Abstract
Ded1 is a DEAD-box RNA helicase with essential roles in translation initiation. It binds to the eukaryotic initiation factor 4F (eIF4F) complex and promotes 48S preinitiation complex assembly and start-site scanning of 5′ untranslated regions of mRNAs. Most prior studies of Ded1 cellular function were conducted in steady-state conditions during nutrient-rich growth. In this work, however, we examine its role in the translational response during target of rapamycin (TOR)C1 inhibition and identify a novel function of Ded1 as a translation repressor. We show that C-terminal mutants of DED1 are defective in down-regulating translation following TORC1 inhibition using rapamycin. Furthermore, following TORC1 inhibition, eIF4G1 normally dissociates from translation complexes and is degraded, and this process is attenuated in mutant cells. Mapping of the functional requirements for Ded1 in this translational response indicates that Ded1 enzymatic activity and interaction with eIF4G1 are required, while homo-oligomerization may be dispensable. Our results are consistent with a model wherein Ded1 stalls translation and specifically removes eIF4G1 from translation preinitiation complexes, thus removing eIF4G1 from the translating mRNA pool and leading to the codegradation of both proteins. Shared features among DED1 orthologues suggest that this role is conserved and may be implicated in pathologies such as oncogenesis.
INTRODUCTION
Gene expression is a central process of the cell, and accordingly, intricate modes of regulation have evolved to ensure faithfulness in the process. In eukaryotic cells, many factors interact with specific mRNAs to facilitate and regulate a variety of processes, including nuclear export, translation, and degradation (Mitchell and Parker, 2014). The composition and structure of the mRNP are thus dynamic throughout its life cycle. A major method of controlling RNA–protein interactions is through the action of RNA helicases, of which DEAD-box proteins are the largest family (for a review, see Linder and Jankowsky, 2011). DEAD-box proteins utilize linked cycles of ATP binding, hydrolysis, and RNA binding to destabilize RNA duplexes, alter RNA–protein interactions, and/or act as ATP-dependent binding proteins.
Ded1 is a DEAD-box RNA helicase in Saccharomyces cerevisiae that has an essential role in translation initiation conserved from yeast to humans (Sharma and Jankowsky, 2014). The human orthologue, DDX3, has been implicated in oncogenesis, most notably in the pediatric brain cancer medulloblastoma, where DDX3 is the second most frequently mutated gene after β-catenin (Northcott et al., 2012; Kool et al., 2014). DDX3 mutations have also been identified in several other cancer types, and DDX3 has been linked to viral replication as well (Valiente-Echeverría et al., 2015; Zhao et al., 2016). However, its function in these pathologies has remained largely unclear, emphasizing the importance of understanding Ded1/DDX3 function and regulation.
Ded1 function in translation is complex. Ded1 activity is necessary for scanning through long and structured 5′ untranslated regions (UTRs) and for 48S preinitiation complex (PIC) assembly (Hilliker et al., 2011; Sen et al., 2015). In addition, our previous work suggests that Ded1 may have a role in translation start site fidelity, and a recent study has presented strong evidence for Ded1 in controlling the use of near-cognate start codons (Aryanpur et al., 2017; Guenther et al., 2018). On the other hand, a repressive function for Ded1 has also been proposed (Hilliker et al., 2011). Ded1 overexpression causes severe growth inhibition, translation repression, and the formation of stalled mRNPs in stress granules (Beckham et al., 2008; Hilliker et al., 2011; Aryanpur et al., 2017). Likewise, high Ded1 levels inhibit 48S PIC assembly in in vitro assays (Hilliker et al., 2011; Aryanpur et al., 2017). Nonetheless, it is currently unclear whether Ded1 has a repressive function in translation when expressed at physiological levels.
The repressive function of overexpressed Ded1 is partially dependent on the Ded1 C-terminal domain, which is a predicted low-complexity sequence that lies outside of the core helicase domains (Sharma and Jankowsky, 2014). Deletion of this domain (amino acids 536–604) substantially rescues growth inhibition on overexpression (Hilliker et al., 2011). Two functions have been described for the domain. First, in vitro assays have identified this domain as essential for Ded1 association with the initiation factor 4F (eIF4F) complex scaffolding protein, eukaryotic initiation factor 4G (eIF4G) (Hilliker et al., 2011; Senissar et al., 2014; Putnam et al., 2015). Second, Ded1 has been shown to homo-oligomerize through the C-terminal domain, which is important for maximal helicase activity in vitro (Putnam et al., 2015). Surprisingly, deletion of the Ded1 C-terminus has not been associated with any deficiencies in growth or translation in vivo. However, these studies have generally focused on favorable growth conditions in rich media rather than cellular stress conditions such as nutrient deprivation or stationary phase culture.
As part of the cellular stress response, new protein synthesis is appropriately regulated with growth rate and cell division to efficiently manage limited resources and to ensure survival (Loewith and Hall, 2011; González and Hall, 2017). A major pathway in eukaryotes involved in this regulation is the target of rapamycin (TOR) pathway, named after the discovery that mutations in TOR conferred resistance to the growth inhibitory effect of the small molecule rapamycin (González and Hall, 2017). TOR can form two functionally different signaling complexes with other proteins (TORC1 and TORC2); however, rapamycin specifically inhibits TORC1 (Loewith and Hall, 2011). When amino acids and nitrogen sources are abundant, TORC1 signaling stimulates cell division and anabolic processes such as new protein, nucleotide, and lipid biosynthesis. Conversely, when nutrients are scarce, a reduction in TORC1 signaling halts cell division and promotes catabolic processes and general inhibition of new protein synthesis. Notably however, while some stresses, such as glucose deprivation, induce sequestration of translation components into cytoplasmic foci termed stress granules (Buchan and Parker, 2009), others, including nitrogen starvation, amino acid starvation, and specific inhibition of TORC1 with rapamycin, result in repression of translation but do not drive stress granule assembly (Simpson and Ashe, 2012). This suggests that general inhibition of translation and stress granule formation can be uncoupled and can be driven by distinct signaling pathways (Mokas et al., 2009). TORC1 signaling has several downstream effects on translation initiation during stress. The most well-conserved mechanism occurs at the level of 48S complex assembly following phosphorylation of eukaryotic initiation factor 2 alpha (eIF2α), which is induced by a wide variety of stresses, including nutrient depletion, DNA damage, and viral infection (Dever et al., 1992; Pakos-Zebrucka et al., 2016; Ryoo and Vasudevan, 2017). In mammals, TORC1 inhibition also enhances the ability of eIF4E binding proteins to repress translation by binding to the cap-binding protein eIF4E, although this mechanism seems to differ in yeast (Raught et al., 2001; Loewith and Hall, 2011; Liu and Qian, 2014; Ryoo and Vasudevan, 2017).
In addition to these two mechanisms, TORC1 inhibition induces the degradation of some translation components by both autophagy and the proteasome (Kelly and Bedwell, 2015; Gonskikh and Polacek, 2017). For example, in S. cerevisiae, eIF4G1 is rapidly degraded by autophagy, whereas other factors such as Dcp2 are degraded via the proteasome (Berset et al., 1998; Kelly and Bedwell, 2015). Other translation factors such as eIF4A and eIF3 remain relatively stable following stress (Berset et al., 1998; Harris et al., 2006). The relatively rapid degradation of eIF4G1 compared with other initiation factors suggests that this may be a regulatory mechanism used to inhibit cap-dependent translation initiation, at least during sustained TORC1 down-regulation. Thus, the tuning of eIF4G1 levels may play an important role in reprogramming gene expression following stress (Rogers et al., 2011; Das and Das, 2016). However, few studies have explored how translation is affected by the degradation of initiation factors such as eIF4G1, and the mechanism by which this occurs and the factors involved are not well understood. Here we take advantage of the genetic tractability of S. cerevisiae to investigate the role of the Ded1 C-terminus in the translational response to TORC1 inhibition and show that Ded1 activity plays an important role in promoting translation repression and adaptation to stress conditions. To carry out this function, Ded1 remodels eIF4G1 in translation complexes and targets it for degradation.
RESULTS
Deletion of the Ded1 C-terminus confers rapamycin resistance
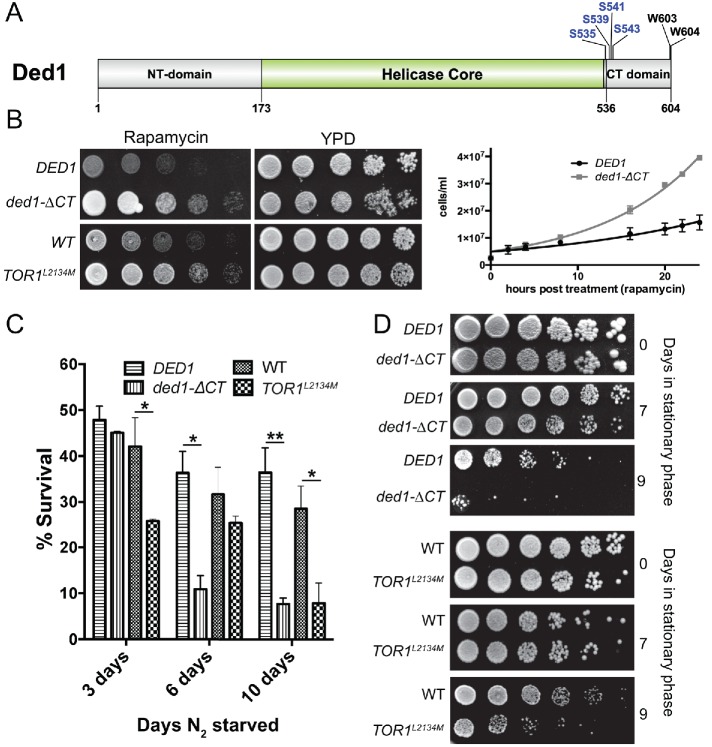
The C-terminal domain of Ded1 is a low complexity sequence that has not been extensively studied (Figure 1A). This domain is necessary for the interaction with eIF4G1 and for self-association and the formation of Ded1 oligomers (Hilliker et al., 2011; Putnam et al., 2015). Although in vitro studies have defined a framework for understanding the effects of these interactions on Ded1 biochemical activity (Putnam et al., 2015; Gao et al., 2016), little is known about how these interactions affect Ded1 cellular function, particularly under different cellular conditions. As previously reported (Hilliker et al., 2011), deletion of the C-terminal region (amino acids 536–604) of DED1 in cells had little effect on growth assays in rich media compared with a wild-type DED1 control (Figure 1B, YPD [yeast extract–peptone–dextrose media]). To test whether this region plays a role in responses to cell stress, we examined growth following TORC1 inhibition by treating cells with rapamycin, a specific inhibitor of TORC1 (Figure 1B). The wild-type DED1 cells showed severe growth inhibition in the presence of rapamycin, a well-known effect of TORC1 inactivation (Loewith and Hall, 2011). Surprisingly, however, deletion of the C-terminal tail of Ded1 (ded1-ΔCT) resulted in significant resistance to rapamycin, as observed via growth on plates and in liquid media (Figure 1B). This effect was comparable to the rapamycin-resistant growth of cells containing a TOR mutant, TOR1L2134M, which has a mutation in its kinase domain that results in a hyperactive enzyme (Takahara and Maeda, 2012) (Figure 1B). These results suggest that Ded1 and the Ded1 C-terminus have roles in suppressing growth in conditions where TORC1 signaling is inactivated and that mutation of this domain results in growth phenotypes similar to a hyperactive TOR mutant.
