Abstract
Mammary tumor cells derived from vitamin D receptor (VDR) knock-out (KO) mice were engineered to stably express wild-type or mutated VDR for characterization of the mechanisms by which 1,25-dihydroxyvitamin D (1,25D), the VDR ligand, mediates growth regulation. Although KO cells were completely resistant to 1,25D, introduction of wild-type human VDR restored gene expression and growth inhibition in response to 1,25D and a variety of structural analogs. Pdgfb, Vegfa and Nfkbi were identified as genomic targets of both human and murine VDR signaling in this cell model. KO cells expressing hVDRs containing point mutations (W286R, R274L) that reduce or abolish ligand binding did not exhibit changes in gene expression or growth in response to physiological doses of 1,25D but did respond to higher doses and more potent analogs. KO cells expressing hVDR with the G46D point mutation, which abrogates VDR binding to DR3 response elements, exhibited partial growth inhibition in response to 1,25D and synthetic vitamin D analogs, providing proof of principle that VDR signaling through alternative genomic or non-genomic mechanisms contributes to vitamin D mediated growth effects in transformed cells. We conclude that the 1,25D-VDR signaling axis that triggers anti-cancer effects is highly conserved between the murine and human systems despite differences in VDR protein, cofactors, and target genes and that these actions are not solely mediated via canonical VDRE signaling.
Keywords: Vitamin D, Breast cancer, Vitamin D analog, Genomic, Non-genomic
Introduction
Both the vitamin D receptor (VDR) and its ligand, 1,25-dihydroxyvitamin D (1,25D), are implicated in regulation of proliferation, apoptosis, differentiation and invasion of breast cancer cells, but the specific molecular mechanisms remain undefined. VDR agonists that display enhanced growth inhibitory potency with minimal calcemic effects provide proof of principle that these distinct actions of VDR can be dissociated, yet the underlying mechanisms remain unclear. As a transcription factor, VDR interacts with structurally distinct vitamin D response elements (VDREs) in target genes, including the consensus direct-repeat with 3 base pair spacer (DR3) which is present in the promoters of many 1,25D-inducible genes, including CYP24 [1]. In other VDR target genes, functional VDREs composed of inverted palindromes with 9-base pair spacers (IP9), everted repeats with 6 or 9-base pair spacers (ER6, ER9), and direct repeats with 4 or 6-base pair spacers (DR4, DR6) have been described [2–7]. In some contexts, VDR binding to IP9–type VDREs correlates with induction of apoptosis, and vitamin D analogs have been reported that preferentially enhance VDR binding to IP9 VDREs [8,9]. The 1,25D-VDR complex can also interact with other transcription factors and induce gene expression via non-VDRE elements. For example, 1,25D promotes VDR binding to Sp1, forming a complex which activates the p27 promoter through Sp1 transcription sites [10]. In addition to genomic effects, 1,25D mediates effects of 1,25D at the plasma membrane and/or in the cytosol through interaction with extra-nuclear VDR or with other proteins such as calcium channels, ERp57, RhoA/ROCK or G-coupled receptors [11–15].
Development of effective VDR modulators as cancer therapeutics requires a more precise understanding of the molecular events by which the natural ligand 1,25D triggers growth inhibition. Although the VDR is required for the anti-proliferative effects of 1,25D, no studies have examined the impact of polymorphic VDRs or mutant VDRs in a cell biology context in general, or in breast cancer in particular. This research gap is directly related to the lack of a model system to differentiate between ligand dependent, ligand independent and DNA independent effects of VDR in a cellular context. Prior attempts to recapitulate VDR-mediated growth inhibitory effects of 1,25D in established human cancer cell lines with low or no detectable VDR protein expression have had mixed results. Stable transfection of VDR into JCA-1 prostate adenocarcinoma cells, which are unresponsive to 1,25D-mediated growth inhibition, increased CYP24 transcription and sensitized cells to growth inhibition by 1,25D [16]. Re-introduction of VDR in C6 rat glioma cells was necessary, but not sufficient, for induction of growth arrest, since some cell lines with detectable VDR protein failed to respond to 1,25D [17]. Most recently, stable expression of VDR in estrogen receptor positive and negative breast cancer cell lines did not confer growth inhibition following treatment with the analog 1α-hydroxyvitamin D5, even though CYP24 gene expression was increased in both cell lines [18]. Therefore, while recapitulation of 1,25D mediated growth arrest in non-responsive cells has been demonstrated via stable transfection of VDR, this has yet to be successfully accomplished in mammary derived cell lines or on a true VDR null background.
The goal of these studies was to create and test a mammary tumor cell model with differential VDR expression from one parental VDR null cell line. In previous work, we established cell lines from mammary tumors generated in wild-type (WT) and VDR knockout (KO) mice and demonstrated proof of principle that the VDR is required for 1,25D mediated growth effects both in vitro and in vivo. Further, we showed that transient transfection of human VDR into the murine VDR null cell line could restore 1,25D-mediated transcription of a heterologous CYP24 promoter [19–21]. In this follow-up work, we have created KO cell lines stably expressing wild-type human VDR (hVDR) or hVDR bearing point mutations that disrupt either ligand binding or DNA binding. These mutations were originally described in patients diagnosed with hereditary vitamin D resistant rickets (HVDRR) type II, indicating that the encoded VDR proteins are deficient in maintenance of calcium homeostasis in vivo. Our data indicate that stable expression of hVDR is necessary and sufficient to confer sensitivity to 1,25D-mediated growth inhibition and apoptosis in VDRKO murine mammary cells and provide new insight into the mechanisms underlying VDR action.
Materials and Methods
Cell Culture:
WT145 and KO240 cells [21] were maintained in DMEM/F12 medium containing 5% charcoal-stripped fetal bovine serum (CSS). Stably transfected cell lines (described below) were maintained in DMEM/F12 medium containing 5% CSS and 500μg/mL hygromycin B. All cells were routinely passaged twice weekly using trypsin/EDTA.
hVDR expression vectors and stable cell selection:
A hygromycin resistance sequence obtained by PCR from pcDNA3.1/Hygro(+) (Life Technologies, Carlsbad, CA) was inserted into the pSG5-hVDR expression vector (gift from Dr. Paul MacDonald, Case Western Reserve University) to generate pSG5-hVDR-hygro. An empty vector control (pSG5-hygro) was generated from pSG5-hVDR-hygro by removal of the hVDR coding sequence. Site directed mutagenesis was used to create the G46D, R274L and W286R mutations to hVDR in this vector (SeqWright, Houston, TX). All inserts were sequenced to confirm that the point mutations were achieved in the absence of any other sequence alterations. Stably expressing cell lines were generated from KO240 cells transfected with 10ng pSG5-hVDR-hygro, pSG5-hVDRG46D-hygro, pSG5-hVDRR274L-hygro, pSG5-hVDRW286R-hygro or pSG5-hygro using TransFast transfection reagent (Promega, Madison, WI) according to manufacturer’s recommendations. 48 hours post-transfection, cells were trypsinized, pooled and incubated in DMEM/F12 medium with 5% CSS and 500μg/mL hygromycin B. After 24 hours, cells were switched to DMEM/F12 with 5% CSS, grown to 80% confluency and passaged. This selection process was repeated twice to generate stable transfectants. Multiple lines stably expressing each wild-type or mutant receptor were generated and characterized.
