Abstract
In this article, we present data on the anticancer activities of green synthesized silver nanoparticles (AgNPs) from ethanolic extracts of fruits (AgNPs-F) and leaves (AgNPs-L) of Annona muricata and standard anticancer drug 5-Fluorouracil (5-FU) on two cancer cell lines, i.e. cervical adenocarcinoma (HeLa cells) and prostate adenocarcinoma (PC3 cells) as well as on an immortalized normal prostate cell line, PNT1A. The cytotoxicity on the cells was determined by measuring the absorbance signal of resazurin dye. It has long been known that metabolically active cells change the resazurin from blue (oxidized) to red (reduced) forms, corresponding to the absorbance signals at a wavelength of 570nm (A570) and 600nm (A600) respectively, from which therefore the effects of any treatments on percentage cell viability/death can be elucidated. The raw data values of the treatments against the HeLa, PC3 and PNT1A cells are shown in the different Tables. Examples of how the data can be analyzed have been illustrated using different growth inhibition curves. The data can be used by academics, students, and researchers working on development of anticancer drugs.
Keywords: Silver nanoparticles (AgNPs), 5FU, HeLa, PC3, PNT1A, Cytotoxicity, Resazurin
Specifications Table
| Subject | Biochemistry, Nanomedicine |
| Specific subject area | Cancer Research |
| Type of data | Tables Graphs |
| How data were acquired | Cell culture (DMEM and RPMI 1640 used as growth media), Inverted microscope (Olympus), microplate reader (Infinite M1000, Tecan) |
| Data format | Raw Analyzed |
| Parameters for data collection | Cells were maintained in appropriate growth media in an incubator at 37 °C, 5% CO2; and 95% humidity. HeLa Cells were grown in DMEM while PC3 and PNT1A were grown in RPMI 1640. Cells passaged 1–2 times a week. Cells were harvested and assayed at 60–75% confluence. |
| Description of data collection | The cytotoxicity was determined by measuring the absorbance signal of resazurin dye on the treated cell lines at 570nm and 600nm in a microplate reader. |
| Data source location | Institution: United States Army Medical Research Directorate - Kenya (USAMRD-K), Department of Emerging Infectious Diseases (DEID), Influenza Laboratory City: Nairobi Country: Kenya |
| Data accessibility | With the article |
Value of the Data
|
1. Data
The raw data of the treatments against the HeLa, PC3 and PNT1A cells are shown in the different Tables described in this section. Each experiment (Exp.) was represented by four independent replicates (Rep.). Table 1, Table 2, Table 3 show the data for cytotoxicity of silver nanoparticles derived from fruits extracts of Annona muricata (AgNPs-F) on HeLa, PC3 and PNT1A cells; Table 4, Table 5, Table 6 show the data for cytotoxicity of silver nanoparticles derived from leaves extracts of Annona muricata (AgNPs-L) on HeLa, PC3 and PNT1A cells; while Table 7, Table 8, Table 9 show the data for cytotoxicity of 5FU on HeLa, PC3 and PNT1A cells. On the other hand, Fig. 1, Fig. 2, Fig. 3 show the growth inhibition curves for cells treated with AgNPs-F, Fig. 4, Fig. 5, Fig. 6 show growth inhibition curves of cells treated with AgNPs-L, while Fig. 7, Fig. 8, Fig. 9 show the growth inhibition curves for cells treated with 5FU.
Table 1.
Data for cytotoxicity of the AgNPs-F against HeLa Cells using the Resazurin metabolic assay.
| Concentration (μg/ml) |
||||||||
|---|---|---|---|---|---|---|---|---|
| Wavelength/nm | Replicates | Blank | 200 | 100 | 50 | 25 | 12.5 | |
| Exp. 1 | A570 | Rep. 1 | 1.4698 | 1.3135 | 1.2295 | 1.2667 | 1.3932 | 1.4757 |
| Rep. 2 | 1.4291 | 0.9098 | 1.2994 | 1.2886 | 1.3688 | 1.4914 | ||
| Rep. 3 | 1.5494 | 1.3617 | 1.2275 | 1.3184 | 1.3802 | 1.4947 | ||
| Rep. 4 | 1.3866 | 0.8855 | 1.3176 | 1.2926 | 1.3883 | 1.5741 | ||
| Mean Absorbance of Reps. | 1.45873 | 1.11763 | 1.2685 | 1.29158 | 1.38263 | 1.50897 | ||
| A600 | Rep. 1 | 0.775 | 1.1489 | 1.0559 | 1.0849 | 0.8845 | 0.646 | |
| Rep. 2 | 0.8197 | 0.8186 | 1.1004 | 1.0854 | 0.9019 | 0.8163 | ||
| Rep. 3 | 0.6554 | 1.1691 | 1.0688 | 1.1144 | 0.8115 | 0.6728 | ||
| Rep. 4 | 0.7578 | 0.7987 | 1.1132 | 1.0988 | 0.9122 | 0.636 | ||
| Mean Absorbance of Reps. | 0.75197 | 0.98383 | 1.08457 | 1.09587 | 0.87753 | 0.69277 | ||
| Net Absorbance (A570-A600) | 0.70676 | 0.1338 | 0.18393 | 0.19571 | 0.5051 | 0.8162 | ||
| % Cell Viability | 100 | 18.9315 | 26.0244 | 27.6911 | 71.4670 | 115 | ||
| Exp. 2 | A570 | Rep. 1 | 1.5427 | 1.2109 | 0.9767 | 1.3123 | 1.323 | 1.444 |
| Rep. 2 | 1.5653 | 0.927 | 1.1317 | 1.1178 | 1.3343 | 1.5014 | ||
| Rep. 3 | 1.4661 | 1.2944 | 1.2287 | 1.4169 | 1.3551 | 1.4673 | ||
| Rep. 4 | 1.4413 | 0.9868 | 1.1181 | 1.0673 | 1.4321 | 1.4468 | ||
| Mean Absorbance of Reps. | 1.50385 | 1.10477 | 1.1138 | 1.22857 | 1.36113 | 1.46488 | ||
| A600 | Rep. 1 | 0.5542 | 1.0557 | 0.8384 | 1.1167 | 0.7809 | 0.6737 | |
| Rep. 2 | 0.4877 | 0.8098 | 0.961 | 0.9482 | 0.8498 | 0.5827 | ||
| Rep. 3 | 0.4531 | 1.0748 | 1.0026 | 1.1551 | 0.8375 | 0.6709 | ||
| Rep. 4 | 0.4348 | 0.8631 | 0.9213 | 0.8983 | 0.7712 | 0.7523 | ||
| Mean Absorbance of Reps. | 0.48245 | 0.95085 | 0.93082 | 1.02958 | 0.80985 | 0.6699 | ||
| Net Absorbance (A570-A600) | 1.0214 | 0.15392 | 0.18298 | 0.19899 | 0.55125 | 0.79498 | ||
| % Cell Viability | 100 | 15.0695 | 17.9146 | 19.4821 | 53.9729 | 77.8324 | ||
| Exp. 3 | A570 | Rep. 1 | 1.446 | 1.313 | 1.2562 | 1.2606 | 1.4306 | 1.4939 |
| Rep. 2 | 1.4997 | 1.1884 | 1.3107 | 1.4239 | 1.4366 | 1.5769 | ||
| Rep. 3 | 1.3913 | 1.1504 | 1.2825 | 1.534 | 1.4756 | 1.5519 | ||
| Rep. 4 | 1.3499 | 1.2011 | 1.3006 | 1.2713 | 1.441 | 1.6484 | ||
| Mean Absorbance of Reps. | 1.42172 | 1.21323 | 1.2875 | 1.37245 | 1.44595 | 1.56777 | ||
| A600 | Rep. 1 | 0.503 | 1.091 | 1.0543 | 1.061 | 0.6198 | 0.4778 | |
| Rep. 2 | 0.3938 | 0.9762 | 1.0516 | 1.1493 | 0.7063 | 0.5043 | ||
| Rep. 3 | 0.3751 | 0.9297 | 1.0299 | 1.2124 | 0.6676 | 0.4227 | ||
| Rep. 4 | 0.4032 | 0.9783 | 1.0403 | 1.027 | 0.778 | 0.5372 | ||
| Mean Absorbance of Reps. | 0.41877 | 0.9938 | 1.04402 | 1.11242 | 0.69292 | 0.4855 | ||
| Net Absorbance (A570-A600) | 1.00295 | 0.21943 | 0.24348 | 0.26003 | 0.75303 | 1.08227 | ||
| % Cell Viability | 100 | 21.8788 | 24.2763 | 25.9265 | 75.0815 | 108 | ||
| % Mean Cell Viability ± SD | 100 ± 0 | 18.6266 ± 3.40 | 22.7384 ± 4.27 | 24.3666 ± 4.32 | 66.8405 ± 11.29 | 100.2775 ± 19.75 | ||
Table 2.
