Abstract
Autophagy is an evolutionarily conserved intracellular lysosomal degradation pathway. It is a multistep process involving de novo formation of double membrane autophagosomes that capture cytosolic constituents (cargo) and eventually fuse with lysosomes wherein the cargo gets degraded and resulting simpler biomolecules get recycled. In addition to their autophagy function, several of the autophagy-related proteins work at the interface of other vesicular trafficking pathways. Hence, development of specific autophagy modulators that do not perturb general endo-lysosomal traffic possesses unique challenges. In this article, we report a novel small molecule EACC that inhibits autophagic flux by blocking autophagosome–lysosome fusion. Strikingly, unlike other late stage inhibitors, EACC does not have any effect on lysosomal properties or on endocytosis-mediated degradation of EGF receptor. EACC affects the translocation of SNAREs Stx17 and SNAP29 on autophagosomes without impeding the completion of autophagosomes. EACC treatment also reduces the interaction of Stx17 with the HOPS subunit VPS33A and the cognate lysosomal R-SNARE VAMP8. Interestingly, this effect of EACC although quite robust is reversible and hence EACC can be used as a tool to study autophagosomal SNARE trafficking. Our results put forward a novel method to block autophagic flux by impeding the action of the autophagosomal SNAREs.
INTRODUCTION
Autophagy is an intracellular catabolic pathway in which double membrane autophagosomes containing cytoplasmic cargo are transported to lysosomes to form a single membrane degradative compartment called autolysosomes. Inside autolysosomes, by the action of lysosomal hydrolases, simpler biomolecules are generated that are recycled back to the cytoplasm for reuse. The rate at which this multistep dynamic process occurs inside cells is referred to as autophagic flux. All these steps are tightly regulated and are constantly occurring inside a cell at a basal rate; however, this basal autophagic flux varies according to cell type and environmental cues. Basal autophagic flux and its appropriate responsiveness to external perturbations are critical to maintain cellular homeostasis. On the other hand, external stress stimuli such as nutrient limitation or starvation lead to an increase in autophagic flux.
Dysfunctional autophagic flux has been associated with several human diseases. Impaired autophagic flux has been associated with neurodegenerative and infectious diseases while excessive autophagy sustains survival of several types of solid tumors. Therefore, pharmacological modulation of autophagy and its application in various disease scenarios has garnered a lot of interest (Mizushima, 2007; Glick et al., 2010; Rubinsztein et al., 2012; Deretic et al., 2013; Nixon, 2013; Singh et al., 2018).
Induction of autophagy is tightly regulated inside cells. The mammalian target of rapamycin (mTOR) senses cellular nutrient status and regulates cell growth. In the case of nutrient limitation, mTOR is deactivated, which leads to dephosphorylation of Unc-51–like autophagy activating kinase1 (ULK1) and allows assimilation of the ULK1 complex. This complex translocates to the phagophore or isolation membrane where it promotes assembly of the second complex comprising BECLIN1, ATG14, p150, and VPS34. Expansion of this isolation membrane requires conjugation of the ATG5-12/ATG16L1 complex that in turn brings LC3-II to autophagosomal membrane. Double membrane autophagosomes capture cytoplasmic cargo by binding to ubiquitinated cargo via the LC3 interacting region (LIR) present in adaptor proteins like SQSTM1/p62. Finally, these autophagosomes should fuse with lysosomes so that the captured cargo can be degraded by action of lysosomal enzymes (Bento et al., 2016).
Autophagosome–lysosome fusion, similar to all vesicle fusion events, involves the action of soluble NSF (N-ethylmaleimide–sensitive factor) attachment protein receptors (SNAREs; Bonifacino and Glick, 2004; Cai et al., 2007). In yeast, autophagosome–vacuole fusion requires SNAREs Vam3 (Qa), Vti1 (Qb), Vam7 (Qc), and R-SNARE YKT6 (Darsow et al., 1997; Sato et al., 1998; Ishihara et al., 2001; Surpin et al., 2003). In mammalian cells, autophagosome–lysosome fusion is orchestrated by the concerted action of autophagosomal Qa-SNARE Syntaxin17 (Stx17), Qbc-SNARE SNAP29, lysosomal R-SNARE VAMP8, homotypic fusion and protein sorting (HOPS) tethering complex, small GTPase RAB7, and accessory proteins like ATG14 (Itakura et al., 2012; Hyttinen et al., 2013; Jiang et al., 2014; Diao et al., 2015).
Stx17 is abundantly present in endoplasmic reticulum and is involved in smooth endoplasmic reticulum membrane trafficking dynamics (Steegmaier et al., 1998, 2000). Stx17 is unique among the Syntaxin family because it possesses a unique C-terminal hairpin structure that is important for its localization to autophagosomes. Interestingly, the translocation of Stx17 occurs only on complete autophagosomes and not on partially formed autophagosomes. This functions as a regulatory step that prevents fusion of incompletely formed autophagosomes with lysosomes (Itakura et al., 2012). Upon its translocation, Stx17 along with its partner SNARE SNAP29 interact with VAMP8 resulting in the formation of a parallel four-helix bundle consisting of Qa, Qbc, and R-SNAREs (Itakura et al., 2012; Guo et al., 2014). This SNARE bundle is stabilized by ATG14 whose role at this step is largely independent of its role in early steps of autophagy (Hamasaki et al., 2013; Diao et al., 2015).
In this article, we report a novel small molecule inhibitor of autophagy EACC that blocks autophagosomal–lysosomal fusion. EACC inhibits autophagic flux by selectively affecting the translocation of Stx17 on autophagosomes. The autophagic pathway and the endocytic pathway both culminate at the lysosomes and share some components of the fusion machinery such as RAB7 and the HOPS complex (Hyttinen et al., 2013; Jiang et al., 2014; Takats et al., 2014). Owing to this, selectively modulating autophagy without perturbing the endo-lysosomal system is difficult. Our investigations into the mechanism of EACC revealed that its action is largely specific to the process of autophagy. Most importantly, the action of EACC is reversible and hence can be used as a tool to study the dynamic recruitment of autophagy-specific SNAREs.
RESULTS
EACC inhibits autophagic flux
Recent reports from our lab described a luciferase-based high-throughput screen for identification of novel small molecule modulators of autophagy (Mishra et al., 2017a,b). Utilizing this assay, we screened 1999 compounds of the Microsource Discovery Systems library and identified EACC as one of the hits. EACC stands for ethyl (2-(5-nitrothiophene-2-carboxamido) thiophene-3-carbonyl) carbamate. To test for its potential to modulate autophagy, EACC was further tested in mammalian systems.
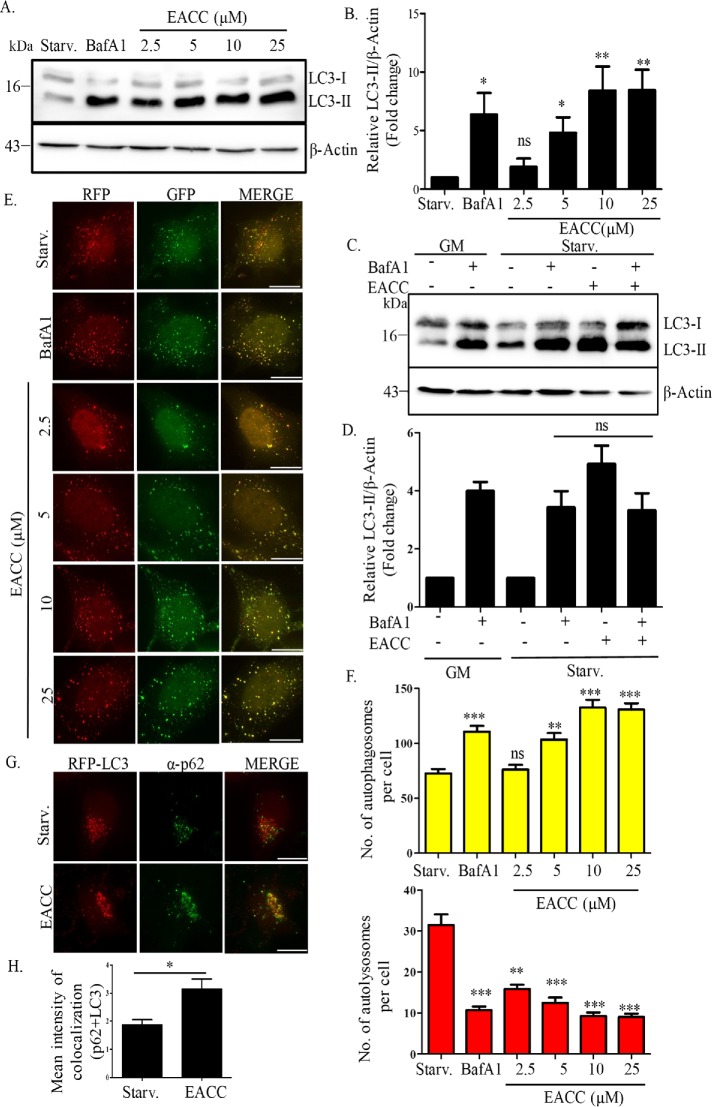
Starvation is a potent physiological inducer of autophagic flux and we wanted to test whether EACC could modulate starvation-induced autophagic flux. We treated HeLa cells with an increasing dose of EACC in starvation conditions (2.5–25 µM) and probed for LC3 expression. An enhanced conversion of LC3 (LC3-I to LC3-II) was seen with increasing dose (Figure 1, A and B). This would indicate either induction or a block in autophagic flux. To address this, we analyzed the accumulation of LC3-II in the presence or absence of a known autophagy inhibitor, bafilomycin A1 (BafA1). An autophagy inducer added along with BafA1 will increase LC3-II levels over and above that of BafA1 alone. On the other hand, in the case of an inhibitor the LC3-II levels will remain unchanged (Mizushima and Yoshimori, 2007; Mizushima et al., 2010). EACC caused an accumulation of LC3-II that was similar to that of BafA1. The combined treatment of BafA1 and EACC did not cause further accumulation of LC3-II, suggesting that EACC is an inhibitor rather than an inducer of autophagic flux (Figure 1, C and D).
