Abstract
Rho-dependent proteins control assembly of the cytokinetic contractile ring, yet it remains unclear how those proteins guide ring closure and how they promote subsequent formation of a stable midbody ring. Citron kinase is one important component required for midbody ring formation but its mechanisms of action and relationship with Rho are controversial. Here, we conduct a structure–function analysis of the Drosophila Citron kinase, Sticky, in Schneider’s S2 cells. We define two separable and redundant RhoGEF/Pebble-dependent inputs into Sticky recruitment to the nascent midbody ring and show that each input is subsequently required for retention at, and for the integrity of, the mature midbody ring. The first input is via an actomyosin-independent interaction between Sticky and Anillin, a key scaffold also required for midbody ring formation. The second input requires the Rho-binding domain of Sticky, whose boundaries we have defined. Collectively, these results show how midbody ring biogenesis depends on the coordinated actions of Sticky, Anillin, and Rho.
INTRODUCTION
In animal cells, cytokinesis follows mitosis by forming a cleavage furrow at the equator of the dividing cell. Furrow ingression is driven at its base by a contractile ring, a dynamic, membrane-anchored assembly of cytoskeletal proteins that constricts via disassembly to reduce the cell equator to a diameter of 1–2 µm (Schroeder, 1972; Carvalho et al., 2009). At this point, the contractile ring has gathered the bundled microtubules of the central spindle, and it must transition into a stable structure called the midbody ring. The midbody ring and associated microtubule-based midbody form a dense structure (Mullins and Biesele, 1977), enriched in many different proteins and lipids (Skop et al., 2004; Atilla-Gokcumen et al., 2014). The contractile ring-to-midbody ring transition is a crucial step in cell division, essential for maintaining the integrity of the intercellular bridge and for setting the stage for the final abscission event that irrevocably separates the cells. The contractile ring-to-midbody ring transition occurs via a maturation process that includes a gradual thinning of the nascent midbody ring and removal of cortical material through internalization and a septin-dependent process that we described and termed “shedding” (El Amine et al., 2013), which involves extrusion and expulsion of material in the form of membranous blebs or tubules (see also Mullins and Biesele, 1977; Renshaw et al., 2014). The transition also involves changes in the actin and microtubule cytoskeleton (Hu et al., 2012; Terry et al., 2018). A complex machinery is involved in organizing the succession of events during cytokinesis, ensuring that they proceed with high fidelity (Fededa and Gerlich, 2012; Green et al., 2012; D’Avino et al., 2015; Glotzer, 2017). Activation of the small molecular weight GTPase RhoA is an early event essential for furrow initiation and contractile ring formation (Bement et al., 2005; Piekny et al., 2005; Basant and Glotzer, 2018). The contractile ring comprises filament-forming cytoskeletal proteins including actin, myosin II, and septins, along with many other accessory proteins that ensure contractile ring contractility, attachment to the plasma membrane, and interaction with central spindle microtubules (Green et al., 2012; D’Avino et al., 2015; Glotzer, 2017). Among these contractile ring proteins is the Citron kinase, named Sticky in Drosophila melanogaster. Originally identified as a Rho-interacting protein (Madaule et al., 1995, 1998; Di Cunto et al., 1998), Citron kinase was initially suspected to play a role in myosin activation and cleavage furrow formation (Madaule et al., 1998, 2000; Yamashiro et al., 2003). However, it has since become clear that the major function and requirement for Citron kinase during cytokinesis is not at the contractile ring per se, but rather in the formation of the midbody ring that occurs after contractile ring closure (D’Avino et al., 2004; Echard et al., 2004; Naim et al., 2004; Dean and Spudich, 2006). Although there is evidence of redundant contributions of Citron kinase/Sticky during furrowing (Madaule et al., 1998; Gai et al., 2011; El Amine et al., 2013), as well as suggestions of a role during abscission (Naim et al., 2004; Gai et al., 2011; Sgro et al., 2016), loss-of-function studies have clearly shown that Citron kinase/Sticky is required for the stabilization of the midbody ring in a variety of cell types (Echard et al., 2004; Gruneberg et al., 2006; Neumann et al., 2010; Gai et al., 2011; Bassi et al., 2013; El Amine et al., 2013; Watanabe et al., 2013; McKenzie et al., 2016). However, precisely how Sticky is regulated and how it acts to stabilize the midbody ring remain unclear. Although Citron kinase is primarily a cortical protein, numerous studies indicate that it can interact with microtubule-associated proteins of the midbody, such as KIF14/Nebbish and kinesin-6/MKLP1/Pavarotti, and that these interactions are important for intracellular bridge stability (Gruneberg et al., 2006; Bassi et al., 2013; Watanabe et al., 2013; McKenzie et al., 2016).
An important and longstanding question concerns the nature of the relationship between Citron kinase/Sticky and the Rho GTPase. The original assumption that Sticky acts as a canonical downstream effector of Rho has recently been challenged by the suggestion that mammalian Citron kinase acts upstream of RhoA to modulate its activity (Gai et al., 2011; Dema et al., 2018). Additionally, there is evidence of Drosophila Sticky interacting with Rho1 via its C-terminal Citron-Nik1 homology (CNH) domain and not via its putative Rho-binding domain (RBD) (Shandala et al., 2004; Bassi et al., 2011; Gai et al., 2011). Thus, the relationship between Citron kinase and Rho remains controversial and unclear.
Here, we show in Drosophila S2 cells that the RhoGEF Pebble (pbl) controls the localization of Sticky to the cleavage furrow through two partially redundant and separable inputs, acting on its central coiled-coil region. One input depends on the highly conserved Sticky Rho-binding homology (RBH) region and likely involves direct binding of Rho1-GTP, while the second input requires interaction with the Rho1-dependent scaffold protein, Anillin, also known as Scraps in Drosophila. Disrupting either the Rho1/Sticky or the Anillin/Sticky interaction led to failures in midbody ring formation, indicating that both inputs are required for Sticky function. Conversely, little to no evidence was found to support a role for the CNH domain in Sticky localization or function during cytokinesis. Collectively, the data lead to the proposal that Rho1 acts on Sticky both directly, and indirectly via Anillin, to recruit Sticky to the contractile ring. In turn, Sticky acts to retain Anillin in the subsequent midbody ring as it forms around the midbody, to which Sticky is also connected. Thus, Rho1 controls Sticky through multiple inputs to coordinate midbody ring formation.
RESULTS
Sticky localization requires Rho1 signaling
To test the requirements for Rho1 signaling in the localization of Sticky, we first depleted the RhoGEF Pbl (the orthologue of mammalian ECT2) in cells expressing Sticky-GFP. We note that many Sticky-derived constructs (Figure 1A) exhibit cytoplasmic punctate structures, presumed to be aggregates, as described for mammalian Citron kinase (Eda et al., 2001; Watanabe et al., 2013). Nevertheless, nonaggregated Sticky-GFP is still robustly recruited to the cell cortex during cytokinesis and, as previously shown, is able to functionally rescue for loss of endogenous Sticky (El Amine et al., 2013, and Figure 1H). Live-cell spinning disk confocal microscopy showed that cells incubated with a double-stranded RNA (dsRNA) targeting the Sticky 3′UTR (see Supplemental Figure S1 for efficacy of different Sticky dsRNAs used in this study) robustly recruited Sticky-GFP to the contractile ring and midbody ring and exhibited some shedding from the nascent midbody ring (Figure 1B and Supplemental Movie 1), similar to that previously described for Anillin (El Amine et al., 2013). However, in cells incubated with pbl dsRNA, cells failed to form furrows, as expected (Somers and Saint, 2003; Hickson et al., 2006), and Sticky-GFP localization to the cell cortex during anaphase/telophase was abolished, indicating that pbl controls Sticky recruitment (Figure 1C). Sticky contains a highly conserved, 29-amino acid RBH region within its central coiled-coil region (residues 1235–1263) that is predicted to bind to Rho-GTP (Bassi et al., 2011). Deletion of the RBH region (Sticky-∆RBH-GFP, Figure 1D) or introduction of a point mutation (L1246N) predicted to abrogate any Rho-GTP binding (Shimizu et al., 2003; Dvorsky et al., 2004; Watanabe et al., 2013; Jungas et al., 2016) did not prevent cortical recruitment of Sticky (Figure 1E). However, live imaging of deplete-and-rescue experiments indicated that these perturbations of the RBH region greatly reduced the ability of Sticky to support cytokinesis, resulting in characteristic failures at the nascent midbody ring stage (Figure 1H, N = 68, **p = 0.004 and N = 87, *p = 0.028 for Sticky-∆RBH and Sticky-L1246N, respectively). Thus, the RhoGEF/Pbl is required for Sticky localization and the RBH region is required for Sticky function, but is dispensable for cortical localization.
FIGURE 1:
Sticky localization requires Rho1 signaling. (A) An illustrated representation of Sticky showing different domains, truncated constructs, and the position of the L1246N mutation (asterisk). (B–G) Representative, high-resolution time-lapse sequences of cells stably expressing mCherry-Tubulin (magenta) and inducibly expressing (in green in the merged top panels, or inverted grayscale in the bottom panels) Sticky-GFP (B, C), Sticky-∆RBH-GFP (D), Sticky-L1246N-GFP (E), Sticky-∆CNH-GFP (F), or Sticky-L1246N-∆CNH-GFP (G) following 3-d incubation with the specified dsRNAs. N represents the number of cells displaying similar patterns of GFP localization at the division plane at t = 00:08:00–00:10:00/total number of cells scored. Data are from two independent experiments. Blue arrowheads highlight the ingressing cleavage furrow, green arrowheads (B) highlight shedding of Sticky-GFP, and dashed lines represent the outline of the cell in the grayscale images. (H) Quantification of the outcome (success or failure) of division attempts scored from low-resolution imaging of cells stably expressing mCherry-Tubulin and induced to express the specified GFP-tagged rescue constructs, following 3-d incubation with sti 3′UTR dsRNA. Data are from two independent experiments. Error bars represent SD between experiments resulting from an unpaired t test; *p < 0.05, **p < 0.01 for significance, and ns = nonsignificance. Times are in h:min:s; scale bars are 5 µm.
Movie 1.
Sticky-GFP is robustly recruited to the contractile ring and midbody ring and exhibits some shedding from the nascent midbody ring. Time-lapse spinning disc confocal microscopy images of Drosophila S2 cell stably expressing Sticky-GFP (green) and mCh-Tubulin (magenta) after treatment with sti 3′UTR dsRNA for 3 days. Frames were taken every 1.8 minutes for 3 hours with a 63x, 1.4 NA objective. A maximum intensity z-projection (extended focus) is shown and video is playing at 5 fps. Video corresponds to Figure 1B. Time is shown in h:min:s. Scale bar is 5 μm.
The C-terminal CNH domain has also been reported to interact with Rho (Shandala et al., 2004; Bassi et al., 2011). However, a construct lacking this domain (Sticky-∆CNH-GFP; residues 1–1450) was indistinguishable from full-length Sticky-GFP in terms of localization to the contractile ring and midbody ring (Figure 1F) and in its ability to support successful cytokinesis on depletion of endogenous Sticky via sti dsRNA 3′UTR (100% success; Figure 1H, N = 65, p = 0.211). A construct combining deletion of the CNH domain with mutation of the RBH region (Sticky-L1246N-∆CNH-GFP) behaved similarly to Sticky-L1246N-GFP (60% vs. 41% success; Figure 1, G and H, N = 59, *p = 0.036). We also tested the ability of the ∆CNH constructs to rescue the more penetrant failures induced by sti dsRNA3 that targets the CNH region (56% vs. 70% success, Supplemental Figure S2F, N = 52, p = 0.053). In this case, partial rescue by Sticky-∆CNH-GFP was observed, in a manner that was abolished by the L1246N mutation (70% vs. 52% success; Supplemental Figure S2, D and F, N = 60, p = 0.052). These results suggest that the CNH domain of Sticky is not essential for cytokinesis in S2 cells. However, given that the rescue observed on induction of Sticky-∆CNH-GFP was incomplete (70% success vs. 56% in cells only expressing mCh-Tubulin, Supplemental Figure S2F), we cannot rule out the possibility that it plays a contributory role herein, or even an essential role in other cell types.
