Abstract
Clinical diagnostic tools requiring direct sample testing cannot be applied to infections deep within the body, and clinically available imaging tools lack specificity. New approaches are needed for early diagnosis and monitoring of bacterial infections and rapid detection of drug-resistant organisms. Molecular imaging allows for longitudinal, noninvasive assessments and can provide key information about infectious processes deep within the body.
INTRODUCTION
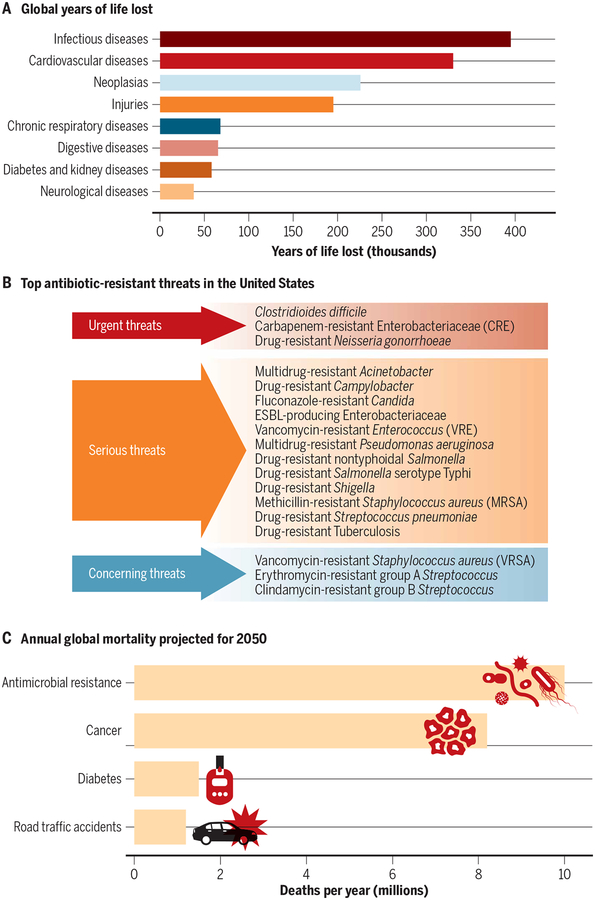
Globally, more than 8 million deaths and 400,000 years of life lost were attributed to infections in 2017, ranking infections third in mortality but first in morbidity among all human diseases (Fig. 1A) (1). Most of these deaths and disability are caused by only a few pathogens, with around 20 bacterial and viral species accounting for two-thirds of deaths from infections (2). For example, Mycobacterium tuberculosis, the causative agent for tuberculosis (TB), is currently the leading infectious cause of death globally (3). Other bacterial pathogens, such as those from the Enterobacteriaceae family (Escherichia coli, Klebsiella spp., Enterobacter sp., and Salmonella sp.), Clostridioides difficile, Staphylococcus aureus, and Pseudomonas aeruginosa, account for the majority of health care–associated infections in the United States (Fig. 1B) (4). The overall annual direct medical costs of health care–associated infections are estimated to be 36 to 45 billion U.S. dollars (USD) per year in the United States alone (5). The alarming rise of antimicrobial drug resistance globally is another major health care challenge. Given current trends, it is estimated that by 2050, drug-resistant infections will become the leading cause of death globally (10 million per year) and surpass those due to cancer (Fig. 1C) (6). A cumulative 100 trillion USD of economic output is estimated to be at risk because of the rise of drug-resistant infections. Although major efforts are under way to curb bacterial infections and antimicrobial drug resistance, there is a need to improve the way we diagnose and treat bacterial infections.
Fig. 1. Global burden of infections.
(A) Global years of life lost due to diseases in 2017. (B) The top 18 antimicrobial drug–resistant threats compiled by the U.S. Centers for Disease Control and Prevention (CDC). Extended spectrum β-lactamase (ESBL) producing Enterobacteriaceae. (C) Annual global mortality projected for 2050. Adapted from the Global Burden of Disease Study (1), U.S. CDC (5) and O’Neill (6).
DIAGNOSIS OF INFECTIONS
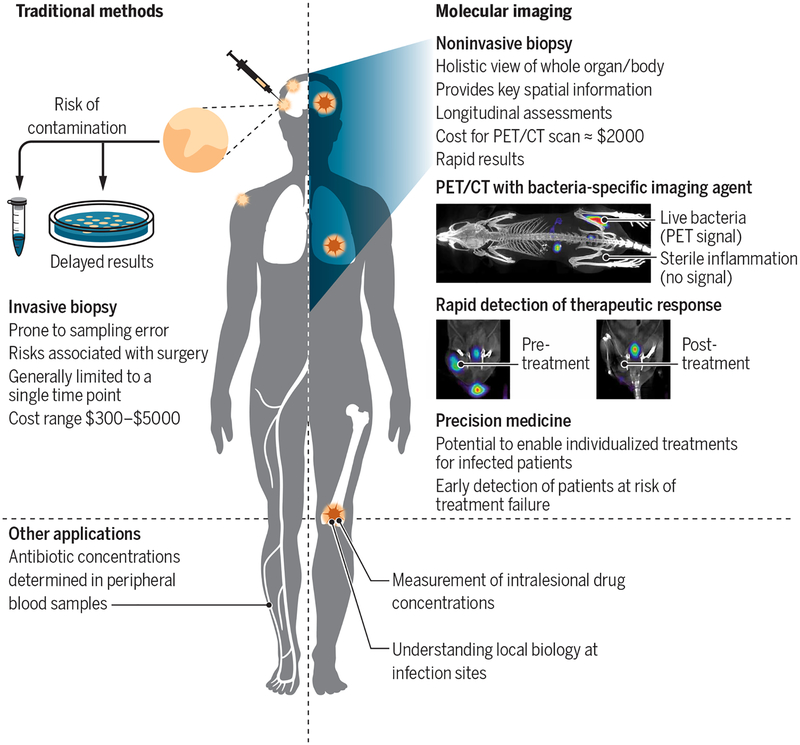
Traditional diagnostic tools used for infectious diseases, such as microscopy, micro-biology, and molecular techniques (nucleic acid amplification and mass spectrometry), require clinical samples (blood, urine, stool, or cerebrospinal fluid) (Fig. 2). Clinical samples may not accurately represent the local biology at infection sites and thus are often nondiagnostic or insensitive for the detection of the bacterial infection (7). Surgical resection or biopsy of infected tissues is often the last resort for establishing a definitive diagnosis of infections deep within the body and is generally limited to the most accessible lesion identified at a single time point. Biopsies introduce risk of contamination, are prone to sampling error, and fail to capture the heterogeneous infectious lesions that often occur in patients, as well as the temporal changes occurring over the course of disease or treatment. M. tuberculosis, anaerobes, and other fastidious organisms are difficult to cultivate ex vivo, which can limit or delay diagnosis. Clinically available imaging tools such as radiography, ultrasonography, computed tomography (CT), and magnetic resonance imaging (MRI) are often incorporated into the diagnostic workup. However, these structural imaging tools are based on changes in anatomy or tissue morphology that are often delayed relative to the molecular events in the disease process, are nonspecific, and may reflect a combination of the infection and the host inflammatory response.