Data for cytotoxicity of the AgNPs-F against PC3 Cells using the Resazurin metabolic assay.
| Concentration (μg/ml) |
||||||||
|---|---|---|---|---|---|---|---|---|
| Wavelength/nm | Replicates | Blank | 200 | 100 | 50 | 25 | 12.5 | |
| Exp. 1 | A570 | Rep. 1 | 1.3333 | 1.1585 | 1.0472 | 1.1885 | 1.2751 | 1.2804 |
| Rep. 2 | 1.2456 | 1.2892 | 1.1179 | 1.1709 | 1.2578 | 1.5277 | ||
| Rep. 3 | 1.2113 | 1.3057 | 1.0965 | 1.2815 | 1.3069 | 1.6487 | ||
| Rep. 4 | 1.1547 | 1.31 | 0.9138 | 1.1196 | 1.2775 | 1.6191 | ||
| Mean Absorbance of Reps. | 1.23623 | 1.26585 | 1.04385 | 1.19013 | 1.27933 | 1.51897 | ||
| A600 | Rep. 1 | 0.8743 | 1.0464 | 1.0163 | 0.9057 | 0.9813 | 0.9847 | |
| Rep. 2 | 0.7974 | 1.1854 | 0.8347 | 0.8893 | 0.9626 | 1.234 | ||
| Rep. 3 | 0.8159 | 1.2455 | 0.8202 | 0.9755 | 0.9999 | 1.3307 | ||
| Rep. 4 | 0.7762 | 1.2624 | 1.003 | 0.8564 | 0.9837 | 1.3069 | ||
| Mean Absorbance of Reps. | 0.81595 | 1.18492 | 0.91855 | 0.90673 | 0.98187 | 1.21408 | ||
| Net Absorbance (A570-A600) | 0.42028 | 0.08092 | 0.1253 | 0.2834 | 0.29745 | 0.3049 | ||
| % Cell Viability | 100 | 19.2552 | 29.8138 | 67.432 | 70.7751 | 72.5477 | ||
| Exp. 2 | A570 | Rep. 1 | 1.1529 | 1.4277 | 1.0416 | 1.1698 | 1.2876 | 1.6169 |
| Rep. 2 | 1.302 | 1.1672 | 1.1424 | 1.1884 | 1.344 | 1.7367 | ||
| Rep. 3 | 1.2233 | 1.2382 | 1.8403 | 1.1754 | 1.2491 | 1.4129 | ||
| Rep. 4 | 1.1472 | 1.1836 | 1.0647 | 1.2895 | 1.34 | 1.4508 | ||
| Mean Absorbance of Reps. | 1.20635 | 1.25417 | 1.27225 | 1.20578 | 1.30518 | 1.55432 | ||
| A600 | Rep. 1 | 0.6568 | 1.7462 | 0.7479 | 0.9004 | 0.9852 | 1.2206 | |
| Rep. 2 | 0.8626 | 1.0341 | 0.8102 | 0.9322 | 1.0424 | 1.2958 | ||
| Rep. 3 | 0.8162 | 1.2045 | 1.7465 | 0.9557 | 0.9652 | 1.0876 | ||
| Rep. 4 | 0.8145 | 1.0109 | 1.0593 | 1.0168 | 1.0288 | 1.1233 | ||
| Mean Absorbance of Reps. | 0.78752 | 1.24893 | 1.09097 | 0.95128 | 1.0054 | 1.18182 | ||
| Net Absorbance (A570-A600) | 0.41883 | 0.00525 | 0.18128 | 0.2545 | 0.29978 | 0.3725 | ||
| % Cell Viability | 100 | 1.2535 | 43.2818 | 60.7652 | 71.5752 | 88.9393 | ||
| Exp. 3 | A570 | Rep. 1 | 1.1465 | 1.1826 | 1.2776 | 1.3187 | 1.4139 | 1.3651 |
| Rep. 2 | 1.1237 | 1.2939 | 1.2806 | 1.0512 | 1.374 | 1.6522 | ||
| Rep. 3 | 1.3081 | 1.2568 | 1.2649 | 1.401 | 1.3359 | 1.2984 | ||
| Rep. 4 | 1.053 | 1.228 | 1.111 | 1.2029 | 1.4066 | 1.7333 | ||
| Mean Absorbance of Reps. | 1.15782 | 1.24033 | 1.23352 | 1.24345 | 1.3826 | 1.51225 | ||
| A600 | Rep. 1 | 0.54 | 1.0571 | 0.9571 | 1.1027 | 1.1306 | 1.0439 | |
| Rep. 2 | 0.6766 | 1.2622 | 0.9634 | 1.0164 | 1.0716 | 1.3143 | ||
| Rep. 3 | 0.928 | 1.1568 | 1.0648 | 1.1281 | 1.0312 | 1.0449 | ||
| Rep. 4 | 0.7152 | 1.1127 | 1.0298 | 0.9907 | 1.0983 | 1.3841 | ||
| Mean Absorbance of Reps. | 0.71495 | 1.1472 | 1.00378 | 1.05948 | 1.08292 | 1.1968 | ||
| Net Absorbance (A570-A600) | 0.44287 | 0.09313 | 0.22975 | 0.18398 | 0.29967 | 0.31545 | ||
| % Cell Viability | 100 | 21.0274 | 51.8769 | 41.5411 | 67.6658 | 71.2278 | ||
| % Mean Cell Viability ± SD | 100 ± 0 | 13.8454 ± 10.94 | 41.6575 ±11.12 |
56.5795 ± 13.44 | 70.0054 ± 2.07 | 77.5716 ± 9.87 | ||
Table 3.
Data for cytotoxicity of the AgNPs-F against PNT1A normal cells using the Resazurin metabolic assay.
| Concentration (μg/ml) |
||||||||
|---|---|---|---|---|---|---|---|---|
| Wavelength/nm | Replicates | Blank | 200 | 100 | 50 | 25 | 12.5 | |
| Exp. 1 | A570 | Rep. 1 | 1.0662 | 1.322 | 1.1746 | 1.3105 | 0.9654 | 1.0308 |
| Rep. 2 | 1.0252 | 1.2622 | 1.1472 | 1.1819 | 1.1266 | 1.146 | ||
| Rep. 3 | 1.0735 | 1.5141 | 1.1129 | 1.0966 | 1.2235 | 2.0434 | ||
| Rep. 4 | 1.0679 | 1.4703 | 1.0964 | 1.2052 | 1.2261 | 1.1985 | ||
| Mean Absorbance of Reps. | 1.0582 | 1.39215 | 1.13278 | 1.19855 | 1.1354 | 1.35468 | ||
| A600 | Rep. 1 | 0.4355 | 0.9898 | 0.7002 | 0.6334 | 0.775 | 0.669 | |
| Rep. 2 | 0.5535 | 0.9617 | 0.7756 | 0.733 | 0.9649 | 0.5 | ||
| Rep. 3 | 0.507 | 1.1626 | 0.7718 | 0.6998 | 1.1027 | 1.3381 | ||
| Rep. 4 | 0.5895 | 1.1143 | 0.9109 | 0.8611 | 1.0304 | 0.8164 | ||
| Mean Absorbance of Reps. | 0.52138 | 1.0571 | 0.78963 | 0.73183 | 0.96825 | 0.83087 | ||
| Net Absorbance (A570-A600) | 0.53682 | 0.33505 | 0.34315 | 0.46673 | 0.16715 | 0.5238 | ||
| % Cell Viability | 100 | 62.4133 | 63.9221 | 86.9417 | 31.1368 | 97.5737 | ||
| Exp. 2 | A570 | Rep. 1 | 1.0403 | 1.0461 | 1.108 | 1.1146 | 1.0012 | 1.053 |
| Rep. 2 | 1.0948 | 1.2727 | 1.0925 | 1.105 | 1.178 | 1.1035 | ||
| Rep. 3 | 1.0239 | 1.1515 | 1.0847 | 1.1341 | 1.2848 | 1.1462 | ||
| Rep. 4 | 1.0595 | 1.3475 | 1.0336 | 1.0617 | 1.2899 | 1.0691 | ||
| Mean Absorbance of Reps. | 1.05463 | 1.20445 | 1.0797 | 1.10385 | 1.18847 | 1.09295 | ||
| A600 | Rep. 1 | 0.6439 | 0.8089 | 0.6201 | 0.6013 | 0.7937 | 0.483 | |
| Rep. 2 | 0.456 | 0.9612 | 0.8254 | 0.7689 | 0.9855 | 0.5061 | ||
| Rep. 3 | 0.6273 | 0.8999 | 0.6621 | 0.8289 | 1.1305 | 0.6187 | ||
| Rep. 4 | 0.4868 | 1.0054 | 0.7801 | 0.8518 | 1.0603 | 0.8104 | ||
| Mean Absorbance of Reps. | 0.5535 | 0.91885 | 0.72193 | 0.76272 | 0.9925 | 0.60455 | ||
| Net Absorbance (A570-A600) | 0.50113 | 0.2856 | 0.35777 | 0.34112 | 0.19598 | 0.4884 | ||
| % Cell Viability | 100 | 56.9918 | 71.3944 | 68.0718 | 39.107 | 97.4607 | ||
| Exp. 3 | A570 | Rep. 1 | 1.4067 | 1.4349 | 1.1762 | 1.2242 | 1.7042 | 1.2268 |
| Rep. 2 | 1.3437 | 1.3423 | 1.2115 | 1.2436 | 1.6892 | 1.3645 | ||
| Rep. 3 | 1.1885 | 1.3744 | 1.1532 | 1.2279 | 1.5748 | 1.3597 | ||
| Rep. 4 | 1.3056 | 1.3265 | 1.1547 | 1.3041 | 1.7545 | 1.3102 | ||
| Mean Absorbance of Reps. | 1.31113 | 1.36953 | 1.1739 | 1.24995 | 1.68068 | 1.3153 | ||
| A600 | Rep. 1 | 0.8095 | 1.128 | 0.8524 | 0.8504 | 0.7139 | 0.9781 | |
| Rep. 2 | 0.5971 | 1.0511 | 0.7117 | 0.7322 | 0.6383 | 0.7012 | ||
| Rep. 3 | 0.5568 | 1.0699 | 0.8382 | 0.9259 | 0.7067 | 0.6284 | ||
| Rep. 4 | 0.6509 | 1.0417 | 0.7584 | 0.9122 | 0.6807 | 0.7825 | ||
| Mean Absorbance of Reps. | 0.65358 | 1.07268 | 0.79018 | 0.85517 | 0.6849 | 0.77255 | ||
| Net Absorbance (A570-A600) | 0.65755 | 0.29685 | 0.38373 | 0.39478 | 0.99578 | 0.54275 | ||
| % Cell Viability | 100 | 45.1449 | 58.3568 | 60.0373 | 151.437 | 82.5412 | ||
| % Mean Cell Viability ± SD | 100 ± 0 | 54.85 ±8.83 |
64.5578 ± 6.54 | 71.6836 ± 13.81 | 73.8936 ± 67.27 | 92.5252 ± 8.65 | ||
Table 4.
