Abstract
We present high-throughput amplicon sequence (HTS) datasets of the microbial metacommunity DNA of the gut tissue and the gut digesta of naturally occurring (n = 3) and laboratory aquaculture (n = 2) green sea urchins, Lytechinus variegatus. The HTS datasets were generated on an Illumina MiSeq by targeting the amplicons of the V4 region of the 16S rRNA gene. After the raw sequences were quality checked and filtered, 88% of the sequence reads were subjected to bioinformatics analyses to generate operation taxonomic units (OTUs), which were then verified for saturation by using rarefaction analysis at a 3% sequence variation. Further, the OTUs were randomly subsampled to the minimum sequence count values. Then, the FASTA-formatted representative sequences of the microbiota were assigned taxonomic identities through multiple databases using the SILVA ACT: Alignment, Classification and Tree Service (www.arb-silva.de/aligner). The HTS datasets of this metagenome can be accessed from the BioSample Submission Portal (https://www.ncbi.nlm.nih.gov/bioproject/) under the BioProject IDs PRJNA291441 and PRJNA326427.
Keywords: Echinoderm, Arcobacter, PhyloToAST, QIIME, MiSeq
Specifications Table
| Subject area | Biology |
| More specific subject area | Metagenomics |
| Type of data | Figures and Tables |
| How data was acquired | Illumina MiSeq platform with 250 paired-end kits. |
| Data format | Raw, analyzed |
| Experimental factors | Laboratory aquaculture (LAB) Lytechinus variegatus (n = 2) were collected from Port Saint Joseph, Florida (29.80° N 85.36° W), and held in the laboratory aquaculture condition, fed with a formulated diet for six months prior to investigation. Naturally occurring (ENV) Lytechinus variegatus (n = 3) were collected from the same location and sample preparation began immediately upon arrival to the University of Alabama at Birmingham (UAB) |
| Experimental features | Targeted high-throughput sequencing of the microbial metacommunity 16S rRNA gene (V4 hypervariable regions) using the Illumina MiSeq with 250 paired-end kits followed by bioinformatics analyses. |
| Data source location | Lytechinus variegatus collected from Port Saint Joseph, Florida, USA (29.80° N 85.36° W) located in the Gulf of Mexico. Lytechinus variegatus were maintained in laboratory aquaculture condition at the UAB Biology Department, 1300 University Blvd., Birmingham, AL 35294, USA. Microbial metacommunity DNA was prepared and sequenced at the UAB Department of Genetics, Heflin Center Genomics Core, School of Medicine, the University of Alabama at Birmingham, 705 South 20th Street, Birmingham, AL 35294, USA. |
| Data accessibility |