Fig. 2. Comparison of traditional methods with molecular imaging.
Traditional diagnostic tools for infectious diseases depend on available clinical samples (blood and urine), with most deep-seated infections requiring surgical biopsies to establish a definitive diagnosis. Microbiological or molecular assays are performed on surgical biopsies; however, some organisms are difficult to cultivate ex vivo or need a long time to grow, which can limit or delay the diagnosis. Clinically available imaging tests [radiographs, ultrasound, computed tomography (CT), and magnetic resonance imaging (MRI)] incorporated into the diagnostic workup are not specific for infection and reflect a combination of infection and the host inflammatory response. Molecular imaging can provide spatial and temporal information about infections and can monitor response to treatment. An example of positron emission tomography (PET)/CT imaging in mice using a bacteria-specific imaging agent, 2-18F-fluorodeoxysorbitol, is shown. The radiopharmaceutical accumulates in the infected muscle but not in the inflamed sterile muscle. Repeat imaging before and after antibiotic treatment can also provide rapid efficacy monitoring, demonstrating a PET signal proportionate to the bacterial burden. Adapted from Weinstein et al. (23).
Molecular imaging tools such as single- photon emission CT (SPECT) or positron emission tomography (PET) provide noninvasive measurements of molecular pathways and are often used in combination with structural imaging (PET/CT or PET/MRI) to provide an anatomic reference. Current PET or SPECT imaging agents such as gallium-67–, indium-111–, and technetium-99 m (99mTc)–radiolabeled leukocytes [white blood cell (WBC) scan] or 18F-fluorodeoxyglucose (18F-FDG) offer increased sensitivity compared to structural imaging tools (8) but are limited as they reflect the physiological changes that are part of the inflammatory process and the host response to infection, as opposed to the infectious pathogen itself. Therefore, they cannot reliably distinguish infections from other diseases. Given that the host responses can be altered in various disease states such as diabetes, cancer, and immunosuppression, this lack of specificity and procedural efforts required for definitive diagnosis remain as major barriers, leading to the indiscriminate use of broad-spectrum antibiotics, development of antimicrobial drug resistance, increased health care costs, and drug toxicity.
Cancer imaging provides an example of the value of specific molecular imaging tools, which can be a road map for infectious diseases. The use of gallium-68 (68Ga)–DOTATATE, a PET radiopharmaceutical that targets somatostatin receptors and is used to evaluate neuroendocrine tumors (NETs), is a recent example of successful translation of molecular imaging affecting patient care. This new molecular imaging agent has caused a paradigm shift in NET cancer management and, along with a related radiotherapeutic agent (lutetium-177—DOTATATE), improved overall survival in patients with these tumors (9). Another example is the use of radiolabeled prostate-specific membrane antigen (PSMA) ligands to improve the sensitivity and specificity of detection of primary and metastatic prostate cancer (10). Similar potential exists for developing molecular imaging tools for infectious diseases to advance clinical care, understand disease pathophysiology, and enable precision medicine approaches for infection management.
BARRIERS TO CLINICAL TRANSLATION
Successful development and translation of infection imaging tools require a comprehensive understanding of bacterial biology and imaging technologies (Table 1). PET is considered the most sensitive among SPECT, PET, MRI, ultrasound, and photoacoustic imaging technologies and will be highlighted in this article.
Table 1.
Key considerations for the development and clinical translation of bacteria-specific imaging agents.