Western Blotting:
Cells were treated 24 hours after plating with 100nM 1,25D or ethanol vehicle, and whole cell lysates were prepared 48 hours later by scraping into Laemmli buffer containing protease and phosphatase inhibitors. Lysates were sonicated, and protein concentration determined with the BCA protein assay (Pierce Biotechnology, Rockford, IL). 50μg of lysate was separated via SDS-PAGE, transferred to nitrocellulose filters, blocked in skim milk, and incubated overnight with primary antibodies including mouse monoclonal VDR clone D-6, 1:100 (Santa Cruz Biotechnology, Santa Cruz, CA), mouse monoclonal CYP24, 1:200 (gift from Cytochroma, Markham, Ontario) and goat polyclonal actin, 1:100 (Santa Cruz Biotechnology). Appropriate horseradish-peroxidase conjugated secondary antibodies (obtained from Amersham Biosciences) were incubated with filters for 1 hour. Specific bands were detected via chemiluminescence (SuperSignal West Dura, Pierce) and exposure to x-ray film. Films were scanned with a flatbed computer scanner.
VDR Transactivation Assay:
Cells were transfected 24 hours after plating with the pGL3–24 hydroxylase luciferase reporter vector (approximately 300bp of the rCYP24 gene promoter containing two DR3 VDRE regions, gift of the late Dr. Jack Omdahl) and a pRL-TK driven luciferase plasmid (Promega) using TransFast transfection reagent. 24 hours post-transfection, cells were treated with 1,25D at the indicated doses or ethanol vehicle. Cells were harvested after 24 hours with Passive Lysis Buffer and luminescence was read via the Dual Luciferase system (Promega). CYP24 reporter luciferase values were corrected for transfection efficiency by normalization against co-transfected pRL-TK luciferase readings. Data are expressed as relative luciferase activity (RLU) of cells treated with vitamin D metabolites/analogs vs. ethanol control treated cells which were set to 100%.
Crystal Violet Cell Growth Assay and Doubling Time:
Cells were seeded in 24-well plates at a density of 2,000 cells per well and treated 24 hours later with the indicated doses of test compounds. 96 hours post-treatment, cells were fixed with 1% glutaraldehyde, stained with 0.1% crystal violet dye and air dried. Dye was resuspended in 0.2% Triton-X100 and absorbance, which is proportional to cell density under these assay conditions, was measured at 590nm. For doubling time, cells were treated and processed as described above daily for five days, and doubling time calculated according to measured absorbance.
Caspase Activity:
Cells were seeded in 150mm dishes (Corning, Corning, NY) at a density of 300,000 cells per dish, and treated 24 hours later with 100nM 1,25D or vehicle control. 48 hours post-treatment, cells were harvested via trypsinization. One million cells per treatment were assayed using Caspase-GLO luminescent activity assays (Promega), according to manufacturer’s recommendations.
MIcroscopy:
Cells were seeded in 2-well chamber slides at a density of 10,000 cells per chamber, and treated 24 hours later with 100nM 1,25D or vehicle control. For visualization of apoptotic morphology cells were fixed after 96 hours, permeabilized with ice cold methanol and incubated with Hoechst 33342 nuclear dye (Life Technologies). For VDR staining, cells were processed similarly after 24 hours treatment and incubated with VDR antibody clone D-6 (Santa Cruz Biotechnology), followed by anti-mouse Alexa Fluor 488 (Life Technologies) secondary antibody and counterstaining nuclei with Hoechst. Coverslips were mounted with anti-fade mounting medium and images were acquired under phase contrast, UV and/or epifluorescence on an Olympus Provis AX70 microscope equipped with a Spot RT Slider digital camera.
RT2 Profiler PCR Array and qPCR Validation:
Cultures were seeded at a density of 500,000 cells/100mm dish and treated 24 hours later with 100nM 1,25D or vehicle control. 12 hours post-treatment, RNA was harvested with the RNeasy Mini Kit (Qiagen, Valencia, CA) and cDNA was synthesized with the TaqMan Reverse Transcription Reagents kit (Applied Biosystems, Foster City, CA). Gene expression analysis was performed in triplicate with the Mouse Cancer Pathway Finder PCR Array (PAMM-033, SA Biosciences, Frederick, MA), and data were analyzed with the PCR Array Data Analysis Software available from SA Biosciences. A subset of genes identified as potential 1,25D/VDR targets from the PCR array were independently analyzed by qPCR using SYBR-green master mix and primers from Invitrogen. WT145 and KOhVDR-F cells were treated the day after plating with 100nm 1,25D and harvested 6, 12 or 24 hrs after treatment. Data were normalized against 18sRNA and expressed as fold-change relative to values from WT145 control cells harvested at 6hr.
Statistical Analysis:
Data are expressed as mean +/− standard error. ANOVA or Student’s t-test was performed using GraphPad Prism software, and means were considered statistically significant when p-values less than 0.05 were obtained. Statistical significance is indicated on all data figures as asterisks.
Results
Effect of re-introduction of hVDR into VDR null murine cells.
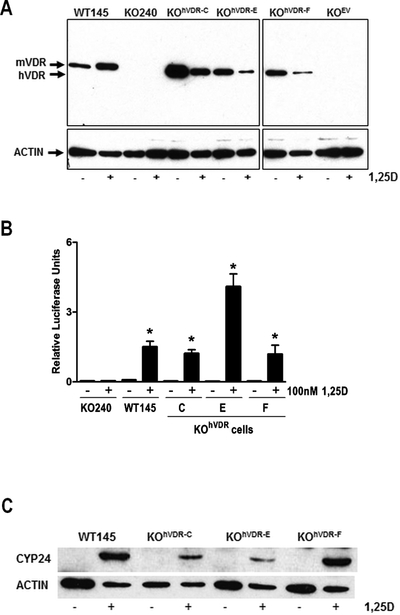
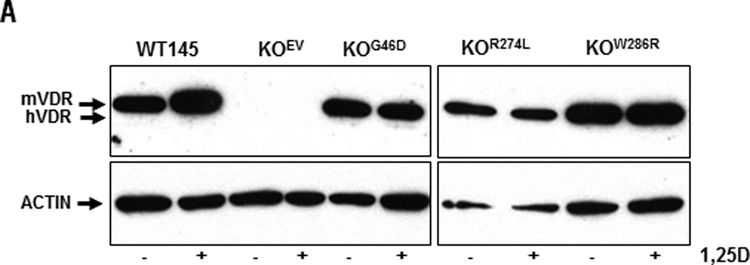
To create a mammary tumor cell model system with differential VDR expression, we stably transfected the hVDR coding sequence into a VDR negative murine cell line (KO240) which we previously established from a VDRKO mouse breast tumor. Three independently derived clonal lines expressing hVDR (designated KOhVDR lines C, E and F) and one line expressing the empty vector (designated KOEV) were characterized in relation to the parental KO240 cells. The responses of these cell lines were also compared to that of WT145 cells, a companion cell line that expresses murine (m) VDR. On western blots (Figure 1A) the VDR antibody identified a 48kDa protein in all three engineered KOhVDR cell lines that migrated just below mVDR (50kDa) in WT145 cells and was not detected in KO240 or KOEV cells. Basal VDR expression was higher in line C than in lines E and F, but roughly comparable to that of WT145 cells. In response to 48h treatment with 100nM, VDR protein was up regulated in WT145 cells but down regulated in all KOhVDR cell lines.
Figure 1. Expression and function of VDR protein in WT145 and engineered VDRKO cell lines.