Raw data corresponding to the 10 samples are available at the NCBI's BioSample database following this link:http://www.ncbi.nlm.nih.gov/sra/?term=sea+urchin+gut+microbiome For the LAB group, the BioProject number is PRJNA291441 and the BioSample IDs are SAMN03944319, SAMN03944320, SAMN03944321, SAMN03944322. For the ENV group, the BioProject number is PRJNA326427 and the BioSample IDs are SAMN05277844, SAMN05277845, SAMN05277846, SAMN05277847, SAMN05277848, SAMN05277849, and SAMN05277850. |
Value of the data
|
1. Data
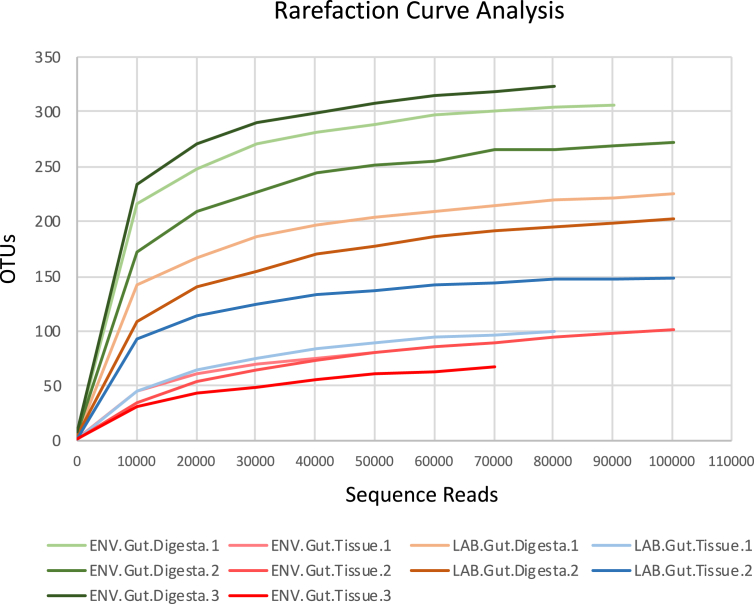
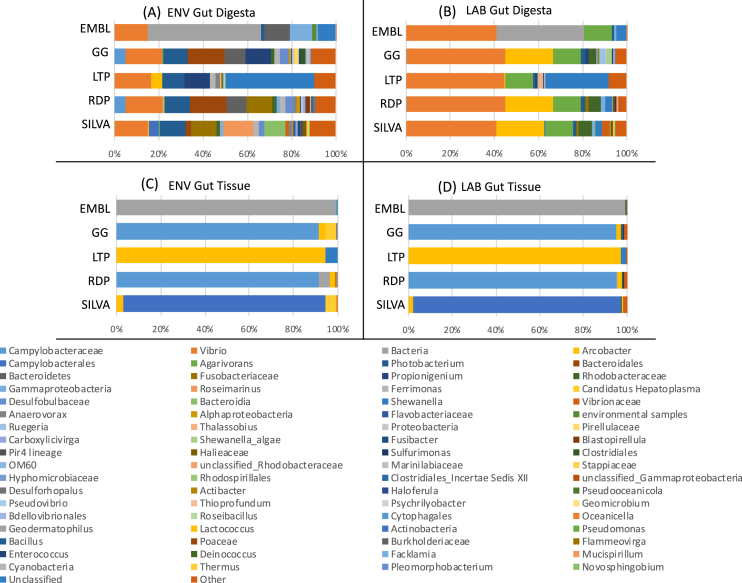
The metagenomic datasets presented in this article describe the microbial community compositions in the gut ecosystem of a marine invertebrate echinoderm of ecological, economic, and scientific importance, Lytechinus variegatus, fed with formulated diet in laboratory aquaculture conditions and from their natural habitat. Fig. 1 describes the OTUs from the quality-checked and filtered HTS data of the 16S rRNA gene and visualized by rarefaction analysis, which indicated that the total quality sequences from each sample are approaching saturation when constructed at a 3% sequence variation. Fig. 2 shows the microbial community profiles of both LAB and ENV gut tissue with a near-exclusive abundance of Epsilonproteobacteria, with the ENV group showing a slightly higher diversity. Table 1 presents the applicability of the HTS datasets for profiling Lytechinus variegatus gut microbiota at the highest possible taxonomic levels following the alignment of the representative sequences to five microbial databases.
Fig. 1.
Rarefaction curve analysis of the HTS data showing the number of OTUs (Y-axis) plotted against total number of sequences (X-axis) per sample. OTUs were determined by using the PhyloToAST (v1.4) taxonomy condensing workflow, which is integrated into QIIME (v1.9.1). Samples were rarefied to the minimum sequence count across all samples for downstream bioinformatics analysis. Data were plotted using Microsoft Excel Software (Seattle, WA, USA).
Fig. 2.
Relative abundance distribution of taxa at the highest resolution determined for the merged biological replicates using multiple taxonomic databases. The FASTA-formatted representative sequences determined by the PhyloToAST (v1.4) workflow integrated into QIIME (v1.9.1) were aligned to multiple databases using the SILVA ACT: Alignment, Classification and Tree Service (www.arb-silva.de/aligner). Taxonomic assignments were performed using the SSU (Small Subunit) category and the Least Common Ancestor (LCA) method with the following databases: SILVA, Ribosomal Database Project (RDP), The All-Species Living Tree (LTP) project, Greengenes (GG), and the European Molecular Biology Laboratory (EMBL). Sequences aligned with a similarity threshold below 70% were discarded. The top 25 taxa from each database were merged based on their common taxonomic assignments at the specific level of classification.
