Abstract
Genome-wide association studies (GWASs) have been performed extensively in diverse populations to identify single nucleotide polymorphisms (SNPs) associated with complex diseases or traits. However, to date, the SNPs identified fail to explain a large proportion of the variance of the traits/diseases. GWASs on type 2 diabetes (T2D) and coronary artery disease (CARD) are generally performed as single-trait studies, rather than analyzing the related traits simultaneously. Despite the extensive evidence suggesting that these two phenotypes share both genetic and environmental risk factors, the shared overlapping genetic biological mechanisms between these traits remain largely unexplored. Here, we adopted a recently developed genetic pleiotropic conditional false discovery rate (cFDR) approach to discover novel loci associated with T2D and CARD by incorporating the summary statistics from existing GWASs of these two traits. Applying the cFDR level of 0.05, 33 loci were identified for T2D and 34 loci for CARD, 9 of which for both. By incorporating pleiotropic effects into a conditional analysis framework, we observed that there is significant pleiotropic enrichment between T2D and CARD. These findings may provide novel insights into the etiology of T2D and CARD, as well as the processes that may influence disease development both individually and jointly.
Keywords: T2D, Type 2 diabetes, Coronary artery disease, Pleiotropic, Conditional FDR
1. Introduction
Genome-wide association studies (GWASs) have successfully identified hundreds of SNPs associated with complex diseases or traits. However, the SNPs identified to date fail to explain a large proportion of the variance and risks involved. Previous studies have suggested that GWAS has the potential to explain a larger proportion of this “missing heritability”1,2 mainly by using enlarged sample sizes.3 However, although acquiring larger sample sizes may increase statistical power, it is often not feasible since the recruiting and genotyping of additional participants is too costly. Therefore, there is a need for analytical methods that can better and more efficiently utilize the information contained in the existing pool of available data for the identification of trait-associated loci. Several of these types of methods have recently been developed4–6 and successfully applied7,8 to identify novel loci for various complex traits.
Pleiotropy is the phenomenon of a single gene affecting two or more phenotypes.9 There is ample evidence to suggest that genetic pleiotropy exists in many correlated diseases and traits, such as bipolar disorder and schizophrenia,10 indicating that related traits may share overlapping genetic mechanisms. Through the incorporation of information regarding genetic pleiotropy, we can improve the detection power of common variants associated with complex diseases or traits by effectively increasing the sample sizes without the need to recruit more individuals. The joint analysis of related phenotypes may reveal novel insights into the common biological mechanisms and overlapping pathophysiological relationships between complex traits.
Andreassen et al.4 developed a genetic-pleiotropy-informed conditional false discovery rate (cFDR) method by leveraging two independent GWASs from associated traits in a conditional analysis. The method has been successfully applied to genetically associated diseases and phenotypes including schizophrenia and bipolar disorder,7 as well as blood pressure and other phenotypes.8 Our group has recently successfully applied the cFDR method to the joint analyses of bone mineral density (BMD) and breast cancer,11 BMD and coronary artery disease (CARD),12 femoral neck (FNK) BMD and height,13 and CARD and birth weight.14 All of these studies improved statistical power through the joint analysis of related traits, and unambiguously demonstrated the utility of the method for improving gene discovery in the identification of potentially novel trait-associated variants.
Type 2 diabetes (T2D) is a long term chronic metabolic disorder mainly characterized by high blood sugar, insulin resistance and relative lack of insulin. Long term exposure to high blood sugar will result multiple complex complications like stroke, diabetic retinopathy and heart disease.15 Epidemiological studies estimate that 422 million people were living with diabetes, with a worldwide prevalence of 8.3% in 2014.15 As the most common complication of T2D, cardiovascular disease is the most primary cause of T2D mortality and mobility.16 The overall prevalence of CARD in diabetic adult individuals was reported as 55% and an estimated 75% of the T2D patients died of cardiovascular disease.17 Heritability studies demonstrate a substantial genetic contribution to T2D risk (h2~40–70%)18 and CARD risk (h2~30–60%).19
Multiple prospective studies suggested that diabetic individuals have 1.5 to threefold increased risk of developing coronary heart disease compared to the nondiabetic individuals.20 What’s more, compared with nondiabetic individuals, the mortality rate of cardiovascular disease is more than twice in men and more than fourfold in women who have diabetes.21 There is strong evidence21,22 that T2D and CARD share primary risk factors such as smoking, hypertension, elevated lipid, dysbetalipoproteinemia and hyperglycemia, also some potential risk factors like obesity, lack of physical activity, cardiovascular family history, gender and age. Although dozens of genetic loci associated with T2D or CARD have been demonstrated by GWASs, these loci can explain at best 10% of the genetic variance for either T2D23 or CARD.24 Considering the high degree of heritability, close relationship and potential pleiotropy between these two phenotypes, we assume those two traits are ideal for the further analyses using the cFDR approach to improve the detection of loci associated with T2D or CARD or both and explore their common etiology.
In this study, we applied the genetic-pleiotropy-informed cFDR method4 on two large and independent GWAS summary statistics of T2D and CARD23,24 to identify novel loci and pleiotropic relationships between T2D and CARD. The purpose of our study is to improve SNP detection for T2D and CARD with these two existent GWASs and gain some novel insights into shared biological mechanisms and overlapping genetic heritability between them.
2. Materials and methods
2.1. GWAS Datasets
The dataset for T2D contains association summary statistics of 12 GWASs of European descent which compromising of 12,171 cases and 56,862 controls.23 The dataset was downloaded from http://www.diagram-consortium.org/downloads.html. The meta-analyses were previously performed by the DIAbetes Genetics Replication And Metaanalysis (DIAGRAM) Consortium. The dataset for CARD contains association summary statistics of 22 GWASs of European descent which comprising of 22,233 cases and 64,762 controls.24 The dataset was downloaded from http://www.cardiogramplusc4d.org/data-downloads. The dataset was conducted by the transatlantic Coronary ARtery Disease Genome-wide Replication and Meta-analysis (CARDloGRAM) Consortium. Both of the datasets consist of the summary statistics for each SNP, providing the p values that have undergone genomic control at the individual study level, and again after meta-analysis. Further details of the samples and methods employed within each group are presented in the corresponding consortium papers.23,24 We further checked the original studies in both GWASs (Table S1), there was one common study between these two GWASs datasets, WTCCC (1926 cases of T2D, and 71.5% × (1926 + 2938) = 3478 cases of CVD25), which makes the rates of CVD in the T2D GWAS and the rates of diabetes in the CVD GWAS are 3% and 5% respectively.