FIGURE 1:
EACC inhibits autophagic flux. (A) HeLa cells were either left untreated or treated with BafA1 (100 nM) or EACC (2.5–25 µM) for 2 h in starvation conditions. Samples were collected and immunoblotted for anti-LC3 and anti–β-actin antibodies. (B) Relative levels of LC3-II:β-actin in untreated vs. treated samples were quantitated for three independent experiments. **, P < 0.01; *, P < 0.05; ns = nonsignificant (two-way ANOVA, replicate means compared with Bonferroni posttest). (C) HeLa cells were either left untreated or pretreated with BafA1 (100 nM) in basal or starvation conditions for 1 h in order to block the autophagic flux. This was followed by treatment with EACC (10 µM) for 2 h. Samples were collected and immunoblotted for anti-LC3 and anti–β-actin antibodies. (D) Relative levels of LC3-II:β-actin in untreated vs. treated samples were quantitated for three independent experiments. ns = nonsignificant. Statistical significance was analyzed by Student’s unpaired t test. (E) HeLa cells transfected with tandem-tagged ptfLC3 (mRFP-GFP-LC3) construct were either left untreated or treated with BafA1 (100 nM) or EACC (2.5–25 µM) for 2 h in starvation conditions. Scale = 10 µm. (F) The autophagosomes (RFP+/GFP+ structures) and autolysosomes (RFP+/GFP− structures) per cell were counted using the cell counter plug-in of ImageJ software. Data shown represent the number of autophagosomes (RFP+/GFP+) and autolysosomes (RFP+/GFP−) as compared with control of a minimum of 45 cells from three independent experiments plotted as mean ± SEM. Statistical significance was analyzed by Student’s unpaired t test. ***, P < 0.001; **, P < 0.01; *, P < 0.05; ns = nonsignificant. (G) Immunostaining with anti-SQSTM1/p62 antibody in RFP-LC3 transfected HeLa cells treated with EACC (10 µM) for 2 h in starvation conditions. Scale = 15 µm. (H) Graph showing the mean intensity of colocalization between p62 and RFP-LC3 in control vs. EACC-treated group. Mean intensity of colocalization was measured using colocalization and analyze plug-ins of ImageJ software. Data shown here represents a minimum of 60 cells from three independent experiments plotted as mean ± SEM. Statistical significance was analyzed by Student’s unpaired t test. *, P < 0.05.
To validate these observations and further dissect the step of autophagic flux affected by EACC, we employed tandem-fluorescent-tagged LC3 reporter, mRFP-GFP-LC3 (Kimura et al., 2007).Using this reporter, while autophagosomes appear yellow, autolysosomes (the fusion product of autophagosomes with lysosomes) are seen as red because the green fluorescence of GFP gets quenched due the acidic nature of lysosomes. HeLa cells transfected with mRFP-GFP-LC3 construct were treated with increasing concentrations of EACC (2.5–25 µM) for 2 h. We saw a significant dose-dependent increase in the number of autophagosomes (mRFP+/GFP+) and a concomitant decrease in the number of autolysosomes (mRFP+/GFP−) (Figure 1, E and F) in EACC-treated cells. Subsequent experiments were carried out at 10 µM concentration for a period of 2 h and at this concentration, cell viability was unaffected even up to a period of 5 h (Supplemental Figure S1A). Next, we assessed the effect of EACC on autophagic adaptor p62/SQSTM1. p62 binds to ubiquitinated cargo via the UBA domain and LC3 via its LC3 interacting region (LIR) region. This step helps in sequestration of cargo in autophagosomes. p62 is degraded by autophagy and hence decreased autophagic flux leads to accumulation of this protein. EACC treatment resulted in increased colocalization between p62 and LC3 suggesting that EACC, while inhibiting autophagic flux, did not affect adaptor loading and LC3 recruitment (Figure 1, G and H). These results together suggest that EACC inhibits autophagic flux most likely at the later stages.
EACC blocks autophagosome–lysosome fusion but does not affect endo-lysosomal function
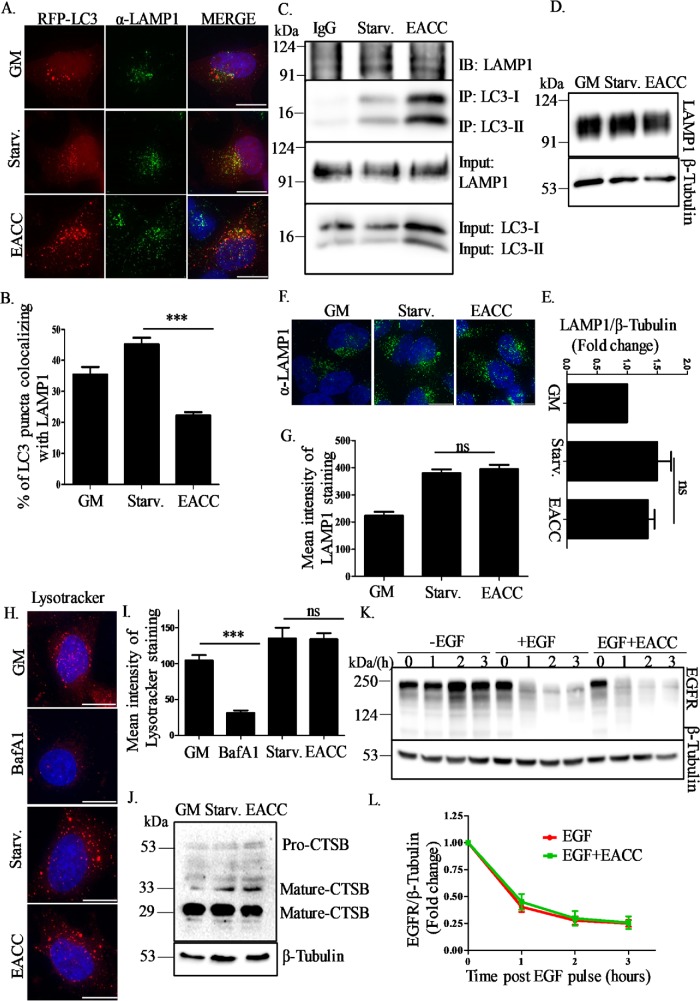
To further understand the inhibitory action of EACC on autophagic flux, we checked the colocalization between the autophagosome marker LC3 and the lysosomal marker, LAMP1. In line with our previous observations, we saw a decrease in the percentage of autolysosomes (RFP-LC3+/LAMP1+) in RFP-LC3 transfected HeLa cells treated with EACC (Figure 2, A and B). A similar decrease in number of autolysosomes was also observed in EACC-treated cells immunostained with LC3 and LAMP1 (Supplemental Figure S2, A and B).
FIGURE 2:
EACC blocks autophagosome–lysosome fusion but does not affect endo-lysosomal function. (A) RFP-LC3 transfected HeLa cells were immunostained with anti-LAMP1 antibody and treated with EACC (10 µM) for 2 h in starvation conditions. Scale = 10 µm. (B) Graph showing percent colocalization between LAMP1 and RFP-LC3 (autolysosomes) in starvation conditions and EACC treatment. The colocalized dots were counted using colocalization and cell counter plug-ins of ImageJ software and plotted with respect to the total number of LC3 puncta. Data shown here represent a minimum of 45 cells from three independent experiments plotted as mean ± SEM. Statistical significance was analyzed by Student’s unpaired t test. ***, P < 0.001. (C) HeLa cells were treated with EACC (10 µM) for 2 h in starvation conditions and immunoprecipitated with anti-LC3 antibody. Anti-mouse IgG was used as an isotype control. The immunoprecipitates were immunoblotted with anti-LAMP1 and anti-LC3 antibodies. (D) HeLa cells were treated with EACC (10 µM) for 2 h in starvation conditions and immunoblotted with anti-LAMP1 and anti–β-tubulin antibodies. (E) Relative levels of LAMP1:β-tubulin in untreated vs. treated samples were quantitated for three independent experiments. Statistical significance was analyzed by Student’s unpaired t test. ns = nonsignificant. (F) HeLa cells were treated with EACC (10 µM) for 2 h in starvation conditions and immunostained with anti-LAMP1 antibody. Scale = 10 µm. (G) Graph represents the mean intensity of LAMP1 staining that was measured using the analyze plug-in in ImageJ. Data shown here represent a minimum of 60 cells from three independent experiments plotted as mean ± SEM. Statistical significance was analyzed by Student’s unpaired t test. ns = nonsignificant. (H) HeLa cells were either treated with BafA1 (100 nM) in basal conditions or EACC (10 µM) in starvation conditions for 2 h. LysoTracker Deep Red (100 nM) was added in the media in the last 15 min of treatment. Cells were fixed and imaged. Scale = 15 µm. (I) Graph showing the mean intensity of LysoTracker staining measured as in D. Data shown here represent a minimum of 45 cells from three independent experiments plotted as mean ± SEM. Statistical significance was analyzed by Student’s unpaired t test. ***, P < 0.001; ns = nonsignificant. (J) Samples collected after EACC treatment were immunoblotted with anti–cathepsin B and anti–β-tubulin antibodies. (K) HeLa cells were serum starved for 3 h and either left untreated or pretreated with EACC before addition of EGF (100 ng/ml) for the indicated time periods. Samples were collected and immunoblotted for anti-EGFR and anti–β-tubulin antibodies. (L) Relative levels of EGFR:β-tubulin in untreated vs. treated samples were quantitated for three independent experiments.
