Abstract
Cells have evolved diverse protein-based machinery to reshape, cut, or fuse their membrane-delimited compartments. Dynamin superfamily proteins are principal components of this machinery and use their ability to hydrolyze GTP and to polymerize into helices and rings to achieve these goals. Nucleotide-binding, hydrolysis, and exchange reactions drive significant conformational changes across the dynamin family, and these changes alter the shape and stability of supramolecular dynamin oligomers, as well as the ability of dynamins to bind receptors and membranes. Mutations that interfere with the conformational repertoire of these enzymes, and hence with membrane fission, exist in several inherited human diseases. Here, we discuss insights from new x-ray crystal structures and cryo-EM reconstructions that have enabled us to infer some of the allosteric dynamics for these proteins. Together, these studies help us to understand how dynamins perform mechanical work, as well as how specific mutants of dynamin family proteins exhibit pathogenic properties.
EUKARYOTIC LIFE AND THE EVOLUTION OF MEMBRANE-BOUND COMPARTMENTS
Membrane-bound cellular compartments are a hallmark of eukaryotic evolution and underlie the efficient division and specialization of work within the cell. Simple compartments feature small vesicles that form at the plasma membrane for the uptake of nutrients and neurotransmitters. More complex compartments include organelles such as mitochondria, which harbor simple genomes that encode some of their own proteins. Each compartment is actively remodeled during endocytosis or organelle division events, respectively. The timing and location of such remodeling activity, in turn, determines the efficiency of critical cellular functions and developmental processes. The endocytic membranes, for example, have to be remodeled and detached from the plasma membrane for neurotransmitter uptake in neurons and are therefore fundamental to nervous system functions (Ferguson and De Camilli, 2012).
Similarly, mitochondrial division requires membrane scission, which ensures the distribution of both catabolic and anabolic capacity and the inheritance of the correct complement of mitochondria by daughter cells (Mishra and Chan, 2014). Endocytic and organelle division events require dedicated protein-based machinery that can remodel and cut membranes. The structure and allosteric domain movements within these remodelers are critical to their activity. We discuss recent developments in this field below.
FISSION GTPASES OF THE DYNAMIN SUPERFAMILY OF PROTEINS
The dynamin superfamily of proteins (DSPs) catalyze membrane fission and fusion reactions during processes ranging from endocytosis in eukaryotes to cell division in certain bacteria (Jimah and Hinshaw, 2019). They play critical roles in organelle homeostasis and as innate immunity effectors that restrict certain pathogens (Bui and Shaw, 2013; Haller et al., 2015). Independent of their membrane binding activity, DSPs also play an increasingly appreciated role as regulators and organizers of both the actin- and microtubule-based cytoskeleta (Strack et al., 2013; Hatch et al., 2014; Zhang et al., 2019).
