Abstract
Mutations of the inositol 5-phosphatase OCRL cause Lowe syndrome (LS), characterized by congenital cataract, low IQ, and defective kidney proximal tubule resorption. A key subset of LS mutants abolishes OCRL’s interactions with endocytic adaptors containing F&H peptide motifs. Converging unbiased methods examining human peptides and the unicellular phagocytic organism Dictyostelium discoideum reveal that, like OCRL, the Dictyostelium OCRL orthologue Dd5P4 binds two proteins closely related to the F&H proteins APPL1 and Ses1/2 (also referred to as IPIP27A/B). In addition, a novel conserved F&H interactor was identified, GxcU (in Dictyostelium) and the Cdc42-GEF FGD1-related F-actin binding protein (Frabin) (in human cells). Examining these proteins in D. discoideum, we find that, like OCRL, Dd5P4 acts at well-conserved and physically distinct endocytic stations. Dd5P4 functions in coordination with F&H proteins to control membrane deformation at multiple stages of endocytosis and suppresses GxcU-mediated activity during fluid-phase micropinocytosis. We also reveal that OCRL/Dd5P4 acts at the contractile vacuole, an exocytic osmoregulatory organelle. We propose F&H peptide-containing proteins may be key modifiers of LS phenotypes.
INTRODUCTION
Phosphoinositide lipids (PIPs) are a group of seven phospholipids that, by reversible phosphorylation of the 3′, 4′, and 5′ positions of their cytosolic inositol ring, help specify membrane function and identity. They achieve this chiefly by recruiting cytosolic proteins, such as adaptor proteins for membrane sorting and deformation complexes, allowing coordinated recruitment and sorting of transmembrane cargoes in the exo/endocytic cycle. As membranes progress through the exo/endocytic pathways, changes in their PIP “signature” signal a change in their identity, thus modifying membrane function. Humans express at least nine enzymes that act on the 5′ position of the lipids PI(4,5)P2 and/or PI(3,4,5)P3. Both OCRL (OCRL1) and its paralog INPP5B (also called OCRL2) contain an N-terminal Pleckstrin homology (PH) domain (Mao et al., 2009), a central 5′-phosphatase catalytic module, an ASH (ASPM, SPD-2, Hydin) domain (Ponting, 2006), which stabilizes a C-terminal catalytically inactive RhoGAP-like domain(Erdmann et al., 2007; Pirruccello et al., 2011). The core structure of the OCRL proteins (5′ phosphatase and ASH-RhoGAP tandem domain) is exceptionally well conserved during evolution. OCRL-like proteins are found in most eukaryotic lineages, including distantly related eukaryotes such as the protist Giardia lamblia (Yichoy et al., 2011), suggesting a significant conserved function.
OCRL acts on the 5-phosphates of PI(4,5)P2 and PI(3,4,5)P3, two PIPs that are typically enriched at the plasma membrane. However, OCRL is a promiscuous interactor of endocytic proteins, which are found on organelles not typically enriched in these substrate lipids. Endocytic interactors include proteins involved in the initial steps of clathrin-mediated endocytosis (Choudhury et al., 2005, 2009; Mao et al., 2009; Taylor et al., 2011), early and late endosomal adaptor proteins (Erdmann et al., 2007; Swan et al., 2010; Pirruccello et al., 2011; Nandez et al., 2014), a wide variety of Rab (Hyvola et al., 2006; Fukuda et al., 2008), and Rho family GTPases (Lichter-Konecki et al., 2006), leading to a distinctive distribution of OCRL-positive intracellular compartments (Erdmann et al., 2007; Williams et al., 2007; Choudhury et al., 2009).
Loss, truncation, or missense mutation of OCRL causes the congenital X-linked disorder, oculo-cerebral renal syndrome of Lowe (Lowe syndrome [LS; OMIM: 309000; Attree et al., 1992]), affecting the brain, eyes, and renal system, or a less severe condition that primarily involves later-onset dysfunction of the renal system, called Dent2 disease (OMIM: 300555; Hoopes et al., 2005). LS and Dent2 disease are caused by a similar spectrum of OCRL (Attree et al., 1992) mutations, including in one family, the same mutation (Hichri et al., 2011). Missense mutations of the ASH-RhoGAP domain provoke the full spectrum of LS symptoms, namely profound defects in PI(4,5)P2 metabolism in patient fibroblasts and the clinical triad of kidney, brain, and ocular disorders. Previously, we have identified that these ASH-RhoGAP mutants are specifically deficient in binding to endocytic proteins containing a helical peptide that we called the F&H motif (for the obligate presence of conserved phenylalanine and histidine residues [Swan et al., 2010; Pirruccello et al., 2011]).
The F&H peptide directly binds a surface unique to the OCRL/INPP5B RhoGAP domain. Previous to this study, we had identified F&H motifs in three endocytic proteins, the very early endocytic protein APPL1 (Erdmann et al., 2007), which is a resident of noncanonical PI3P-negative, Rab5-positive endosomes (Zoncu et al., 2009), and a pair of later, endosomal adaptors Ses1 and Ses2 (Swan et al., 2010), otherwise known as IPIP27A/B (Noakes et al., 2011), which are resident on canonical PI3P-positive endosomes. Binding of APPL1 and Ses1/2 to OCRL is mutually exclusive and occurs at different endosomal stations defined by the absence or presence of PI3P, respectively (Swan et al., 2010), suggesting formation of OCRL-F&H adaptor protein complexes is subject to regulation by other endocytic factors.
The function of F&H motif-dependent interactions of OCRL and INPP5B and their contribution to LS pathology remain elusive. In different systems, fluid-phase endocytosis (Oltrabella et al., 2015) and efficient ciliary traffic (Coon et al., 2012) require the interaction of OCRL with F&H proteins, suggesting their relevance to LS phenotypes. Ses2/IPIP27B recruits OCRL to sort cation-independent mannose-6-phosphate receptor (Billcliff et al., 2016), a retromer cargo, demonstrating a requirement for OCRL-driven PI(4,5)P2 dephosphorylation via F&H proteins in late endocytic traffic. The F&H binding patch is conserved in most species that express an OCRL-like protein, including a variety of protists and other unicellular eukaryotes, including Dictyostelium discoideum, Trypanosoma brucei (Pirruccello et al., 2011), Albugo candida (CCI48278.1, an Oomycete), and Cyanidioschyzon merolae (XP_005535444.1, a red alga), suggesting that interactions with F&H peptides are critical to OCRL function. In the above organisms, no orthologues of APPL1 and Ses1/2 could be identified by primary amino acid sequence similarity or domain structure predictions. We hypothesized that critical functions of OCRL and INPP5B were mediated by yet-unknown F&H peptide-containing binding partners that may have human orthologues, which may help to explain cellular dysfunction in LS.
This study aimed to identify ancestral F&H interactors to gain functional insight into the F&H peptide-binding interface of OCRL/INPP5B and to help understand OCRL dysfunction in LS/Dent2 disease. We utilized the unicellular organism D. discoideum, where the presence of a single OCRL/INPP5B orthologue (Dd5P4, which has a predicted F&H-motif-binding surface) avoids complications of vertebrate models that express two, partially redundant, enzymes (Janne et al., 1998; Bernard and Nussbaum, 2010), OCRL and INPP5B. Importantly, we expected that the study of Dd5P4 would allow us to assess the role of F&H domain binding to OCRL/Dd5P4 in a cellular context where orthologues of APPL1 and Ses1/2 did not appear to be present.
Strikingly, our studies showed that not only a network of F&H peptide interactors of OCRL/INPP5B but also the subcellular localization of these interactors are strongly conserved, suggesting that OCRL-driven hydrolysis of PI(4,5)P2 or PI(3,4,5)P3 on endolysosomal organelles is a widely conserved feature of membrane trafficking. We isolated three D. discoideum proteins with a bona fide F&H motif. Two are related in domain structure to APPL1 and Ses1/2, in spite of divergent amino acid sequence: an APPL1-like early endocytic BAR domain protein and a Ses1/2-like PH domain protein. A third interactor with a novel domain structure is a Rho family GEF, GxcU, orthologous to mammalian FGD1-related F-actin binding protein (Frabin), which we find also has a conserved F&H motif, suggesting its relevance to LS/Dent pathology. We find that overexpression of GxcU leads to endocytic defects that are enhanced by Dd5P4 loss, suggesting Dd5P4 represses GxcU activity. We additionally reveal that both Dd5P4 and the APPL1-like protein are recruited to the contractile vacuole (CV) membrane of D. discoideum upon their kiss-and-run exocytic water discharge, providing new evidence for a role of OCRL-APPL1 partnership in membrane remodeling in the early endocytic pathway.
RESULTS
Peptide array experiments confirm F&H motif and identify Frabin/FGD4 as a potential F&H motif-containing OCRL interactor
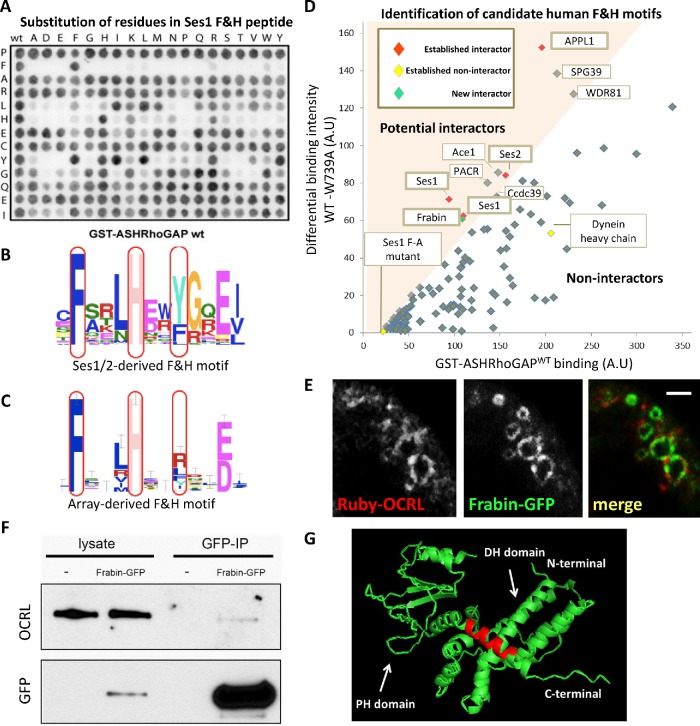
To search for potential new F&H motif interactors of human OCRL, we first defined the F&H interaction consensus via peptide microarray experiments to then search for proteins harboring this consensus in the human proteome (Figure 1 and Supplemental Figure S1). Each residue of the F&H peptide of human Ses1 (13 amino acids) was systematically substituted and immobilized on nitrocellulose membranes. These membranes were then probed in an overlay assay with a GST fusion of the ASHRhoGAP domain of human OCRL (Figure 1A), or of the same domain harboring a tryptophan to alanine mutation at amino acid 739 (i.e., a tryptophan critically required for F&H motif binding) (Pirruccello et al., 2011) to exclude nonspecific interactors. We found that this assay validated the F&H motif deduced by evolutionary conservation of residues in APPL1 and Ses1/2 (Swan et al., 2010) (Figure 1, B and C). Interaction with OCRL in the overlay assay was absolutely dependent on the presence of F and H residues at positions 2 and 6 of the motif as previously predicted. The peptide array assay, however, appears to favor an APPL1-like mode of binding (Erdmann et al., 2007; Pirruccello et al., 2011), which requires only 11 amino acids of the 13 amino acid F&H helical peptide (Figure 1, A, C, and D). The array-derived motif also includes either an R or a hydrophobic residue as opposed to the bulky hydrophobic residue at position 9 defined by conservation (Figure 1B), generating a slightly less selective F&H motif than that derived by evolutionary conservation.