To dissect the effect of EACC on LC3-LAMP1 interaction endogenous immunoprecipitation (IP) was employed. Control and EACC-treated lysates were subjected to IP using LC3 antibody. We observed that in EACC-treated lysates, the levels of LC3-II were significantly high as compared with control in both LC3 input as well as immunoprecipitates, but the levels of LAMP1 in the LC3 IP as detected by immunoblotting remains unchanged indicating decreased interaction between LC3 and LAMP1 (Figure 2C).
Autophagic flux inhibition can also be achieved by affecting lysosomal function. As other commonly used late stage autophagy inhibitors (chloroquine and BafA1) affect lysosomal function, we investigated whether the effect of EACC on autophagic flux impinged on lysosomes and related pathways. To test this, we checked the expression of LAMP1 in the presence or absence of EACC. HeLa cells treated with EACC were immunoblotted with LAMP1 antibody. There was no significant change in the LAMP1 expression in control versus treated cells (Figure 2, D and E). We also did not see any obvious difference in lysosomal positioning or LAMP1 signal intensity in EACC-treated cells (Figure 2, F and G). Although the overall levels of lysosomes remain unchanged, we wondered whether there was loss of acidification of lysosomes that stalls all fusion events as seen in chloroquine and BafA1 treatments. To test the effect of EACC on lysosomal acidification, we used LysoTracker Deep Red, which preferably accumulates in acidic compartments. The intensity of LysoTracker staining was diminished in BafA1-treated cells but remained unchanged in EACC-treated cells suggesting that EACC does not affect lysosomal pH (Figure 2, H and I).
Next, we investigated whether the EACC-treated lysosomes harbored functional hydrolases. We checked the expression and processing of cathepsin B (CTSB), a lysosomal cysteine protease that is cleaved inside the lysosomes to release a proteolytically active mature form. EACC treatment did not impede the conversion of procathepsin B to mature cathepsin B (Figure 2J).
Finally, we tested whether these lysosomes received and processed endocytic pathway cargo upon EACC treatment by performing epidermal growth factor receptor (EGFR) degradation assay.
Upon EGF treatment, EGF bound to EGFR gets internalized via endocytosis and gets degraded in lysosomes. Hence, the temporal decrease in levels of EGFR after EGF pulse is indicative of endocytic trafficking of the receptor to the lysosomes. We found that the rate of EGFR degradation with time followed a comparable trend in treated versus untreated cells (Figure 2, K and L).
These results clearly indicate that EACC prevents autophagosome–lysosome fusion without affecting lysosomes and other vesicular trafficking pathways in general.
EACC does not affect early autophagic events
Our results so far suggest that EACC selectively affected autophagic flux. So, our next approach was to narrow down to the step of autophagy at which EACC acts.
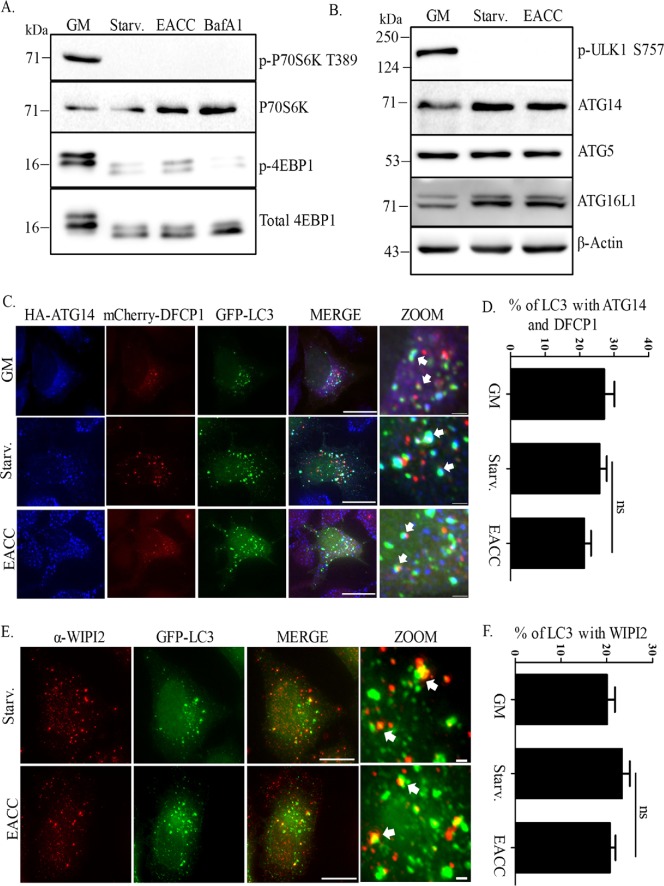
First, we tested the effect of EACC on mTOR signaling. In nutrient starvation conditions, mTOR is inhibited, which allows induction of autophagy. The status of mTOR can be predicted by the phosphorylation status of its substrates P70S6 kinase and eukaryotic translation initiation factor 4E (eIF4E)–binding protein 1 (4EBP1). HeLa cells treated with EACC were immunoblotted for phospho-P70S6 kinase and phospho-4EBP1. Loss of phosphorylation of these substrates suggested that mTOR is inhibited in EACC-treated cells similar to that of control (Figure 3A). Active mTOR phosphorylates ULK1 at serine 757 and shuts down autophagy, whereas in starvation, inhibition of mTOR activity leads to dephosphorylation of ULK1 at 757 position and induction of autophagy. Unaltered dephosphorylation events of mTOR substrates and ULK1 in the presence of EACC suggested that the early signaling events that lead to starvation-mediated induction of autophagy is not perturbed (Figure 3, A and B). We also checked whether the massive accumulation of LC3-II upon EACC treatment is dependent on enhanced transcription or translation (Supplemental Figure S3, A–D).
FIGURE 3:
EACC does not affect early autophagic events. (A) HeLa cells were either left untreated or treated with BafA1 (100 nM) or EACC (10 µM) for 2 h in starvation conditions. Samples were collected and immunoblotted with anti–phospho-P70S6K (T389), anti-P70S6K, anti–phospho-4EBP1, and anti-4EBP1 antibodies. (B) Samples collected after EACC treatment were immunoblotted with anti–phospho-ULK1 (S757), anti-ATG14, anti-ATG5, anti-ATG16L1, and anti–β-actin antibodies. (C) HeLa cells cotransfected with mCherry-DFCP1, GFP-LC3, and HA-ATG14 were either left untreated or treated with EACC and immunostained with anti-HA antibody. Scale = 15 µm, 1 µm. (D) Graph showing the percent of LC3 puncta colocalizing with DFCP1 and ATG14. This population represents immature autophagosomes. The colocalized dots were counted and plotted as in Figure 2B. Data shown here represent a minimum of 50 cells from three independent experiments plotted as mean ± SEM. Statistical significance was analyzed by Student’s unpaired t test. ns = nonsignificant. (E) GFP-LC3 transfected HeLa cells were treated with EACC (10 µM) for 2 h in starvation conditions and immunostained with anti-WIPI2 antibody. Scale = 15 µm. (F) Graph showing the percent of LC3 puncta colocalizing with WIPI2. This population represents omegasomes. The analysis was done similarly as in D. Data shown here represent a minimum of 45 cells from three independent experiments plotted as mean ± SEM. Statistical significance was analyzed by Student’s unpaired t test. ns = nonsignificant.
Furthermore, relative levels of proteins involved in early and middle stages of the autophagy pathway such as ATG14, ATG5, and ATG16L1 were not changed upon EACC treatment (Figure 3B).
As mTOR-mediated control of autophagy was unaltered, we next investigated the effect of EACC on molecular events that lead to autophagosome biogenesis. Isolation membrane or phagophore formation upon autophagy induction is characterized by the presence of phosphatidylinositol-3-phosphate (PI3P) generated by vacuolar protein sorting 34 (VPS34) kinase complex activity. This local increase in PI3P is recognized by PI3P-binding proteins like double FYVE-domain–containing protein 1 (DFCP1) and WD repeat domain phosphoinositide-interacting protein 2 (WIPI2). In addition, this nascent membrane is also marked by ATG14 (Itakura and Mizushima, 2010; Hamasaki et al., 2013). Triple colocalization results between ATG14, DFCP1, and LC3 in control and EACC-treated cells suggested that there was no decrease in number of autophagosome biogenesis sites (Figure 3, C and D).