The dataset for attention-deficit/hyperactivity disorder (ADHD) contains association summary statistics of European descent which compromising of 5415 individuals (2064 trios, 896 cases and 2455 controls),26 the dataset was downloaded from https://www.med.unc.edu/pgc/results-and-downloads/data-use-agreement-forms/ADHD_data_download_agreement. The dataset for major depressive disorder (MDD) contains association summary statistics of 18,759 independent and unrelated subjects of European ancestry (9240 MDD cases and 9519 controls),27 The dataset was downloaded from https://www.med.unc.edu/pgc/results-and-downloads/data-use-agreement-forms/MDD_data_download_agreement. Both meta-analyses were previously performed by the Major Depressive Disorder Working Group of the Psychiatric GWAS Consortium (PGC).
2.2. Conditional false discovery rate
The cFDR approach is well-established now and has been widely applied by many other groups4,7,8,28,29 and our group.12–14,30 We briefly summarize this cFDR approach as follows: after the data preparation processing as indicated in the previous papers, we computed the conditional empirical cumulative distribution functions (cdfs) of the corrected p-values for the x axis in conditional QQplot. Empirical cdfs for T2D SNP p-values were conditioned on nominal p-values in CARD, and vice versa. For each nominal p-value, an estimate of the cFDR was obtained from the conditional empirical cdfs. Using this cFDR approach, we obtained two cFDR tables–cFDR result for T2D conditioned on CARD and vice versa. Using these tables we identified loci associated with T2D and CARD (cFDR <0.05), respectively. Then a conjunction method was used to find SNPs significantly associated with both T2D and CARD. Specifically, we took the maximum of those two cFDR values above as our conjunction FDR.
2.3. Conditional QQ and enrichment plots for assessing pleiotropic enrichment
To assess the pleiotropic enrichment of SNP association compared to that expected under the null hypothesis, we presented conditional QQ plots based on different levels of significance of the conditional phenotype. The QQplots show the observed distribution of p-values plotted against the expected distribution of p-values under the null hypothesis. We plotted the QQ curve for the quantiles of nominal –log10(p)-values obtained from GWAS summary statistics for association of the subset of SNPs that are below each significance threshold in the conditional trait. The nominal –log10(p)-values are plotted on the y-axis and the empirical quantiles (cdfs) of the nominal p-values are plotted on the x-axis. Pleiotropic enrichment is expressed as the degree of leftward shift from the expected null line, and as the p values of the conditional phenotypes decrease, earlier leftward shift from the null line will persist.
In order to check the pleiotropic enrichment and provide a baseline that can be used to confirm novel findings, we also generated conditional QQ plots for two traits that are unlikely to be correlated with T2D and CARD, ADHD and MDD, as “control traits.”
2.4. Conditional Manhattan plots for localizing genetic variants
To demonstrate the localization of the SNPs associated with T2D conditional on their significance on CARD, and the reverse, we present conditional Manhattan plots. The plots present the relationship between all SNPs within an LD block and their chromosomal locations. The 22 chromosomal locations are plotted on the x-axis, and the –logi0(FDR) T2D values conditional on CARD are plotted on the y-axis and vice versa for CARD. Any SNP with a –log10(FDR) value >1.3 (FDR < 0.05) was deemed to be significantly associated with the principal phenotype. We also present a conjunction Manhattan plot to demonstrate the locations of the common pleiotropic genetic variants associated with both phenotypes.
2.5. Functional annotation and gene enrichment analysis
In order to evaluate the biological functions of the individual trait associated loci identified by cFDR and pleiotropic loci identified by conjunction FDR, we performed functional annotation and gene enrichment analysis using the gene ontology (GO) terms database (http://geneontology.org/.).31 All significant genes identified by cFDR and conjunction FDR in our study were annotated and characterized based on three main categories: biological processes, cellular component and molecular functions. This analysis provided comprehensive biological information, allowing us to partially validate our findings by determining specific genes that are enriched in T2D-and CARD-related GO terms.
2.6. Protein-protein interaction network
In order to detect interactions and associations of the T2D-associated and CARD-associated genes respectively, protein-protein interaction analyses were conducted by searching the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database (http://string-db.org/). The STRING database comprises known and predicted associations from curated databases or high-throughput experiments, and also with other associations derived from text mining, co-expression, and protein homology.32
3. Results
3.1. Assessment of pleiotropic enrichment
As an intuitive illustration, we present the data as conditional Q-Q plots (Fig. 1) to graphically assess the pleiotropic enrichment of SNPs of the principal phenotype successively conditioning on various strengths of associations with the conditional phenotype. Under the global null hypothesis, the theoretical distribution of p-values is expected to lie approximately on the diagonal line of the Q-Q plots. Enrichment of genetic associations is indicated as a leftward deflection from the null line as the principal phenotype is successively conditioned on increasing strength of associations with conditional phenotype. The degree of deflection between curves provides important information about the degree of pleiotropy between the two phenotypes. Larger deflection is considered as a greater enrichment of pleiotropic genes between the two phenotypes.
Fig. 1.
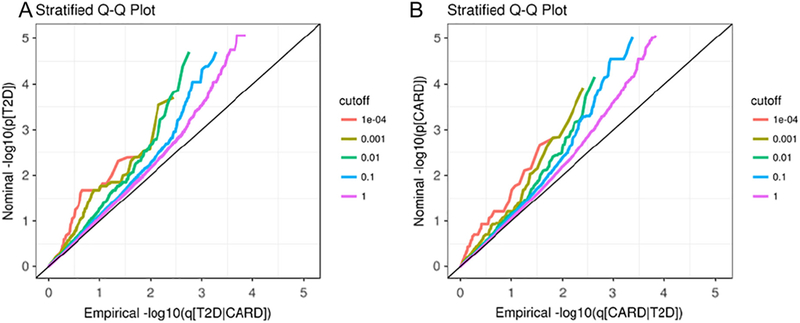
Stratified QQ plots. Stratified QQ plots of nominal versus empirical –log10 p-values in T2D (A) as a function of significance of the association with CARD, and in CARD (B) as a function of significance of the association with T2D. The purple line with slope of zero represents all SNPs.
