Abstract
Marine amniotes, a polyphyletic group, provide an excellent opportunity for studying convergent evolution. Their sense of smell tends to degenerate, but this process has not been explored by comparing fully aquatic species with their amphibious relatives in an evolutionary context. Here, we sequenced the genomes of fully aquatic and amphibious sea snakes and identified repertoires of chemosensory receptor genes involved in olfaction. Snakes possess large numbers of the olfactory receptor (OR) genes and the type-2 vomeronasal receptor (V2R) genes, and expression profiling in the olfactory tissues suggests that snakes use the ORs in the main olfactory system (MOS) and the V2Rs in the vomeronasal system (VNS). The number of OR genes has decreased in sea snakes, and fully aquatic species lost MOS which is responsible for detecting airborne odours. By contrast, sea snakes including fully aquatic species retain a number of V2R genes and a well-developed VNS for smelling underwater. This study suggests that the sense of smell also degenerated in sea snakes, particularly in fully aquatic species, but their residual olfactory capability is distinct from that of other fully aquatic amniotes. Amphibious species show an intermediate status between terrestrial and fully aquatic snakes, implying their importance in understanding the process of aquatic adaptation.
Keywords: OR, V1R, V2R, TAAR, amphibious, fully aquatic
1. Background
Shifts between terrestrial and aquatic lifestyles are among the most striking types of evolutionary transitions in the history of life. Vertebrates invaded land during the Devonian to the Carboniferous in two steps: first, they became amphibious (i.e. both aquatic and terrestrial habitats are required) with the emergence of tetrapods, and then they adapted for terrestriality with the emergence of amniotes [1]. Among groups containing mostly terrestrial amniotes, there are several groups which readapted to the aquatic habitat independently from each other. Amniotes are also suggested to have reinvaded water from land with two major steps: they become amphibious prior to the completion of aquatic invasion. For example, all extant cetaceans are fully aquatic, but their intermediate ancestors from the Early Eocene were amphibious [2] (figure 1). Marine elapids (Suborder Serpentes, Order Squamata, Class Reptilia), collectively called sea snakes, consist of two monophyletic clades: Laticaudini and Hydrophiini. Laticaudins are oviparous and lay eggs on land, whereas hydrophiins are viviparous and spend all their life in water. Both groups have a paddle-shaped tail adapted to aquatic locomotion, but laticaudins retain enlarged ventrals required for terrestrial locomotion which hydrophiins lost [5]. Although recent studies have suggested that laticaudins and hydrophiins adapted to the marine habitat independently, these two clades are phylogenetically close to each other with a divergence time of approximately 12–20 Ma [6–9]. Thus, sea snakes provide an excellent study system of aquatic adaptation because phylogenetically closely related fully aquatic and amphibious species can be compared directly.
Figure 1.
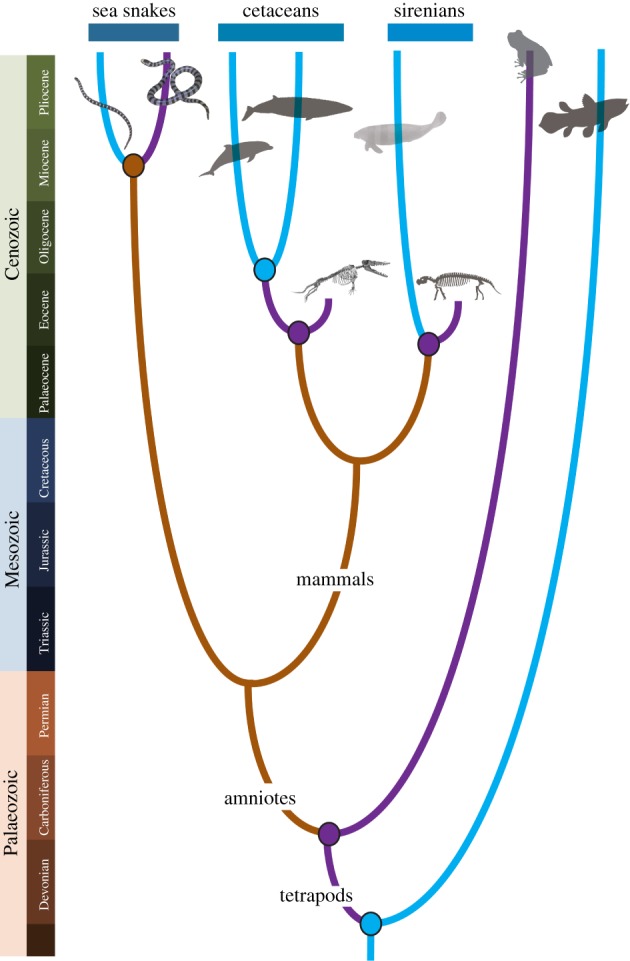
A schematic view of the evolution of terrestrial adaptation of vertebrates and three major groups of extant fully aquatic amniotes. Branch colour indicates representative lifestyle in each branch (brown, terrestrial; purple, amphibious; blue, fully aquatic), and circles in ancestral nodes represent lifestyles at these points in evolution. Extinct amphibious species are also shown for cetaceans (Amburocetus [3]) and sirenians (Pezosiren [4]). (Online version in colour.)
Aquatic amniotes offer a valuable opportunity for studying convergent evolution because evolutionary hypotheses of specific adaptation can be tested for multiple aquatic groups that independently migrated from land to water [10,11]. One of the most remarkable differences between terrestrial and aquatic vertebrates involves the sense of smell. Broad taxa of vertebrates detect odourants mainly using four major groups of G-protein-coupled receptors (GPCRs) encoded by different multigene families: olfactory receptors (ORs), trace amine-associated receptors (TAARs), and two types of vomeronasal receptors (V1Rs and V2Rs) [12]. It has been hypothesized that olfactory GPCRs are functionally divided into two groups: receptors for airborne molecules and those for water-soluble molecules [12,13]. The OR gene repertoire changed drastically in our ancestors during their transition from water to land, and amphibians show an intermediate form. Modern anurans retain mostly ancestral OR gene subfamilies for detecting water-soluble molecules which amniotes lost, but they also share newly diverged OR gene subfamilies with amniotes which are thought to detect airborne odourants [14–16]. Aquatic tadpoles (larvae of amphibians) possess olfactory organs for smelling underwater, but extreme remodelling occurs during metamorphosis to meet the requirement of the adult lifestyle, and adult anurans develop a so-called ‘air nose’ for smelling in the air [17], in which the newly diverged OR genes are expressed [18]. The OR genes possessed by terrestrial amniotes are prone to secondary loss from the genomes of aquatic amniotes [19–23], and extant toothed whales possess no olfactory nervous systems [24]. Baleen whales, the other group of extant cetaceans, still possess a functional olfactory system, but anatomical, histological, and genomic studies suggest that they cannot smell underwater and their olfactory capability is highly limited [25–28]. However, olfactory capabilities of non-cetacean aquatic amniotes remain largely elusive. Furthermore, no extensive studies on genomes of amphibious amniotes have ever been compared and contrasted with those of fully aquatic relatives, in spite of their importance upon aquatic adaptation. Here, we sequenced and assembled the genomes of fully aquatic and amphibious sea snakes. Olfactory GPCR genes were identified in each genome assembly, and the expression profiling of these receptors was performed. Our present study explores the genomic traces of evolution of olfaction in sea snakes and provides new perspectives on the aquatic adaptation of amniotes.