Anillin contributes to the Rho-dependent cortical recruitment of Sticky
We previously showed that Sticky and Anillin cooperate during formation of the stable midbody ring, and in particular that Sticky acts to retain Anillin at the nascent midbody ring (El Amine et al., 2013). We therefore wished to test whether Anillin, itself a Rho-dependent protein (Hickson and O’Farrell, 2008b; Piekny and Glotzer, 2008; Sun et al., 2015), contributes to the cortical localization of Sticky. Depletion of Anillin using different, efficacious dsRNAs (Supplemental Figure S4) that block complete closure of the contractile ring did not prevent the cortical recruitment of Sticky-GFP, indicating that Anillin is dispensable for Sticky localization (Figure 2B, arrowheads at 00:03:00 and 00:09:00). However, Anillin depletion did prevent the cortical recruitment of Sticky-L1246N-GFP (Figure 2C and Supplemental Movie 2) and Sticky-∆RBH-GFP (Figure 2D), albeit Sticky-∆CNH-GFP was still recruited following Anillin depletion (Figure 2E, arrowheads at 00:05:00 and 00:15:00). These results indicate that Sticky recruitment to the contractile ring involves at least two inputs: one dependent on Anillin and the other dependent on Sticky’s RBH region with no major contribution from the CNH domain.
FIGURE 2:
Anillin contributes to the Rho1-dependent cortical recruitment of Sticky. (A) Illustrated representation of Sticky showing different domains and Sticky-CC2a. (B–I) Representative, high-resolution time-lapse sequences of cells stably expressing mCherry-Tubulin (magenta) and inducibly expressing (in green in the merged top panels, or inverted grayscale in the bottom panels) Sticky-GFP (B), Sticky-L1246N-GFP (C), Sticky-∆RBH-GFP (D), Sticky-∆CNH-GFP (E), or Sticky-CC2a-GFP (F–I) following 3-d incubation with the specified dsRNAs. N represents the number of cells displaying similar patterns of GFP localization at the division plane at t = 00:08:00-00:10:00/total number of cells scored. Data are from two independent experiments. Blue arrowheads highlight the ingressing cleavage furrow and dashed lines represent the outline of the cell in the grayscale images. (J) Immunofluorescence micrographs of S2 cells induced to express Sticky-CC2a-GFP treated for 3 d with the indicated dsRNAs, fixed, and stained with anti-Rho1, anti-Tubulin antibodies, and Hoechst DNA stain. White arrowheads highlight the ingressed cleavage furrow. Times are h:min:s; scale bars are 5 µm.
Movie 2.
Depletion of Anillin prevents the cortical recruitment of Sticky-L1246N-GFP Time-lapse spinning disc confocal microscopy images of Drosophila S2 cell stably expressing Sticky-L1246N-GFP (green) and mCh-Tubulin (magenta) after treatment with control anillin dsRNA1 for 3 days. Frames were taken every minute 3 for 3 hours with a 63x, 1.4 NA objective. A maximum intensity z-projection (extended focus) is shown and video is playing at 5 fps. Video corresponds to Figure 2C. Time is shown in h:min:s. Scale bar is 5 μm.
Bassi et al. (2011) used bioinformatic approaches to subdivide the central coiled-coil region of Sticky into three subregions, CC1, CC2a, and CC2b, and showed that CC2a (residues 774–1227) was sufficient for cortical localization. In agreement, we found that Sticky-CC2a-GFP (residues 774–1228) was robustly recruited to the contractile ring and nascent midbody ring (Figure 2F, arrowheads at 00:09:00 and 00:24:00). The localization pattern of Sticky-CC2a-GFP remained unchanged on RNA interference (RNAi)-mediated depletion of endogenous Sticky (sti dsRNA3; Figure 2G, arrowheads at 00:08:00 and 00:56:00), but it was abolished by depletion of Anillin (Figure 2H). RNAi of Pbl also prevented the recruitment of Sticky-CC2a-GFP, confirming that CC2a recruitment is RhoGEF-dependent (pbl dsRNA; Figure 2I). Immunofluorescence analysis of fixed S2 cells depleted of endogenous Sticky (sti dsRNA3) revealed colocalization of Sticky-CC2a-GFP and endogenous Rho1 at the nascent midbody ring stage, providing further support that the recruitment of Sticky-CC2a is Rho1-dependent (Figure 2J).
The coiled-coil domain of Sticky contains two Pbl-dependent inputs
The above results suggested that Anillin contributes to Sticky localization via the CC2a region, while the RBH domain of Sticky, which resides within the CC2b region, also plays a role. To further test this notion, we closely examined the behavior of a construct containing both CC2a and CC2b (residues 774–1370) (Figure 3A) using live-cell imaging of mitotic cells. In cells treated with a control lacI dsRNA, Sticky-CC2a-CC2b-GFP was robustly recruited to the contractile ring and nascent midbody ring (Figure 3B). This recruitment was also seen on depletion of endogenous Sticky using sti dsRNA3 that targets the mRNA sequence encoding part of the CNH domain (Supplemental Figure S1A), although the construct was unable to rescue cytokinesis failures at the midbody stage (Figure 3C). As with full-length Sticky-GFP, Pbl depletion abolished the equatorial recruitment of Sticky-CC2a-CC2b-GFP (Figure 3D), whereas Anillin depletion did not (Figure 3E). Introduction of the L1246N mutation within the RBH domain did not prevent the cortical recruitment of Sticky-CC2a-CC2b-GFP (Figure 3F), unless Anillin was simultaneously depleted (Figure 3G). These observations confirm that the coiled-coil region of Sticky responds to two Pbl-dependent inputs: one via the CC2a region involving Anillin and one via the RBH of Sticky within the CC2b region presumably involving Rho binding. Furthermore, since both Anillin depletion and mutation of the RBH region are required to abrogate localization, we conclude that these inputs act in a partially redundant manner to recruit Sticky to the cortex.
FIGURE 3:
The coiled-coil domain of Sticky contains two Pbl-dependent inputs. (A) Illustrated representation of Sticky showing different domains and Sticky-CC2a-CC2b with and without the L1246N mutation (asterisk). (B–G) Representative, high-resolution time-lapse sequences of cells stably expressing mCherry-Tubulin (magenta) and inducibly expressing (in green in the merged top panels, or inverted grayscale in the bottom panels) Sticky-CC2a-CC2b-GFP (B–E) or Sticky-CC2a-CC2b-L1246N-GFP (F, G) following 3-d incubation with the specified dsRNAs. N represents the number of cells displaying similar patterns of GFP localization at the division plane at t = 00:08:00–00:10:00/total number of cells scored. Data are from two independent experiments. Blue arrowheads highlight the ingressing cleavage furrow and dashed lines indicate the outline of the cell in the grayscale images. Times are h:min:s; scale bars are 5 µm.
Defining the minimal Anillin-dependent input to Sticky localization
We sought to define the minimal requirements for each of these two inputs (CC2a and CC2b) and determine whether they could both be physically separated from one another. A series of constructs was generated in which the CC2a region (residues 774–1228) was progressively truncated by 50-residue increments from either the N- or the C-terminus (Figure 4, A and B). These fragments were fused to GFP and expressed in S2 cells, and their ability to localize to the furrow cortex was assessed by live-imaging time-lapse microscopy. Truncation of the N-terminus did not prevent cortical recruitment until residues 974–1024 were deleted (Figure 4A); the construct Sticky1024–1228-GFP and smaller constructs remained cytoplasmic during furrow ingression. Truncation of the C-terminus did not prevent cortical recruitment until residues 1078–1128 were deleted (Figure 4B); the construct Sticky774–1078–GFP and smaller truncations were no longer recruited to the cortex. Thus, we conclude that residues 974–1128 are required for the cortical localization of Sticky’s CC2a domain. However, we noted that some of the shorter constructs (Sticky774–1078-GFP, Sticky774–1028-GFP, and Sticky774-978–GFP) that did not localize to the cortex nonetheless exhibited a later midbody localization, which might indicate additional interactions within the CC2a region (Figure 4B).
FIGURE 4:
Defining the minimal Anillin-dependent input to Sticky localization. (A, B) Illustrations on the left indicate the series of N-terminal (A) and C-terminal (B) truncations of Sticky-CC2a generated and, on the right, high-resolution time-lapse sequences of the corresponding, representative localization patterns observed for the indicated truncations fused to GFP (inverted grayscale images of the GFP channel are shown). Horizontal black lines separate the illustrated depictions of the constructs that localized to the contractile ring/nascent midbody ring (above the line) from those that did not (below the line), while the light blue shaded region represents the Sticky-miniCC2a974-1128 region defined by this truncation analysis. (C–E) Time-lapse sequence of representative cells stably expressing mCherry-Tubulin (magenta) and inducibly Sticky-miniCC2a974-1128-GFP (green in top panel and inverted grayscale in bottom panel) incubated for 3 d with control lacI dsRNA (C), anillin dsRNA2 (D), or sticky dsRNA3 (E). Times are h:min:s; scale bars are 5 µm. (F) Quantification from high-resolution imaging of the percentage of cells that showed cortical recruitment vs. no cortical recruitment of Sticky-miniCC2a974-1128-GFP during anaphase/telophase following indicated RNAi treatment. (G) Quantification of the outcome (success or failure) of division attempts scored from low-resolution imaging of cells stably expressing mCherry-Tubulin and induced to express Sticky-miniCC2a974-1128-GFP following 3-d incubation with sti dsRNA3. Data are from three independent experiments. Total number of cells analyzed (N) for each condition is indicated on respective bars. Error bars represent SD between experiments resulting from an unpaired t test with ns = nonsignificant.
We next tested whether the residues 974–1128 were sufficient for cortical localization by generating a copper-inducible Sticky974–1128-GFP construct and expressing it in S2 cells. This construct, named “miniCC2a,” was robustly recruited to the contractile ring and nascent midbody ring in 100% of control cells observed via live imaging (Figure 4, C and F, and Supplemental Movie 3, N = 100), while 100% of Anillin-depleted cells failed to recruit Sticky-miniCC2a974–1128-GFP to the furrow cortex (Figure 4, D and F, N = 120, and Supplemental Movie 4). In cells depleted of endogenous Sticky (dsRNA3), Sticky-miniCC2a974–1128-GFP was still recruited in 100% of cases (N = 78), but failed to rescue cytokinesis since 61% of these division attempts failed (N = 108) compared with 56% in the uninduced controls (N = 80, Figure 4, E–G, p = 0.467). We further noted that in control cells expressing endogenous Sticky, Sticky-miniCC2a974-1128-GFP could localize along the length of the intercellular bridge at the nascent midbody ring stage, but was specifically excluded from mature midbody rings (e.g., Figure 4C), suggesting that Sticky-miniCC2a974-1128-GFP lacks elements required for the formation of the midbody ring and/or its retention at the midbody ring. Given that a Protein A-CC2a774–1227 fusion was reported to pull down actin and myosin from cell extracts (Bassi et al., 2011), we tested for potential contributions of actomyosin in recruiting Sticky-miniCC2a974-1128-GFP. Treatment with Latrunculin A (LatA), which prevents actin polymerization (Coue et al., 1987; Ayscough et al., 1997, 1998), specifically disrupts cortical F-actin. However, LatA does not prevent the recruitment during anaphase of Rho1, Anillin, septins, or myosin II, which rather assemble into cortical structures at the equatorial membrane (Hickson and O’Farrell, 2008b). Hence, in order to test whether the recruitment of miniCC2a was actin-dependent, cells were treated with LatA and recruitment of miniCC2a was assayed. Persistent cortical localization of Sticky-miniCC2a974–1128-GFP in 1 µg/ml LatA indicates that its recruitment is actin-independent (Supplemental Figure S3B). Recruitment of Sticky-miniCC2a974–1128-GFP was also unaffected, in the presence or absence of LatA, by depletion of Rho-kinase (Rok; Supplemental Figure S3F) or myosin heavy chain (Zipper; Supplemental Figure S3G), indicating that recruitment is independent of myosin II. Depletion of kinesin 6/Pavarotti or Kif14/Nebbish, which can also bind to the CC1 region of Sticky during cytokinesis (Bassi et al., 2013), had no effect on Sticky-miniCC2a974–1128-GFP recruitment either (Supplemental Figure S3, H and I). Indeed, of all candidates tested, only Anillin or RhoGEF/pbl RNAi diminished cortical recruitment of Sticky-miniCC2a974–1128-GFP, which were assayed both in the presence and in the absence of LatA (Figure 4D and Supplemental Figure S3, D and E). We therefore conclude that residues 974–1128 of Sticky are sufficient for cortical recruitment to the equatorial cortex in a manner that specifically requires RhoGEF/Pbl and Anillin, but does not require Rok, actomyosin, kinesin-6/Pav, or Kif14/Nebbish.