Endocytic dynamin knockout mice display neuronal dysfunction and, depending on the dynamin isoform, either are embryonic-lethal or have a lifespan limited to days or hours (Park et al., 2013). Similarly, knockout mice for the mitochondrial fission dynamin DRP1 (dynamin-related protein 1) are embryonic-lethal (Wakabayashi et al., 2009). In cells, dynamin knockouts result in clathrin-coated pits stuck to the plasma membrane, characterized by long tubules projecting into the cell (Ferguson et al., 2009). DRP1 down-regulation similarly results in undivided and hyperelongated mitochondria (Otsuga et al., 1998). In this review, we focus on comparing the plasma membrane-active endocytic, or “modern,” dynamins with the mitochondrial and peroxisomal membrane-active “ancient” or dynamin-related proteins to highlight our current understanding of their macromolecular motions, their assembly determinants, and the structural basis of their membrane fission function (Ramachandran and Schmid, 2018)
FISSION DYNAMIN STRUCTURES INCLUDE TWO HINGES
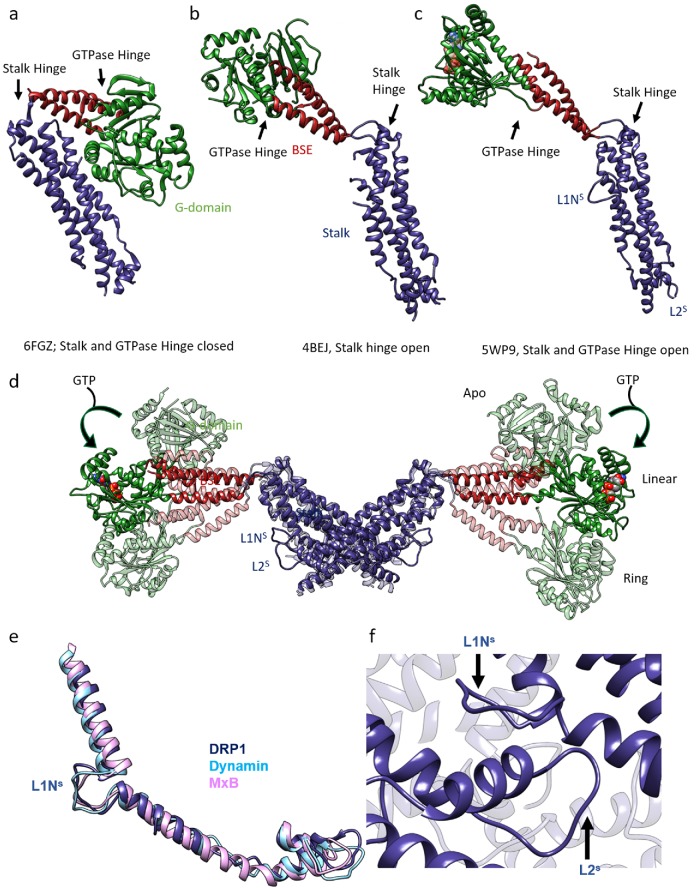
Both the modern and ancient dynamin family proteins considered here are “large” GTPases that utilize the energy of nucleotide hydrolysis to fission membranes. Structural elements include the highly conserved N-terminal GTPase (G) domain that binds GTP with low affinity (Km ∼ 15 µM) and hydrolyzes it with a high basal hydrolysis rate (Kcat ∼ 0.3 s–1 for unpolymerized modern dynamins) that can be stimulated ∼1000-fold upon exposure to lipids and subsequent oligomerization (Stowell et al., 1999). The G-domain is followed by a three-helix bundle called the bundle signaling element (BSE) connected to the G-domain via a flexible hinge (hereafter referred to as the GTPase hinge). A second hinge (hereafter referred to as the Stalk hinge) connects the BSE to a four-helix bundle called the stalk (Figure 1, a–c). All known fission dynamins are obligate dimers, and the dimeric interface (interface-2) resides within the four-helix stalk region (Figure 1d). Additional interactions at the tips of the stalk regions (including interface 1 at the “top” of the stalk and interface 3 at the “bottom” of the stalk) promote assembly of dynamins into tetramers and higher-order oligomers (Gao et al., 2010; Faelber et al., 2011; Ford et al., 2011). Indeed, all dynamins purified to date purify as dimers and/or tetrameric oligomers, and their structures have revealed a range of open versus closed states for the two hinges (Faelber et al., 2011; Ford et al., 2011; Gao et al., 2011; Reubold et al., 2015; Bohuszewicz and Low, 2018; Kalia et al., 2018; Kong et al., 2018).
FIGURE 1:
Structural rearrangements within the DRP1 molecules upon nucleotide binding: (a) Cyanidioschyzon merolae Dnm1 crystal structure depicts a closed state of a DRP1 molecule. (b) Crystal structure of human DRP1 shows an open GTPase hinge from chain (A) where both the loops that constitute the GTPase hinge were ordered. (c) Cryo-EM structure of DRP1 determined with nucleotide and the MID49 receptor. The G-domain is rotated and loops L1NS and L2S are stabilized and visible. (d) Overlay of DRP1 dimers in the apo state (4BEJ, light shade, Fröhlich et al., 2013), the linear cryo-EM structure (5WP9, solid color), and the ring model (light shade, bent downward), as seen from Kalia et al. (2018), showing the range of movements exhibited by the G-BSE region relative to the stalk. The stalk is kept constant. (e, f) L1NS and L2S stabilization and interaction in dynamin family members: (e) Overlay of the region of the stalk that contains L1NS and L2S—from DRP1 cryo-EM structure (PDB ID: 5WP9), dynamin-3 crystal structure (PDB ID: 53AF), and MxB cryo-EM structure (PDB ID: 5UOT)—to depict the structural conservation of the region across dynamin family members. In each case, L1NS and L2S are stabilized by interdynamin and/or dynamin–receptor contacts. (f) L1NS and L2S mediate interdynamin contacts in the DRP1 cryo-EM structure (PDB ID: 5WP9).