2. Results
(a). Sea snake genome assemblies
We sequenced and assembled the genomes of two hydrophiins (Hydrophis melanocephalus and Emydocephalus ijimae) and two laticaudins (Laticauda laticaudata and L. colubrina). These genome assemblies are estimated to contain at least 90% of all protein-coding genes (including those recognized as ‘Fragmented’ by BUSCO [29,30] or ‘Partial’ by CEGMA [31], see Materials and methods) based on completeness assessments using a one-to-one reference orthologue set (electronic supplementary material, table S1).
Different methodologies were employed for performing de novo assembly of the genomes of four snakes (see Material and methods for details). We assembled the genome of L. colubrina based on the linked-read sequencing technology [32,33]. It is known that this method allows us to generate relatively long genome sequences of diploid species with a single library for short-read sequencing [34]. Among the four species sequenced in this study, the L. colubrina assembly shows the largest scaffold N50 length (3.1 Mb) and completeness score of one-to-one orthologue coverage (electronic supplementary material, table S1). However, the proportion of truncated genes in the L. colubrina genome assembly does not differ greatly in comparison with other assemblies (electronic supplementary material, table S2). Consistently, the contig N50 lengths do not largely vary between the assemblies of the four species (electronic supplementary material, table S1).
(b). Olfactory GPCR gene repertoires in snake genomes
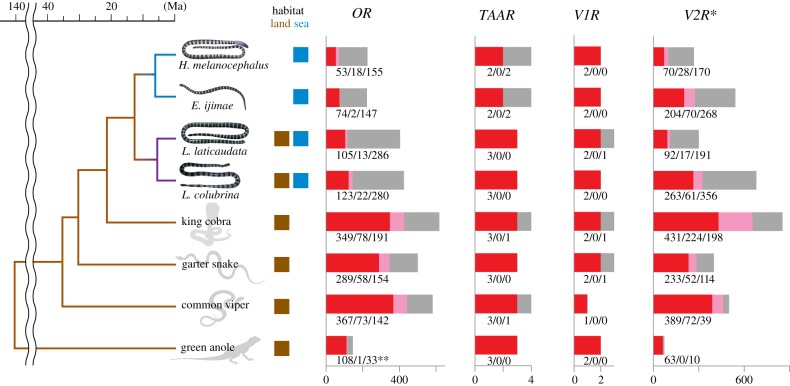
We identified the olfactory GPCR genes in the genome assemblies of sea snakes and their terrestrial relatives. Snakes possess large numbers of ORs and V2Rs, which vary between species (mean numbers of intact ORs and V2Rs are 194 ± 136 and 240 ± 136 (mean ± s.d., calculated using all snake species shown in figure 2), respectively), whereas the numbers of TAARs and V1Rs are small and comparable across species (snakes possess only two or three intact TAARs and two intact V1Rs, as described below) (figure 2).
Figure 2.
Phylogenetic relationship of squamates analysed in this study, and the numbers of olfactory GPCR genes identified in the genome assemblies of these species. Red, pink, and grey bars indicate the numbers of intact genes, truncated genes, and pseudogenes, respectively. Approximate divergence time follows Sanders et al. [6–8] and Kim et al. [35]. *Only the third exon of the V2R genes was identified and analysed. **The OR gene repertoire of a green anole is taken from Vandewege et al. [36]. (Online version in colour.)
Sea snakes possess a smaller number of intact OR genes with higher proportions of pseudogenes (electronic supplementary material, table S2) compared with terrestrial snakes (mean number of intact ORs of hydrophiins: 63.5 ± 14.9, laticaudins: 114 ± 12.7, terrestrial snakes: 335 ± 40.8 (mean ± s.d.)) mainly due to massive loss of the OR genes in the sea snake lineages (figure 3a). They also possess a relatively small number of intact V2Rs, but the numbers of intact V2R genes vary greatly between species (mean number of intact V2Rs of hydrophiins: 137 ± 95, laticaudins: 177.5 ± 121, terrestrial snakes: 351 ± 104 (mean ± s.d.)), and massive gain of the V2R genes is observed in two species of sea snakes, E. ijimae and L. colubrina (figure 3b). Snakes possess two intact TAARs (TAAR1 and TAAR5). In addition, terrestrial snakes and laticaudins possess one more intact TAAR gene (TAAR2, which is pseudogenized in the hydrophiin genomes). All snakes including sea snakes (except for the common viper) possess two intact V1Rs, the ancV1R [38] and a V1R gene which is not orthologous to the mammalian V1Rs (Squamata-V1R; electronic supplementary material, figure S1).
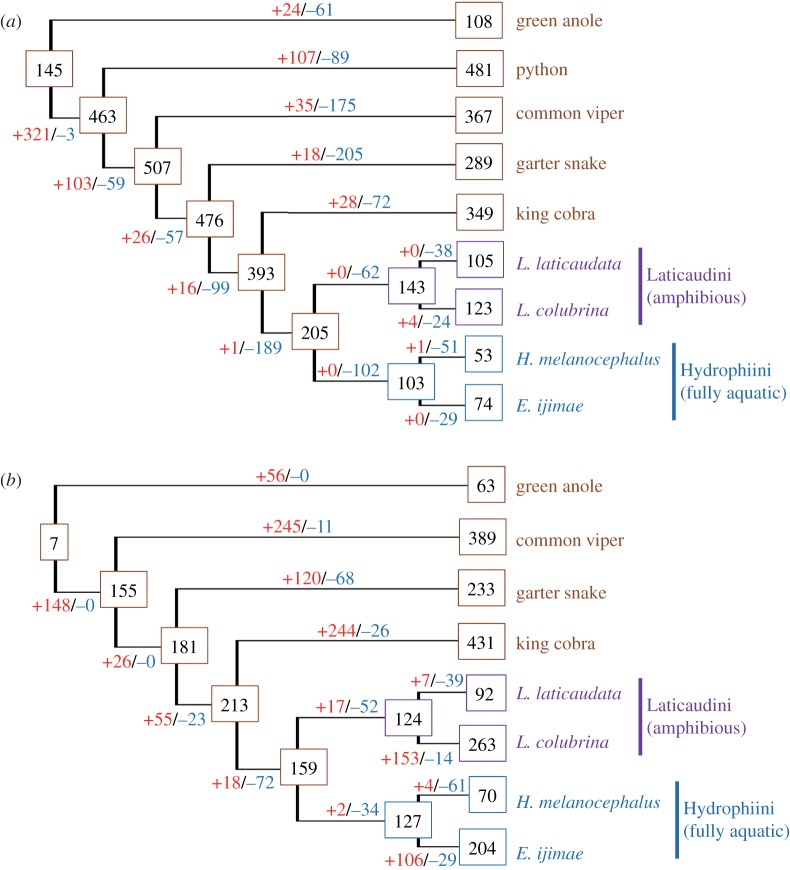
Figure 3.
Evolution of the gain and loss of OR and V2R genes in snakes. Evolutionary changes in the number of intact OR (a) and V2R (b) genes are estimated using the reconciled tree method [37]. Python intact ORs identified by Vandewege et al. [36] were included in the dataset for this calculation. (Online version in colour.)