Data for cytotoxicity of the AgNPs-L against HeLa Cells using the Resazurin metabolic assay.
| Concentration (μg/ml) |
||||||||
|---|---|---|---|---|---|---|---|---|
| Wavelength/nm | Replicates | Blank | 200 | 100 | 50 | 25 | 12.5 | |
| Exp. 1 | A570 | Rep. 1 | 0.9300 | 0.8548 | 1.0364 | 0.9733 | 0.9654 | 0.9224 |
| Rep. 2 | 1.3646 | 1.0182 | 1.2764 | 0.9654 | 1.1266 | 1.1541 | ||
| Rep. 3 | 1.2094 | 1.3914 | 1.3338 | 1.2995 | 1.2235 | 1.1968 | ||
| Rep. 4 | 1.3059 | 1.1796 | 1.2899 | 1.1952 | 1.2261 | 1.2324 | ||
| Mean Absorbance of Reps. | 1.20247 | 1.111 | 1.23412 | 1.10835 | 1.1354 | 1.12642 | ||
| A600 | Rep. 1 | 0.6142 | 0.8395 | 0.8456 | 0.7411 | 0.775 | 0.5721 | |
| Rep. 2 | 0.9294 | 0.9751 | 1.0388 | 0.6971 | 0.9649 | 0.6962 | ||
| Rep. 3 | 1.0003 | 1.4323 | 1.0919 | 1.1155 | 1.1027 | 0.9274 | ||
| Rep. 4 | 0.8611 | 1.1857 | 1.0725 | 0.9207 | 1.0304 | 0.9128 | ||
| Mean Absorbance of Reps. | 0.85125 | 1.10815 | 1.0122 | 0.8686 | 0.96825 | 0.77713 | ||
| Net Absorbance (A570-A600) | 0.35122 | 0.00285 | 0.22192 | 0.23975 | 0.16715 | 0.3493 | ||
| % Cell Viability | 100 | 0.81144 | 63.186 | 68.2611 | 47.5906 | 99.4519 | ||
| Exp. 2 | A570 | Rep. 1 | 0.9551 | 0.8854 | 1.0429 | 1.0087 | 1.0012 | 0.9341 |
| Rep. 2 | 1.4089 | 1.0653 | 1.2939 | 1.006 | 1.178 | 1.1937 | ||
| Rep. 3 | 1.2444 | 1.4537 | 1.362 | 1.3543 | 1.2848 | 1.2442 | ||
| Rep. 4 | 1.3292 | 1.2382 | 1.3149 | 1.2499 | 1.2899 | 1.264 | ||
| Mean Absorbance of Reps. | 1.2344 | 1.16065 | 1.25343 | 1.15473 | 1.18847 | 1.159 | ||
| A600 | Rep. 1 | 0.6179 | 0.8533 | 0.8723 | 0.7601 | 0.7937 | 0.5722 | |
| Rep. 2 | 0.9504 | 0.9948 | 1.0855 | 0.7129 | 0.9855 | 0.712 | ||
| Rep. 3 | 1.0168 | 1.4649 | 1.1522 | 1.1398 | 1.1305 | 0.9507 | ||
| Rep. 4 | 0.8705 | 1.2156 | 1.1315 | 0.945 | 1.0603 | 0.9185 | ||
| Mean Absorbance of Reps. | 0.8639 | 1.13215 | 1.06038 | 0.88945 | 0.9925 | 0.78835 | ||
| Net Absorbance (A570-A600) | 0.3705 | 0.0285 | 0.19305 | 0.26528 | 0.19598 | 0.37065 | ||
| % Cell Viability | 100 | 7.6923 | 52.1053 | 71.5992 | 52.8947 | 100.04 | ||
| Exp. 3 | A570 | Rep. 1 | 1.618 | 1.4486 | 1.4134 | 1.4438 | 1.7042 | 1.5772 |
| Rep. 2 | 1.6132 | 1.3799 | 1.3523 | 1.3666 | 1.6892 | 1.5764 | ||
| Rep. 3 | 1.5871 | 1.3893 | 1.3497 | 1.3177 | 1.5748 | 1.4538 | ||
| Rep. 4 | 2.2674 | 1.3500 | 1.3508 | 1.5011 | 1.7545 | 1.4657 | ||
| Mean Absorbance of Reps. | 1.77143 | 1.39195 | 1.36655 | 1.4073 | 1.68068 | 1.51828 | ||
| A600 | Rep. 1 | 0.7961 | 1.0479 | 1.2882 | 1.1222 | 0.7139 | 0.7698 | |
| Rep. 2 | 0.8412 | 1.0662 | 1.1898 | 1.0908 | 0.6383 | 0.8527 | ||
| Rep. 3 | 0.8130 | 0.9942 | 1.1778 | 1.1227 | 0.7067 | 0.8380 | ||
| Rep. 4 | 1.0122 | 0.9290 | 1.1759 | 0.9830 | 0.6807 | 0.9100 | ||
| Mean Absorbance of Reps. | 0.86563 | 1.00932 | 1.20793 | 1.07968 | 0.6849 | 0.84263 | ||
| Net Absorbance (A570-A600) | 0.9058 | 0.38263 | 0.15863 | 0.32762 | 0.99578 | 0.67565 | ||
| % Cell Viability | 100 | 42.2417 | 17.5121 | 36.1697 | 109.933 | 74.5915 | ||
| % Mean Cell Viability ± SD | 100 ± 0 | 16.9151 ± 22.2 | 44.2678 ± 23.82 | 58.6767 ± 19.56 | 70.1395 ± 34.56 | 91.3613 ± 14.53 | ||
Table 5.
Data for cytotoxicity of the AgNPs-L against PC3 Cells using the Resazurin metabolic assay.