| Challenges | Solutions |
|---|---|
| Discovery | |
| Bacterial target selection | Target conserved pathways |
| Use clinical strains, drug-resistant strains, and bacteria in different metabolic states | |
| Dosage selection | Evaluate the agent using physiologically relevant assays |
| Use nano- to picomolar doses in vitro | |
| Bacterial specificity | Use eukaryotic cells and heat-inactivated bacteria as specificity controls |
| Select agents showing >100× favorable accumulation in bacteria | |
| Preclinical animal studies | |
| Model selection | Use multiple clinically relevant animal models |
| Quantify bacterial burden at infection site at the time of imaging | |
| Control selection | Use sterile inflammation controls within the same animal model |
| Agent pharmacokinetics | Monitor clearance, half-life, and tissue penetration of agents |
| Consider protein binding characteristics and host metabolism | |
| Use kinetic modeling to evaluate blood pool effects due to leaky vasculature | |
| Clinical studies | |
| Patient selection | Prioritize diseases with clear clinical needs and appropriate bacterial burden |
| Enroll patients with confirmed diagnosis of infection using current gold standard | |
| Empiric antibiotic use with variable residual bacterial burden | Select patients who have received limited antibiotic treatment |
| Specificity determination | Include patients with inflammatory or oncologic conditions as noninfectious disease controls |
Target selection
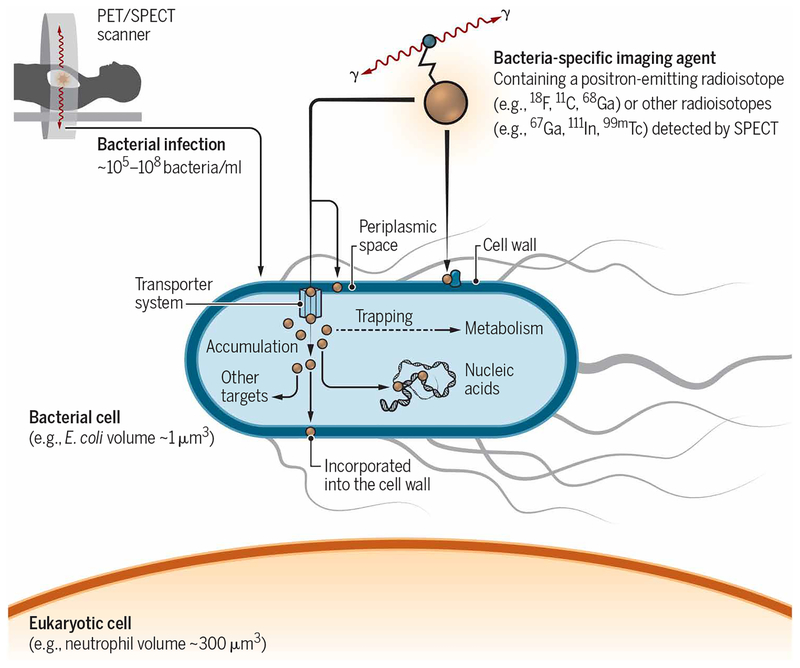
Bacteria (prokaryotes) are evolutionarily and phylogenetically distinct from eukaryotic cells/mammals. Fundamental biochemical differences in metabolism, proteins, and cell wall components between bacteria and mammalian cells provide opportunities for target selection to develop bacteria-specific imaging agents (Fig. 3). Thus far, target selection has focused on antibiotics or antimicrobial peptides. While being nontoxic to human cells, antibiotics and antimicrobial peptides specifically target and kill (or disable) bacteria at high potency. However, although extensively evaluated as bacteria-specific agents, many radiolabeled antibiotics or antimicrobial peptides may not be ideal candidates for bacteria-specific imaging (11) unless the mechanism of bacterial accumulation is orders of magnitude greater for the bacteria than for human tissues. The failure of 99mTc-ciprofloxacin is one such example. Although promising as a bacteria-specific imaging agent in an initial clinical study (12), subsequent studies demonstrated variability and ultimately an inability to reliably differentiate infection from sterile inflammatory processes (13). In contrast, a PET agent based on the broad-spectrum antibiotic trimethoprim (TMP), 18F-FPTMP, has over 30,000-fold selectivity for its bacterial target over the human homolog and has shown promise in animal models (14). The use of bacteria-specific radiolabeled antibodies has also been attempted in preclinical studies, with encouraging results. However, their use may be limited because of an inability to differentiate “live” versus “dead” bacteria and decreased penetration into necrotic lesions.
Fig. 3. Bacteria-specific imaging tools.
PET and other molecular imaging modalities can detect the accumulation of an imaging agent at diseased sites with very high sensitivity. Fundamental biochemical differences between bacterial and human (mammalian) metabolism offer distinct targeting opportunities for developing bacteria-specific imaging tools. The relative volume of a single bacterium and the target density of the proposed agent need to be considered to achieve adequate target-to-background contrast.
An alternative approach is to develop bacteria-specific radiolabeled molecular imaging agents based on prokaryotic metabolism (15). Selective metabolism of small molecules (mostly sugars) has been used to identify and differentiate bacteria in the clinical micro-biology laboratory (16). Small molecules selectively metabolized by pathways expressed only in bacteria (or in a specific class/species of bacteria) could not only be exploited to accurately differentiate bacterial infections from noninfectious processes but also provide information about the causative bacterial class/species (15). Because the efficacy spectrum of many antibiotics is dependent on the class of bacteria (Gram-positive or Gram- negative organisms), this distinction will allow narrowing of the antimicrobial regimen. The advantages of metabolized substrates include amplification via enzymatic turnover and opportunities for specific retention in the cell wall or other macromolecules leading to substantial bacterial accumulation over the background host tissues. Radiolabeled analogs of small molecules easily penetrate diseased tissues, are rapidly cleared from nontarget tissues, show increased stability due to covalent bonding of the radiolabel, and are generally cost effective and simple to produce. Recently developed bacteria- specific metabolic imaging agents include the following: 11C-para-aminobenzoic acid (PABA) and 2-18F-PABA, which target the bacterial folate pathway (17, 18); 18F-labeled maltohexaose and 6-18F-fluoromaltotriose, which are taken up via the maltodextrin transporter in bacteria (19, 20); radio-analogs of d–amino acids that are incorporated into the bacterial cell wall (21); siderophore-derived agents (22); and 2-18F-fluorodeoxysorbitol (18F-FDS), synthesized from 18F-FDG and used to specifically localize infections due to Gram-negative Enterobacteriaceae, the most common cause of bacterial infections in humans (23).