The conditional Q-Q plot for T2D conditional on CARD (A in Fig. 1) shows some enrichment across varying significance thresholds for CARD. The presence of leftward shift when restricting the analysis to include the SNPs that have more significant associations with T2D indicates an increase in the number of true associations for a given CARD p-value. Similar enrichment is observed for CARD given T2D (B in Fig. 1), as there appears to be a similar departure pattern between the different curves. These earlier deflections from the null line indicate a great proportion of true associations for any given T2D nominal p-value.
On the other hand, as negative controls, the conditional Q-Qplots for T2D given nominal p-values of association with ADHD (A in Fig. S1), CARD given nominal p-values of association with ADHD (C in Fig. S1), T2D given nominal p-values of association with MDD (A in Fig. S2), and CARD given nominal p-values of association with MDD (C in Fig. S2) all show no enrichment, and vice versa.
3.2. T2D loci identified with cFDR
Conditional on their association with CARD, we identified 33 significant SNPs (cFDR <0.05) for T2D variation (A in Fig. 2 and Table 1), which were mapped to 13 different chromosomes (1,3, 5–12,15–17) and annotated to 37 genes. In the original meta-analysis for T2D GWAS,23 16 SNPs had p-values smaller than 1 × 10−5 while 6 of them reached the standard genome-wide significance of 5 × 10−8. We confirmed 11 SNPs that were reported in the original T2D GWAS analysis23 and previous T2D related GWASs.33,34 Another 7 SNPs that were reported to be associated with T2D-related traits were also confirmed in our analysis.35,36 The remaining 15 SNPs were not previously reported in the original T2D GWAS23 and the previous studies did not show their significance for T2D, while 2 SNPs of them showed high LD (r2 > 0.6) with the T2D-associated SNPs reported previously. For the 37 genes these 33 SNPs annotated to, 16 of them were newly detected compared to the original T2D23 and previous T2D-related studies. The details are provided in Table S1. Of the detected loci for T2D, most of the genes were enriched in T2D-related terms “positive regulation of fatty acid oxidation”, and “white fat cell differentiation”. GO term enrichment analysis results are detailed in Table 2.
Fig. 2.
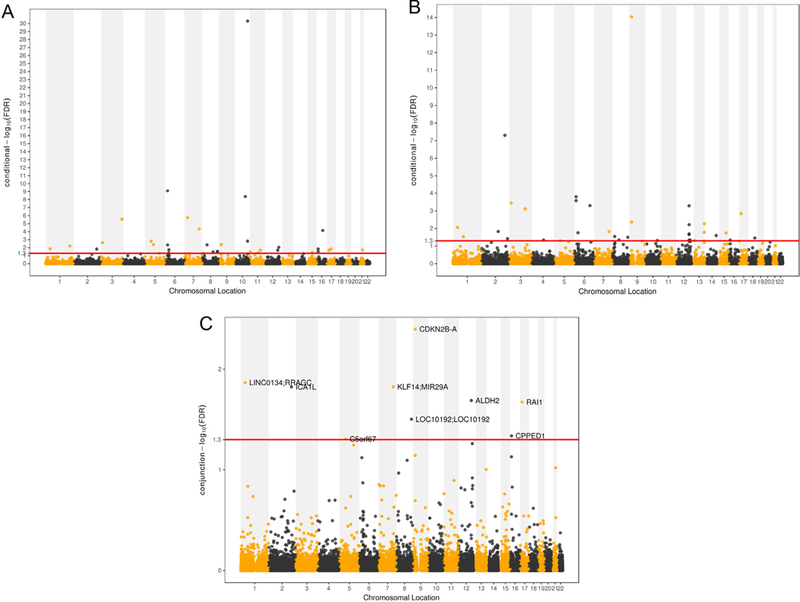
Conditional Manhattan plot of conditional –log10 FDR values for A) T2D given CARD (T2D|CARD), B) CARD given T2D (CARD|T2D), C) T2D and CARD. The red line marks the conditional –log10 FDR value of 1.3 corresponds to a cFDR <0.05.
Table 1.
Conditional FDR value for T2D given the CARD (cFDR <.05).