Modern dynamins also have a pleckstrin homology (PH) domain that interacts with the phospholipid headgroups of phosphatidylinositol (4,5) bisphosphate (PI(4,5)P2) for membrane localization (Ferguson et al., 1994; Zheng et al., 1996). While the PH domain is absent from ancient dynamins such as DNM1/DRP1, these enzymes harbor an ∼100–amino acid unstructured region in its place, the B-insert, which binds to other anionic lipids such as phosphatidic acid (Adachi et al., 2016) and cardiolipin on mitochondrial membranes (Bustillo-Zabalbeitia et al., 2014; Francy et al., 2017). Fission dynamins utilize these membrane-binding elements to form polymeric assemblies that perform mechanical work on the underlying membrane, and these assemblies on lipid templates are often helical or ringlike oligomers that can encircle and constrict membrane tubes of variable dimensions in vitro (Marks et al., 2001; Roux et al., 2006; Mears et al., 2011; Kalia et al., 2015; Kong et al., 2018).
CONFORMATION-DEPENDENT REMODELING OF MEMBRANES BY DSPs
Remodeling of membranes by DSPs was first suggested by Drosophila shibire mutants that became paralyzed at nonpermissive temperatures. Electron micrographs of sections through their synapses revealed unreleased synaptic vesicles, the necks of which immunostained positive for dynamin (Koenig and Ikeda, 1989; Takei et al., 1995). Similarly, assemblies of DRP1 have been observed to constrict and catalyze fission at the mitochondria (Labrousse et al., 1999; Legesse-Miller et al., 2003). Consistent with membrane remodeling in vivo, assemblies of purified DSPs over a diverse range of templates such as liposomes (Sweitzer and Hinshaw, 1998; Takei et al., 1998), microtubules (Shpetner and Vallee, 1989), and lipid nanotubes (Stowell et al., 1999; Marks et al., 2001) have also been described in vitro. In particular, modern dynamins are known to tubulate lipid vesicles containing PI(4,5)P2, and these tubules can be further constricted, and even vesiculated, upon addition of GTP in the reaction mixtures (Roux et al., 2006; Pucadyil and Schmid, 2008). PI(4,5)P2 seems to be important in particular to stimulate GTPase activity of PH domain–containing dynamins, since additional amounts of other anionic lipids on lipid templates have little or no effect on GTPase activity (Stowell et al., 1999). Ancient dynamins such as DRP1, on the other hand, are known to tubulate liposomes that have an artificially high, up to 100% content of anionic lipids such as phosphatidylserine (PS), suggesting a more nonspecific interaction between the B-insert and the underlying phospholipids in the membrane (Lackner et al., 2009; Mears et al., 2011). In addition to the lipid content, the tension in the underlying membrane is also considered an important determinant for fission, and some reports indicate that tension on lipid templates is needed to proceed toward complete fission, and loss of such membrane tension in cultured cells results in trapped clathrin–coated pits at late stages of endocytosis (Roux et al., 2006; Morlot et al., 2012).
Because the crystal structures of nearly full-length recombinant DSPs do not contain nucleotides, membranes, or binding partners, how nucleotide binding affects membrane interaction and structural dynamics of full-length dynamins has remained an area of active study. In particular, nucleotide-based changes within the G-domain, and how these changes remodel the relationship and orientations of the G-BSE and the BSE-stalk regions via the flexible hinges are important details that may explain dynamin assembly properties in detail. Moreover, how interfaces within dynamin dimers or tetramers may change to accommodate membranes and binding partners is an important structural question. In the next sections, we discuss structural details that illustrate the differences in dynamin conformations during this conversion from the cytosolic apo state to the membrane- or receptor-bound and nucleotide-activated states.