Screening of candidate bacteria- specific imaging agents requires rigorous preclinical testing and should be physiologically relevant. In vivo tissue concentrations achieved after intravenous injection of traditional PET agents are in the nano- to picomolar range of the parent compound; therefore, 18F-labeled PET agents should be incubated at nano- to picomolar concentrations for 2 hours in vitro to simulate in vivo conditions and to match the physical (radiological) half-life (15). Bacteria under stress from host immunological responses or antibiotic treatments exhibit altered metabolic expression; thus, when selecting targets, it is critical to select highly conserved pathways expressed by bacteria in a wide range of microbial milieus. Molecules requiring enzymatic activation based on adenosine triphosphate (ATP) may not fare well in clinical infections such as chronic joint infections where bacterial populations are predominantly metabolically slow or inactive. However, some bacteria-specific imaging agents such as PABA, which is metabolized via the bacterial folate pathway, are not affected by the bacterial growth phase (15).
Achieving sufficient tissue contrast
To achieve adequate sensitivity for bacterial infections, the relative volume of a bacterium and the target density of the proposed agent need to be considered. The average volume of a single pathogenic bacterium ranges from ~0.5 μm3 for cocci such as S. aureus to ~3 μm3 for bacilli such as E. coli or M. tuberculosis, which is smaller than the volume of a typical mammalian host cell (~100 to 1000 μm3) (24). Selectivity (100 to 2000 times) favoring bacterial accumulation over the background signal in host tissues is therefore required to adequately visualize bacteria.
Acute bacterial infections are associated with high bacterial burdens [an average of 2 × 108 CFU/ml in soft tissue and peritoneal infections (25) and 107 to 109 mycobacteria in cavitary TB lesions in humans (26)], although only limited data exist due to the difficulties of direct tissue measurements. However, bacterial burden at infection sites can also be inferred from other observations: A cutoff of 105 CFU/ml in clinical samples such as bronchoalveolar lavage fluid or urine originating from infected tissues is recommended for determining a true infection (27), suggesting that much higher bacterial concentrations exist at the source of the infection sites. Similarly, rifampin failure occurs when used as monotherapy due to the development of rifampin-resistant bacteria (28). Given that in vitro studies suggest that 1 in 108 bacteria is rifampin-resistant due to random mutants (29), bacterial burdens at infection sites during active infections are likely to be at least 108 or higher. Therefore, ~105 CFU/ml would be a promising threshold for acute infections. However, even higher agent selectivity would be needed for detecting lower bacterial densities (<105 CFU/ml), such as those encountered in partially treated or chronic infections.
Clinical studies
A major challenge to patient recruitment for initial validation studies is the widespread clinical practice of administering antibiotics empirically to patients with suspected infection before a confirmatory diagnosis is established. Procedural delays or the time required for definitive diagnosis using microbiological culture leads to a variable residual bacterial burden, potentially affecting the sensitivity of imaging tests.
Complementary anatomic imaging findings that allow orthogonal measurement of the lesion do not usually exist for bacterial infections. Unlike a typical solid tumor detected by anatomic imaging that has a measurable component of cancer cells in the lesion [which correlates with a molecular imaging agent, enabling tracking of treatment response over time (30)], infectious lesions detected by traditional imaging may be sterile and instead represent host inflammatory responses rather than true infection. In these false-positive situations, a pathogen-specific imaging technique might inappropriately be considered to have sensitivity inferior to that of traditional anatomic and nuclear medicine techniques (18F-FDG, PET, and WBC scan).
Patient selection
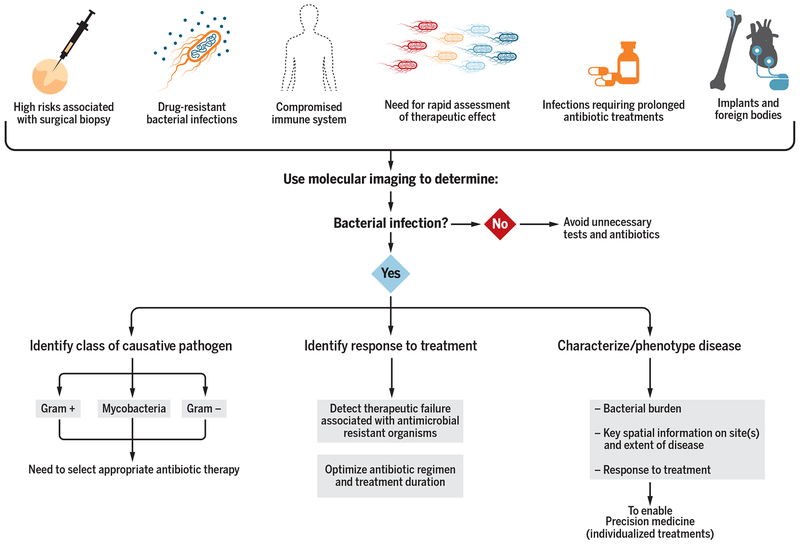
Molecular imaging could complement traditional clinical tools (Fig. 4). This applies to scenarios in which available clinical samples (blood and urine) would be insensitive, high risk, or impractical (biopsy for brain infection); to infections with drug-resistant bacterial strains, where the risks (and costs) of empiric second- or third-line antibiotic treatment are high; and for patients with compromised immune systems (fever and neutropenia due to cancer chemotherapy, HIV/AIDS, and organ transplant), for whom routine imaging to detect disease foci is already an established clinical practice. Similarly, molecular tools could address the need for rapid assessment of therapeutic effect, could help optimize treatment and establish end points in patients requiring prolonged antibiotic duration (months to years), and could be useful for patients with implants and foreign bodies for which traditional imaging tools may be limited.
Fig. 4. Molecular imaging to complement traditional tools in select patient populations.
Molecular imaging can be useful for several patient populations and could be easily incorporated into the current clinical workflows to address the relevant clinical question(s) at hand.
