaitis M., Felix V., Feng G., Kibbe W.A. Disease ontology: a backbone for disease semantic integration. Nucleic Acids Res. 2012;40(D1):D940–D946. doi: 10.1093/nar/gkr972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Parkinson H., Kapushesky M., Shojatalab M., Abeygunawardena N., Coulson R., Farne A., Holloway E., Kolesnykov N., Lilja P., Lukk M. ArrayExpress—a public database of microarray experiments and gene expression profiles. Nucleic Acids Res. 2007;35(Suppl. 1):D747–D750. doi: 10.1093/nar/gkl995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang Y., Li H., Hou S., Hu B., Liu J., Wang J. The noncoding RNA expression profile and the effect of lncRNA AK126698 on cisplatin resistance in non-small-cell lung cancer cell. PLoS ONE. 2013;8:e65309. doi: 10.1371/journal.pone.0065309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen X. KATZLDA: KATZ measure for the lncRNA-disease association prediction. Sci. Rep. 2015;5:16840. doi: 10.1038/srep16840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen X., Yang J.R., Guan N.N., Li J.Q. GRMDA: graph regression for miRNA-disease association prediction. Front. Physiol. 2018;9:92. doi: 10.3389/fphys.2018.00092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang D., Wang J., Lu M., Song F., Cui Q. Inferring the human microRNA functional similarity and functional network based on microRNA-associated diseases. Bioinformatics. 2010;26:1644–1650. doi: 10.1093/bioinformatics/btq241. [DOI] [PubMed] [Google Scholar]
- 69.Li G., Luo J., Xiao Q., Liang C., Ding P. Prediction of microRNA–disease associations with a Kronecker kernel matrix dimension reduction model. RSC Advances. 2018;8:4377–4385. [Google Scholar]
- 70.Adomavicius G., Tuzhilin A. Toward the next generation of recommender systems: a survey of the state-of-the-art and possible extensions. IEEE Trans. Knowl. Data Eng. 2005;17(6):734–749. [Google Scholar]
- 71.Debnath S., Ganguly N., Mitra P. Feature weighting in content based recommendation system using social network analysis. Proceedings of the 17th International Conference on World Wide Web. ACM. 2008:1041–1042. [Google Scholar]