FIGURE 1:
Overlay assay detects new F&H candidate proteins. (A) Peptide overlay experiments confirm the established consensus sequence for F&H peptides. Each residue was systematically substituted in the Ses1 F&H peptide (vertical: the original residue in the peptide, horizontal: substituted residue) immobilized on nitrocellulose and probed with GST-hOCRL-ASHRhoGAP or the non-F&H binding GST-hOCRL-ASHRhoGAPW739A to control for nonspecific binding. (B) F&H consensus derived from sequence conservation of F&H peptides in Ses1/2 proteins. (C) F&H consensus derived from peptide overlay. (D) Naturally occurring human peptides that fall within the F&H consensus derived in C were immobilized on nitrocellulose and probed with WT GST-hOCRL-ASHRhoGAP (D, x-axis) and control as before. WT binding intensity is plotted against the difference (y-axis, arbitrary units) in intensity of binding to WT and W739A control. We defined candidate interactors as peptides in the top left quadrant of the graph (i.e., peptides whose binding to GST-hOCRL-ASHRhoGAP is reduced by over 50% by the W739A mutation). All known F&H peptide interactors (red) were isolated by this analysis, including two identical Ses1 peptides. Peptides determined as noninteractors by ITC are excluded (yellow) (Supplemental Figure S1B). The Frabin F&H peptide (green) was isolated as an interactor in this assay. (E) Ruby-OCRL and Frabin-GFP are found together on pinosomal compartments in Cos7; see Supplemental Movie S1. Scale bar: 3 μm. (F) Full length Frabin-GFP immunoprecipitations isolated a weak but specific interaction with endogenous OCRL in Cos7 cells. (G) Predicted structure of the Frabin DH-PH module shows the Frabin F&H peptide (red) is found on an extended alpha helix of the Frabin PH1 domain.
Movie S1.
Supplementary Movie 1, related to Figure 1: Ruby-OCRL (red) and Frabin-GFP (green) are found together on pinosomal compartments in Cos7. Images 0.25Hz. Scale Bar 3μm.
Peptides in the human proteome that fit the peptide-array F&H consensus were synthesized and immobilized on nitrocellulose membranes and probed as above with GST-OCRL-ASHRhoGAP and GST-OCRL-ASHRhoGAPW739A to isolate candidate F&H proteins in humans. Peptides predicted to be inaccessible to cytosolic proteins (i.e., peptides known to be buried in folded domains, transmembrane regions, or extracellular domains) were excluded from the analysis. Peptides whose binding to GST-ASHRhoGAP is diminished by at least 50% by W739A mutation (colored region in Figure 1D) were considered to be candidate F&H peptides.
This screen identified known F&H peptide interactors Ses1, Ses2, and APPL1, as expected, and excluded peptides that we had previously determined using isothermal titration calorimetry (ITC) to be nonspecific (Swan et al., 2010; Pirruccello et al., 2011) (Supplemental Figure S1), confirming the specificity of the analysis. It also identified additional potential interactors: PACR, Ace1, Ccdc39, SPG39, WDR81, and Frabin (Figure 1D). PACR was excluded on closer inspection as key residues are buried in the plasma membrane (McCulloch et al., 2001). Ace1, Ccdc39, and SPG39 were also excluded as, based on Western blotting on GST-ASHRhoGAP pull downs in HKC cells (Supplemental Figure S1), full-length proteins did not show a specific interaction with the F&H interaction surface of OCRL. WDR81 has a peptide that does not fit in the consensus defined by the conservation of Ses1/2 F&H peptides (Figure 1B), but fits with the looser consensus (including an R residue at position 9) defined in Figure 1C by peptide array. This low-stringency F&H motif is lost in WDR81 in lower vertebrates such as Gallus gallus and Danio rerio, suggesting that it is not a conserved interactor of OCRL via this interface.
Our final candidate, Frabin/FGD4, is a Cdc42-GEF in the Dbl homology (DH) family of GEFs, consisting of a highly affine actin binding peptide region(Banerjee et al., 2009), the DH-PH tandem GEF domain followed by two lipid binding domains: the PI3P-binding FYVE domain(Catimel et al., 2013) and a second PH domain, which binds a broad spectrum of phosphoinositides including PI(3)P and PI(4,5)P2 (Catimel et al., 2013). As we had no antibody that recognized endogenous Frabin, we examined the interaction of Frabin-GFP with endogenous OCRL. Accordingly, a pool of Frabin-GFP was detected along with OCRL on spontaneously generated pinosomes in Cos7 cells (Figure 1E). It remained unclear whether this partial colocalization was due to a direct interaction or to the ability of both proteins to independently associate with pinosomes. However, when Cos7 cells expressing Frabin-GFP were subjected to anti-GFP immunoprecipitation, a weak, albeit specific, coprecipitation with OCRL was observed (Figure 1F). Modeling of Frabin using PHYRE2 (Kelley et al., 2015) to thread the Frabin DH-PH sequence on the known structure of Vav (PDB:3BJI) suggests that the F&H peptide sits on an extended helix of the PH domain (Figure 1G), which may be concealed by the neighboring FYVE domain when Frabin is inactive. The full-length Frabin-OCRL interaction may thus be regulated and only transient, despite the interaction of the Frabin F&H peptide with OCRL being strong.
Inspection of databases revealed that the F&H binding patch of OCRL is present in species in which APPL1, Ses1/2, and Frabin orthologues cannot be identified by primary amino acid similarity, suggesting that the original interactor of the OCRL-F&H interface had not yet been identified. Attempts to use unbiased bioinformatics approaches to identify conserved proteins harboring F&H peptides failed, due largely to the large number of peptides that fit the putative F&H consensus and poor sequence conservation for domains such as PH or BAR domains. Thus, we turned to an ancient unicellular organism, D. discoideum to isolate potential additional evolutionary conserved OCRL F&H interface interactors and to understand the function of this interface.
Identification of three F&H motif-dependent interactors of Dd5P4
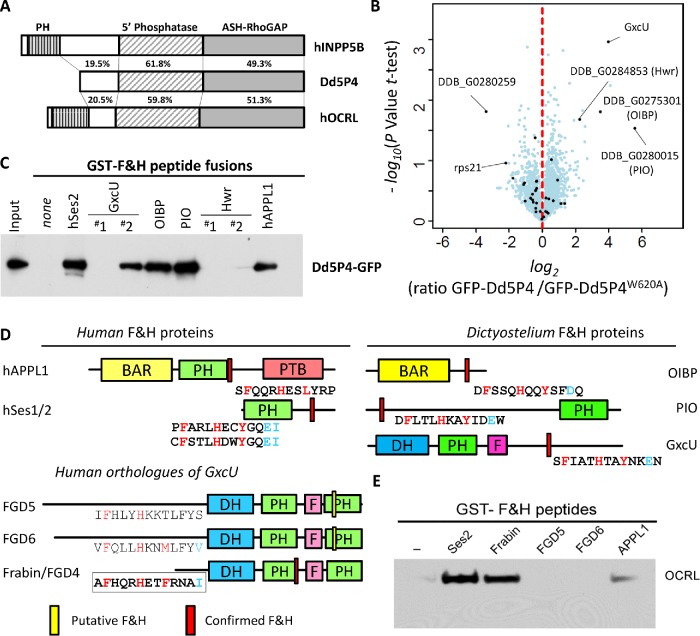
The single OCRL-like protein of D. discoideum, Dd5P4, has the same overall architecture as mammalian OCRL (Loovers et al., 2003, 2007), except that it lacks the N-terminal PH domain (Mao et al., 2009) and any identifiable clathrin coat binding motif (Figure 2A and Supplemental Figure S2). Importantly, the F&H motif binding surface of the RhoGAP-like domain is conserved. Dd5P4 knockout cells (Dd5P4- mutants) can be partially rescued by expression of human OCRL (Loovers et al., 2007), demonstrating that Dd5P4 and OCRL have well-conserved functions.
FIGURE 2:
The F&H interface specifically interacts with three proteins in D. discoideum. (A) Comparison of human INPP5B/OCRL and Dd5P4 amino acid sequences. (B) Analysis of Dd5P4-; GFP-Dd5P4WT vs. Dd5P4-; GFP-Dd5P4W620A immunoprecipitates shows enrichment of four possible interactors of the F&H surface of Dd5P4. Points labeled in black are proteins that were also enriched in an analysis of WT vs. Dd5P4-; GFP-Dd5P4WT (Supplemental Figure S3). (C) All possible F&H peptides in the four putative F&H-surface interactors were produced as GST fusions and purified on beads. These beads were exposed to lysates of D. discoideum expressing GFP-Dd5P4WT. (D) Domain structure of F&H interactors in D. discoideum and humans with validated interacting peptides shown below. DH, Dbl Homology domain; PH, Phox Homology homain; F, FYVE domain; BAR, Bin-Amphiphysin-Rvs domain; PTB, phosphotyrosine binding domain. The location of validated F&H peptide consensus is highlighted in red, unsuccessful F&H motifs are highlighted in yellow. (E) GST and GST-F&H peptides were exposed to HKC cell lysate. A specific interaction with endogenous OCRL was found with the known interactors Ses2 and APPL1.
To identify F&H interactors of Dd5P4 in D. discoideum, in Dd5P4-cells we expressed GFP fusions of wild-type (WT) Dd5P4 and of a mutant Dd5P4 harboring a tryptophan to alanine substitution at amino acid position 620, which corresponds to mammalian W739 required for F&H motif binding(Pirruccello et al., 2011) (Supplemental Figure S2). We then analyzed triplicate GFP immunoprecipitates from untransfected WT cells alongside GFP-Dd5P4 and GFP-Dd5P4W620A transfected Dd5P4-knockout cells by mass spectrometry. We found four proteins that were specifically enriched in precipitates from GFP-Dd5P4-expressing cells versus negative control precipitates, but not in precipitates from GFP-Dd5P4W620A-expressing cells (Figure 2B; for comparison against GFP alone, Supplemental Figure S3). These four proteins were identified: three uncharacterized gene products, DDB_G0275301, DDB_G0280015, and DDB_G0284853, and the putative Rho-GEF GxcU.
To determine whether there was a direct F&H peptide-mediated interaction with Dd5P4-GFP, we made GST fusions of potential F&H peptides of each protein, performed pull downs from lysates of Dd5P4- cells expressing Dd5P4-GFP, and probed the blot with anti-GFP antibodies. To be certain that we captured any possible interaction, we selected as our test F&H peptide any 13 amino acid stretch in the four proteins that contained the conserved F&H residues, including peptides that did not fit to either of our consensus motifs (Figure 2C and Supplemental Figure S1). GST fusions of the F&H peptides of human Ses2 and APPL1 were also included. Figure 2C shows that Dd5P4-GFP bound the F&H motifs of human APPL1 and Ses2, confirming conservation of the F&H interaction between humans and D. discoideum. GST fusions of the putative F&H peptides of three of the four candidate D. discoideum interactors (DDB_G0275301, DDB_G0280015, and GxcU) bound Dd5P4, while two low-stringency F&H peptides of protein DDB_G0284853, a mycBP2/Highwire-like protein, did not (Figure 2C; Supplemental Figure S1C). We thus conclude that GxcU, DDB_G0275301 (which we called OCRL-Interacting Bar domain Protein [OIBP]), and DDB_G0280015 (which we called PH domain Interacting with OCRL [PIO]) are bona fide direct interactors of the Dd5P4 F&H binding surface. Examining RNA expression profiles (Van Driessche et al., 2002), all three F&H-containing proteins are well expressed in cells under nondifferentiation conditions, suggesting that all three proteins are available to interact competitively with Dd5P4 at the same time.