| RSID | ROLE | GENE | CHR | SNP type | Gene type | P.valueA | cFDR.AcB |
|---|---|---|---|---|---|---|---|
| rs10787472 | Intronic | TCF7L2 | chr10 | Confirmed | Confirmed | 1.10E-35 | 4.83E-31 |
| rs6906327 | Intronic | CDKAL1 | chr6 | Confirmed | Confirmed | 3.10E-14 | 7.92E-10 |
| rs7911264 | Intergenic | KIF11,HHEX | chr10 | Confirmed | Confirmed | 4.50E-13 | 4.24E-09 |
| rs849135 | Intronic | JAZF1 | chr7 | Confirmed | Confirmed | 3.40E-10 | 1.79E-06 |
| rs4481184 | Intronic | IGF2BP2 | chr3 | Confirmed | Confirmed | 3.20E-10 | 2.83E-06 |
| rs11979110 | Intergenic | KLF14,MIR29A | chr7 | HDL(24097068) | Confirmed, novel | 1.00E-07 | 4.66E-05 |
| rs9940128 | Intronic | FTO | chr16 | Confirmed | Confirmed | 1.10E-08 | 7.15E-05 |
| rs70797H | Intronic | TCF7L2 | chr10 | Novel | Confirmed | 2.50E-07 | 0.001524 |
| rs3843467 | Intronic | C5orf67 | chr5 | HDL (24097068) | Novel | 2.50E-06 | 0.001541 |
| rs2881654 | Intronic | PPARG | chr3 | T2D(24509480) | Confirmed | 1.70E-07 | 0.002381 |
| rs10965212 | ncRNA_intronic | CDKN2B-AS1 | chr9 | CARD (28530674) | Confirmed | 0.004 | 0.004 |
| rs6885904 | ncRNA_intronic | ZBED3-AS1 | chr5 | Novel | Confirmed | 1.10E-06 | 0.004178 |
| rs516946 | Intronic | ANK1 | chr8 | T2D (22885922) | Confirmed | 7.30E-07 | 0.0044 |
| rs4712540 | Intronic | CDKAL1 | chr6 | Confirmed | Confirmed | 5.80E-06 | 0.00453 |
| rs340835 | Intronic | PROX1 | chr1 | Fasting glucose (22885924) | Confirmed | 1.10E-06 | 0.006194 |
| rs7965349 | Intronic | OASL | chr12 | LD (0.817 rs7957197 T2D) | Novel | 2.00E-05 | 0.00894 |
| rs11211039 | Intergenic | LINC01343,RRAGC | chr1 | Novel | Novel, novel | 2.00E-04 | 0.0136 |
| rs4430796 | Intronic | HNF1B | chr17 | T2D (26551672) | Confirmed | 2.40E-06 | 0.014504 |
| rs4780476 | Intronic | CPPED1 | chr16 | Novel | Novel | 4.10E-05 | 0.014514 |
| rs4510208 | Intronic | ICA1L | chr2 | CARD (26343387) | Novel | 0.015 | 0.015 |
| rs3892710 | Intergenic | HLA-DQB1,HLA-DQA2 | chr6 | CARD (21971053) | Novel, novel | 8.70E-06 | 0.018475 |
| rs7280071 | Intronic | RUNX1 | chr21 | Novel | Novel | 4.40E-05 | 0.01881 |
| rs10744777 | Intronic | ALDH2 | chr12 | CARD (23202125) | CARD (23364394) | 0.0041 | 0.0205 |
| rs1876602 | Intergenic | MTNR1B,SLC36A4 | chr11 | Novel | Confirmed, novel | 4.10E-05 | 0.020739 |
| rs3818717 | Exonic | RAI1 | chr17 | Novel | CARD (24262325) | 0.0049 | 0.021233 |
| rs13275988 | Intergenic | LOC101927798,LOC101927822 | chr8 | Novel | Novel, novel | 0.00029 | 0.02987 |
| rs1878016 | Intronic | KCNQ3 | chr8 | Novel | CARD (23870195) | 1.30E-05 | 0.030072 |
| rs7l93741 | Intronic | CPPED1 | chr16 | Novel | Novel | 0.00014 | 0.031267 |
| rs163177 | Intronic | KCNQ1 | chr11 | T2D (26551672) | Confirmed | 4.80E-05 | 0.0318 |
| rs6991067 | Intronic | INTS8 | chr8 | LD (0.604 rs896854 T2D) | Confirmed | 0.00015 | 0.033188 |
| rs1723839! | Intergenic | C2CD4B,MIR8067 | chr15 | Novel | Confirmed, novel | 2.90E-05 | 0.039832 |
| rs3l30931 | UTR5 | POU5F1 | chr6 | Novel | Confirmed | 1.80E-05 | 0.043902 |
| rs1783598 | Intronic | FCHSD2 | chr11 | Novel | Novel | 3.50E-05 | 0.043995 |
Notes:
SNP type means whether SNPs identified in our study compared to the original T22D GWAS and previous studies are Novel or Confirmed or associated with T2D-related traits (trait (PMID)) or in high LD with T2D-associated loci.
Gene type means whether genes identified in our study compared to the original T2D GWAS and previous studies are Novel or Confirmed.
P.valueA is the p value of T2D, A is T2D.
cFDRAcB is the cFDR value of T2D conditioned on CARD, B is CARD.
Table 2.
Functional Term Enrichment Analysis.
| Pathway ID | Pathway description | Count in gene set | False discovery rate |
|---|---|---|---|
| T2D GO:0002504 | Antigen processing and presentation of peptide or polysaccharide antigen via MHC class II | 14 | 5.533 |
| GO:0061008 | Hepaticobiliary system development | 7 | 4.363 |
| GO:0001889 | Liver development | 7 | 4.381 |
| GO:0030855 | Epithelial cell differentiation | 10 | 3.249 |
| GO:0070365 | Hepatocyte differentiation | 5 | 7.310 |
| GO:0019904 | Protein domain specific binding | 18 | 3.425 |
| GO:0016055 | Wnt signaling pathway | 10 | 3.842 |
| GO:0048713 | Regulation of oligodendrocyte differentiation | 5 | 5.359 |
| GO:0002674 | Negative regulation of acute inflammatory response | 4 | 6.895 |
| GO:0046321 | Positive regulation of fatty acid oxidation | 4 | 6.648 |
| GO:0050872 | White fat cell differentiation | 4 | 6.436 |
| GO:0060214 | Endocardium formation | 3 | 7.895 |
| GO:0070309 | Lens fiber cell morphogenesis | 3 | 7.895 |
| GO:2000977 | Regulation of forebrain neuron differentiation | 3 | 8.158 |
| GO:0042611 | MHC protein complex | 13 | 4.868 |
| GO:0042613 | MHC class II protein complex | 13 | 6.530 |
| GO:0098796 | Membrane protein complex | 17 | 2.256 |
| GO:0098797 | Plasma membrane protein complex | 17 | 3.084 |
| CARD GO:0019904 | Protein domain specific binding | 15 | 3.208 |
| GO:0004465 | Lipoprotein lipase activity | 4 | 7.942 |
| GO:0017129 | Triglyceride binding | 4 | 8.620 |
| GO:0019433 | Triglyceride catabolic process | 4 | 6.418 |
| T2D and CARD GO:0019904 | Protein domain specific binding | 14 | 5.173 |
| GO:0040015 | Negative regulation of multicellular organism growth | 3 | 7.890 |
3.3. CARD gene loci identified with cFDR
Conditional on their association with T2D, we identified 34 significant SNPs (cFDR <0.05) for CARD variation (B in Fig. 2 and Table 3), which were located on 17 chromosomes (1–10,12–18) and annotated to 43 genes. In the original meta-analysis for CARD GWAS,24 18 SNPs had p-values smaller than 1 × 10−5 while 5 of them reached the standard genome-wide significance of 5 × 10−8. We confirmed 13 SNPs that were reported in the original CARD GWAS analysis24 and previous CARD related GWASs.37,38 Another 5 SNPs that were reported to be associated with CARD-related traits were also confirmed in our analysis.36,39 The other 16 SNPs were not previously reported in the original CARD GWAS24 and the previous studies did not show their significance for CARD, and none of the novel SNPs showed high LD (r2 > 0.6) with the CARD-associated SNPs reported previously. For the 43 genes these 34 SNPs annotated to, there were 24 of them were newly detected compared to the original CARD24 and previous CARD-related studies. The details are provided in Table S2. Of the detected loci for CARD, some of the genes were enriched in CARD-related terms “protein domain specific binding” and “lipoprotein lipase activity”. GO term enrichment analysis are detailed in Table 2.