(c). Expression of the olfactory GPCR genes
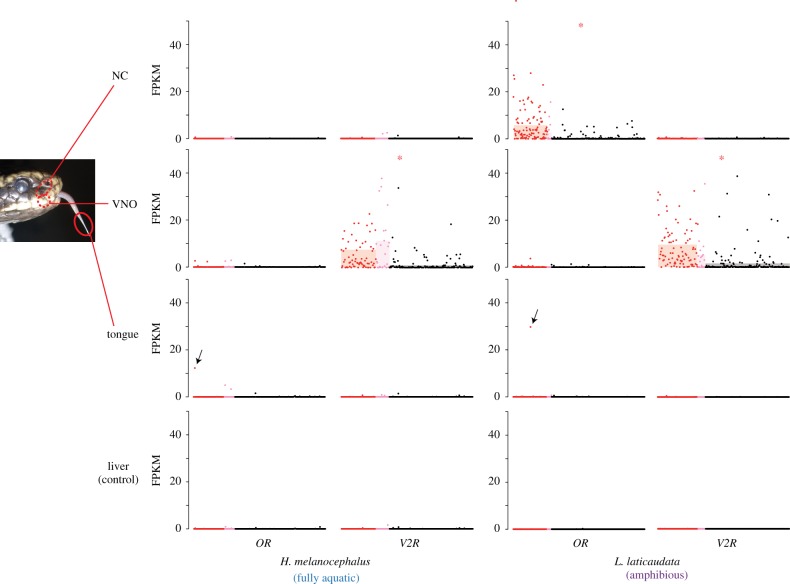
There are two anatomically distinctive olfactory systems in terrestrial snakes: the main olfactory system (MOS) and the vomeronasal system (VNS). The olfactory epithelium of MOS (i.e., the main olfactory epithelium) is located in the nasal cavity (NC) and that of VNS (vomeronasal epithelium) is located in the vomeronasal organ (VNO) [39]. The snake tongue also plays a role in VNS by delivering chemicals to the VNO [39]. We performed transcriptome sequencing with RNA-seq on these potential olfactory organs of L. laticaudata and H. melanocephalus. The expression pattern of these genes in L. laticaudata suggests that most of the ORs are used in MOS, while V2Rs function in VNS (figure 4). It is noted that some pseudogenes are also expressed in the olfactory organs, coinciding with a previous report [40]. Expression levels of V1Rs and TAARs suggest that the ancV1R is expressed in the VNO of both species, and the TAAR2, which is pseudogenized in the hydrophiin genomes, is expressed in the NC of L. laticaudata (electronic supplementary material, table S3).
Figure 4.
Expression levels of the OR and V2R genes in the three potential olfactory organs and the liver. Each dot represents a single OR/V2R gene identified in this study, and the y-axis shows normalized gene expression levels in FPKM values. Red, pink, and black dots represent intact genes, truncated genes, and pseudogenes, respectively. The mean FPKM values of intact, truncated, and pseudogenes in each organ are shown as bars in the background. Difference of the mean FPKM values of intact OR/V2R genes between each chemosensory organ and a control (liver) is calculated, and chemosensory organs with obviously (greater than 1) and significantly (p < 0.01, paired t-test) larger FPKM values compared to the control are shown with asterisks (see electronic supplementary material, table S5 for details). Arrows indicate an intact OR gene expressed in the tongue. Approximate position of each organ in a fully aquatic hydrophiin (H. melanocephalus) is also shown. (Online version in colour.)
An intact OR gene is expressed in the tongue in each snake species (figure 4, indicated by arrows). The arrowed gene of Hydrophis and that of Laticauda is orthologous to each other. All squamates investigated in this study (except for Emydocephalus) possess one-to-one orthologues of this OR gene (electronic supplementary material, figure S2).
3. Discussion
(a). Evolution of the main olfactory system
The repertoires and expression pattern of the olfactory GPCR genes shown in this study suggest that snakes mainly use ORs in MOS and V2Rs in VNS. It has been widely considered that VNS is the predominant chemosensory system in snakes, being more responsible than MOS for their sense of smell [39,41]. However, in our results, the numbers of intact ORs and V2Rs are almost comparable among terrestrial snakes (figure 2), implying that terrestrial snakes potentially detect and discriminate as many chemicals using MOS as using VNS.
Sea snakes possess an apparently smaller number of intact OR genes compared with terrestrial snakes, and our phylogenetic analysis suggests that it is mainly because of massive loss of the OR genes in the sea snake lineages after the king cobra–sea snakes split (figure 3a). Although the most recent common ancestor of hydrophiins and laticaudins, which lived on land, was also estimated to possess a smaller number (205) of intact OR genes than other terrestrial snakes (figure 3a), it is possible that hydrophiins and laticaudins lost OR orthologues in their unique lineages independently. In any case, massive loss of OR genes was also confirmed in both hydrophiin and laticaudin lineages after the Hydrophiini–Laticaudini split, coinciding with their transition from land to water. Amphibious carnivorans (pinnipeds and otters) and fully aquatic cetaceans were also estimated to have lost a large number of intact OR genes when they migrated from land to water [23,26], showing a remarkable case of convergent evolution on becoming aquatic. Although both hydrophiins and laticaudins possess fewer intact OR genes, their expression patterns are different from each other. Most of the OR genes possessed by L. laticaudata are expressed in the NC, while those possessed by H. melanocephalus are not (figure 4). This contrast indicates that most of the OR genes possessed by hydrophiins do not have an olfactory function and that hydrophiins lost a functional MOS. The loss of MOS in hydrophiins is also supported by the evolution of the TAAR genes—the TAAR2 gene, which is expressed in the NC of laticaudins (electronic supplementary material, table S3), is pseudogenized in the hydrophiin genomes. Histological studies showed that a relative size of the olfactory region in the NC is highly reduced in sea snakes, and in particular, that of hydrophiins lacks an external nasal gland which lubricates the olfactory epithelium [42,43]. The role of MOS is poorly understood in snakes, but these findings suggest that MOS became less useful for snakes to sense the surrounding environment on becoming aquatic, and it was completely lost in fully aquatic hydrophiins, probably because the snake MOS functions only in the air.
(b). Evolution of the vomeronasal system
The evolutionary pattern of the gain and loss of V2R genes differs from that of ORs in snakes. Although sea snakes possess a relatively small number of intact V2Rs, the numbers of intact V2R genes vary largely even among hydrophiins and laticaudins (figure 2). Snakes are known to have a pair of well-developed VNOs [39,42], and fully aquatic hydrophiins are no exception [42–44]. Most of the intact V2Rs are expressed in the VNO even in the case of hydrophiins (figure 4). The snake VNO is linked to the oral cavity, and snakes deliver odour molecules to their VNOs through tongue-flicking [41]. Underwater tongue-flicking is widely observed among squamates [42] including hydrophiins [42,45], and Hydrophis can distinguish fish species solely by tongue-flicking [46]. All these pieces of evidence strongly suggest that the hydrophiin VNS is functional, and that sea snakes can smell underwater through tongue-flicking. A recent study reported that the presence/absence of a V1R gene named ancV1R corresponds to the presence/absence of the functional VNO among tetrapods [38]. We found an intact V1R gene orthologous to the ancV1R in each snake genome (electronic supplementary material, figure S1), and expression of this V1R in the VNO is confirmed in hydrophiins (electronic supplementary material, table S3), supporting the suggestion that the hydrophiin VNS is functional. Gene duplication and loss of the V2R genes are more frequent than that of OR genes, and massive gain of the V2Rs is observed even in two species of sea snakes, E. ijimae and L. colubrina (figure 3b). Previous studies showed that Emydocephalus relies heavily on VNS for foraging [45], while not only olfaction but also vision plays an important role for Hydrophis to find prey [46]. This implies that Emydocephalus relies more on olfaction than Hydrophis, which is consistent with our results that Emydocephalus possesses a larger number of intact V2Rs than Hydrophis. Olfactory capabilities through VNS may differ largely between snake species including sea snakes.