Molecular imaging could help address relevant clinical questions. For example, an otherwise healthy 15-year-old presenting with osteomyelitis would be treated with an empiric antibiotic regimen targeting both methicillin-resistant (MRSA) and drug-sensitive (MSSA) bacteria when blood cultures do not yield a pathogen and biopsy is not performed. A molecular imaging agent that could rapidly assess response to oxacillin/methicillin would be cost effective and useful in streamlining antibiotic treatment. Conversely, determining the causative bacterial species would be the clinically relevant question to answer for an otherwise healthy 3-year-old with septic arthritis/osteomyelitis, as both S. aureus (Gram-positive bacteria) and Kingella kingae (Gram-negative bacteria) are common in this patient demographic. A molecular imaging agent that distinguishes Gram-positive from Gram-negative infections may allow the tailoring of antibiotic regimens to target the appropriate pathogen.
Many deep-seated infections are mono-microbial (31). However, even for polymicrobial infections, a general bacteria-specific molecular imaging agent (PABA, TMP, or maltotriose) capable of identifying multiple species could specifically detect and localize infection sites and also allow rapid assessments of therapeutic response to treatment. Imaging also provides detailed spatial information and could discriminate true infections from the normal flora inhabiting the body. For example, co-registration of PET with anatomic imaging (CT/MRI) could distinguish intraluminal normal gut flora from extraluminal sites, such as abdominal infections (peritonitis and intra-abdominal abscesses).
Molecular imaging is often assumed to be restricted to resource-rich settings. Developing countries, where infections are still the leading cause of mortality, are important sites for innovation and implementation of molecular imaging for infections. Over the past decade, developing countries have witnessed considerable growth in the installation and use of advanced imaging (32). Moreover, imaging costs are substantially lower in developing nations than in the United States (33). 18F PET agents can be transported locally usually within a 2- to 3-hour travel radius, and some radio-isotopes such as 68Ga can be produced without the need of a cyclotron and hold promise for use in remote areas (34).
Radiation risks
Another barrier to molecular imaging is the perceived risk of radiation exposure from radioligands and CT performed in conjunction with PET or SPECT. The use of PET/MRI could reduce radiation exposure by eliminating the need for CT (35), although MRI may require anesthesia in young children due to its longer acquisition time. Technological advancements have lowered radiation exposure due to CT. The effective dose for each chest CT can be ≤0.5 mSv [a dose equivalent to 2 months of natural background radiation, one screening mammography, or four trans-Atlantic airplane round trips (8)] and is performed in a few seconds, precluding the need for sedation (36). Unlike 18F-FDG, which is retained by many tissues, many bacteria-specific imaging agents in development are inert in mammalian tissues and therefore are rapidly eliminated from the body, substantially reducing radiation exposure. Technical advances such as iterative reconstruction algorithms and time-of-flight PET systems have also enabled lower radiation exposures. More recently, total-body PET, with an overall >40-fold gain in effective sensitivity, can generate equivalent image quality with less injected radioactivity than conventional PET, thus potentially expanding the use of this technology in children, in whom radiation dose concerns have impeded its use (37). In addition, total-body PET will offer simultaneous whole-body analysis with four to five times more sensitivity and enable kinetic time profiling of bacteria-specific molecular imaging agents. This will provide a means to identify quantitative “kinetic signatures” of bacterial uptake and retention, which could prove to be much more accurate than the static PET imaging currently used by conventional scanners. Last, the mortality risks for patients with serious infections, especially due to antimicrobial drug–resistant bacteria, are considerably higher than the often theoretical risks of radiation, the risk of mortality with many infections due to multidrug-resistant (MDR) bacteria is similar or higher than 5-year mortality risks due to common cancers (8). Although molecular imaging techniques are used in the management of many cancers, they are usually avoided in infectious diseases, and so, a pragmatic approach is needed to surpass this bias.
Lack of research funding
Governmental and industrial support for developing molecular imaging for infectious diseases is not commensurate with the burden of disease due to bacterial infections. The U.S. Department of Health awarded 1378 projects and 1.97 billion USD in total costs for the development of PET-based approaches to study cancer from 1985 to 2018 (source: National Institutes of Health reporter; see Supplementary Materials and Methods and data file S1). In contrast, over the same period, only 26 grants and 39.61 million USD in total costs were awarded for the development of novel PET approaches to image bacterial infections. Cancer research also gets a boost from initiatives such as “Cancer Moonshot” and “Stand Up to Cancer,” receiving billions of dollars in public and industry funding. The number of publications in this area reflects this large discrepancy in government funding. The field of oncology has about 2324 publications listed on PubMed by the National Center for Biotechnology Information (NCBI) pertaining to the use of PET agents for cancer, excluding the use of 18F-FDG. In contrast, the field of infectious diseases had only 51 publications in the same time period (1985–2018) pertaining to the development of novel PET agents for diagnosing bacterial infection. In addition, the Molecular Imaging and Contrast Agent Database (MICAD) maintained by NCBI currently lists 5359 novel PET agents that were developed for oncology versus only 13 listed for imaging bacterial infection.
UNDERSTANDING PATHOGENESIS AND PRECISION MEDICINE APPROACHES
Much of our understanding of infection micro-environments stems from biopsy and tissue resection. Molecular imaging could help develop new platforms for basic research by providing detailed spatial and temporal information about local microenvironments supporting bacterial survival. Multimodality imaging could simultaneously visualize several different processes (bacterial burden, antibiotic exposure, and local milieu) and allow integration of cross-species data from animals to humans. This will facilitate the study of pathogenesis in patients in an unprecedented fashion that is not feasible with traditional ex vivo tools. Imaging using dual or multi-agent PET studies could also provide accurate data on the classes of bacteria causing the infections as well as the diversity of microbiota.