The three interactors of Dd5P4 have domain homologies to APPL1, Ses1/2, and Frabin
Remarkably, despite there being no identifiable amino acid sequence homology, modeling (Kelley et al., 2015) of the domain structures of the uncharacterized D. discoideum F&H motif interactors (OIBP, PIO, and GxcU) suggested these to contain signature domains of those of the well-characterized F&H partners of OCRL: APPL1 (a BAR domain) and Ses1/2 (a PH domain with extended PxxP motifs), respectively. GxcU is a DH-PH domain Rho GEF with similar domain structure Frabin, which we had identified as a human candidate interactor by peptide array (Figure 2D).
Using Phyre2 (Kelley et al., 2015) to model D. discoideum sequences onto crystal structures deposited in the PDB database, OIBP (amino acids 37–228) displayed a high structural homology with the BAR domain portion of the APPL1 paralogue APPL2 (PDB:4H8S, 98.8% confidence) in spite of a low (12%) sequence identity. The only published study referring to D. discoideum OIBP (Journet et al., 2012) reported that this protein was purified in association with macropinosomes, indicating a possible role in endocytosis/macropinocytosis, a function that is conserved with mammalian APPL1/2 (Zoncu et al., 2009; Swan et al., 2010; Bohdanowicz et al., 2012).
PIO is a 741 amino acid protein with a predicted PH domain. Since PIO’s PH domain contains a series of long unstructured peptide loops that interfere with accurate tertiary structure prediction, to assess its structural "neighbors" we made structure predictions by searching only the portions of the PH domain of PIO conserved in the closely related species Dictyostelium purpureum (XP_003295168.1) (Supplemental Figure S4) (Schaap et al., 2006). This PH domain falls loosely into the same structural family as the PH domain of PRKD3 (PDB:2D9Z 99.2% confidence, 18% sequence similarity) and of PKCD2 (PDB:2COA; 99.2% confidence, 17% sequence similarity), as does the PH domain of human Ses1 (99.9% model confidence, 17% sequence similarity). No function has previously been assigned to this protein.
GxcU is one of four D. discoideum proteins (GxcU, GxcV, GxcW, and GxcX) with a domain structure similar to that of Frabin (Vlahou and Rivero, 2006): a Dbl-Rho family GEF domain, followed by a PH domain and a FYVE domain. GxcU is largely unstudied, but knockout strains show that this gene is not essential for life (Wang et al., 2013). Frabin is one of seven closely related so-called FGD (faciogenital dysplasia) proteins, FGD1-6 and FGD1-related Cdc42-GEF/FARP2, thought to function as Cdc42 GEFs. Three FGD proteins are associated with disease: FGD1 is mutated in Aarskog-Scott syndrome (Pasteris et al., 1994), FGD4/Frabin mutations cause the peripheral neuropathy Charcot Marie Tooth 4H (Delague et al., 2007), and mutations of FGD6 are a risk factor for macular degeneration (Huang et al., 2016). Expanding on our peptide array analysis, we examined all putative F&H motifs in the FGD family regardless of possible presence in folded domains, isolating F&H peptides in Frabin/FGD4, FGD5 (a protein specific to haemopoetic stem cells [Gazit et al., 2014]), and FGD6 (Figure 2D).
Pull downs from mouse brain extracts using GST fusions of the putative 13 amino acid F&H motif peptides of each of the three FGD proteins, and of Ses2 and APPL1 as positive controls, revealed that the F&H peptide of Frabin, but not that of FGD5 and FGD6, specifically enriched endogenous OCRL (Figure 2E). The F&H motif captured for Frabin was the same as that independently identified by peptide array (Figure 1D).
Two unbiased methods, peptide array on human proteins and mass spectrometry in Dictyostelium, converge on a common set of conserved F&H-peptide harboring OCRL interactors, APPL1, Ses1/2, and Frabin. We hypothesize that distant homologues of at least one of these proteins exist in all cells that express an OCRL protein harboring this patch.
F&H motif-containing proteins are functionally similar in D. discoideum and mammals
We next examined the properties of our newly identified F&H binding partners in D. discoideum. PIO-GFP fusion proteins were located to compartments of the late-acidic/postlysosomal phase (Figure 3). PIO-GFP colocalized with the fluid-phase marker TRITC-dextran (a fluid-phase dye taken up in the phagosomal-lysosomal system and excreted via postlysosomes) in organelles labeled by 30 min of fluid phase dye uptake by micropinocytosis, suggesting a function in the endo-lysosomal system. Figure 3 shows PIO-GFP localization in both WT and Dd5P4- cells exposed for 30 min to TRITC-Dextran. There is very little colocalization between PIO-GFP and dextran label at earlier timepoints (Figure 3A, 10 min dextran pulse) and strong but not complete colocalization at 30 min dye uptake (Figure 3, B–D), suggesting that PIO-GFP labels organelles at the transition between late acidic lysosomes to postlysosomes. This is reminiscent of the localization of Ses1/2 proteins at late endosomal stages in mammals (Swan et al., 2010; Noakes et al., 2011).
FIGURE 3:
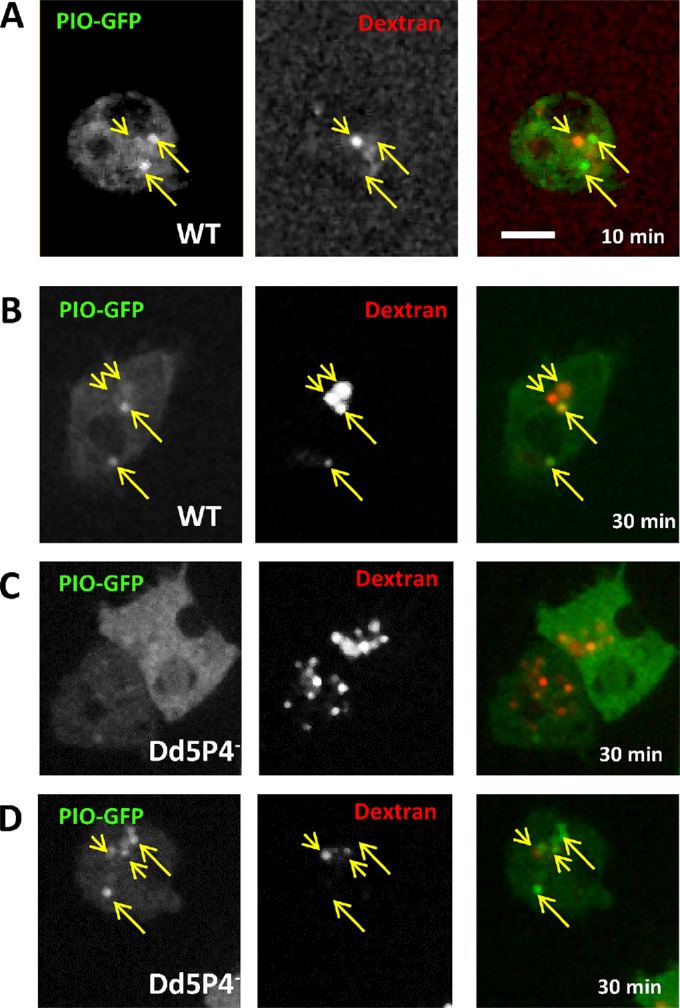
PIO-GFP labels a late lysosomal station. (A) In WT cells, PIO-GFP (arrows) does not colocalize with early lysosomal intermediates labeled by 10 min of fluid-phase dextran uptake (arrowheads). (B) At 30 min, fluid phase dextran (red) marks some, but not all, PIO-GFP positive compartments. (C, D) There is no significant change in PIO-GFP localization in Dd5P4- cells: two examples of PIO-GFP expressing Dd5P4- cells (one cytosolic, one punctate) labeled for 30 min with TRITC-dextran. Scale bar: 5 μm.
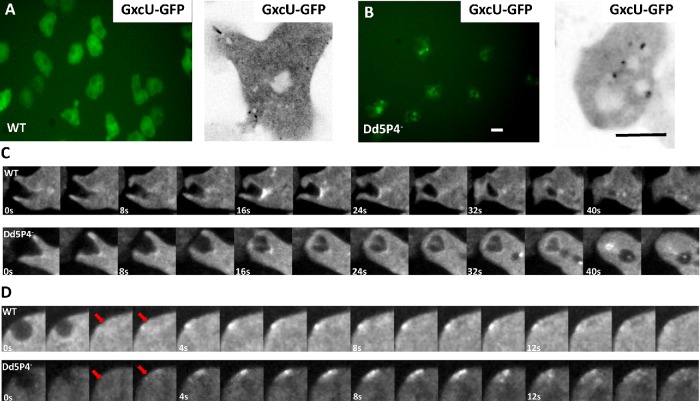
Due to a long, low complexity AT-rich stretch in the D. discoideum GxcU DNA sequence, it was not possible to clone or to synthetize the D. discoideum GxcU cDNA as a GFP fusion. Instead, we synthetized the sequence of the GxcU orthologue from the closely related amoeba Dictyostelium intermedium (Schaap et al., 2006) (see Supplemental Figure S5 for sequence comparison) and expressed it as a fusion protein. Overall, GxcU-GFP appears more punctate in Dd5P4- cells (Figure 4, A and B). GxcU-GFP appears on several subcellular structures, including sites of macropinosome formation (Figure 4C) and on rare occasions at the plasma membrane some seconds after CV exocytosis events (for a more complete description of CV, please see below) (Figure 4D), suggesting that GxcU participates in endocytic events. GxcU-GFP recruitment was evident at the base of the forming macropinocytic cup (Figure 4C), which would indicate an early endocytic function, and several seconds after CV fusion (Figure 4C) (i.e., later than the appearance of OIBP-GFP on these organelles [Figure 5C]). A role in coordinating actin-dependent remodeling of the plasma membrane at endocytic sites has also been reported for Frabin (Nakanishi and Takai, 2008; Horn et al., 2012). Recruitment of the GxcU-GFP fusion protein appeared delayed in the Dd5P4- mutant (Figure 4C).
FIGURE 4:
GxcU-GFP labels several endocytic intermediates. Wide-field microscopy (left panel) and confocal microscopy (right panel) of GxcU-GFP expressing cells. (A) WT cells expressing GFP-GxcU. (B) GxcU-GFP is more punctate in Dd5P4- cells. Scale bars in B: 10 µm. (C) GxcU-GFP can be detected on phagosomes in both WT (top) and Dd5P4- (bottom) cells. Gallery of images 0.25 Hz (see Supplemental Movies S2 and S3). (D) GxcU-GFP is also (rarely) detectible several seconds after CV collapse (site of CV collapse marked by red arrows) in both WT and Dd5P4- cells, gallery at 1 Hz.
FIGURE 5:
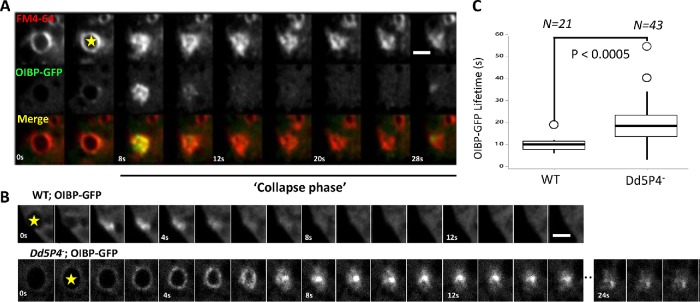
F&H interactions contribute to membrane reformation kinetics at the CV. (A) OIBP-GFP in wild-type cells. Expression of OIBP-GFP is largely cytosolic except for endocytic structures associated with CVs, labeled in by FM4-64 (red). Shown is a gallery of images showing CV fusing en face, image rate 0.25 Hz. Yellow Star, CV beginning to collapse (see Supplemental Movie S4). (B) Gallery showing OIBP-GFP recruitment to osmotically triggered CV fusion events imaged at 1 Hz. Top panel, WT cells; bottom panel, Dd5P4- cells. Scale bars in A and B: 4 μm. (C) Lifetime of OIBP-GFP fluorescence during CV collapse (start of the collapse indicated by yellow stars in A and B) is approximately twice as long in Dd5P4- cells. N = 21 WT events, N = 43 Dd5P4- events, two-sided Student’s t test: P < 0.0005.