As mentioned above, WIPI2 is an effector of mammalian PI3Ps that is recruited to omegasomes and marks the sites of autophagosome biogenesis. We looked at the colocalization between LC3 and WIPI2 and the results again suggest that the number of sites of autophagosome biogenesis (omegasomes) is unaffected upon EACC treatment (Figure 3, E and F). A similar trend was also observed in EACC-treated cells immunostained with LC3 and WIPI antibodies (Supplemental Figure S3E).
Developing autophagosomes undergo expansion of the phagophore and are characterized by localization of the ATG5-12/16 complex. In HeLa cells transfected with RFP-LC3, the colocalization between ATG5 and LC3 as well as ATG16L1 and LC3 that represents expanding phagophores also was comparable to that of control (Supplemental Figure S3, F–I).
As shown earlier, we also tested whether cargo recognition was affected by EACC. The colocalization analysis of the autophagy adaptor p62/SQSTM1 with the autophagosome membrane marker LC3 showed increased association between these proteins (Figure 1, G and H). Taken together, these results indicate that signaling events leading to autophagy induction, the number of autophagosome biogenesis sites, expansion of the phagophore, and cargo recognition remain unaltered in the presence of EACC. Thus, it is likely that the autophagic flux inhibition due to EACC may be affecting some downstream steps.
EACC inhibits autophagy by preventing SNARE Stx17 loading on autophagosomes
As all our previous observations suggested that autophagosome formation is unaffected upon EACC treatment, we next tested whether these accumulated autophagosomes have the molecular machinery required for fusion with lysosomes. Elegant studies by Noburu Mizushima’s group identified Stx17 as an autophagosomal SNARE that translocates to autophagosomes and interacts with SNAP29 and endo/lysosomal SNARE VAMP8 with the help of a multisubunit tethering complex like HOPS. Depletion of Stx17 blocked autophagic flux by inhibiting fusion of autophagosomes with lysosomes (Itakura et al., 2012).
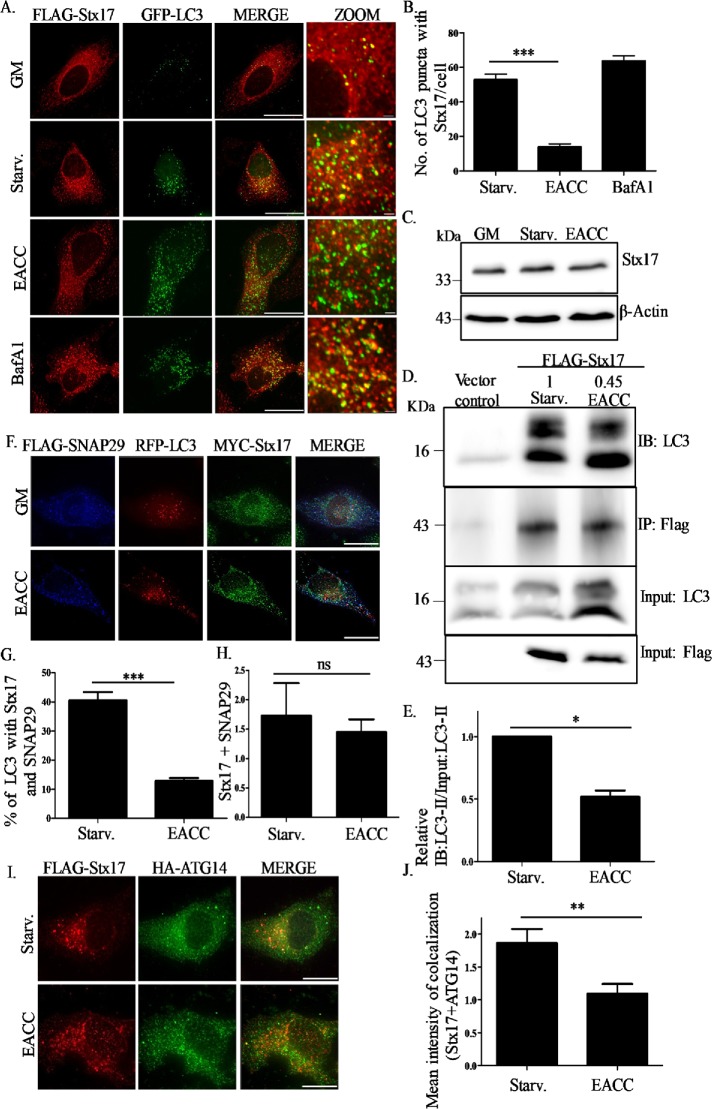
In HeLa cells cotransfected with FLAG-Stx17 and GFP-LC3, we quantitated the colocalization between Stx17 and LC3. Similar to previous reports, in basal conditions, Stx17 depicted a reticulate pattern suggesting ER/mitochondrial localization. Upon induction of autophagy, the Stx17 staining pattern became punctate and showed significantly increased colocalization with LC3. This colocalization increased further in the presence of BafA1 because this treatment blocks fusion by affecting lysosomal pH but does not affect autophagosomal SNARE assembly (Itakura et al., 2012). Interestingly, upon EACC treatment the colocalization between Stx17 and LC3 reduced significantly (Figure 4, A and B).
FIGURE 4:
EACC inhibits autophagy by preventing SNARE Stx17 loading on autophagosomes. (A) HeLa cells cotransfected with FLAG-Stx17 and GFP-LC3 were treated with BafA1 (100 nM) or EACC (10 µM) for 2 h in starvation conditions and immunostained with anti-FLAG antibody. Scale = 15 µm, 1 µm. (B) Graph represents the number of colocalized dots of FLAG-Stx17 and GFP-LC3. The colocalized dots were counted as mentioned in Figure 2B. Data shown here represent a minimum of 45 cells from three independent experiments plotted as mean ± SEM. Statistical significance was analyzed by Student’s unpaired t test. ***, P < 0.001. (C) Samples from EACC- or BafA1-treated HeLa cells were immunoblotted with anti-Stx17 and anti–β-actin antibodies. (D) Co-IP analysis of interaction between FLAG-Stx17 and endogenous LC3B in HeLa cells either left untreated or treated with EACC. Relative levels of LC3B-II in untreated and EACC-treated cells are mentioned. (E) Data indicate mean ± SEM of relative levels of LC3B-II in FLAG-Stx17 IP normalized to input LC3B-II from three independent experiments. Statistical significance was analyzed by Student’s paired t test. *, P < 0.05. (F) HeLa cells cotransfected with MYC-Stx17, RFP-LC3, and FLAG-SNAP29 were either left untreated or treated with EACC and immunostained with anti-FLAG and anti-MYC antibodies. Scale = 15 µm. (G) Graph represents the percentage of LC3 puncta colocalizing with Stx17 and SNAP-29. The colocalized dots were counted as mentioned in Figure 2B. Data shown here represent a minimum of 45 cells from three independent experiments plotted as mean ± SEM. Statistical significance was analyzed by Student’s unpaired t test. ***, P < 0.001. (H) Graph showing the mean intensity of colocalization between FLAG-SNAP29 and MYC-Stx17 measured as explained in Figure 1H. Data shown here represent a minimum of 45 cells from three independent experiments plotted as mean ± SEM. Statistical significance was analyzed by Student’s unpaired t test. ns = nonsignificant. (I) HeLa cells cotransfected with FLAG-Stx17 and HA-ATG14 were treated with EACC and immunostained with anti-FLAG and anti-HA antibodies. Scale = 15 µm. (J) Graph showing the mean intensity of colocalization between FLAG-Stx17 and HA-ATG14 measured as explained in Figure 1H. Data shown here represent a minimum of 30 cells from three independent experiments plotted as mean ± SEM. Statistical significance was analyzed by Student’s unpaired t test. **, P < 0.01.
A recent report showed that the pathogenic bacterium Legionella pneumophila can block autophagy by degrading Stx17 (Arasaki et al., 2017). However, the presence of EACC did not affect levels of Stx17 expression (Figure 4C).
To further dissect the effect of EACC on LC3-Stx17 interaction we performed coimmunoprecipitation (Co-IP) analysis. In HeLa cells, transfected with either FLAG-Stx17 or an empty vector and either left untreated or treated with EACC, we probed for the levels of LC3-II. The relative levels of LC3-II in FLAG-Stx17 IP (after normalizing it to input LC3) were reduced upon EACC treatment (Figure 4, D and E).
Stx17 is a Qa SNARE that partners with Qbc-SNARE SNAP29 (Itakura et al., 2012; Guo et al., 2014). The autophagosomes having both SNAP-29 and Stx17 were fewer in EACC-treated cells as compared with control (Figure 4, F and G). Interestingly, the colocalization of the partners SNARE Stx17 and SNAP29 was largely unaffected post EACC treatment (Figure 4H). In addition to these SNAREs, ATG14 also participates in autophagosome–lysosome fusion, where it binds to the SNARE domain of Stx17 and stabilizes the Stx17-SNAP29 complex on autophagosomes. This function of ATG14 is independent of its role in autophagosome biogenesis (Hamasaki et al., 2013; Diao et al., 2015). EACC treatment reduced ATG14 and Stx17 colocalization (Figure 4, I and J).