We found an OR gene expressed in the tongue. This gene is conserved among snake species, implying that the function of this OR is evolutionarily maintained and important for snakes. Unlike most of the other OR genes, the expression of this gene is confirmed even in hydrophiins (figure 4). Snakes do not use the tongue as a gustatory organ because it lacks taste buds [41]. Still, the tongue may be used as a chemosensory organ which enables efficient tongue-flicking of snakes. Further studies are required for testing this hypothesis.
(c). Olfactory capabilities of sea snakes
In this study, we investigated the molecular basis of snake olfaction and showed the presence of VNS but the absence of MOS in fully aquatic hydrophiins. Although hydrophiins cannot smell in the air using MOS, they smell underwater using VNS. To our knowledge, hydrophiins are the only vertebrates which possess a functional VNS without the presence of MOS. The functional VNS is absent from all extant fully aquatic mammals (cetaceans and sirenians) though their terrestrial relatives (artiodactyls and terrestrial afrotherians) have it [47], indicating that VNS is required only on land in most mammals. However, squamates are understood to smell underwater using VNS. This may be because V2Rs, which are abundant in diverse aquatic vertebrates and putatively detect water-soluble molecules, are predominant in squamate genomes over V1Rs, which have diversified in mammals after their terrestrial invasion to detect odourants on land [48–52]. Modern anurans, particularly in the larval form, also use VNS for smelling underwater [17] via the VNOs in which V2Rs are predominantly expressed [52–54].
The olfactory capability of amphibious laticaudins is speculated to be an intermediate between terrestrial snakes and hydrophiins: they still possess a functional MOS, but their OR gene repertoire has largely degenerated. This is consistent with our assumption that amphibious species are intermediates between fully terrestrial and fully aquatic. Amphibious mammals also tend to show an intermediate status between fully terrestrial and fully aquatic mammals. For example, the majority of pinnipeds retain putatively functional VNS, while some species such as harbour seals lack it [55]. Careful interpretation is required for amphibious species when studying convergent evolution among marine amniotes.
(d). Comparison among fully aquatic amniotes
Our results show that hydrophiins, which adapted to water independently from aquatic mammals, also profoundly reduced olfaction. However, the residual olfactory abilities are very different between fully aquatic mammals and hydrophiins. Baleen whales smell in the air using a highly degenerated but functional MOS [25,28]. Little is known about olfactory capacities of sirenians, but they also possess a putatively functional though degenerated MOS [47,56], and their olfactory anatomy suggests that they smell in the air, not underwater [56]. On the other hand, hydrophiins possess well-developed VNOs, and behavioural studies suggest that they smell underwater using VNS [45,46] (table 1). The well-developed underwater-functional VNS of sea snakes is derived from the V2R-predominant well-developed snake VNS, and the difference of the olfactory capabilities between hydrophiins and fully aquatic mammals is explained by the difference of the olfactory capabilities between their terrestrial ancestors. Underwater olfaction might have been favoured by natural selection if it was adaptive for whales and sirenians to survive in water, but they have never acquired underwater-functional olfactory systems. This shows a striking contrast with the terrestrial adaptation of vertebrates. Tetrapods modified their olfactory organs and generated novel OR gene subfamilies for smelling in the air when they migrated from water to land [15,17]. In addition, adults of secondarily aquatic pipid frogs acquired an additional olfactory epithelium called ‘water nose’ for smelling underwater [52,57–59]. But no amniotes are known to have modified their olfactory organs for sensing the newly invaded environment upon aquatic adaptation. Histological and genomic studies imply that whales lack innate avoidance behaviour against predator odours probably because their predators cannot be detected by smelling in the air [26,27]. Amniotes belonging to various taxa have to adapt themselves to handle similar problems inflicted by their new environment upon aquatic adaptation. However, not only the ecological demands but also phylogenetic backgrounds play important roles in the formation of sensory modalities in this process.
Table 1.
Olfactory capabilities of extant fully aquatic amniotes.
| MOS | VNS | references | ||
|---|---|---|---|---|
| Cetacea | Odontoceti | absent | absent | [24,47] |
| Mysticeti | present, smelling in the air | absent | [25–28] | |
| Sirenia | present, smelling in the air | absent | [47,56] | |
| Hydrophiini | absent | present, smelling underwatera | [42–46], this study |
aIt remains unknown whether hydrophiins use the VNS for smelling in the air or not.
4. Material and methods
(a). Specimens
The following specimens were used for DNA/RNA extraction. Specimens used for genome sequencing were not used for RNA extraction in order to save all internal/external organs for future studies. All specimens used in the study are deposited in the Zoological Collection of the Kyoto University Museum (KUZ) with the specimen vouchers shown below.
Laticauda laticaudata Linnaeus 1758 (blue-lipped sea krait, Laticaudini)
-
1.
Specimen voucher: KUZ R72402; sex: male; locality: Okinawa Island, Japan (genome sequencing; electronic supplementary material, figure S3A).
-
2.
Specimen voucher: KUZ R68692; sex: female; locality: Okinawa Island, Japan (RNA sequencing).
Hydrophis melanocephalus Gray 1849 (slender-necked sea snake, Hydrophiini)
-
1.
Specimen voucher: KUZ R72403; sex: male; locality: Okinawa Island, Japan (genome sequencing; electronic supplementary material, figure S3B).
-
2.
Specimen voucher: KUZ R73056; sex: female; locality: Okinawa Island, Japan (RNA sequencing).
Laticauda colubrina Schnieder 1799 (yellow-lipped sea krait, Laticaudini)
Specimen voucher: KUZ R77260; sex: male; locality: Ishigaki Island, Japan (genome sequencing).
Emydocephalus ijimae Stejneger 1898 (turtlehead sea snake, Hydrophiini)
Specimen voucher: KUZ R72604; sex: male; locality: Okinawa Island, Japan (genome sequencing).