Movie S2.
Supplementary Movie 2, related to Figure 4: GxcU-GFP can be detected on phagosomes in WT cells. Images 0.25Hz. Scale Bar 15μm.
Movie S3.
Supplementary Movie 3, related to Figure 4: GxcU-GFP can be detected on phagosomes in Dd5P4-cells. Images 0.25Hz . Scale Bar 15μm.
Movie S4.
Supplementary Movie 4, related to Figure 5: Recruitment of a C-terminal GFP fusion of OIBP (green). Contractile vacuoles are labelled by FM4-64 (red). Images 0.25Hz. Scale Bar 10μm.
A C-terminal GFP fusion of OIBP was almost completely cytosolic except for brief and intense ‘flashes’ of recruitment, lasting only a few seconds, near the plasma membrane (Figure 5, A and B, and Supplemental Movie S4). As we could see no colocalization of this signal with TRITC-dextran (i.e., the endolysosomal system), we examined another organelle, the CV. CVs act to excrete excess fluid to prevent osmotic rupture and are found in many water and soil dwellers. In D. discoideum, CVs are a reticulum of bladders and connecting tubules that fill with excess water, undergo rapid kiss-and-run fusion to disgorge their contents, and locally recycle CV membrane without mixing with the plasma membrane by a striking process of tubulation, fragmentation, and reformation of the CV organelle (Clarke et al., 2002; Gerisch et al., 2002; Heuser, 2006) (here indicated as collapse phase). The lipophilic dye FM4-64 preferentially incorporates in CV membranes (Heuser et al., 1993; Nishihara et al., 2007). Colabeling of the intensely OIBP-GFP-labeled structures with FM4-64 showed that these flashes occurred at the moment at which CVs open to the extracellular medium during the discharge and collapse phase (Figure 5, A and B). This observation suggests that OIBP-GFP is recruited as the CV membrane undergoes scission from the plasma membrane and is reformed by tubulation and fragmentation. This localization fits very well with the site of action in mammalian cells of APPL1, a protein with curvature inducer/sensor properties due to its BAR domain (McMahon and Boucrot, 2015), which is recruited to endocytic membranes at the earliest stages of endocytosis in mammalian cells (Zoncu et al., 2009). When this OIBP-GFP construct was expressed in Dd5P4- cells, its localization was essentially unchanged (as is the case for its mammalian orthologue APPL1 in OCRL mutant patient fibroblasts [Swan et al., 2010]). However, when we examined the OIBP-GFP flash at CV exocytic pores, we found that the lifetime of these events had approximately doubled in Dd5P4- mutants (Figure 5, B and C).
Novel function of Dd5P4 in CV fusion/endocytosis is modulated by F&H peptide interactions
Having determined that the F&H proteins of D. discoideum recapitulate the localization of their mammalian counterparts, we turned to characterize Dd5P4 recruitment and function on these organelles. Complementation of a Dd5P4- line with Dd5P4 fused to GFP at either the N- or the C-terminus rescued previously characterized growth deficits (Schaap et al., 2006) in these mutants, indicating that the fusion proteins were functional (Supplemental Figure S5A). We also successfully rescued the Dd5P4- growth deficit with the F&H interface mutant GFP-Dd5P4W620A, suggesting that either sufficient overexpression of Dd5P4W620A is enough to overcome any defects in efficient targeting of Dd5P4 activity to membranes or, as we had previously observed in human patient fibroblasts (Pirruccello et al., 2011), loss of the F&H interface led to loss of OCRL-like activity from very specific membrane subcompartments and not a generalized deficit in OCRL-mediated traffic.
Given the presence of GxcU-GFP and PIO-GFP in the micropinocytic system and the functional requirement for Dd5P4 for micropinocytosis/nutrient uptake (Supplemental Figure S5A), we reexamined the subcellular recruitment (Loovers et al., 2007) of fluorescently tagged Dd5P4. Despite an established function of Dd5P4 in phagocytosis (Loovers et al., 2007), and despite the recruitment of mammalian OCRL to endocytic vesicles engulfing pathogens (Weber et al., 2009), neither published studies (Loovers et al., 2007) nor our own study with N- or C-terminal GFP-tagged Dd5P4 fusions detected a clear accumulation of Dd5P4 on macropinocytic intermediates (labeled by TRITC-dextran in Supplemental Figure S5B). We hypothesize that GFP-tagged fusion proteins are likely to be expressed at higher levels than endogenous Dd5P4, and the excess cytosolic fluorescence may mask the presence of a macropinosome-associated pool of the protein.
Dd5P4- cells expressing either GFP-Dd5P4WT or GFP-Dd5P4W620A exhibited marked transient recruitment of GFP fluorescence to organelles that appeared to fuse with the plasma membrane. We excluded postlysosomes (Gopaldass et al., 2012; King et al., 2013) as the organelles in question, as GFP-Dd5P4WT and GFP-Dd5P4W620A fluorescence segregated away from postlysosomal organelles when cells had been preloaded overnight with TRITC-dextran (Supplemental Figure S5B). FM4-64 dye labeling showed both GFP-Dd5P4WT and GFP-Dd5P4W620A were recruited to CVs in the moments before its fusion and collapse at the plasma membrane (Figure 6D). We then explored the functional requirement for Dd5P4 and its F&H interface on this organelle.
FIGURE 6:
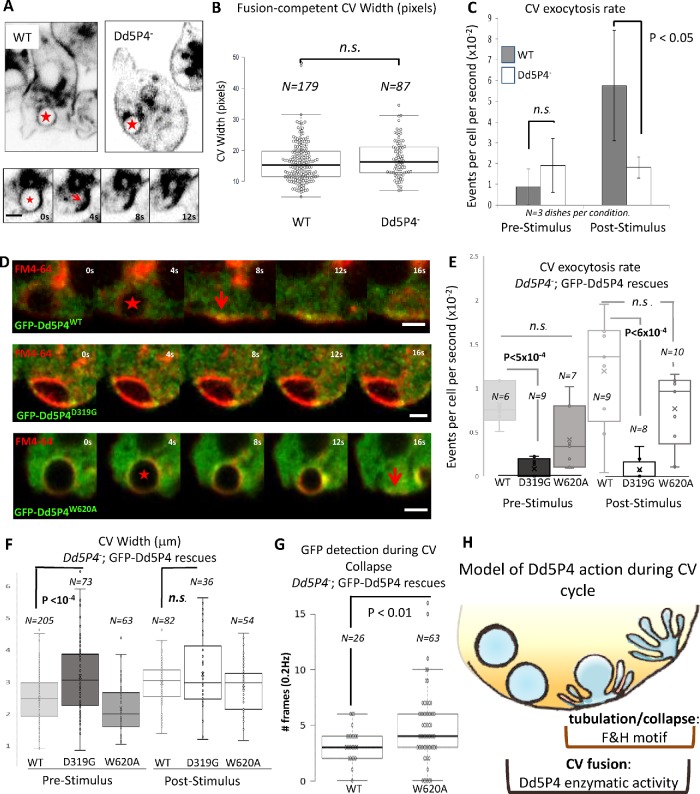
CV dynamics in Dd5P4 mutants and rescues. (A) FM4-64 dye labeling of CV membranes in WT and Dd5P4- cells show CV distribution is similar. Bottom panel: CV collapse event (frames from 0.25 Hz movie) quantified in C and E. The star indicates a CV about to collapse; the arrow indicates the collapsed CV membrane. Scale bar: 2 μm. (B) The size at the widest point of CVs undergoing fusion is unchanged by Dd5P4 loss: N = 179 (WT), N = 87 (Dd5P4-). (C) The rate of CV fusion triggered by 1:1 osmotic stimulus is strongly reduced, but not abolished, in Dd5P4- mutants. Average of three movies per condition, SD, two-sided Student’s t test. (D) Recruitment of GFP-Dd5P4 constructs to CVs marked by FM4-64. GFP-Dd5P4WT and GFP-Dd5P4W620A are recruited to CVs at the moment that the CV starts to collapse (marked by a red star) and persists at the moment of full collapse (arrow). GFP-Dd5P4D319G is recruited in punctae to the CV but do not appear to spread over the CV surface, CVs appear enlarged. Movie: 0.25 Hz. Scale bars: 2 μm. (E) FM4-46 labeling shows that Dd5P4-; GFP-Dd5P4D319G cells exhibit significantly reduced CV fusion/collapse in both basal and osmotically stimulated conditions. Dd5P4-; GFP-Dd5P4W620A cells show a similar rate of CV fusions as Dd5P4-; GFP-Dd5P4WT. The number of movies per condition are indicated on graph. Student’s t test. See Supplemental Movies S7 and S8 for live imaging of GFP signal. (F) Cells in basal conditions show significantly larger CV diameters in Dd5P4-; GFP-Dd5P4D319G cells. (G) GFP signal in collapse phase lasts longer in Dd5P4-; GFP-Dd5P4W620A cells than in cells reexpressing GFP-Dd5P4WT. (H) Model of Dd5P4 action during CV cycle.
When we imaged FM4-64-labeled cells, we identified frequent CV discharges (see Figure 6A for examples of fusion events showing a full CV, collapse of the organelle, and local membrane tubulation). The size of CVs that underwent exocytosis was unchanged between WT and Dd5P4- cells (Figure 6B), suggesting that CV biogenesis was grossly normal. Additionally, basal CV fusion, unstimulated by osmotic pressure, occurred at the same rate in both WT and Dd5P4- mutant strains. However, 1:1 dilution of the imaging medium with distilled water caused an increase in water pumping and of the CV exocytic rate in WT (Figure 6C; Supplemental Movie S5), whereas the exocytic rate of CVs did not change from the basal rate in the Dd5P4- mutants (Supplemental Movie S6).
Movie S5.
Supplementary Movie 5, related to Figure 6: 1:1 dilution of imaging medium with distilled water caused an increase CV exocytosis rate in WT. Images 0.2 Hz. Scale Bar 15μm.
Movie S6.
Supplementary Movie 6, related to Figure 6: 1:1 dilution of imaging medium with distilled water in Dd5P4- mutants does not change CV exocytosis rate. Images 0.2 Hz. Scale Bar 15μm.
Movie S7.
Supplementary Movie 7, related to Figure 6: the collapse/fusion phase of CV exocytosis in Dd5P4- mutants re-expressing GFP-Dd5P4. Images 0.2Hz. Scale Bar 15μm.
Movie S8.
Supplementary Movie 8, related to Figure 6: the collapse/fusion phase of CV exocytosis in Dd5P4- mutants re-expressing F&H mutant GFP-Dd5P4W620A. Images 0.2Hz. Scale Bar 15μm.
