Abstract
A previously developed and centrally validated MammaPrint® (MP) and BluePrint® (BP) targeted RNA next-generation sequencing (NGS) kit was implemented and validated in two large academic European hospitals. Additionally, breast cancer molecular subtypes by MP and BP RNA sequencing were compared with immunohistochemistry (IHC). Patients with early breast cancer diagnosed at University Hospitals Leuven and Curie Institute Paris were prospectively included between September 2017 and January 2018. Formalin-fixed paraffin-embedded tissue sections were analyzed with MP and BP NGS technology at the beta sites and with both NGS and microarray technology at Agendia. Raw NGS data generated on Illumina MiSeq instruments at the beta sites were interpreted and compared with NGS and microarray data at Agendia. MP and BP NGS molecular subtypes were compared to surrogate IHC breast cancer subtypes. Equivalence of MP and BP indices was determined by Pearson's correlation coefficient. Acceptable limits were defined a priori, based on microarray data generated at Agendia between 2012 and 2016. The concordance, the Negative Percent Agreement and the Positive Percent Agreement were calculated based on the contingency tables and had to be equal to or higher than 90%. Out of 124 included samples, 48% were MP Low and 52% High Risk with microarray. Molecular subtypes were BP luminal, HER2 or basal in 82%, 8% and 10% respectively. Concordance between MP microarray at Agendia and MP NGS at the beta sites was 91.1%. Concordance of MP High and Low Risk classification between NGS at the beta sites and NGS at Agendia was 93.9%. Concordance of MP and BP molecular subtyping using NGS at the beta sites and microarray at Agendia was 89.5%. Concordance between MP and BP NGS subtyping, and IHC was 71.8% and 76.6%, for two IHC surrogate models. The MP/BP NGS kit was successfully validated in a decentralized setting.
Introduction
Currently, MammaPrint® (MP) and BluePrint® (BP) tests are performed centrally at Agendia in two laboratories, one in Europe and one in the United States of America (USA). MP and BP tests are cDNA microarray-based, with MP providing a 70-gene prognostic signature for distant recurrence (Low Risk versus High Risk) and BP a 80-gene signature for molecular subtyping of breast cancer (Luminal-, HER2-, and Basal-type) [1], [2]. Both assays can be performed on total RNA extracted from formalin-fixed paraffin-embedded (FFPE) breast cancer tissue [1], [3]. The clinical utility of MP has been validated in a large prospective and randomized international phase III controlled clinical trial; the Microarray in Node-negative and 1–3 positive lymph nodes Disease may Avoid Chemo Therapy trial (MINDACT) [4]. The test is mainly used to guide the decision to administer or avoid adjuvant chemotherapy in patients with early breast cancer carrying estrogen receptor positive (ERpos) and human epidermal growth factor receptor 2 non-amplified (HER2neg) tumors. Low Risk corresponds with 10% or less risk of distant recurrences in 10 years without any adjuvant systemic treatment and patients with Low Risk classified tumor could therefore potentially omit chemotherapy from their treatment. Patients with High Risk classification have a risk of distant recurrences in 10 years without any adjuvant systemic treatment of 29% and should be treated with chemotherapy to reduce their risk of developing recurrences [1], [4], [5], [6], [7], [8], [9], [10]. Data from MINDACT are only validated for five to 6 years of follow-up [4].
Several factors have impeded the broad use of MP and other similar tests in many countries including: the high cost, the risk of overconsumption in absence of a defined target population and the lack of a decentralized test that can be integrated in existing workflows in molecular diagnostic laboratories. Other commercially available multi-gene signatures that can be used in a decentralized setting lack validation of clinical utility and sometimes require investments in laboratory equipment to implement the test locally [4], [11], [12], [13].
Recently, molecular diagnostics by means of next-generation sequencing (NGS) technology has entered the clinic and many laboratories have now installed NGS platforms to answer diagnostic questions. Therefore, NGS technology might be the preferred testing platform to exploit gene expression multi-gene signatures such as MP and BP in a decentralized setting, thereby increasing accessibility to the test.
A targeted RNA — from FFPE tissue — based NGS kit for the implementation of MP and BP in a decentralized setting was recently developed by Agendia, in collaboration with Agilent Technologies (Santa Clara, California). The NGS kit has been validated by demonstrating equivalence between results generated with the standard microarray-based test and with the MP/BP NGS kit in the two central laboratories of Agendia located in Amsterdam, The Netherlands and Irvine, CA, USA. The validation of the NGS kit also included reproducibility between the two laboratories, stability over time and the clinical validity [14].
The MP/BP NGS kit development also included a preliminary validation in a decentralized setting where a set of 15 FFPE RNA samples previously isolated at Agendia were processed with the Agendia NGS kit in three independent laboratories: Agendia Amsterdam, University Hospitals Leuven (UHL) and Curie Institute Paris (CIP). A preliminary validation study aiming at assessing the competence of both beta sites already showed a high reproducibility and concordance between results obtained at Agendia (Amsterdam), UHL and CIP on RNA extracted from a set of 15 patients [14]. However, a larger prospective validation study aiming at the full introduction of the test in the daily routine of a diagnostic laboratory was needed.
Therefore, we prospectively validated the MP/BP NGS kit for FFPE tissue samples in two large tertiary academic hospitals in Europe using routine diagnostic tissue samples obtained from the two test sites. Additionally, we aimed at comparing breast cancer subtypes by molecular MP and BP targeted RNA sequencing with immunohistochemistry (IHC).
Materials and Methods
In this study, women between 30 and 91 years old with primary operable breast cancer diagnosed at the Multidisciplinary Breast Center in UHL and at CIP were prospectively and consecutively included between September 2017 and January 2018. Patients with bilateral breast cancer and with more than three positive lymph nodes were excluded. There were no restrictions on estrogen receptor (ER), progesterone receptor (PR) and human epidermal growth factor receptor 2 (HER2) status.
This study was approved by the Ethics Committee of UHL and CIP, and all patients gave written informed consent. All patient sample data were anonymized in accordance with national ethical guidelines; study samples had Institutional Review Board approvals.