Table 3.
Conditional FDR value for CARD given the T2D (cFDR <.05).
| RSID | ROLE | GENE | CHR | SNP type | Gene type | P.valueB | cFDR.BcA |
|---|---|---|---|---|---|---|---|
| rs10965212 | ncRNA_intronic | CDKN2B-AS1 | chr9 | Confirmed | Confirmed | 1.37E-17 | 9.10E-15 |
| rs4510208 | Intronic | ICA1L | chr2 | Confirmed | Confirmed | 4.29E-11 | 4.97E-08 |
| rs9381462 | Intronic | PHACTR1 | chr6 | Confirmed | Confirmed | 5.13E-09 | 0.000157 |
| rs2876303 | Intronic | PHACTR1 | chr6 | Confirmed | Confirmed | 9.10E-09 | 0.000264 |
| rs7651039 | Intronic | BTD | chr3 | Confirmed | Confirmed | 1.85E-08 | 0.000357 |
| rs1029212 | ncRNA_intronic | LINC01312,TARID | chr6 | CARD (28530674) | Confirmed | 6.23E-08 | 0.000504 |
| rs10744777 | Intronic | ALDH2 | chr12 | CARD (23202125) | Confirmed | 1.52E-06 | 0.000519 |
| rs2347252 | Intronic | MRAS | chr3 | CARD (23202125) | Confirmed | 9.83E-08 | 0.000775 |
| rs3818717 | Exonic | RAI1 | chr17 | Novel | Confirmed | 5.16E-06 | 0.001424 |
| rs1011970 | ncRNA_intronic | CDKN2B-AS1 | chr9 | Novel | Confirmed | 6.37E-06 | 0.004352 |
| rs4773144 | Intronic | COL4A2 | chr13 | CARD (23202125) | Confirmed | 4.15E-07 | 0.005338 |
| rsll066301 | Intronic | PTPN11 | chr12 | Total cholesterol (24097068) | Novel | 5.20E-07 | 0.006047 |
| rs11211039 | Intergenic | LINC01343,RRAGC | chr1 | Novel | Novel, novel | 0.00012 | 0.008633 |
| rs2252641 | ncRNA_intronic | TEX41 | chr2 | CARD (23202125) | Confirmed | 1.37E-05 | 0.014755 |
| rs11979110 | Intergenic | KLF14,MIR29A | chr7 | HDL(20686565) | Novel, novel | 0.002137 | 0.014959 |
| rs9515203 | Intronic | COL4A2 | chr13 | CARD (23202125) | Confirmed | 3.42E-05 | 0.016238 |
| rs2523414 | ncRNA_exonic | LOC554223 | chr6 | Novel | Novel | 2.83E-05 | 0.017416 |
| rs4539564 | Intergenic | ADAMTS7,MORF4L1 | chr15 | CARD (23202125) | Confirmed | 9.46E-06 | 0.017974 |
| rs17696736 | Intronic | NAA25 | chr12 | Novel | Confirmed | 4.12E-06 | 0.020291 |
| rs7970490 | Intronic | CUX2 | chr12 | Novel | Confirmed | 2.18E-05 | 0.021993 |
| rs2146238 | Intronic | CYP46A1 | chr14 | Novel | Novel | 2.61E-06 | 0.025067 |
| rs894210 | Intergenic | LPL,SLC18A1 | chr8 | Triglycerides (24097068) | Lipid (24386095), Triglycerides (24886709) | 6.93E-05 | 0.028257 |
| rs4415546 | Intergenic | ZNF326,BARHL2 | chr1 | Novel | Novel, novel | 5.56E-06 | 0.028883 |
| rs13275988 | Intergenic | LOC101927798,LOC101927822 | chr8 | Novel | Novel, novel | 0.000647 | 0.031365 |
| rs8089632 | Intergenic | MALT1,ZNF532 | chr18 | Novel | Novel, novel | 1.11E-05 | 0.034666 |
| rs6713510 | ncRNA_intronic | LOC646736 | chr2 | T2D (26551672) | Novel | 9.77E-05 | 0.038066 |
| rs6474069 | ncRNA_intronic | LINC00968,LOC101929415 | chr8 | Novel | Novel, novel | 9.83E-06 | 0.045441 |
| rs4699748 | Intergenic | ADH1C,ADH7 | chr4 | Novel | Novel, novel | 7.84E-05 | 0.04562 |
| rs2708081 | Intronic | OASL | chr12 | CARD (28530674) | Confirmed | 0.000189 | 0.045881 |
| rs4780476 | Intronic | CPPED1 | chr16 | Novel | Novel | 0.001434 | 0.045891 |
| rs10774625 | Intronic | ATXN2 | chr12 | Novel | Blood pressure (19430483) | 7.19E-06 | 0.04631 |
| rs9581678 | Intergenic | CDK8,WASF3 | chr13 | Novel | Novel, novel | 1.47E-05 | 0.047478 |
| rs7902587 | Intergenic | OBFC1,SLK | chr10 | Novel | Novel, novel | 7.07E-05 | 0.048121 |
| rs3843467 | Intronic | C5orf67 | chr5 | Triglycerides (24097068) | Novel | 0.00705 | 0.049348 |
Notes:
SNP type means whether SNPs identified in our study compared to the original CARD GWAS and previous studies are Novel or Confirmed or associated with CARD-related traits (trait (PMID)).