Table 1.
Statistical analysis of the representative sequences aligned to multiple databases using the SILVA ACT: Alignment, Classification and Tree Service (www.arb-silva.de/aligner). Taxonomic assignments were performed using the SSU (Small Subunit) category and the Least Common Ancestor (LCA) method with the following databases: SILVA, Ribosomal Database Project (RDP), The All-Species Living Tree (LTP) project, Greengenes (GG), and the European Molecular Biology Laboratory (EMBL). Sequences aligned at a similarity threshold below 70% were discarded. For each database, the total number of uniquely assigned sequences were determined, and the fraction of those assignments to the family and the genus level were listed.
| Level | SILVA | RDP | LTP | GG | EMBL |
|---|---|---|---|---|---|
| Family | 234 | 193 | 128 | 219 | 18 |
| 82.98% | 80.08% | 100.00% | 76.04% | 54.55% | |
| Genus | 167 | 147 | 121 | 132 | 12 |
| 59.22% | 61.00% | 94.53% | 45.83% | 36.36% | |
| Total Unique | 282 | 241 | 128 | 288 | 33 |
2. Experimental design, materials and methods
2.1. Sample description
The sea urchins were collected from Saint Joseph Bay Aquatic Preserve of the U.S. Gulf of Mexico (29.80° N 85.36° W). For the laboratory aquaculture (LAB) group [1], adult sea urchins (n = 2) were kept in a recirculating saltwater tank system for six months, and fed a formulated feed ad libitum once every 24–48 h that consisted of 6% lipid, 28% protein, and 36% carbohydrate relative percentages [2]. The aquaria were maintained at 22 ± 2 °C with a pH of 8.2 ± 0.2 and salinity of 32 ± 1 ppt. For the naturally occurring (ENV) group [3], adult sea urchins (n = 3) were collected from within the same 1 m2 area and transported to the laboratory at the University of Alabama at Birmingham (UAB) for sample collection. Water conditions were recorded as 20 ± 2 °C with a pH of 7.8 ± 0.2 and salinity of 28 ± 1 ppt. For both groups, the Illumina MiSeq high throughput-sequencing (HTS) platform was used with the 250 bp paired-end kits targeting the V4 hypervariable region [4], [5]. The paired-end raw sequence data were demultiplexed and formatted into FASTQ files [6]. The raw data were deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) under BioProject #PRJNA291441 and #PRJNA326427 for the LAB and ENV group, respectively. The paired-end sequence data for the gut microbial communities can be accessed under the following NCBI BioSample Ids: SAMN03944319 - SAMN03944322 (LAB group) and SAMN05277845 - SAMN05277850 (ENV group). Subgroups for the laboratory-fed group are as follows: LAB.Gut.Tissue (n = 2), LAB.Gut.Digesta (n = 2), ENV.Gut.Tissue (n = 3), and ENV.Gut.Digesta (n = 3).
2.2. Quality assessment and filtering
The raw and demultiplexed paired-end sequence datasets were initially assessed by FastQC [7], and only reads showing 80% of bases at a Q score of >33 were retained by using the “fastx_trimmer” command from the FASTX Toolkit [5], [8] and merged using USEARCH [9]. Paired-end reads with <50 base overlap and/or >20 mismatching nucleotides were filtered from the analysis, and chimeric sequences were removed using USEARCH [9].