Reexpression of GFP-Dd5P4WT in Dd5P4- mutants rescued the osmotically triggered component of CV exocytosis observed in these mutants (Supplemental Figure S5C). GFP-Dd5P4W620A showed localization on CVs similar to GFP-Dd5P4WT, speaking against a role of F&H-containing proteins in the recruitment of Dd5P4 to the CV membrane. Comparing the reexpression of GFP-Dd5P4WT in Dd5P4- mutants with that of GFP-Dd5P4W620A or the catalytic mutant GFP-Dd5P4D319G (Figure 6D), we noted that the catalytic mutant exerted a dominant effect, almost completely abolishing CV cycling (Figure 6E) in both resting and osmotically stimulated conditions. Additionally, we observed that CV diameter (FM4-64 staining) increased in resting cells expressing GFP-Dd5P4D319G (Figure 6, D, quantified in F). Interestingly, whereas WT and W620A Dd5P4 appeared to be recruited to the entire CV membrane at about the time when CV starts decreasing in size due to CV discharge (stars in Figure 6D) and persists during the tubulation/collapse phase (arrowheads in Figure 6D), GFP-Dd5P4D319G appeared punctate and did not spread across the CV membrane upon arrival at a FM4-64-labeled organelle. We did not capture any events where GFP-Dd5P4D319G was recruited to a collapsed/tubulating CV. We therefore analyzed only the kinetics of GFP recruitment in Dd5P4-;GFP-Dd5P4WT, or Dd5P4-; GFP-Dd5P4W620A CV fusion events. There was no significant difference in the lifetime of the GFP signal on precollapse CVs between the two fusion proteins (Supplemental Figure S5D), as in general both fluorescent proteins were recruited when the CV started to discharge and collapse (a few CVs took some time to collapse after GFP-Dd5P4 recruitment as shown in Supplemental Figure S5D). We found that GFP-Dd5P4W620A lingered for a significantly longer time than GFP-Dd5P4WT during the process of CV collapse/tubulation (Figure 6G), suggesting defects in the process by which membranes tubulate and reform after CVs discharge their content. Taken together, this indicates that catalytic activity is necessary for Dd5P4-mediated CV fusion and that F&H peptide interactions in D. discoideum are not essential for recruitment of Dd5P4 to CV membranes, but are necessary for efficient dynamics/remodeling of these membranes during fusion and subsequent reendocytosis of CV membranes (model in Figure 6H).
Dd5P4 loss-of-function deficit in fluid phase dye uptake is phenocopied and enhanced by GxcU overexpression
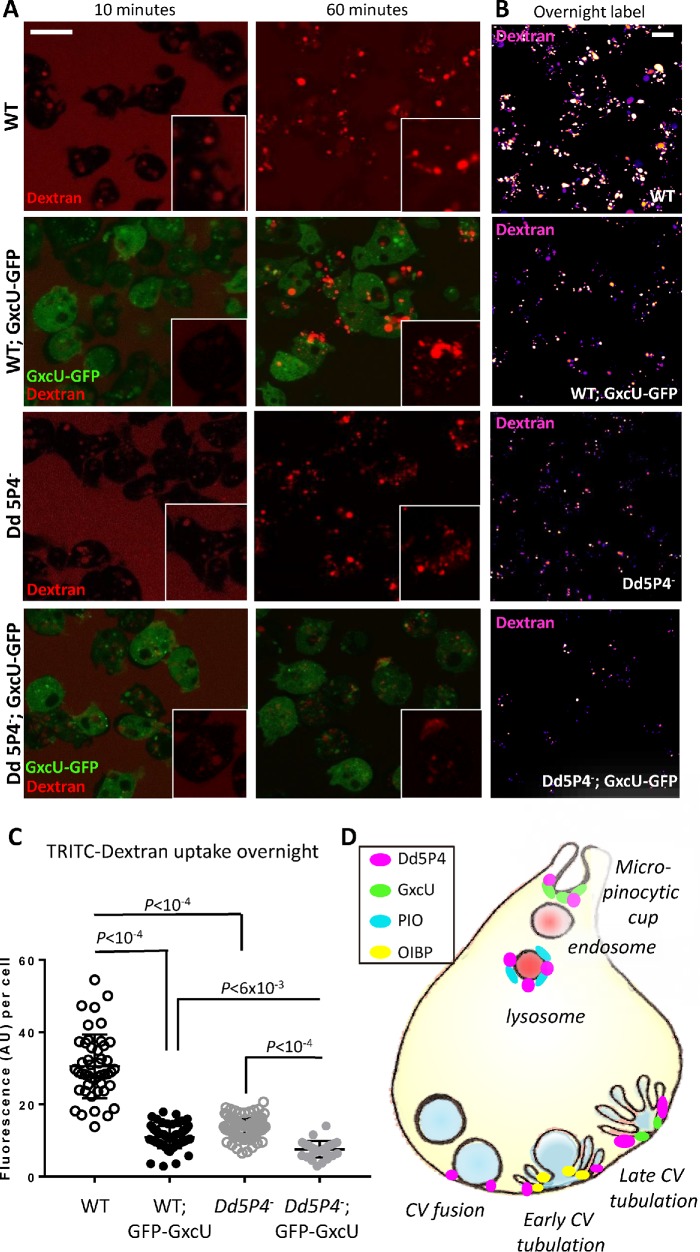
Loss of Dd5P4 has been shown to cause deficits in fluid-phase micropinocytosis. Neither PIO-GFP nor OIBP-GFP appeared to be recruited to early micropinocytic intermediates, but GxcU-GFP had a wide distribution on organelles including micropinosomes (Figure 4), suggesting the two proteins may interact in this process. Overexpression of GxcU-GFP inhibited uptake of TRITC-dextran (Figure 7, A and B), over all the timescales measured (10 min, 60 min, and overnight application of labeled dextran to the culture medium), as observed with Dd5P4- mutants. Both Dd5P4- mutants and GFP-GxcU overexpressors exhibited numerous small dye-positive endolysosomal structures rather than the large lysosomes/postlysosomes found in WT cells, suggestive of a defect in membrane traffic along multiple parts of the endo/lysosomal pathway (see inset images at 60 min uptake in Figure 7A). GxcU-GFP overexpression in Dd5P4- evoked a profound deficit in dye uptake, suggesting that Dd5P4 activity opposes the action of overexpressed GxcU (Figure 7, B and C). Cumulatively, these data suggest that OCRL and its F&H protein interactors each act defined points of the membrane cycle as illustrated in Figure 7D.
FIGURE 7:
GxcU-GFP overexpression and Dd5P4 loss show similar effects in micropinocytic dye uptake and subsequent membrane trafficking. (A) The timed application of TRITC-dextran (red) shows defects in initial micropinocytic uptake (10 min) and subsequent membrane trafficking (60 min) in both Dd5P4- cells and GxcU-GFP overexpressors, which is enhanced by GxcU-GFP overexpression in the Dd5P4- background. Individual cells are shown the inset. (B) Deficits in dye uptake are not resolved by longer exposure to dye (overnight labeling). TRITC-dextran label is shown in false color scale for ease of comparison. Scale bars in A and B: 8 μm. (C) GFP-GxcU overexpression enhances dye uptake defects caused by Dd5P4 loss. Fluorescence per cell after overnight TRITC-dextran uptake, ANOVA. (D) Localization of Dd5P4 and F&H motif-containing interactors.
DISCUSSION
Previous studies have shown that interactions of F&H-peptide-containing proteins with OCRL/INPP5B participate in the localization and function of these inositol 5-phosphatases on intracellular membranes (Noakes et al., 2011; Pirruccello et al., 2011). Loss of the F&H binding surface in mammals very subtly relocalizes OCRL, whereas loss of the OCRL Rab binding interface (Pirruccello et al., 2011; Luo et al., 2012) almost completely delocalizes the protein, suggesting that the F&H interface, as we see in D. discoideum, has a specialized function in membrane traffic, rather than a generalized function recruiting OCRL. The F&H motif-containing OCRL/INPP5B interactors APPL1 and Ses1/2 (IPIP27A/B) were thought to appear later in evolution than OCRL, as searches of databases had detected APPL1 only in vertebrates (Erdmann et al., 2007) and Ses/CG12393 in Drosophila and other insects, but not in unicellular organisms (Swan et al., 2010). In contrast, OCRL/INPP5B-like proteins are widely present in unicellular organisms. Amino acids critically required for the F&H interacting surface of OCRL (Pirruccello et al., 2011) were highly conserved even in these ancestral OCRL species, suggestive of the presence of yet-to-be-identified F&H interactors in these evolutionary older organisms.
We tested this possibility in the social amoeba D. discoideum, which expresses a single OCRL orthologue, Dd5P4, displaying an F&H interacting surface. Loss of Dd5P4 is not lethal (Loovers et al., 2003), but leads to quantifiable defects in macropinocytosis and growth (Loovers et al., 2007), which can be rescued by expression of GFP-fusions of Dd5P4 and of human OCRL, suggesting closely conserved function and subcellular targeting (Loovers et al., 2007).
To our surprise, we found that the identity of the F&H proteins that interact with OCRL is as well conserved as the F&H surface itself: in both humans and amoeba, OCRL uses the F&H interface to couple to three proteins, a BAR-domain protein present at the earliest stages of endocytosis (APPL1 in humans and OIBP in D. discoideum), a PH-domain containing protein on late lysosomes/postlysosomes (Ses1/2 in humans and PIO in D. discoideum), and a novel interactor GxcU/ Frabin, a Rho family GEF, which in humans is associated with the disorder Charcot Marie Tooth Type 4. Live imaging of GxcU and Frabin showed that both proteins appear at multiple early and later stages of endocytosis. Our data suggest that there is a specific requirement of OCRL phosphatase activity coordinated by different F&H proteins at well-delineated endocytic stations.
We also established a novel function for OCRL-like phosphatases in membrane trafficking: in addition to known phenotypes of Dd5P4 loss in D. discoideum (Loovers et al., 2003, 2007; Weber et al., 2009; Hilbi et al., 2011) (reduced growth and a defect in both micropinocytosis and phagocytosis), we identified a novel phenotype in axenic cells: Dd5P4 controls the rate of fusion and endocytic tubulation/recapture of an osmoregulatory organelle, the CV. CV exocytosis is thought to be a kiss-and-run process, involving minimal mixing of the CV membrane with the plasma membrane (Essid et al., 2012).
Notably, we found that Dd5P4 has two separable functions in the CV discharge process: CV fusion, which did not depend on F&H peptide interactions but was blocked by overexpression of a catalytically inactive Dd5P4 fusion protein, and tubulation/reformation (which we indicate as collapse phase), whose kinetic was influenced by F&H-mediated interactions. We measured extended kinetics of the CV collapse phase in GFP-Dd5P4W602A expressing mutants (where the CV fusion rate defects in Dd5P4 knockouts is restored), and the extended kinetics of the OIBP-GFP fusion protein in this same stage in Dd5P4-.
How Dd5P4 interferes with CV fusion is yet unclear, although several functions of OCRL/Dd5P4 might be implicated. Disturbances of the actin cytoskeleton cause inappropriate mixing of CV and plasma membranes (Heuser, 2006), leading to CV-resident proton pumps being found at the plasma membrane. Similarly, ‘spillover’ of the transmembrane protein Dajumin, responsible for the biogenesis of CVs, is retrieved from the plasma membrane via clathrin/AP2 (Macro et al., 2012), where mammalian OCRL is also known to act. Thus it is possible that Dd5P4 could have several functions on this organelle, for example, regulating underlying actin and/or PI(4,5)P2 at the CV fusion site, or in trafficking of receptors that are necessary for the CV fusion/reformation cycle. Our images of GFP-Dd5P4D319G show that this mutant protein remains clustered in punctae upon arriving to docked CVs, whereas GFP-Dd5P4WT and GFP-Dd5P4W620A arrive at CVs prior to fusion and then spread over the CV membrane, coincident with the CV starting to fuse and decrease in size. Previous work (Essid et al., 2012; Parkinson et al., 2014) suggests that during this phase of the CV cycle (docking and fusion-ready), CVs are labeled by Drainin and Rab8 and undergo ‘ring to patch’ transition to allow full fusion. In the absence of the ring to patch transition (i.e., in P2XA mutant CVs [Parkinson et al., 2014]), CV fusion becomes inefficient, as we have seen in Dd5P4- mutants. In contrast, the more severe phenotype of GFP-Dd5P4D319G suggests that this mutant may sequester factors such as Rab8GTP. Indeed, the swollen CV phenotype found in GFP-Dd5P4D319G is reminiscent of the Rab8DN and drainin- phenotypes previously published (Essid et al., 2012).