A. Whole cell lysates of WT145, KO240, KOhVDR-C, KOhVDR-E, KOhVDR-F and KOEV cultures treated for 48h with 100nM 1,25D or vehicle were immunoblotted with antibodies directed against VDR (top) and actin (bottom). Blot is representative of three independent preparations for each cell line. Arrows indicate position of murine (m) and human (h) VDR proteins at 50kDa and 48kDa respectively. B. CYP24 reporter gene activity in KO240, WT145, KOhVDR-C, KOhVDR-E, and KOhVDR-F cells treated with vehicle or 100nM 1,25D for 24h. Data were normalized for transfection efficiency measured by co-transfected pRL-TK and are expressed as fold increase in relative luciferase activity (RLU) of 1,25D treated versus vehicle treated for each cell line. Bars represent mean ± SEM of four-six values. Asterisks indicate significant difference from respective control. C. Whole cell lysates of WT145, KOhVDR-C, KOhVDR-E, and KOhVDR-F cells treated for 48h with 100nM 1,25D or vehicle were immunoblotted with antibodies directed against CYP24 (top) and actin (bottom); blot is representative of three independent experiments.
Since 1,25D binding is known to stabilize the VDR, the ligand-induced down regulation of VDR protein in all three 1,25D treated KOhVDR cell lines was unexpected. We further investigated this phenomenon in single cell clones isolated from the KOhVDR-F parental line which behaved similarly to the parental cell line with respect to 1,25D mediated growth inhibition (representative doubling times are shown in Supplemental Table 1). Single cell clones were selected in order to reduce the heterogeneity of VDR expression in the mass cultures which we hypothesized could lead to apparent VDR down-regulation secondary to outgrowth of cells lacking VDR. In contrast to this expectation, 1,25D treatment down-regulated the steady state expression of hVDR protein in both newly isolated clones (Supplemental Figure 1A). In both clones, 1,25D treatment also reduced hVDR transcript levels as measured by real-time PCR (Supplemental Figure 1B). We also considered the possibility that 1,25D treatment might inhibit the SV40 promoter which drives VDR expression in this model. However, 1,25D did not suppress the activity of the SV40 promoter as measured in a luciferase reporter assay (Supplemental Figure 1C). Thus, the underlying basis for ligand induced down regulation of hVDR transcript and protein when expressed in the context of a VDRKO murine cell background remains unclear.
Given the unusual down regulation of VDR protein in the engineered cell lines, we sought to compare VDR function in the KOhVDR cell lines to that in the WT145 cells using induction of a known VDR target gene (CYP24) as readout. In transient transfection assays, 1,25D significantly induced the rat CYP24 promoter luciferase construct in WT145 cells and in the KOhVDR cell lines but was without effect in KOEV cells (Figure 1B). Western blot analysis indicated that the CYP24 protein was not detectable in any of the cell lines in the absence of 1,25D but was induced in all three KOhVDR cell lines and in WT145 cells (Figure 1C). No induction of CYP24 protein was detected in KO240 or KOEV cells (Supplemental Figure 2). These data confirm that hVDR exerts functional transcriptional activity on both endogenous and exogenous promoters when stably expressed in murine VDR negative cells.
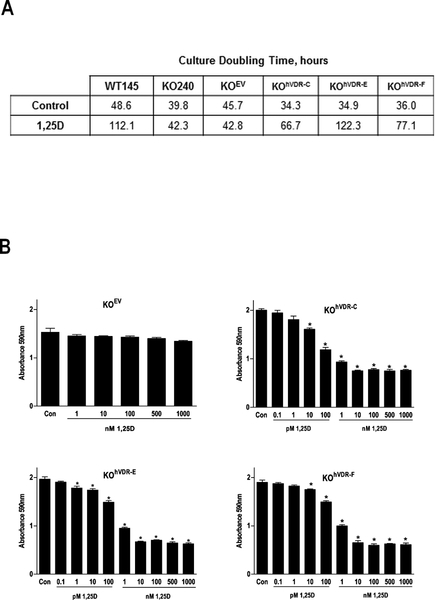
We next examined the impact of VDR on the growth of the KOhVDR cell lines in the presence and absence of 1,25D. Cells treated with ethanol vehicle or 100nM 1,25D were harvested daily for analysis of cell density and doubling times were calculated (Figure 2A). In the absence of 1,25D, the doubling times of the six cell lines ranged from 34–48h and did not correlate with VDR expression. Comparison of doubling times for KO240 cells and KOEV cells (40h and 46h respectively) to that of the hVDR expressing derivatives (range 34–36h) indicated that introduction of VDR in the absence of 1,25D did not slow cell growth (Figure 2B). This observation argues against a ligand independent effect of VDR on cell growth kinetics. However, in the presence of 100nM 1,25D, doubling times of WT145 cells and all three KOhVDR cell lines (but not KO240 or KOEV cells) were significantly increased (between 2 and 3 fold). Dose response assays (Figure 2B) indicated that 96 hour treatment with concentrations of 1,25D as low as 10pM reduced cell denisty of all three KOhVDR cell lines. Under the same conditions, 1,25D had no effect on growth of KOEV cells even at concentrations as high as 1μM. These data indicate that stable expression of VDR confers cellular sensitivity to 1,25D mediated growth inhibition at physiologically relevant concentrations.
Figure 2. Expression of human VDR in murine VDRKO cells confers 1,25D mediated growth inhibition.
A. Doubling times for WT145, KO240, KOEV, KOhVDR-C, KOhVDR-E and KOhVDR-F cell lines treated with 100nM 1,25D or vehicle were calculated from crystal violet stained cultures harvested for five consecutive days. B. Dose dependent effects of 1,25D in KOEV, KOhVDR-C, KOhVDR-E and KOhVDR-F cultures on cell density after 96 h treatment. Data are expressed as absorbance of crystal violet dye, which is proportional to cell density under the conditions used. Data points represent mean ± SEM of four values; bars with asterisks are statistically different from vehicle control.
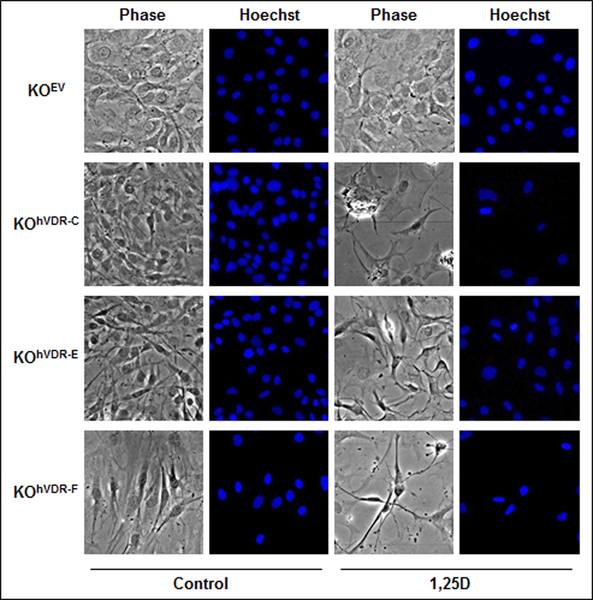
Phase contrast microscopy was used to assess the impact of VDR expression on the morphology of the KO240 derivative cell lines in the presence and absence of 100nM 1,25D. As shown in Figure 3, these tumor-derived cell lines exhibit fibroblastic morphology indicative of an epithelial-mesenchymal transition. In basal media, no consistent changes in morphology were observed after stable expression of hVDR in the VDR-negative cell lines. In contrast, hVDR-expressing cell lines exposed to 100nM 1,25D for 96 hours exhibited morphological changes indicative of apoptosis including cell shrinkage, pyknotic nuclei and reduced adherence. The morphology of KOEV cells was not altered by treatment with 100nM 1,25D.
Figure 3. 1,25D induces apoptotic morphology in KOhVDR cell lines, but not in KOEV cells.