FIGURE 1:
Ded1 has a role in growth regulation and viability following cellular stress. (A) Ded1 domain structure. The carboxy-terminal domain (CT domain) lies outside of the helicase core domain. It contains TORC1-dependent phosphoserines. Two conserved tryptophans at the C-terminal tail are also shown. (B) Wild-type DED1, ded1-ΔCT, WT, and TOR1L2134M cells were serially diluted onto rich media ± rapamycin (200 ng/ml) and incubated at 30°C (left). Wild-type DED1 or ded1-ΔCT cells were grown in rich liquid media ± rapamycin (200 ng/ml) at 30°C, and cell concentration was measured via OD600 at different time points in a 24-h window. Data points represent the mean of four independent experiments and were fit to simple logarithmic curves (right). (C) The strains described in B were starved of amino acids and ammonium sulfate for 3, 6, and 10 d and then plated on rich media to assay for survival. Survival percentages are normalized for the plating efficiency of each strain. The data show the mean and SEM of three to five independent experiments; 3 d, *p < 0.05 TOR1L2134M vs. WT; 6 d, *p < 0.05 ded1-ΔCT vs. wild-type DED1; 10 d, **p < 0.01 ded1-ΔCT vs. wild-type DED1; 10 d, *p < 0.05 TOR1L2134M vs. WT. (D) The strains described in B were grown to stationary phase in minimal media and were incubated at 30°C for either 7 or 9 d before being serially diluted and plated on rich media.
We next asked whether the C-terminal domain had a similar repressive function during natural stresses in which TORC1 signaling is inhibited. Two such conditions in which TORC1 signaling is attenuated are nitrogen deprivation and stationary phase growth. After depriving cells of nitrogen, the ded1-ΔCT cells showed a decreased ability to survive compared with controls (Figure 1C). Similarly, the ded1-ΔCT mutant also showed a decreased ability to survive prolonged stationary phase (Figure 1D). TOR1L2134M cells showed similar impaired survival after both nitrogen deprivation and prolonged stationary phase compared with its respective wild-type control (Figure 1, C and D). Both the rapamycin resistance and impaired survivability following nutrient starvation suggest an important role for the Ded1 C-terminus in the cellular changes that occur during long-term nutrient stress and inhibition of TORC1. The differing phenotypes in these conditions likely reflect negative consequences to an improper cellular response to these natural stresses, in contrast to rapamycin treatment in rich media. Taken together, these results suggest that Ded1 and the C-terminal domain could be involved in cellular reprogramming during stress conditions.
The Ded1 C-terminal deletion shows impaired translation down-regulation following rapamycin treatment
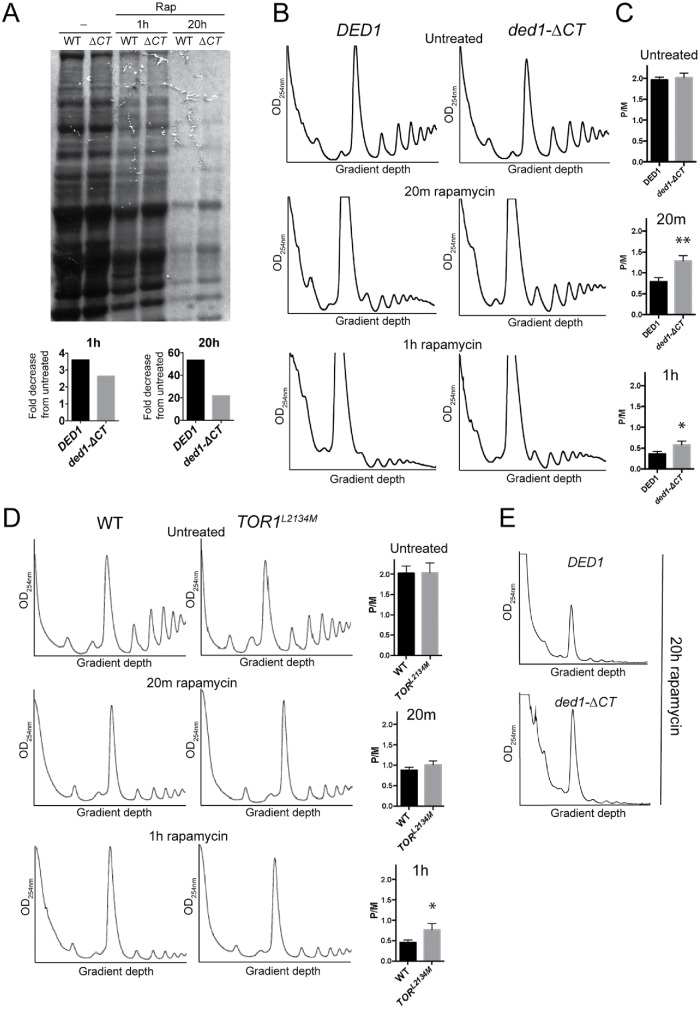
Ded1 has been described as having a role in both promoting (Chuang et al., 1997; Hilliker et al., 2011; Aryanpur et al., 2017) and repressing (Beckham et al., 2008; Hilliker et al., 2011; Aryanpur et al., 2017) translation, and these roles correlate with the activity level of Ded1(Aryanpur et al., 2017). Ded1 activity is essential for translation initiation; however, above a certain threshold Ded1 becomes inhibitory toward translation (Hilliker et al., 2011; Aryanpur et al., 2017). High levels of Ded1 overexpression thus result in severe growth inhibition and translation down-regulation. This effect is at least partially dependent on the Ded1 C-terminus as deletion of the C-terminus partially rescues the observed growth inhibition (Hilliker et al., 2011). As Ded1 can both activate and repress translation, and we observed rapamycin-resistant growth in the ded1-ΔCT mutant, we hypothesized that Ded1 has a role in translation down-regulation following TORC1 inhibition. To test this, we measured new protein synthesis after rapamycin treatment using 35S-methionine (35S-Met) labeling. In wild-type DED1 cells, there was a 3.6-fold reduction in new protein synthesis after 1 h of rapamycin treatment, and a 53-fold reduction was observed after 20 h of rapamycin treatment (Figure 2A). Consistent with a role for Ded1 in translation down-regulation, this reduction in protein synthesis was attenuated in ded1-ΔCT cells. Specifically, in ded1-ΔCT, there was a 2.6-fold reduction in new protein synthesis after 1 h of rapamycin treatment and a 22-fold reduction after 20 h of rapamycin treatment, indicating that ded1-ΔCT cells showed ∼40% more new protein synthesis than wild-type cells at 1 h and ∼220% more after 20 h of rapamycin treatment (Figure 2A). This result shows that compared with wild-type DED1, ded1-ΔCT cells are partially defective in down-regulating protein translation when TORC1 is inhibited. To further test whether Ded1 affects translation down-regulation following rapamycin treatment, we analyzed polyribosomal profiles via sucrose density sedimentation and quantified the relative polysome to 80S monosome levels (P/M ratio). Although the P/M ratio was similar for untreated wild-type DED1 and ded1-ΔCT cells, translation was significantly less down-regulated in the mutant cells compared with controls after 20 min and 1 h of rapamycin treatment (Figure 2, B and C). A similar, though smaller, effect was observed for the TOR1L2134M mutant compared with its respective wild-type control after rapamycin treatment (Figure 2D). These data suggest that Ded1 and its C-terminal domain may have a novel function in translation repression following rapamycin treatment. Long-term rapamycin treatment (20 h) resulted in massive translation down-regulation and loss of ribosomes in both DED1 and and ded1-ΔCT cells (Figure 2E). However, the monosome peak was significantly elevated in ded1-ΔCT cells, indicating higher ribosome levels and possibly continued low-level translation (Heyer and Moore, 2016). Consistent with higher ribosome levels, we also observed elevated levels of ribosomal RNA in ded1-ΔCT cells after rapamycin treatment (Supplemental Figure S1A). Together, these data suggest that Ded1 C-terminus has a role in repressing translation during inhibition of TORC1 signaling.
FIGURE 2:
Ded1 has a role in repressing translation following rapamycin treatment. (A) DED1 and ded1-∆CT cells were grown in the presence of rapamycin for 0, 1, or 20 h. 35S-Methionine (200 µCi ) was added to the medium 1 h prior to harvest. SDS–PAGE and autoradiography of the samples were performed to show incorporation of 35S-Met into new proteins, and the total signal in each lane was quantified to calculate fold difference of ded1-∆CT compared with DED1 (numbers below lanes), or fold decrease from untreated (bar graphs). (B) Cultures of the indicated strains were grown in the presence of rapamycin for the indicated time, and polysome profiles were generated by subjecting cell lysates to 7–47% sucrose density centrifugation and OD254 analysis. A representative result is shown. (C) The monosome/polysome (P/M) ratio in B was determined by comparing the sum of the areas of the polysome peaks to the area of the monosome peak. Each P/M ratio shown is the mean and SEM from of three to four independent trials. **p < 0.01 vs. wild-type DED1, *p < 0.05 vs. wild-type DED1. (D) WT and TOR1L2134M strains were grown in the presence of rapamycin for the indicated time, and polysome profiles were generated as in B. A representative trace is shown with the P/M ratio (mean and SEM of three independent trials) shown to the right. *p < 0.05 vs. WT. (E) Representative traces of polysome profiles from the indicated strains after 20 h of rapamycin treatment.