| Concentration (μg/ml) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Wavelength/nm | Replicates | Blank | 200 | 100 | 50 | 25 | 12.5 | ||
| Exp. 1 | A570 | Rep. 1 | 1.0656 | 1.0033 | 0.8548 | 1.0472 | 1.1885 | 1.2751 | |
| Rep. 2 | 1.0504 | 0.8598 | 1.0182 | 1.1179 | 1.1709 | 1.2578 | |||
| Rep. 3 | 0.9836 | 1.1382 | 1.3914 | 1.0965 | 1.2815 | 1.3069 | |||
| Rep. 4 | 0.9948 | 0.9712 | 1.1796 | 0.9138 | 1.1196 | 1.2775 | |||
| Mean Absorbance of Reps. | 1.0236 | 0.99312 | 1.111 | 1.04385 | 1.19013 | 1.27933 | |||
| A600 | Rep. 1 | 0.6045 | 1.0854 | 0.8395 | 1.0163 | 0.9057 | 0.9813 | ||
| Rep. 2 | 0.6152 | 0.8481 | 0.9751 | 0.8347 | 0.8893 | 0.9626 | |||
| Rep. 3 | 0.6472 | 1.1427 | 1.4323 | 0.8202 | 0.9755 | 0.9999 | |||
| Rep. 4 | 0.6218 | 1.0194 | 1.1857 | 1.003 | 0.8564 | 0.9837 | |||
| Mean Absorbance of Reps. | 0.62217 | 1.0239 | 1.10815 | 0.91855 | 0.90673 | 0.98187 | |||
| Net Absorbance (A570-A600) | 0.40143 | −0.0307 | 0.00285 | 0.1253 | 0.2834 | 0.29745 | |||
| % Cell Viability | 100 | −7.6664 | 0.70997 | 31.2138 | 70.5985 | 74.0985 | |||
| Exp. 2 | A570 | Rep. 1 | 0.9578 | 0.9334 | 0.8854 | 1.0416 | 1.1698 | 1.2876 | |
| Rep. 2 | 1.1571 | 1.0754 | 1.0653 | 1.1424 | 1.1884 | 1.344 | |||
| Rep. 3 | 1.1137 | 1.0846 | 1.4537 | 1.8403 | 1.1754 | 1.2491 | |||
| Rep. 4 | 1.0617 | 0.9629 | 1.2382 | 1.0647 | 1.2895 | 1.34 | |||
| Mean Absorbance of Reps. | 1.07257 | 1.01407 | 1.16065 | 1.27225 | 1.20578 | 1.30518 | |||
| A600 | Rep. 1 | 0.5302 | 0.9295 | 0.8533 | 0.7479 | 0.9004 | 0.9852 | ||
| Rep. 2 | 0.8297 | 0.8506 | 0.9948 | 0.8102 | 0.9322 | 1.0424 | |||
| Rep. 3 | 0.764 | 1.1326 | 1.4649 | 1.7465 | 0.9557 | 0.9652 | |||
| Rep. 4 | 0.83 | 0.9381 | 1.2156 | 1.0593 | 1.0168 | 1.0288 | |||
| Mean Absorbance of Reps. | 0.73847 | 0.9627 | 1.13215 | 1.09097 | 0.95128 | 1.0054 | |||
| Net Absorbance (A570-A600) | 0.3341 | 0.05138 | 0.0285 | 0.18128 | 0.2545 | 0.29978 | |||
| % Cell Viability | 100 | 15.3771 | 8.53037 | 54.2577 | 76.1748 | 89.7261 | |||
| Exp. 3 | A570 | Rep. 1 | 1.323 | 1.132 | 1.4486 | 1.2776 | 1.3187 | 1.4139 | |
| Rep. 2 | 1.2966 | 1.2185 | 1.3799 | 1.2806 | 1.0512 | 1.374 | |||
| Rep. 3 | 1.2871 | 1.1995 | 1.3893 | 1.2649 | 1.401 | 1.3359 | |||
| Rep. 4 | 1.209 | 1.1804 | 1.35 | 1.111 | 1.2029 | 1.4066 | |||
| Mean Absorbance of Reps. | 1.27892 | 1.1826 | 1.39195 | 1.23352 | 1.24345 | 1.3826 | |||
| A600 | Rep. 1 | 1.0241 | 1.0888 | 1.0479 | 0.9571 | 1.1027 | 1.1306 | ||
| Rep. 2 | 0.9343 | 1.0871 | 1.0662 | 0.9634 | 1.0164 | 1.0716 | |||
| Rep. 3 | 0.9466 | 1.1122 | 0.9942 | 1.0648 | 1.1281 | 1.0312 | |||
| Rep. 4 | 1.0884 | 1.2177 | 0.929 | 1.0298 | 0.9907 | 1.0983 | |||
| Mean Absorbance of Reps. | 0.99835 | 1.12645 | 1.00932 | 1.00378 | 1.05948 | 1.08292 | |||
| Net Absorbance (A570-A600) | 0.28057 | 0.05615 | 0.38263 | 0.22975 | 0.18398 | 0.29967 | |||
| % Cell Viability | 100 | 20.0125 | 136.372 | 81.8854 | 65.5707 | 106.807 | |||
| % Mean Cell Viability ± SD | 100 ± 0 | 9.24106 ± 7.16 | 48.5374 ± 76.16 | 55.7856 ± 25.37 | 70.7813 ± 5.3 | 90.2107 ± 16.36 | |||
Table 6.
Data for cytotoxicity of the AgNPs-L against PNT1A normal cells using the Resazurin metabolic assay.
| Concentration (μg/ml) |
||||||||
|---|---|---|---|---|---|---|---|---|
| Wavelength/nm | Replicates | Blank | 200 | 100 | 50 | 25 | 12.5 | |
| Exp. 1 | A570 | Rep. 1 | 1.2407 | 1.0866 | 1.2406 | 1.3137 | 1.2693 | 0.9282 |
| Rep. 2 | 1.1232 | 0.8972 | 1.28 | 1.2804 | 1.2856 | 0.9183 | ||
| Rep. 3 | 1.269 | 1.5388 | 1.1828 | 1.2567 | 1.069 | 0.9439 | ||
| Rep. 4 | 1.2601 | 1.2797 | 1.2009 | 1.303 | 1.235 | 0.8617 | ||
| Mean Absorbance of Reps. | 1.22325 | 1.20057 | 1.22607 | 1.28845 | 1.21472 | 0.91302 | ||
| A600 | Rep. 1 | 0.7743 | 0.746 | 0.9653 | 1.0537 | 0.9954 | 0.4771 | |
| Rep. 2 | 0.6654 | 0.9776 | 1.0465 | 1.0191 | 1.0113 | 1.0198 | ||
| Rep. 3 | 0.7117 | 0.95 | 0.9685 | 1.0086 | 0.8496 | 0.7099 | ||
| Rep. 4 | 0.9448 | 1.0352 | 0.9474 | 1.0497 | 0.9955 | 1.0908 | ||
| Mean Absorbance of Reps. | 0.77405 | 0.9272 | 0.98192 | 1.03277 | 0.96295 | 0.8244 | ||
| Net Absorbance (A570-A600) | 0.4492 | 0.27338 | 0.24415 | 0.25568 | 0.25177 | 0.08862 | ||
| % Cell Viability | 100 | 60.8582 | 54.3522 | 56.9179 | 56.0496 | 19.7295 | ||
| Exp. 2 | A570 | Rep. 1 | 1.1515 | 1.1619 | 1.1257 | 1.1935 | 1.1548 | 0.9842 |
| Rep. 2 | 1.211 | 1.3064 | 1.2002 | 1.1932 | 1.1526 | 1.3729 | ||
| Rep. 3 | 1.0892 | 1.382 | 1.4051 | 1.2978 | 1.2415 | 1.2624 | ||
| Rep. 4 | 1.2657 | 1.1699 | 1.1682 | 1.3704 | 1.2996 | 1.2985 | ||
| Mean Absorbance of Reps. | 1.17935 | 1.25505 | 1.2248 | 1.26372 | 1.21213 | 1.2295 | ||
| A600 | Rep. 1 | 0.5864 | 0.9968 | 0.8734 | 0.9459 | 0.9092 | 0.4771 | |
| Rep. 2 | 0.7193 | 0.9433 | 0.9986 | 0.9383 | 0.8934 | 1.0198 | ||
| Rep. 3 | 0.71 | 1.2182 | 1.1052 | 1.0181 | 0.9764 | 0.7099 | ||
| Rep. 4 | 0.8115 | 1.2238 | 0.9416 | 1.0979 | 1.0063 | 1.0908 | ||
| Mean Absorbance of Reps. | 0.7068 | 1.09552 | 0.9797 | 1.00005 | 0.94633 | 0.8244 | ||
| Net Absorbance (A570-A600) | 0.47255 | 0.15952 | 0.2451 | 0.26367 | 0.2658 | 0.4051 | ||
| % Cell Viability | 100 | 33.7583 | 51.8675 | 55.7983 | 56.248 | 85.7264 | ||
| Exp. 3 | A570 | Rep. 1 | 1.1275 | 1.035 | 1.2563 | 1.3947 | 1.2913 | 1.1149 |
| Rep. 2 | 1.2802 | 1.156 | 1.1997 | 1.4059 | 1.3251 | 1.246 | ||
| Rep. 3 | 1.2155 | 1.1991 | 1.2531 | 1.0914 | 1.3846 | 1.2462 | ||
| Rep. 4 | 1.2674 | 1.2178 | 1.2982 | 1.4632 | 1.3368 | 1.2599 | ||
| Mean Absorbance of Reps. | 1.22265 | 1.15198 | 1.25183 | 1.3388 | 1.33445 | 1.21675 | ||
| A600 | Rep. 1 | 0.8274 | 0.746 | 0.9999 | 1.1287 | 1.0471 | 0.9102 | |
| Rep. 2 | 0.6811 | 0.9776 | 0.979 | 1.1563 | 1.0674 | 0.623 | ||
| Rep. 3 | 0.6737 | 0.95 | 1.0378 | 0.8926 | 1.0979 | 0.9252 | ||
| Rep. 4 | 0.7668 | 1.0352 | 1.0329 | 1.1917 | 1.0738 | 0.9023 | ||
| Mean Absorbance of Reps. | 0.73725 | 0.9272 | 1.0124 | 1.09232 | 1.07155 | 0.84018 | ||
| Net Absorbance (A570-A600) | 0.4854 | 0.22478 | 0.23943 | 0.24648 | 0.2629 | 0.37657 | ||
| % Cell Viability | 100 | 46.3072 | 49.3253 | 50.7777 | 54.1615 | 77.5803 | ||
| % Mean Cell Viability ± SDs | 100 ± 0 | 46.9746 ± 13.56 | 51.8483 ± 2.51 | 54.498 ± 3.23 | 55.4864 ± 1.15 | 61.0121 ± 35.98 | ||
Table 7.