(b). DNA extraction, sequencing, and performing de novo assembly
DNA was extracted manually, following the methods of Blin & Stafford [60] with modifications, from muscle tissues of specimens KUZR72402, KUZR72403, and KUZR72604. Whole-genome shotgun (WGS) sequences were generated using Illumina platforms. Paired-end libraries were prepared using the TruSeq DNA PCR-free Sample Prep Kit (Illumina) (specimen KUZR72604) and the TruSeq Nano DNA Sample Prep Kit (Illumina) (KUZR72402 and KUZR72043). Mate-pair libraries were prepared following Tatsumi et al. [61] having a size range of 6–10 kb with a peak of around 7 kb. A PacBio RS II sequencer was also employed for sequencing the L. laticaudata genome using the PacBio DNA Template Prep Kit 1.0 (Pacific Biosciences). The details of sequencing results are provided in electronic supplementary material, table S4, and k-mer frequency spectrum of the WGS reads of each specimen is shown in electronic supplementary material, figure S4. Platanus_trim and Platanus_internal_trim [62] were employed to trim low-quality regions and adapters of paired-end and mate-pair sequences, respectively, with default parameters, except for reads of L. colubrina. Regarding PacBio long-reads, the following filtering criteria were used to obtain subreads from the polymerase reads: minimum subread length 50 and minimum polymerase read quality 0.75. For the L. colubrina (specimen KUZR77260), high-molecular weight (HMW) DNA was extracted from the liver using Nuclei PURE Prep Kit (Sigma). Using the Chromium System (10xGenomics), a linked-read library was constructed from 1.25 ng of HMW DNA of 50 kb or longer. The library was sequenced on an Illumina HiSeq platform and then processed using Supernova v1.2.2 [32,33] with default settings. Assembling the L. laticaudata genome: trimmed paired-end reads were used to construct contig assembly using the PLATANUS v. 1.2.4 [62] with a step size of k-mer extension set at 1. Scaffolds were constructed based on this contig assembly using the Redundans v0.12c [63]. Finally, the PacBio subreads were merged, and gap-closing was performed using the pbjelly software in the PBSuite package v. 15.8.24 [64,65]. The H. melanocephalus genome: PLATANUS v. 1.2.4 [62] was employed for contig assembling, scaffolding, and gap-closing with a step size of k-mer extension set to be 1. Only paired-end reads were used for contig assembling, and then, mate-pair reads were added for scaffolding and gap-closing. The E. ijimae genome: SOAPdenovo2 [66] was employed for contig assembling, scaffolding, and gap-closing with a k-mer set to be 81. Completeness of these genome assemblies was evaluated using CEGMA v. 2.5 [31] and BUSCO v. 3 [29,30] referring to the orthologue set CVG [67].
In addition to these sea snake genome assemblies, genome assemblies of terrestrial snakes closely related to sea snakes (king cobra Ophiophagus hannah (Elapidae, GenBank accession no. GCA_000516915.1), garter snake Thamnophis sirtalis (Colubridae, GCA_001077635.2), and common viper Vipera berus (Viperidae, GCA_000800605.1)) and a green anole Anolis carolinensis (Iguanidae, GCA_000090745.2) were obtained from the GenBank FTP server.
(c). Identification of the olfactory GPCR genes
(i). OR genes
The OR genes were searched against the genome assemblies of seven snake species (four sea snake genomes assembled in this study, and three terrestrial snake genomes retrieved from the GenBank) using the tblastn program in the BLAST+ v. 2.6.0 package [68] with the cut-off E-value of 1 × 10−5. Deduced amino acid sequences of all intact OR genes of the green anole and the western clawed frog identified by Niimura [69] were used as queries. Each sequence thus obtained was searched against the GenBank protein database using the blastx program and was discarded if its best hit was not an OR. A sequence was judged to be a non-functional pseudogene if the sequence was interrupted by premature stop codon(s) and/or frame shift(s), or it lacked five or more consecutive amino acids including a trans-membrane domain. If a sequence was interrupted by contig-gap(s) although it was not judged to be a pseudogene, it was labelled as ‘truncated’. The OR gene repertoires of the anole and the python were retrieved from the dataset provided by Vandewege et al. [36].
(ii). TAAR and V1R genes
Essentially, a uniform method employed to find the OR genes was used to identify intronless TAAR and V1R genes from the genome assemblies of seven snakes and a green anole with the following amino acid sequences as queries: TAAR, all intact mammal TAARs identified by Kishida et al. [26]; V1R, all intact vertebrate V1Rs identified by Zapilko & Korsching [51].
(iii). V2R genes
Only the longest exon (third exon, approx. 800 bp) of V2R genes was analysed in this study because V2Rs are multi-exon genes and it is difficult to identify all exons derived from a V2R gene if exons are scattered in two or more scaffolds. Using deduced amino acid sequences of the third exon of all intact vertebrate V2R genes identified by Shi & Zhang [48] as queries, V2R sequences were searched and identified based on the same method employed to identify OR genes.
(d). RNA extraction, sequencing, and expression analyses
Total RNA was extracted from three potential olfactory organs (the VNO, the NC, and the tongue) and the liver (as a negative control) of specimens KUZR68692 (L. laticaudata) and KUZR73056 (H. melanocephalus) using the RNeasy Mini Kit (Qiagen) and following the manufacturer's guidelines. Regarding the NC, we sampled entire tissues around the NC broadly from both specimens for RNA extraction because the olfactory epithelia of both specimens were hard to locate exactly. It is noted that this might cause reduction in the estimated expression level of chemosensory receptors due to contamination of non-olfactory tissues. The extracted RNA was used to construct paired-end sequencing libraries using the TruSeq RNA Sample Prep Kit v. 2 (Illumina), and these libraries were sequenced using an Illumina HiSeq platform (2 × 101 bp). As a result, the following sizes of RNA-seq reads were obtained: [KUZR68692] VNO 5.19 Gb, NC 5.27 Gb, tongue 5.91 Gb, and liver 5.24 Gb; [KUZR73056] VNO 4.73 Gb, NC 5.74 Gb, tongue 5.38 Gb, and liver 8.46 Gb. Low-quality sequences and adapters were removed using the Trimmomatic v. 0.36 [70] with the following parameters: ILLUMINACLIP:TruSeq3-PE-2.fa: 2:30:10, LEADING: 20, TRAILING: 20, SLIDINGWINDOW: 5:25, and MINLEN: 36. Trimmed RNA-seq reads were mapped to the conspecific genome assembly using HISAT2 [71] v. 2.1.0 with default parameters. Gene expression levels were quantified with fragments per kilobase of exon per million mapped fragments (FPKM) values using Cufflinks [72,73] v. 2.2.1 after the removal of duplicated reads.
(e). Phylogenetic analyses
Deduced amino acid sequences were aligned using the l-ins-i program in the MAFFT package v. 7.266 [74], and gap sites were excluded from further analyses. Trees were inferred using the neighbour-joining method [75] based on the Poisson-corrected distance matrices. Evolutionary changes in the number of OR/V2R genes were inferred using the reconciled tree method [37]. Amniote ORs are clearly classified into two classes: class I and class II [76]. All intact OR genes were classified into 35 clades identified by Niimura & Nei (a class I clade and 34 class II clades) [37] based on sequence similarities, and a calculation was performed for each clade separately. Eight human class I ORs (OR51Q1, OR51G1, OR51L1, OR51I1, OR52K1, OR52H1, OR52B4, and OR56A1) retrieved from the HORDE database [77] build #44 were used as outgroups for class II OR trees; 16 human class II ORs (OR1C1, OR1Q1, OR2C1, OR5F1, OR5J2, OR5P3, OR6B2, OR6N1, OR7D4, OR8D2, OR8U1, OR9Q2, OR10A3, OR10K1, OR11H4, and OR13D1) for a class I OR tree. Vandewege et al. [36] reported the OR gene repertoire of a python, a phylogenetically distant snake species which possesses a much larger number of intact OR genes (481) than that of any snakes investigated in this study. We included the python intact ORs identified by them for this analysis to estimate the ancestral OR gene repertoires thoroughly. V2R genes were classified into two clades (families C and non-C [49]) based on a phylogenetic tree using green anole Tas1Rs (Tas1R1, GenBank accession no. XM_016998922; Tas1R2, XM_008124605; Tas1R3, XM_003228934) as outgroups, and the evolutionary gains and losses of non-C V2Rs were calculated using the family-C V2Rs as outgroups. Because all snakes possess exactly one family-C V2R, we concluded that the number of family-C V2R did not change through the evolution of snakes. Bootstrap values were obtained by 500 resamplings, and a bootstrap value of 70% was used as a threshold for reconciliation. Truncated genes and pseudogenes were excluded from this calculation. Phylogenetic trees of class I and class II OR genes are shown in electronic supplementary material, figures S5 and S2, respectively, and changes in the number of class I and class II OR genes are shown in electronic supplementary material, figure S6.