Rapamycin resistance of ded1-ΔCT is not a result of TORC1 reactivation or eIF2α phosphorylation, but is linked to eIF4G1
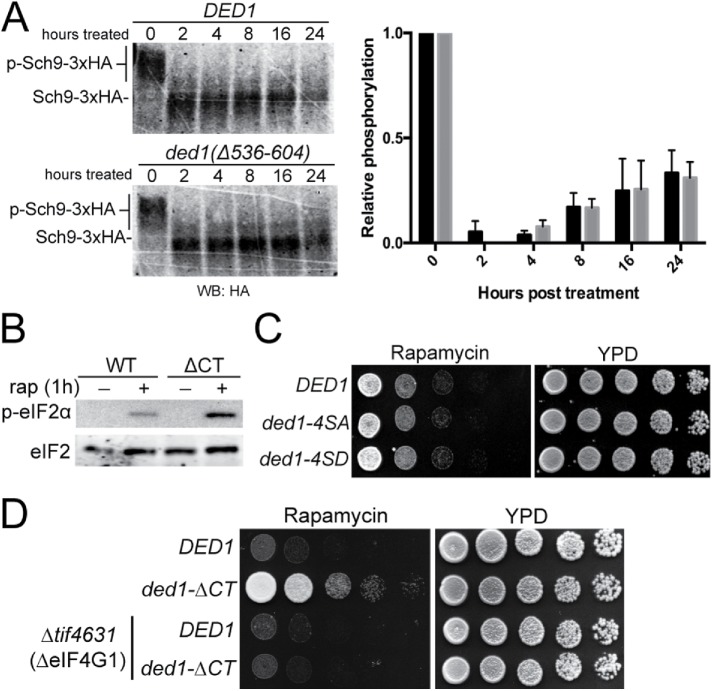
Next, we sought to determine how Ded1 affects the translational response to TORC1 inactivation. To test whether the ded1-ΔCT rapamycin resistance phenotype was due to effects on TORC1 activity or rapamycin efficacy, we examined phosphorylation of Sch9, a direct downstream target of TORC1 in S. cerevisiae. Sch9 is phosphorylated by TORC1 in conditions of nutrient abundance and is rapidly dephosphorylated when TORC1 is deactivated during nutrient stress (Urban et al., 2007; Loewith and Hall, 2011). We used a chemical cleavage band-shift assay (Urban et al., 2007) to assay the phosphorylation level of Sch9 over a 24-h window following rapamycin treatment (Figure 3A). Both wild-type and ded1-ΔCT strains showed similar initial losses of Sch9 phosphorylation following treatment with rapamycin. Over a 24-h period, the relative phosphorylation of Sch9 slowly recovered to ∼40% in both wild-type and mutant cells. This suggested that the rapamycin resistance phenotype observed in the ded1-ΔCT mutant was not driven by the loss of rapamycin efficacy or a result of TORC1 reactivation after long-term treatment, but by a different mechanism.
FIGURE 3:
Ded1 phosphorylation and/or misregulation of downstream TORC1 signaling are not the cause of ded1-∆CT rapamycin resistance. (A) Sch9::3xHA cells harboring DED1 or ded1-∆CT were subjected to a time course of rapamycin treatment, NTCB chemical cleavage of cell extracts, and blotted for with 12CA5 antibody (∝HA) to visualize the band-shift. A representative blot is shown along with a graph of the means from three independent trials. (B) DED1 or ded1-∆CT cells were treated for 1 h with 200 ng/ml rapamycin, and cell extracts were prepared and blotted with antibodies against eIF2α, p-eIF2α, and PGK1. (C) Cells containing DED1, ded1-4SA (S535/S539/541/543A: phospho-deficient), or ded1-4SD (S535/539/541/543D: phospho-mimetic) were serially diluted onto rich media ± rapamycin (200 ng/ml) and incubated at 30°C. (D) DED1 or ded1-∆CT, plasmids were shuffled into WT or tif4631∆ (eIF4G1-null) strains. Cells were serially diluted onto YPD ± rapamycin (200 ng/ml) and incubated at 30°C.
A well-characterized mechanism of translation down-regulation during stress is phosphorylation of eIF2α. Rapamycin treatment and other stresses cause rapid phosphorylation of eIF2α, which contributes to down-regulation of translation of most mRNAs (Liu and Qian, 2014; Pakos-Zebrucka et al., 2016). It is possible that deletion of the Ded1 C-terminus could either directly or indirectly reduce the phosphorylation of eIF2α by an unknown mechanism, which then would result in impaired translation down-regulation and increased protein synthesis. To test whether the rapamycin-resistant phenotype in ded1-ΔCT cells was a result of impaired eIF2α phosphorylation, we assayed the phosphorylation level of eIF2α after 1 h of rapamycin treatment in wild-type and ded1-ΔCT mutant cells (Figure 3B). Phosphorylation of eIF2α was not impaired in ded1-ΔCT, and surprisingly, levels of phosphorylated eIF2α were slightly higher compared with the wild type, perhaps reflecting a feedback mechanism as a result of higher translation levels. In any case, this result supports the hypothesis that the rapamycin resistance phenotypes in ded1 mutants are due to a direct effect on translation and not a result of impaired phosphorylation of eIF2α.
Notably, phosphoproteomic analyses have identified four serine residues (S535, S539, S541, S543) in Ded1 near the beginning of the C-terminal domain that are phosphorylated in stress conditions where TORC1 signaling is inhibited; however, the functionality of these sites is unknown (Figure 1A) (Albuquerque et al., 2008; Huber et al., 2009; Soulard et al., 2010). Therefore, we next asked whether these phosphosites were required for rapamycin-resistant growth. We made a phosphorylation-incapable Ded1, where these putative phosphosites were mutated to alanine (ded1-4SA), and a phospho-mimetic Ded1, where they were mutated to aspartic acid (ded1-4SD). In both phosphorylation-incapable and phospho-mimetic ded1 mutants, no growth phenotypes were observed in the presence of rapamycin or when treated with vehicle control (Figure 3C). From this result, we conclude that these phosphorylation sites are not critical for rapamycin-resistant growth; however, we cannot rule out they serve a biological function under specific, as-yet-undefined conditions.
Since the Ded1 C-terminus has been shown to physically associate with eIF4G1 (Hilliker et al., 2011; Senissar et al., 2014; Putnam et al., 2015), we hypothesized that the disruption of this interaction may contribute to the rapamycin growth resistance of ded1-ΔCT cells. First utilizing a genetic approach, we constructed a TIF4631 (eIF4G1) deletion strain with either DED1 or ded1-ΔCT to test how the absence of eIF4G1 affected rapamycin-resistant growth. Supporting the idea that the rapamycin resistance in ded1-ΔCT cells is mediated by eIF4G1, deletion of eIF4G1 in ded1-ΔCT cells significantly suppressed rapamycin resistance growth, suggesting that this phenotype is dependent on cellular levels of eIF4G1 (Figure 3D). Taken together, these data suggest that the rapamycin-resistant phenotypes of ded1-ΔCT are not due to indirect effects (e.g., on TORC1 activity or eIF2 phosphorylation), but rather are mediated through Ded1’s interaction with eIF4G1.
Ded1 specifically affects the polysome distribution and degradation of eIF4G1 following rapamycin treatment
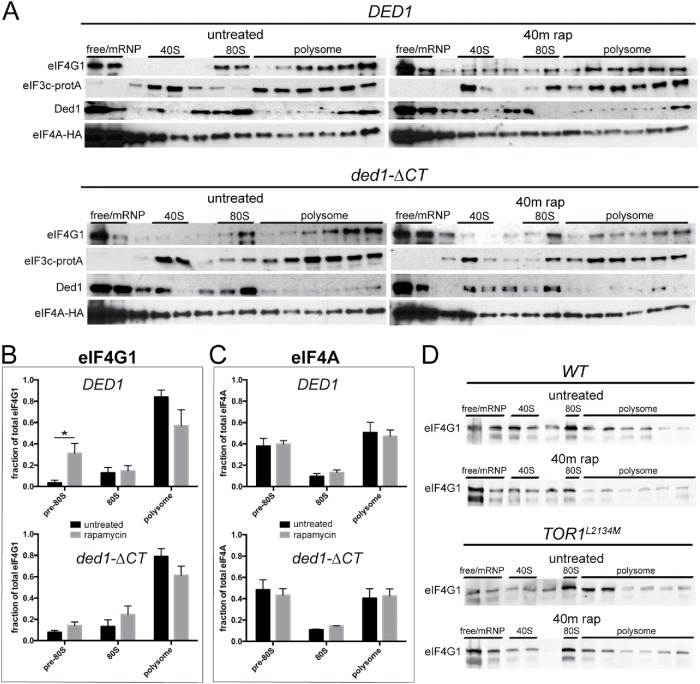
To further examine the interaction between Ded1 and eIF4G1 during TORC1 inhibition, we analyzed in vivo translation complexes. In normal conditions, Ded1 promotes PIC formation (Hilliker et al., 2011; Aryanpur et al., 2017); therefore, we hypothesized that during TORC1 inhibition Ded1 exerts its repressive role through stalling and/or disruption of PICs. If this were the case, we would expect to observe an altered association of translation initiation factors with these complexes. To visualize these changes following rapamycin treatment, we utilized an established protocol for cross-linking translation complexes (Valášek et al., 2007), minimizing the formaldehyde treatment to halt translation and prevent polysome runoff while preventing aberrant cross-linking of initiation factors and maintaining bona fide physical interactions. We then performed sucrose density sedimentation and fractionation, followed by Western blotting for translation initiation factors (Valášek et al., 2007). In wild-type cells, eIF4G1 shifted from the polysome fractions to 80S and pre-80S ribosomal fractions after 40 min of rapamycin treatment (Figure 4A). Quantitation of the initiation complex-associated eIF4G1 distribution showed a significant shift from 3 to 31% of total eIF4G1 into pre-80S fractions following rapamycin treatment (Figure 4B). Interestingly, the distribution of initiation factors eIF4A (Tif1) and eIF3c (Nip1) did not significantly change (Figure 4, A and C), suggesting that a mechanism specific to eIF4G1 alters its association with active translation complexes. In ded1-ΔCT cells, the shift of eIF4G1 into pre-80S fractions was much reduced and not significant (8–14%) after rapamycin treatment. Likewise, the distributions of eIF3c and eIF4A still remained unchanged (Figure 4, B and C). Similar to ded1-ΔCT cells, in TOR1L2134M hyperactive mutant cells, more eIF4G1 was retained in polysome fractions and less shifted to pre-80S fractions compared with its respective wild-type strain after rapamycin treatment (Figure 4D). These results suggest that Ded1 modulates the association of eIF4G1 with pre-80S translation complexes during TORC1 inhibition.