Data for cytotoxicity of 5FU against HeLa Cells using the Resazurin metabolic assay.
| Concentration (μg/ml) |
||||||||
|---|---|---|---|---|---|---|---|---|
| Wavelength/nm | Replicates | Blank | 200 | 100 | 50 | 25 | 12.5 | |
| Exp. 1 | A570 | Rep. 1 | 0.9300 | 0.9917 | 1.2199 | 0.8006 | 1.0193 | 1.0169 |
| Rep. 2 | 1.3646 | 1.186 | 1.2638 | 1.1729 | 1.1181 | 1.207 | ||
| Rep. 3 | 1.2094 | 1.0287 | 1.1894 | 1.1752 | 1.0646 | 1.1992 | ||
| Rep. 4 | 1.3059 | 1.171 | 1.1563 | 1.0809 | 1.1307 | 1.1797 | ||
| Mean Absorbance of Reps. | 1.20247 | 1.09435 | 1.20735 | 1.0574 | 1.08318 | 1.1507 | ||
| A600 | Rep. 1 | 0.6142 | 0.8624 | 1.127 | 0.699 | 0.9253 | 0.8917 | |
| Rep. 2 | 0.9294 | 1.1043 | 1.1901 | 1.0642 | 1.074 | 1.1148 | ||
| Rep. 3 | 1.0003 | 1.0022 | 1.1349 | 1.1609 | 1.0259 | 1.0741 | ||
| Rep. 4 | 0.8611 | 1.144 | 1.102 | 1.0754 | 1.0492 | 1.1133 | ||
| Mean Absorbance of Reps. | 0.85125 | 1.02823 | 1.1385 | 0.99988 | 1.0186 | 1.04847 | ||
| Net Absorbance (A570-A600) | 0.35122 | 0.06612 | 0.06885 | 0.05752 | 0.06457 | 0.10223 | ||
| % Cell Viability | 100 | 18.827 | 19.6028 | 16.3784 | 18.3856 | 29.1053 | ||
| Exp. 2 | A570 | Rep. 1 | 0.9551 | 1.017 | 1.2367 | 0.8372 | 1.0507 | 1.0592 |
| Rep. 2 | 1.4089 | 1.2117 | 1.3115 | 1.2111 | 1.1562 | 1.2348 | ||
| Rep. 3 | 1.2444 | 1.0983 | 1.2719 | 1.2354 | 1.12 | 1.2559 | ||
| Rep. 4 | 1.3292 | 1.2225 | 1.213 | 1.1333 | 1.1685 | 1.2467 | ||
| Mean Absorbance of Reps. | 1.2344 | 1.13737 | 1.25828 | 1.10425 | 1.12385 | 1.19915 | ||
| A600 | Rep. 1 | 0.6179 | 0.8865 | 1.1511 | 0.7214 | 0.9486 | 0.9195 | |
| Rep. 2 | 0.9504 | 1.1268 | 1.2173 | 1.0868 | 1.0954 | 1.1367 | ||
| Rep. 3 | 1.0168 | 1.0383 | 1.1735 | 1.2021 | 1.0573 | 1.1011 | ||
| Rep. 4 | 0.8705 | 1.1749 | 1.1333 | 1.1027 | 1.0712 | 1.1387 | ||
| Mean Absorbance of Reps. | 0.8639 | 1.05663 | 1.1688 | 1.02825 | 1.04312 | 1.074 | ||
| Net Absorbance (A570-A600) | 0.3705 | 0.08075 | 0.08948 | 0.076 | 0.08072 | 0.12515 | ||
| % Cell Viability | 100 | 21.7949 | 24.1498 | 20.5128 | 21.7881 | 33.7787 | ||
| Exp. 3 | A570 | Rep. 1 | 1.618 | 1.3869 | 1.4109 | 1.4461 | 1.5886 | 1.51 |
| Rep. 2 | 1.6132 | 1.2718 | 1.4172 | 1.5096 | 1.4624 | 1.5024 | ||
| Rep. 3 | 1.5871 | 1.306 | 1.4215 | 1.5138 | 1.374 | 1.4649 | ||
| Rep. 4 | 2.2674 | 1.2012 | 1.393 | 1.3669 | 1.3833 | 1.4478 | ||
| Mean Absorbance of Reps. | 1.77143 | 1.29147 | 1.41065 | 1.4591 | 1.45207 | 1.48128 | ||
| A600 | Rep. 1 | 0.7961 | 0.9932 | 0.9462 | 1.0964 | 0.7275 | 0.9649 | |
| Rep. 2 | 0.8412 | 0.8292 | 1.0772 | 1.0013 | 0.9073 | 0.8436 | ||
| Rep. 3 | 0.813 | 0.9733 | 1.0002 | 0.8611 | 1.0423 | 0.9563 | ||
| Rep. 4 | 1.0122 | 0.9924 | 1.1114 | 0.9739 | 0.9566 | 0.9414 | ||
| Mean Absorbance of Reps. | 0.86563 | 0.94703 | 1.03375 | 0.98318 | 0.90843 | 0.92655 | ||
| Net Absorbance (A570-A600) | 0.9058 | 0.34445 | 0.3769 | 0.47592 | 0.54365 | 0.55473 | ||
| % Cell Viability | 100 | 38.0272 | 41.6096 | 52.542 | 60.0188 | 61.2414 | ||
| Mean % Cell Viability ± SD | 100 ± 0 | 26.2163 ± 10.33 | 28.4541 ± 11.62 | 29.811 ± 19.79 | 33.3975 ± 23.11 | 41.3751 ± 17.36 | ||
Table 8.
Data for cytotoxicity of 5FU against PC3 Cells using the Resazurin metabolic assay.
| Concentration (μg/ml) |
||||||||
|---|---|---|---|---|---|---|---|---|
| Wavelength/nm | Replicates | Blank | 200 | 100 | 50 | 25 | 12.5 | |
| Exp. 1 | A570 | Rep. 1 | 1.3333 | 1.2295 | 1.4598 | 1.3509 | 1.5099 | 1.3809 |
| Rep. 2 | 1.2456 | 1.2565 | 1.3126 | 1.2195 | 1.3586 | 1.259 | ||
| Rep. 3 | 1.2113 | 1.2528 | 1.4194 | 1.2509 | 1.454 | 1.277 | ||
| Rep. 4 | 1.1547 | 1.219 | 1.3856 | 1.2766 | 1.3622 | 1.2752 | ||
| Mean Absorbance of Reps. | 1.23623 | 1.23945 | 1.39435 | 1.27448 | 1.42118 | 1.29802 | ||
| A600 | Rep. 1 | 0.8743 | 1.1243 | 1.3535 | 1.0984 | 1.3006 | 1.1187 | |
| Rep. 2 | 0.7974 | 1.058 | 1.1941 | 1.0904 | 1.1127 | 0.9825 | ||
| Rep. 3 | 0.8159 | 1.101 | 0.969 | 0.9938 | 1.2125 | 1.0203 | ||
| Rep. 4 | 0.7762 | 1.0578 | 1.0978 | 1.0803 | 1.1213 | 1.0756 | ||
| Mean Absorbance of Reps. | 0.81595 | 1.08527 | 1.1536 | 1.06572 | 1.18677 | 1.04928 | ||
| Net Absorbance (A570-A600) | 0.42028 | 0.15418 | 0.24075 | 0.20875 | 0.2344 | 0.24875 | ||
| % Cell Viability | 100 | 36.6843 | 57.2839 | 49.6699 | 55.773 | 59.1874 | ||
| Exp. 2 | A570 | Rep. 1 | 1.1529 | 1.1256 | 1.0922 | 1.189 | 1.1088 | 1.2155 |
| Rep. 2 | 1.302 | 1.0736 | 2.0665 | 1.1893 | 1.2698 | 1.1701 | ||
| Rep. 3 | 1.2233 | 1.0479 | 1.2537 | 1.1769 | 1.2981 | 1.1123 | ||
| Rep. 4 | 1.1472 | 1.0659 | 1.0555 | 1.1281 | 1.1668 | 1.1998 | ||
| Mean Absorbance of Reps. | 1.20635 | 1.07825 | 1.36698 | 1.17083 | 1.21088 | 1.17443 | ||
| A600 | Rep. 1 | 0.6568 | 0.8058 | 0.6521 | 0.8702 | 0.7814 | 0.8249 | |
| Rep. 2 | 0.8626 | 0.8734 | 2.0876 | 0.8702 | 0.8966 | 0.8882 | ||
| Rep. 3 | 0.8162 | 0.7755 | 0.9804 | 1.0262 | 0.9785 | 0.7266 | ||
| Rep. 4 | 0.8145 | 0.9318 | 0.9941 | 0.8094 | 0.8577 | 1.0238 | ||
| Mean Absorbance of Reps. | 0.78752 | 0.84663 | 1.17855 | 0.894 | 0.87855 | 0.86587 | ||
| Net Absorbance (A570-A600) | 0.41883 | 0.23162 | 0.18843 | 0.27682 | 0.33232 | 0.30855 | ||
| % Cell Viability | 100 | 55.3035 | 44.989 | 66.0956 | 79.347 | 73.6704 | ||
| Exp. 3 | A570 | Rep. 1 | 1.1465 | 1.0529 | 0.9879 | 1.4099 | 1.0342 | 1.215 |
| Rep. 2 | 1.1237 | 1.1887 | 1.251 | 1.2178 | 1.274 | 1.2613 | ||
| Rep. 3 | 1.3081 | 1.3231 | 1.2296 | 1.2199 | 1.3973 | 1.3347 | ||
| Rep. 4 | 1.053 | 1.2915 | 1.3369 | 1.3105 | 1.3249 | 1.3175 | ||
| Mean Absorbance of Reps. | 1.15782 | 1.21405 | 1.20135 | 1.28953 | 1.2576 | 1.28212 | ||
| A600 | Rep. 1 | 0.54 | 0.7176 | 0.7193 | 1.195 | 0.7924 | 0.9112 | |
| Rep. 2 | 0.6766 | 0.8633 | 1.1052 | 0.8249 | 1.0303 | 1.0017 | ||
| Rep. 3 | 0.928 | 1.2248 | 0.9405 | 1.1032 | 1.1282 | 1.0617 | ||
| Rep. 4 | 0.7152 | 1.019 | 1.0162 | 1.0546 | 1.0763 | 1.039 | ||
| Mean Absorbance of Reps. | 0.71495 | 0.95618 | 0.9453 | 1.04442 | 1.0068 | 1.0034 | ||
| Net Absorbance (A570-A600) | 0.44287 | 0.25787 | 0.25605 | 0.2451 | 0.2508 | 0.27872 | ||
| % Cell Viability | 100 | 58.2275 | 57.8154 | 55.3429 | 56.63 | 62.9354 | ||
| Mean % Cell Viability ± SD | 100 ± 0 | 50.0718 ± 11.69 | 53.3628 ± 7.26 | 57.0361 ± 8.34 | 63.9167 ± 13.37 | 65.2644 ± 7.52 | ||
Table 9.