Overall, all these results suggest that EACC renders autophagosomes fusion incompetent by preventing Stx17 translocation onto autophagosomes.
EACC does not affect RABs, tethers, and lysosomal SNARE but prevents their interaction with LC3 and Stx17
Apart from SNAREs, autophagosome–lysosome fusion also requires accessory proteins like small GTPase RAB7 and multisubunit tethering complex HOPS.
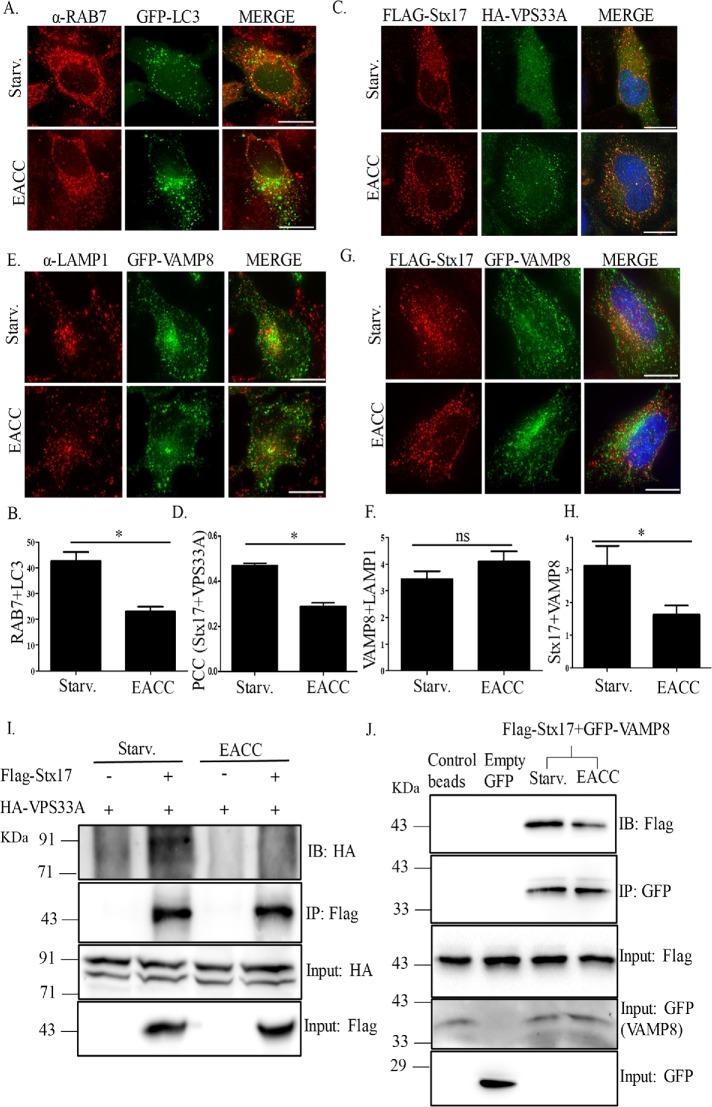
Through its interaction with Stx17 and LC3, RAB7 is required for autolysosome formation (Hyttinen et al., 2013). Although control cells showed significant association of RAB7 with LC3, EACC treatment revealed decreased RAB7 and LC3 colocalization, reiterating that EACC renders autophagosomes fusion incompetent (Figure 5, A and B).
FIGURE 5:
EACC does not affect RABs, tethers, and lysosomal SNARE but prevents their interaction with LC3 and Stx17. (A) GFP-LC3 transfected HeLa cells were treated with EACC and immunostained with anti-RAB7 antibody. Scale = 10 µm. (B) Graph represents the number of LC3 puncta colocalizing with RAB7. The colocalized dots were counted as in Figure 2B. Data shown here represent a minimum of 45 cells from three independent experiments plotted as mean ± SEM. Statistical significance was analyzed by Student’s unpaired t test. *, P < 0.05. (C) HeLa cells cotransfected with FLAG-Stx17 and HA-VPS33A were either left untreated or treated with EACC (10 µM) for 2 h. Scale = 10 µm. (D) Graph showing Pearson’s correlation coefficient (PCC) between Stx17 and VPS33A. PCC was measured using SoftWoRx software from DeltaVision. Data shown here represent a minimum of 45 cells from three independent experiments plotted as mean ± SEM. Statistical significance was analyzed by Student’s unpaired t test. *, P < 0.05. (E) GFP-VAMP8 transfected HeLa cells were immunostained with anti-LAMP1 antibody. Scale = 10 µm. (F) Graph representing the mean intensity of colocalization between LAMP1 and VAMP8. The mean intensity of colocalized dots was measured as in Figure 1H. Data shown here represent a minimum of 45 cells from three independent experiments plotted as mean ± SEM. Statistical significance was analyzed by Student’s unpaired t test. ns = nonsignificant. (G) HeLa cells cotransfected with FLAG-Stx17 and GFP-VAMP8 were either left untreated or treated with EACC. Scale = 10 µm. (H) Graph representing the mean intensity of colocalization between Stx17 and VAMP8. The mean intensity of colocalization was measured as in Figure 1H. Data shown here represent a minimum of 45 cells from three independent experiments plotted as mean ± SEM. Statistical significance was analyzed by Student’s unpaired t test. *, P < 0.05. (I) HeLa cells transfected with FLAG-Stx17 and HA-VPS33A or only HA-VPS33A were either left untreated or treated with EACC. IP was performed using FLAG-tagged magnetic beads and the levels of HA-VPS33A and FLAG-Stx17 were checked by immunoblotting. (J) HeLa cells transfected with FLAG-Stx17 and GFP-VAMP8 or FLAG-Stx17 and empty GFP vector were either left untreated or treated with EACC. IP was performed using control agarose beads or GFP-Trap beads and the levels of GFP-VAMP8 and FLAG-Stx17 were checked by immunoblotting.
Multisubunit tethering complex HOPS through its interaction with Stx17 promotes autophagosome–lysosome fusion (Jiang et al., 2014; Takats et al., 2014). We addressed whether this interaction was altered in the presence of EACC by colocalization analysis between HOPS-specific subunit VPS33A and Stx17. In HeLa cells cotransfected with FLAG-Stx17 and HA-VPS33A, we calculated Pearson’s correlation coefficient (PCC) between Stx17 and VPS33A. Colocalization between Stx17 and VPS33A decreased in EACC treatment (Figure 5, C and D).
Although EACC treatment resulted in accumulation of fusion-incompetent autophagosomes, we further tested whether the lysosomes were competent to receive incoming vesicles for fusion. As shown in Figure 2, K and L, unperturbed EGFR degradation hinted at unaltered lysosomal competence in the presence of EACC. We further investigated the status of the fusion machinery on lysosomes, in particular the v-SNARE VAMP8 required for autolysosome formation (Itakura et al., 2012). In HeLa cells transfected with GFP-VAMP8 and immunostained for LAMP1, there was no apparent change in VAMP8 and LAMP1 association after EACC treatment as compared with control (Figure 5, E and F). Because EACC prevents Stx17 translocation onto autophagosome and blocks autophagosome–lysosome fusion, as expected, treatment with EACC in cells cotransfected with FLAG-Stx17 and GFP-VAMP8 showed decreased colocalization between Stx17 and VAMP8 (Figure 5, G and H).
To further consolidate these findings, using immunoprecipitation assays we checked the effect of EACC on Stx17-HOPS and Stx17-VAMP8 interactions. Cells cotransfected with HA-VPS33A and FLAG-Stx17 were either left untreated or treated with EACC and subjected to IP using FLAG-tagged magnetic beads. We observed that in EACC-treated lysates, the levels of HA-VPS33A were reduced in FLAG-Stx17 immunoprecipitates (Figure 5I).
We also performed a Co-IP using GFP-Trap beads in cells cotransfected with GFP-VAMP8 and FLAG-Stx17. EACC treatment reduced the levels of FLAG-Stx17 in GFP-VAMP8 Co-IP (Figure 5J).
Taken together, these results suggest that EACC treatment renders autophagosomes “fusion incompetent” by preventing Stx17 translocation onto them. It also reduces Stx17 interaction with tethers (VPS33A) and the lysosomal R-SNARE VAMP8. All these factors collectively prevent autophagosome–lysosome fusion and block autophagic flux.
The action of EACC is reversible
Thus far, EACC appears to inhibit autophagic flux by accumulating fusion-incompetent (Stx17-negative) autophagosomes. We wondered whether this effect of EACC is reversible. Toward this, we carried out EACC washout experiments and followed autophagic flux and loading of Stx17 onto autophagosomes.