Movie 3.
Sticky-miniCC2a974-1128-GFP is robustly recruited to the contractile ring and midbody ring and exhibits shedding from the midbody ring. Time-lapse spinning disc confocal microscopy images of Drosophila S2 cell stably expressing Sticky-miniCC2a974-1128-GFP (green) and mCh-Tubulin (magenta) after treatment with control lacI dsRNA for 3 days. Frames were taken every 2.4 minutes for 3 hours and 50 minutes with a 63x, 1.4 NA objective. A maximum intensity z-projection (extended focus) is shown and video is playing at 6 fps. Video corresponds to Figure 4C. Time is shown in h:min:s. Scale bar is 5 μm.
Movie 4.
Sticky-miniCC2a974-1128-GFP fails to be recruited to the contractile ring and midbody ring upon Anillin depletion. Time-lapse spinning disc confocal microscopy images of Drosophila S2 cell stably expressing Sticky-miniCC2a974-1128-GFP (green) and mCh-Tubulin (magenta) after treatment with anil dsRNA2 for 3 days. Frames were taken every 1.8 minutes for 3 hours with a 63x, 1.4 NA objective. A maximum intensity z-projection (extended focus) is shown and video is playing at 5 fps. Video corresponds to Figure 4D. Time is shown in h:min:s and scale bar is 5 μm.
Mapping the RBD-dependent contribution to Sticky localization
Our analysis of the L1246N mutation in the context of full-length Sticky (Figures 1E and 2C) and Sticky-CC2a-CC2b (Figure 3) indicated the involvement of the RBH domain in Sticky localization. However, Bassi et al. (2011) had shown that a construct comprising only residues 1228–1386, which harbors the Rho-binding homology domain and which they termed CC2b, was not recruited to the cortex. We confirmed this finding using Sticky-CC2b-GFP (residues 1229–1370; Figure 5A), but also generated an additional series of constructs that extended this construct in ∼50-residue increments in the N-terminal direction. These extended constructs now localized to the contractile ring (Figure 5, B, D, and F) in a manner that was inhibited by inclusion of the L1246N mutation (Figure 5, C, E, and G), indicating that they depended on a functional RBH domain. The only constructs that still localized to the contractile ring when harboring the L1246N mutation were the ones that also included the entire Anillin-dependent miniCC2a974–1128 residues (Figure 5, H and I). Importantly, Sticky residues 1174–1370, which we refer to here as “maxiCC2b,” promoted contractile ring localization without any overlapping amino acid residues with the miniCC2a (residues 974–1128) region, confirming that these two inputs are physically separable, albeit directly adjacent.
FIGURE 5:
Mapping the RBD-dependent contribution to Sticky localization. (A–I) High-resolution time-lapse sequences of the representative localization patterns observed for the Sticky-CC2a-CC2b-GFP N-terminal truncations, and L1246N mutants are shown on the left and depicted in the illustrations on the right. Inverted grayscale confocal images of the GFP channel are shown. Only Sticky-CC2b1229-1370-GFP failed to localize to the cell cortex (A), whereas the longer GFP constructs were recruited to the cortex (B, D, F, H) in a manner that was abolished by the L1246N mutation (C, E, G), except for when the full miniCC2a974-1128 sequence was also present (I). The yellow shaded area of the illustrations represents the minimal RBH-dependent sequence (residues 1174–1370) required for cortical recruitment of maxiCC2b that is distinct from the miniCC2a sequence (residues 974–1128; light blue shading). (J–M) Representative, high-resolution time-lapse sequences of cells transiently expressing mCherry-Tubulin (magenta) and inducibly expressing Sticky-maxiCC2b1174-1370-GFP (green in the merged panels on the left; inverted grayscale in the panels on the right) following 3-d incubation with the specified dsRNAs. Blue arrowheads highlight cortical recruitment at the CR and dashed lines indicate the outline of the cell in the grayscale images. Times are h:min:s; scale bars are 5 µm.
We further confirmed that Sticky-maxiCC2b1174–1370-GFP (Figure 5J) was recruited independently of endogenous Sticky (Figure 5K) and Anillin (Figure 5L). However, as expected, pbl RNAi blocked equatorial enrichment of Sticky-maxiCC2b1174–1370-GFP (Figure 5M). Collectively, these results support the conclusion that the CC2 region of Sticky contains at least two separable Rho-dependent inputs that contribute to furrow localization: one through residues 974–1128 (miniCC2a; Rho1- and Anillin-dependent) and another one through residues 1174–1370 (maxiCC2b; Rho1-dependent, but Anillin-independent).
The localization of miniCC2a specifically requires the N-terminal domain (NTD) of Anillin
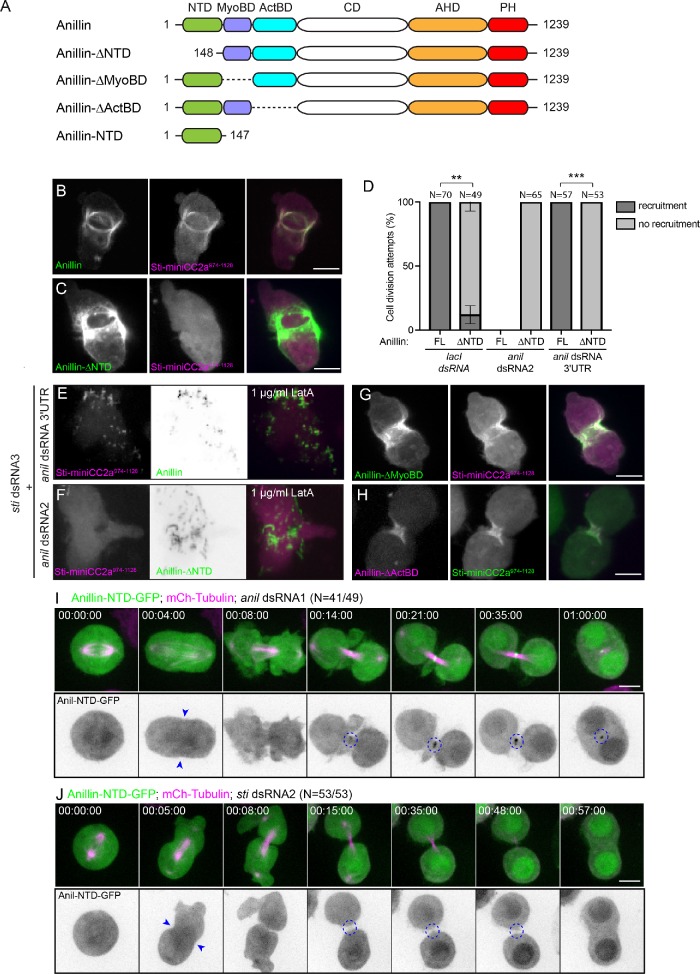
We sought to further define how Anillin contributes to Sticky localization. Anillin is a multidomain, Rho-dependent scaffold protein (Hickson and O’Farrell, 2008a,b; Piekny and Glotzer, 2008). We previously showed that Sticky acts to retain the N-terminal half of Anillin (residues 1–802), which lacks the C-terminal Anillin homology (AH) and Pleckstrin homology (PH) domains, but is sufficient to form a midbody ring-like structure (El Amine et al., 2013). We therefore hypothesized that Anillin would require sequences within its N-terminal half to recruit miniCC2a, even though the Rho-dependent recruitment of Anillin occurs via its C-terminal AH-PH region (Piekny and Glotzer, 2008; Kechad et al., 2012; Sun et al., 2015). To test this hypothesis, we generated stable cell lines coexpressing Sticky-miniCC2a974–1128-mCherry and various Anillin truncations fused to GFP (Figure 6A) and examined their localization patterns in live cells depleted of endogenous Anillin. While 100% of cells expressing full-length Anillin-GFP robustly recruited miniCC2a to the contractile ring (Figure 6, B and D, N = 70), cells expressing Anillin lacking its NTD of 147 amino acids (Anillin-∆NTD-GFP) failed to recruit miniCC2a in 95% of division attempts (Figure 6, C and D, N = 49, **p = 0.0032). On depletion of endogenous Anillin by RNAi, Anillin-∆NTD failed to recruit miniCC2a in 100% of division attempts (Figure 6D, N = 65 for anil dsRNA2 and N = 53 for anil dsRNA 3′UTR, ***p = 0.00012). We next wished to examine the recruitment of miniCC2a to the Rho1-, Anillin-, and septin-dependent structures that form, on disruption of the actin cytoskeleton with LatA, at the equatorial membrane during anaphase (Hickson and O’Farrell, 2008b). Anillin-GFP recruited miniCC2a to these structures (Figure 6E and Supplemental Movie 5) following LatA treatment, while Anillin-∆NTD-GFP did not (Figure 6F, Supplemental Movie 6), confirming that F-actin is not required for the Anillin-dependent recruitment of miniCC2a. Constructs lacking Anillin’s N-terminal myosin binding (Anillin-∆MyoBD-GFP) or actin-binding (Anillin-∆ActBD-GFP) domain were still able to recruit miniCC2a to the cleavage furrow (Figure 6, G and H). Thus, the NTD of Anillin is specifically required for the recruitment of miniCC2a to the cleavage furrow, presumably reflecting an interaction between Anillin and Sticky. Consistent with this notion, a fragment of Anillin comprising only the NTD fused to GFP was recruited to the nascent midbody ring in Anillin-depleted cells (anil dsRNA1; Figure 6I), while this recruitment was lost following Sticky depletion (sti dsRNA2; Figure 6J).
FIGURE 6:
The localization of Sticky-miniCC2a974-1128 specifically requires the Anillin NTD. (A) Illustrated representation of Anillin showing different domains and truncated constructs. (B, C) Stills from time-lapse sequences of cells midanaphase, inducibly expressing Sticky-miniCC2a974-1128-mCherry (middle panels, magenta in merged) and (B) Anillin-GFP or (C) Anillin-∆NTD-GFP (left panels, green in merged). (D) Quantification from time-lapse recordings of the percentage of cells that showed cortical recruitment vs. no cortical recruitment of Sticky-miniCC2a974-1128-mCherry during anaphase/telophase in cells coexpressing either Anillin-GFP (full-length, FL) or Anillin-∆NTD (∆NTD) following 3-d incubation with the specified dsRNAs. N values indicate the total number of cells scored for recruitment per condition and data are from three independent experiments. Error bars represent SD between experiments; **p < 0.01; ***p < 0.001 for significance in an unpaired t test. (E, F) Stills from time-lapse sequences of cells midanaphase inducibly expressing Sticky-miniCC2a974-1128mCherry (left panels, magenta in merged) and (E) Anillin-GFP or (F) Anillin-∆NTD-GFP (grayscale middle panel and green in merged) after a 3-d incubation with sti and anil dsRNAs as indicated, and following pretreatment with 1 µg/ml LatA. (G, H) Stills from time-lapse sequences of cells midanaphase, inducibly expressing (G) Sticky-miniCC2a974-1128-mCherry (middle panel, magenta in merged) and Anillin-∆MyoBD-GFP (left panel, green in merged) or (H) Sticky-miniCC2a974-1128-GFP (middle panel, green in merged) and Anillin-∆ActBD-mCherry (left panel, magenta in merged). (I, J) Stills from time-lapse recordings of cells expressing mCherry-Tubulin (magenta in merged top panels) and Anillin-NTD-GFP (green in top merged panels; grayscale in bottom panels showing inverted images) progressing through cytokinesis following a 3-d incubation with anil dsRNA1 (I) or sti dsRNA2 (J). Blue arrowheads highlight the ingressing furrow; dashed circles highlight the nascent MR. Times are h:min:s; scale bars are 5 µm.
Movie 5.
Anillin-GFP recruits Sticky-miniCC2a974-1128-mCherry to LatA-induced structures. Time-lapse spinning disc confocal microscopy images of Drosophila S2 cell stably expressing Anillin-GFP (green) and Sticky-miniCC2a974-1128-mCherry (magenta) treated with anil dsRNA 3′UTR and sti dsRNA3 for 3 days, followed by treatment with 1 μg/ml of LatA. Frames were taken every minute for 2 hours and 10 minutes with a 63x, 1.4 NA objective. A maximum intensity z-projection (extended focus) is shown and video is playing at 5 fps. Video corresponds to Figure 6E. Time is shown in h:min:s and scale bar is 5 μm.