KOEV, KOhVDR-C, KOhVDR-E and KOhVDR-F cells were treated with 100nM 1,25D or vehicle for 96h. Cells were incubated with Hoechst dye and identical fields were photographed under phase contrast and UV fluorescence to visualize nuclear morphology.
In previous studies we demonstrated that 1,25D induces caspase activity in both human and murine derived breast cancer cells. To explore whether apoptosis was triggered by 1,25D in the hVDR-expressing murine cells, the activity of several caspases was measured in WT145, KO240 and KOhVDR cell lines treated with 100nM 1,25D for 48h (Supplemental Table 1). This data confirmed our previous finding that 1,25D activates caspase 3 activity in WT145 cells but not in KO240 cells [21]. With respect to the KOhVDR cell lines, 1,25D increased the activity of several caspases in KOhVDR lines C and F but not in line E. Further analysis would be necessary to determine whether 1,25D alters caspase activity in KOhVDR-E with different kinetics or at different doses. Statistical analysis of the pooled data from KOhVDR-D and KOhVDR-F cell lines indicated that 1,25D significantly increased caspase 3 activity through hVDR in the murine cells. Although there were trends for 1,25D to enhance the activity of caspases 2 and 9 in WT145 cells and in the KOhVDR cell lines, the differences were not statistically significant.
Response of hVDR expressing cells to 25-hydroxvitamin D and synthetic VDR agonists.
Both WT145 and KO240 cells express CYP27B1, the mitochondrial hydroxylase that converts 25-hydroxyvitamin D (25D) to 1,25D [21]. The presence of active CYP27B1 confers sensitivity to 25D mediated growth inhibition in WT145 cells but not in KO240 cells. We therefore assessed whether expression of hVDR in the KO240 cellular context would enable growth inhibitory responses to 25D. Cell density in the presence and absence of increasing doses of 25D was compared in KOEV, KOhVDR-C, KOhVDR-E and KOhVDR-F cells after 96 hours. As shown in Figure 4, 25D inhibited growth of KOhVDR-F, KOhVDR-C and KOhVDR-E cells at 500nM but had no effect on growth of KOEV cells at any concentration tested. The high concentration of 25D required to impact growth of these cell lines likely reflects the presence of DBP (which impedes uptake of 25D into cells lacking megalin and cubilin) in the serum-containing media [22].
Figure 4. KOhVDR cells are growth inhibited by 25D.
Crystal violet growth assay in KOEV, KOhVDR-C, KOhVDR-E and KOhVDR-F cells treated for 96 hours with 25D at indicated concentrations or vehicle control. Data are expressed as absorbance of crystal violet dye, which is proportional to cell density under the conditions used. Data points represent mean ± SEM of four values. For all graphs, bars with asterisks are significantly different from control.
Synthetic vitamin D analogs with more potent anti-proliferative effects than 1,25D have been developed as potential therapeutics, but their mechanism of action remains poorly understood. We previously reported [23] that growth of KO240 cells was unaffected in the presence of nanomolar concentrations of synthetic vitamin D analogs developed by LEO Pharmaceuticals (Ballerup, Denmark). Here we examined whether introduction of functional VDR into KO240 cells was permissive for conferring sensitivity to these analogs. Dose response assays to the analog EB1089 (Figure 5A) indicated that the growth of all three KOhVDR cell lines was inhibited at concentrations as low as 1nM. Dose response studies indicated that the growth of all three KOhVDR cell lines was also reduced in response to the 1,25D analogs CB1093, MC1288, and KH1230. Figure 5B shows the relative cell density of KOhVDR cells treated with low (1nM) and high (1 μM) concentrations of these analogs compared to vehicle. None of the synthetic vitamin D analogs significantly altered growth of the control KOEV cells, even at concentration as high as1μM (Figure 5A,B). Though there were slight differences between cell lines in their growth inhibitory responses to the synthetic analogs, all were maximally inhibited at the lowest concentration tested (1nM). Collectively, these data indicate that stable expression of hVDR into VDR null murine cells is permissive for mediating growth inhibition in response to both natural (1,25D and 25D) and synthetic (EB1089, CB1093, MC1288, and KH1230) vitamin D compounds.
Figure 5. Vitamin D analogs induce growth arrest in KOhVDR cells.
A. Cell density was measured by crystal violet staining in KOEV, KOhVDR-C, KOhVDR-E and KOhVDR-F cultures after treatment for 96h with EB1089 at indicated concentrations or vehicle control. Data points represent mean ± SEM of four values, and bars with asterisks are significantly different from control. B. Crystal violet growth assays were conducted in the indicated cell lines after treatment for 96h with CB1093, MC1288, or KH1230 at 1nM or 1μM concentration. Data represent mean of four values and are expressed as percentage of vehicle treated cells which were set to 100%. Asterisks signify significant difference from control.
Analysis of gene expression changes in response to 1,25D.
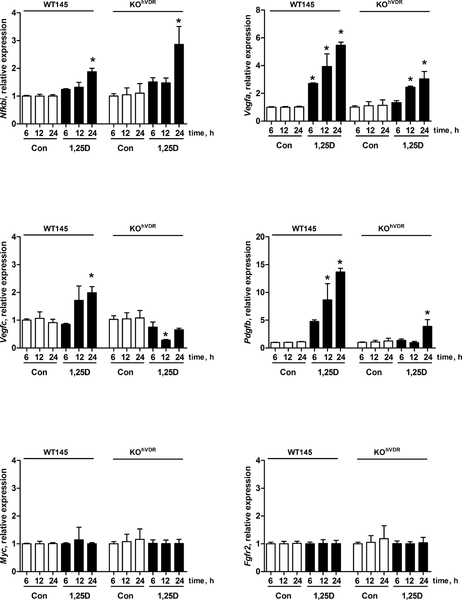
To gain insight into potential downstream pathway(s) altered by the 1,25D/VDR complex, we used focused PCR Arrays to profile the expression of 84 genes representative of six biological pathways involved in transformation and tumorigenesis (Mouse Cancer Pathway Finder PCR Array #PAMM-033, SA Biosciences, Frederick, MD). We sought to identify genes that were commonly altered by 1,25D in WT145 cells and KOhVDR-F cells since growth of both cell lines was inhibited by 1,25D treatment. We conducted the PCR array analysis after 12h of treatment with 100nM 1,25D in order to identify early mediators of the response. Of the 84 genes represented on the PCR array, nine were altered two-fold or more in WT145 cells (c-Myc, Vegfa, Nfkbia, Tnfrsf1a, c-Jun, Pdgfb, Fgfr2, Tnf, Syk), and 21 were altered two-fold or more in KOhVDR-F cells (Vegfc, Pdgfb, Vegfa, Cdkn2a, Igf1, Egfr, Fos, Ifnb, Kiss1, Itga4, Tert, Cdh1, Cdk4, Muc1, Angpt1, Fas, Pik3r1, Fgf1, Itga2, Casp8, Mmp9). From these lists we chose Myc, Nfkbi, Fgfr2, Pdgfb, Vegfa and Vegfc for follow-up by qPCR in WT145 and KOhVDR-F cell lines treated with 1,25D for up to 24h. Of these genes, neither Myc nor Fgfr2 were regulated by 1,25D in either cell line (Figure 6). Pdgfb, Vegfa and Nfkbi were up-regulated by 1,25D in both WT145 and KOhVDR-F cells, but (with the exception of Nfkbi) the induction of these genes by 1,25D was more robust and consistent in WT145 cells expressing endogenous murine VDR than in KOhVDR-F cells expressing ectopic human VDR. Vegfc was induced in WT145 cells but not in KOhVDR-F cells. These differences may reflect species specificity in gene regulation by mVDR (in WT145 cells) versus hVDR (in KOhVDR-F cells) or inherent differences in the genomes of the cell lines themselves.