NUCLEOTIDE-INDUCED STRUCTURAL CHANGES
Soluble-state dynamin structures
Early hints that the membrane fission activity of DSPs depended on conformational dynamics came from loss-of-function mutants that displayed defective oligomerization properties (Gao et al., 2010). Because polymerization of dynamins hinders crystallization, these polymerization-incompetent mutants were sought and used to determine crystal structures of nearly full-length molecules (PDB ID’s: 3SNH, 3ZVR, 4BEJ, and 53AF) (Faelber et al., 2011; Ford et al., 2011; Fröhlich et al., 2013; Reubold et al., 2015). The winning constructs typically carried mutations in the stalk region as well as deletions of the PRD or the B-insert. These structures revealed a closed GTPase hinge, such that the G-domain packed closely with the BSE. The Stalk hinge in these structures showed flexibility within the asymmetric units of the crystal, in that some asymmetric units showed more compaction of the G-BSE than the stalk. This flexibility suggests that the Stalk hinge samples a range of different conformations even in a defined nucleotide-free state (Faelber et al., 2011; Fröhlich et al., 2013).
A recent 7 Å–resolution crystal structure of the mitochondrial fission dynamin from Cyanidioschyzon merolae (CmDnm1, PDB ID: 6FGZ) shows a new and wholly compacted state of the molecule (Bohuszewicz and Low, 2018). The apo-state tetramer contains a stalk interface 2 and a new interface 5 that mediates contacts between two adjacent stalk dimers. The authors argue that this is the fully closed, unassembled state of the molecule, which must open to form 15- to 18-membered rings around membranes—as observed in vitro at higher protein concentrations. Remarkably, both the Stalk hinge and the GTPase hinge are closed in this DRP1 ortholog structure (Figure 1a). The structure reinforces the importance of interface 2 in the dynamin superfamily, since this interface buries the largest surface area and occurs in this and all other structures of fission dynamins. Also, an overall resemblance of the structure to the fusion dynamins speaks to a shared evolutionary ancestry across diverse dynamin family members (Low et al., 2009; Bohuszewicz et al., 2016).
Nucleotide-bound minimal constructs
By contrast with the nucleotide-free crystal structures described above, x-ray structures of nucleotide-bound dynamins have primarily been studied in the context of minimized, engineered constructs that consist only of a G-domain or a fusion of the G-domain and an engineered BSE domain. For both endocytic and the mitochondrial dynamins, multiple crystal structures exist for these constructs in the GDP (PDB ID: 5D3Q), GMPPCP (PDB ID: 3ZYC, 3W6O), and GDP-AlF4 (PDB ID: 2 × 2E, 3W6P) bound states (Chappie et al., 2010, 2011; Anand et al., 2016). Importantly, all these structures reveal a dimeric assembly state formed via a direct G–G interaction, which is understood to stabilize the catalytic residues for nucleotide hydrolysis, including the main-chain carbon atoms that coordinate a hydrolyzing water molecule and a cation that participates in charge compensation in the transition state. Subtle changes in nucleotide coordination within the binding pocket, due to the nonhydrolyzsable methylene in GMPPCP, revealed a crucial change in the GMPPCP-bound structure: while the rest of the catalytic site was similar to the GDP–AlF4 state, the BSE in the GMPPCP state was in the open conformation, rotated ∼69° away from the G-domain (Chappie et al., 2011). This rotation depends on a conserved proline in this region (P294 in human dynamin-1) and leads to an opening of the GTPase hinge. The opening observed in these minimal constructs left open the question as to what happens to the Stalk hinge upon nucleotide binding, since the stalk and its associated hinge were both missing from these constructs. Cryo-EM structures have also suggested a G–G contact between adjacent rungs of dynamin helices, corroborating the idea of trans-G domain dimerization. Biochemical measurements support this idea, and dynamin-1, for example, shows dramatically increased GTPase activity with G–G dimerization—a phenomenon described as “assembly-stimulated GTP hydrolysis” (Chappie et al., 2011; Chappie and Dyda, 2013; Mears et al., 2011).