FIGURE 4:
The Ded1 C-terminus has a role in remodeling eIF4G1 from translation complexes following rapamycin treatment. (A) DED1 and ded1-ΔCT cells were grown in the presence of rapamycin for 40 min, after which formaldehyde cross-linking was performed. Polysome profiles were generated as in Figure 2, and fractions were collected for Western blotting using antibodies against Tif4631 (eIF4G1), Ded1, HA (Tif1/eIF4A), and rabbit IgG for Protein-A (Nip1/eIF3c). Representative results for the distributions of each protein are shown. (B, C) Densitometry was performed on eIF4G1 (B) and eIF4A (C) from A. The figure depicts the distribution as a percentage of the total protein in each subfraction. The averages of five independent experiments are shown. *p < 0.05 vs. untreated fraction. (D) WT and TOR1L2134M cells were treated with rapamycin for 40 min, and cells were analyzed as in A using antibodies against Tif4631 (eIF4G1).
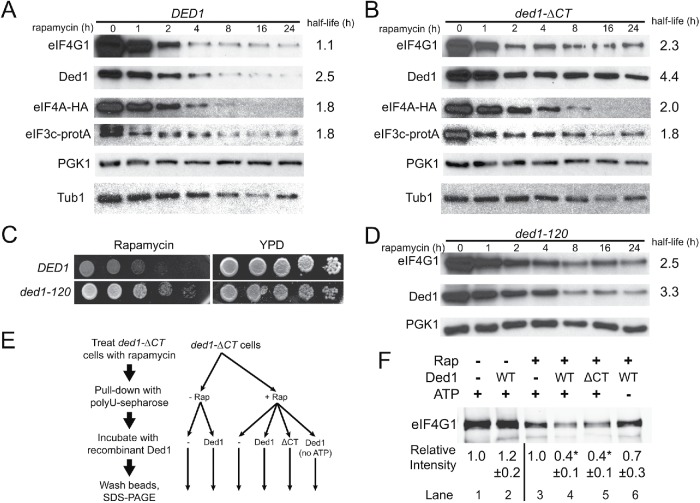
The cellular abundance of eIF4G1 has been shown to decrease via autophagic degradation following inhibition of TORC1 during nitrogen starvation and rapamycin treatment (Berset et al., 1998; Kelly and Bedwell, 2015). Higher cellular levels of eIF4G1 are correlated with higher levels of translation and cell growth (Park et al., 2011; Sen et al., 2015). Since ded1-ΔCT cells display an altered association of eIF4G1 with translation complexes, we hypothesized that these effects may lead to inefficient degradation of eIF4G1 (higher cellular levels of eIF4G1) and contribute to the rapamycin resistance phenotype. To further test this possibility, we monitored initiation factor levels during a rapamycin time course in both wild-type and ded1-ΔCT cells. Levels of eIF4G1 were decreased following rapamycin treatment with a half-life of just over 1 h, but this loss of protein was substantially reduced in the ded1-ΔCT mutant, with an increased half-life of 2.3 h (Figure 5, A and B). Consistent with these effects, blotting of polysome fractions after prolonged (18 h) rapamycin treatment revealed virtually undetectable levels of eIF4G1 in wild-type cells but higher levels and continued association with translation complexes in ded1-ΔCT mutants (Supplemental Figure S1B). In contrast, degradation of the initiation factors eIF4A and eIF3c was similar in both wild-type and mutant cells (Figure 5, A and B), suggesting that the Ded1 C-terminus has a specific effect on eIF4G1 degradation following rapamycin treatment.
FIGURE 5:
The Ded1 C-terminus and enzymatic activity are required for eIF4G1 remodeling and degradation. (A) DED1 cells were subjected to a time course of rapamycin treatment, and cell extracts were prepared and blotted for the indicated translation initiation factors (and PGK1 and Tub1 as a loading controls) as in Figure 3. Half-lives were determined via curve fitting, with 95% confidence intervals of 0.8–1.7 h (4G1), 2.3–2.7 (Ded1), 1.4–2.5 (4A), and 1.3–2.9 (3c). (B) Time course as in A for ded1-∆CT cells. Half-life 95% confidence intervals were 1.7–3.7 (4G1), 3.7–5.3 (Ded1), 1.6–2.7 (4A), and 1.1–5.7 (3c). The K values for eIF4G1 and Ded1 for B were significantly different from A by an extra sum-of-squares F test, while eIF4A and eIF3c were not. (C) DED1 and ded1-120 cells were serially diluted onto rich media ± rapamycin (200 ng/ml) and incubated at 30°C. (D) Time course as in A for ded1-120 cells, blotting for the indicated proteins. Half-life 95% confidence intervals were 1.9–3.8 (4G1) and 2.8–4.2 (Ded1), and the K values for eIF4G1 and Ded1 were significantly different from A by an extra sum-of-squares F test. E Diagram showing experimental workflow for F. Cells were incubated ± rapamycin for 40 min and then pull downs with polyuridine sepharose were performed to isolate RNA-binding proteins. Pull downs were then incubated with 2 μM recombinant Ded1 or ded1-∆CT protein ± 2 mM ATP. (F) Blot of eIF4G1 levels from pull downs described in E. Densitometry was performed and normalized to eIF4G1 levels without added Ded1 separately for rapamycin samples and untreated controls. Values represent the mean of four to five independent experiments for each condition. *p < 0.01 vs. no Ded1 control by Student’s t test.
Like eIF4G1, Ded1 was also degraded on rapamycin treatment but to a lesser extent in the ded1-ΔCT mutant with the half-life increasing from 2.5 to 4.4 h (Figure 5, A and B). Since Ded1 itself is a key activator of translation and cell growth, we asked whether the elevated levels of the protein in mutant cells following rapamycin treatment were contributing to the increased translation levels and growth rate. To test this, we moderately overexpressed DED1 on a high-copy plasmid. Cells containing this construct showed elevated levels of Ded1 following rapamycin treatment similar to the ded1-ΔCT, but these cells did not show a rapamycin-resistant growth phenotype (Supplemental Figure S1, C and D). In fact, they appeared to be slightly more sensitive to rapamycin than cells containing empty plasmid, indicating that the resistance phenotype observed in the ded1-ΔCT mutant was not due to the presence of more Ded1 protein in the cell. Additionally, when ded1-ΔCT was coexpressed in cells also expressing wild-type DED1, the cells did not become resistant to rapamycin, indicating that the ded1-ΔCT phenotype is recessive and reflects a loss of function (unpublished data).
Collectively, the data support a model wherein Ded1 has a novel function during cellular stress, specifically remodeling eIF4G1-containing translation complexes, which leads to degradation of eIF4G1 and down-regulation of translation. This model suggests the catalytic activity of Ded1 would be required for the rapamycin-resistant growth phenotype and the efficient degradation of eIF4G1 following rapamycin treatment. To test this, we used cells containing a ded1 mutation (ded1-120) that is known to have defects in activity in vivo (Chuang et al., 1997). The ded1-120 strain is cold-sensitive; however, it has minor growth and translation defects even at a permissive temperature of 30°C (Chuang et al., 1997; Bolger and Wente, 2011). In growth assays (at 30°C), ded1-120 cells showed nearly identical resistance to rapamycin as the ded1-ΔCT mutant, suggesting that Ded1 enzymatic activity is required for resistance to rapamycin-induced growth inhibition (Figure 5C). We next asked whether Ded1 enzymatic activity was involved in eIF4G1 degradation following rapamycin treatment. Again, similar to the ded1-ΔCT mutant, the ded1-120 strain showed increased abundance of eIF4G1 following rapamycin treatment compared with wild-type cells (Figure 5D), linking Ded1 activity to the loss of eIF4G1 following TORC1 inhibition.
To further test our model that Ded1 remodels eIF4G1 following TORC1 inhibition, we performed an in vitro remodeling experiment (Figure 5E). Extracts were generated from ded1-ΔCT cells (used to maximize eIF4G1 levels) with or without rapamycin treatment for 40 min. RNA-binding proteins, including eIF4G1, were then isolated using polyuridine-sepharose, and after washing, the beads were treated with recombinant Ded1 for 1 h. After further washes, remaining protein was eluted from the beads by sample buffer and immunoblotted for eIF4G1 (Figure 5F). In the pull downs from untreated cells, eIF4G1 levels were unaffected by incubation with Ded1 (lanes 1 and 2). Strikingly, however, in pull downs from rapamycin-treated cells, the amount of eIF4G1 remaining after Ded1 incubation was reduced by 60% (lanes 3 and 4). When ATP was omitted from the Ded1 incubation step, the remodeling of eIF4G1 was markedly reduced (lane 6), consistent with this effect being a result of Ded1 enzymatic activity. Interestingly, addition of recombinant ded1-ΔCT protein had a similar effect to the wild-type Ded1 (lane 5). This may be due to regulation in vivo that is not reflected in the experimental setup with isolated pull downs and recombinant proteins. Taken together, these results suggest that the C-terminus of Ded1 and Ded1 enzymatic activity are required for rapamycin-induced remodeling of eIF4G1 from initiation complexes, leading to eIF4G1 degradation and translation down-regulation.
Conserved critical residues in the Ded1 C-terminus are required for its function in stress, interaction with eIF4G1, and efficient oligomerization
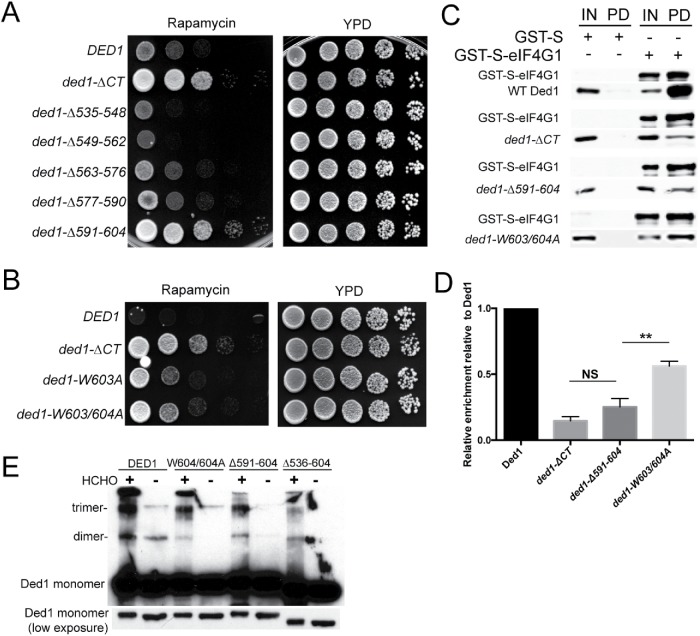
To better understand the mechanism of Ded1’s role in translation down-regulation during cellular stress, we constructed a series of successive 14 amino acid deletions within the Ded1 C-terminus to narrow down the important amino acid(s) required for the rapamycin resistance phenotype. Deletion of the last 14 amino acids (ded1-Δ591–604) phenocopied the rapamycin resistance seen in the full deletion of the entire C-terminus (ΔCT), while the other deletions had no effect (Figure 6A). The last 14 amino acids in the Ded1 C-terminus comprise a low complexity sequence that is mostly poorly conserved in Ded1 orthologues (Supplemental Figure S2, A and B). However, these orthologues all share the striking similarity in having two conserved tryptophans near the C-terminus (Figure 1A). Thus, we hypothesized that these tryptophans are important for Ded1 function during conditions of cellular stress. To test this, we mutated one or both of these tryptophan residues to alanine and performed growth assays in the presence of rapamycin. Mutation of these tryptophans (ded1-W603/604A) largely, but not completely, phenocopied the rapamycin resistance phenotype seen in ded1-ΔCT and ded1-Δ591–604 (Figure 6B). These results suggest that the C-terminal tryptophans in Ded1 are key residues required for active translation down-regulation and growth inhibition following rapamycin treatment, and that their function has been conserved in evolution.