The laboratory workflow in a decentralized and centralized setting can be found in Supplementary Figure 1. At the beta testing site, tumor block selection was done by a board certified pathologist (AVS and GF). A minimum of 30% viable tumor cells was required for RNA extraction according to standard Agendia procedures and the MINDACT study [4], [9]; tumor enrichment by scraping of the marked tumor area was only necessary in case of tumor cell percentage less than 30%. Tumor cell percentage was assessed at Agendia and assessed at the beta sites by board certified pathologists (AVS, GF, JW and CF). For each selected formalin-fixed, paraffin-embedded (FFPE) block 20 serial tissue sections of 5 μm were cut; 10 tissue slides were kept at the beta testing site, deparaffinization was performed with Histo-Clear in UHL and xylene in CIP. RNA extraction was performed with the AllPrep kit (Qiagen) or RNeasy FFPE kit (Qiagen) at UHL and with the RNeasy FFPE kit (Qiagen) at CIP. The other 10 tissue slides were sent to the Agendia laboratory for deparaffinization with xylene and RNA extraction with the RNeasy FFPE kit (Qiagen). At UHL, the even numbered FFPE slides were kept internally and the uneven slides were sent to Agendia laboratories to minimize intra-tumor heterogeneity. At CIP, the alternation of the FFPE slides was done only from the second batch onwards. The laboratory procedure was performed by two different operators at UHL, a single operator at CIP and two different operators at Agendia.
After RNA extraction, library preparation and sequencing set-up was performed following the standard procedure of the MP/BP NGS kit [14]. A minimum of 100 ng of RNA was needed to proceed with capturing of the targeted genes using a specifically developed bait library, pooling of captured libraries and performing the sequencing. Per MiSeq run, 15 to 16 samples were pooled at UHL, 12 to 21 samples were pooled at CIP, and 7 to 18 samples were pooled at Agendia. Targeted RNA sequencing of the 70 MP and 80 BP signature genes RNA baits with a size of 120 base pairs (bp) was performed on Illumina MiSeq instruments using the V3 150 cycles kit and a 150 bp single-end protocol. The size of the capture library was 0.9 Mega base (Mb). On average 1.5 million reads were obtained per sample, which corresponds to approximately 225 Mb per sample. During the process, quality assessments were performed on RNA and on the prepared sequencing library. The raw NGS data (FASTQ files) generated at the beta test sites were sent through a secure file transfer protocol server to Agendia for analysis and interpretation following the NGS Agendia standard pipeline [14]. Each sample was processed at the beta site and at the Agendia laboratories with the same NGS kit and procedure, and results were compared to the gold standard microarray MP and BP results. Additionally, all beta site samples were analyzed at Agendia with the MP and BP microarray diagnostic test as part of the standard diagnostic workflow [1]. Briefly, after isolation and DNase treatment of the RNA, the sample was amplified and labeled. The labeled material was then hybridized on a diagnostic customized microarray. After data acquisition and normalization, standardized custom developed software was used to obtain the MP and BP test results. The MP microarray and NGS test results are reported as an index (I) that corresponds to High Risk (I ≤ 0.000) or Low Risk (I > 0.000). BP microarray and NGS test results are also reported as an index and each sample results in three indices, one index for each molecular subtype (Luminal-, HER2- and basal-type index). The final test result is the highest index among the three indices [2]. By combining the MP and the BP test results, luminal-type tumors can be further stratified in Luminal A-type (BP luminal-type and MP Low Risk) and Luminal B-type (BP luminal-type and MP High Risk). The procedure is publicly available in the Standard Operating Procedures at the Agendia website [15].
The beta site MP and BP NGS results were compared both to the Agendia MP and BP microarray results and to the Agendia MP and BP NGS results. Furthermore, molecular subtypes obtained by the MP/BP NGS kit were compared to tumor surrogate intrinsic molecular subtypes obtained by IHC.
Surrogate intrinsic breast cancer subtypes based on IHC were defined as follows: ERpos (with nuclear ER expression in at least 10% of the tumor cells) and HER2neg tumors were defined as Luminal-like. Luminal-like tumors were further stratified depending on level of Ki-67 and progesterone receptor (PR) expression, into Luminal A-like and Luminal B-like according to the classification described by Prat et al. and by Maisonneuve et al. as described in Table 1 [16], [17].
Table 1.
Definition of surrogate Luminal A- and Luminal B-like tumors depending on Ki-67 and PR expression by immunohistochemistry. While Prat et al. define the two luminal subtypes primarily depending on PR expression levels with a fixed Ki-67 cut-off, Maisonneuve et al. propose a variable Ki-67 cut-off depending on PR expression levels. The % indicates the percentage of positive tumor cells.
When Ki-67% was not available at UHL or CIP, tumors with grade 1 or 2 were classified as Luminal A-like and tumors with grade 3 as Luminal B-like according to a classification described earlier by Brouckaert et al. [18]. Tumors in which HER2 was amplified (with or without confirmation by in situ hybridization in case of score 2+ and/or 3+ by IHC) were defined either as Luminal B-like (HER2pos) in case of ERpos, or as HER2pos in case of ERneg. ERneg, PRneg and HER2neg tumors were assigned to the Basal-like subtype. The IHC-based surrogate subtypes were then compared to the molecular subtypes obtained by BP (Luminal-, HER2- and Basal-type) based on NGS at the beta sites and microarray technology in the central laboratory.
Statistical Analysis
The analyses of the data were performed using the MATLAB® (The MathWorks, Inc., Natick, Massachusetts, United States) software version R2012a. Visualization of data was performed using the MATLAB® (The MathWorks) software version R2012a, Microsoft PowerPoint and Excel 2016 MSO (16.0.9330.2124). Equivalence of MP and BP indices was determined by a Pearson's correlation coefficient for assessment of the degree of linear correlation, and a Passing-Bablok regression analysis to obtain the regression equation. Scatterplots were used to visually examine the existence of any constant bias in the difference of measurements between samples analyzed with targeted RNA sequencing and microarray technologies. Acceptable limits were defined a priori, based on microarray data generated at Agendia between 2012 and 2016. MP and BP test outcomes (MP: High/Low Risk, BP: Luminal-, Basal- and HER2-type) assessed using targeted RNA sequencing technology were compared with the standard microarray diagnostic outcomes using a contingency table. Based on the contingency table we measured the concordance for both MP and BP, the Negative Percent Agreement (NPA) and the Positive Percent Agreement (PPA) for MP only. Concordance, NPA and PPA had to be equal to or higher than 90%.