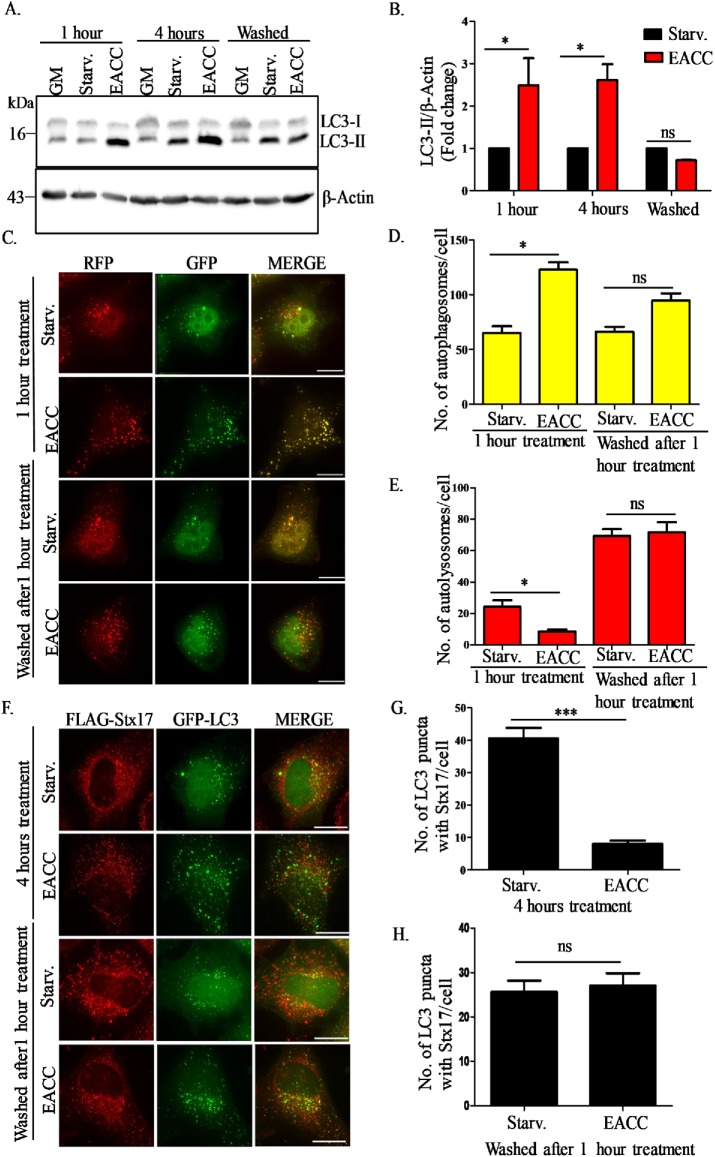
We divided EACC-treated cells into three subgroups. In the first group, cells in starvation media were treated with EACC for 1 h and lysates were collected. In the second group, after a similar treatment with EACC for 1 h, cells were washed with Dulbecco phosphate buffered saline (DPBS) and kept in starvation medium without EACC for 3 h and lysates were collected. In the third group, the treatment with EACC was allowed to go on for 4 h and lysates were collected after that. All the lysates were probed for LC3-II expression. The robust accumulation of LC3-II in EACC treatment was seen as early as 1 h. Interestingly, this accumulation of LC3-II was abrogated after washing out EACC (Figure 6, A and B).
FIGURE 6:
The action of EACC is reversible. (A) We divided EACC-treated cells into three subgroups. In the first group, cells in starvation media were treated with EACC for 1 h and lysates were collected. In the second group, after a similar treatment with EACC for 1 h, cells were washed with DPBS and kept in starvation medium without EACC for 3 h and lysates were collected. In the third group, the treatment with EACC was allowed to go on for 4 h and lysates were collected after that. All the lysates were probed for LC3B-II expression. (B) Relative levels of LC3-II:β-actin in untreated vs. treated samples were quantitated for three independent experiments. *, P < 0.05; ns = nonsignificant (two-way ANOVA, replicate means compared with Bonferroni posttest). (C) HeLa cells were transfected with tandem-tagged mRFP-GFP-LC3 construct for 48 h and treatment was carried out as explained above in A. Scale: 15 µm. (D, E) The autophagosomes (RFP+/GFP+ structures) and autolysosomes (RFP+/GFP− structures) per cell in various treatment conditions were counted as mentioned in Figure 1F. Data shown represent the number of autophagosomes (RFP+/GFP+) and autolysosomes (RFP+/GFP−) for a minimum of 45 cells from three independent experiments plotted as mean ± SEM. Statistical significance was analyzed by Student’s unpaired t test. *, P < 0.05; ns = nonsignificant. (F) HeLa cells transfected with FLAG-Stx17 and GFP-LC3 were treated with EACC (10 µM) as explained above and immunostained with anti-FLAG antibody. Scale: 10 µm. (G, H) Graph represents the number of LC3 puncta colocalizing with Stx17. The colocalized dots were counted as mentioned in Figure 2B. Data shown here represent a minimum of 45 cells from three independent experiments plotted as mean ± SEM. Statistical significance was analyzed by Student’s unpaired t test. ***, P < 0.001; ns = nonsignificant.
We next corroborated our immunoblotting-based results by utilizing the tandem-tagged mRFP-GFP-LC3 construct. HeLa cells transfected with mRFP-GFP-LC3 were treated with EACC in a similar manner as explained above. After 1-h treatment, we saw a significant increase in the number of autophagosomes (mRFP+/GFP+) and a concomitant decrease in the number of autolysosomes (mRFP+/GFP−) as compared with control. After washing out EACC, the autophagosome and autolysosome numbers became comparable to that of control (Figure 6, C–E). Taken together these results suggest that the block in autophagosome–lysosome fusion can be reversed by washing out EACC.
We have shown that EACC inhibits translocation of Stx17 to autophagosomes. So, next we tested whether the localization of SNARE Stx17 can be restored after washing out EACC. In HeLa cells transfected with FLAG-Stx17 and RFP-LC3, we quantitated the number of Stx17+ autophagosomes before and after EACC washout. There were very few LC3+/Stx17+ puncta in cells treated with EACC for 4 h. On the other hand, the number of LC3+/Stx17+ puncta was higher in cells in which EACC was washed out after 1 h (Figure 6, F–H).
In summary, EACC is a reversible inhibitor of autophagosome–lysosome fusion and mechanistically, it acts by preventing translocation of Stx17 onto autophagosomes and decreasing its interaction with the HOPS subunit VPS33A and the lysosomal R-SNARE VAMP8.
DISCUSSION
In this article, we report a novel small molecule EACC that can block autophagic flux in a previously unreported manner. EACC inhibits the translocation of autophagosome-specific SNARE Stx17 thereby blocking autophagosome–lysosome fusion.
We show that EACC causes a massive accumulation of LC3-II over and above that of starvation-induced autophagy. Using various experimental approaches, we deduce that the increase in LC3-II is due to a block in autophagic flux rather than autophagy induction. Additionally, upon probing each step in the process of autophagy, we narrow down the action of EACC to the penultimate step of autophagic flux, that is, fusion of autophagosomes with lysosomes resulting in accumulation of autophagosomes. By further systematic analysis of stage-specific components of autophagy and lysosomal machinery, we conclude that EACC selectively renders autophagosome “fusion incompetent”’ but does not affect the ability of lysosomes to fuse with other incoming traffic.
It is suggested that the fusion step proceeds temporally by first loading Stx17 on autophagosomes followed by SNAP29 recruitment. This Qabc SNARE complex is stabilized by ATG14. Subsequently, successful fusion ensues when SNARE pairing (Qa Stx17, Qbc SNAP29, and the lysosomal R-SNARE VAMP8) is promoted by small GTPase RAB7 and tethering complex HOPS (Itakura et al., 2012; Guo et al., 2014; Jiang et al., 2014; Takats et al., 2014; Diao et al., 2015).
The striking feature of EACC-mediated block of autophagic flux is impaired Stx17 loading onto autophagosomes. To the best of our knowledge, there is no other report suggesting any chemical modulator of autophagy that can selectively prevent Stx17 translocation thereby rendering autophagosomes “fusion incompetent.” The exact mechanism by which Stx17 is translocated onto complete autophagosomes is not very clear. A recent report suggested that Stx17 recruitment to autophagosomes occurs via interaction with a small GTPase IRGM and mammalian ATG8 proteins (Kumar et al., 2018). Although we have not checked whether EACC can affect interaction between Stx17 and IRGM, we propose that identification of Stx17-binding partners in the presence or absence of EACC could give a clue regarding the target of EACC as well as help in identification of any other accessory factors that might be involved in Stx17 recruitment on autophagosomes. Furthermore, we also showed that the action of EACC is reversible. The block in autophagic flux is eliminated after washing out EACC because Stx17 is now able to translocate to autophagosomes and participate in further fusion events. Hence, due to the reversible nature of its action, EACC can be used as a useful tool to study Stx17 trafficking.
To determine the rate of autophagic flux, lysosomal inhibitors like BafA1 and chloroquine are routinely used. Unfortunately, these treatments are not ideal as they not only can impair lysosomal function but impede all other lysosomal pathways including the endo-lysosomal trafficking. Our results also show that the action of EACC is specific to autophagosomes and it does not affect lysosomal pH, function, or endocytic trafficking. It also does not affect the localization of lysosomal SNAREs or RABs. Additionally, even the well-known early inhibitors of autophagy such as wortmannin and 3-methyl adenine are promiscuous as they block all phosphatidylinositol 3-kinase–dependent signaling pathways thereby resulting in a plethora of side effects. In such scenarios, inhibiting Stx17 translocation by using EACC, which leads to a specific block in autophagy, might be a cleaner way to perform autophagic flux experiments. In fact, silencing Stx17 expression is recommended as a desired attribute for selectively inhibiting autophagic flux (Hegedus et al., 2013). In conclusion, molecules like EACC can fill the lacuna that exists in the field due to lack of specific autophagy inhibitors.