Gene type means whether genes identified in our study compared to the original CARD GWAS and previous studies are Novel or Confirmed.
P.valueB is the p value of CARD, B is CARD.
cFDRBcA is the cFDR value of CARD conditioned on T2D, A is T2D.
3.4. Pleiotropic gene loci for both T2D and CARD
The conjunction FDR analysis detected 9 independent pleiotropic loci that were significantly (conjunction FDR < 0.05) associated with both traits (C in Fig. 2 and Table 4). Of the 9 identified pleiotropic variants, three SNPs rs10965212 (CDKN2B-AS1), rs4510208 (ICA1L) and rs10744777 (ALDH2) were reported to be significant for CARD in the original CARD GWAS24 or previous CARD GWAS.38 The other two SNPs (rs11979110 and rs3843467) were previously reported to be associated with high density lipoprotein (HDL) and triglycerides.36 The remaining four SNPs were not previously reported in the original T2D and CARD related GWASs and in the previous studies they were not significant for either T2D or CARD. For the 12 genes those pleiotropic SNPs annotated to, we found six of them (CDKN2B-AS1, ICA1L, ALDH, RAI1, C5orf67 and KLF14) were reported by T2D or CARD related GWAS. The other six genes were not identified by any T2D or CAD related GWAS. For the SNPs that were annotated to these 6 genes, one SNP was located in the intronic regions of gene CPPED1, the rest of the SNPs were all located in intergenic regions of the genes. Detailed information were shown in Table 2. Of the detected 9 pleiotropic loci, most of the genes were enriched in T2D and CARD related terms “protein domain specific binding” and “negative regulation of multicellular organism growth”. Detailed information of GO term analysis is given in Table 2.
Table 4.
Conjunction FDR: Pleiotropic Loci in T2D and CARD (cFDR<0.05).
| RSID | ROLE | GENE | CHR | P.valueA | P.valueB | cFDR.AcB | cFDR.BcA | conjunction FDR |
|---|---|---|---|---|---|---|---|---|
| rs10965212 | ncRNA_intronic | CDKN2B-AS1 | chr9 | 0.004 | 1.37E-17 | 0.004 | 9.10E-15 | 0.004 |
| rs11211039 | Intergenic | LINC01343,RRAGC | chr1 | 2.00E-04 | 0.00012 | 0.0136 | 0.0086328 | 0.0136 |
| rs11979110 | Intergenic | KLF14,MIR29A | chr7 | 1.00E-07 | 0.002137 | 4.66E-05 | 0.014959 | 0.014959 |
| rs4510208 | Intronic | ICA1L | chr2 | 0.015 | 4.29E-11 | 0.015 | 4.97E-08 | 0.015 |
| rs10744777 | Intronic | ALDH2 | chr12 | 0.0041 | 1.52E-06 | 0.0205 | 0.00051908 | 0.0205 |
| rs3818717 | Exonic | RAI1 | chr17 | 0.0049 | 5.16E-06 | 0.021233 | 0.00142416 | 0.02123333 |
| rs13275988 | Intergenic | LOC101927798,LOC101927822 | chr8 | 0.00029 | 0.000647 | 0.02987 | 0.03136495 | 0.03136495 |
| rs4780476 | Intronic | CPPED1 | chr16 | 4.10E-05 | 0.001434 | 0.014514 | 0.0458912 | 0.0458912 |
| rs3843467 | Intronic | C5orf67 | chr5 | 2.50E-06 | 0.00705 | 0.001541 | 0.0493479 | 0.0493479 |
Notes:
P.valueA is the p value of T2D.
P.valueB is the p value of CARD.
3.5. Protein-protein interaction network
The 37 identified T2D-associated genes were retrieved from the STRING database. Only 18 genes, including 3 novel genes, were annotated in this database. The 18 genes were clearly enriched in two clusters: TCF7L2 and HLA (Fig. S3). Three novel genes OASL, HLA-DQA2 and HLA-DQB1, respectively encoding 2′−5′-oligoadenylate synthetase like, major histocompatibility complex, class II, DQ alpha 2 and major histocompatibility complex, class II, DQ beta 1, were directly connected with the HLA cluster.
The 43 identified CARD-associated genes were retrieved from the STRING database. Only 4 genes, including 2 novel genes, were annotated in this database. The 4 genes were clearly enriched into two clusters: ALDH2 and LPL (Fig. S4). Two novel genes, ADH7 and CDK8, those respectively encoding alcohol dehydrogenase class 4 mu/sigma chains and cyclin-dependent kinase 8, were directly connected to the two clusters.
4. Discussion
In our study, two independent GWASs with summary statistic p values were combined to explore the pleiotropic enrichment of SNPs that are associated with T2D and CARD. Compared to the conventional standard single phenotype GWAS, simultaneously analyzing multiple related traits allows for the increased discovery of trait-associated variants without requiring additional larger datasets for individual trait. By leveraging the power of two different GWAS datasets from T2D and CARD, we discovered 33 loci for T2D and 34 loci for CARD. Using the standard GWAS significance in the datasets, only 6 for T2D and 5 for CARD were significant. Most of the genes have not been reported to show borderline significance with T2D and CARD respectively, as detailed in Tables S1 and S2. Adopting the genetic pleiotropic-informed cFDR method, we found 9 novel genes associated with both T2D and CARD. These novel findings may enable us to further dissect the overlapping genetic mechanisms between these two related phenotypes. The improved detection of novel susceptibility loci with genetic pleiotropy may lead us to a better understanding of common etiology between disorders and have a significant impact on the clinical treatment and prevention of related complex human diseases.