Figure 6. Effects of 1,25D on gene expression in WT and KOhVDR cells.
RNA was isolated from WT145 and KOhVDR-F cells treated with 100nM 1,25D for 6, 12 or 24 h. Gene expression was analyzed by real time PCR using primers against the murine Myc, Fgfr2, Nfkbi, Pdgfb, Vegfa and Vegfc. Data are mean ± SEM of three independent cell harvests, each analyzed in duplicate. Values are expressed as fold-change relative to vehicle treated WT145 cells harvested at 6h. Asterisks signify significant difference between control and 1,25D treated values for each cell line at the indicated time point.
Characterization of KO240 cell lines expressing mutant hVDR proteins.
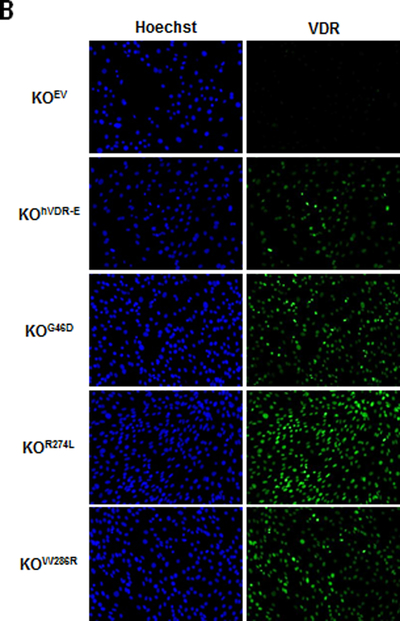
To probe structure-function relationships between hVDR and growth inhibition, we created KO cell lines stably expressing hVDRs containing point mutations that either abolish DNA binding to DR3-type VDREs (G46D) or impair ligand binding (R274L, W286R). Western blotting and immunofluorescent microscopy was used to confirm the expression of mutant VDRs in these newly created stable cell lines. On western blots, the VDR antibody identified a 48kDa protein in lysates of KOG46D, KOR274L and KOW286R stable cell lines but not in KOEV cells (Figure 7A). There was no evidence for truncated products of any of the mutant VDR proteins (data not shown). When protein loading was taken into account, the level of expression for the ectopic VDRs in the stable cell lines was approximately equal to that of endogenous wild-type VDR in the WT145 cells. Comparative VDR western blots in the WT145 cells and several engineered KO cell lines (Supplemental Figure 3) indicate that the level of VDR expressed in the KOG46D cells was roughly equivalent to that achieved in KOhVDR-E and KOhVDR-F cell lines. Immunofluorescence was used to verify that the majority of cells in each population retained VDR expression after continued passage and that VDR expression was equivalent to that achieved in KO cell lines expressing non-mutated hVDR such as KOhVDR-E (Figure 7B).
Figure 7. Expression of LBD and DBD mutant VDR proteins in KO240 cells.
A. Whole cell lysates of WT145, KOEV, KOG46D, KOR274L and KOW286R cells treated for 48 hours with 100nM 1,25D or vehicle were immunoblotted with antibodies directed against VDR (top) and actin (bottom) as loading control. Blot is representative of two independent whole cell lysate preps for each cell line. Arrows indicate position of murine (m) and human (h) VDR proteins at 50kDa and 48kDa respectively. B. KOEV, KOhVDR-E, KOG46D, KOR274L, and KOW286R cells treated with 100nM 1,25D for 48h were fixed and incubated with Hoechst and VDR antibody to confirm that the majority of cells in each stable cell line express VDR.
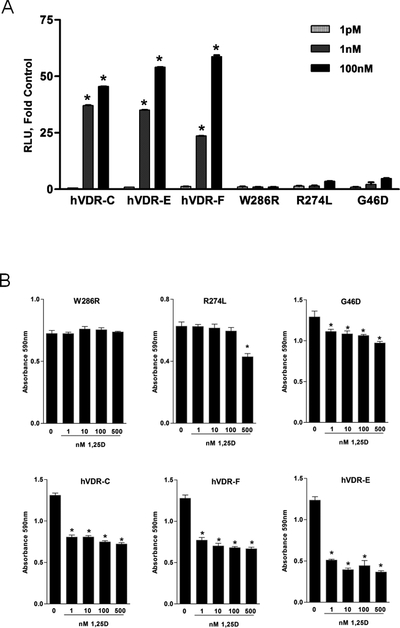
To assess transcriptional activity of mutant VDRs, transient transfection assays were conducted with the VDR-responsive CYP24 luciferase construct. CYP24 promoter activity was not induced by 1,25D in KOW286R cells and slightly but not significantly elevated by 1,25D in KOG46D and KOR274L cells (1.4–2 fold at 1nM and 3–4 fold at 100nM, Figure 8A). In contrast, 1,25D up-regulated CYP24 promoter activity between 23–37 fold at 1nM and 45–60 fold at 100nM through the wild-type VDR in the KOhVDR cell lines. Thus, the transcriptional activity of the KOG46D and KOR274L hVDR mutant proteins was more than 10-fold less than that of wild-type hVDR even at supra-physiological (μM) concentrations and neither exhibited significant transcriptional activity at physiological (1nM) concentrations.
Figure 8. Transcriptional activity and growth responses of KO240 cells expressing mutant VDR proteins.
A. CYP24 reporter gene activity in KOhVDR-F, KOhVDR-C, KOhVDR-E, KOW286R, KOR274L and KOG46D cells treated with vehicle or indicated doses of 1,25D for 24 hours. Data were normalized for transfection efficiency measured by co-transfected pRL-TK, and are expressed as fold increase in relative luciferase activity (RLU) of 1,25D treated versus vehicle treated for each cell line. Bars represent mean ± SEM of four-six values. Asterisks indicate significant difference from respective control. B. Crystal violet growth assays in KOW286R, KOR274L, KOG46D, KOhVDR-C, KOhVDR-Fand KOhVDR-E cells treated for 96h with vehicle or 1,25D at the indicated doses. Asterisks signify significant difference from control.
Growth assays were conducted to determine whether any of the transcriptionally inactive mutant VDR proteins could mediate growth inhibitory effects of 1,25D (Figure 8B) or vitamin D analogs (Figure 9). Growth of the KOW286R cell line was unaffected by 1,25D up to 1μM and even the highest potency analogs had minimal effect at this concentration. In the KOR274L cell line, dose dependent growth reduction (25% decrease) was observed at concentrations above 500nM 1,25D. At 1μM, 1,25D and synthetic analogs reduced growth of KOR274L cells from 40–55%. Of the three mutant receptors, the KOG46D cell line was the most sensitive to growth inhibition by vitamin D compounds, with dose-dependent reduction in cell density beginning at 1nM 1,25D (Figure 8B). In replicate assays, 1nM 1,25D inhibited growth of the KOG46D cells from 14–28% of control, which was consistently less than that observed in any of the KOhVDR cell lines (range 34–37% of control). Interestingly, cells expressing the G46D mutant were less responsive to higher concentrations of both 1,25D and synthetic analogs than cells expressing the R274L mutation, indicating different mechanisms of action for the two mutants.