2.3. Taxonomic distribution and alpha diversity
The merged sequence data was analyzed using Quantitative Insights into Microbial Ecology (QIIME; v1.9.1) along with Phylogenetic Tools for Analysis of Species-level Taxa (PhylotoAST; v1.4.0) [10], [11]. The initial OTUs were clustered at a 97% similarity through UCLUST in QIIME (v1.9.1) [9], and representative sequences were established by the “most_abundant” option. Then, OTUs with <0.0005% average abundance across all samples were filtered. The redundant OTUs were merged by using the “condense_workflow.py” command through PhyloToAST (v1.4.0) [11]. The OTUs per sample were plotted against the filtered sequence read counts as rarefaction curves, and the data was subsampled to the minimum value using “single_rarefaction.py” in QIIME (v1.9.1). The representative sequences were then assigned taxonomy using the SILVA ACT: Alignment, Classification and Tree Service (www.arb-silva.de/aligner), which utilizes the SILVA Incremental Aligner (SINA; v1.2.11) to align rRNA gene sequences and classify based on Least Common Ancestor (LCA) methods [12]. For this, the SSU (Small Sub-Unit) option selected at a minimum similarity of 0.7 with 20 neighbors per query sequence, and the databases selected were as follows: SILVA database [13], Ribosomal Database Project (RDP) [14], All-Species Living Tree (LTP) project [15], Greengenes (GG) [16], [17], and European Molecular Biology Laboratory (EMBL) [18]. Biological replicates were validated and merged according to their sub-group assignment based on significant Analysis of Similarity (ANOSIM) [19] and Adonis [20] measurements (p = 0.001) using the weighted Unifrac distances [21] calculated for each sample. The top 25 taxa at the highest resolution from each database were combined and plotted as relative abundance graphs using Microsoft Excel Software (Seattle, WA, USA). The taxonomic data derived from each of the five databases is summarized in Table 1 showing the total number of OTUs that were assigned a taxonomy, including the proportion that was resolved to the family and the genus level.
Acknowledgments
We would like to thank Dr. Peter Eipers of the Department of Cell, Developmental and Integrative Biology, and Dr. Michael Crowley of the Heflin Center for Genomic Sciences at the University of Alabama at Birmingham (UAB), for their assistance in high-throughput sequencing. Graduate Research Assistant funding to J.A.H. was provided from grant support to C.D.M by the UAB School of Medicine. The following are acknowledged for their support of the Microbiome Resource at the University of Alabama at Birmingham: School of Medicine, Comprehensive Cancer Center (P30AR050948; C.D.M.), Center for Clinical Translational Science (UL1TR000165; C.D.M.), UAB Microbiome Center (C.D.M.), and Heflin Center. Animal husbandry supported in part by NIH P30DK056336 (S.A.W.).
Contributor Information
Joseph A. Hakim, Email: joe21@uab.edu.
Casey D. Morrow, Email: caseym@uab.edu.
Asim K. Bej, Email: abej@uab.edu.
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Hakim J.A., Koo H., Dennis L.N., Kumar R., Ptacek T., Morrow C.D., Lefkowitz E.J., Powell M.L., Bej A.K., Watts S.A. An abundance of Epsilonproteobacteria revealed in the gut microbiome of the laboratory cultured sea urchin, Lytechinus variegatus. Front. Microbiol. 2015;6:1047. doi: 10.3389/fmicb.2015.01047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hammer H., Hammer B., Watts S., Lawrence A., Lawrence J. The effect of dietary protein and carbohydrate concentration on the biochemical composition and gametogenic condition of the sea urchin Lytechinus variegatus. J. Exp. Mar. Biol. Ecol. 2006;334(1):109–121. [Google Scholar]