FIGURE 6:
Conserved critical residues in the Ded1 C-terminus are required for its function in stress, interaction with eIF4G1, and efficient oligomerization. (A) Cells containing DED1, ded1-∆CT, or one of a series of 14 amino acid C-terminal deletions were serially diluted onto rich media ± rapamycin (200 ng/ml) and incubated at 30°C. (B) DED1, ded1-∆CT, ded1-W604A, or ded1-W603/604A cells were serially diluted onto rich media ± rapamycin (200 ng/ml), and incubated at 30°C. (C) Bacterially expressed GST-S-eIF4G1 or GST-S was enriched using a glutathione sepharose resin, and affinity chromatography was performed with the addition of recombinant Ded1, ded1-∆CT, ded1-∆591-604, or ded1W603/604A. The bound proteins were analyzed via Western blotting using antibodies to Ded1 and eIF4G1. Input (IN), pull down (PD). (D) Quantitation of the results in C. The data show the mean and SEM of three independent experiments. **p < 0.01 vs. ded1-∆591-604. (E) Recombinant Ded1, ded1-∆CT, ded1-∆591-604, and ded1-W603/604A were cross-linked with formaldehyde or left untreated and visualized via Western blotting with anti-Ded1 antibody. Mobilities of the Ded1 monomer, dimer and trimer are indicated. Note that a lower exposure of Ded1 monomer is also shown to demonstrate equal protein loading.
The Ded1 C-terminus has been implicated in two cellular functions: interaction with eIF4G1 and homo-oligomerization. We hypothesized that the C-terminal tryptophans are required for one or both of these functions. We first analyzed the effects of the C-terminal tryptophans on Ded1 association with eIF4G1. We performed pull downs with bacterially expressed GST-S-eIF4G1 bound to glutathione sepharose and incubated with recombinant purified WT Ded1, ded1-ΔCT, ded1-Δ591-604, or ded1-W603/604A. Full-length Ded1 pulled down with GST-S-eIF4G1, whereas deletion of the entire C-terminus (ded1-ΔCT) reduced this association by 85% (Figure 6C). Likewise, the ded1-Δ591–604 mutant showed a 75% reduction in binding to GST-S-eIF4G1. However, the ded1-W603/604A mutant bound GST-S-eIF4G1 significantly better than ded1-Δ591–604 or ded1-ΔCT, although still less well than wild-type Ded1, showing a 44% reduction in binding to GST-S-eIF4G1 compared with WT Ded1. These results suggest that the last 14 amino acids in the Ded1 C-terminus are critical for the physical interaction with eIF4G1. Furthermore, since deletion of these amino acids phenocopies the ded1-ΔCT strain (Figure 6A), we hypothesize that the interaction between eIF4G1 and Ded1 is a determinant of growth and survival during cell stress. We posit that the C-terminal tryptophans have a novel role in this function of Ded1 since mutating them, but not adjacent amino acids, induced phenotypes (unpublished data).
We next asked whether homo-oligomerization was affected in any of the Ded1 C-terminal mutants. To test this, we performed an in vitro oligomerization assay by cross-linking recombinant wild-type Ded1 or C-terminal mutants with formaldehyde (Figure 6D). Consistent with previous studies, dimerization and trimerization were observed with wild-type Ded1 and were severely reduced in the ded1-ΔCT mutant (Putnam et al., 2015). A smaller reduction in oligomerization was observed in the ded1-Δ591–604 and ded1-W603/604A mutants. The magnitude of the effect on oligomerization appeared to correlate with the size of the disruption of the C-terminus, since the ded1-W603/604A mutant showed the smallest reduction in oligomerization. This suggests that the rapamycin resistance phenotype is, at best, only partially linked to a defect in oligomerization, since deletion of the last 14 amino acids (ded1-Δ591–604) fully phenocopied the entire deletion of the C-terminus (ded1-ΔCT) in growth assays (Figure 6A). We conclude that the function of the Ded1 C-terminus in the TORC1 stress response is likely linked to its physical association with eIF4G1, although oligomerization may also play a role through affecting Ded1 catalytic activity (Putnam et al., 2015).
DISCUSSION
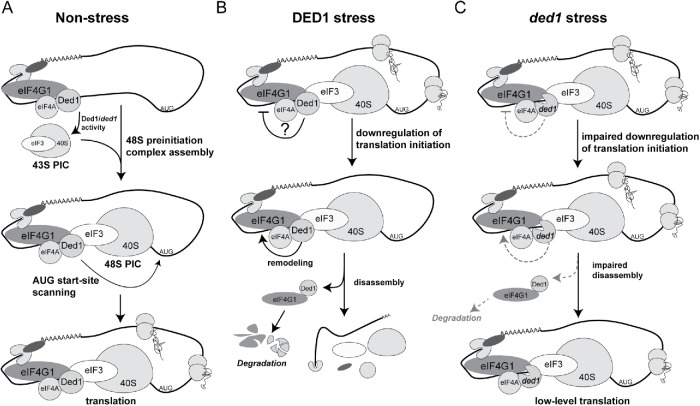
In this work, we have defined a repressive function for Ded1 in conditions that affect the TORC1 signaling pathway. We have shown that the Ded1 C-terminus is required for growth inhibition and efficient translation down-regulation following rapamycin treatment, and furthermore, that this domain serves a function in survival following long-term nutrient deprivation (Figure 1, C and D). Hilliker et al. (2011) have previously shown that this domain contributes to growth repression following Ded1 overexpression, and we and others have speculated that Ded1, as a “gatekeeper” of translation, may also possess the ability to repress translation at physiological levels (Hilliker et al., 2011; Aryanpur et al., 2017). This work has elucidated a repressive function of Ded1 at physiological levels in the context of inhibition of the TORC1 signaling pathway. Our data suggest the following model for Ded1 function (Figure 7). In steady-state growth conditions, Ded1 promotes initiation through PIC assembly and start site scanning as previously described (Sen et al., 2015; Aryanpur et al., 2017; Gupta et al., 2018) (Figure 7A). During stress conditions, however, Ded1 represses translation, possibly first inhibiting 48S PIC assembly. This is followed by the enzymatic removal of eIF4G1 from translation complexes and the codegradation of both eIF4G1 and Ded1 (Figure 7B). In ded1 mutant cells, this remodeling and degradation of eIF4G1 is attenuated, resulting in continued eIF4G1 association with translation complexes and sustained translation during TORC1 inhibition (Figure 7C). In support of our model, we show genetic and functional data indicating that the Ded1 C-terminus and enzymatic activity are required for growth inhibition and translation repression following TORC1 inhibition (Figures 1, 2, and 5). We demonstrate that this is at least partially due to enzymatic removal of eIF4G1 from translation complexes and subsequent degradation (Figures 4 and 5). Furthermore, our finding that Ded1 possesses the ability to dissociate eIF4G1 from RNA in vitro is consistent with active remodeling of translation complexes as proposed by our model (Figure 5, E and F). Surprisingly, both recombinant Ded1 and ded1-ΔCT were able to dissociate eIF4G1 from RNA, but only from extracts treated with rapamycin. This suggests that inhibition of TORC1 activity modulates the ability of Ded1 to dissociate eIF4G1 from RNA. We speculate that the ability of ded1-ΔCT to still affect eIF4G1 association with RNA derives from the excess of protein used in this assay, although it is possible that this effect represents a lack of dependence on the C-terminus for this specific Ded1 function. Additionally, the remodeling of eIF4G1 requires ATP, which suggests that it is dependent on Ded1 enzymatic activity. In summary, our work has helped define a unique function of Ded1 in translation repression following TORC1 inhibition.
FIGURE 7:
Speculative model for Ded1 in the translational stress response. (A) In nonstress conditions, Ded1 stimulates translation by promoting assembly of the 48S PIC. (B) In stress, WT Ded1 may be directly involved in two steps of translation repression. First, Ded1 may stall the assembly of the 48S PIC. In a second step, Ded1 promotes the removal of eIF4G1 from stalled translation PICs, which leads to the codegradation of both factors. (C) In cellular stress ded1 mutants (ded1-120/ded1-ΔCT) are not effective in stalling 48S PIC assembly. The removal and degradation of eIF4G1 from PICs is also impaired, as this step requires Ded1 enzymatic activity and/or a functional interaction between the Ded1 C-terminus and eIF4G1. Thus, eIF4G1 is retained in translation complexes and helps sustain translational activity.
A key outcome of this work is elucidation of the functional requirements for Ded1’s role in the cellular response during TORC1 inhibition. We first noted rapamycin-resistant phenotypes in a C-terminal deletion mutant of DED1 (Figure 1), suggesting that interaction with eIF4G1 and/or Ded1 oligomerization is required for normal translation down-regulation. We were able to map the rapamycin resistance phenotype to a function of the last 14 amino acids of Ded1 (amino acids 591–604) and even further to two highly conserved C-terminal tryptophans (Figure 6). Interestingly, the ded1-Δ591–604 mutant phenocopied ded1-ΔCT in rapamycin-resistant growth, while it also reduced eIF4G1 binding to a nearly equal degree as the larger deletion. On the other hand, we observed a severe oligomerization defect in ded1-ΔCT and only a minor defect in ded1-Δ591–604. Thus, the stress phenotypes we observed directly correlate with eIF4G1 binding rather than oligomerization, suggesting that eIF4G1 binding is likely the more critical interaction. Likewise, we observed incomplete resistance to rapamycin in the ded1-W603/604A mutant, commensurate with the partial reduction we observed in eIF4G1 binding (Figure 6). These findings fit with the effects on eIF4G1 shown in Figures 3, 4, and 5, although it remains possible that oligomerization also plays a role in these processes.