DRP1 cryo-EM structure with MID49 and GMPPCP
As noted above for the visualization of G–G dimers in a reconstruction of dynamin-1 (Chappie et al., 2011), cryo-EM has begun to reveal how nucleotide binding and hydrolysis lead to allosteric protein motions within full-length dynamins (Kalia et al., 2018; Kong et al., 2018). For DRP1, guanine nucleotide binding induces a 90° rotation of the G-domain via the GTPase hinge while orchestrating an ∼40-Å movement of the G-domain toward the stalk via the Stalk hinge (Kalia et al., 2018; Figure 1d). In the structure, both hinges are open, leading to 1) exposure of two of the four interfaces where DRP1 binds with its receptor MID49 or MID51 and 2) copolymerization of DRP1 with MID49/51, leading to a vast and highly avid DRP1 assembly network on mitochondrial membranes. Perhaps coassembly of DRP1 and MID49/51 into these long and approximately linear polymers on the outer surfaces of mitochondria facilitates the recruitment of sufficient copies of DRP1 to appreciably decorate the much larger surface around a mitochondrial tubule, as compared with the thin plasma membrane necks that “modern” dynamins sever. Similar linear polymers have been observed for the assembly of DRP1 with MFF, but structural details of that assembly are unknown (Clinton et al., 2016).
Part of the interaction network with the MID49/MID51 receptors included the conserved loops that interrupt the alpha helices of the stalk. The DRP1 L1NS and L2S loops involved in these receptor interfaces determine the geometry of assembly for other dynamin-family oligomers (Reubold et al., 2015; Alvarez et al., 2017). The conserved L1NS loop is a 12-residue interruption of the first stalk helix and the site of several disease alleles that correlate with elongated mitochondria. These mutations include missense substitutions of the deeply conserved residues G362 and G363. A prior x-ray crystallography study established that this loop composes part of the intramolecular PH-domain binding site for a soluble conformation of endocytic dynamin tetramers (Reubold et al., 2015). Indeed, the disease-associated G362D mutant of DRP1 failed to coassemble with MID49 and displayed both altered assembly and conformational properties (Kalia et al. 2018). These observations are consistent with the phenotypes associated with this mutation: elongated mitochondria in cells and a defect in DRP1 recruitment to mitochondria (Sheffer et al., 2016; Vanstone et al., 2016).
The entire backbone of the DRP1 L1NS loop was resolved in the cryo-EM structure with MiD49, while the same region in a crystal lattice was mostly disordered (Fröhlich et al., 2013). The stability can be explained by the multivalent contacts that the loop made both with the receptor and with the L2s loop of the next molecule over within the linear oligomer (Figure 1, e and f). This loop was observed to make similar critical contacts within the helical oligomer studied in a recent cryo-EM structure of MxB (Alvarez et al., 2017) and the lattices of the dynamin-3 crystal structure (Reubold et al., 2015; Figure 1e). Interestingly, only one residue change (G362D) within this loop was sufficient to break the interaction of DRP1 with MiD49 and was also able—via yet unknown allosteric motions—to shift the polymer architecture from linear strings to closed rings that compose twelve dimers of DRP1 in a new conformation (Kalia et al., 2018).
Interestingly, wild-type DRP1 could also form closed rings, but only through a path-dependent reaction that depended on the formation of a linear oligomer first with MID49. Nucleotide exchange from GMPPCP to GTP then drove the linear oligomer to dissociate from MID49/51 and to curl into closed rings. Complete hydrolysis of the GTP in solution led to complete disassembly of the rings. L1Ns also maintains critical intermolecular contacts in the recent dynamin-on-lipid cryo-EM structure discussed below (Kong et al., 2018). Based on its location, we predict that this loop must undergo conformational changes that underlie the structural basis of the linear filament to ring (or helix) conversion. Finally, while L1NS is vital for the allostery that mediates the filament to ring formation, it may not be the only determinant of such changes. We also noted that a G-domain mutant—D221A, which is remote from the L1NS loop—also induces DRP1 to form rings with GMPPCP and MID49, rather than linear filaments. Further structural studies of the closed ring state of DRP1 will help explain the short- and long-range allosteric motions that are responsible for DRP1 curling into closed rings (Srinivasan et al., 2016).