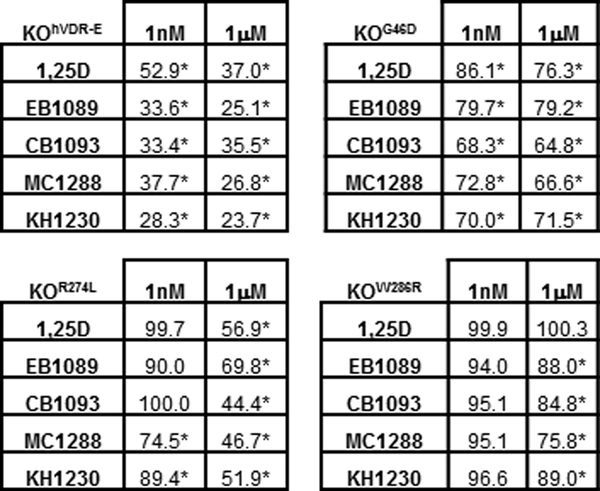
Figure 9. Effects of 1,25D analogs on growth of KO240 cells expressing mutant VDR proteins.
Crystal violet growth assays in KOhVDR-E, KOG46D, KOR274L and KOW286R cells treated for 96h with vehicle or 1,25D, EB1089, CB1093, MC1288, or KH1230 at indicated concentrations. Data represent mean of four values expressed as percentage of control absorbance which was set to 100%. Asterisks signify significant difference from vehicle treated control. The data shown for KOhVDR-E cells is reproduced from Figure 5 to facilitate comparison.
Discussion
The goal of these studies was to develop a model system to dissect the impact of specific functional domains of the VDR on breast cancer cell growth and gene expression. Towards this goal, we stably expressed wild-type VDR and VDRs containing point mutations which abolished the functionality of either the DNA- or ligand- binding domains in KO240 murine mammary tumor cells which are resistant to the growth inhibitory and apoptotic effects of 1,25D [19]. We show restoration of 1,25D mediated growth arrest, apoptosis and gene expression in KOhVDR cell lines engineered to stably express wild-type VDR protein. Despite variability in the responses of individual clonal cell lines, all hVDR-expressing lines demonstrated high levels of VDR protein, robust transcriptional activity and dose-dependent growth arrest similar to the matched WT145 cell line that endogenously expresses mVDR. These data demonstrate that the 1,25D-VDR signaling axis that triggers anti-cancer effects is highly conserved between the murine and human systems despite differences in VDR protein, nuclear cofactors, and target gene promoters.
Although VDR is clearly required for 1,25D mediated growth arrest, comparison of the three clonal cell lines did not indicate strict correlation between the level of VDR expression and the biological response to 1,25D. For example, KOhVDR-F cells exhibited the lowest VDR expression but was the most sensitive to 1,25D with respect to CYP24 induction and growth arrest (growth reduction was triggered in response to 1pM 1,25D in KOhVDR-F cells as compared to 10pM for KOhVDR-E cells and 100pM for KOhVDR-C cells). Similar trends in responses were observed for growth inhibition of the individual hVDR lines treated with synthetic vitamin D analogs. Despite these differences, maximal growth inhibition of approximately 50% was achieved at 10nM for 1,25D and 60–75% at 1nM for the analogs in all hVDR-expressing cell lines. Furthermore, all cell lines exhibited morphological features of apoptosis when treated with 100nM 1,25D. These data suggest that the percentage of VDR positive cells rather than the absolute level of cellular VDR expression in a heterogenous tumor would be the best predictor of therapeutic response to endogenous or exogenous vitamin D compounds.
Since both the endogenous mVDR in WT145 cells and the hVDR in the engineered KO240 cell lines successfully triggered growth arrest and apoptosis, we reasoned that a set of common target genes might mediate these responses. Using a PCR array focused on cancer pathways, we identified nine genes in WT145 cells and 21 genes in KOhVDR-F cells that were altered two-fold or more in response to 100nM 1,25D. Of six genes chosen for qPCR follow-up, three (Pdgfb, Vegfa and Nfkbi) were confirmed to be commonly induced by 1,25D in cells expressing either mVDR or hVDR. Potential VDR binding sites have been identified in the promoter regions of Nfkbi and Pdgfb in both the human and mouse genomes, suggesting they may be direct VDR target genes [24]. Nfkbi encodes the NfκB inhibitor IkB-α, and it’s up-regulation by 1,25D is consistent with reports that NfκB signaling is enhanced in VDRKO cells [25] and is inhibited by 1,25D in breast cancer cells [26]. Since NfκB is pro-inflammatory and has been associated with cancer progression, induction of its major cellular inhibitor IkB-α could contribute to the anti-cancer actions of 1,25D. Induction of Pdgfb and Vegfa, which code for growth factors that stimulate mitogenesis and angiogenesis in several cell types, is seemingly incongruent with the growth inhibitory effects of 1,25D we observed in our model system. However, Vegfa was also identified as a 1,25D inducible gene in association with growth inhibition of squamous carcinoma cells [24] and in some contexts Pdgfb has been linked to tissue remodeling, cellular differentiation and morphogenesis. For example, loss of the tumor suppressor PTEN during prostate cancer progression is associated with a ligand switch from Pdgfb in normal tissue to Pdgfd in tumors [27]. Additional mechanistic studies and more comprehensive genomic profiling will be necessary to integrate the effects reported here with the multiple actions of 1,25D that mediate growth inhibition and apoptosis in this model system.
A major application of our model system is structure-function analysis of VDR mutations in the context of growth control by vitamin D compounds. As proof of principle of this approach, we developed cell lines expressing hVDRs bearing natural point mutations (G46D, R274L and W286R) that have been identified in patients with hVDRR. It is well accepted that these VDR mutations are defective in mediating the calcemic effects of vitamin D, but none have been studied for their anti-cancer actions. We found that all three mutant hVDR proteins were expressed comparably to wild-type hVDR in the KO cell background yet had negligible transactivation activity on the CYP24 reporter which is driven by DR3-type response elements [28].
The W286R mutation was of considerable mechanistic interest for our studies since it appears to retain some non-canonical functions of VDR. For example, HVDRR patients with mutations at W286 do not develop alopecia [29] suggesting that this mutant receptor retains the ability to functionally interact with co-factors such as β-catenin to regulate hair cycling. Furthermore, the W286R mutant can mediate rapid non-genomic effects on calcium signaling [30] indicating that the transient, low affinity binding of 1,25D to the mutant receptor (which cannot drive genomic signaling) is sufficient for rapid responses. Our data clearly demonstrates that expression of the W286R mutation had no impact on cell growth kinetics in the presence or absence of 1,25D, arguing against a role for ligand independent actions or rapid, non-genomic signaling in the anti-cancer actions of VDR in this model.
Like HVDRR patients with W286 mutations, those with mutations in R274 do not exhibit alopecia indicating that this residue is not essential for hair cycle maintenance. The R274 residue in the hVDR ligand binding domain makes direct contact with the 1α-hydroxyl group on 1,25D. Functional analyses indicated that missense mutations at this site decrease activity 100–1000 fold compared to wild-type VDR [31,32]. Consistent with these reports, our data indicate that expression of the R274L mutant receptor in KO cells did not confer significant transcriptional activity or growth inhibition in response to 1,25D at concentrations up to 100nM. However, at supra-physiological concentrations (500nM) which presumably overcome the reduced affinity of the mutant ligand binding domain, 1,25D inhibited growth of the KOR274L cells. These data indicate a close correlation between ligand binding, transcriptional activity and growth inhibition by the 1,25D/VDR complex and do not support a ligand independent effect of VDR on cell growth kinetics.