We have identified a cellular context (TORC1 inhibition) in which Ded1 activity represses translation at physiological levels (not overexpressed). However, an important question raised from this work is how this activity is modulated. Specifically, what is the “switch” that changes Ded1 from a translation activator in normal conditions to a translation repressor during cellular stress? We hypothesize that Ded1 activity is modulated by a currently unidentified factor during TORC1 inhibition. Interestingly, in our in vitro remodeling experiments, Ded1 was only able to remove eIF4G1 from RNA in cell extracts that had been treated with rapamycin (Figure 5F). This suggests that on inhibition of TORC1 signaling, there are modifications to the components in the extract that affect Ded1 function. These could include posttranslational modifications that directly modulate protein function and/or a change in expression levels of a regulatory factor. Moderate overexpression of wild-type DED1 or mutants had no effect on rapamycin-resistant growth, indicating that the switch is not caused by modulation of Ded1 levels themselves (Supplemental Figure S2C). Notably, mutations in GLE1, a regulator of Ded1 activity (Bolger and Wente, 2011; Aryanpur et al., 2017), also did not have any effect (unpublished data). Furthermore, our data suggest that the previously identified phosphorylation sites are not critical since mutation of these sites did not show any phenotypes in the conditions we tested (Figure 3C). A different posttranslational modification or phosphorylation of another translation factor could still play a role, however. One possibility is that the repressive function of Ded1 is indirectly stimulated by the well-established stress response pathway of eIF2α phosphorylation through the inhibition of ternary complex formation and the rate of PIC assembly. As shown in Figure 3B, eIF2α phosphorylation is not reduced in ded1-ΔCT cells. Further studies will be required to determine whether Ded1 is downstream of or a parallel supplement to eIF2α phosphorylation.
Kelly and Bedwell (2015) previously have shown that eIF4G1, along with eRF3 and Pab1, are among a small subset of translation factors that are degraded by autophagy following nitrogen deprivation (Kelly and Bedwell, 2015). In this work, we have identified Ded1 as another translation factor that is degraded during TORC1 inhibition. Our data indicate that degradation of eIF4G1 is linked to physical interaction with Ded1 (Figure 5, A and B), suggesting that they may follow the same autophagic degradation program. Interestingly, both Ded1 and eIF4G accumulate in stress granules following induction with sodium azide or glucose withdrawal (Brengues and Parker, 2007; Beckham et al., 2008; Hilliker et al., 2011; Jain et al., 2016), and work by Buchan et al. (2013) has shown that stress granule components, including Ded1, can be degraded via autophagic pathways in a process termed granulophagy (Buchan et al., 2013). Thus, although rapamycin does not promote stress granule assembly, both cellular stress conditions still result in autophagic degradation of Ded1 and eIF4G. Furthermore, parallel to the remodeling activity of Ded1 that we propose in TORC1 inhibition, Ded1 also has been suggested to play a role in stress granule assembly. Therefore, despite differences in mechanism depending on the conditions, it appears that Ded1 and eIF4G may play parallel roles in multiple responses to cellular stress. We speculate that during TORC1 inhibition, the degradation of Ded1 and eIFG1 is a regulated mechanism of gene reprogramming, affecting cap-dependent translation and/or specific subclasses of genes that have different dependence on eIF4G1 and Ded1 for efficient translation (Park et al., 2011; Sen et al., 2015; Costello et al., 2017; Gupta et al., 2018). Our data also do not rule out the possibility that on TORC1 inhibition, Ded1 and eIF4G still form small stress granule-like structures or microaggregates that are not distinguishable as such by standard microscopy techniques. Such microgranules could potentially link these cellular stress response mechanisms, but examination of their dynamics and/or abundance would likely be technically challenging.
Currently, it is unknown whether the translation repression function of Ded1 affects all mRNAs or a subset of mRNAs. In nonstress conditions, Ded1 activity has been shown to promote translation of mRNAs with long 5′ UTRs and cap distal secondary structure (Sen et al., 2015, 2016). In addition, a recent study showed that Ded1 normally prevents translation initiation from near cognate start codons associated with RNA secondary structure (Guenther et al., 2018); thus mRNAs with these sites may be more affected by changes in Ded1 activity. Guenther et al. (2018) go on to suggest that a lower abundance of Ded1 during meiosis may result in the use of alternative start-codons as a part of the meiotic gene expression program. Given the degradation of Ded1 that we have observed (Figure 5A), a similar effect may occur in TOR-inhibited cells. Furthermore, our work has shown that during stress, Ded1 represses translation by removing eIF4G1 from the pool of actively translating mRNAs, ultimately leading to its degradation. Therefore, mRNAs that are dependent on eIF4G1 may also be preferentially affected. In support of this idea is evidence that a subclass of mRNAs that are dependent on eIF4G1 have the highest translation efficiencies and encode the most abundant cellular proteins (Park et al., 2011; Sen et al., 2016). This is also in agreement with the observation that translation of the most abundant proteins in ded1-ΔCT cells was affected in the 35S-Met labeling experiment. Testing these hypotheses will be a priority for future studies on Ded1 function in the cellular stress response.
The TORC1 signaling pathway is a master regulator of cell growth and metabolism (Loewith and Hall, 2011; González and Hall, 2017), so it is not surprising that mutations affecting TORC1 signaling have been implicated in cancer (Zhao et al., 2016). Here we have shown that mutations in Ded1 make cells partially refractory to regulation by the TORC1 signaling pathway. Mutations in the Ded1 human orthologue, DDX3, are found frequently in cancer (Northcott et al., 2012; Kool et al., 2014; Zhao et al., 2016); however, an understanding of how DDX3 contributes to cancer is lacking and often contradictory (Oh et al., 2016; He et al., 2018). A recent paper by Oh et al. (2016) presented evidence that cancer-associated DDX3 mutations can impair the translation of some mRNAs while selectively preserving translation on other mRNAs during stress. Thus, it is possible that alterations in DDX3 in cancer may promote TOR-independent growth. However, most of the cancer-associated mutations in DDX3 are located outside of the C-terminus and nearer to the conserved helicase motifs, suggesting that perturbation of the RNA-dependent ATPase activity of DDX3 and not of the C-terminus is critical in oncogenesis. On the other hand, our observation that the ded1-120 mutant also exhibits rapamycin-resistant growth suggests that cancer mutations affecting the helicase core regions of DDX3 could also potentially promote TOR-independent growth. At the least, the implication that Ded1 function can be modulated by different cellular conditions reveals another dimension that must be considered when investigating the biological consequences of disease-associated mutations in DDX3. Our findings may thus provide further context for functional studies that aim to understand the complex role of Ded1/DDX3 in cancer.
MATERIALS AND METHODS
Yeast strains and plasmids
Yeast strains and plasmids used are listed in Supplemental Tables S1 and S2. Strains containing different ded1 mutants were created by plasmid shuffle starting with the strain SWY4093 (ded1::KAN +pDED1). TBY101 and 103 and TBY120 and 131 were generated by integrating a sequence encoding a C-terminal 3x-HA tag marked with TRP1 at the SCH9 and TIF1 locus, respectively. TBY134-137 were created by integrating a sequence encoding a protein conferring resistance hygromycin B into the TIF4631 locus of SWY4093. Plasmid shuffle was then used to create the different ded1 tif4631 double mutant strains. pTB136 was created by cutting pRP2044 with ApaI and SacI and cloning into pSW3619 after digestion with the same enzymes. DED1 mutant plasmids were created by site-directed mutagenesis of either DED1 or pSW3619. pTB111-115 were created by amplifying pSW3619 using PCR primers designed to exclude the targeted 14 amino acid region, followed by blunt end ligation. Bacterial expression plasmids (pTB6, pTB160, pTB161) were created by PCR amplification of pRP2044, pTB115, and pTB124 respectively, with primers flanked with NdeI or EcoRI cut sites. The PCR products were then cut with NdeI and EcoRI and ligated into SW3576, which was cut with the same enzymes.
Growth assays
Yeast growth assays were performed by serial dilution as previously described (Aryanpur et al., 2017). Images of growth on rapamycin plates were taken after 5 d for W303 background strains and after 3 d for S288C background strains to ensure spots were not oversaturated with colonies (growth of S288C background strains was inherently less sensitive to rapamycin). Rapamycin (Santa Cruz Biotechnology) was dissolved in dimethylsulfoxide (DMSO) and was used at concentrations of 200 ng/ml in YPD plates and 100 ng/ml in selective auxotrophy plates. Rapamycin growth curves were generated by diluting log-phase cells to an optical density (OD) of 0.1 in liquid YPD on which rapamycin was added to a concentration of 200 ng/ml. Cultures were incubated at 30°C on a shaker and optical density measurements were taken every 2–6 h and converted to cells per milliliter.
Survival assays
Nitrogen starvation survival assays were performed by first growing cells to mid–log phase in SD media + amino acids and ammonium sulfate. The cells were spun down, washed 2×, resuspended in SD media–amino acids and ammonium sulfate to an OD of 0.2, and incubated at 30°C on a shaker. Cultures were incubated for 3, 6, and 10 d, after which time they were spun down, washed, and resuspended to a concentration of 5 × 107 cells/ml. The cells were serially diluted and plated on YPD. The number of yeast colonies was counted on each plate, from which the average number of viable yeast per milliliter of original culture was calculated. The percent survival was calculated by normalizing to the plating efficiencies for each strain, which were virtually identical (unpublished data). An unpaired t test was used to determine significance. The stationary phase survival assay was performed by growing cells to mid–log phase in liquid SD media and allowing them to grow to stationary phase at 30°C on a shaker. After 0, 7, and 9 d of post-mid-log growth, serial dilution growth assays were performed as described previously.
Polysome preparation
Sucrose density gradients and cell extracts were prepared as previously described (Aryanpur et al., 2017). For the rapamycin time-course polysomes, cells were grown to mid–log phase in YPD, and rapamycin was added to a concentration of 200 ng/ml. Cells were harvested before the addition of rapamycin and after 20 min and 1 h of rapamycin treatment. Polysome to monosome ratios were determined by comparing the area under the curve for the sum of the polyribosome peaks to the sum of the 80S peak in ImageJ (National Institutes of Health). In samples where rapamycin was added, the absorbance reading of the top portion of the monosome peak exceeded the maximum measurable by the fractionator. Thus, the true monosome area is slightly larger than the measured one, and the P/M ratio is a maximum value. A paired t test was used to determine significance. Note that the difference between the wild type and the mutant is still significant despite this limitation.