Movie 6.
Anillin-ΔNTD-GFP fails to recruit Sticky-miniCC2a974-1128-mCherry to LatA-induced structures. Time-lapse spinning disc confocal microscopy images of Drosophila S2 cell stably expressing Anillin-ΔNTD-GFP (green) and Sticky-miniCC2a974-1128-mCherry (magenta) treated with anil dsRNA 2 and sti dsRNA3 for 3 days, followed by treatment with 1 μg/ml of LatA. Frames were taken every minute for 1 hour and 40 minutes with a 63x, 1.4 NA objective. A maximum intensity z-projection (extended focus) is shown and video is playing at 5 fps. Video corresponds to stills shown in Figure 6F. Stillswere rotated by 90? clockwise. Time is shown in h:min:s and scale bar is 5 μm.
A physical interaction between the Anillin NTD and Sticky-miniCC2a
Biochemical experiments were employed to test the hypothesis that the Anillin NTD physically interacts with the miniCC2a region of Sticky. GST fusion proteins were expressed and purified from Escherichia coli and used in pull-down assays. Unlike GST alone, GST-Anillin-NTD effectively pulled down endogenous Sticky from untransfected S2 cell lysate (Figure 7A). GST-Anillin-NTD also specifically pulled down miniCC2a-GFP from cell lysate expressing this construct (Figure 7B). Reciprocally, a GST-miniCC2a fusion protein, but not GST alone, effectively pulled down endogenous Anillin from untransfected S2 cell lysate (Figure 7C). Further, GST-miniCC2a pulled down Anillin-NTD-GFP from transfected cell lysate (Figure 7D), but did not pull down Anillin-∆NTD-GFP (Figure 7E). These data lead to the conclusion that the miniCC2a region (residues 974–1128) of Sticky physically interacts, directly or indirectly, with the NTD of Anillin (residues 1–147). This supports the observations regarding the recruitment of Sticky-miniCC2a to the contractile ring and nascent midbody ring during live-cell imaging presented in Figures 4 and 6.
Figure 7:
A physical interaction between the Anillin NTD and Sticky-miniCC2a974-1128. (A, B) GST-pull-down experiments in which GST-Anillin-NTD was used to pull down endogenous Sticky (A) or Sticky-miniCC2a974-1128-GFP (B) from S2 cell whole cell extracts (WCE). Pull-down fractions (P) and supernatants (S) were separated by SDS–PAGE, transferred to membranes and immunoblotted with antibodies against Sticky (A) or GFP (B). (C–E) Analogous blots for which GST-Sticky-miniCC2a974-1128 was used to pull down endogenous Anillin (C), Anillin-NTD-GFP (D), or Anillin-∆NTD-GFP (E) from S2 cell whole cell extracts (WCE). Membranes were immunoblotted with antibodies against Anillin (C) or GFP (D, E). Ponceau S-stained membranes prior to blotting are shown (bottom panel) (A–E) and, in A and C, prior to being cut into two for separate immunoblotting with alpha-Tubulin antibody. The position of GST and GST-fusion proteins is indicated on the Ponceau S-stained membranes. The GST was run off the bottom of the gels shown in C–E. Both the GST and GST-fusion protein were run off the bottom of the gel in A.
The Anillin NTD and Sticky-miniCC2a domains are each required for a successful contractile ring-to-midbody ring transition
Given that Anillin-∆NTD-GFP had failed to recruit (Figure 6C) and interact with (Figure 7E) the miniCC2a fragment of Sticky, we tested the ability of Anillin-∆NTD-GFP to support cytokinesis in rescue experiments. Stably transfected cells were treated with control dsRNA or one of two different dsRNAs targeting endogenous Anillin, induced to express Anillin-∆NTD-GFP, and scored for the outcome of division attempt by low-resolution live-imaging 3 d post-RNAi. In total, 95% of control depleted cells (lacI RNAi) successfully divided (Figure 8A, N = 314 for lacI dsRNA-induced cells and N = 203 for lacI dsRNA-uninduced cells, p = 0.714). Conversely, 50% of cells depleted of endogenous Anillin failed cytokinesis (Figure 8A, N = 238 for anil dsRNA2-induced cells, ***p = 0.0006 and N = 175 for anil dsRNA 3′UTR-induced cells, *p = 0.024). This indicates that Anillin requires its 147 amino acid NTD for faithful cytokinesis. High-resolution imaging of these cells also revealed evidence of shedding of Anillin-∆NTD-GFP (Figure 8, B–D, and Supplemental Movie 7), although cells often failed cytokinesis with an internal Anillin-∆NTD-positive structure (Figure 8, C and D, and Supplemental Movie 7). This suggests that the normal retention of Anillin at the midbody ring does not solely depend on the NTD, and other mechanisms must exist. As an additional test of the role of the Anillin NTD, we examined the behaviors of Anillin-NTD-GFP and Anillin-∆NTD-mCherry coexpressed in the same cells. This confirmed specific retention of Anillin-NTD at the midbody ring with concomitant shedding of Anillin-∆NTD (Figure 8E and Supplemental Movie 8).
FIGURE 8:
The Anillin NTD and Sticky miniCC2a974-1128 domains are each required for a successful contractile ring-to-midbody ring transition. (A) Quantification of the outcomes (success or failure) of division attempts scored from low-resolution time-lapse imaging of cells stably expressing mCherry-Tubulin and inducibly expressing Anillin-∆NTD-GFP following 3-d incubation with the specified dsRNAs. N values indicate the total number of cells scored per condition and data are from three independent experiments. Error bars represent SD between experiments; *p < 0.05, ***p < 0.001 for significance and ns = nonsignificant in an unpaired t test. (B, D) High-resolution time-lapse sequences of representative examples of the most prevalent phenotypes observed for cells expressing mCherry-Tubulin (magenta) and Anillin-∆NTD-GFP (green in top panels, inverted grayscale in bottom panels) progressing through cytokinesis, following 3-d incubation with lacI (B) or anil (D) dsRNAs. Insets are magnifications of the boxed regions, and dashed white boxes indicate the region shown in the insets. Blue arrowheads highlight shed particles of Anillin-∆NTD-GFP, and dashed lines indicate the outline of the cell in the grayscale images. (C) Scatter plot of the phenotypes observed by high-resolution imaging of the Anillin RNAi condition. The relative times of success (abscission) or failure (binucleation) of individual cells are plotted, and color-coding reflects whether or not shedding of Anillin-∆NTD-GFP was observed in relation to whether or not an internalized Anillin-∆NTD-GFP-positive structure was observed. (E) High-resolution time-lapse sequence of a cell coexpressing Anillin-NTD-GFP (green) and Anillin-∆NTD-mCherry (magenta) following a 3-d Anillin RNAi treatment. The bottom panels represent separated channels of the dashed boxed region in the top panels. Dashed circles highlight the nascent MR enriched in Anillin-NTD-GFP; arrowheads highlight shed particles enriched in Anillin-∆NTD-mCherry. (F) Top, an illustrated representation of Sticky and Sticky-∆miniCC2a. Bottom, quantification of the outcomes (success or failure) of division attempts in the percentage scored from low-resolution time-lapse imaging of cells stably expressing mCherry-Tubulin and inducibly expressing Sticky-∆miniCC2a-GFP following 3-d incubation with the specified dsRNAs. N values indicate the total number of cells scored per condition and data are from three independent experiments. Error bars represent SD between experiments; ns = nonsignificance in an unpaired t test. (G–I) High-resolution time-lapse sequences of the most prevalent phenotypes observed for cells transiently expressing mCherry-Tubulin (magenta) and inducibly expressing Sticky-∆miniCC2a-GFP (G, H) or Sticky-∆miniCC2a-L1246N-GFP (I, green in top panels, inverted grayscale in bottom panels) progressing through cytokinesis following 3-d incubation with lacI (G, I) or sti (H) dsRNAs. Insets are magnifications of the boxed regions. Arrowheads in G highlight the nascent midbody and shed particles of Sticky-∆miniCC2a-GFP. Times are h:min:s; scale bars are 5 µm. (J) Scatter plots of the quantification of localization of Sticky-GFP, Sticky-∆miniCC2a-GFP and Sticky-∆miniCC2a-L1246N-GFP to the nascent midbody ring following incubation with lacI or sti dsRNAs by measuring fluorescence intensity at 20 min after anaphase onset for all cells analyzed from high-resolution time-lapse sequences acquired after a 3-d incubation with the specified dsRNAs. N values indicate the total number of cells scored per condition and data are from three independent experiments. Data are displayed as median with the interquartile range; *p ≤ 0.05, ***p < 0.0001 for significance and ns = nonsignificant in a Mann-Whitney U test.
Movie 7.
Partial shedding from the midbody ring and subsequent internalization of Anillin-ΔNTD-GFP in a cell that fails cytokinesis after depletion of Anillin.Time-lapse spinning disc confocal microscopy images of Drosophila S2 cell stably expressing Anillin-ΔNTD-GFP (green) and mCh-Tubulin (magenta) treated with anil dsRNA2 for 3 days. Frames were taken every 1.6 minutes for 2 hours and 56 minutes with a 63x, 1.4 NA objective. A maximum intensity z- projection (extended focus) is shown and video is playing at 5 fps. Video corresponds to Figure 8D. Time is shown in h:min:s and scale bar is 5 μm.
Movie 8.
Retention of Anillin-NTD-GFP at the midbody ring and shedding of Anillin-ΔNTD-mCherry after depletion of Anillin. Time-lapse spinning disc confocal microscopy images of Drosophila S2 cell stably expressing Anillin-NTD-GFP (green) and Anillin-ΔNTD-mCherry (magenta) treated with anil dsRNA 3′UTR for 3 days. Frames were taken every 4 minutes for 3 hours with a 63x, 1.4 NA objective. A maximum intensity z-projection (extended focus) is shown and video is playing at 5 fps. Video corresponds to Figure 8E. Time is shown in h:min:s and scale bar is 5 μm.
Similar deplete-and-rescue experiments were performed to test the requirement for the miniCC2a domain for the localization and function of Sticky. Cells were induced to express Sticky-∆miniCC2a-GFP and the outcome of division attempts was scored through low-resolution live imaging. In control lacI dsRNA-treated cells, 90% of divisions were successful (Figure 8F, N = 52 for lacI dsRNA-uninduced cells, N = 94 for lacI dsRNA-induced cells, p = 0.473), while 95% of cells treated for 3 d with Sticky 3′UTR dsRNAs failed cytokinesis (Figure 8F, N = 131 for Sti dsRNA 3′UTR-uninduced cells and N = 96 for Sti dsRNA 3′UTR-induced cells, p = 0.366). Thus, amino acids 974–1128 of Sticky, composing the miniCC2a sequence, are required for cytokinesis.
High-resolution imaging revealed that in the control lacI dsRNA-treated cells, Sticky-∆miniCC2a-GFP localized to the midbody region after contractile ring closure, although this localization was significantly weaker compared with Sticky-GFP (Figure 8, G and J, N = 21 for Sticky-GFP and N = 18 for Sticky-∆miniCC2a-GFP, p ≤ 0.0001). However, in Sticky-depleted cells, Sticky-∆miniCC2a-GFP localization was still detectable, albeit barely (Figure 8, H and J, N = 19, p ≤ 0.0001, and Supplemental Movie 9). This suggests that the presence of endogenous, full-length Sticky can contribute to Sticky-∆miniCC2a-GFP localization. We observed that Sticky-GFP intensity at the midbody ring was also slightly decreased in Sticky-depleted cells (Figure 8J, N = 25, p = 0.019). We also introduced the L1246N mutation into Sticky-∆miniCC2a-GFP, which abolished any localization of this construct at the division site (Figure 8I, N = 53), regardless of whether or not endogenous Sticky was depleted (Figure 8J, N = 25 for lacI-depleted cells and N = 23 for sti-depleted cells, p ≤ 0.0001). This further supports the conclusion that both miniCC2a and the RBH-containing maxiCC2b contribute to Sticky localization and in a partially redundant manner. However, the particularly dramatic decrease observed in the localization of Sticky-∆miniCC2a suggests that miniCC2a constitutes the major recruitment mechanism.