- 3.Hakim J.A., Koo H., Kumar R., Lefkowitz E.J., Morrow C.D., Powell M.L., Watts S.A., Bej A.K. The gut microbiome of the sea urchin, Lytechinus variegatus, from its natural habitat demonstrates selective attributes of microbial taxa and predictive metabolic profiles. FEMS Microbiol. Ecol. 2016;92(9):fiw146. doi: 10.1093/femsec/fiw146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kozich J.J., Westcott S.L., Baxter N.T., Highlander S.K., Schloss P.D. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 2013;79(17):5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kumar R., Eipers P., Little R.B., Crowley M., Crossman D.K., Lefkowitz E.J., Morrow C.D. Getting started with microbiome analysis: sample acquisition to bioinformatics. Curr. Protoc. Hum. Genet. 2014;82(1):18.8.1–18.8.29. doi: 10.1002/0471142905.hg1808s82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cock P.J., Fields C.J., Goto N., Heuer M.L., Rice P.M. The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants. Nucleic Acids Res. 2009;38(6):1767–1771. doi: 10.1093/nar/gkp1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andrews S. 2010. FastQC: a Quality Control Tool for High Throughput Sequence Data.https://www.bioinformatics.babraham.ac.uk/projects/fastqc [cited 2019]. Available from: [Google Scholar]
- 8.Gordon A., Hannon G. 2010. Fastx-toolkit. FASTQ/A Short-Reads Pre-processing Tools.http://hannonlab.cshl.edu/fastx_toolkit [cited 2019]. Available from: [Google Scholar]
- 9.Edgar R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26(19):2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 10.Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K., Fierer N., Peña A.G., Goodrich J.K., Gordon J.I., Huttley G.A., Kelley S.T., Knights D., Koenig J.E., Ley R.E., Lozupone C.A., McDonald D., Muegge B.D., Pirrung M., Reeder J., Sevinsky J.R., Turnbaugh P.J., Walters W.A., Widmann J., Yatsunenko T., Zaneveld J., Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7(5):335. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dabdoub S.M., Fellows M.L., Paropkari A.D., Mason M.R., Huja S.S., Tsigarida A.A., Kumar P.S. PhyloToAST: bioinformatics tools for species-level analysis and visualization of complex microbial datasets. Sci. Rep. 2016;6:29123. doi: 10.1038/srep29123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pruesse E., Peplies J., Glöckner F.O. SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics. 2012;28(14):1823–1829. doi: 10.1093/bioinformatics/bts252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., Peplies J., Glöckner F.O. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2012;41(D1):D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cole J.R., Wang Q., Fish J.A., Chai B., McGarrell D.M., Sun Y., Brown C.T., Porras-Alfaro A., Kuske C.R., Tiedje J.M. Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2013;42(D1):D633–D642. doi: 10.1093/nar/gkt1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yilmaz P., Parfrey L.W., Yarza P., Gerken J., Pruesse E., Quast C., Schweer T., Peplies J., Ludwig W., Glöckner F.O. The SILVA and “all-species living tree project (LTP)” taxonomic frameworks. Nucleic Acids Res. 2013;42(D1):D643–D648. doi: 10.1093/nar/gkt1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DeSantis T.Z., Hugenholtz P., Larsen N., Rojas M., Brodie E.L., Keller K., Huber T., Dalevi D., Hu P., Andersen G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006;72(7):5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McDonald D., Price M.N., Goodrich J., Nawrocki E.P., DeSantis T.Z., Probst A., Andersen G.L., Knight R., Hugenholtz P. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012;6(3):610. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kanz C., Aldebert P., Althorpe N., Baker W., Baldwin A., Bates K., Browne P., van den Broek A., Castro M., Cochrane G., Duggan K., Eberhardt R., Faruque N., Gamble J., Garcia Diez F., Harte N., Kulikova T., Lin Q., Lombard V., Lopez R., Mancuso R., McHale M., Nardone F., Silventoinen V., Sobhany S., Stoehr P., Tuli M.A., Tzouvara K., Vaughan R., Wu D., Zhu W., Apweiler R. The EMBL nucleotide sequence database. Nucleic Acids Res. 2005;33(suppl_1):D29–D33. doi: 10.1093/nar/gki098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clarke K.R. Non-parametric multivariate analyses of changes in community structure. Austral Ecol. 1993;18(1):117–143. [Google Scholar]
- 20.Oksanen J., Kindt R., Legendre P., O'Hara B., Simpson G.L., Solymos P., Stevens M.H.H., Wagner H. The vegan package. Community Ecology Package. 2007;10:631–637. [Google Scholar]
- 21.Lozupone C., Lladser M.E., Knights D., Stombaugh J., Knight R. UniFrac: an effective distance metric for microbial community comparison. ISME J. 2011;5(2):169. doi: 10.1038/ismej.2010.133. [DOI] [PMC free article] [PubMed] [Google Scholar]