While there is no direct analogue of this organelle in mammals, the CV is decorated by the small GTPase Rab8 prior to its exocyst-directed fusion with the plasma membrane (Essid et al., 2012). Rab8 is a strong interactor of OCRL in mammals (Hou et al., 2011; Hagemann et al., 2012), where the interaction is responsible for membrane traffic to the primary cilium (Coon et al., 2012). Interestingly, both CV fusion in D. discoideum (Essid et al., 2012) and growth of cilia involve fusion via the exocyst complex (Mukherjee et al., 2014), suggesting OCRL may serve an analogous function in both contexts.
The PH domain protein PIO, like its higher-organism counterpart Ses1/2, is a late endocytic protein, only present on endocytic organelles that are reached by fluid-phase endocytic tracer 30 min or more after ingestion (Figure 3), consistent with a late endosomal or lysosomal function. The localization of the newly identified OCRL/Dd5P4 interactor, GxcU/Frabin, was particularly intriguing. In human cells, Frabin-GFP has a similar multistage recruitment pattern to both early and late endosomal and macropinocytic membranes (Kim et al., 2002; Hayakawa et al., 2004; Brooks et al., 2017) (Figure 2; Supplemental Movie S1). In D. discoideum, GxcU-GFP was recruited to several distinct stages of the endocytic pathway (Figure 4), both at what would be considered very early (at the base of the macropinocytic crown) and at the site of reforming CVs, several seconds after the timepoint when the other F&H interactor, OIBP-GFP, appears (e.g., Figure 5A). While the target of the GxcU GEF activity is not known, the localization of GxcU to macropinosomes is highly reminiscent of the localization of active Rac1 (Marinovic et al., 2016; Rivero and Xiong, 2016). Interestingly, overexpression of GxcU-GFP phenocopied Dd5P4 loss in terms of fluid-phase dye uptake and processing and enhanced the Dd5P4- mutant phenotype (Figure 7), suggesting that Dd5P4 may be a repressor of GxcU function.
In summary, our findings reveal that the network of F&H peptide interactors of OCRL is more evolutionary conserved than expected based on previous studies. Furthermore, in all cases that we examined, selective disruption of the F&H interface of OCRL/Dd5P4 does not significantly impair its localization, but impacts downstream reactions. In D. discoideum, F&H mutant Dd5P4 localizes to the same subcompartments as WT Dd5P4, but where we made kinetic measurements, we found that the lack of F&H interactions affected the kinetics of the trafficking step under study. Likewise, F&H-containing interactors of Dd5P4 were recruited to membranes for longer periods in Dd5P4- mutants than in WT, suggesting that coordinated activity of OCRL and F&H adaptors is necessary for efficient membrane progression.
Our study gives weight to the possibility of functionally relevant amounts of PI(4,5)P2 present in the endocytic system under normal conditions. Recent research suggests that PI(4,5)P2 and/or OCRL catalytic activity has a critical role in membrane traffic separate from its role at the plasma membrane, including the transition from early to late endosomes (Vicinanza et al., 2011; Sun et al., 2013; Henmi et al., 2016; Muriel et al., 2016) and the transition from endosome to trans-Golgi (Choudhury et al., 2005; Billcliff et al., 2016). The presence of conserved, endomembrane-specific interactors for OCRL argues against a function of OCRL solely as a cellular ‘housekeeper’ and suggests instead that OCRL-directed PI(4,5)P2 on endosomes is assisted and directed by interaction with F&H proteins.
Fibroblasts from OCRL-mutated LS or Dent2 patients are equally deficient in PI(4,5)P2 phosphatase activity (Montjean et al., 2015); however, Dent2 disease fibroblasts manifest milder phenotypes (e.g., the degree of defective actin stress fiber formation, abnormal alpha-actinin staining, and shortened primary cilia [Montjean et al., 2015]) than their LS counterparts, suggesting the presence of a modifier gene on an autosome. The OCRL paralog INPP5B has been excluded as a phenotype modifier in Lowe/Dent2 patients (Montjean et al., 2015). Given that the F&H interaction is so well conserved and appears to change the kinetics of OCRL-dependent membrane trafficking steps, F&H proteins should be considered as highly likely candidates for modifiers of the LS phenotype.
MATERIALS AND METHODS
Plasmids
Frabin-GFP was a kind gift of Valerie Delague (Delague et al., 2007). Ruby-OCRL was created by replacing the EGFP fluorophore with Ruby in pEGFP-OCRL (Swan et al., 2010). Dd5P4 cDNA was a kind gift of Peter van Haastert (Loovers et al., 2007). D. discoideum genomic DNA was isolated as previously described and the loci for DDB_G0275301/OIBP and DDB_G0280015/PIO were amplified by PCR. The D. intermedium GxcU cDNA from the genomic locus AJWI01001082.1 was codon optimized and synthetized (IDT-DNA) to minimize recombination (protein sequence is shown in Supplemental Figure S4). GFP fusion constructs were subcloned into pDM317 and pDM323 for expression as N- and C-terminal GFP fusions, respectively. Site-directed mutagenesis (Stratagene XL II kit) was used to introduce the W620A and D319G substitutions to Dd5P4 cDNA. See Table 1 for more details.
TABLE 1:
Constructs and Dictyostelium cell lines used in this article.
| Dictyostelium strains used | Plasmids used for transformation | Reference |
|---|---|---|
| AX3 (WT) | ||
| dd5P4 knockout strain | Loovers et al., 2003, 2007 | |
| GFP-Dd5P4/dd5P4 KO | See below | |
| Dd5P4-GFP/dd5P4 KO | See below | |
| GFP-Dd5P4W620A/dd5P4 KO | See below | |
| GFP-Dd5P4D319G/dd5P4 KO | See below | |
| OIBP-GFP/dd5P4 KO | See below | |
| PIO-GFP/dd5P4 KO | See below | |
| GxcU-GFP/dd5P4 KO | See below |
| Plasmids generated for this work | Insert | Plasmid and reference |
|---|---|---|
| Dd5P4-GFP | DDB_G0267462 | pDM323 (Veltman et al., 2009) |
| GFP-Dd5P4 | DDB_G0267462 | pDM317 (Veltman et al., 2009) |
| GFP-Dd5P4W620A | DDB_G0267462 with a tryptophan to alanine substitution at amino acid position 620 | pDM317 (Veltman et al., 2009) |
| GFP-Dd5P4D319G | DDB_G0267462 with an aspartate to glutamine substitution at amino acid 319 | pDM317 (Veltman et al., 2009) |
| GxcU-GFP | GxcU (D. intermedium) | pDM323 (Veltman et al., 2009) |
| OIBP-GFP | DDB_G0275301 | pDM323 (Veltman et al., 2009) |
| PIO-GFP | DDB_G0280015 | pDM323 (Veltman et al., 2009) |
GST fusion proteins of F&H peptides were made by annealing oligos of the following sequences to allow cloning into pGex6P-1: OIBP peptide: DFSSQHQQYSFDQ*, PIO peptide: DFLTLHKAYIDEW*, GxcU peptide #1: SFQLIHPIKSFTL*, GxcU peptide #2: SFIATHTAYNKEN*, MycBP2 ortholgoue #1: GFVYIHCNLGLFK*, MycBP2 ortholgoue #2: CFGTTHFCDTCHD*, human proteins FGD4/Frabin: AFHQRHETFRNAI*, Fgd5: IFHLYHKKTLFYS*, FGD6: VFQLLHKNMLFYV*. GST fusions of APPL1 and Ses2 F&H peptides were previously described (Swan et al., 2010).
Dictyostelium knockout and transgenic lines
Knockout lines and overexpressors are described in Table 1. Knockout and fusion protein-expressing strains were maintained under antibiotic selection, 10 and 5 µg/ml G418, respectively.
GST pull down
D. discoideum lysates were prepared in lysis buffer (20 mM Tris, pH 8, 10% [vol/vol] glycerol, 100 mM NaCl, 1 mM EDTA, and 1 mM DTT), divided into equal aliquots, and exposed to bacterially expressed GST or GST F&H peptides immobilized on 40 μl of glutathione Sepharose (GE Healthcare Lifesciences). GFP-Dd5P4 was then detected in Western blot by anti-GFP antibodies (Abcam 290). HKC (a human proximal tubule cell line) cells were lysed and exposed to GST fusion proteins of human F&H peptides, or GST fusions of OCRL ASHRhoGAP WT and W739A (Pirruccello et al., 2011). Blots were probed with antibodies to Ace1 (EPR2757, Novus Biologicals), Ccdc39 (HPA035364, Sigma) and SPG39 (14261-1-AP, Proteintech). Endogenous OCRL was detected in Western blot using anti-OCRL antibodies (Erdmann et al., 2007).
Immunoprecipitation
Three 15-cm-diameter dishes per D. discoideum sample were grown to 80% confluency in full medium (HL5 medium supplemented with glucose vitamins and trace elements; Formedium, UK), harvested by scraping and centrifugation, and lysed in lysis buffer (20 mM Tris, pH 8, 10% [vol/vol] glycerol, 100 mM NaCl, 1 mM EDTA, and 1 mM DTT). GFP-immunopreciptations were performed in triplicate on 10 μl GFP-trap beads (Chromotek) per sample, washed extensively, and frozen for mass spectrometric analysis. Frabin-GFP was expressed in Cos7 cells and immunoprecipated on GFP trap beads as above. Frabin-GFP was detected in Western blot by anti-GFP antibody (Abcam 290). Endogenous OCRL was detected in Western blot using mouse anti-human OCRL (Erdmann et al., 2007).
ITC
Calorimetry was performed on purified GST-OCRL-ASHRhoGAP as previously described (Pirruccello et al., 2011). Dynein heavy chain putative F&H peptide (KFRRQHEQLRAVI) and a peptide with an alanine in the critical Phe residue (KARRQHEQLRAVI) were tested.
Mass spectrometry
Proteins were reduced, alkylated, and digested overnight at 37°C with trypsin directly on the agarose beads. The resulting peptide mixture was removed from the beads and acidified by the addition of trifluoroacetic acid. Peptides were desalted following the protocol for StageTip purification (Rappsilber et al., 2007). Samples were eluted with 60 ml of buffer B (80% ACN, 0.1% formic acid in H2O) and reduced in a Vacufuge plus (Eppendorf) to a final volume of 3 µl. Buffer A (2 µl) (0.1% formic acid in H2O) was added, and the resulting 5 µl was injected through high-performance liquid chromatography.
Analysis of the peptide mixture was performed as described previously (Frohlich et al., 2013). Briefly, peptides were separated on 15-cm columns (New Objectives) with a 75-mm inner diameter and packed in-house with 1.9 mm C18 resin (Dr. Maisch GmbH). Peptides were eluted at a constant flow rate of 250 nl for 95 min with a linear acetonitrile gradient from 5 to 30% and directly sprayed into an online coupled Q Exactive Mass Spectrometer (Thermo Scientific). The mass spectrometer was operated in a data-dependent mode to automatically switch between full-scan MS and up to 10 data-dependent MS/MS scans. Maximum injection time for MS scans was 20 ms with a target value of 3,000,000 at a resolution of 70,000 at mass/charge ratio (m/z) of 200. The 10 most intense multiple charged ions (z ≥ 2) from the survey scan were selected for MS/MS scans. Peptides were fragmented with higher-energy collision dissociation (38) with normalized collision energies of 25. Target values for MS/MS were set to 100,000 with a maximum injection time of 120 ms at a resolution of 17,500 atm/z of 200. Dynamic exclusion of sequenced peptides was set to 35 s. MaxQuant (version1.5.3.30) with its integrated Andromeda search algorithm was used to analyze the resulting MS and MS/MS spectra (Cox and Mann, 2008; Cox et al., 2011). Peak lists were searched against local databases for D. discoideum (downloaded from dictyBase). Carbamidomethylation of cysteine was set as fixed and methionine oxidation and N-terminal acetylation as variable modifications. Maximum mass deviation was 6 ppm for MS peaks and 20 ppm for MS/MS peaks with a maximum of two missed cleavages allowed and a minimum peptide length of six amino acids. Label-free quantitation was performed using the QUBIC software package as described previously (Hubner et al., 2010). All calculations and plots were done with the R software package (http://r-project.org/).