Movie 9.
Sticky-ΔminiCC2a-GFP is weakly localized to the late contractile ring/nascent midbody ring after depletion of Sticky. Time-lapse spinning disc confocal microscopy images of Drosophila S2 cell transiently expressing Sticky-ΔminiCC2a-GFP (Δ974-1128; green) and mCh-Tubulin (magenta) treated with stidsRNA 3′UTR for 3 days. Frames were taken every 5 minutes for 3 hours with a 63x, 1.4 NA objective. A maximum intensity z-projection (extended focus) is shown and video is playing at 4 fps. Video corresponds to Figure 8H. Time is shown in h:min:s and scale bar is 5 μm.
The Sticky RBH domain is required for midbody ring maturation
Interfering with the interaction between the Sticky miniCC2a region and the Anillin NTD led to a failure of the contractile ring-to-midbody ring transition. We further explored the consequences of perturbing the RBH-dependent input in the context of full-length Sticky. Despite localizing to the nascent midbody ring, neither Sticky-∆RBH-GFP nor Sticky-L1246N-GFP was able to rescue for loss of endogenous Sticky (Figure 1H), with cells failing at the nascent midbody ring stage. On close inspection of time-lapse sequences, we noted that Sticky-L1246N-GFP was significantly less well recruited to the nascent midbody ring than Sticky-GFP, irrespective of the presence or absence of endogenous Sticky (Figure 9A, N = 21 for Sticky-GFP and N = 26 for Sticky-L1246N-GFP for lacI-depleted cells; N = 25 for Sticky-GFP and N = 28 for Sticky-L1246N-GFP for sti-depleted cells, p ≤ 0.0001 in both cases). Nonetheless, regardless of whether the initial recruitment to the nascent midbody ring was weak or strong, the retention at the maturing midbody ring of Sticky-L1246N-GFP appeared markedly impaired compared with that of Sticky-GFP. The persistence of Sticky-GFP versus Sticky-L1246N-GFP signals at the mature midbody ring was therefore measured over time in high-resolution imaging experiments that were performed in parallel, blind as to the presence or absence of the mutation, and without RNAi (so as not to induce cytokinesis failure). This revealed that while Sticky-GFP signal remained detectable at the mature midbody ring for 249 ± 50 min (mean ± SD, Figure 9B, N = 31), Sticky-L1246N-GFP was prematurely lost at 132 ± 56 min (mean ± SD, Figure 9B, N = 29, p = 0.0002). Remarkably, under these same conditions, Sticky-L1246N-GFP also exhibited an increased propensity to be shed than did wild-type Sticky-GFP (Figure 9, C and D). Sticky-GFP was only rarely observed to shed from the nascent midbody ring and in a limited manner (Figure 9C), while Sticky-L1246N-GFP exhibited frequent and extensive shedding (Figure 9D, arrowheads, and Supplemental Movie 10). We previously reported that Sticky is required to form a midbody ring that retains Anillin and protects it from septin-dependent shedding at the nascent midbody ring, since Sticky depletion enhanced shedding and prevented midbody ring formation (El Amine et al., 2013). The L1246N mutant described here appears compromised in its ability to incorporate into a stable midbody ring structure and to resist shedding. We therefore conclude that the RBH domain of Sticky, which makes only a partially redundant contribution to the initial recruitment of Sticky, is required for successful cytokinesis through a role in Sticky retention and stable, mature midbody ring formation.
FIGURE 9:
The Sticky RBH domain promotes its retention at the stable midbody ring. (A) Scatter plot of the quantification of localization of Sticky-GFP and Sticky-L1246N -GFP to the nascent midbody ring by measuring fluorescence intensity at 20 min after anaphase onset for all cells analyzed from high-resolution time-lapse sequences acquired after a 3-d incubation with the specified dsRNAs. N values indicate the total number of cells scored per condition and data are from three independent experiments. Data are displayed as median with the interquartile range; *p = 0.0195 and ***p < 0.0001 for significance in a Mann-Whitney U test. (B) Scatter plot of the relative times (in minutes) of the disappearance of detectable GFP signal from the mature MR of individual cells expressing Sticky-GFP or Sticky-L1246N-GFP scored in a blind manner using high-resolution time-lapse sequences acquired in parallel and without RNAi. N values indicate the total number of cells scored per condition and data are from two independent experiments. Error bars represent SD between cells; ***p < 0.001 for significance obtained in an unpaired t test. (C, D) High-resolution time-lapse sequences of cells transiently expressing mCherry-Tubulin (magenta) and inducibly expressing Sticky-GFP (green in C) or Sticky-L1246N-GFP (green in D) progressing through cytokinesis. Bottom panels represent magnifications of the dashed boxed regions. Arrowheads highlight shed particles. Times are h:min:s; scale bars are 5 µm. (E) Model showing proposed, cooperative interactions between Rho1, Sticky and Anillin at play during recruitment to the contractile ring/nascent midbody ring and retention at the mature midbody ring.
Movie 10.
Sticky-L1246N-GFP is poorly retained and shed from the nascent midbody ring. Time-lapse spinning disc confocal microscopy images of Drosophila S2 cell transiently expressing Sticky-L1246N-GFP (green) and mCh-Tubulin (magenta). Frames were taken every 4 minutes for 3 hours with a 63x, 1.4 NA objective. A maximum intensity z-projection (extended focus) is shown and video is playing at 5 fps. Video corresponds to Figure 9D. Time is shown in h:min:s and scale bar is 5 μm.
DISCUSSION
We report a detailed structure–function analysis that helps clarify the controversial relationship between the Citron kinase, Sticky, and the small GTPase Rho. The work defines two adjacent, Rho-dependent inputs that both contribute to the cortical recruitment and retention of Sticky during midbody ring formation. One input requires interaction with Anillin, itself a Rho-dependent protein (Hickson and O’Farrell, 2008b; Piekny and Glotzer, 2008; Sun et al., 2015), while the other requires the RBH domain of Sticky, presumably via a direct interaction with Rho1-GTP. Although these two inputs are experimentally separable, they function together as a unit with each playing essential roles in ensuring the error-free transition to a stable, mature midbody ring.
Interactions of Sticky with Rho
Mammalian Citron kinase was first identified through a yeast two-hybrid screen for RhoA-GTP binding proteins (Madaule et al., 1995, 1998). It was thus assumed that Citron kinase, and its Drosophila orthologue Sticky, would act as canonical Rho effectors, that is, direct Rho-GTP binding would drive its recruitment to the equatorial cortex and activate its kinase domain in much the same way as the related Rho-dependent kinase Rok is thought to be regulated (Di Cunto et al., 1998; Madaule et al., 1998; Madaule et al., 2000; Eda et al., 2001; Shandala et al., 2004; Thumkeo et al., 2013). However, the assumption that Citron kinase/Sticky acts as a canonical Rho-effector at the midbody ring has been called into question on the basis of several observations, as discussed in D’Avino (2017). In particular, Gai et al. (2011) showed that inhibiting RhoA with cell-permeant C3 exoenzyme during late cytokinesis of HeLa cells led to a loss of Anillin from the intercellular bridge, but not a loss of Citron kinase, whereas depletion of Citron kinase led to a loss of both RhoA and Anillin. This was interpreted to signify that Citron kinase is upstream of RhoA, unlike Anillin, which is downstream of RhoA. Sticky was shown only to bind to Rho1 via its C-terminal CNH domain and independently of Rho1 nucleotide status (Bassi et al., 2011), while Sticky localization was shown to not require its putative RBD (Bassi et al., 2011). Indeed, the structure–function analysis of Bassi et al. (2011) failed to identify a role for the RBH domain of Sticky, but the only fragment examined (CC2b, residues 1228–1386) lacked essential residues for it to act as a functional RBD. Indeed, the mapping analysis presented here demonstrates that the minimal RBD rather resides within residues 1174–1370 (maxiCC2b). The present study also clearly shows that the RBD of Sticky is not only functional in that it contributes to Sticky recruitment to the contractile ring but also essential, since the Sticky-L1246N mutant predicted to no longer bind Rho-GTP is unable to form a stable midbody ring that can resist the shedding and disintegration that otherwise occurs. A similar conclusion was reached by Watanabe et al. (2013), who found that the coiled-coil domain of mammalian Citron kinase was largely able to rescue the loss of Citron kinase in a manner that was sensitive to the analogous mutation.
As for the reports of Rho1 interacting with the C-terminal CNH domain of Sticky (Shandala et al., 2004; Bassi et al., 2011), we failed to find any clear functional consequences during cytokinesis for such an interaction based on deletion of the CNH domain in depletion-and-rescue experiments. This argues that the CNH domain is dispensable for cytokinesis of S2 cells, although it may be important in other cell types.
A conserved Rho1/Anillin/Sticky complex is required for midbody ring formation
Bassi et al. (2011) identified residues 774–1227 (CC2a) as being sufficient for cortical recruitment of Sticky and showed that GST-CC2a pulled down actin and myosin from cell lysate. This led to the proposal that the CC2a region localized to the cortex through interactions with actin and myosin filaments. We have further mapped the minimal region required for cortical localization to residues 974–1128 (miniCC2a) and found that its recruitment was uniquely sensitive to Anillin depletion, with persistent recruitment observed on loss of F-actin (LatA treatment) or myosin II (Rok or MHC/Zipper depletion). We show that the NTD of Anillin is necessary for the cortical recruitment of Sticky-miniCC2a, in the presence or absence of F-actin (Figure 6), and both necessary and sufficient for the interaction between Anillin and Sticky-miniCC2a (Figure 7). Accordingly, we define the miniCC2a region (residues 974–1128) as the Anillin-binding domain (ABD) of Sticky, and conclude that the recruitment of Sticky via this ABD occurs through interaction with the Anillin NTD, independently of actomyosin. Coimmunoprecipitation of Anillin and Citron kinase from HeLa cells has also been reported (Gai et al., 2011). Despite poor sequence conservation from flies to mammals within the Anillin NTD, truncation analysis also strongly implicated the mammalian Anillin NTD in the interaction. While the interacting region of Citron kinase mapped to more than one region within the C-terminus (Gai et al., 2011), one of these regions (comprising residues 955–1388 of murine Citron kinase) contains the sequence most homologous to Sticky-miniCC2a. Thus, an Anillin-Citron kinase interaction appears to be evolutionarily conserved from flies to mammals. The Anillin-Sticky interaction described here may be direct or indirect, but regardless of the nature of this interaction, essential contributions from Rok, myosin II, F-actin, kinesin-6/Pavarotti, KIF14/Nebbish can be excluded on the basis of persistent colocalization of Anillin and the ABD of Sticky (miniCC2a) following specific depletions and LatA treatment.
Regarding the importance of the Anillin-Sticky interaction to midbody ring formation, we previously showed that depletion of Sticky leads to a complete loss of Anillin at the normal time of midbody ring formation, through mechanisms that include septin-dependent membrane shedding (El Amine et al., 2013). This indicated that Sticky is required to retain Anillin at the mature midbody ring and counteract septin-dependent removal mechanisms. We therefore hypothesized that disrupting the interaction between the Anillin NTD and the Sticky miniCC2a region might lead to a phenotype similar to that observed following Sticky depletion (El Amine et al., 2013). However, in cells expressing endogenous Sticky, deletion of the Anillin NTD, while blocking cytokinesis, often did not result in complete loss of Anillin via shedding (Figure 8, C and D). Shedding of Anillin-∆NTD was frequently observed, but some Anillin-∆NTD was nonetheless still retained in internal structures after furrow regression (Figure 8D). These were not persistent midbody ring-like structures like those observed on Anillin-∆C-terminus expression (Kechad et al., 2012; El Amine et al., 2013), but they appeared rather transient and disintegrated over time. However, their existence nevertheless suggests that the interaction between Sticky and Anillin NTD is not the only mechanism through which Anillin is retained during midbody ring formation. Further studies will be required to define these additional mechanisms.