Antibiotic drug development
Dosing recommendations for antibiotics continue to be developed based on plasma concentrations and historic measures of efficacy. Plasma pharmacokinetics (PK)—essential in the developmental pipeline for new antibiotics—do not always correlate with intralesional PK. Direct measurement cannot be achieved in humans except in rare circumstances and is generally limited to sampling a single, accessible lesion. Molecular imaging with radiolabeled antibiotics can measure in situ biodistribution of antibiotics simultaneously in multiple organ systems/compartments in patients and animal models, without biopsy- related artifacts or alterations to tissue physiology (7, 38).
Understanding local biology at infection sites
Bacteria are known to adapt to their local milieu (hypoxia or nutrient starvation) and develop a quiescent state (“dormancy”), which can successfully evade antibiotics and immune responses for months to decades. Extended antibiotic courses are required to kill this subpopulation of dormant bacteria, although treatments could also be improved by immunotherapies that could alter the local milieu to prevent this bacterial adaptation (39). Specific molecular imaging approaches that measure the local biology could provide insights into pathogenesis and mechanism(s) of novel immunologic therapeutics and could also be used as biomarkers to measure the efficacy and monitor responses to treatments.
Precision medicine
Current antibiotic treatment strategies are designed for efficacy (>90%) at a population level but ignore inter- and intrasubject heterogeneity. Although much shorter treatments could cure >70% of patients, accurate tools to identify at-risk patients requiring longer treatments are lacking. Such tools could decrease the inappropriate use of antimicrobials that contributes to the rise of MDR bacteria. By providing information about the bacterial burden, the location and extent of disease, and response to treatment, molecular imaging using bacteria-specific imaging agents could be a major advance toward developing precision medicine for infectious diseases to accurately phenotype and identify at-risk patients at the time of diagnosis and to optimize individualized therapeutic approaches.
CHALLENGES AND OPPORTUNITIES
Bacterial infections remain a major threat to human health. The rampant increase in antimicrobial drug–resistant bacteria and patient populations susceptible to acquiring infections pose additional challenges to health care. Rather than focusing on previously known targets, unbiased, high-throughput screening approaches that exploit the distinct and highly conserved biochemical pathways in prokaryotes are needed to discover candidate molecules that could be developed as novel bacteria-specific imaging agents (15). High-quality preclinical research is needed to understand the specificity and sensitivity of promising candidates. A detailed understanding of the mechanism(s) of bacterial specificity and accumulation should be sought. Because there is heterogeneity in the presentation of infectious lesions in patients, evaluation of promising imaging agents should be performed in canonical animal models with subsequent and prompt validation in human studies.
The use of empiric antibiotics hinders selection of appropriate patient populations for initial clinical studies and can affect the sensitivity of imaging testing, necessitating stringent proof-of-principle clinical studies in patients with a confirmed diagnosis and those who have received limited duration of antibiotic treatment. Study sites in the developing world should be considered for the recruitment of eligible patients. Where safe and feasible, therapeutic trials using experimental infections in human volunteers, which are growing in acceptance (40), may also be useful.
Molecular imaging techniques are used routinely in the management of many cancers but are avoided in infectious diseases due to perceived risks of radiation, although the morbidity and mortality due to many antimicrobial drug–resistant bacterial infections are similar to or higher than the 5-year mortality risks for common cancers. Advancements in imaging technologies and PK characteristics of many bacteria-specific imaging agents allow for lower radiation exposure. This scientific knowledge needs to be disseminated to patients and their treating clinicians, and the general public needs to be made aware of the threats posed by bacterial infections and antimicrobial drug resistance.
Molecular imaging has the potential to improve diagnostics and to provide prognostic information for patients with bacterial infections and is also broadly applicable to other classes of infectious pathogens. However, the use of novel molecular imaging modalities in infectious diseases lags behind its use in cancer and other fields. A multi-disciplinary approach involving microbiologists, molecular imaging scientists, infectious diseases physicians, and radiologists is necessary to address the key challenges preventing the translation of bacteria-specific imaging tracers. Open sharing of data from research studies, including negative results, is encouraged. Because current research funding for developing molecular imaging tools for bacterial infections is not commensurate with the burden of disease, substantially increased and sustained support for basic and translational research is needed to develop and translate novel diagnostic tools for bacterial infections.
Supplementary Material
Acknowledgments:
We thank M. Pomper (Johns Hopkins Hospital), D. Mankoff (Hospital of the University of Pennsylvania), and S. Gambhir (Stanford University Medical Center) for comments.
Funding: This work was funded by the NIH (Director’s Transformative Research Award R01-EB020539, R01-HL131829, R01-EB025985, and R56-AI145435 to S.K.J. and Director’s Early Independence Award DP5-OD26386 to M.A.S.), the Department of Defense’s Congressionally Directed Medical Research Programs PR-171338P1 to S.K.J., the Burroughs Wellcome Fund Career Award for Medical Scientists to M.A.S., and Johns Hopkins All Children’s Hospital Foundation Institutional Grant Program to E.W.T.
Footnotes
Competing interests: S.K.J. received consulting fees from Mediso Medical Imaging Systems Ltd., unrelated to this work. A.A.O. and S.K.J. are co-inventors on pending patent US20150250906A1 on bacteria-specific labeled substrates as imaging biomarkers, filed by Johns Hopkins University. M.A.S. is a co-inventor on pending patent US20180104365A1 filed by the University of Pennsylvania on radiotracer derivatives of TMP for medical imaging and is a cofounder of Vellum Biosciences. G.G. is an inventor on US20160303259A1 and US20140314671, both of which are held by Stanford University and cover the 6-18F-fluoromaltose and 6″−18F-fluoromaltotriose PET tracers.
Data and materials availability: All data associated with this study are present in the paper or the Supplementary Materials.
SUPPLEMENTARY MATERIALS
stm.sciencemag.org/cgi/content/full/11/508/eaax8251/DC1
Materials and Methods
Data file S1. Primary data.