In contrast to cells expressing R274L or W286R, cells expressing hVDR with the G46D mutation exhibited partial growth inhibition at physiological concentrations of 1,25D (1nM). In replicate assays, growth inhibition by 1nM 1,25D averaged 22% in cells expressing hVDRG46D as compared to 35% in the three cell lines expressing wild-type hVDR. Mutations of VDR at G46, located in the DNA binding domain, do not affect binding of ligand or co-factors but abrogate binding to DR3-type VDREs. At 1nM, a concentration which mediated growth inhibition in KOG46D cells, 1,25D had no significant effect on CYP24 reporter gene activity. In contrast, cells expressing wild-type hVDR exhibited 20-fold induction of CYP24 reporter gene activity in response to 1nM 1,25D. Furthermore, HVDRR patients with mutations at G46 exhibit alopecia indicating that mutations at this residue abrogate the ligand independent actions of VDR in hair cycling. Collectively, these data suggest that a percentage of the growth inhibition triggered by 1,25D is potentially mediated through mechanisms that are distinct from the actions of VDR in the hair cycle and do not require VDR binding to DR3-type VDREs. This suggestion is supported by comparison of cells expressing hVDRG46D to those expressing hVDRR274L which as discussed above reduces ligand binding but does not affect DNA binding [32]. Cells expressing hVDRR274L are not arrested at physiological concentrations of 1,25D, but do respond to concentrations > 500nM and are more sensitive to 1uM concentrations of synthetic vitamin D analogs than cells expressing hVDRG46D.
In summary, these data suggest that the ligand binding mutant hVDRR274L mimics the growth inhibitory signaling of wild-type hVDR when ligand concentrations are high enough to overcome the weak affinity associated with the mutation, whereas the DNA binding mutant hVDRG46D responds to physiological concentrations of the ligand but mediates a subset of wild-type hVDR actions since its binding to DR3 sequences is compromised. Furthermore, the hVDRW286R mutant lacks the ability to trigger growth inhibition despite being competent in mediating rapid responses in some contexts [30]. Thus, we conclude that 1,25D mediates growth arrest in cancer cells via multiple novel pathways which can be mechanistically dissociated at the level of the VDR.
Our model system will be useful in identifying the specific mechanisms by which hVDRG46D elicits growth arrest in response to 1,25D and analogs. Laboratory and in silico approaches support the existence of cis regulatory elements distinct from the consensus DR3 that mediate VDR transcriptional regulation including IP9, ER6, ER4, DR4 and DR6 elements [2–7]. Of note, some of these alternate VDREs function in VDR target genes that are not involved in calcium homeostasis such as p21 [6] and c-fos [2] and correlate with induction of apoptosis by 1,25D [8]. Furthermore, VDR bound to the synthetic analog EB1089 exhibits increased affinity for IP9 VDREs as compared to DR3 VDREs [9]. Although genome wide ChIP-Seq approaches in human cells have confirmed enrichment of consensus DR3 elements at VDR binding sites, up to 60% of VDR binding is at sites lacking DR3 sequences and likely involves interactions between VDR and novel co-factors such as β-catenin and TCF-4 [33,34]. Interactions between ligand-bound VDR and cytoplasmic signaling proteins such as PP1c/PP2Ac phosphatases, RHO-A kinase and p38MAP kinases could also contribute to control of cell proliferation by 1,25D [15,35]. Currently, it is not clear whether ligand bound hVDRG46D mimics any of these alternative genomic or non-genomic hVDR actions or whether these mechanisms are relevant for growth control by 1,25D in our model system. The cell lines described here comprise a valuable model system for further mechanistic study of these diverse actions of VDR in relation to the anti-cancer actions of vitamin D.
Supplementary Material
Acknowledgements.
The help of Dr. Jamie Keith in preparation of expression plasmids is greatly appreciated.
Grant support:
This work was funded by National Institutes of Health RO1CA69700 to JW, Department of Defense IDEA award DAMD-17-03-1-0359 to JW and Breast Cancer Research Program Dissertation Fellowship W81XWH-06-1-0439 to MEK.
Definitions:
- 1,25D
1,25-dihydroxyvitamin D
- 25D
25-hydroxyvitamin D
- CSS
charcoal stripped fetal bovine serum
- DBD
DNA binding domain
- DR3/4/6
direct repeat with 3, 4, or 6 base-pair spacer
- ER6/9
everted repeat with 6 or 9 base-pair spacer
- EV
empty vector
- G46D
natural occurring mutation of position 46 in hVDR from G to D
- h
human
- HVDRR
hereditary vitamin D resistant rickets
- IP9
inverted palindrome with 9 base-pair spacer
- KO
knock-out
- LBD
ligand binding domain
- m
mouse
- μM
micromolar
- nM
nanomolar
- pM
picomolar
- r
rat
- R274L
natural occurring mutation of position 274 in hVDR from R to L
- RLU
relative luciferase units
- VDR
vitamin D receptor
- VDRE
vitamin D response element
- W286R
natural occurring mutation of position 286 in hVDR from W to R
- WT
wild-type
References
- 1.Haussler MR, Jurutka PW, Mizwicki M, Norman AW. Vitamin D receptor (VDR)-mediated actions of 1alpha,25(OH)vitamin D: genomic and non-genomic mechanisms. Best Pract Res Clin Endocrinol Metab 2011;25(4):543–559. [DOI] [PubMed] [Google Scholar]
- 2.Schrader M, Kahlen J-P, Carlberg C. Functional characterization of a novel type of 1a,25-dihydroxyvitamin D3 response element identified in the mouse c-fos promoter. Biochemical and Biophysical Research Communications 1997;230:646–651. [DOI] [PubMed] [Google Scholar]
- 3.Thummel KE, Brimer C, Yasuda K et al. Transcriptional control of intestinal cytochrome P-4503A by 1alpha,25-dihydroxy vitamin D3. Mol Pharmacol 2001;60(6):1399–1406. [DOI] [PubMed] [Google Scholar]
- 4.Quack M, Carlberg C. Ligand-triggered stabilization of vitamin D receptor/retinoid X receptor heterodimer conformations on DR4-type response elements. J Mol Biol 2000;296(3):743–756. [DOI] [PubMed] [Google Scholar]
- 5.Xie Z, Bikle DD. Cloning of the human phospholipase C-gamma1 promoter and identification of a DR6-type vitamin D-responsive element. J Biol Chem 1997;272(10):6573–6577. [DOI] [PubMed] [Google Scholar]
- 6.Saramaki A, Banwell CM, Campbell MJ, Carlberg C. Regulation of the human p21(waf1/cip1) gene promoter via multiple binding sites for p53 and the vitamin D3 receptor. Nucleic Acids Res 2006;34(2):543–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thompson PD, Jurutka PW, Whitfield GK et al. Liganded VDR induces CYP3A4 in small intestinal and colon cancer cells via DR3 and ER6 vitamin D responsive elements. Biochem Biophys Res Commun 2002;299:730–738. [DOI] [PubMed] [Google Scholar]