Results
Patient Samples
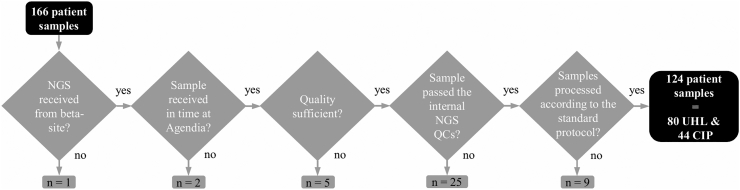
For this beta testing study, 124 samples from patients diagnosed at UHL and CIP between September 2017 and January 2018 were included for the analysis. The sample inclusion flowchart can be found in Figure 1 and patient and tumor characteristics can be found in Supplementary Table 1.
Figure 1.
Patient sample inclusion diagram. Patient samples need to successfully pass different stages and quality controls before they can be included in the final analysis. Abbreviations: NGS, next-generation sequencing; QC, quality control; UHL, University Hospitals Leuven; CIP, Curie Institut Paris.
Equivalence Between Beta Site NGS and Agendia Microarray Results
Out of the 124 patient samples, 60 samples (48.4%) were MP Low Risk and 64 samples (51.6%) were MP High Risk according to microarray. The comparison between MP microarray indices assessed at Agendia central laboratory and MP NGS indices at the beta sites are shown in the scatterplot of Figure 2. MP NGS indices showed high correlation with the matching microarray indices (Pearson's r = 0.96), as confirmed by the Passing-Bablok regression analysis (Supplementary Table 2). The concordance between risk results obtained by MP microarray from Agendia and by MP NGS from the beta sites was 91.1% with an NPA of 93.8% (60/64, 95% CI: 85.0–97.5) and a PPA of 88.3% (53/60, 95% CI: 77.8–94.2) (Table 2). For concordance and NPA, the pre-defined criteria were met. There were 11 samples (8.9%) that showed discrepancies in MP risk categories between microarray and NGS (Table 2). Those samples had indices very close to the classification threshold with microarray and/or NGS. The comparison between MP microarray indices assessed at Agendia Amsterdam central laboratory and MP NGS at the beta sites separately is shown in Supplementary Figure 2. The MP risk classification concordance between Agendia microarray and UHL NGS and CIP NGS, NPA and PPA for each beta site separately can be found in Supplementary Table 3 and 4. When we excluded all samples with indices very close to the classification threshold, the concordance between risk results obtained by MP microarray from Agendia and by NGS from the beta sites was 99.0% with an NPA of 98.1% (53/54, 95% CI: 90.2–99.7) and a PPA of 100.0%, and by NGS from each beta site separately was 98.4% for UHL with an NPA of 96.8% (30/31, 95% CI: 83.8–99.4) and a PPA of 100.0% and 100.0% for CIP (data not shown).
Figure 2.
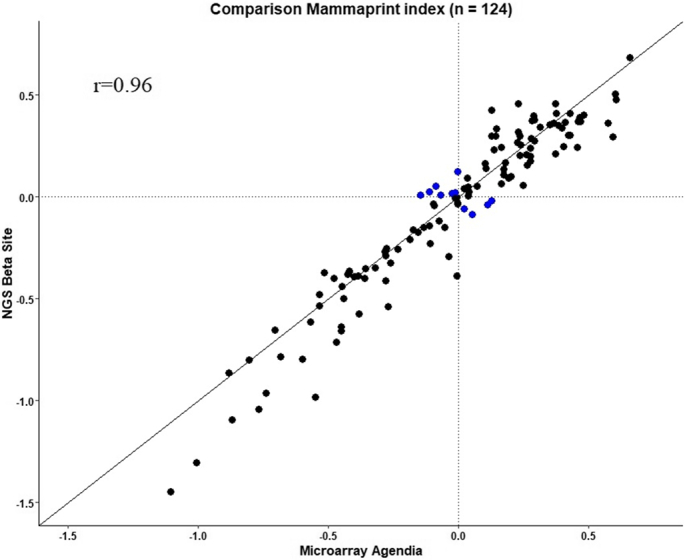
MammaPrint (MP) microarray indices assessed at Agendia Amsterdam central laboratory in comparison to MP NGS indices at the beta sites. The comparison shows equivalence between the two technologies (Pearson's r = 0.96). The x-axis reports the MP microarray index, the y-axis reports the MP NGS index. Each dot represents a single breast cancer sample for which total RNA underwent microarray and NGS laboratory processing and analysis. The blue dots represent the discordant cases with indices close to the classification threshold.
Table 2.
Comparison of test outcomes from MammaPrint (High/Low Risk) between microarray assessed at Agendia Amsterdam central laboratory and NGS assessed at the beta sites. These results show a Negative Predictive Agreement (Low Risk) of 93.8% (60/64, 95% CI: 85.0–97.5), a Positive Predictive Agreement (High Risk) of 88.3% (53/60, 95% CI: 77.8–94.2), a concordance of 91.1% and a Cohen's kappa of 0.82.
| Microarray |
||||
|---|---|---|---|---|
| High Risk | Low Risk | Total | ||
| NGS beta sites | High Risk | 53 | 4 | 57 |
| Low Risk | 7 | 60 | 67 | |
| Total | 60 | 64 | 124 | |
Next, we assessed the potential clinical impact of the patients with a false-negative NGS results obtained by the beta sites (n = 8; respectively 5 with MA and NGS Agendia, 2 with only MA Agendia and 1 with only NGS Agendia). A detailed summary of the false-negative cases is provided in Supplementary Table 5. After clinic-pathological review, we found that most likely in 6 of the 8 discordant cases the MP test would have not be requested by the clinician because of the clear-cut clinical management of these patients. Two of them were spared from chemotherapy because of comorbidities and the older age (D-UHL1/2); two received anti-HER2 therapy in combination with chemo-endocrine therapy because of HER2 gene amplification (D-UHL3/4). The inclusion of HER2pos cases was dictated by the validation of the BluePrint signature. The fifth one was a clinically High Risk lobular carcinoma with high tumor volume and two positive lymph nodes for which major international guidelines would recommend the use of chemotherapy (D-UHL5). The sixth case was a small screening detected well differentiated ERpos/HER2neg tubular carcinoma with negative sentinel-biopsy procedure in a postmenopausal woman (D-UHL6). Interestingly, in the latter the MP NGS index at the beta site and at Agendia showed a positive value outside the 5.0% zone around the 0 with low genomic risk. In the two remaining cases (D-UHL7, D-CIP1), the clinical impact of the false-negative outcome is less clear because of the young age of both, the presence of borderline MP index in the MA results of one of them and, the presence of a nodal micrometastasis despite a small grade 1 breast carcinoma of no special type in the other.