REFERENCES AND NOTES
- 1.GBD 2017 Causes of Death Collaborators, Global, regional, and national age-sex-specific mortality for 282 causes of death in 195 countries and territories, 1980–2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet 392, 1736–1788 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dye C, After 2015: Infectious diseases in a new era of health and development. Philos. Trans. R. Soc. London B Biol. Sci 369, 20130426 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World Health Organization, Global Tuberculosis Report 2018 (World Health Organization, 2018); www.who.int/gho/tb/en/. [Google Scholar]
- 4.Magill SS, Edwards JR, Bamberg W, Beldavs ZG, Dumyati G, Kainer MA, Lynfield R, Maloney M, McAllister-Hollod L, Nadle J, Ray SM, Thompson DL, Wilson LE, Fridkin SK; Emerging Infections Program Healthcare-Associated Infections; Antimicrobial Use Prevalence Survey Team, Multistate point-prevalence survey of health care–associated infections. N. Engl. J. Med 370, 1198–1208 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention, The Direct Medical Costs of Healthcare-Associated Infections in U.S. Hospitals and the Benefits of Prevention (Centers for Disease Control and Prevention, 2009); www.cdc.gov/hai/pdfs/hai/scott_costpaper.pdf. [Google Scholar]
- 6.O’Neill J, Tackling drug-resistant infections globally: Final report and recommendations (Wellcome Trust and UK Government, 2016); https://amr-review.org/sites/default/files/160525_Final%20paper_with%20cover.pdf. [Google Scholar]
- 7.Tucker EW, Guglieri-Lopez B, Ordonez AA, Ritchie B, Klunk MH, Sharma R, Chang YS, Sanchez-Bautista J, Frey S, Lodge MA, Rowe SP, Holt DP, Gobburu JVS, Peloquin CA, Mathews WB, Dannals RF, Pardo CA, Kannan S, Ivaturi VD, Jain SK, Noninvasive 11C-rifampin positron emission tomography reveals drug biodistribution in tuberculous meningitis. Sci. Transl. Med 10, eaau0965 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jain SK, The promise of molecular imaging in the study and treatment of infectious diseases. Mol. Imaging Biol 19, 341–347 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Strosberg J, El-Haddad G, Wolin E, Hendifar A, Yao J, Chasen B, Mittra E, Kunz PL, Kulke MH, Jacene H, Bushnell D, O’Dorisio TM, Baum RP, Kulkarni HR, Caplin M, Lebtahi R, Hobday T, Delpassand E, Van Cutsem E, Benson A, Srirajaskanthan R, Pavel M, Mora J, Berlin J, Grande E, Reed N, Seregni E, Öberg K, Lopera Sierra M, Santoro P, Thevenet T, Erion JL, Ruszniewski P, Kwekkeboom D, Krenning E; NETTER-1 Trial Investigators, Phase 3 trial of 177Lu-Dotatate for midgut neuroendocrine tumors. N. Engl. J. Med 376, 125–135 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rowe SP, Gorin MA, Pomper MG, Imaging of prostate-specific membrane antigen with small-molecule PET radiotracers: From the bench to advanced clinical applications. Annu. Rev. Med 70, 461–477 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Welling M, Stokkel M, Balter J, Sarda-Mantel L, Meulemans A, Le Guludec D, The many roads to infection imaging. Eur. J. Nucl. Med. Mol. Imaging 35, 848–849 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vinjamuri S, Hall AV, Solanki KK, Bomanji J, Siraj Q, O’Shaughnessy E, Das SS, Britton KE, Comparison of 99mTc infecton imaging with radiolabelled white-cell imaging in the evaluation of bacterial infection. Lancet 347, 233–235 (1996). [DOI] [PubMed] [Google Scholar]
- 13.Sarda L, Cremieux A-C, Lebellec Y, Meulemans A, Lebtahi R, Hayem G, Génin R, Delahaye N, Huten D, Le Guludec D, Inability of 99mTc-ciprofloxacin scintigraphy to discriminate between septic and sterile osteoarticular diseases. J. Nucl. Med 44, 920–926 (2003). [PubMed] [Google Scholar]
- 14.Sellmyer MA, Lee I, Hou C, Weng C-C, Li S, Lieberman BP, Zeng C, Mankoff DA, Mach RH, Bacterial infection imaging with [18F]fluoropropyl-trimethoprim. Proc. Natl. Acad. Sci. U.S.A 114, 8372–8377 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ordonez AA, Weinstein EA, Bambarger LE, Saini V, Chang YS, DeMarco VP, Klunk MH, Urbanowski ME, Moulton KL, Murawski AM, Pokkali S, Kalinda AS, Jain SK, A systematic approach for developing bacteria-specific imaging tracers. J. Nucl. Med 58, 144–150 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mac Faddin JF, Biochemical Tests for Identification of Medical Bacteria (Williams & Wilkins Baltimore, 1976). [Google Scholar]
- 17.Mutch CA, Ordonez AA, Qin H, Parker M, Bambarger LE, Villanueva-Meyer JE, Blecha J, Carroll V, Taglang C, Flavell R, Sriram R, VanBrocklin H, Rosenberg O, Ohliger MA, Jain SK, Neumann KD, Wilson DM, [11C]Para-aminobenzoic acid: A positron emission tomography tracer targeting bacteria-specific metabolism. ACS Infect. Dis 4, 1067–1072 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Z, Ordonez AA, Wang H, Li Y, Gogarty KR, Weinstein EA, Daryaee F, Merino J, Yoon GE, Kalinda AS, Mease RC, Iuliano JN, Smith-Jones PM, Jain SK, Tonge PJ, Positron emission tomography imaging with 2-[18F]F- p-aminobenzoic acid detects Staphylococcus aureus infections and monitors drug response. ACS Infect. Dis 4, 1635–1644 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gowrishankar G, Namavari M, Jouannot EB, Hoehne A, Reeves