Supplementary Material
Acknowledgements
We are grateful to Prof. Tsutomu Hikida for helping dissection of snakes; Takahide Sasai, Koji Mochida, and members of Tropical Biosphere Research Center, University of the Ryukyus for supporting sampling and fixation of specimens; the technical staff at Latobatory of Phyloinformatics in RIKEN BDR for obtaining mate-pair reads; the technical staffs at Cognitive Genomics Research Group in ExCELLS, NINE for constructing the linked-read library; Dr Rob Ogden for checking the English of the text and reviewers of this manuscript for helpful comments. Computations were partially performed on the NIG supercomputer at ROIS National Institute of Genetics.
Data accessibility
Specimens: Zoological Collection of the Kyoto University Museum (KUZ) with specimen vouchers KUZ R68692, R72402, R72403, R72604, R73056, and R77260. Sequencing data and assembled genome sequences: GenBank BioProject accessions PRJDB7221 (E. ijimae genome sequencing), PRJDB7226 (L. laticaudata genome sequencing), PRJDB7271 (H. melanocephalus genome sequencing), PRJDB7284 (L. colubrina genome sequencing), PRJDB7257 (L. laticaudata RNA-seq), and PRJDB7258 (H. melanocephalus RNA-seq). The locus of each gene (electronic supplementary material, tables S6–S13) and amino acid sequences of intact olfactory GPCRs (electronic supplementary material, data S1) identified in this study is available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.t8sm4m6 [78].
Authors' contributions
T.K. designed this study, sequenced and assembled genomes of sea snakes, analysed data, and wrote the paper. T.K. and M.T. sampled, dissected, and fixed specimens. Y.G. and S.T. sequenced and assembled the L. colubrna genome. S.K. and K.T. participated in the H. melanocephalus genome sequencing. T.K., Y.G., and S.K. finalized the manuscript.
Competing interests
We declare we have no competing interests.
Funding
This work was supported by JSPS KAKENHI (grant nos. 15K07184 and 18K06378 to T.K., grant no. 16H06531 to Y.G.) and the Collaborative Research of Tropical Biosphere Research Center, University of the Ryukyus.
References
- 1.Benton MJ. 2008. The history of life. New York, NY: Oxford University Press. [Google Scholar]
- 2.Thewissen JGM, Cooper LN, George JC, Bajpai S. 2009. From land to water: the origin of whales, dolphins, and porpoises. Evol. Educ. Outreach 2, 272–288. ( 10.1007/s12052-009-0135-2) [DOI] [Google Scholar]
- 3.Thewissen JGM, Hussain ST, Arif M. 1994. Fossil evidence for the origin of aquatic locomotion in archaeocete whales. Science 263, 210–212. ( 10.1126/science.263.5144.210) [DOI] [PubMed] [Google Scholar]
- 4.Domning DP. 2001. The earliest known fully quadrupedal sirenian. Nature 413, 625–627. ( 10.1038/35098072) [DOI] [PubMed] [Google Scholar]
- 5.Heatwole H. 1999. Sea snakes. Sydney, New South Wales: University of New South Wales Press. [Google Scholar]
- 6.Sanders KL, Mumpuni LMS. 2010. Uncoupling ecological innovation and speciation in sea snakes (Elapidae, Hydrophiinae, Hydrophiini). J. Evol. Biol. 23, 2685–2693. ( 10.1111/j.1420-9101.2010.02131.x) [DOI] [PubMed] [Google Scholar]
- 7.Sanders KL, Lee MS, Leys R, Foster R, Keogh JS. 2008. Molecular phylogeny and divergence dates for Australasian elapids and sea snakes (Hydrophiinae): evidence from seven genes for rapid evolutionary radiations. J. Evol. Biol. 21, 682–695. ( 10.1111/j.1420-9101.2008.01525.x) [DOI] [PubMed] [Google Scholar]
- 8.Sanders KL, Lee MS. 2008. Molecular evidence for a rapid late-Miocene radiation of Australasian venomous snakes (Elapidae, Colubroidea). Mol. Phylogenet. Evol. 46, 1165–1173. ( 10.1016/j.ympev.2007.11.013) [DOI] [PubMed] [Google Scholar]
- 9.Rasmussen AR, Murphy JC, Ompi M, Gibbons JW, Uetz P. 2011. Marine reptiles. PLoS ONE 6, e27373 ( 10.1371/journal.pone.0027373) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thewissen JGM, Nummela S. 2008. Introduction: on becoming aquatic. In Sensory evolution on the threshold: adaptations in secondarily aquatic vertebrates (eds Thewissen JGM, Nummela S), pp. 1–25. Berkeley, CA: University of California Press. [Google Scholar]
- 11.Foote AD, et al. 2015. Convergent evolution of the genomes of marine mammals. Nat. Genet. 47, 272–275. ( 10.1038/ng.3198) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nei M, Niimura Y, Nozawa M. 2008. The evolution of animal chemosensory receptor gene repertoires: roles of chance and necessity. Nat. Rev. Genet. 9, 951–963. ( 10.1038/nrg2480) [DOI] [PubMed] [Google Scholar]
- 13.Korsching S. 2016. Aquatic olfaction. In Chemosensory transduction (eds Zufall F, Munger SD), pp. 81–100. New York, NY: Academic Press. [Google Scholar]
- 14.Niimura Y, Nei M. 2005. Evolutionary dynamics of olfactory receptor genes in fishes and tetrapods. Proc. Natl Acad. Sci. USA 102, 6039–6044. ( 10.1073/pnas.0501922102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Niimura Y. 2009. Evolutionary dynamics of olfactory receptor genes in chordates: interaction between environments and genomic contents. Hum. Genom. 4, 107–118. ( 10.1186/1479-7364-4-2-107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mezler M, Fleischer J, Breer H. 2001. Characteristic features and ligand specificity of the two olfactory receptor classes from Xenopus laevis. J. Exp. Biol. 204, 2987–2997. [DOI] [PubMed] [Google Scholar]
- 17.Reiss JO, Eisthen HL. 2008. Comparative anatomy and physiology of chemical senses in amphibians. In Sensory evolution on the threshold: adaptations in secondarily aquatic vertebrates (eds Thewissen JGM, Nummela S), pp. 43–63. Berkeley, CA: University of California Press. [Google Scholar]
- 18.Freitag J, Krieger J, Strotmann J, Breer H. 1995. Two classes of olfactory receptors in Xenopus laevis. Neuron 15, 1383–1392. ( 10.1016/0896-6273(95)90016-0) [DOI] [PubMed] [Google Scholar]
- 19.Hayden S, Bekaert M, Crider TA, Mariani S, Murphy WJ, Teeling EC. 2010. Ecological adaptation determines functional mammalian olfactory subgenomes. Genome Res. 20, 1–9. ( 10.1101/gr.099416.109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kishida T, Hikida T. 2010. Degeneration patterns of the olfactory receptor genes in sea snakes. J. Evol. Biol. 23, 302–310. ( 10.1111/j.1420-9101.2009.01899.x) [DOI] [PubMed] [Google Scholar]
- 21.Kishida T, Kubota S, Shirayama Y, Fukami H. 2007. The olfactory receptor gene repertoires in secondary-adapted marine vertebrates: evidence for reduction of the functional proportions in cetaceans. Biol. Lett. 3, 428–430. ( 10.1098/rsbl.2007.0191) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McGowen MR, Gatesy J, Wildman DE. 2014. Molecular evolution tracks macroevolutionary transitions in Cetacea. Trends Ecol. Evol. 29, 336–346. ( 10.1016/j.tree.2014.04.001) [DOI] [PubMed] [Google Scholar]