Data for cytotoxicity of 5FU against PNT1A normal cells using the Resazurin metabolic assay.
| Concentration (μg/ml) |
||||||||
|---|---|---|---|---|---|---|---|---|
| Wavelength/nm | Replicates | Blank | 200 | 100 | 50 | 25 | 12.5 | |
| Exp. 1 | A570 | Rep. 1 | 1.0662 | 0.9282 | 1.0414 | 1.085 | 1.0312 | 1.0644 |
| Rep. 2 | 1.0252 | 0.9183 | 1.1152 | 1.0759 | 1.0472 | 1.1821 | ||
| Rep. 3 | 1.0735 | 0.9439 | 1.2742 | 1.1065 | 1.1835 | 1.2094 | ||
| Rep. 4 | 1.0679 | 0.8617 | 0.9168 | 1.1691 | 1.0481 | 1.1196 | ||
| Mean Absorbance of Reps. | 1.0582 | 0.91302 | 1.0869 | 1.10913 | 1.0775 | 1.14388 | ||
| A600 | Rep. 1 | 0.4355 | 0.4771 | 0.47 | 0.482 | 0.5063 | 0.5058 | |
| Rep. 2 | 0.5535 | 1.0198 | 0.675 | 0.6988 | 0.6371 | 0.6727 | ||
| Rep. 3 | 0.507 | 0.7099 | 0.8579 | 0.6031 | 0.852 | 0.7457 | ||
| Rep. 4 | 0.5895 | 1.0908 | 0.5706 | 0.7968 | 0.6933 | 0.7201 | ||
| Mean Absorbance of Reps. | 0.52138 | 0.8244 | 0.64338 | 0.64518 | 0.67217 | 0.66107 | ||
| Net Absorbance (A570-A600) | 0.53682 | 0.08862 | 0.44352 | 0.46395 | 0.40533 | 0.4828 | ||
| % Cell Viability | 100 | 16.5091 | 82.62 | 86.4248 | 75.5041 | 89.9362 | ||
| Exp. 2 | A570 | Rep. 1 | 1.0403 | 0.9842 | 0.9956 | 1.0839 | 1.0614 | 1.0555 |
| Rep. 2 | 1.0948 | 1.3729 | 1.0657 | 1.088 | 0.9943 | 0.9717 | ||
| Rep. 3 | 1.0239 | 1.2624 | 0.9841 | 1.0846 | 1.0802 | 1.1183 | ||
| Rep. 4 | 1.0595 | 1.2985 | 1.0744 | 1.0832 | 1.07 | 1.1113 | ||
| Mean Absorbance of Reps. | 1.05463 | 1.2295 | 1.02995 | 1.08492 | 1.05148 | 1.0642 | ||
| A600 | Rep. 1 | 0.6439 | 0.4771 | 0.5945 | 0.4893 | 0.4823 | 0.4356 | |
| Rep. 2 | 0.456 | 1.0198 | 0.5494 | 0.6673 | 0.6906 | 0.5797 | ||
| Rep. 3 | 0.6273 | 0.7099 | 0.701 | 0.6897 | 0.633 | 0.5618 | ||
| Rep. 4 | 0.4868 | 1.0908 | 0.6393 | 0.6875 | 0.6322 | 0.6472 | ||
| Mean Absorbance of Reps. | 0.5535 | 0.8244 | 0.62105 | 0.63345 | 0.60953 | 0.55607 | ||
| Net Absorbance (A570-A600) | 0.50113 | 0.4051 | 0.4089 | 0.45147 | 0.44195 | 0.50812 | ||
| % Cell Viability | 100 | 80.8381 | 81.5964 | 90.0923 | 88.1916 | 101.397 | ||
| Exp. 3 | A570 | Rep. 1 | 1.4067 | 1.1149 | 1.2754 | 1.2814 | 1.2513 | 1.3242 |
| Rep. 2 | 1.3437 | 1.246 | 1.1233 | 1.2189 | 1.2208 | 1.216 | ||
| Rep. 3 | 1.1885 | 1.2462 | 1.1105 | 1.1816 | 1.2943 | 1.1357 | ||
| Rep. 4 | 1.3056 | 1.2599 | 1.1501 | 1.0069 | 1.2266 | 1.1305 | ||
| Mean Absorbance of Reps. | 1.31113 | 1.21675 | 1.16482 | 1.1722 | 1.24825 | 1.2016 | ||
| A600 | Rep. 1 | 0.8095 | 0.9102 | 0.8707 | 1.0449 | 0.9541 | 0.7732 | |
| Rep. 2 | 0.5971 | 0.623 | 0.6806 | 0.7104 | 0.6601 | 0.741 | ||
| Rep. 3 | 0.5568 | 0.9252 | 0.7461 | 0.708 | 0.6388 | 0.5442 | ||
| Rep. 4 | 0.6509 | 0.9023 | 0.7008 | 0.6725 | 0.6796 | 0.66 | ||
| Mean Absorbance of Reps. | 0.65358 | 0.84018 | 0.74955 | 0.78395 | 0.73315 | 0.6796 | ||
| Net Absorbance (A570-A600) | 0.65755 | 0.37657 | 0.41527 | 0.38825 | 0.5151 | 0.522 | ||
| % Cell Viability | 100 | 57.2694 | 63.1549 | 59.0449 | 78.3362 | 79.3856 | ||
| % Mean Cell Viability ± SD | 100 ± 0 | 51.5389 ± 32.55 | 75.7904 ± 10.95 | 78.5207 ± 16.97 | 80.6773 ± 6.66 | 90.2396 ± 11.0 | ||
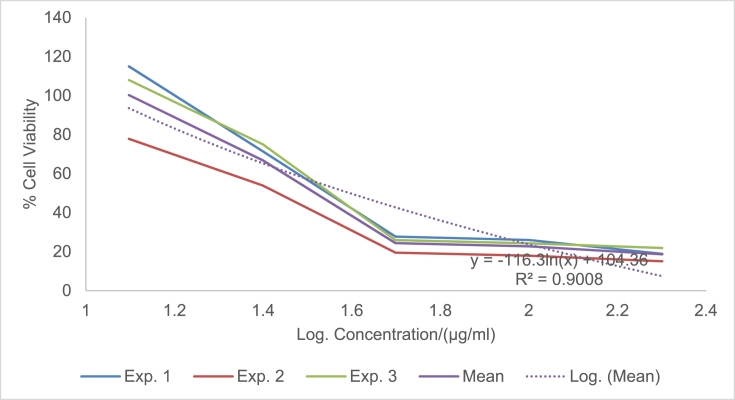
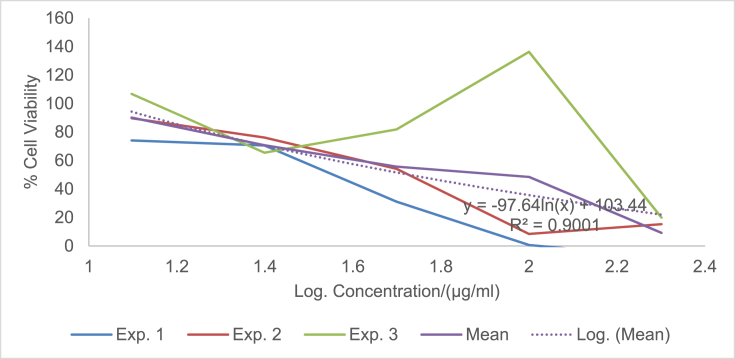
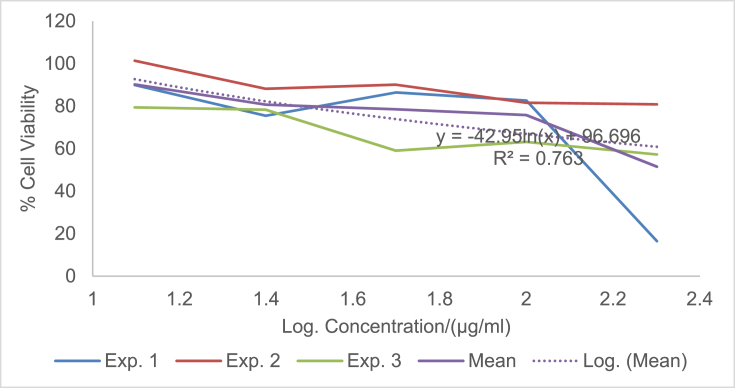
Fig. 1.