Rho-dependent control of midbody ring formation
To summarize our findings, we propose a model (Figure 9E and Supplemental Figure S5) in which Pbl-activated Rho1 directly recruits Anillin via its C-terminus, as shown previously (Hickson and O’Farrell, 2008b; Piekny and Glotzer, 2008; Sun et al., 2015), and Sticky via its RBD. At the same time, Anillin and Sticky also promote each other’s recruitment via an interaction between their NTD and Anillin-binding domain (ABD, that is, miniCC2a), respectively. That this mutual support works both ways is evident from the observations that Anillin can recruit Sticky-miniCC2a and Sticky-L1246N (this study), while Sticky can recruit Anillin-∆C-terminus (El Amine et al., 2013) and Anillin-NTD (this study). In addition to these two Rho-dependent inputs providing partially redundant contributions to the recruitment of Sticky to the nascent midbody ring, they also appear to be essential for Sticky (and therefore Anillin) retention at the mature midbody ring. This is particularly clear for the RBD input, given the observed premature loss of Sticky-L1246N. The two Rho-dependent inputs that we have described, and their shifting importance from recruitment to retention during the transition from contractile ring to midbody ring, can help reconcile the confusion in the literature regarding the relationship between Sticky and Rho. According to the proposed model, Sticky can be considered as both “downstream” of Rho during its recruitment to the contractile ring/ nascent midbody ring and “upstream” of Rho during its retention at the mature midbody ring. However, in both cases interaction with Rho-GTP (either directly or indirectly via Anillin) is presumably required, so it does not seem unreasonable to consider Sticky as a conventional Rho effector protein that acts downstream of Rho activation.
In considering the overall role of Sticky, it is important to note that the N-terminal portion of the coiled-coil domain (CC1) is also likely essential for midbody ring formation. Constructs expressing the CC1 domain alone localize to the microtubules at the center of the midbody (Bassi et al., 2011, 2013, and our unpublished observations). Furthermore, the CC1 domain can bind to microtubule-associated proteins and motors (Bassi et al., 2013; Watanabe et al., 2013), suggesting that it links midbody ring-localized Sticky to the underlying microtubules of the midbody. It will therefore be important in future work to determine how the CC1-dependent activities of Citron kinase are coordinated with the Rho-dependent activities described here to promote formation of a stable midbody ring.
In summary, we have defined two inputs into the cortical recruitment of Sticky, both dependent on Rho and one via Anillin. These inputs also appear to be crucial for stable midbody ring formation. Overall, the results provide strong indications that Rho, the master regulator of the actomyosin contractile ring, also controls biogenesis of the subsequent midbody ring through its coordinated actions on Citron kinase and Anillin.
MATERIALS AND METHODS
Molecular biology and cloning
All constructs, except for Sticky-∆miniCC2a-L1246N, were generated by PCR amplification of the ORF, without stop codons followed by TOPO-cloning into pENTR-D-TOPO (Invitrogen, #K-2400) and then by Gateway recombination using LR Clonase into appropriate destination vectors (Drosophila Gateway Vector Collection; T. Murphy, Carnegie Institution for Science, Washington, DC). The sticky (CG10522) ORF was PCR amplified from clone RE26327 (Drosophila Gene Collection release 2 collection; Drosophila Genomics Resource Center). For all cloning into Gateway destination vectors, initial PCR amplification was carried out with respective forward primers that included the 5′ CACCATG sequence (Table 1). The Sticky-L1246N point mutant was generated by site-directed mutagenesis (see Table 1). Sticky-∆RBH lacks the 29 amino acids (residues ∆1235–1263) that form the conserved RBH domain, and Sticky-∆RBH lacks the last C-terminal 404 amino acids (residues 1–1450). Sticky-CC2a encodes amino acids 774–1228, Sticky-CC2a-CC2b encodes amino acids 774–1370, and Sticky-miniCC2a encodes amino acids 974–1128. Sticky-∆miniCC2a encodes amino acids 1–973 and 1129–1854 and therefore lacks amino acids 974–1128 and was generated by PCR by overlap extension followed by cloning into pENTR-D-TOPO and subsequent recombination into metallothionein promoter (pMT)-WG. Sticky-∆miniCC2a-L1246N was generated by first digesting Sticky-L1246N-pENTR and Sticky-∆miniCC2a-pMT plasmids with MfeI and BsaAI restriction enzymes (New England Biolabs, #R0589 and #R0531) for 2 h at 37°C followed by treatment of the Sticky-L1246N-pENTR backbone with 1 µl of calf alkaline phosphatase for 30 min at 37°C and gel extraction and purification of digested plasmids. The digested plasmid backbone and insert were ligated at 16°C overnight in a ratio of 1:5 followed by transformation into competent E. coli cells (DH5α strain) and screening for positive colonies. The remaining Sticky constructs have encoded amino acids indicated in the relevant figure illustrations. All Anillin constructs derive from clone LD23793 (Berkeley Drosophila Genome Project Gold collection; Drosophila Genomics Resource Center) that encodes the CG2092-RB polypeptide. Anillin full length, Anillin-∆MyoBD, and Anillin-∆ActBD have been described previously (El Amine et al., 2013). Anillin-∆NTD encodes amino acids 148–1239 and lacks the first 147 amino acids ,while Anillin-NTD encodes the first 147 amino acids only. All constructs were verified by sequencing.
TABLE 1:
Primers.
| Sticky-455 F: 5′-CACCATGATTTCCGCTACCACCGATGAA-3′ |
| Sticky-774 F: 5′-CACCATGCCTGGATCTTTGACCGAACTG-3′ |
| Sticky-783 R: 5′-AATGGCATTCAGTTCGGTCAA-3′ |
| Sticky-1229 F: 5′-CACCATGTACGTGCAGCGGGACATTAAA-3′ |
| Sticky-1228 R: 5′-GAACTGCTCCTTCTCGGCCAA-3′ |
| Sticky-1370 R: 5′-CGTCGTCTTCAGCTCCACCTG-3′ |
| Sticky-1450 R: 5′-GCTAAGATCATCGGCTGACGG-3′ |
| Sticky-824 F: 5′-CACCATGGAGCAGAGTCTTTCACCCACG-3′ |
| Sticky-874 F: 5′-CACCATGACAGCGAATCTATCGCTCTGG-3′ |
| Sticky-924 F: 5′-CACCATGTCACAGGAGGAAACTCGCCAG-3′ |
| Sticky-974 F: 5′-CACCATGTTGGCCAATGTGCACAGATTA-3′ |
| Sticky-1024 F: 5′-CACCATGGACTCTTGTTTGGTCTTACAG-3′ |
| Sticky-1074 F: 5′-CACCATGCAACTTGATACCCTTCATGAG-3′ |
| Sticky-1124 F: 5′-CACCATGCTCAAGGAGCAGCAGAAGAAG-3′ |
| Sticky-1174 F: 5′-CACCATGGTTAGTTTGAAGGAGGAAAAT-3′ |
| Sticky-828 R: 5′-TGAAAGACTCTGCTCGTTGAA-3′ |
| Sticky-878 R: 5′-CGATAGATTCGCTGTACGCGC-3′ |
| Sticky-928 R: 5′-AGTTTCCTCCTGTGACGTCTT-3′ |
| Sticky-978 R: 5′-GTGCACATTGGCCAAGTGCTC-3′ |
| Sticky-1028 R: 5′-GACCAAACAAGAGTCATTCGC-3′ |
| Sticky-1078 R: 5′-AAATCTCAACTTGATACCCTT-3′ |
| Sticky-1128 R: 5′-CTGCTGCTCCTTGAGATTGAG-3′ |
| Sticky-1178 R: 5′-CTCCTTCAAACTAACCATTTC-3′ |
| Sticky -L1246N F: 5′-GCGCAGCACAAAAAGaacATTGACTACCTTCAG-3′ |
| Sticky-L1246N R: 5′-CTGAAGGTAGTCAATgttCTTTTTGTGCTGCGC-3′ |
| Sticky-∆miniCC2a F: 5′-ATAATCGAGCACAAGAAGCTGGTGGCGCAGCAG-3′ |
| Sticky-∆miniCC2a R: 5′-CACCAGCTTCTTGTGCTCGATTATCTCCGACTT-3′ |
| Sticky-∆RBH F: 5′-CAGCGGGACATTACATTGGCGGACAAACTTTTC-3′ |
| Sticky-∆RBH R: 5′-GTCCGCCAATGTAATGTCCCGCTGCACGTAGAA-3′ |
| Sticky-∆CNH F: 5′-CACCATGCCACCCAAGATGGAGCCG-3′ |
| Sticky-∆CNH R: 5′-GCTAAGATCATCGGCTGACGG-3′ |
| Primers used to generate DNA templates to produce dsRNA. Note that the 5′ 8–base pair sequence GGGCGGGT is an anchor sequence allowing for a second PCR amplification using a universal T7 primer, sequence 5′-TAATACGACTCACTATAGGGAGACCACGGGCGGGT-3′: |
| lacI dsRNA F: 5′-GGGCGGGTTGGTGGTGTCGATGGTAGAA-3′ |
| lacI dsRNA R: 5′-GGGCGGGTCGGTATCGTCGTATCCCACT-3′ |
| pbl dsRNA F: 5′-GGGCGGGTATGGAAATGGAGACCATTGAA-3′ |
| pbl dsRNA R: 5′-GGGCGGGTCAGATAGTCCATCTCATTGGC-3′ |
| sti dsRNA1 F: 5′-GGGCGGGTGTGAAACCGTTGGTGATATGC-3′ |
| sti dsRNA1 R: 5′-GGGCGGGTTTCAACCTCTGGAAGTTATCG-3′ |
| sti dsRNA2 F: 5′-GGGCGGGTATGGAGCCGATTAGCGTGCGC-3′ |
| sti dsRNA2 R: 5′-GGGCGGGTCTGTGAGCTCGGCCAGATAGA-3′ |
| sti dsRNA3 F: 5′-GGGCGGGTGTCACATCCATATTCTTTACG-3′ |
| sti dsRNA3 R: 5′-GGGCGGGTCGAGCCCAAAGTCACAGAAGT-3′ |
| sti dsRNA 3′UTR F: 5′-GGGCGGGTGCATATAGATGAAGGATATAT-3′ |
| sti dsRNA 3′UTR R: 5′-GGGCGGGTTTTAAAATATTATGCGACTTC-3′ |
| anil dsRNA1 F: 5′-GGGCGGGTTAGAAATCTATGGCATGTTGGC-3′ |
| anil dsRNA1 R: 5′-GGGCGGGTGAGAAAACTGTTAACAACCCGC-3′ |
| anil dsRNA2 F: 5′-GGGCGGGTATGGACCCGTTTACTCAGC-3′ |
| anil dsRNA2 R: 5′-GGGCGGGTTGGTTCCGCCTCCAGCAGGG-3′ |
| anil dsRNA3 F: 5′-GGGCGGGTATGGACCCGTTTACTCAGC-3′ |
| anil dsRNA3 R: 5′-GGGCGGGTTCGACTGGACAAATGCGGTTTC-3′ |
| anil dsRNA 3′UTR F: 5′-GGGCGGGTGGAACCACCCACTGACCCGCT-3′ |
| anil dsRNA 3′UTR R: 5′-GGGCGGGTGCGAGTCATCCTAAATTAAATG-3′ |
| rok dsRNA F: 5′-GGGCGGGTGAACGCCAACCGATCCGATGC-3′ |
| rok dsRNA R: 5′-GGGCGGGTGATACTCGCTTCTGGTACACG-3′ |
| zipper dsRNA F: 5′-GGGCGGGTCCTAAAGCCACTGACAAGACG-3′ |
| zipper dsRNA R: 5′-GGGCGGGTCGGTACAAGTTCGAGTCAAGC-3′ |
| pavarotti dsRNA F: 5′-GGGCGGGTAAATCCGTAACGAAACTAACCG-3′ |
| pavarotti dsRNA R: 5′-GGGCGGGTACAACTGCTCTTGGCAGATACC-3′ |
| nebbish dsRNA F: 5′-GGGCGGGTAGCAACTTCGCTCCGAATTGG-3′ |
| nebbish dsRNA R: 5′-GGGCGGGTCAGGAGACCAAATTCATCTGG-3′ |
| T7 anchor primer :5′-TAATACGACTCACTATAGGGAGACCACGGGCGGGT-3′ |
Cell culture and RNAi
Wild-type Drosophila S2 cells (University of California, San Francisco, origin) were grown in Schneider’s medium (Invitrogen/Thermo Fisher Scientific, #21720001) supplemented with 10% (vol/vol) heat-inactivated fetal bovine serum (FBS; U.S. origin, Invitrogen/Thermo Fisher Scientific, #16140071), 50,000 U of penicillin, and 50 mg/ml Streptomycin at 25°C in ambient CO2. To generate stable cell lines, wild-type S2 cells were seeded at a density of 7.5 × 105 cells/ml and incubated at 25°C overnight before being cotransfected with 1.0 µg of each respective plasmid and 0.25 µg of pCoHygro (Invitrogen) using FuGENE HD (Promega, #E2311) transfection reagent. Starting from 48 h after transfection, resistant clones were selected in 0.4 mg/ml Hygromycin B (Invitrogen, #10687010) for 4 wk. After selection, cells were grown without Hygromycin B in medium supplemented with 10% FBS for at least 1 wk prior to use. For transient DNA transfections, wild-type S2 cells were plated at a density of 1.0 × 105 cells/ml a day prior to transfection. Each relevant plasmid (1.0 µg) was transfected using FuGENE HD transfection reagent and per reaction, a 3:1 ratio of FuGENE HD:DNA was diluted in serum-free Schneider’s medium in a total volume of 100 μl and incubated at room temperature for 20 min prior to addition to the cells in normal growth medium.