For BP subtype outcomes based on microarray, the patient samples had a BP luminal, HER2 or basal subtype in 82.3% (102/124), 8.1% (10/124) and 9.7% (12/124), respectively. The concordance between subtype results obtained by BP microarray from Agendia and by BP NGS from the beta sites was 98.4% (122/124). Two patient samples were classified as BP Luminal-type by NGS at the beta site and as BP HER2-type by microarray at Agendia (Table 4C). In those samples, HER2 was amplified with 2+ and 3+ by IHC with confirmation of in situ hybridization.
Table 4.
Comparison of molecular subtyping using MP and BP tests (Luminal A for Low Risk MP and Luminal B for High Risk) between NGS Beta Site and IHC according to Prat et al. (n = 124) (A), between NGS Beta Site and IHC according to Maisonneuve et al. (n = 124) (B), between NGS Beta Site and microarray Agendia (n = 124) (C), and between NGS Beta Site and NGS Agendia (n = 114∗) (D). These results show a concordance of 71.8% (89/124) (A), 76.6% (95/124) (B), 89.5% (111/124) (C) and 93.9% (107/114) (D).
| A. |
|||||
|---|---|---|---|---|---|
| MP/BP NGS Beta Site |
|||||
| IHC | Luminal A | Luminal B | HER2 | Basal | Total |
| Luminal A-like | 52 | 18 | 0 | 0 | 70 |
| Luminal B-like, HER2-negative |
10 | 11 | 0 | 1 | 22 |
| Luminal B-like, HER2-positive |
5 | 7 | 5 | 0 | 17 |
| HER2-positive | 0 | 0 | 3 | 0 | 3 |
| Triple negative | 0 | 1 | 0 | 11 | 12 |
| Total | 67 | 37 | 8 | 12 | 124 |
| B. | |||||
| Luminal A-like | 53 | 13 | 0 | 0 | 66 |
| Luminal A-like | 53 | 13 | 0 | 0 | 66 |
| Luminal B-like, HER2-negative |
9 | 16 | 0 | 1 | 26 |
| Luminal B-like, HER2-positive |
5 | 7 | 5 | 0 | 17 |
| HER2-positive | 0 | 0 | 3 | 0 | 3 |
| Triple negative | 0 | 1 | 0 | 11 | 12 |
| Total | 67 | 37 | 8 | 12 | 124 |
| C. | |||||
| Microarray | Luminal A | Luminal B | HER2 | Basal | Total |
| Luminal A | 60 | 4 | 0 | 0 | 64 |
| Luminal B | 7 | 31 | 0 | 0 | 38 |
| HER2 | 0 | 2 | 8 | 0 | 10 |
| Basal | 0 | 0 | 0 | 12 | 12 |
| Total | 67 | 37 | 8 | 12 | 124 |
| D. | |||||
| NGS Agendia | Luminal A | Luminal B | HER2 | Basal | Total |
| Luminal A | 58 | 1 | 0 | 0 | 59 |
| Luminal B | 6 | 31 | 0 | 0 | 37 |
| HER2 | 0 | 0 | 7 | 0 | 7 |
| Basal | 0 | 0 | 0 | 11 | 11 |
| Total | 64 | 32 | 7 | 11 | 114∗ |
For 10 samples, no NGS results were obtained at Agendia.
Equivalence Between Beta Site and Agendia NGS Results
Out of the 124 samples, 114 led to NGS results both at Agendia and at the beta sites. The comparison between MP NGS indices assessed at Agendia Amsterdam central laboratory and MP NGS at the beta sites is shown in Figure 3. MP NGS indices generated at the beta site showed near perfect correlation with the matching Agendia NGS indices (Pearson's r = 0.96) as confirmed by the Passing-Bablok regression analysis (Supplementary Table 2). The MP risk classification concordance between Agendia NGS and NGS at the beta sites was 93.9%; NPA of 98.3% (58/59, 95% CI: 91.0–99.7) and PPA of 89.1% (49/55, 95% CI: 78.2–94.9) (Table 3). For concordance and NPA, the pre-defined criteria were met. Seven samples show discordant MP classification results (Table 3) between the beta site and Agendia. These samples had indices very close to the classification threshold. The comparison between MP NGS indices assessed at Agendia Amsterdam central laboratory and MP NGS at the beta sites separately is shown in Supplementary Figure 3. The MP risk classification concordance between Agendia NGS and UHL NGS and CIP NGS, NPA and PPA can be found in Supplementary Table 6 and 7. When we excluded all samples with indices very close to the classification threshold, the concordance between risk results obtained by MP NGS from Agendia and by MP NGS from the beta sites was 98.9% with an NPA of 98.0% (50/51, 95% CI: 89.7–99.6) and a PPA of 100.0%, and by NGS from each beta site separately was 100.0% for UHL and 97.1% for CIP with an NPA of 95.5% (21/22, 95% CI: 78.2–99.2) and a PPA of 100.0% (data not shown). For BP, the results from NGS Agendia versus NGS at the beta sites gave a 100.0% concordance.
Figure 3.
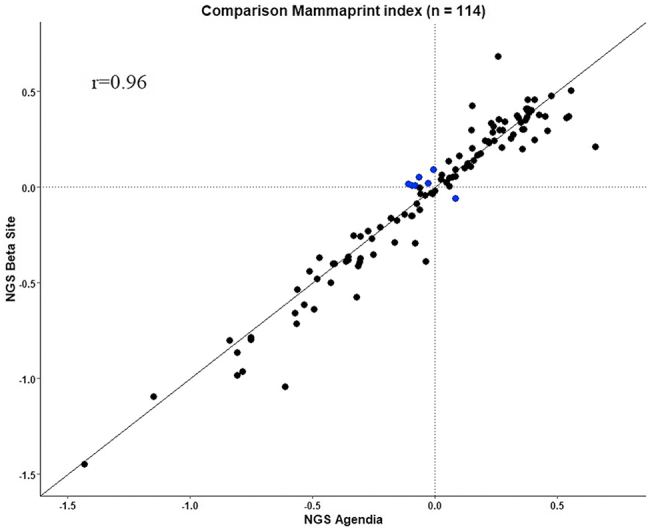
MammaPrint (MP) NGS indices assessed at Agendia Amsterdam central laboratory in comparison to MP NGS at the beta sites. The comparison shows equivalence between MP NGS performed at Agendia (x-axis) and MP NGS at the beta sites (y-axis) (Pearson's r = 0.96) (n = 114). Each dot represents a single breast cancer sample for which total RNA underwent NGS or microarray laboratory processing and analysis. Out of the 124 samples processed on NGS, 10 samples lacked NGS results at Agendia. The blue dots represent the discordant cases with indices close to the classification threshold.