Another important aspect of this structure is the L2S loop (residues 402–409 in human DRP1). This loop resides at the tip of the stalk (Figure 1e) and mediates contact between dynamin tetramers by participating in the formation of stalk interface-3. Previous structures (PDB ID’s 5UOT, 53AF) have also shown this loop in a similar configuration (Reubold et al., 2015; Alvarez et al., 2017). Similarly to L1NS, the reason for the stabilization of this loop in these structures is the interaction with L1NS of a neighboring dynamin (Figure 1, d–f). This loop also harbors a disease allele, Arg403, and patients harboring the R403C mutation show symptoms that include epilepsy and encephalopathy, possibly due to defective DRP1 oligomerization (Fahrner et al., 2016).
These in vitro observations raise new questions about DRP1-based mitochondrial constriction: 1) Are the DRP1 rings sufficient to constrict mitochondria? In the Kalia et al. (2018) study, the rings formed by the wild-type protein after disengaging from the MID49/51 receptors ranged in size from 11 to 16 dimers of DRP1, and, interestingly, some reports have indicated that efficient membrane fission could be driven by oligomers of human dynamin-2 corresponding to this size (Cocucci et al., 2014). 2) How are the receptors modified or removed to enable DRP1 to disengage and curl up into rings? If the MID49/51 receptors are always around, a linear, rather than a ring state of DRP1, will be favored—precluding constriction and most likely explaining the overexpression phenotype for transgenic MID49/51 receptors (Zhao et al., 2011). Interestingly, MARCH-V, a ubiquitin ligase that ubiquitylates MID49, supports mitochondrial division (Xu et al., 2016), which suggests that ubiquitination of receptors may be a step in the division process.
New views of dynamin-1 around a lipid tubule
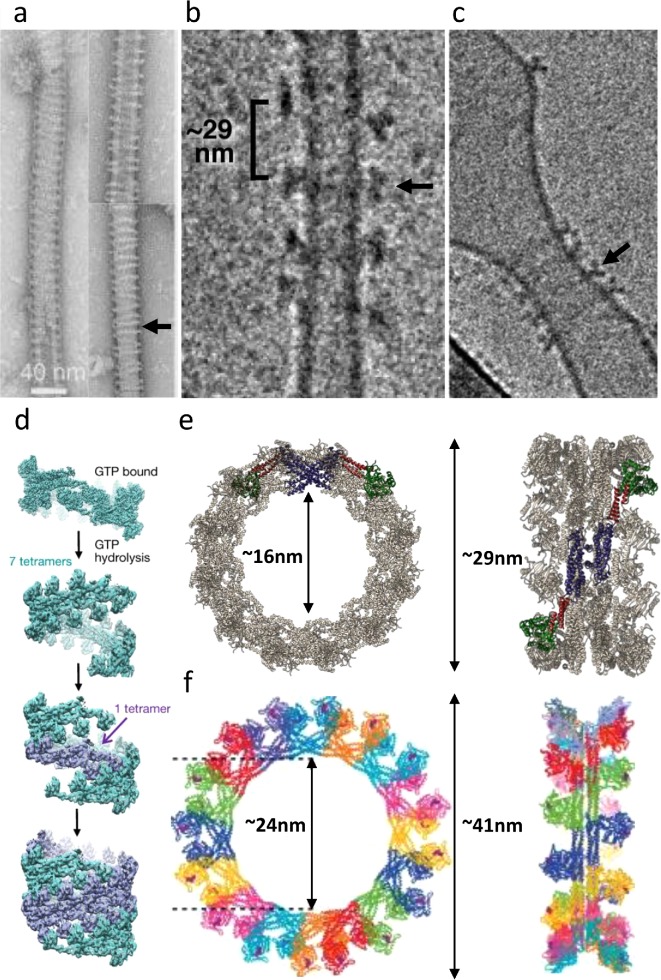
A recent study has also reported high-resolution structures of helical assemblies of endocytic dynamin on a lipid template (Kong et al., 2018). While low-resolution structures of dynamins around membrane cylinders were known (Chappie et al., 2011; Mears et al., 2011; Sundborger et al., 2014), this study reports near–atomic resolution snapshots of dynamin bound to lipid tubules in a GMPPCP bound state. The nucleotide-based conformational changes here closely resemble those seen in the case of the minimal constructs with GMPPCP (Chappie et al., 2011) and the DRP1 study mentioned above (Kalia et al., 2018). While G–G dimerization observed in this study strengthens the concept of assembly-mediated hydrolysis for certain members of the dynamin family, fundamental questions remain as to the steps that follow and lead to complete fission. While the initial assembly appears to be a one-start helix, the addition of GTP leads to nucleotide hydrolysis and a conversion into a pitch-expanded two-start helix with greater constriction, perhaps due to the force exerted on the membrane via the PH domains (Figure 2, a and b, Marks et al., 2001; Figure 2d, Kong et al., 2018).