- 23.Beichman AC, et al. In press Aquatic adaptation and depleted diversity: a deep dive into the genomes of the sea otter and giant otter. Mol. Biol. Evol. ( 10.1093/molbev/msz101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oelschlager HHA, Oelschlager JS. 2008. Brain. In Encyclopedia of marine mammals, 2nd edn (eds Perrin WF, Wursig B, Thewissen JGM), pp. 134–149. Amsterdam, The Netherlands: Elsevier. [Google Scholar]
- 25.Thewissen JGM, George J, Rosa C, Kishida T. 2011. Olfaction and brain size in the bowhead whale (Balaena mysticetus). Mar. Mamm. Sci. 27, 282–294. ( 10.1111/j.1748-7692.2010.00406.x) [DOI] [Google Scholar]
- 26.Kishida T, Thewissen JGM, Hayakawa T, Imai H, Agata K. 2015. Aquatic adaptation and the evolution of smell and taste in whales. Zool. Lett. 1, 9 ( 10.1186/s40851-014-0002-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kishida T, Thewissen JGM, Usip S, Suydam RS, George JC. 2015. Organization and distribution of glomeruli in the bowhead whale olfactory bulb. PeerJ 3, e897 ( 10.7717/peerj.897) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kishida T, Thewissen JGM. 2012. Evolutionary changes of the importance of olfaction in cetaceans based on the olfactory marker protein gene. Gene 492, 349–353. ( 10.1016/j.gene.2011.11.013) [DOI] [PubMed] [Google Scholar]
- 29.Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212. ( 10.1093/bioinformatics/btv351) [DOI] [PubMed] [Google Scholar]
- 30.Waterhouse RM, Seppey M, Simão FA, Manni M, Ioannidis P, Klioutchnikov G, Kriventseva EV, Zdobnov EM. 2017. BUSCO applications from quality assessments to gene prediction and phylogenomics. Mol. Biol. Evol. 35, 543–548. ( 10.1093/molbev/msx319) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parra G, Bradnam K, Ning Z, Keane T, Korf I. 2009. Assessing the gene space in draft genomes. Nucleic Acids Res. 37, 289–297. ( 10.1093/nar/gkn916) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marks P, et al. 2017. Resolving the full spectrum of human genome variation using linked-reads. bioRxiv. ( 10.1101/230946) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng GXY, et al. 2016. Haplotyping germline and cancer genomes with high-throughput linked-read sequencing. Nat. Biotechnol. 34, 303 ( 10.1038/nbt.3432) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weisenfeld NI, Kumar V, Shah P, Church DM, Jaffe DB. 2017. Direct determination of diploid genome sequences. Genome Res. 27, 757–767. ( 10.1101/gr.214874.116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim I, Park J, Suk H, Bae H, Min M, Tsai T, Park D. 2018. Phylogenetic relationships of three representative sea krait species (genus Laticauda; Elapidae; Serpentes) based on 13 mitochondrial genes. Mitochondrial DNA Part A 29, 772–777. ( 10.1080/24701394.2017.1357710) [DOI] [PubMed] [Google Scholar]
- 36.Vandewege MW, Mangum SF, Gabaldón T, Castoe TA, Ray DA, Hoffmann FG. 2016. Contrasting patterns of evolutionary diversification in the olfactory repertoires of reptile and bird genomes. Genome Biol. Evol. 8, 470–480. ( 10.1093/gbe/evw013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Niimura Y, Nei M. 2007. Extensive gains and losses of olfactory receptor genes in mammalian evolution. PLoS ONE 2, e708 ( 10.1371/journal.pone.0000708) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suzuki H, et al. 2018. A single pheromone receptor gene conserved across 400 million years of vertebrate evolution. Mol. Biol. Evol. 35, 2928–2939. ( 10.1093/molbev/msy186) [DOI] [PubMed] [Google Scholar]
- 39.Schwenk K. 1995. Of tongues and noses: chemoreception in lizards and snakes. Trends Ecol. Evol. 10, 7–12. ( 10.1016/S0169-5347(00)88953-3) [DOI] [PubMed] [Google Scholar]
- 40.Zhang X, De la Cruz O, Pinto JM, Nicolae D, Firestein S, Gilad Y.. 2007. Characterizing the expression of the human olfactory receptor gene family using a novel DNA microarray. Genome Biol. 8, R86 ( 10.1186/gb-2007-8-5-r86) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Halpern M. 1992. Nasal chemical senses in reptiles. In Biology of the reptilia (eds Gans C, Crews D), pp. 423–523. Chicago, IL: The University of Chicago Press. [Google Scholar]
- 42.Schwenk K. 2008. Comparative anatomy and physiology of chemical senses in nonavian aquatic reptiles. In Sensory evolution on the threshold: adaptations in secondarily aquatic vertebrates (eds Thewissen JGM, Nummela S), pp. 65–81. Berkeley, CA: University of California Press. [Google Scholar]
- 43.Gabe M, Saint Girons H. 1976. Contribution a la morphologie comparee des fosses nasales et de leurs annexes chez les lepidosoriens. Mém. Mus. Natl. Hist. Nat. Ser. A 98, 1–87. [Google Scholar]
- 44.Hibbard E. 1975. Eyes and other sense organs of sea snakes. In The biology of sea snakes (ed. Dunson WA.), pp. 355–382. Baltimore, MD: University Park Press. [Google Scholar]
- 45.Shine R, Bonnet X, Elphick M, Barrott E. 2004. A novel foraging mode in snakes: browsing by the sea snake Emydocephalus annulatus (Serpentes, Hydrophiidae). Funct. Ecol. 18, 16–24. ( 10.1046/j.0269-8463.2004.00803.x) [DOI] [Google Scholar]
- 46.Kutsuma R, Sasai T, Kishida T. 2018. How snakes find prey underwater: sea snakes use visual and chemical cues for foraging. Zool. Sci. 35, 483–486. ( 10.2108/zs180059) [DOI] [PubMed] [Google Scholar]
- 47.Pihlström H. 2008. Comparative anatomy and physiology of chemical senses in aquatic mammals. In Sensory evolution on the threshold: adaptations in secondarily aquatic vertebrates (eds Thewissen JGM, Nummela S), pp. 95–109. Berkeley, CA: University of California Press. [Google Scholar]