MATERIALS AND METHODS
Cell culture
HeLa cells were maintained in growth medium composed of DMEM (Sigma-Aldrich; D5648) supplemented with 3.7 g/l sodium bicarbonate (Sigma-Aldrich; S5761) plus 10% fetal bovine serum (Life Technologies; 10270-106) and 100 U/ml penicillin and streptomycin (Life Technologies; 15140-122) at 5% CO2 and 37°C. Autophagy was induced by washing cells with DPBS (Sigma-Aldrich; D5773) and incubating them in starvation media/Earle’s balanced salt solution (Sigma-Aldrich; E7510) made to 1× and supplemented with 2.2 g/l sodium bicarbonate for 2 h until otherwise stated.
Plasmids
Plasmids used in the study were as follows: ptfLC3 (mRFP-GFP-LC3; Addgene plasmid #21074) and pmRFP-LC3 (Addgene plasmid #21075) were gifts from Tamotsu Yoshimori (Osaka University). FLAG-Stx17 (Addgene plasmid #45911) and FLAG-SNAP29 (Addgene plasmid#45915) were gifts from Noburu Mizushima (The University of Tokyo). GFP-VAMP8 was a gift from Thierry Galli (Institute of Psychiatry and Neuroscience of Paris [IPNP]) (Addgene plasmid #42311; Paumet et al., 2000), mCherry-DFCP1 was a gift from Do-Hyung Kim (University of Minnesota) (Addgene plasmid #86746; Kim et al., 2015), and HA-hATG14 was a gift from Noburu Mizushima (Addgene plasmid #24294; Itakura et al., 2008). Plasmid-containing HA-VPS33A was a kind gift from Mahak Sharma, IISER Mohali. Myc-Stx17 plasmid was a kind gift from Viktor Korolchuk, Newcastle University. GFP-LC3 plasmid was generated in the lab by excising out mRFP fragment from mRFP-GFP-LC3 plasmid.
Antibodies and reagents
The following primary antibodies were used: LC3 (MBL; M152-3), LC3B (Sigma-Aldrich; L7543), β-actin (CST; 4970), LAMP1 (CST; 9091), p62/SQSTM1 (Abcam; ab56416), β-tubulin (DHSB; E7), cathepsin B (Cloud Clone; PAC964Hu01), EGFR (Santa Cruz Biotechnology; sc-03), phospho-P70S6K (T389; CST; 9234), total P70S6K (CST; 9202), phospho-4EBP1 (CST; 2855), total 4EBP1 (CST; 9452), phospho-ULK1 (S757; CST; 6888), ATG14 (CST; 5504), ATG5 (CST; 12994), ATG16L1 (CST; 8089), WIPI2 (Abcam; ab105459), Stx17 (Sigma-Aldrich; HPA001204), FLAG (Sigma-Aldrich; F3165), FLAG (Sigma-Aldrich; F1804), HA (CST; 3724), Myc (Abcam; ab9106), RAB7 (CST; 9367), GFP (Roche; 11814460001), mouse immunoglobulin G (IgG) (Genei, IGP3). Secondary antibodies used were goat anti-mouse IgG (H+L) HRP (horseradish peroxidase) conjugate (Bio-Rad; 1721011), goat anti-rabbit IgG (H+L) HRP conjugate (Biorad; 1706515). Fluorescent secondary antibodies used were Atto 633 (goat anti-rabbit IgG; Sigma-Aldrich; 41176), Atto 488 (goat anti-rabbit IgG; Sigma-Aldrich; 41057), Alexa 647 (goat anti-mouse IgG; Invitrogen; A21236), Alexa 405 (goat anti-mouse IgG; Invitrogen; A31556).
Chemicals used in this study were EACC (Life Chemicals; F1358-0554), bafilomycin A1 (Sigma-Aldrich; B1793), actinomycin D (Sigma-Aldrich; A1410), cycloheximide (Sigma-Aldrich; C7698), and EGF (Thermo Fisher Scientific; PHG0311L). LysoTracker Deep Red (L12492) was purchased from Thermo Fisher Scientific.
Immunoprecipitation
For immunoprecipitation assays, cells were lysed in lysis buffer (20 mM Tris-HCl, pH 7.2, 2 mM MgCl2, 150 mM NaCl, 0.5% NP-40) supplemented with protease inhibitor/phosphatase inhibitor cocktails for 30 min at 4°C and centrifuged at 13,000 × g for 15 min. Protein (500 μg to 1 mg) was incubated with specific primary antibody at 4°C (2 h to overnight) on a tube rotator followed by incubation with protein G dynabeads (Invitrogen; #10004D) for 2 h at 4°C. The beads were washed three times with ice-cold PBS and the proteins were eluted from washed beads by boiling for 5 min in 2× Laemmli sample buffer and processed for immunoblot analysis.
For immunoprecipitation with GFP-Trap beads (Chromotek), cells were lysed in lysis buffer recommended by the manufacturer (10 mM Tris-HCl, pH 7.2, 0.5 mM EDTA, 150 mM NaCl, 0.5% NP-40) supplemented with protease inhibitor/phosphatase inhibitor cocktails for 30 min at 4°C and centrifuged at 13,000 × g for 15 min. Protein (1 mg) from the supernatant was used, and immunoprecipitation was performed by following the manufacturer’s instructions.
Immunoblotting
Following appropriate treatments, cells were washed with ice-cold PBS. Cells were then lysed in 100 μl of sample buffer (10% wt/vol SDS, 10 mM dithiothreitol, 20% vol/vol glycerol, 0.2 M Tris-HCl, pH 6.8, 0.05% wt/vol bromophenol blue) and then collected using a rubber cell scraper. The lysates were boiled at 99°C for 15 min and stored at −20°C. Immunoblotting was performed using standard methods.
Blots were incubated overnight with the above-mentioned primary antibodies. Secondary antibody used at 1:10,000 was goat anti-mouse (Bio-Rad; #172-1011) or goat anti-rabbit antibody (Bio-Rad; #172-1019) conjugated to HRP. Blots were developed by using enhanced chemiluminescence (ECL) substrate (Bio-Rad; #170-5061) and images captured using auto capture or series capture program in a Gel documentation system (Syngene G-Box; UK). ImageJ software (National Institutes of Health [NIH]) was used for quantitation of band intensities.
Immunofluorescence
An appropriate number of cells were plated on top of coverslips placed in 60-mm cell culture dishes for transfection. The following day, transfection was done on a 60-mm dish with HeLa cells at 60–70% confluency. Cells were transfected using 5 μl of lipofectamine 2000 (Invitrogen; 11668-019) and 2.5 μg of DNA (2:1 ratio) diluted in 100 μl of OPTI-MEM (Invitrogen; 31985-070) separately. At 48 h posttransfection, cells were either left untreated or treatment with EACC was done for 2 h. Starvation was induced by treating cells with Earle’s balanced salt solution (EBSS). After treatment, cells were fixed in 4% paraformaldehyde and permeabilized using 0.25% Triton X-100. Overnight incubation with primary antibody was done at 4°C. Excess antibody was washed with PBS and coverslips were incubated with appropriate fluorescent secondary antibody. The coverslips were mounted with Vectashield antifade reagent (H-1000/ H-1200; Vector Laboratories). Imaging for HeLa cells was carried out using a DeltaVision microscope, GE (Olympus 60X/1.42, Plan ApoN, excitation and emission filter Cy5, FITC, DAPI, and TRITC, polychroic Quad).
CellTiter-Glo cell viability assay
Toxicity of the compound was monitored by CellTiter-Glo cell viability assay (Promega; G7570). HeLa cells were counted and equal numbers (1500 cells/well) were plated in a 384-well plate in growth medium. The following day, different concentrations of EACC ranging from 100 nM to 100 μM were mixed in starvation media, added onto the cells, and incubated for 5 h. After 5 h, CellTiter-Glo Reagent was added to each well and luminescence measured using Varioskan Flash (Thermo Fisher Scientific).
EGFR trafficking
HeLa cells were plated on six-well plates and allowed to attach. The following day, cells were washed with PBS and then starved in DMEM (serum-free media) for 3 h. Pretreatment with EACC was carried out for 1 h, following which cells were pulsed with 100 ng/ml EGF and samples were collected at 0, 1-, 2-, and 3-h intervals.
Colocalization analysis and mean intensity calculation
ImageJ software (NIH) was used to calculate the mean intensity of staining or mean intensity of colocalization. Images were opened using the split channel plug-in. In the case of colocalization, a colocalization plug-in in the analyze tool was used to obtain the colocalized area between two channels as a separate window. The intensity was measured using the analysis measure plug-in in analysis tools. A cell counter plug-in was used to count the colocalized structures.
Statistical analysis and image preparation
Statistical analysis was performed using GraphPad Prism (GraphPad Software). Statistical analyses were performed by comparing the means using the paired/unpaired Student t test or two-way analysis of variance (ANOVA) followed by the Bonferroni posttest to compare replicate means by row. Images were prepared using SoftWoRx software (GE Healthcare). Some fluorescent MIP images had their brightness and contrast modified equally in control and treatment conditions just for the purpose of visualization.