RNAi in S2 cells was performed using long (250–700 base pairs) dsRNA. DNA templates were generated in a two-step PCR amplification from cDNA. In the first round of PCR, gene-specific primers (Table 1) were used that included a 5′ 8–base pair anchor sequence (GGGCGGGT). In a second round PCR, a universal T7 primer containing the anchor (5′-TAATACGACTCACTATAGGGAGACCACGGGCGGGT-3′) was used to generate gene-specific PCR templates that included the T7 promoter sequence at each 5′ end. RNAs were subsequently generated by in vitro transcription overnight at 37°C using RiboMAX transcription kits (Promega). After RQ1 DNase digestion of the template cDNA, the resulting RNAs were then precipitated in ethanol, resuspended in nuclease-free water, denatured at 95°C, and slowly cooled to room temperature to allow the RNA to anneal and form dsRNAs, which were further verified by agarose gel electrophoresis and quantified.
For RNAi experiments, S2 cells were plated in plastic, flat-bottomed 96-well dishes (Falcon) and incubated with 1 µg/ml relevant dsRNA for 3–6 d (indicated in the figure legends). For analyses after 6-d RNAi, cells were split one in three into fresh complete medium 3 d after (the first round of dsRNA treatment) followed by the addition of 1µg/ml fresh dsRNA and incubation for a further 3 d. RNAi experiments in stable cell lines were only carried out after the 4-wk selection period, and in the case of transient transfections, dsRNA incubation was initiated 2 d after transfection. Expression of relevant constructs under control of the pMT plasmids was induced by the addition of 0.1 mM CuSO4 24 h prior to imaging, or 20 min after the addition of dsRNA, in the case of rescue experiments. One hour prior to live imaging, cells were transferred into 200 µl of fresh medium in an eight-well chamber slide (Nunc Lab-Tek II, Thermo Fisher Scientific). Live imaging was carried out after 3 d for sticky, anillin, pbl, pavarotti, and nebbish RNAi and after 7 d for zipper and rok RNAi. In the case of codepletion experiments, these incubation times were maintained, but individual dsRNA incubations in the same experiment were performed in the presence of the lacI dsRNA to control for potential effects of combining dsRNAs. For LatA experiments, LatA (Calbiochem/EMD Millipore) was gently pipetted to the cells at a final concentration of 1 µg/ml immediately prior to the start of image acquisition.
Live-cell microscopy
Time-lapse imaging of Drosophila S2 cells in Schneider’s medium was performed at room temperature using a spinning-disk confocal system (UltraVIEW Vox; Perkin Elmer-Cetus) comprising a scanning unit (CSU-X1; Yokogawa America), a charge-coupled device camera (ORCA-R2; Hamamatsu Photonics), fitted to an inverted microscope (DMI6000 B; Leica Microsystems), and equipped with a motorized piezoelectric stage (Applied Scientific Instrumentation). Image acquisition was controlled using Volocity version 6.3 (Perkin Elmer-Cetus) and was performed in emission discrimination mode. Low-resolution time-lapse imaging was performed using a Plan Apochromat 40 × 0.85 NA air objective with camera binning set to 2 × 2, and 10-µm Z-stacks were acquired with an optical spacing of 1 µm at 4- to 5-min intervals overnight. High-resolution imaging was performed using a Plan Apochromat 63 × oil immersion objectives NA 1.4 with camera binning set to 2 × 2, and 10-µm Z-stacks were acquired with an optical spacing of 0.5 µm at 1- to 2-min intervals, unless otherwise stated in the figure legends.
Immunofluorescence microscopy
Cells expressing Sticky-CC2a-GFP and mCherry-Tubulin were transferred to a 96-well glass-bottomed plate (Whatman) 2 h before fixation for 5 min in 4% formaldehyde in phosphate-buffered saline (PBS). After permeabilization and blocking for 1 h in PBS containing 0.1% Triton X-100 (PTX buffer) and 5% normal goat serum, cells were incubated with primary antibody at 4°C overnight (concentrated mouse anti-Rho1 mAb p1D9 used at 1:1000), washed with PTX buffer, and incubated for 1 h with Alexa Fluor 647–conjugated goat anti-mouse antibody (1:500; Molecular Probes) and Hoechst 33258 (1:1000). Cells were washed in PTX and mounted in Fluoromount-G (SouthernBiotech). Images were acquired using the UltraVIEW Vox system described in the preceding section using a Plan Apochromat 63 × 1.4 NA oil immersion objective without camera binning and in emission discrimination mode.
Image processing and statistical analysis
At least three independent repeats were carried out for each experiment (unless otherwise indicated in the figure legends). Statistical analyses were conducted comparing independent experiments with use of GraphPad Prism 6-7 software (GraphPad Software). Gaussian distribution of data was assessed using the D’Agostino-Pearson test. Means were compared using an unpaired Student’s t test to analyze data with a normal distribution and medians were compared using the Mann-Whitney U test to analyze data that were not normally distributed. The total number of cells quantified (N value), p values, and significance level are indicated in the respective figure legend and in the main text. Fluorescence intensity measurements at the nascent MR were carried out at 20 min postanaphase onset for all cells and all conditions (construct and dsRNA treatment) by drawing a fixed-size ROI around the nascent MR. The average background fluorescence intensity in the green channel at that specific time-point was determined by recording intensity values (A.U.) within the fixed-size ROI placed at three random positions in the field of view, which excluded the cell being analyzed and also other neighboring cells. The background-subtracted fluorescence intensity value for each cell analyzed per condition is displayed on the scatter plot (related to Figures 8J and 9A). All measurements were made using Volocity Analysis 6.3. Images were processed for publication using Photoshop CS6 (Adobe) or ImageJ (National Institutes of Health) and assembled as figures using Illustrator CS6 (Adobe). Video files were exported from Volocity as QuickTime videos (Apple).
Recombinant protein expression
GST-tagged proteins were expressed in BL21 E. coli cells. A 5-ml culture of cells was grown overnight in LB with ampicillin at 37°C with shaking (220 rpm) and used to inoculate 500 ml of fresh LB+Amp, and cells were grown at 37°C with shaking until an optical density of 0.6 at A600nm. Recombinant protein expression was then induced by the addition of 0.15 mM of isopropyl b-d-1-thiogalactopyranoside and further incubation at 37°C for 4 h with shaking. Bacterial cells were harvested by centrifugation at 6000 × g at 4°C followed by resuspension of the pellet in lysis buffer (20 mM HEPES, pH 7.4, 0.2 M NaCl, 1 mM EDTA, 5 mM MgCl2, 0.1% Triton X-100, 1 mM dithiothreitol, 1 mg/ml lysozyme, 0.2 mg/ml Pefabloc), supplemented with complete protease inhibitors (Roche), and lysed at 4°C on ice for 1.5 h (with intermittent resuspension). Bacterial lysates were then sonicated on ice for 5–10 min using a cycle alternating between 30 s of pulse and 30 s of rest at 50% amplitude. Sonicated lysate was cleared by centrifugation at 12,000 × g for 20 min at 4°C and then applied to prewashed glutathione sepharose beads (GE healthcare) overnight at 4°C with rotation. GST-tagged proteins bound to beads were separated from the unbound fraction by centrifugation at 4500 rpm at 4°C for 15 min followed by three washes in HMNT lysis buffer (20 mM HEPES, pH 7.4, 10 mM MgCl2, 0.1 M NaCl, 0.5% Triton X-100).
Whole cell extract preparation
Wild-type S2 cells or cells expressing the GFP-tagged protein of interest were cultured in T75 culture flasks for 7 d and induced with 0.1 mM of CuSO4 48 h before harvesting by centrifugation at 1500 rpm for 5 min. Cell pellets were then washed twice in ice-cold PBS prior to lysis in HMNT buffer (500 µl/T75 flask) (20 mM HEPES, pH 7.4, 10 mM MgCl2, 0.1 M NaCl, 0.5% Triton X-100 and supplemented with a protease inhibitor tablet) by incubating on ice for 45 min with vortexing every 5 min. Extracts were clarified by centrifugation at 13,200 rpm for 20 min at 4°C in a microfuge. For protein knockdown experiments, wild-type S2 cells were grown in eight wells of a 96-well plate per condition and treated with relevant dsRNA for 3 d before pooling the contents of the eight wells and harvesting by centrifugation at 1500 rpm for 5 min and extracted in HMNT buffer (100 µl/8 wells) as above. The concentration of protein on beads and whole cell extracts were measured using the BCA assay (Pierce BCA protein assay kit; Thermo Fisher Scientific).
GST pull-down assays
The S2 whole cell extract (500 µl at 3 mg/ml per pull down) was cleared by two rounds of ultracentrifugation at 55,000 rpm for 15 min at 4°C followed by incubation with GST-tagged bait protein, or GST alone, bound to glutathione-Sepharose beads (500 µg/pull down) overnight at 4°C on a rotating platform. After binding, beads were centrifuged at 5000 rpm for 5 min at 4°C to collect the unbound fraction. Beads were then washed three times with cold HMNT, and the bound fraction was eluted by incubating the beads in 2× SDS sample buffer, boiling at 100°C for 5 min and centrifugation at 13,200 rpm for 5 min.
Immunoblotting
Equal amounts of protein from different samples were subjected to SDS–PAGE (6–10% polyacrylamide gels) and transferred to Amersham Protran nitrocellulose membrane (0.45 µm pore size) using constant current (300 mA) for 2–3 h. Membranes were stained with Ponceau S, digitally scanned, then blocked with 5% milk in PBS containing 0.15% Tween-20 (PBS-T; blocking solution) for 1 h at room temperature. Where appropriate, membranes were cut at the relevant molecular weight marker and incubated in blocking solution containing primary antibody overnight at 4°C on a rocking platform. Membranes were washed three times for 5 min in PBS-T and incubated in horseradish peroxidase–conjugated donkey anti-rabbit, anti-mouse secondary antibody (1:4000; GE Healthcare), or alpaca anti-chicken (1:4000; Immune Biosolutions) in blocking solution for 1 h at room temperature on a rocking platform. Membranes were then washed three times in PBS-T followed by a final wash in PBS before signal development in Clarity Western ECL substrate (Bio-Rad laboratories) and visualization on a ChemiDoc MP imager.
Antibodies
Polyclonal antibodies against Drosophila melanogaster Anillin were generated by Immune Biosolutions (Sherbrooke, Quebec, Canada) in chickens against amino acids 299–802 of Anillin fused to GST followed by two rounds of affinity purification of the antibodies, first against the immunogen and then against an N-terminal GST tagged- and C-terminal His tagged-Anillin fusion protein (GST-Anillin257–298-His) to remove anti-GST antibodies. The purified antibody was used for immunoblotting at 1:2500 and for validation of the Anillin dsRNAs in Supplemental Figure S4. Other polyclonal antibodies used include: rabbit anti-Sticky (used for immunoblotting at 1:3000) raised in rabbits against a fragment of the Sticky protein (amino acids 531–742) fused to a C-terminal 6xHis tag (Paolo D’Avino, University of Cambridge, Cambridge, UK; D’Avino et al., 2004) and rabbit anti-GFP (used for immunoblotting at 1:3000; A-6455, Invitrogen). Monoclonal antibodies used include mouse anti-tubulin antibody (used for immunoblotting at 1:2500; Clone DM1A, Sigma-Aldrich) and concentrated mouse anti-Rho1 antibody (used for immunofluorescence at 1:1000; clone p1D9, Developmental Studies Hybridoma Bank).