The cFDR approach was adopted here to account for some of the missing heritability between traits or diseases. This method employs the idea that a variant with significant effects in two associated phenotypes is more likely to be a true effect, and therefore has a higher probability of being detected in multiple independent studies. This technique allows for an increase in effective sample size and therefore a sub-sequent increase in power to detect true associations for more variants with small to moderate effect sizes which are often easily ignored in the standard single phenotype GWAS. In addition, the genetic enrichment presented in conditional Q-Q plots conveys that the decreased cFDR value for a given nominal p value greatly increases power to detect true association effects. When initially implementing the cFDR method, Andreassen et al.7 demonstrated one advantage of this model-free empirical cdf approach is for the avoidance of bias in conditional FDR estimates from model misspecification, and they made a comparison of traditional unconditional FDR and cFDR methods, and found that the latter resulted in an increase of 15–20 times the number of SNPs under the same FDR threshold of 0.05.7
Our cFDR analysis identified 9 pleiotropic signals, which supported the close relationship and shared genetic determination between these two traits. These 9 pleiotropic SNPs were annotated to 12 genes. Five genes CDKN2B-AS1, ICA1L, ALDH, C5orf67 and RAH were frequently reported and replicated in previous CARD related studies. The implementation of cFDR method in our study not only furnishes another empirical validation for the cFDR method to successfully detect novel and known disease associated genetic variants, but also shows the practicability of improved discovery of novel susceptibility loci using existing GWASs summary results. Six genes (CDKN2B-AS1, ICA1L, ALDH, RAH, C5orf67 and KLF14) thatwere associated with either T2D or CARD in previous studies but not with both were detected as pleiotropic loci in this analysis. Furthermore, seven novel genes are worth noting because no previous study has reported associations with either T2D or CARD for them. For the SNPs that were annotated to these 6 genes, one SNP was located in the intronic regions of gene CPPED1, the rest SNPs were all located in intergenic regions of the genes. As examples, we will discuss gene CPPED1 in the following for their potential functional relevance and significance.
The SNP rs4780476 is located at the intronic region of gene CPPED1. A study reported that the expression of CPPED1 decreased after weight reduction in subcutaneous adipose tissue.40 Moreover, CPPED1 knockdown experiment demonstrated that CPPED1 knockdown with small interfering RNA increased expression of genes involved in glucose metabolism and improved insulin-stimulated glucose uptake, which suggests the potential of CPPED1 knockdown in the treatment of obesity-related phenotypes such as T2D.40 We assume that this gene might be involved in certain processes that are significant in the development of T2D and CARD, however, more future studies are expected to explore the exact mechanisms of the novel gene we identified.
Our study presents several strengths. First, the statistical power is increased through the cFDR method by leveraging two large GWAS datasets, providing an increase in effective sample size. Although a meta-analysis of the same data would offer a similar gain, a meta-analysis only allows for more powerful detection of loci with the same direction of allelic effects in the phenotypes,41 whereas the cFDR method allows for detecting loci regardless of their effect directions. Secondly, we consider two traits that are unlikely to be correlated with T2D and CARD, ADHD and MDD, and generate conditional QQ plots with respect to these “control traits.” This “control traits” enrichment analysis provides an alternative way to examine pleiotropic enrichment and provides a baseline that can be used to statistically partially validate the novel findings in our study. Our study may also have some limitations. First, we could not provide information about the effect estimates of pleiotropic loci on the phenotypes due to a lack of detailed individual-study-level data. However, we can infer this information from the summary beta values in the original GWAS study. This cFDR approach cannot distinguish between vertical and horizontal pleiotropy of the pleiotropic signals, although this question might be partially addressed in future summary-based Mendelian Randomization (SMR)42,43 study. Second, it is likely that some of our cFDR results may be overstated due to overlapping samples although the model-free approach is able to neutralize this overestimation of the conservative cFDR estimate.4,7,8 Alternative approaches may be applied to check whether novel loci could still be identified in order to further confirm novel findings in our study or to furnish an empirical comparison of the relative performance of alternative methods, a topic we wish to pursue in the future with comprehensive theoretical and simulation approaches.
In summary, by incorporating pleiotropic effects of two closely related traits into a conditional analysis framework, we observed significant pleiotropic enrichment between T2D and CARD, supporting the improved statistical power of the method. We identified several novel pleiotropic loci of potential functional significance for T2D and CARD in our analysis, and the results may provide us with novel insights into the shared genetic influences between these two disorders.
Supplementary Material
Research in context.
T2D and CARD share primary risk factors such as smoking, hypertension, elevated lipid, dysbetalipoproteinemia and hyperglycemia, also some potential risk factors like obesity, lack of physical activity, cardiovascular family history, gender and age. We found additional common variants associated with T2D and CARD. We found 9 pleiotropic loci associated with both T2D and CARD. These findings may provide novel insights into the etiology of T2D and CARD, as well as the processes that may influence disease development both individually and jointly
Acknowledgments
Chang Qing Sun took responsibility for the contents of this article as he conceived and initiated this project, provided advice on experimental design, oversaw the implementation of the statistical method, and revised/finalized the manuscript. We appreciate the support from Zhengzhou University Key scientific research projects of universities in Henan Henan Provice [19A330005] in providing necessary support for this project.
Footnotes
Disclosures: The authors declare no competing financial interests.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jdiacomp.2018.09.003.