35S-Met incorporation
Cells were grown to mid–log phase and then diluted to an OD600 of 0.1. Rapamycin was added at a concentration of 200 ng/ml and the cultures were incubated for 1 h and 20 h on a 30°C shaker. Three OD units of cells were then spun down and resuspended in 1 ml of supernatant on which 75 μCi 35S-Met was added, followed by 1-h incubation at 30°C with shaking. Cells were spun down and washed with water before lysis in 1.85 M NaOH and 7.4% β-mercaptoethanol. Proteins were precipitated with trichloroacetic acid, resuspended in SDS sample buffer, and separated by SDS–PAGE. The polyacrylamide gel was fixed and dried using a vacuum drier before autoradiographic detection of 35S-Met incorporated proteins. In each lane of the gel, the total 35S-Met signal was quantitated using ImageJ.
Formaldehyde cross-linking and fractionation
Formaldehyde cross-linking and polysome fractionation were performed as described with a few modifications (Valášek et al., 2007). Briefly, after 40 min of rapamycin treatment (200 ng/ml), formaldehyde was added to the cultures to a final concentration of 0.25%, followed by incubation on ice for 15 min, before quenching with 0.5 mM glycine. Cell extracts were prepared by first lysing cells by vortexing with glass beads (3 × 2 min, with 30 cooling on ice) and then spinning twice for 5 min at 13,000 rpm at 4°C. Polysome profiling and fractionation was performed as previously described. Protein was precipitated from each fraction by the addition of 0.8 ml of ice-cold ethanol to 0.5 ml of each fraction, vortexing, and incubation overnight at –20°C. The samples were spun at full speed at 4°C, washed once with 75% ethanol, and the pellets resuspended in 0.1 ml sample buffer (100 mM Tris-HCl,15% glycerol, 5% SDS, 100 mM dithiothreitol [DTT]).
Western blotting of polysome fractions
Fraction samples were heated to 95°C prior to loading onto gel, separated by SDS–PAGE, and blotted with specific antibodies toward Ded1 (VU318), eIF4G1 (gift from R. Parker, University of Colorado–Boulder), and anti-HA for eIF4A (Novus). eIF3C-ProtA was detected with rabbit, anti-mouse IgG (Invitrogen). HRP-conjugated secondary antibodies were used for chemiluminescent detection of bands. Band intensity was measured using ImageJ software. Quantitation of eIF4G1 and eIF4A-HA in different subfractions (pre-80S, 80S, and polysome) was achieved by first calculating the percentage of the total intensity for each individual fraction and summing the percentages for each subfraction. The mRNP fractions (first two) were not included in the total intensity because the signal was beyond the linear range of detection. A paired t test was used to determine significance.
Translation factor time courses
Cells were cultured in YPD, and rapamycin was added to a concentration of 200 ng/ml. At the indicated time points posttreatment an aliquot of each culture was taken (normalizing to the same number of cells) for harvest. Briefly, cells were lysed in 1.85 M NaOH and 7.4% β-mercaptoethanol, proteins were precipitated with trichloroacetic acid, and samples were resuspended in SDS sample buffer. Samples were separated by SDS–PAGE and blotted for the indicated proteins as described previously. Specific antibodies were used to detect PGK1 (Invitrogen) and Tubulin (Novus). Phospho-eIF2α (Ser51) antibody was purchased from Cell Signaling Technology. The eIF2α anti-sera was a gift from Tom Dever (National Institutes of Health). Band detection was accomplished with LI-COR secondary antibodies, in conjunction with an Odyssey infrared imager. For half-life calculations, bands from three to eight independent trials were quantified via densitometry and degradation curves were fitted using the 0- to 8-h time points in Graphpad Prism. Similarity of the derived K values (half-life = ln(2)/K) for each protein in wild-type and mutant strains was assessed by an extra sum-of-squares F test with a P value threshold of 0.05. Half-life values are shown with 95% confidence intervals listed in the figure legends.
In vitro GST-pull downs
Rosetta cells (Novagen) containing the pET41a(+)-GST-S or pET41a(+)-GST-S-eIF4G1-His plasmid (gift from E. Jankowsky, Case Western Reserve University) were induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) overnight at 15°C. Cells were lysed by sonication and were spun down to clear cell debris. The supernatants were incubated with glutathione-sepharose beads for 4 h at 4°C and then washed 3× with RIPA buffer (150 nM NaCl, 1% Nonidet-P40, 0.1% SDS, 50 mM Tris-HCl; pH 7.4). Protein-bound beads (50 μl ) were used for the different Ded1p pull downs. Purified, recombinant WT or mutant Ded1p was then added to each pull down to a final concentration of 1 μM, along with 5 mg/ml bovine serum albumin. The pull downs were incubated overnight at 4°C on a rotator after which they were washed 4× with RIPA buffer and resuspended in SDS sample buffer. Proteins were visualized by Western blotting for eIF4G1 and Ded1 as previously described. Quantitation was performed using ImageJ. WT and mutant Ded1p enrichment values were calculated by first normalizing the signal to eIF4G1. The relative enrichment was determined as the ratio between the normalized pull down and input values. These values were normalized to WT recombinant Ded1, which was set to 1.
Remodeling assay
For the Ded1/eIF4G1 remodeling assay, ded1-ΔCT cells were grown to mid–log and incubated in the presence or absence of rapamycin for 40 min. Extracts were generated via bead beater as in Aryanpur et al. (2017) and incubated with poly(U)-sepharose resin (GE Healthcare) at 4°C for 2 h. Beads were washed, then resuspended in buffer containing 20 mM HEPES (pH 7.5), 50 mM NaCl, 2 mM MgCl2, and 10% glycerol. Recombinant Ded1 or ded1-ΔCT protein was added in the presence or absence of 2 mM ATP, and the samples were incubated for 1 h at 37°C. Beads were then washed again, resuspended in sample buffer, and immunoblotted for the presence of eIF4G1 as above. Bands were quantified via densitometry and presented as the mean of 4–5 independent trials. Significance was assessed via Student’s t test.
Oligomerization assay
WT Ded1p and mutant ded1p were purified as described previously (Aryanpur et al., 2017). In vitro oligomerization of Ded1p was assayed as described in Putnam et al. (2015). Briefly, purified recombinant Ded1p (1 μM), ded1-ΔCT (1 μM), ded1-Δ591-604 (1 μM), or ded1-W603/604A (1 μM) were incubated in helicase reaction buffer (40 mM Tris-HCl, pH 8.0), 50 mM NaCl, 8.3% (vol/vol) glycerol, 0.01% (wt/vol) IGEPAL CA 630, 2 mM DTT, and 0.5 mM MgCl2) for 1 h at 19°C. Formaldehyde was added to a final concentration of 1% vol/vol, and the reactions were incubated at room temperature for 30 min. Reactions were quenched with 0.5 mM glycine (pH 6.8) and samples were diluted in sample buffer (100 mM Tris, pH 6.8, 24% glycerol, 8% SDS, 0.02%, 0.2 mM DTT) prior to Western blotting (αDed1).
Sch9-3xHA band-shift assay
Band-shift measurements were performed using a modified version of the protocol developed by Urban and Loewith (Urban et al., 2007; Hughes-Hallett et al., 2014). Briefly, at the indicated time points after rapamycin treatment (200 ng/ml), 47 ml of culture was mixed with 3 ml of 100% trichloroacetic acid and incubated on ice for 1 h. The cultures were spun down, washed with ddH2O and then acetone, and cells were disrupted by sonication, followed by centrifugation at 8000 rpm for 30 s. The pellets were then subjected to bead beating (6 × 1 min) in urea buffer (6M urea, 50 mM Tris-HCl, pH 7.5, 5 mM EDTA, 1 mM PMSF, 5 mM NaF, 5 mM NaN3, 5 mM NaH2PO4, 5 mM p-nitrophenylphosphate, 5 mM β-glycerophosphate, 1% SDS) supplemented with additional protease and phosphatase inhibitors. The lysate was heated at 65°C for 10 min and cleared by centrifugation at 15,000 rpm for 7 min. Chemical cleavage of the extracts was accomplished by adding nitro-5-thiocyanatobenzoic acid (NTCB) for 12–16 h at room temperature (1 mM NTCB in 100 mM CHES, pH 10.5). Western blotting of the samples was performed with an anti-HA antibody (Novus), LI-COR secondary antibodies, on an Odyssey infrared imager. Quantitation of Sch9 phosphorylation was performed using a MATLAB script written by James Hughes-Hallett (Hughes Hallett et al., 2014).
RNA gel electrophoresis
Cultures were grown to mid–log phase in YPD and diluted to an OD600 of 0.1. Rapamycin was added to a concentration of 200 ng/ml, and the cultures were incubated for 18 h at 30°C with shaking. RNA was isolated by lysing cells with TRIZOL, chloroform extraction, followed by isopropyl alcohol precipitation. The resuspended RNA samples were mixed with RNA sample loading buffer, separated on a formaldehyde-agarose gel, and visualized by a UV gel imager.
Supplementary Material
Acknowledgments
We thank Roy Parker, Angela Hilliker, Eckhard Jankowsky, and Tom Dever for reagents and members of the Bolger and J.R. Buchan laboratories for helpful advice and discussions. This work was supported by the American Cancer Society (RSG-13-263-01-RMC to T.A.B.) and the Arizona Biomedical Research Commission (ADHS14-082993 to T.A.B.).
Abbreviations used:
- DTT
dithiothreitol
- eIF2α
eukaryotic initiation factor 2 alpha
- eIF4F
initiation factor 4F
- eIF4G
eukaryotic initiation factor 4G
- 35S-Met
35S-methionine
- NTCB
nitro-5-thiocyanatobenzoic acid
- OD
optical density
- TOR
target of rapamycin
- UTR
untranslated regions
- YPD
yeast extract–peptone–dextrose media.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E18-11-0702) on May 29, 2019.