Table 3.
Comparison of test outcomes from MP (High/Low Risk) between NGS assessed at Agendia Amsterdam central laboratory and NGS assessed at the beta sites (n = 114∗). These results show a Negative Predictive Agreement (Low Risk) of 98.3% (58/59, 95% CI: 91.0–99.7), a Positive Predictive Agreement (High Risk) of 89.1% (49/55, 95% CI: 78.2–94.9), a concordance of 93.9% and a Cohen's kappa of 0.88.
| NGS Agendia |
||||
|---|---|---|---|---|
| High Risk | Low Risk | Total | ||
| NGS Beta Sites | High Risk | 49 | 1 | 50 |
| Low Risk | 6 | 58 | 64 | |
| Total | 55 | 59 | 114∗ | |
For 10 samples, no NGS results were obtained at Agendia.
Comparison Between IHC and MP/BP Molecular Subtyping
The concordance between IHC, and MP and BP NGS molecular subtyping according to Prat was 71.8% (89/124) (Table 4A), and according to Maisonneuve et al. was 76.6% (95/124) (Table 4B). The concordance for molecular subtyping between NGS at the beta sites and microarray with the addition of MP to further stratify Luminal-type into Luminal A-type and Luminal B-type was 89.5% (111/124) (Table 4C) and between NGS at the beta sites and NGS at Agendia was 93.9% (107/114) (Table 4D).
MP/BP NGS subtyping identified less Low Risk Luminal A tumors compared to IHC (54.0% (67/124) versus 56.5% (70/124)) according to Prat et al. but more Low Risk Luminal A tumors compared to IHC (54.0% (67/124) versus 53.2% (66/124)) according to Maisonneuve et al. Notably, the concordance was excellent for triple-negative and, to a lesser extent for HER2 driven tumors (Luminal B-like-HER2 positive and HER2+) (Table 4A and Table 4B).
Discussion
The current MP and BP gold standard FFPE microarray test was recently successfully translated to a targeted RNA NGS based test at Agendia laboratories [14]. A high concordance was already observed between results obtained by microarray performed centrally at Agendia and NGS performed at UHL and CIP on a small sample set of which RNA was previously isolated at Agendia (n = 15) [14]. However, a larger prospective validation study where samples are processed locally obtained from tissue onwards with the NGS kit was required.
In this beta testing study, we validated the Next-Generation RNA sequencing-based MP/BP NGS kit in UHL and CIP and we demonstrated reproducibility of the MP and BP NGS test between two independent decentralized laboratories and Agendia central laboratories. The concordance rates between microarray, the gold standard technology performed centrally at Agendia laboratories, and NGS performed at UHL and at CIP met the pre-determined 90% concordance, NPA and PPA for both MP and BP. Furthermore, the comparison between results generated with the NGS technology at Agendia and at the beta sites also showed a high concordance, which highlights the robustness of the NGS test. However, for MP, the PPA for the comparison between microarray performed centrally and NGS performed at the beta sites did not reach 90%. Notably, the samples that were discordant in the MP classification results (Low Risk versus High Risk) between microarray at Agendia and NGS at both beta sites and between NGS at Agendia and NGS at each beta site had indices very close to the classification threshold.
We looked in more detail at 8 false-negative cases, discrepant Low Risk according to decentral NGS versus High Risk according to central laboratory because of their major potential clinical impact that might result in undertreatment of the patients. Beside standard clinic-pathological parameters, well known tumor-associated histopathological features may provide prognostic information and therefore influence the results of multi-gene signatures, such as a higher amount of stromal tumor infiltrating lymphocytes (TILs) or intra-tumoral morphological heterogeneity [19]. Histopathological review showed lymph-vessel invasion (LVI) in five cases, intra-tumoral morphological heterogeneity in one case, a relative increase in stromal TILs in two other cases (up to 20–30%), and a heterogeneous HER2 gene amplification with a mixture of HER2pos and HER2neg tumor cells in two cases. Most remarkably one tubular carcinoma was called high risk by MA analysis, while in our opinion the case was correctly classified as low risk by both NGS at Agendia and UHL (Supplementary Table 5). Nevertheless, morphological features such as LVI, which is a well-established poor prognostic factor in breast cancer, are not always linked to specific molecular features [20]. Therefore, we strongly believe that the results obtained by the multi-gene signatures must be interpreted in the light of all clinic-pathological features of each patient and should also be put in the correct clinical perspective. In this regard, we speculate that the clinical impact of the eight false-negative results in our study would have been negligible in the correct clinical setting where only Luminal HER2neg tumors with uncertain indication for chemotherapy would have been tested. Indeed, in our opinion 6 out of 8 patients would have not been tested either for the well-established clinical management, age, or comorbidities. In the remaining two cases the clinical impact remains uncertain, however the younger age of both patients in combination with MP indices in the equivocal region should advocate careful examination of all features in the context of a multi-disciplinary oncological meeting.