Supplementary Material
Acknowledgments
We thank past and present members of the Hickson lab for many useful discussions and Jean-Claude Labbé for critical reading of the manuscript. We thank Silvana Jananji for purifying the proteins used to generate the Anillin antibody. We thank Paolo D’Avino for providing the rabbit anti-Sticky antibody and Emile Levy for access to a ChemiDoc system. N.E-a. thanks the Fondation CHU Sainte-Justine and the Département Pathologie et Biologie Cellulaire, Université de Montréal, for graduate studentships. G.R.X.H. thanks the Fonds de Recherche du Québec, Santé (FRQS), for research scholarships. This work was supported by grants from Canadian Institutes for Health Research (CIHR) (MOP-97788), the Natural Sciences and Engineering Research Council (NSERC) (RGPIN-2014-05083), the Canadian Fund for Innovation (CFI), and the Cole Foundation to G.R.X.H.
Abbreviations used:
- CNH
Citron-Nik1 homology
- pbl
pebble
- PBS
phosphate-buffered saline
- pMT
metallothionein promoter.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E19-04-0194) on June 5, 2019.
REFERENCES
- Atilla-Gokcumen GE, Muro E, Relat-Goberna J, Sasse S, Bedigian A, Coughlin ML, Garcia-Manyes S, Eggert US. (2014). Dividing cells regulate their lipid composition and localization. Cell , 428–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayscough K. (1998). Use of latrunculin-A, an actin monomer-binding drug. Method Enzymol , 18–25. [DOI] [PubMed] [Google Scholar]
- Ayscough KR, Stryker J, Pokala N, Sanders M, Crews P, Drubin DG. (1997). High rates of actin filament turnover in budding yeast and roles for actin in establishment and maintenance of cell polarity revealed using the actin inhibitor latrunculin-A. J Cell Biol , 399–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basant A, Glotzer M. (2018). Spatiotemporal regulation of RhoA during cytokinesis. Curr Biol , R570–R580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassi ZI, Audusseau M, Riparbelli MG, Callaini G, D’Avino PP. (2013). Citron kinase controls a molecular network required for midbody formation in cytokinesis. Proc Natl Acad Sci USA , 9782–9787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassi ZI, Verbrugghe KJ, Capalbo L, Gregory S, Montembault E, Glover DM, D’Avino PP. (2011). Sticky/Citron kinase maintains proper RhoA localization at the cleavage site during cytokinesis. J Cell Biol , 595–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bement WM, Benink HA, von Dassow G. (2005). A microtubule-dependent zone of active RhoA during cleavage plane specification. J Cell Biol , 91–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho A, Desai A, Oegema K. (2009). Structural memory in the contractile ring makes the duration of cytokinesis independent of cell size. Cell , 926–937. [DOI] [PubMed] [Google Scholar]
- Coue M, Brenner SL, Spector I, Korn ED. (1987). Inhibition of actin polymerization by latrunculin A. FEBS Lett , 316–318. [DOI] [PubMed] [Google Scholar]
- D’Avino PP. (2017). Citron kinase—renaissance of a neglected mitotic kinase. J Cell Sci , 1701–1708. [DOI] [PubMed] [Google Scholar]
- D’Avino PP, Giansanti MG, Petronczki M. (2015). Cytokinesis in animal cells. Cold Spring Harb Perspect Biol , a015834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Avino PP, Savoian MS, Glover DM. (2004). Mutations in sticky lead to defective organization of the contractile ring during cytokinesis and are enhanced by Rho and suppressed by Rac. J Cell Biol , 61–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean SO, Spudich JA. (2006). Rho kinase’s role in myosin recruitment to the equatorial cortex of mitotic Drosophila S2 cells is for myosin regulatory light chain phosphorylation. Plos One , e131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dema A, Macaluso F, Sgro F, Berto GE, Bianchi FT, Chiotto AA, Pallavicini G, Di Cunto F, Gai M. (2018). Citron kinase-dependent F-actin maintenance at midbody secondary ingression sites mediates abscission. J Cell Sci , jcs209080. [DOI] [PubMed] [Google Scholar]
- Di Cunto F, Calautti E, Hsiao J, Ong L, Topley G, Turco E, Dotto GP. (1998). Citron rho-interacting kinase, a novel tissue-specific ser/thr kinase encompassing the Rho-Rac-binding protein Citron. J Biol Chem , 29706–29711. [DOI] [PubMed] [Google Scholar]
- Dvorsky R, Blumenstein L, Vetter IR, Ahmadian MR. (2004). Structural insights into the interaction of ROCKI with the switch regions of RhoA. J Biol Chem , 7098–7104. [DOI] [PubMed] [Google Scholar]
- Echard A, Hickson GRX, Foley E, O’Farrell PH. (2004). Terminal cytokinesis events uncovered after an RNAi screen. Curr Biol , 1685–1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eda M, Yonemura S, Kato T, Watanabe N, Ishizaki T, Madaule P, Narumiya S. (2001). Rho-dependent transfer of Citron-kinase to the cleavage furrow of dividing cells. J Cell Sci , 3273–3284. [DOI] [PubMed] [Google Scholar]
- El Amine N, Kechad A, Jananji S, Hickson GR. (2013). Opposing actions of septins and Sticky on Anillin promote the transition from contractile to midbody ring. J Cell Biol , 487–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fededa JP, Gerlich DW. (2012). Molecular control of animal cell cytokinesis. Nat Cell Biol , 440–447. [DOI] [PubMed] [Google Scholar]
- Gai M, Camera P, Dema A, Bianchi F, Berto G, Scarpa E, Germena G, Di Cunto F. (2011). Citron kinase controls abscission through RhoA and anillin. Mol Biol Cell , 3768–3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glotzer M. (2017). Cytokinesis in metazoa and fungi. Cold Spring Harb Perspect Biol , a022342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RA, Paluch E, Oegema K. (2012). Cytokinesis in animal cells. Annu Rev Cell Dev Biol , 29–58. [DOI] [PubMed] [Google Scholar]
- Gruneberg U, Neef R, Li X, Chan EH, Chalamalasetty RB, Nigg EA, Barr FA. (2006). KIF14 and citron kinase act together to promote efficient cytokinesis. J Cell Biol , 363–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickson GR, Echard A, O’Farrell PH. (2006). Rho-kinase controls cell shape changes during cytokinesis. Curr Biol , 359–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickson GR, O’Farrell PH. (2008a). Anillin: a pivotal organizer of the cytokinetic machinery. Biochem Soc Trans , 439–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickson GR, O’Farrell PH. (2008b). Rho-dependent control of anillin behavior during cytokinesis. J Cell Biol , 285–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu CK, Coughlin M, Mitchison TJ. (2012). Midbody assembly and its regulation during cytokinesis. Mol Biol Cell , 1024–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungas T, Perchey RT, Fawal M, Callot C, Froment C, Burlet-Schiltz O, Besson A, Davy A. (2016). Eph-mediated tyrosine phosphorylation of citron kinase controls abscission. J Cell Biol , 555–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kechad A, Jananji S, Ruella Y, Hickson GRX. (2012). Anillin acts as a bifunctional linker coordinating midbody ring biogenesis during cytokinesis. Curr Biol , 197–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madaule P, Eda M, Watanabe N, Fujisawa K, Matsuoka T, Bito H, Ishizaki T, Narumiya S. (1998). Role of citron kinase as a target of the small GTPase Rho in cytokinesis. Nature , 491–494. [DOI] [PubMed] [Google Scholar]
- Madaule P, Furuyashiki T, Eda M, Bito H, Ishizaki T, Narumiya S. (2000). Citron, a Rho target that affects contractility during cytokinesis. Microsc Res Techniq , 123–126. [DOI] [PubMed] [Google Scholar]
- Madaule P, Furuyashiki T, Reid T, Ishizaki T, Watanabe G, Morii N, Narumiya S. (1995). A novel partner for the GTP-bound forms of rho and rac. FEBS Lett , 243–248. [DOI] [PubMed] [Google Scholar]
- McKenzie C, Bassi ZI, Debski J, Gottardo M, Callaini G, Dadlez M, D’Avino PP. (2016). Cross-regulation between Aurora B and Citron kinase controls midbody architecture in cytokinesis. Open Biology , 160019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins JM, Biesele JJ. (1977). Terminal phase of cytokinesis in D-98s cells. J Cell Biol , 672–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naim V, Imarisio S, Di Cunto F, Gatti M, Bonaccorsi S. (2004). Drosophila citron kinase is required for the final steps of cytokinesis. Mol Biol Cell , 5053–5063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann B, Walter T, Heriche JK, Bulkescher J, Erfle H, Conrad C, Rogers P, Poser I, Held M, Liebel U, et al. (2010). Phenotypic profiling of the human genome by time-lapse microscopy reveals cell division genes. Nature , 721–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piekny A, Werner M, Glotzer M. (2005). Cytokinesis: welcome to the Rho zone. Trends Cell Biol , 651–658. [DOI] [PubMed] [Google Scholar]
- Piekny AJ, Glotzer M. (2008). Anillin is a scaffold protein that links RhoA, actin, and myosin during cytokinesis. Curr Biol , 30–36. [DOI] [PubMed] [Google Scholar]
- Renshaw MJ, Liu J, Lavoie BD, Wilde A. (2014). Anillin-dependent organization of septin filaments promotes intercellular bridge elongation and Chmp4B targeting to the abscission site. Open Biol , 130190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder TE. (1972). The contractile ring. II. Determining its brief existence, volumetric changes, and vital role in cleaving Arbacia eggs. J Cell Biol , 419–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sgro F, Bianchi FT, Falcone M, Pallavicini G, Gai M, Chiotto AMA, Berto GE, Turco E, Chang YJ, Huttner WB, et al. (2016). Tissue-specific control of midbody microtubule stability by Citron kinase through modulation of TUBB3 phosphorylation. Cell Death Differ , 801–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shandala T, Gregory SL, Dalton HE, Smallhorn M, Saint R. (2004). Citron kinase is an essential effector of the Pbl-activated Rho signalling pathway in Drosophila melanogaster. Development , 5053–5063. [DOI] [PubMed] [Google Scholar]
- Shimizu T, Ihara K, Maesaki R, Amano M, Kaibuchi K, Hakoshima T. (2003). Parallel coiled-coil association of the RhoA-binding domain in Rho-kinase. J Biol Chem , 46046–46051. [DOI] [PubMed] [Google Scholar]
- Skop AR, Liu H, Yates J, 3rd, Meyer BJ, Heald R. (2004). Dissection of the mammalian midbody proteome reveals conserved cytokinesis mechanisms. Science , 61–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers WG, Saint R. (2003). A RhoGEF and Rho family GTPase-activating protein complex links the contractile ring to cortical microtubules at the onset of cytokinesis. Dev Cell , 29–39. [DOI] [PubMed] [Google Scholar]
- Sun LF, Guan RF, Lee IJ, Liu YJ, Chen MR, Wang JW, Wu JQ, Chen ZC. (2015). Mechanistic insights into the anchorage of the contractile ring by Anillin and Mid1. Dev Cell , 413–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry SJ, Dona F, Osenberg P, Carlton JG, Eggert US. (2018). Capping protein regulates actin dynamics during cytokinetic midbody maturation. Proc Natl Acad Sci USA , 2138–2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thumkeo D, Watanabe S, Narumiya S. (2013). Physiological roles of Rho and Rho effectors in mammals. Eur J Cell Biol , 303–315. [DOI] [PubMed] [Google Scholar]
- Watanabe S, De Zan T, Ishizaki T, Narumiya S. (2013). Citron kinase mediates transition from constriction to abscission through its coiled-coil domain. J Cell Sci , 1773–1784. [DOI] [PubMed] [Google Scholar]
- Yamashiro S, Totsukawa G, Yamakita Y, Sasaki Y, Madaule P, Ishizaki T, Narumiya S, Matsumura F. (2003). Citron kinase, a Rho-dependent kinase, induces di-phosphorylation of regulatory light chain of myosin II. Mol Biol Cell , 1745–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.