A graph showing the Cytotoxicity of AgNPs-F against HeLa Cells using the Resazurin Metabolic Assay.
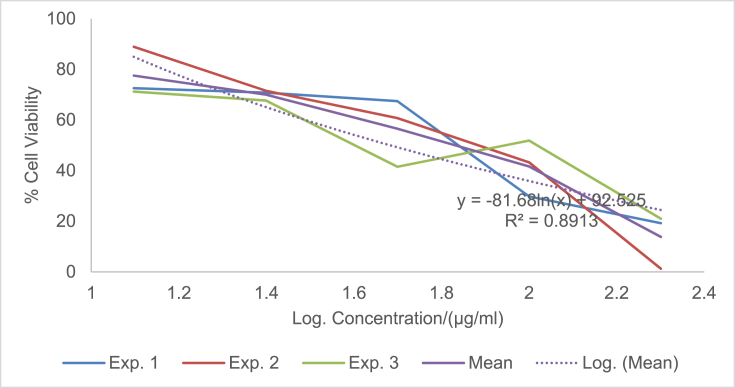
Fig. 2.
A graph showing the Cytotoxicity of AgNPs-F against PC3 Cells using the Resazurin Metabolic Assay.
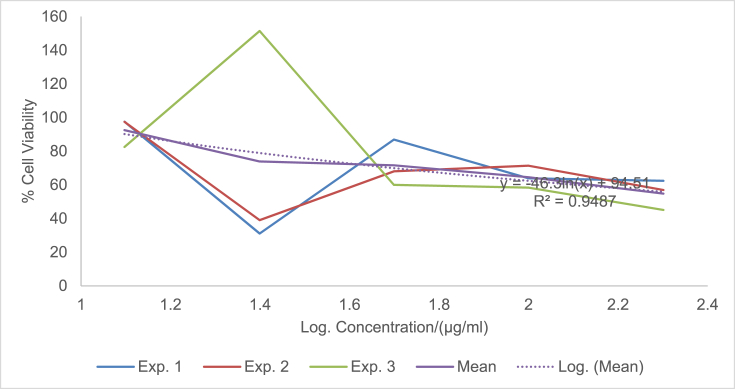
Fig. 3.
A graph showing the Cytotoxicity of AgNPs-F against PNT1A Cells using the Resazurin Metabolic Assay.
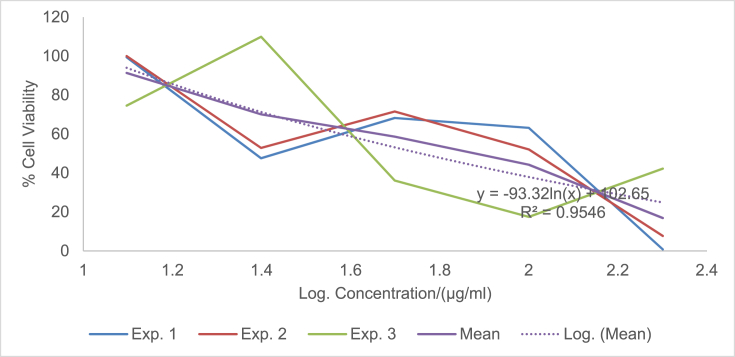
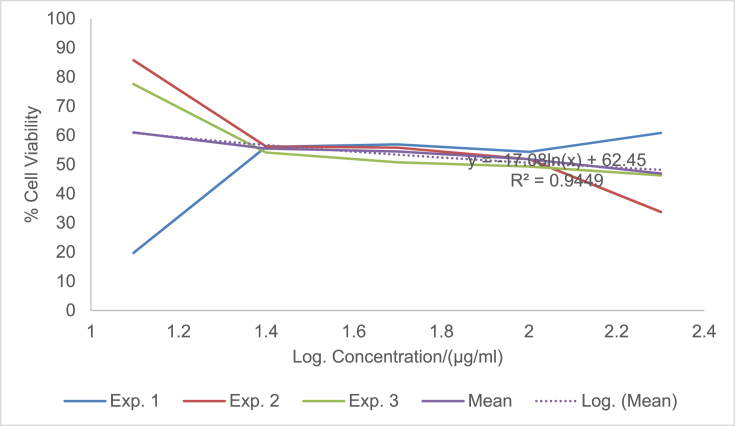
Fig. 4.
A graph showing the Cytotoxicity of AgNPs-L against HeLa Cells using the Resazurin Metabolic Assay.
Fig. 5.
A graph showing the Cytotoxicity of AgNPs-L against PC3 Cells using the Resazurin Metabolic Assay.
Fig. 6.
A graph showing the Cytotoxicity of AgNPs-L against PNT1A Cells using the Resazurin Metabolic Assay.
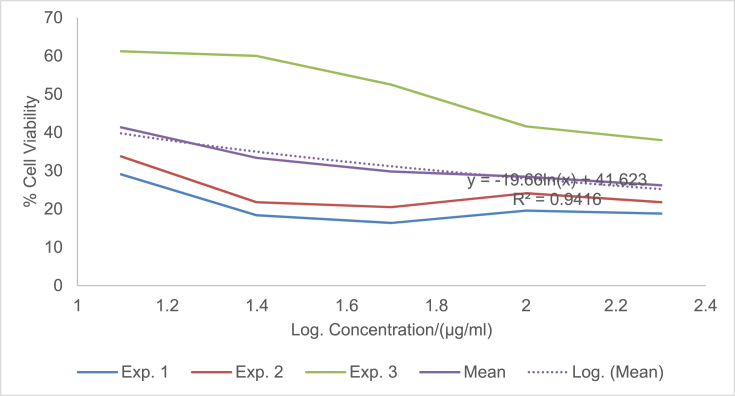
Fig. 7.
A graph showing the Cytotoxicity of 5FU against HeLa Cells using the Resazurin Assay.
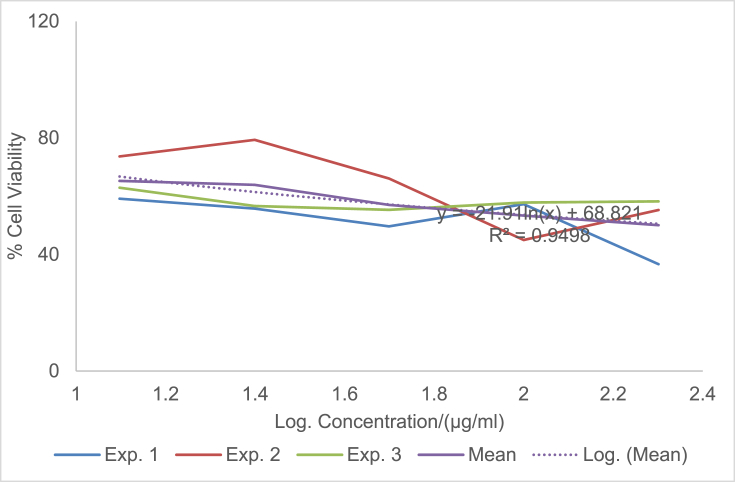
Fig. 8.