The robustness of the MP and BP NGS kit was further assessed by an additional experiment performed at UHL in which samples falling in the region with MP index between +0.057 and− 0.057, and just outside this region were re-run in triplicate (n = 5). The triple experiment confirmed in all cases the initial results highlighting the reproducibility of the test (data not shown). These findings are consistent with those described by Sapino et al. [1], where the PPA of MP indices measured in FFPE material within this region was about 61% as compared to results obtained from fresh frozen material. This discrepancy is due to the assay variability, to tumor heterogeneity and possibly to intrinsic morphological features of the tumor. Physicians who use MP results to supplement clinic-pathological parameters in their decision making should be notified about the decreased analytical accuracy of the test in these cases. Because of the reduced analytical accuracy of the region close to the classification threshold, one could speculate to administer chemotherapy regardless of the MP risk category in order to avoid undertreatment. However, we anticipate that most cases with MP index in the 5% region around the 0 will probably show a discrepant clinical-versus-molecular risk profile (see also false-negative case 7 and 8 in our series in Supplementary Table 5), a category for which the added value of using MP is uncertain in those patients with clinicalLOW/molecularHIGH risk, according to the results of the MINDACT trial [4]. Therefore, in absence of more solid evidence coming from larger series of patients or from clinical trials, we would suggest to thoroughly discuss the cases with a MP index in the region around the classification threshold in the context of a well-informed multidisciplinary oncological meeting. Next, we would like also to acknowledge that this statement is based on an observation made on a relatively small series of cases, in a study that did not aim at recording the impact of MP-NGS on clinical decisions.
There were some differences in the laboratory procedure used at the three sites (Agendia, UHL, CIP). For deparaffinization, Agendia used xylene whereas UHL used the less toxic solvent Histo-Clear. In CIP, nine samples were excluded from the validation set as another deparaffinization buffer (Qiagen deparaffinization solution) than xylene or Histo-Clear was used which resulted in lower quality data. In light of the results obtained during the beta testing, the instructions for use of the MP/BP NGS kit have been adjusted by adding xylene or Histo-Clear as recommended buffers for deparaffinization. For RNA extraction, the RNeasy FFPE kit was used in Agendia and CIP and the AllPrep FFPE kit was used in UHL. We observed lower RNA yield from FFPE with the AllPrep kit as compared to the RNeasy kit [21], [22]. In UHL, two operators performed the laboratory procedure and we observed only a small difference in results between an experienced and a less experienced operator (data not shown). This indicates the robustness of the NGS test. However, as the laboratory protocol to perform the MP and BP NGS tests is a delicate multi-step procedure, previous experience with NGS tests is advisable and on-site training is crucial.
When we compared MP/BP NGS molecular subtyping with IHC surrogate subtypes, we observed a discordance of 28.2% (Prat classification) and 23.4% (Maisonneuve classification) compared to 10.5% when comparing NGS at the beta sites with microarray. The discordance of about 25% between IHC and NGS is in line with previous findings where IHC surrogate subtypes were compared to molecular subtyping based on microarray underlining the complementarity of genomic testing in early breast cancer [23], [24], [25]. Molecular characterization by genomic analysis is supposed to better capture the real biologic behavior of breast carcinomas than IHC, because it measures the expression level of a larger number of genes. Despite the observations that in the neo-adjuvant setting HER2-enriched and Basal subtypes seem to predict a higher pathological complete response rate in IHCER+/HER2- breast carcinomas, to the best of our knowledge there are no randomized prospective clinical trials that show benefit from anti-HER2 therapy based on molecular subtype [26]. Therefore, the structural implementation of molecular subtyping in the daily clinical routine remains largely controversial. Major international guidelines as well as expert panels do not advocate the use of molecular subtyping by genomic analysis for treatment decision making and recommend the use of IHC for ER and HER2 for the identification of therapeutically relevant subtypes. Nevertheless, the information obtained by molecular subtyping seems to provide clinically relevant information beyond the current surrogate classification [26], [27], [28], [29], [30], [31].
Importantly, we observed a high concordance of 98.4% between NGS at the beta sites and microarray BP molecular subtyping (luminal-, HER2- and basal-types). Two patient samples were classified as BP Luminal-type by NGS at the beta site and as BP HER2-type by microarray at Agendia. For those samples, the HER2 and the luminal BP indices were close to each other. Both samples were ERpos; HER2 amplification was confirmed in both cases by in situ hybridization with scores of 2+ and 3+ by IHC respectively.
In this beta testing study, patients were consecutively included which led to the inclusion of patients with different histological subtypes. The MP and BP test is only validated for invasive ductal and invasive lobular carcinoma [32], [33], [34], [35]. The majority of tumors in this study had an invasive ductal or invasive lobular histological subtype. There were only eight patient samples (8/124 = 6.5%) with other histological subtypes. Interestingly, one tumor sample with a special histologic subtype showed MP index within the region around the 0, suggesting that other morphological features might be associated to the equivocal results. Additionally, there was one patient with six positive lymph nodes who did not meet the standard MP inclusion criteria [4] that was included.
In summary, we report here that the MP/BP NGS kit was successfully validated in a decentralized setting, showing high concordance between results obtained at two independent laboratories and at Agendia. We observed a high concordance between NGS and microarray molecular subtyping suggesting a successful translation of the MP and BP microarray test to a MP and BP NGS test in a decentralized setting. The MP/BP NGS kit, the first FFPE targeted RNA sequencing based multi-gene signature for early breast cancer, is straightforward to implement in a decentralized setting and is compatible with existing infrastructure. The availability of such a MP/BP NGS kit will increase accessibility to the MP and BP tests offering an in-house solution for physicians and patients without compromising the level of accuracy and clinical utility.
Declaration of competing interest
Lorenza Mittempergher, Leonie JMJ Delahaye, Mireille HJ Snel, Anke T Witteveen, Sari Neijenhuis and Annuska M Glas are employed by Agendia, the commercial entity that markets the 70-gene signature as MammaPrint and the 80-gene signature as BluePrint. Annuska M Glas is named inventor of the 80-gene signature as BluePrint.
Laurence Slembrouck received financial support from Agendia.
All remaining authors have declared no conflict of interest.
Acknowledgements
We would like to thank Gijs van Hest and Geert Van der Borght from University Hospitals Leuven for cutting the tissue sections from the selected formalin-fixed paraffin-embedded block of each patient from University Hospitals Leuven and their excellent technical skills.
Footnotes
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.tranon.2019.08.008.