R, Hardy J, Gambhir SS, Investigation of 6-[18F]-fluoromaltose as a novel PET tracer for imaging bacterial infection. PLOS ONE 9, e107951 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ning X, Seo W, Lee S, Takemiya K, Rafi M, Feng X, Weiss D, Wang X, Williams L, Camp VM, Eugene M, Taylor WR, Goodman M, Murthy N, PET imaging of bacterial infections with fluorine-18-labeled maltohexaose. Angew. Chem. Int. Ed. Engl 53, 14096–14101 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neumann KD, Villanueva-Meyer JE, Mutch CA, Flavell RR, Blecha JE, Kwak T, Sriram R, VanBrocklin HF, Rosenberg OS, Ohliger MA, Wilson DM, Imaging active infection in vivo using D-amino acid derived PET radiotracers. Sci. Rep 7, 7903 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Petrik M, Umlaufova E, Raclavsky V, Palyzova A, Havlicek V, Haas H, Novy Z, Dolezal D, Hajduch M, Decristoforo C, Imaging of Pseudomonas aeruginosa infection with Ga-68 labelled pyoverdine for positron emission tomography. Sci. Rep 8, 15698 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weinstein EA, Ordonez AA, DeMarco VP, Murawski AM, Pokkali S, MacDonald EM, Klunk M, Mease RC, Pomper MG, Jain SK, Imaging Enterobacteriaceae infection in vivo with 18F-fluorodeoxysorbitol positron emission tomography. Sci. Transl. Med 6, 259ra146 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levin PA, Angert ER, Small but mighty: Cell size and bacteria. Cold Spring Harb. Perspect. Biol 7, a019216 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.König C, Simmen HP, Blaser J, Bacterial concentrations in pus and infected peritoneal fluid—Implications for bactericidal activity of antibiotics. J. Antimicrob. Chemother 42, 227–232 (1998). [DOI] [PubMed] [Google Scholar]
- 26.Canetti G, Present aspects of bacterial resistance in tuberculosis. Am. Rev. Respir. Dis 92, 687–703 (1965). [DOI] [PubMed] [Google Scholar]
- 27.American Thoracic Society; Infectious Diseases Society of America, Guidelines for the management of adults with hospital-acquired, ventilator-associated, and healthcare-associated pneumonia. Am. J. Respir. Crit. Care Med 171, 388–416 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Vall-Spinosa A, Lester W, Moulding T, Davidson PT, McClatchy JK, Rifampin in the treatment of drug-resistant Mycobacterium tuberculosis infections. N. Engl. J. Med 283, 616–621 (1970). [DOI] [PubMed] [Google Scholar]
- 29.O’Neill AJ, Cove JH, Chopra I, Mutation frequencies for resistance to fusidic acid and rifampicin in Staphylococcus aureus. J. Antimicrob. Chemother 47, 647–650 (2001). [DOI] [PubMed] [Google Scholar]
- 30.Wahl RL, Jacene H, Kasamon Y, Lodge MA, From RECIST to PERCIST: Evolving considerations for PET response criteria in solid tumors. J. Nucl. Med 50 (suppl. 1), 122S–150S (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsou TP, Lee PI, Lu CY, Chang LY, Huang LM, Chen JM, Hsueh PR, Lee CY, Microbiology and epidemiology of brain abscess and subdural empyema in a medical center: A 10-year experience. J. Microbiol. Immunol. Infect 42, 405–412 (2009). [PubMed] [Google Scholar]
- 32.Jankharia GR, Commentary—Radiology in India: The next decade. Indian J. Radiol. Imaging 18, 189–191 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rigamonti D, Liem L, Sampath P, Knoller N, Namaguchi Y, Schreibman DL, Sloan MA, Wolf A, Zeidman S, Spinal epidural abscess: Contemporary trends in etiology, evaluation, and management. Surg. Neurol 52, 189–196; discussion 197 (1999). [DOI] [PubMed] [Google Scholar]
- 34.Ebenhan T, Zeevaart JR, Venter JD, Govender T, Kruger GH, Jarvis NV, Sathekge MM, Preclinical evaluation of 68Ga-labeled 1,4,7-triazacyclononane-1,4,7-triacetic acid-ubiquicidin as a radioligand for PET infection imaging. J. Nucl. Med 55, 308–314 (2014). [DOI] [PubMed] [Google Scholar]
- 35.Ehman EC, Johnson GB, Villanueva-Meyer JE, Cha S, Leynes AP, Larson PEZ, Hope TA, PET/MRI: Where might it replace PET/CT? J. Magn. Reson. Imaging 46, 1247–1262 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salazar-Austin N, Ordonez AA, Hsu AJ, Benson JE, Mahesh M, Menachery E, Razeq JH, Salfinger M, Starke JR, Milstone AM, Parrish N, Nuermberger EL, Jain SK, Extensively drug-resistant tuberculosis in a young child after travel to India. Lancet Infect. Dis 15, 1485–1491 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cherry SR, Badawi RD, Karp JS, Moses WW, Price P, Jones T, Total-body imaging: Transforming the role of positron emission tomography. Sci. Transl. Med 9, eaaf6169 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ordonez AA, Bambarger LE, Jain SK, Weinstein EA, Biodistribution and pharmacokinetics of antimicrobials, in Imaging Infections: From Bench to Bedside, Jain SK, Ed. (Springer International Publishing, 2017), pp. 209–222. [Google Scholar]
- 39.Kaufmann SHE, Dorhoi A, Hotchkiss RS, Bartenschlager R, Host-directed therapies for bacterial and viral infections. Nat. Rev. Drug Discov 17, 35–56 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roestenberg M, Hoogerwerf MA, Ferreira DM, Mordmüller B, Yazdanbakhsh M, Experimental infection of human volunteers. Lancet Infect. Dis 18, E312–E322 (2018). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.