- 48.Shi P, Zhang J. 2007. Comparative genomic analysis identifies an evolutionary shift of vomeronasal receptor gene repertoires in the vertebrate transition from water to land. Genome Res. 17, 166–174. ( 10.1101/gr.6040007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brykczynska U, Tzika AC, Rodriguez I, Milinkovitch MC. 2013. Contrasted evolution of the vomeronasal receptor repertoires in mammals and squamate reptiles. Genome Biol. Evol. 5, 389–401. ( 10.1093/gbe/evt013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saraiva LR, Korsching SI. 2007. A novel olfactory receptor gene family in teleost fish. Genome Res. 17, 1448–1457. ( 10.1101/gr.6553207) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zapilko V, Korsching SI. 2016. Tetrapod V1R-like ora genes in an early-diverging ray-finned fish species: the canonical six ora gene repertoire of teleost fish resulted from gene loss in a larger ancestral repertoire. BMC Genomics 17, 83 ( 10.1186/s12864-016-2399-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Syed AS, Sansone A, Hassenklöver T, Manzini I, Korsching SI. 2017. Coordinated shift of olfactory amino acid responses and V2R expression to an amphibian water nose during metamorphosis. Cell. Mol. Life Sci. 74, 1711–1719. ( 10.1007/s00018-016-2437-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Syed AS, Sansone A, Nadler W, Manzini I, Korsching SI. 2013. Ancestral amphibian v2rs are expressed in the main olfactory epithelium. Proc. Natl Acad. Sci. USA 93, 13 321–13 326. ( 10.1073/pnas.1302088110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hagino-Yamagishi K, Moriya K, Kubo H, Wakabayashi Y, Isobe N, Saito S, Ichikawa M, Yazaki K. 2004. Expression of vomeronasal receptor genes in Xenopus laevis. J. Comp. Neurol. 472, 246–256. ( 10.1002/cne.20073) [DOI] [PubMed] [Google Scholar]
- 55.Meisami E, Bhatnagar KP. 1998. Structure and diversity in mammalian accessory olfactory bulb. Microsc. Res. Tech. 43, 476–499. () [DOI] [PubMed] [Google Scholar]
- 56.Mackay-Sim A, Duvall D, Graves BM. 1985. The West Indian manatee (Trichechus manatus) lacks a vomeronasal organ. Brain Behav. Evol. 27, 186–194. ( 10.1159/000118729) [DOI] [PubMed] [Google Scholar]
- 57.Dittrich K, Kuttler J, Hassenklöver T, Manzini I. 2016. Metamorphic remodeling of the olfactory organ of the African clawed frog, Xenopus laevis. J. Comp. Neurol. 524, 986–998. ( 10.1002/cne.23887) [DOI] [PubMed] [Google Scholar]
- 58.Higgs DM, Burd GD. 2001. Neuronal turnover in the Xenopus laevis olfactory epithelium during metamorphosis. J. Comp. Neurol. 433, 124–130. ( 10.1002/cne.1130) [DOI] [PubMed] [Google Scholar]
- 59.Hansen A, Reiss JO, Gentry CL, Burd GD. 1998. Ultrastructure of the olfactory organ in the clawed frog, Xenopus laevis, during larval development and metamorphosis. J. Comp. Neurol. 398, 273–288. () [DOI] [PubMed] [Google Scholar]
- 60.Blin N, Stafford DW. 1976. A general method for isolation of high molecular weight DNA from eukaryotes. Nucleic Acids Res. 3, 2303–2308. ( 10.1093/nar/3.9.2303) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tatsumi K, Nishimura O, Itomi K, Tanegashima C, Kuraku S. 2015. Optimization and cost-saving in tagmentation-based mate-pair library preparation and sequencing. Biotechniques 58, 253–257. ( 10.2144/000114288) [DOI] [PubMed] [Google Scholar]
- 62.Kajitani R, et al. 2014. Efficient de novo assembly of highly heterozygous genomes from whole-genome shotgun short reads. Genome Res. 24, 1384–1395. ( 10.1101/gr.170720.113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pryszcz LP, Gabaldón T. 2016. Redundans: an assembly pipeline for highly heterozygous genomes. Nucleic Acids Res. 44, e113 ( 10.1093/nar/gkw294) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.English AC, et al. 2012. Mind the gap: upgrading genomes with Pacific Biosciences RS long-read sequencing technology. PLoS ONE 7, e47768 ( 10.1371/journal.pone.0047768) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.English AC, Salerno WJ, Reid JG. 2014. PBHoney: identifying genomic variants via long-read discordance and interrupted mapping. BMC Bioinformatics 15, 180 ( 10.1186/1471-2105-15-180) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Luo R, et al. 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaScience 1, 18 ( 10.1186/2047-217X-1-18) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hara Y, Tatsumi K, Yoshida M, Kajikawa E, Kiyonari H, Kuraku S. 2015. Optimizing and benchmarking de novo transcriptome sequencing: from library preparation to assembly evaluation. BMC Genomics 16, 977 ( 10.1186/s12864-015-2007-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. ( 10.1093/nar/25.17.3389) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Niimura Y. 2009. On the origin and evolution of vertebrate olfactory receptor genes: comparative genome analysis among 23 chordate species. Genome Biol. Evol. 1, 34–44. ( 10.1093/gbe/evp003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. ( 10.1093/bioinformatics/btu170) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kim D, Langmead B, Salzberg SL. 2015. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. ( 10.1038/nmeth.3317) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L.. 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515. ( 10.1038/nbt.1621) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Roberts A, Trapnell C, Donaghey J, Rinn JL, Pachter L. 2011. Improving RNA-Seq expression estimates by correcting for fragment bias. Genome Biol. 12, R22 ( 10.1186/gb-2011-12-3-r22) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. ( 10.1093/molbev/mst010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. [DOI] [PubMed] [Google Scholar]
- 76.Niimura Y, Nei M. 2006. Evolutionary dynamics of olfactory and other chemosensory receptor genes in vertebrates. J. Hum. Genet. 51, 505–517. ( 10.1007/s10038-006-0391-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Glusman G, Yanai I, Rubin I, Lancet D. 2001. The complete human olfactory subgenome. Genome Res. 11, 685–702. ( 10.1101/gr.171001) [DOI] [PubMed] [Google Scholar]
- 78.Kishida T, Go Y, Tatsumoto S, Tatsumi K, Kuraku S, Toda M. 2019. Data from: Loss of olfaction in sea snakes provides new perspectives on the aquatic adaptation of amniotes Dryad Digital Repository. ( 10.5061/dryad.t8sm4m6) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Kishida T, Go Y, Tatsumoto S, Tatsumi K, Kuraku S, Toda M. 2019. Data from: Loss of olfaction in sea snakes provides new perspectives on the aquatic adaptation of amniotes Dryad Digital Repository. ( 10.5061/dryad.t8sm4m6) [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
Specimens: Zoological Collection of the Kyoto University Museum (KUZ) with specimen vouchers KUZ R68692, R72402, R72403, R72604, R73056, and R77260. Sequencing data and assembled genome sequences: GenBank BioProject accessions PRJDB7221 (E. ijimae genome sequencing), PRJDB7226 (L. laticaudata genome sequencing), PRJDB7271 (H. melanocephalus genome sequencing), PRJDB7284 (L. colubrina genome sequencing), PRJDB7257 (L. laticaudata RNA-seq), and PRJDB7258 (H. melanocephalus RNA-seq). The locus of each gene (electronic supplementary material, tables S6–S13) and amino acid sequences of intact olfactory GPCRs (electronic supplementary material, data S1) identified in this study is available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.t8sm4m6 [78].