Appendix A. Supplementary data
Supplementary material
References
- 1.Sapino A, Roepman P, Linn SC, Snel MH, Delahaye LJ, van den Akker J, Glas AM, Simon IM, Barth N, de Snoo FA. 2014 American Society for Investigative Pathology and the Association for Molecular Pathology. Vol. 16. Published by Elsevier Inc; United States: 2014. MammaPrint molecular diagnostics on formalin-fixed, paraffin-embedded tissue, In: J Mol Diagn. pp. 190–197. [DOI] [PubMed] [Google Scholar]
- 2.Krijgsman O, Roepman P, Zwart W, Carroll JS, Tian S, de Snoo FA, Bender RA, Bernards R, Glas AM. A diagnostic gene profile for molecular subtyping of breast cancer associated with treatment response. Breast Cancer Res Treat. 2012;133:37–47. doi: 10.1007/s10549-011-1683-z. [DOI] [PubMed] [Google Scholar]
- 3.Whitworth P, Beitsch P, Mislowsky A, Pellicane JV, Nash C, Murray M, Lee LA, Dul CL, Rotkis M, Baron P. Chemosensitivity and endocrine sensitivity in clinical luminal breast cancer patients in the prospective Neoadjuvant Breast Registry Symphony Trial (NBRST) predicted by molecular subtyping. Ann Surg Oncol. 2017;24:669–675. doi: 10.1245/s10434-016-5600-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cardoso F, van't Veer LJ, Bogaerts J, Slaets L, Viale G, Delaloge S, Pierga JY, Brain E, Causeret S, DeLorenzi M. 70-gene signature as an aid to treatment decisions in early-stage breast cancer. N. Engl. J. Med. 2016;375:717–729. doi: 10.1056/NEJMoa1602253. [DOI] [PubMed] [Google Scholar]
- 5.van 't Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT. Nature. Vol. 415. 2002. Gene expression profiling predicts clinical outcome of breast cancer; pp. 530–536. [England] [DOI] [PubMed] [Google Scholar]
- 6.Drukker CA, Bueno-de-Mesquita JM, Retel VP, van Harten WH, van Tinteren H, Wesseling J, Roumen RM, Knauer M, van 't Veer LJ, Sonke GS. A prospective evaluation of a breast cancer prognosis signature in the observational RASTER study. Int J Cancer. 2013;133:929–936. doi: 10.1002/ijc.28082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lancet. Vol. 365. 2005. Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials; pp. 1687–1717. [England] [DOI] [PubMed] [Google Scholar]
- 8.Ross JS, Fletcher JA, Linette GP, Stec J, Clark E, Ayers M, Symmans WF, Pusztai L, Bloom KJ. The Her-2/neu gene and protein in breast cancer 2003: biomarker and target of therapy. Oncologist. 2003;8:307–325. doi: 10.1634/theoncologist.8-4-307. [DOI] [PubMed] [Google Scholar]
- 9.Delahaye LJ, Wehkamp D, Floore AN, Bernards R, van 't Veer LJ, Glas AM. Performance characteristics of the MammaPrint((R)) breast cancer diagnostic gene signature. Per Med. 2013;10:801–811. doi: 10.2217/pme.13.88. [DOI] [PubMed] [Google Scholar]
- 10.Glas AM, Floore A, Delahaye LJ, Witteveen AT, Pover RC, Bakx N, Lahti-Domenici JS, Bruinsma TJ, Warmoes MO, Bernards R. Converting a breast cancer microarray signature into a high-throughput diagnostic test. BMC Genomics. 2006;7:278. doi: 10.1186/1471-2164-7-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sparano JA, Gray RJ, Makower DF, Pritchard KI, Albain KS, Hayes DF, Geyer CE, Jr., Dees EC, Perez EA, Olson JA., Jr. Prospective Validation of a 21-Gene Expression Assay in Breast Cancer. N Engl J Med. 2015;373:2005–2014. doi: 10.1056/NEJMoa1510764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sparano JA, Gray RJ, Makower DF, Pritchard KI, Albain KS, Hayes DF, Geyer CE, Jr., Dees EC, Goetz MP, Olson JA., Jr. Adjuvant Chemotherapy Guided by a 21-Gene Expression Assay in Breast Cancer. N Engl J Med. 2018;379:111–121. doi: 10.1056/NEJMoa1804710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chia SK, Bramwell VH, Tu D, Shepherd LE, Jiang S, Vickery T, Mardis E, Leung S, Ung K, Pritchard KI. A 50-gene intrinsic subtype classifier for prognosis and prediction of benefit from adjuvant tamoxifen. Clin Cancer Res. 2012;18:4465–4472. doi: 10.1158/1078-0432.CCR-12-0286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mittempergher L, Delahaye LJ, Witteveen A, Spangler JB, Hassenmahomed F, Mee S, Mahmoudi S, Chen J, Bao S, Snel MH. J Mol Diagn. 2019. 2019. MammaPrint and BluePrint Molecular Diagnostics using targeted RNA Next-Generation Sequencing technology. Published by Elsevier Inc.: United States. [DOI] [PubMed] [Google Scholar]
- 15.Agendia NV. MammaPrint and BluePrint Breast Cancer Recurrence and Molecular Subtyping Kit - Package Insert. 2019. https://www.agendia.com/diagnostic-products/resources.html
- 16.Prat A, Cheang MC, Martin M, Parker JS, Carrasco E, Caballero R, Tyldesley S, Gelmon K, Bernard PS, Nielsen TO. Prognostic significance of progesterone receptor-positive tumor cells within immunohistochemically defined luminal A breast cancer. J Clin Oncol. 2013;31:203–209. doi: 10.1200/JCO.2012.43.4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maisonneuve P, Disalvatore D, Rotmensz N, Curigliano G, Colleoni M, Dellapasqua S, Pruneri G, Mastropasqua MG, Luini A, Bassi F. Proposed new clinicopathological surrogate definitions of luminal A and luminal B (HER2-negative) intrinsic breast cancer subtypes. Breast Cancer Res. 2014;16:R65. doi: 10.1186/bcr3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brouckaert O, Laenen A, Vanderhaegen J, Wildiers H, Leunen K, Amant F, Berteloot P, Smeets A, Paridaens R, Christiaens MR. Ann Oncol. Vol. 23. 2012. Applying the 2011 St Gallen panel of prognostic markers on a large single hospital cohort of consecutively treated primary operable breast cancers; pp. 2578–2584. [England] [DOI] [PubMed] [Google Scholar]