Measurement of D. discoideum growth curves
Growth curves were measured as previously described (Loovers et al., 2007).
Microscopy
D. discoideum cells were habituated to SIH medium (Formedium UK) for 2 d with a media change 12 h and then 1 h prior to imaging and imaged on one of three microscopes (PerkinElmer spinning disk, 3i-Marianas spinning disk, and Zeiss LSM800). For dye-uptake experiments, 7 KDa TRITC-dextran was added to cells at a final concentration of 1 μg/μl for the time indicated. Cos7 cells were transfected using Amaxa nucleofection.
CV labeling
Cells were habituated for 2 d before imaging to SIH culture medium as described above, resuspended, and plated on 35-mm glass-bottomed imaging dishes (Mattek) and prelabeled for 1 h with FM4-64 dye (1 μg/μl FM4-64 [Life Technologies] in DMSO at a dilution of 1:2000). Cells were then imaged to establish a basal rate of fusion (events/second/cell) and stimulated for 1 h (or as indicated) by the addition of 1:1 distilled water to measure fusion rates.
Peptide arrays
Peptide microarrays were synthesized on cellulose membranes by the SPOT synthesis protocol (Frank, 2002) using a MultiPep Spot synthesizer (Abimed, Langenfeld, Germany) as previously described (Panni et al., 2011). After synthesis, membranes were first washed with ethanol (10 min) and with phosphate-buffered saline (PBS) (30 min). After overnight incubation at 4°C with blocking buffer (5% bovine serum albumin in PBS), the membranes were rinsed (10 min) with PBS and incubated with 1 μg/ml the GST-hOCRL-ASHRhoGAPWT and the nonbinding mutant control GST-hOCRL-ASHRhoGAPW739A fusion proteins, in blocking buffer, for 1 h at room temperature (RT). Next the membranes were incubated with anti-GST antibody (PHARMACIA) at a concentration of 1 μg/ml for 2 h at RT. Finally the membranes were incubated with a secondary peroxidase–conjugated anti-goat antibody (0.2 μg/ml in blocking buffer) for 1 h at RT, followed by washing three times with PBS. Quantification of peptide-bound domain was obtained by an electrochemiluminescence assay (Bio-Rad) read on a Lumi-Imager instrument (Roche Diagnostics, Basel, Switzerland).
Supplementary Material
Acknowledgments
L.E.S. was supported for this project by the Wellcome Trust (105616/Z/14/Z), the Medical Research Council (MRC/N010035/1), and Lowe Syndrome Association USA. P.D.C. was supported by NIH grants (DA018343; DK082700; NS036251), the Lowe Syndrome Trust, the Lowe Syndrome Association USA, and the Kavli Institute for Neuroscience. T.S.’s laboratory is supported by multiple grants from the Swiss National Science Foundation. T.S. is a member of iGE3 ( www.ige3.unige.ch). G.C. was supported by the Italian Association for Cancer Research Grant N (IG 2013 N.14135). M.S. is supported by North West Cancer Research (CR1081).
Abbreviations used:
- ASH
ASPM, SPD-2, Hydin
- CV
contractile vacuole
- FGD
faciogenital dysplasia
- Frabin
FGD1- related F- actin binding protein
- DH
Dbl homology
- ITC
isothermal titration calorimetry
- LS
Lowe syndrome
- OIBP
OCRL- Interacting Bar domain Protein
- PBS
phosphate-buffered saline
- PH
Pleckstrin homology
- PIO
PH domain Interacting with OCRL
- PIP
phosphoinositide lipid
- RT
room temperature
- WT
wild type.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E18-08-0510) on June 19, 2019.
REFERENCES
- Attree O, Olivos IM, Okabe I, Bailey LC, Nelson DL, Lewis RA, McInnes RR, Nussbaum RL. (1992). The Lowe’s oculocerebrorenal syndrome gene encodes a protein highly homologous to inositol polyphosphate-5-phosphatase. Nature , 239–242. [DOI] [PubMed] [Google Scholar]
- Banerjee J, Fischer CC, Wedegaertner PB. (2009). The amino acid motif L/IIxxFE defines a novel actin-binding sequence in PDZ-RhoGEF. Biochemistry , 8032–8043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard DJ, Nussbaum RL. (2010). X-inactivation analysis of embryonic lethality in Ocrl wt/-; Inpp5b-/- mice. Mamm Genome , 186–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billcliff PG, Noakes CJ, Mehta ZB, Yan G, Mak L, Woscholski R, Lowe M. (2016). OCRL1 engages with the F-BAR protein pacsin 2 to promote biogenesis of membrane-trafficking intermediates. Mol Biol Cell , 90–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohdanowicz M, Balkin DM, De Camilli P, Grinstein S. (2012). Recruitment of OCRL and Inpp5B to phagosomes by Rab5 and APPL1 depletes phosphoinositides and attenuates Akt signaling. Mol Biol Cell , 176–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks AB, Humphreys D, Singh V, Davidson AC, Arden SD, Buss F, Koronakis V. (2017). MYO6 is targeted by Salmonella virulence effectors to trigger PI3-kinase signaling and pathogen invasion into host cells. Proc Natl Acad Sci USA , 3915–3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catimel B, Kapp E, Yin MX, Gregory M, Wong LS, Condron M, Church N, Kershaw N, Holmes AB, Burgess AW. (2013). The PI(3)P interactome from a colon cancer cell. J Proteomics , 35–51. [DOI] [PubMed] [Google Scholar]
- Choudhury R, Diao A, Zhang F, Eisenberg E, Saint-Pol A, Williams C, Konstantakopoulos A, Lucocq J, Johannes L, Rabouille C, et al (2005). Lowe syndrome protein OCRL1 interacts with clathrin and regulates protein trafficking between endosomes and the trans-Golgi network. Mol Biol Cell , 3467–3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury R, Noakes CJ, McKenzie E, Kox C, Lowe M. (2009). Differential clathrin binding and subcellular localization of OCRL1 splice isoforms. J Biol Chem , 9965–9973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke M, Kohler J, Arana Q, Liu T, Heuser J, Gerisch G. (2002). Dynamics of the vacuolar H(+)-ATPase in the contractile vacuole complex and the endosomal pathway of Dictyostelium cells. J Cell Sci , 2893–2905. [DOI] [PubMed] [Google Scholar]
- Coon BG, Hernandez V, Madhivanan K, Mukherjee D, Hanna CB, Barinaga-Rementeria Ramirez I, Lowe M, Beales PL, Aguilar RC. (2012). The Lowe syndrome protein OCRL1 is involved in primary cilia assembly. Hum Mol Genet , 1835–1847. [DOI] [PubMed] [Google Scholar]
- Cox J, Mann M. (2008). MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol , 1367–1372. [DOI] [PubMed] [Google Scholar]
- Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M. (2011). Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res , 1794–1805. [DOI] [PubMed] [Google Scholar]
- Delague V, Jacquier A, Hamadouche T, Poitelon Y, Baudot C, Boccaccio I, Chouery E, Chaouch M, Kassouri N, Jabbour R, et al (2007). Mutations in FGD4 encoding the Rho GDP/GTP exchange factor FRABIN cause autosomal recessive Charcot-Marie-Tooth type 4H. Am J Hum Genet , 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann KS, Mao Y, McCrea HJ, Zoncu R, Lee S, Paradise S, Modregger J, Biemesderfer D, Toomre D, De Camilli P. (2007). A role of the Lowe syndrome protein OCRL in early steps of the endocytic pathway. Dev Cell , 377–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essid M, Gopaldass N, Yoshida K, Merrifield C, Soldati T. (2012). Rab8a regulates the exocyst-mediated kiss-and-run discharge of the Dictyostelium contractile vacuole. Mol Biol Cell , 1267–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank R. (2002). The SPOT-synthesis technique. Synthetic peptide arrays on membrane supports–principles and applications. J Immunol Methods , 13–26. [DOI] [PubMed] [Google Scholar]
- Frohlich F, Christiano R, Walther TC. (2013). Native SILAC: metabolic labeling of proteins in prototroph microorganisms based on lysine synthesis regulation. Mol Cell Proteomics , 1995–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda M, Kanno E, Ishibashi K, Itoh T. (2008). Large scale screening for novel rab effectors reveals unexpected broad Rab binding specificity. Mol Cell Proteomics , 1031–1042. [DOI] [PubMed] [Google Scholar]
- Gazit R, Mandal PK, Ebina W, Ben-Zvi A, Nombela-Arrieta C, Silberstein LE, Rossi DJ. (2014). Fgd5 identifies hematopoietic stem cells in the murine bone marrow. J Exp Med , 1315–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerisch G, Heuser J, Clarke M. (2002). Tubular-vesicular transformation in the contractile vacuole system of Dictyostelium. Cell Biol Int , 845–852. [DOI] [PubMed] [Google Scholar]
- Gopaldass N, Patel D, Kratzke R, Dieckmann R, Hausherr S, Hagedorn M, Monroy R, Kruger J, Neuhaus EM, Hoffmann E, et al (2012). Dynamin A, Myosin IB and Abp1 couple phagosome maturation to F-actin binding. Traffic , 120–130. [DOI] [PubMed] [Google Scholar]
- Hagemann N, Hou X, Goody RS, Itzen A, Erdmann KS. (2012). Crystal structure of the Rab binding domain of OCRL1 in complex with Rab8 and functional implications of the OCRL1/Rab8 module for Lowe syndrome. Small GTPases , 107–110. [DOI] [PubMed] [Google Scholar]
- Hayakawa A, Hayes SJ, Lawe DC, Sudharshan E, Tuft R, Fogarty K, Lambright D, Corvera S. (2004). Structural basis for endosomal targeting by FYVE domains. J Biol Chem , 5958–5966. [DOI] [PubMed] [Google Scholar]
- Henmi Y, Morikawa Y, Oe N, Ikeda N, Fujita A, Takei K, Minogue S, Tanabe K. (2016). PtdIns4KIIalpha generates endosomal PtdIns(4)P and is required for receptor sorting at early endosomes. Mol Biol Cell , 990–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuser J. (2006). Evidence for recycling of contractile vacuole membrane during osmoregulation in Dictyostelium amoebae—a tribute to Gunther Gerisch. Eur J Cell Biol , 859–871. [DOI] [PubMed] [Google Scholar]
- Heuser J, Zhu Q, Clarke M. (1993). Proton pumps populate the contractile vacuoles of Dictyostelium amoebae. J Cell Biol , 1311–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hichri H, Rendu J, Monnier N, Coutton C, Dorseuil O, Poussou RV, Baujat G, Blanchard A, Nobili F, Ranchin B, et al (2011). From Lowe syndrome to Dent disease: correlations between mutations of the OCRL1 gene and clinical and biochemical phenotypes. Hum Mutat , 379–388. [DOI] [PubMed] [Google Scholar]
- Hilbi H, Weber S, Finsel I. (2011). Anchors for effectors: subversion of phosphoinositide lipids by legionella. Front Microbiol , 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoopes RR, Jr, Shrimpton AE, Knohl SJ, Hueber P, Hoppe B, Matyus J, Simckes A, Tasic V, Toenshoff B, Suchy SF, et al (2005). Dent Disease with mutations in OCRL1. Am J Hum Genet , 260–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn M, Baumann R, Pereira JA, Sidiropoulos PN, Somandin C, Welzl H, Stendel C, Luhmann T, Wessig C, Toyka KV, et al (2012). Myelin is dependent on the Charcot-Marie-Tooth Type 4H disease culprit protein FRABIN/FGD4 in Schwann cells. Brain , 3567–3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X, Hagemann N, Schoebel S, Blankenfeldt W, Goody RS, Erdmann KS, Itzen A. (2011). A structural basis for Lowe syndrome caused by mutations in the Rab-binding domain of OCRL1. EMBO J , 1659–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Zhang H, Cheng CY, Wen F, Tam PO, Zhao P, Chen H, Li Z, Chen L, Tai Z, et al (2016). A missense variant in FGD6 confers increased risk of polypoidal choroidal vasculopathy. Nat Genet , 640–647. [DOI] [PubMed] [Google Scholar]