Supplementary Material
Acknowledgments
We acknowledge Robert Damoiseaux, Director, Molecular Shared Screening Resource, UCLA and Bryan France for their support in the primary screening of the Microsource library. We also thank Mahak Sharma (IISER Mohali), G. Subba Rao (Indian Institute of Science), Tapas Kundu (JNCASR), and Sovan Sarkar (University of Birmingham) for sharing reagents and providing expert suggestions. We acknowledge Piyush Mishra, Sunaina Singh, Irine Maria Abraham, and Prashanta Kumar Panda for their scientific advice and critical reading of the manuscript. This work was supported by a Wellcome Trust/DBT India Alliance Intermediate Fellowship (500159-Z-09-Z), the DST Science and Engineering Research Board (SERB) (Grant no. EMR/2015/001946), the DBT-JNCASR Partnership Programme in Biology (BT/INF/22/SP27679/2018), and JNCASR intramural funds to R.M. S.V. acknowledges the support of a JNCASR doctoral fellowship. Application of EACC in autophagy modulation has been awarded an Indian patent, and an international patent application has been filed.
Abbreviations used:
- BafA1
bafilomycin A1
- Co-IP
coimmunoprecipitation
- DPBS
Dulbecco phosphate buffered saline
- EACC
ethyl (2-(5-nitrothiophene-2-carboxamido) thiophene-3-carbonyl) carbamate
- ECL
enhanced chemiluminescence
- EGF
epidermal growth factor
- EGFR
epidermal growth factor receptor
- GM
growth medium
- HRP
horseradish peroxidase
- IB
immunoblotting
- IP
Immunoprecipitation
- IRGM
immunity related GTPase M
- MIP
maximum intensity projection
- PCC
Pearson’s correlation coefficient
- Starv.
starvation medium.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E18-08-0482) on June 12, 2019.
REFERENCES
- Arasaki K, Mikami Y, Shames SR, Inoue H, Wakana Y, Tagaya M. (2017). Legionella effector Lpg1137 shuts down ER-mitochondria communication through cleavage of syntaxin 17. Nat Commun , 15406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bento CF, Renna M, Ghislat G, Puri C, Ashkenazi A, Vicinanza M, Menzies FM, Rubinsztein DC. (2016). Mammalian autophagy: how does it work? Annu Rev Biochem , 685–713. [DOI] [PubMed] [Google Scholar]
- Bonifacino JS, Glick BS. (2004). The mechanisms of vesicle budding and fusion. Cell , 153–166. [DOI] [PubMed] [Google Scholar]
- Cai H, Reinisch K, Ferro-Novick S. (2007). Coats, tethers, Rabs, and SNAREs work together to mediate the intracellular destination of a transport vesicle. Dev Cell , 671–682. [DOI] [PubMed] [Google Scholar]
- Darsow T, Rieder SE, Emr SD. (1997). A multispecificity syntaxin homologue, Vam3p, essential for autophagic and biosynthetic protein transport to the vacuole. J Cell Biol , 517–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V, Saitoh T, Akira S. (2013). Autophagy in infection, inflammation and immunity. Nat Rev Immunol , 722–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao J, Liu R, Rong Y, Zhao M, Zhang J, Lai Y, Zhou Q, Wilz LM, Li J, Vivona S, et al (2015). ATG14 promotes membrane tethering and fusion of autophagosomes to endolysosomes. Nature , 563–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick D, Barth S, Macleod KF. (2010). Autophagy: cellular and molecular mechanisms. J Pathol , 3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo B, Liang Q, Li L, Hu Z, Wu F, Zhang P, Ma Y, Zhao B, Kovacs AL, Zhang Z, et al (2014). O-GlcNAc-modification of SNAP-29 regulates autophagosome maturation. Nat Cell Biol , 1215–1226. [DOI] [PubMed] [Google Scholar]
- Hamasaki M, Furuta N, Matsuda A, Nezu A, Yamamoto A, Fujita N, Oomori H, Noda T, Haraguchi T, Hiraoka Y, et al (2013). Autophagosomes form at ER-mitochondria contact sites. Nature , 389–393. [DOI] [PubMed] [Google Scholar]
- Hegedus K, Takats S, Kovacs AL, Juhasz G. (2013). Evolutionarily conserved role and physiological relevance of a STX17/Syx17 (syntaxin 17)-containing SNARE complex in autophagosome fusion with endosomes and lysosomes. Autophagy , 1642–1646. [DOI] [PubMed] [Google Scholar]
- Hyttinen JM, Niittykoski M, Salminen A, Kaarniranta K. (2013). Maturation of autophagosomes and endosomes: a key role for Rab7. Biochim Biophys Acta , 503–510. [DOI] [PubMed] [Google Scholar]
- Ishihara N, Hamasaki M, Yokota S, Suzuki K, Kamada Y, Kihara A, Yoshimori T, Noda T, Ohsumi Y. (2001). Autophagosome requires specific early Sec proteins for its formation and NSF/SNARE for vacuolar fusion. Mol Biol Cell , 3690–3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E, Kishi C, Inoue K, Mizushima N. (2008). Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell , 5360–5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E, Kishi-Itakura C, Mizushima N. (2012). The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell , 1256–1269. [DOI] [PubMed] [Google Scholar]
- Itakura E, Mizushima N. (2010). Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy , 764–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang P, Nishimura T, Sakamaki Y, Itakura E, Hatta T, Natsume T, Mizushima N. (2014). The HOPS complex mediates autophagosome-lysosome fusion through interaction with syntaxin 17. Mol Biol Cell , 1327–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YM, Jung CH, Seo M, Kim EK, Park JM, Bae SS, Kim DH. (2015). mTORC1 phosphorylates UVRAG to negatively regulate autophagosome and endosome maturation. Mol Cell , 207–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S, Noda T, Yoshimori T. (2007). Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy , 452–460. [DOI] [PubMed] [Google Scholar]
- Kumar S, Jain A, Farzam F, Jia J, Gu Y, Choi SW, Mudd MH, Claude-Taupin A, Wester MJ, Lidke KA, et al (2018). Mechanism of Stx17 recruitment to autophagosomes via IRGM and mammalian Atg8 proteins. J Cell Biol , 997–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra P, Dauphinee AN, Ward C, Sarkar S, Gunawardena A, Manjithaya R. (2017a). Discovery of pan autophagy inhibitors through a high-throughput screen highlights macroautophagy as an evolutionarily conserved process across 3 eukaryotic kingdoms. Autophagy , 1556–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra P, Rai S, Manjithaya R. (2017b). A novel dual luciferase based high throughput assay to monitor autophagy in real time in yeast S. cerevisiae. Biochem Biophys Rep , 138–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N. (2007). Autophagy: process and function. Genes Dev , 2861–2873. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T. (2007). How to interpret LC3 immunoblotting. Autophagy , 542–545. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, Levine B. (2010). Methods in mammalian autophagy research. Cell , 313–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon RA. (2013). The role of autophagy in neurodegenerative disease. Nat Med , 983–997. [DOI] [PubMed] [Google Scholar]
- Paumet F, Le Mao J, Martin S, Galli T, David B, Blank U, Roa M. (2000). Soluble NSF attachment protein receptors (SNAREs) in RBL-2H3 mast cells: functional role of syntaxin 4 in exocytosis and identification of a vesicle-associated membrane protein 8-containing secretory compartment. J Immunol , 5850–5857. [DOI] [PubMed] [Google Scholar]
- Rubinsztein DC, Codogno P, Levine B. (2012). Autophagy modulation as a potential therapeutic target for diverse diseases. Nat Rev Drug Discov , 709–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato TK, Darsow T, Emr SD. (1998). Vam7p, a SNAP-25-like molecule, and Vam3p, a syntaxin homolog, function together in yeast vacuolar protein trafficking. Mol Cell Biol , 5308–5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SS, Vats S, Chia AY, Tan TZ, Deng S, Ong MS, Arfuso F, Yap CT, Goh BC, Sethi G, et al (2018). Dual role of autophagy in hallmarks of cancer. Oncogene , 1142–1158. [DOI] [PubMed] [Google Scholar]
- Steegmaier M, Oorschot V, Klumperman J, Scheller RH. (2000). Syntaxin 17 is abundant in steroidogenic cells and implicated in smooth endoplasmic reticulum membrane dynamics. Mol Biol Cell , 2719–2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steegmaier M, Yang B, Yoo JS, Huang B, Shen M, Yu S, Luo Y, Scheller RH. (1998). Three novel proteins of the syntaxin/SNAP-25 family. J Biol Chem , 34171–34179. [DOI] [PubMed] [Google Scholar]
- Surpin M, Zheng H, Morita MT, Saito C, Avila E, Blakeslee JJ, Bandyopadhyay A, Kovaleva V, Carter D, Murphy A, et al (2003). The VTI family of SNARE proteins is necessary for plant viability and mediates different protein transport pathways. Plant Cell , 2885–2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takats S, Pircs K, Nagy P, Varga A, Karpati M, Hegedus K, Kramer H, Kovacs AL, Sass M, Juhasz G. (2014). Interaction of the HOPS complex with Syntaxin 17 mediates autophagosome clearance in Drosophila. Mol Biol Cell , 1338–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.