- 8.Danielsson C, Mathiasen IS, James SY et al. Sensitive induction of apoptosis in breast cancer cells by a novel 1,25-dihydroxyvitamin D3 analogue shows relation to promoter selectivity. J Cell Biochem 1997;66(4):552–562. [DOI] [PubMed] [Google Scholar]
- 9.Nayeri S, Danielsson C, Kahlen JP et al. The anti-proliferative effect of vitamin D3 analogues is not mediated by inhibition of the AP-1 pathway, but may be related to promoter selectivity. Oncogene 1995;11(9):1853–1858. [PubMed] [Google Scholar]
- 10.Huang Y-C, Chen J-Y, Hung W-C. Vitamin D3 receptor/Sp1 complex is required for the induction of p27Kip1 expression by vitamin D3. Oncogene 2004;23:4856–4861. [DOI] [PubMed] [Google Scholar]
- 11.Rochel N, Moras D. Ligand binding domain of vitamin D receptors. Curr Top Med Chem 2006;6:1229–1241. [DOI] [PubMed] [Google Scholar]
- 12.Capiati DA, Rossi AM, Picotto G, Benassati S, Boland RL. Inhibition of serum-stimulated mitogen activated protein kinase by 1alpha,25(OH)2-vitamin D3 in MCF-7 breast cancer cells. J Cell Biochem 2004;93(2):384–397. [DOI] [PubMed] [Google Scholar]
- 13.Huhtakangas JA, Olivera CJ, Bishop JE, Zanello LP, Norman AW. The vitamin D receptor is present in caveolae-enriched plasma membranes and binds 1 alpha,25(OH)2-vitamin D3 in vivo and in vitro. Mol Endocrinol 2004;18(11):2660–2671. [DOI] [PubMed] [Google Scholar]
- 14.Zanello LP, Norman AW. Rapid modulation of osteoblast ion channel responses by 1alpha,25(OH)2-vitamin D3 requires the presence of a functional vitamin D nuclear receptor. Proc Natl Acad Sci U S A 2004;101(6):1589–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ordonez-Moran P, Alvarez-Diaz S, Valle N, Larriba MJ, Bonilla F, Munoz A. The effects of 1,25-dihydroxyvitamin D3 on colon cancer cells depend on RhoA-ROCK-p38MAPK-MSK signaling. J Steroid Biochem Mol Biol 2010;121(1–2):355–361. [DOI] [PubMed] [Google Scholar]
- 16.Hedlund TE, Moffatt KA, Miller GJ. Stable expression of the nuclear vitamin D receptor in the human prostatic carcinoma cell line JCA-1: evidence that the antiproliferative effects of 1 alpha, 25-dihydroxyvitamin D3 are mediated exclusively through the genomic signaling pathway. Endocrinology 1996;137(5):1554–1561. [DOI] [PubMed] [Google Scholar]
- 17.Davoust N, Wion D, Chevalier G, Garabedian M, Brachet P, Couez D. Vitamin D receptor stable transfection restores the susceptibility to 1,25-dihydroxyvitamin D3 cytotoxicity in a rat glioma resistant clone. J Neurosci Res 1998;52(2):210–219. [DOI] [PubMed] [Google Scholar]
- 18.Peng X, Jhaveri P, Hussain-Hakimjee EA, Mehta RG. Overexpression of ER and VDR is not sufficient to make ER-negative MDA-MB231 breast cancer cells responsive to 1alpha-hydroxyvitamin D5. Carcinogenesis 2007;28(5):1000–1007. [DOI] [PubMed] [Google Scholar]
- 19.Zinser GM, McEleney K, Welsh J. Characterization of mammary tumor cell lines from wild type and vitamin D3 receptor knockout mice. Mol Cell Endocrinol 2003;200(1–2):67–80. [DOI] [PubMed] [Google Scholar]
- 20.Valrance ME, Brunet AH, Welsh J. Vitamin D receptor-dependent inhibition of mammary tumor growth by EB1089 and ultraviolet radiation in vivo. Endocrinology 2007;148(10):4887–4894. [DOI] [PubMed] [Google Scholar]
- 21.Valrance ME, Brunet AH, Acosta A, Welsh J. Dissociation of growth arrest and CYP24 induction by VDR ligands in mammary tumor cells. J Cell Biochem 2007. [DOI] [PubMed] [Google Scholar]
- 22.Rowling MJ, Kemmis CM, Taffany DA, Welsh J. Megalin-mediated endocytosis of vitamin D binding protein correlates with 25-hydroxycholecalciferol actions in human mammary cells. J Nutr 2006;136(11):2754–2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Valrance ME, Welsh J. Breast cancer cell regulation by high-dose Vitamin D compounds in the absence of nuclear vitamin D receptor. J Steroid Biochem Mol Biol 2004;89–90(1–5):221–225. [DOI] [PubMed] [Google Scholar]
- 24.Wang TT, Tavera-Mendoza LE, Laperriere D et al. Large-scale in silico and microarray-based identification of direct 1,25-dihydroxyvitamin D3 target genes. Mol Endocrinol 2005;19(11):2685–2695. [DOI] [PubMed] [Google Scholar]
- 25.Sun J, Kong J, Duan Y et al. Increased NF-kappaB activity in fibroblasts lacking the vitamin D receptor. Am J Physiol Endocrinol Metab 2006;291(2):E315–322. [DOI] [PubMed] [Google Scholar]
- 26.Tse AK, Zhu GY, Wan CK, Shen XL, Yu ZL, Fong WF. 1alpha,25-Dihydroxyvitamin D3 inhibits transcriptional potential of nuclear factor kappa B in breast cancer cells. Molecular immunology 2010;47(9):1728–1738. [DOI] [PubMed] [Google Scholar]
- 27.Conley-LaComb MK, Huang W, Wang S et al. PTEN regulates PDGF ligand switch for beta-PDGFR signaling in prostate cancer. Am J Pathol 2012;180(3):1017–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matilainen JM, Malinen M, Turunen MM, Carlberg C, Vaisanen S. The number of vitamin D receptor binding sites defines the different vitamin D responsiveness of the CYP24 gene in malignant and normal mammary cells. J Biol Chem 2010;285(31):24174–24183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nguyen TM, Adiceam P, Kottler ML et al. Tryptophan missense mutation in the ligand-binding domain of the vitamin D receptor causes severe resistance to 1,25-dihydroxyvitamin D. J Bone Miner Res 2002;17(9):1728–1737. [DOI] [PubMed] [Google Scholar]
- 30.Nguyen TM, Lieberherr M, Fritsch J et al. The rapid effects of 1,25-dihydroxyvitamin D3 require the vitamin D receptor and influence 24-hydroxylase activity: studies in human skin fibroblasts bearing vitamin D receptor mutations. Journal of Biological Chemistry 2004;279(9):7591–7591. [DOI] [PubMed] [Google Scholar]
- 31.Aljubeh JM, Wang J, Al-Remeithi SS, Malloy PJ, Feldman D. Report of two unrelated patients with hereditary vitamin D resistant rickets due to the same novel mutation in the vitamin D receptor. Journal of pediatric endocrinology & metabolism : JPEM 2011;24(9–10):793–799. [DOI] [PubMed] [Google Scholar]
- 32.Kristjansson K, Rut AR, Hewison M, O’Riordan JL, Hughes MR. Two mutations in the hormone binding domain of the vitamin D receptor cause tissue resistance to 1,25 dihydroxyvitamin D3. J Clin Invest 1993;92(1):12–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carlberg C, Seuter S, Heikkinen S. The first genome-wide view of vitamin D receptor locations and their mechanistic implications. Anticancer Res 2012;32(1):271–282. [PubMed] [Google Scholar]
- 34.Meyer MB, Goetsch PD, Pike JW. VDR/RXR and TCF4/beta-catenin cistromes in colonic cells of colorectal tumor origin: impact on c-FOS and c-MYC gene expression. Mol Endocrinol 2012;26(1):37–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bettoun DJ, Buck DW, Lu J, Khalifa B, Chin WW, Nagpal S. A vitamin D receptor-Ser/Thr phosphatase-p70 S6 kinase complex and modulation of its enzymatic activities by the ligand. J Biol Chem 2002;277(28):24847–24850. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.