- 19.Acs G, Esposito NN, Kiluk J, Loftus L, Laronga C. Mod Pathol. Vol. 25. 2012. A mitotically active, cellular tumor stroma and/or inflammatory cells associated with tumor cells may contribute to intermediate or high Oncotype DX Recurrence Scores in low-grade invasive breast carcinomas; pp. 556–566. [United States] [DOI] [PubMed] [Google Scholar]
- 20.Heng YJ, Lester SC, Tse GM, Factor RE, Allison KH, Collins LC, Chen YY, Jensen KC, Johnson NB, Jeong JC. The molecular basis of breast cancer pathological phenotypes. J Pathol. 2017;241:375–391. doi: 10.1002/path.4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kresse SH, Namlos HM, Lorenz S, Berner JM, Myklebost O, Bjerkehagen B, Zepeda LA. Evaluation of commercial DNA and RNA extraction methods for high-throughput sequencing of FFPE samples. PLoS One. 2018;13 doi: 10.1371/journal.pone.0197456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Patel PG, Selvarajah S, Guerard KP, Bartlett JMS, Lapointe J, Berman DM, Okello JBA, Park PC. Reliability and performance of commercial RNA and DNA extraction kits for FFPE tissue cores. PLoS One. 2017;12 doi: 10.1371/journal.pone.0179732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Viale G, de Snoo FA, Slaets L, Bogaerts J, van 't Veer L, Rutgers EJ, Piccart-Gebhart MJ, Stork-Sloots L, Glas A, Russo L. Breast Cancer Res Treat. Vol. 167. 2018. Immunohistochemical versus molecular (BluePrint and MammaPrint) subtyping of breast carcinoma. Outcome results from the EORTC 10041/BIG 3-04 MINDACT trial; pp. 123–131. [Netherlands] [DOI] [PubMed] [Google Scholar]
- 24.Whitworth P, Stork-Sloots L, de Snoo FA, Richards P, Rotkis M, Beatty J, Mislowsky A, Pellicane JV, Nguyen B, Lee L. Chemosensitivity predicted by BluePrint 80-gene functional subtype and MammaPrint in the Prospective Neoadjuvant Breast Registry Symphony Trial (NBRST) Ann Surg Oncol. 2014;21:3261–3267. doi: 10.1245/s10434-014-3908-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beitsch P, Whitworth P, Baron P, Rotkis MC, Mislowsky AM, Richards PD, Murray MK, Pellicane JV, Dul CL, Nash CH. Ann Surg Oncol. Vol. 24. 2017. Pertuzumab/Trastuzumab/CT Versus Trastuzumab/CT Therapy for HER2+ Breast Cancer: Results from the Prospective Neoadjuvant Breast Registry Symphony Trial (NBRST) pp. 2539–2546. [United States] [DOI] [PubMed] [Google Scholar]
- 26.Llombart-Cussac A, Cortes J, Pare L, Galvan P, Bermejo B, Martinez N, Vidal M, Pernas S, Lopez R, Munoz M. Lancet Oncol. Vol. 18. 2017. Elsevier Ltd; 2017. HER2-enriched subtype as a predictor of pathological complete response following trastuzumab and lapatinib without chemotherapy in early-stage HER2-positive breast cancer (PAMELA): an open-label, single-group, multicentre, phase 2 trial; pp. 545–554. England. [DOI] [PubMed] [Google Scholar]
- 27.Cardoso F, Kyriakides S, Ohno S, Penault-Llorca F, Poortmans P, Rubio IT, Zackrisson S, Senkus E. Ann Oncol. England. vol. 30. 2019. Early breast cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up; pp. 1194–1220. [DOI] [PubMed] [Google Scholar]
- 28.Guiu S, Michiels S, Andre F, Cortes J, Denkert C, Di Leo A, Hennessy BT, Sorlie T, Sotiriou C, Turner N. Ann Oncol. Vol. 23. 2012. Molecular subclasses of breast cancer: how do we define them? The IMPAKT 2012 Working Group Statement; pp. 2997–3006. [England] [DOI] [PubMed] [Google Scholar]
- 29.Prat A, Pineda E, Adamo B, Galvan P, Fernandez A, Gaba L, Diez M, Viladot M, Arance A, Munoz M. Breast. Vol. 24 Suppl 2. 2015 The Authors. 2015. Clinical implications of the intrinsic molecular subtypes of breast cancer; pp. S26–S35. Published by Elsevier Ltd.: Netherlands. [DOI] [PubMed] [Google Scholar]
- 30.Cejalvo JM, Pascual T, Fernandez-Martinez A, Braso-Maristany F, Gomis RR, Perou CM, Munoz M, Prat A. Cancer Treat Rev. Vol. 67. 2018 The Author(s) 2018. Clinical implications of the non-luminal intrinsic subtypes in hormone receptor-positive breast cancer; pp. 63–70. Published by Elsevier Ltd.: Netherlands. [DOI] [PubMed] [Google Scholar]
- 31.National Comprehensive Cancer Network Breast Cancer (Version 2.2019) 2019. https://www.nccn.org/professionals/physician_gls/pdf/breast.pdf
- 32.van de Vijver MJ, He YD, van't Veer LJ, Dai H, Hart AA, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C, Marton MJ. N Engl J Med. Vol. 347. 2002. Massachusetts Medical Society; United States: 2002. A gene-expression signature as a predictor of survival in breast cancer; pp. 1999–2009. [DOI] [PubMed] [Google Scholar]
- 33.Buyse M, Loi S, van't Veer L, Viale G, Delorenzi M, Glas AM, d'Assignies MS, Bergh J, Lidereau R, Ellis P. J Natl Cancer Inst. Vol. 98. 2006. Validation and clinical utility of a 70-gene prognostic signature for women with node-negative breast cancer; pp. 1183–1192. [United States] [DOI] [PubMed] [Google Scholar]
- 34.Mook S, Schmidt MK, Viale G, Pruneri G, Eekhout I, Floore A, Glas AM, Bogaerts J, Cardoso F, Piccart-Gebhart MJ. The 70-gene prognosis-signature predicts disease outcome in breast cancer patients with 1-3 positive lymph nodes in an independent validation study. Breast Cancer Res Treat. 2009;116:295–302. doi: 10.1007/s10549-008-0130-2. [DOI] [PubMed] [Google Scholar]
- 35.Beumer IJ, Persoon M, Witteveen A, Dreezen C, Chin SF, sammut SJ, Snel M, Caldas C, Linn S, van 't Veer LJ. Prognostic Value of MammaPrint((R)) in Invasive Lobular Breast Cancer. Biomark Insights. 2016;11:139–146. doi: 10.4137/BMI.S38435. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material