- Hubner NC, Bird AW, Cox J, Splettstoesser B, Bandilla P, Poser I, Hyman A, Mann M. (2010). Quantitative proteomics combined with BAC TransgeneOmics reveals in vivo protein interactions. J Cell Biol , 739–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyvola N, Diao A, McKenzie E, Skippen A, Cockcroft S, Lowe M. (2006). Membrane targeting and activation of the Lowe syndrome protein OCRL1 by rab GTPases. EMBO J , 3750–3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janne PA, Suchy SF, Bernard D, MacDonald M, Crawley J, Grinberg A, Wynshaw-Boris A, Westphal H, Nussbaum RL. (1998). Functional overlap between murine Inpp5b and Ocrl1 may explain why deficiency of the murine ortholog for OCRL1 does not cause Lowe syndrome in mice. J Clin Invest , 2042–2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Journet A, Klein G, Brugiere S, Vandenbrouck Y, Chapel A, Kieffer S, Bruley C, Masselon C, Aubry L. (2012). Investigating the macropinocytic proteome of Dictyostelium amoebae by high-resolution mass spectrometry. Proteomics , 241–245. [DOI] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. (2015). The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc , 845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Ikeda W, Nakanishi H, Tanaka Y, Takekuni K, Itoh S, Monden M, Takai Y. (2002). Association of frabin with specific actin and membrane structures. Genes Cells , 413–420. [DOI] [PubMed] [Google Scholar]
- King JS, Gueho A, Hagedorn M, Gopaldass N, Leuba F, Soldati T, Insall RH. (2013). WASH is required for lysosomal recycling and efficient autophagic and phagocytic digestion. Mol Biol Cell , 2714–2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichter-Konecki U, Farber LW, Cronin JS, Suchy SF, Nussbaum RL. (2006). The effect of missense mutations in the RhoGAP-homology domain on ocrl1 function. Mol Genet Metab , 121–128. [DOI] [PubMed] [Google Scholar]
- Loovers HM, Kortholt A, de Groote H, Whitty L, Nussbaum RL, van Haastert PJ. (2007). Regulation of phagocytosis in Dictyostelium by the inositol 5-phosphatase OCRL homolog Dd5P4. Traffic , 618–628. [DOI] [PubMed] [Google Scholar]
- Loovers HM, Veenstra K, Snippe H, Pesesse X, Erneux C, van Haastert PJ. (2003). A diverse family of inositol 5-phosphatases playing a role in growth and development in Dictyostelium discoideum. J Biol Chem , 5652–5658. [DOI] [PubMed] [Google Scholar]
- Luo N, West CC, Murga-Zamalloa CA, Sun L, Anderson RM, Wells CD, Weinreb RN, Travers JB, Khanna H, Sun Y. (2012). OCRL localizes to the primary cilium: a new role for cilia in Lowe syndrome. Hum Mol Genet , 3333–3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macro L, Jaiswal JK, Simon SM. (2012). Dynamics of clathrin-mediated endocytosis and its requirement for organelle biogenesis in Dictyostelium. J Cell Sci , 5721–5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y, Balkin DM, Zoncu R, Erdmann KS, Tomasini L, Hu F, Jin MM, Hodsdon ME, De Camilli P. (2009). A PH domain within OCRL bridges clathrin-mediated membrane trafficking to phosphoinositide metabolism. EMBO J , 1831–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinovic M, Sostar M, Filic V, Antolovic V, Weber I. (2016). Quantitative imaging of Rac1 activity in Dictyostelium cells with a fluorescently labelled GTPase-binding domain from DPAKa kinase. Histochem Cell Biol , 267–279. [DOI] [PubMed] [Google Scholar]
- McCulloch DA, Lutz EM, Johnson MS, Robertson DN, MacKenzie CJ, Holland PJ, Mitchell R. (2001). ADP-ribosylation factor-dependent phospholipase D activation by VPAC receptors and a PAC(1) receptor splice variant. Mol Pharmacol , 1523–1532. [DOI] [PubMed] [Google Scholar]
- McMahon HT, Boucrot E. (2015). Membrane curvature at a glance. J Cell Sci , 1065–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montjean R, Aoidi R, Desbois P, Rucci J, Trichet M, Salomon R, Rendu J, Faure J, Lunardi J, Gacon G, et al (2015). OCRL-mutated fibroblasts from patients with Dent-2 disease exhibit INPP5B-independent phenotypic variability relatively to Lowe syndrome cells. Hum Mol Genet , 994–1006. [DOI] [PubMed] [Google Scholar]
- Mukherjee D, Sen A, Aguilar RC. (2014). RhoGTPase-binding proteins, the exocyst complex and polarized vesicle trafficking. Small GTPases , e28453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muriel O, Tomas A, Scott CC, Gruenberg J. (2016). Moesin and cortactin control actin-dependent multivesicular endosome biogenesis. Mol Biol Cell , 3305–3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi H, Takai Y. (2008). Frabin and other related Cdc42-specific guanine nucleotide exchange factors couple the actin cytoskeleton with the plasma membrane. J Cell Mol Med , 1169–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandez R, Balkin DM, Messa M, Liang L, Paradise S, Czapla H, Hein MY, Duncan JS, Mann M, De Camilli P. (2014). A role of OCRL in clathrin-coated pit dynamics and uncoating revealed by studies of Lowe syndrome cells. Elife , e02975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishihara E, Shimmen T, Sonobe S. (2007). New aspects of membrane dynamics of Amoeba proteus contractile vacuole revealed by vital staining with FM 4–64. Protoplasma , 25–30. [DOI] [PubMed] [Google Scholar]
- Noakes CJ, Lee G, Lowe M. (2011). The PH domain proteins IPIP27A and B link OCRL1 to receptor recycling in the endocytic pathway. Mol Biol Cell , 606–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oltrabella F, Pietka G, Ramirez IB, Mironov A, Starborg T, Drummond IA, Hinchliffe KA, Lowe M. (2015). The Lowe syndrome protein OCRL1 is required for endocytosis in the zebrafish pronephric tubule. PLoS Genet , e1005058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panni S, Montecchi-Palazzi L, Kiemer L, Cabibbo A, Paoluzi S, Santonico E, Landgraf C, Volkmer-Engert R, Bachi A, Castagnoli L, Cesareni G. (2011). Combining peptide recognition specificity and context information for the prediction of the 14-3-3-mediated interactome in S. cerevisiae and H. sapiens. Proteomics , 128–143. [DOI] [PubMed] [Google Scholar]
- Parkinson K, Baines AE, Keller T, Gruenheit N, Bragg L, North RA, Thompson CR. (2014). Calcium-dependent regulation of Rab activation and vesicle fusion by an intracellular P2X ion channel. Nat Cell Biol , 87–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasteris NG, Cadle A, Logie LJ, Porteous ME, Schwartz CE, Stevenson RE, Glover TW, Wilroy RS, Gorski JL. (1994). Isolation and characterization of the faciogenital dysplasia (Aarskog-Scott syndrome) gene: a putative Rho/Rac guanine nucleotide exchange factor. Cell , 669–678. [DOI] [PubMed] [Google Scholar]
- Pirruccello M, Swan LE, Folta-Stogniew E, De Camilli P. (2011). Recognition of the F&H motif by the Lowe syndrome protein OCRL. Nat Struct Mol Biol , 789–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting CP. (2006). A novel domain suggests a ciliary function for ASPM, a brain size determining gene. Bioinformatics , 1031–1035. [DOI] [PubMed] [Google Scholar]
- Rappsilber J, Mann M, Ishihama Y. (2007). Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc , 1896–1906. [DOI] [PubMed] [Google Scholar]
- Rivero F, Xiong H. (2016). Rho Signaling in Dictyostelium discoideum. Int Rev Cell Mol Biol , 61–181. [DOI] [PubMed] [Google Scholar]
- Schaap P, Winckler T, Nelson M, Alvarez-Curto E, Elgie B, Hagiwara H, Cavender J, Milano-Curto A, Rozen DE, Dingermann T, et al (2006). Molecular phylogeny and evolution of morphology in the social amoebas. Science , 661–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Hedman AC, Tan X, Schill NJ, Anderson RA. (2013). Endosomal type Igamma PIP 5-kinase controls EGF receptor lysosomal sorting. Dev Cell , 144–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swan LE, Tomasini L, Pirruccello M, Lunardi J, De Camilli P. (2010). Two closely related endocytic proteins that share a common OCRL-binding motif with APPL1. Proc Natl Acad Sci USA , 3511–3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MJ, Perrais D, Merrifield CJ. (2011). A high precision survey of the molecular dynamics of mammalian clathrin-mediated endocytosis. PLoS Biol , e1000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Driessche N, Shaw C, Katoh M, Morio T, Sucgang R, Ibarra M, Kuwayama H, Saito T, Urushihara H, Maeda M, et al (2002). A transcriptional profile of multicellular development in Dictyostelium discoideum. Development , 1543–1552. [DOI] [PubMed] [Google Scholar]
- Veltman DM, Akar G, Bosgraaf L, Van Haastert PJ. (2009). A new set of small, extrachromosomal expression vectors for Dictyostelium discoideum. Plasmid , 110–118. [DOI] [PubMed] [Google Scholar]
- Vicinanza M, Di Campli A, Polishchuk E, Santoro M, Di Tullio G, Godi A, Levtchenko E, De Leo MG, Polishchuk R, Sandoval L, et al (2011). OCRL controls trafficking through early endosomes via PtdIns4,5P(2)-dependent regulation of endosomal actin. EMBO J , 4970–4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlahou G, Rivero F. (2006). Rho GTPase signaling in Dictyostelium discoideum: insights from the genome. Eur J Cell Biol , 947–959. [DOI] [PubMed] [Google Scholar]
- Wang Y, Senoo H, Sesaki H, Iijima M. (2013). Rho GTPases orient directional sensing in chemotaxis. Proc Natl Acad Sci USA , E4723–E4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber SS, Ragaz C, Hilbi H. (2009). The inositol polyphosphate 5-phosphatase OCRL1 restricts intracellular growth of Legionella, localizes to the replicative vacuole and binds to the bacterial effector LpnE. Cell Microbiol , 442–460. [DOI] [PubMed] [Google Scholar]
- Williams C, Choudhury R, McKenzie E, Lowe M. (2007). Targeting of the type II inositol polyphosphate 5-phosphatase INPP5B to the early secretory pathway. J Cell Sci , 3941–3951. [DOI] [PubMed] [Google Scholar]
- Yichoy M, Duarte TT, De Chatterjee A, Mendez TL, Aguilera KY, Roy D, Roychowdhury S, Aley SB, Das S. (2011). Lipid metabolism in Giardia: a post-genomic perspective. Parasitology , 267–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoncu R, Perera RM, Balkin DM, Pirruccello M, Toomre D, De Camilli P. (2009). A phosphoinositide switch controls the maturation and signaling properties of APPL endosomes. Cell , 1110–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.