References
- 1.Yang J, Benyamin B, McEvoy BP, et al. Common SNPs explain a large proportion of the heritability for human height. Nat Genet 2010;42:565–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yoo YJ, Pinnaduwage D, Waggott D, Bull SB, Sun L. Genome-wide association analyses of north American rheumatoid arthritis consortium and Framingham heart study data utilizing genome-wide linkage results. BMC Proc 2009;3:S103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stahl EA, Wegmann D, Trynka G, et al. Bayesian inference analyses of the polygenic architecture of rheumatoid arthritis. Nat Genet 2012;44:483–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andreassen OA, Djurovic S, Thompson WK, et al. Improved detection of common variants associated with schizophrenia by leveraging pleiotropy with cardiovascular-disease risk factors.AmJ Hum Genet 2013;92:197–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cichonska A, Rousu J, Marttinen P, et al. metaCCA: summary statistics-based multivariate meta-analysis of genome-wide association studies using canonical correlation analysis. Bioinformatics 2016;32(13):1981–9, 10.1093/bioinformatics/btw052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chung D, Yang C, Li C, Gelernter J, Zhao H. GPA: a statistical approach to prioritizing GWAS results by integrating pleiotropy and annotation. PLoS Genet 2014;10, e1004787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andreassen OA, Thompson WK, Schork AJ, et al. Improved detection of common variants associated with schizophrenia and bipolar disorder using pleiotropy-informed conditional false discovery rate. PLoS Genet 2013;9, e1003455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andreassen OA, McEvoy LK, Thompson WK, et al. Identifying common genetic variants in blood pressure due to polygenic pleiotropy with associated phenotypes. Hypertension 2014;63:819–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stearns FW. One hundred years of pleiotropy: a retrospective. Genetics 2010;186: 767–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Consortium C-DGotPG. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet 2013;381:1371–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peng C, Lou HL, Liu F, et al. Enhanced identification of potential pleiotropic genetic variants for bone mineral density and breast cancer. Calcif Tissue Int 2017;101(5): 489–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peng C, Shen J, Lin X, et al. Genetic sharing with coronary artery disease identifies potential novel loci for bone mineral density. Bone 2017;103:70–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greenbaum J, Wu K, Zhang L, Shen H, Zhang J, Deng HW. Increased detection of genetic loci associated with risk predictors of osteoporotic fracture using a pleiotropic cFDR method. Bone 2017;99:62–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zeng CP, Chen YC, Lin X, et al. Increased identification of novel variants in type 2 diabetes, birth weight and their pleiotropic loci. J Diabetes 2016;9(10):898–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Organization WH. Global Report on Diabetes. 2016:88. [Google Scholar]
- 16.Diabetes mellitus: a major risk factor for cardiovascular disease A joint editorial statement by the American Diabetes Association; the National Heart, Lung, and Blood Institute; the juvenile diabetes foundation international; the National Institute of Diabetes and Digestive and Kidney Diseases; and the American Heart AssociationCirculation 1999;100:1132–3. [DOI] [PubMed]
- 17.Hammoud T, Tanguay J-F, Bourassa MG. Management of coronary artery disease: therapeutic options in patients with diabetes. J Am Coll Cardiol 2000;36:355–65. [DOI] [PubMed] [Google Scholar]
- 18.Almgren P, Lehtovirta M, Isomaa B, et al. Heritability and familiality of type 2 diabetes and related quantitative traits in the Botnia Study. Diabetologia 2011;54:2811–9. [DOI] [PubMed] [Google Scholar]
- 19.Marenberg ME, Risch N, Berkman LF, Floderus B, de Faire U. Genetic susceptibility to death from coronary heart disease in a study of twins. N Engl J Med 1994;330:1041–6. [DOI] [PubMed] [Google Scholar]
- 20.López-Jaramillo P, Pradilla LP, Lahera V, Sieger FAS, Rueda-Clausen CF, Márquez GA. A randomized, double blind, cross-over, placebo-controlled clinical trial to assess the effects of Candesartan on the insulin sensitivity on non diabetic, non hypertense subjects with dysglyce mia and abdominal obesity. “ARAMIA”. Trials 2006;7:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kannel WB. Lipids, diabetes, and coronary heart disease: insights from the Framingham Study. Am Heart J 1985;110:1100–7. [DOI] [PubMed] [Google Scholar]
- 22.Norhammar A, Schenck-Gustafsson K. Type 2 diabetes and cardiovascular disease in women. Diabetologia 2013;56:1–9. [DOI] [PubMed] [Google Scholar]
- 23.Morris AP, Voight BF, Teslovich TM, et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet 2012;44:981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schunkert H, Konig IR, Kathiresan S, et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet 2011; 43,333–U153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controlsNature 2007;447:661–78. [DOI] [PMC free article] [PubMed]
- 26.Neale BM, Medland SE, Ripke S, et al. Meta-analysis of genome-wide association studies of attention-deficit/hyperactivity disorder. J Am Acad Child Adolesc Psychiatry 2010;49:884–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Major Depressive Disorder Working Group of the Psychiatric GC. A mega-analysis of genome-wide association studies for major depressive disorder. Mol Psychiatry 2013;18, 10.1038/mp.2012.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Le Hellard S, Wang Y, Witoelar A, et al. Identification of gene loci that overlap between schizophrenia and educational attainment. Schizophr Bull 2017;43:654–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smeland OB, Wang Y, Lo MT, et al. Identification of genetic loci shared between schizophrenia and the Big Five personality traits. 2017;7:2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Q, Wu KH, He JY, et al. Novel common variants associated with obesity and type 2 diabetes detected using a cFDR method. Sci Rep 2017;7,16397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Consortium. GO. Gene ontology consortium: going forward. Nucleic Acids Res 2015;43:D1049–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Franceschini A, Szklarczyk D, Frankild S, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res 2013;41: D808–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mahajan A, Go MJ, Zhang W, et al. Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat Genet 2014;46:234–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gaulton KJ, Ferreira T, Lee Y, et al. Genetic fine mapping and genomic annotation defines causal mechanisms at type 2 diabetes susceptibility loci. Nat Genet 2015;47: 1415–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scott RA, Lagou V, Welch RP, et al. Large-scale association analyses identify new loci influencing glycemic traits and provide insight into the underlying biological pathways. Nat Genet 2012;44:991–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Willer CJ, Schmidt EM, Sengupta S, et al. Discovery and refinement of loci associated with lipid levels. Nat Genet 2013;45:1274–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Howson JMM, Zhao W, Barnes DR, et al. Fifteen new risk loci for coronary artery disease highlight arterial-wall-specific mechanisms. Nat Genet 2017;49:1113–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deloukas P, Kanoni S, Willenborg C, et al. Large-scale association analysis identifies new risk loci for coronary artery disease. Nat Genet 2013;45:25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Teslovich TM, Musunuru K, Smith AV, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature 2010;466:707–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vaittinen M, Kaminska D, Kakela P, et al. Downregulation of CPPED1 expression improves glucose metabolism in vitro in adipocytes. Diabetes 2013;62:3747–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zeggini E, Scott LJ, Saxena R, et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet 2008;40:638–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu Z, Zhang F, Hu H, et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat Genet 2016;48:481–7. [DOI] [PubMed] [Google Scholar]
- 43.Pavlides JM, Zhu Z, Gratten J, McRae AF, Wray NR, Yang J. Predicting gene targets from integrative analyses of summary data from GWAS and eQTL studies for 28 human complex traits. Genome Med 2016;8:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