A graph showing the Cytotoxicity of 5FU against PC3 Cells using the Resazurin Metabolic Assay.
Fig. 9.
A graph showing the Cytotoxicity of 5FU against PNT1A Cells using the Resazurin Metabolic Assay.
1.1. Data for cytotoxicity of AgNPs-F on HeLa, PC3 and PNT1A cells
.
1.2. Data for cytotoxicity of AgNPs-L on HeLa, PC3 and PNT1A cells
.
1.3. Data for cytotoxicity of 5FU on HeLa, PC3 and PNT1A cells
.
2. Experimental design, materials, and methods
2.1. Chemicals and reagents
All chemicals and reagents were procured from certified suppliers and were of the highest analytical standard. DMEM, RPMI1640, Penicillin/Streptomycin, Non-Essential Amino Acids, Trypsin-EDTA, and Resazurin were obtained from Solarbio (China). FCS, 5FU, Phosphate buffered saline (PBS) and Dimethyl Sulfoxide (DMSO) were obtained from Sigma Aldrich (Germany).
2.2. The silver nanoparticles
Previously prepared and characterized AgNPs from ethanolic extracts of fruits and leaves of Annona muricata were used for the study from which the current data was obtained [1], [2]. AgNPs-F used had an absorption maximum at 427 nm and were stable under different pH, Temperature and storage conditions. The AgNPs-F had an average crystalline size of 60.12 nm, a polydispersity index of 0.1235 and were spherical in nature. The functional groups responsible for the formation of the AgNPs included; Alkanes and alkyls, aldehydes and esters, nitro groups, alcohol groups, amines, amides, alkenes, acids and alkyl halides [1], [2]. On the other hand, AgNPs-L used had an absorption maximum at 429 nm and were stable under different pH, Temperature and storage conditions. The AgNPs-L had an average crystalline size of 87.36 nm, a polydispersity index of 0.16 and were spherical in nature. The functional groups responsible for the formation of the AgNPs included; Alkanes and alkyls, aldehydes and esters, nitro groups, alcohol groups, amines, amides, alkenes, acids and alkyl halides [1].
2.3. Cell lines
The HeLa and PC-3 cells were Cervical and Prostate adenocarcinomas respectively. On the other hand, the PNT1A cells were normal immortalized prostate cells. HeLa, PC3, and PNT1A were sourced from the European collection of Animal Cell Cultures (ECACC). All cells were adherent.
2.4. Cell culture
The cells were grown separately in appropriate media (HeLa in DMEM; PC3 and PNT1A in RPMI 1640) containing l-Glutamine and supplemented with 10% batch tested inactivated fetal calf serum (FCS), 1% Penicillin/Streptomycin, and 1% Non-essential amino acids. The cells were kept an incubator at 37 °C, 5% CO2; and 95% humidity. Cells were Trypsinized and passaged 1–2 times a week and were harvested and used for the assays during their logarithmic growth phase at about 60–75% confluence.
2.5. Preparation of the AgNPs solutions, 5-FU and blanks in media
AgNPs-F and AgNPs-L stock solutions (10mg/ml) were prepared by dispersing them in 0.5% DMSO in culture media. Briefly, 100mg of the AgNPs were dispersed in 10 mL of culture medium (containing Dimethyl Sulfoxide (DMSO) of 0.5%v/v). Required treatment concentrations of (200, 100, 50, 25, and 12.5 μg/mL were then made by dilutions of the stock solutions using the formula C1V1 C2V2. To prepare the standard anticancer treatment regimen of 5-FU, a stock solution was prepared as above and then diluted with culture media to desired concentrations ranging from 12.5 to 200 μg/mL. The final concentration of dimethyl sulfoxide (DMSO) in each cell culture did not exceed 1% v/v to keep the cytotoxicity of DMSO low [3].
2.6. Measurement of the anticancer activities of the AgNPs and 5FU using the Resazurin Assay
The effects of the AgNPs on each of the cell lines' viability and death was determined using the Resazurin (7-hydroxy-10-oxido- phenoxazin-10-ium-3-one) assay as previously described [4], [5], [6], [7]. Exponentially growing cells were harvested, washed and seeded in 96 well plates containing 0.5 × 104Cells/well and incubated with 100 μL per well culture media and allowed to attach overnight. Seeding media was then removed from each of the plates. The attached cultured cells were then treated by adding of 100 μL of the treatments at concentrations of 200, 100, 50, 25, and 12.5μg/mL (in culture media). In addition, the DMSO alone in media was added to another set of cells as the solvent control blank (DMSO = 0.5%v/v). Standard drug 5-FU was used as a reference drug for cancer as positive control. The treated cells were then incubated in a humified CO2 incubator at 37 °C. 24 Hours from the start of the incubation, 20 μl resazurin at a concentration of 0.15mg/ml in PBS was added to each of the wells and then incubated at 37 °C for an additional 4 hours. After 4 hours from the addition of resazurin, the plates containing the treated cells were then retrieved from the incubator and the absorbance signal was quickly measured at 570/600nm (excitation/emission wavelengths), using a microplate reader (Infinite M1000, Tecan). Each treatment was read in at least four replicates.
2.7. How the data can be analyzed
The presented data can be analyzed by determining the percentage cell viability using the formula: % Viability = (Net absorbance of treated samples/Net absorbance of blank) ×100. The effect of the samples on the proliferation of the cell lines can then be expressed in form of graphs of percentage cell viability against logarithm of concentration as shown in Fig. 1, Fig. 2, Fig. 3, Fig. 4, Fig. 5, Fig. 6, Fig. 7, Fig. 8, Fig. 9 under the data section above. Fifty percent of inhibitory concentration (IC50) or cytotoxic concentration (CC50) of each of the treatments can then be calculated from the growth inhibition curves.
Research clearance and registration
The study from which the current data was obtained was cleared by the PAUSTI board of examiners (MB400-0007/17), The Uganda National Council for Science and Technology (NS 43ES) as well as the Jomo Kenyatta University of Agriculture and Technology Institutional Ethics Review Committee (Ref. no: JKU/2/4/896B).
Acknowledgments
The authors would wish to thank the Pan African University for the funding that allowed the study from which this data was obtained to be carried out. They further thank Prof Wallace D Bulimo, Janet Majanja, Meshack Wedagu, Silvanos Mukunzi, Samwel Symekher, Rachel Achilla, Agnes Gathemia, Tiffany H Wandera, and the entire team at the United States Army Medical Research Directorate - Kenya at the Department of Emerging Infectious Diseases (USAMRD-K DEID) Influenza Laboratory in Nairobi for their selfless support rendered that ensured the success of this work to be achieved.
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Gavamukulya Y., Elshemy H.A., Meroka A.M., Madivoli E.S., Maina E.N., Wamunyokoli F., Magoma G. Advances in green nanobiotechnology: data for synthesis and characterization of silver nanoparticles from ethanolic extracts of fruits and leaves of Annona muricata. Data Br. 2019;25:104194. doi: 10.1016/j.dib.2019.104194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gavamukulya Y., Maina E.N., Meroka A.M., Madivoli E.S., El-Shemy H.A., Wamunyokoli F., Magoma G. Green synthesis and characterization of highly stable silver nanoparticles from ethanolic extracts of fruits of Annona muricata. J. Inorg. Organomet. Polym. Mater. 2019 doi: 10.1007/s10904-019-01262-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abdel-Hamid N.M., Tabl G.A., El-Bolkiny Y.E., Zeina W.O. In vitro antitumor efficacy of Kochiaindica extract on human hepatocellular carcinoma cell line with or without 5-fluorouracil. Hepatoma Res. 2017;3:149. doi: 10.20517/2394-5079.2016.50. [DOI] [Google Scholar]
- 4.Riss T.L., Moravec R.A., Niles A.L., Duellman S., Benink H.A., Worzella T.J., Minor L. In: Assay Guid. Man. Sittampalam G.S., Coussens N.P., Brimacombe K., et al., editors. Eli Lilly & Company and the National Center for Advancing Translational Sciences; Bethesda (MD): 2013. Cell viability assays; pp. 785–796. [Internet] [DOI] [Google Scholar]
- 5.Vega-Avila E., Pugsley M.K. An overview of colorimetric assay methods used to assess survival or proliferation of mammalian cells. Proc. West. Pharmacol. Soc. 2011;54:10–14. [PubMed] [Google Scholar]
- 6.Borra R.C., Lotufo M.A., Gagioti S.M., Barros F. de M., Andrade P.M. A simple method to measure cell viability in proliferation and cytotoxicity assays. Braz. Oral Res. 2009;23:255–262. doi: 10.1590/S1806-83242009000300006. [DOI] [PubMed] [Google Scholar]
- 7.Kuete V., Wabo H.K., Eyong K.O., Feussi M.T., Wiench B., Krusche B., Tane P., Folefoc G.N., Efferth T. Anticancer activities of six selected natural compounds of some Cameroonian medicinal plants. PLoS One. 2011;6:4–10. doi: 10.1371/journal.pone.0021762. [DOI] [PMC free article] [PubMed] [Google Scholar]