REFERENCES
- Albuquerque CP, Smolka MB, Payne SH, Bafna V, Eng J, Zhou H. (2008). A multidimensional chromatography technology for in-depth phosphoproteome analysis. Mol Cell Proteomics , 1389–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aryanpur PP, Regan CA, Collins JM, Mittelmeier TM, Renner DM, Vergara AM, Brown NP, Bolger TA. (2017). Gle1 regulates RNA binding of the DEAD-box helicase Ded1 in its complex role in translation initiation. Mol Cell Biol , e00139-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckham C, Hilliker A, Cziko A-M, Noueiry A, Ramaswami M, Parker R. (2008). The DEAD-box RNA helicase Ded1p affects and accumulates in Saccharomyces cerevisiae P-bodies. Mol Biol Cell , 984–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berset C, Trachsel H, Altmann M. (1998). The TOR (target of rapamycin) signal transduction pathway regulates the stability of translation initiation factor eIF4G in the yeast Saccharomyces cerevisiae. Biochemistry , 4264–4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger TA, Wente SR. (2011). Gle1 is a multifunctional DEAD-box protein regulator that modulates Ded1 in translation initiation. J Biol Chem , 39750–39759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brengues M, Parker R. (2007). Accumulation of polyadenylated mRNA, Pab1p, eIF4E, and eIF4G with P-bodies in Saccharomyces cerevisiae. Mol Biol Cell , 2592–2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, Kolaitis R-M, Taylor JP, Parker R. (2013). Eukaryotic stress granules are cleared by autophagy and Cdc48 / VCP function. Cell , 1461–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, Parker R. (2009). Eukaryotic stress granules: the ins and outs of translation. Mol Cell , 932–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang RY, Weaver PL, Liu Z, Chang TH. (1997). Requirements of the DEAD-box protein Ded1p for messenger RNA translation. Science , 1468–1471. [DOI] [PubMed] [Google Scholar]
- Costello JL, Kershaw CJ, Castelli LM, Talavera D, Rowe W, Sims PFG, Ashe MP, Grant CM, Hubbard SJ, Pavitt GD. (2017). Dynamic changes in eIF4F-mRNA interactions revealed by global analyses of environmental stress responses. Genome Biol , 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Das B. (2016). eIF4G-an integrator of mRNA metabolism? FEMS Yeast Res , 87–93. [DOI] [PubMed] [Google Scholar]
- Dever TE, Feng L, Wek RC, Cigan AM, Donahue TF, Hinnebusch AG. (1992). Phosphorylation of initiation factor 2α by protein kinase GCN2 mediates gene-specific translational control of GCN4 in yeast. Cell , 585–596. [DOI] [PubMed] [Google Scholar]
- Gao Z, Putnam AA, Bowers HA, Guenther UP, Ye X, Kindsfather A, Hilliker AK, Jankowsky E. (2016). Coupling between the DEAD-box RNA helicases Ded1p and eIF4A. Elife , e16408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonskikh Y, Polacek N. (2017). Alterations of the translation apparatus during aging and stress response. Mech Ageing Dev , 30–36. [DOI] [PubMed] [Google Scholar]
- González A, Hall MN. (2017). Nutrient sensing and TOR signaling in yeast and mammals. EMBO J , 397–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther UP, Weinberg DE, Zubradt MM, Tedeschi FA, Stawicki BN, Zagore LL, Brar GA, Licatalosi DD, Bartel DP, Weissman JS, et al. (2018). The helicase Ded1p controls use of near-cognate translation initiation codons in 5′ UTRs. Nature , 130–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta N, Lorsch JR, Hinnebusch AG. (2018). Yeast Ded1 promotes 48S translation pre-initiation complex assembly in an mRNA-specific and eIF4F-dependent manner. Elife , e38892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris TE, Chi A, Shabanowitz J, Hunt DF, Rhoads RE, Lawrence JC. (2006). mTOR-dependent stimulation of the association of eIF4G and eIF3 by insulin. EMBO J , 1659–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Zhang D, Yang Y, Wang X, Zhao X, Zhang P, Zhu H, Xu N, Liang S. (2018). A double-edged function of DDX3, as an oncogene or tumor suppressor, in cancer progression (Review). Oncol Rep , 883–892. [DOI] [PubMed] [Google Scholar]
- Heyer EE, Moore MJ. (2016). Redefining the translational status of 80S monosomes. Cell , 757–769. [DOI] [PubMed] [Google Scholar]
- Hilliker A, Gao Z, Jankowsky E, Parker R. (2011). The DEAD-box protein Ded1 modulates translation by the formation and resolution of an eIF4F-mRNA complex. Mol Cell , 962–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber A, Bodenmiller B, Uotila A, Stahl M, Wanka S, Gerrits B, Aebersold R, Loewith R. (2009). Characterization of the rapamycin-sensitive phosphoproteome reveals that Sch9 is a central coordinator of protein synthesis. Genes Dev , 1929–1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes Hallett JE, Luo X, Capaldi AP. (2014). State transitions in the TORC1 signaling pathway and information processing in Saccharomyces cerevisiae. Genetics , 773–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S, Wheeler JR, Walters RW, Agrawal A, Barsic A, Parker R. (2016). ATPase-modulated stress granules contain a diverse proteome and substructure. Cell , 487–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly SP, Bedwell DM. (2015). Both the autophagy and proteasomal pathways facilitate the Ubp3p-dependent depletion of a subset of translation and RNA turnover factors during nitrogen starvation in Saccharomyces cerevisiae. RNA , 898–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kool M, Jones DTW, Jager N, Northcott PA, Pugh TJ, Hovestadt V, Piro RM, Esparza LA, Markant SL, Remke M, et al. (2014). Genome sequencing of SHH medulloblastoma predicts genotype-related response to smoothened inhibition. Cancer Cell , 393–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder P, Jankowsky E. (2011). From unwinding to clamping—the DEAD box RNA helicase family. Nat Rev Mol Cell Biol , 505–516. [DOI] [PubMed] [Google Scholar]
- Liu B, Qian SB. (2014). Translational reprogramming in cellular stress response. Wiley Interdiscip Rev RNA , 301–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewith R, Hall MN. (2011). Target of rapamycin (TOR) in nutrient signaling and growth control. Genetics , 1177–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell SF, Parker R. (2014). Principles and properties of eukaryotic mRNPs. Mol Cell , 547–558. [DOI] [PubMed] [Google Scholar]
- Mokas S, Mills JR, Garreau C, Fournier M-J, Robert F, Arya P, Kaufman RJ, Pelletier J, Mazroui R. (2009). Uncoupling stress granule assembly and translation initiation inhibition. Mol Biol Cell , 2673–2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northcott PA, Jones DT, Kool M, Robinson GW, Gilbertson RJ, Cho YJ, Pomeroy SL, Korshunov A, Lichter P, Taylor MD, et al. (2012). Medulloblastomics: The end of the beginning. Nat Rev Cancer , 818–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S, Flynn RA, Floor SN, Purzner J, Martin L, Do BT, Schubert S, Vaka D, Morrissy S, Li Y, et al. (2016). Medulloblastoma-associated DDX3 variant selectively alters the translational response to stress. Oncotarget , 28169–28182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pakos-Zebrucka K, Koryga I, Mnich K, Ljujic M, Samali A, Gorman AM. (2016). The integrated stress response. EMBO Rep , 1374–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park EH, Zhang F, Warringer J, Sunnerhagen P, Hinnebusch AG. (2011). Depletion of eIF4G from yeast cells narrows the range of translational efficiencies genome-wide. BMC Genomics . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam AA, Gao Z, Liu F, Jia H, Yang Q, Jankowsky E. (2015). Division of labor in an oligomer of the DEAD-box RNA helicase Ded1p. Mol Cell , 541–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raught B, Gingras A-C, Sonenberg N. (2001). The target of rapamycin (TOR) proteins. Proc Natl Acad Sci USA , 7037–7044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers AN, Chen D, McColl G, Czerwieniec G, Felkey K, Gibson BW, Hubbard A, Melov S, Lithgow GJ, Kapahi P. (2011). Life span extension via eIF4G inhibition is mediated by posttranscriptional remodeling of stress response gene expression in C. elegans. Cell Metab , 55–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryoo HD, Vasudevan D. (2017). Two distinct nodes of translational inhibition in the integrated stress response. BMB Rep , 539–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen ND, Zhou F, Harris MS, Ingolia NT, Hinnebusch AG. (2016). eIF4B stimulates translation of long mRNAs with structured 5′ UTRs and low closed-loop potential but weak dependence on eIF4G. Proc Natl Acad Sci USA , 10464–10472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen ND, Zhou F, Ingolia NT, Hinnebusch AG. (2015). Genome-wide analysis of translational efficiency reveals distinct but overlapping functions of yeast DEAD-box RNA helicases Ded1 and eIF4A. Genome Res , 1196–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senissar M, Le Saux A, Belgareh-Touze N, Adam CC, Banroques J, Tanner NK, Belgareh-Touzé N, Adam CC, Banroques J, Tanner NK. (2014). The DEAD-box helicase Ded1 from yeast is an mRNP cap-associated protein that shuttles between the cytoplasm and nucleus. Nucleic Acids Res , 10005–10022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma D, Jankowsky E. (2014). The Ded1/DDX3 subfamily of DEAD-box RNA helicases. Crit Rev Biochem Mol Biol , 343–360. [DOI] [PubMed] [Google Scholar]
- Simpson CE, Ashe MP. (2012). Adaptation to stress in yeast: to translate or not? Biochem Soc Trans , 794–799. [DOI] [PubMed] [Google Scholar]
- Soulard A, Cremonesi A, Moes S, Schutz F, Jeno P, Hall MN. (2010). The rapamycin-sensitive phosphoproteome reveals that TOR controls protein kinase a toward some but not all substrates. Mol Biol Cell , 3475–3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahara T, Maeda T. (2012). Transient sequestration of TORC1 into stress granules during heat stress. Mol Cell , 242–252. [DOI] [PubMed] [Google Scholar]
- Urban J, Soulard A, Huber A, Lippman S, Mukhopadhyay D, Deloche O, Wanke V, Anrather D, Ammerer G, Riezman H, et al. (2007). Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol Cell , 663–674. [DOI] [PubMed] [Google Scholar]
- Valášek L, Szamecz B, Hinnebusch AG, Nielsen KH. (2007). In vivo stabilization of preinitiation complexes by formaldehyde cross-linking. Methods Enzymol , 163–183. [DOI] [PubMed] [Google Scholar]
- Valiente-Echeverría F, Hermoso MA, Soto-Rifo R. (2015). RNA helicase DDX3: At the crossroad of viral replication and antiviral immunity. Rev Med Virol , 286–299. [DOI] [PubMed] [Google Scholar]
- Zhao L, Mao Y, Zhou J, Zhao Y, Cao Y, Chen X. (2016). Multifunctional DDX3: dual roles in various cancer development and its related signaling pathways. Am J Cancer Res , 387–402. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.