FIGURE 2:
Dynamin helices and rings. (a) Left: A continuous dynamin-1 helix arranged on lipid nanotubes, in the presence of GTP-γS (Marks et al., 2001). Reproduced with permission from Nature Publishing Group. Right: same as left after incubation with 500 µM GTP for 30 min. Note the increase in the helical pitch (arrows) and possible discontinuous helices/rings. (b) DRP1 helical polymers incubated with GTP, leading to an increase in helical pitch (Mears et al., 2011). Reproduced with permission from Nature Publishing Group. These structures (arrow) could represent helices with a very elongated pitch, or rings of DRP1. (c) Rings of MxA observed on a lipid template, as seen in Kochs et al. (2005); Haller et al. (2010). Reproduced with permission from Academic Press and ASBMB. (d) A model for dynamin-1–based membrane fission, as described by Kong et al. (2018). Reproduced with permission from Nature Publishing Group. (e) Models for rings of DRP1 (Kalia et al., 2018) and (f) of MxA (Haller et al., 2010). Reproduced with permission from ASBMB.
Interestingly, the monomers within the dimeric subunit of this helical oligomer exhibit an asymmetry in the GTPase hinge. While one monomer appeared to have a completely open GTPase hinge, another displayed a kink in this area (residues 292–294). Consequently, the PH domain of the bent dynamin was also better defined in the structure, leading the authors to propose that the bent dynamin is stabilized by a transfer of force to the lipid membrane. Indeed, in terms of the secondary structure, mutants that increase the helical propensity of this region, and thereby prevent kinking, display reduced transferrin uptake in cell cultures, consistent with decreased dynamin-mediated endocytosis. Because such kinking may underlie a mechanism for the relay of the force generated by polymeric assembly via allosteric coupling between the G-BSE and PH domains upon nucleotide binding and hydrolysis, this may be a way to generate torque for membrane fission, as has been hypothesized elsewhere (Pannuzzo et al., 2018). Overall, these observations are consistent with previous biophysical and crystallographic predictions regarding the conformational opening of the PH domains with respect to the stalk and the involvement of L1NS and the L2S loops in maintaining inter-dynamin contacts (Reubold et al., 2015; Srinivasan et al., 2016).
Helices versus closed rings
The steps leading to complete membrane fission by DSPs have been a subject of intense study. GTP exposure to preassembled dynamin polymers results in constricted assemblies—a two-start helix for dynamin and rings for DRP1. Given the differences in the states of the polymers in the dynamin-1 vs. DRP1 structures, what is the role of G–G contacts in DRP1-based constriction? While no G–G contacts were seen within DRP1 rings, “stacked” or adjacent rings could form G–G contacts in trans on lipid surfaces, leading to assembly-stimulated hydrolysis of GTP and generation of torque on the underlying membrane. Alternatively, the oligomeric state could be different in the absence of a membrane template and, perhaps, become helical when assembled on a lipid surface. While DRP1 helical oligomers have not been observed over a “mitochondrial” lipid mix, pure DOPS liposomes can be tubulated by DRP1, which forms loosely organized helical assemblies around such membranes (Lackner et al., 2009; Mears et al., 2011). Intriguingly, it has been demonstrated that incubation of such mixtures with GTP leads to structures that resemble rings, or at least helical oligomers, with a very large pitch or interrung distance (Figure 2b; Mears et al., 2011).
Moreover, rings have been observed on lipid templates in the case of the antiviral dynamin MxA (Figure 2, c and f; Kochs et al., 2005; Haller et al., 2010) and even dynamin-1 on lipid nanotubes, after these proteins were incubated with lipid templates and GTP (Figure 2, a and b; Marks et al., 2001). Such observations suggest that closed rings may be an overlooked state of dynamin-based, or at least DRP1-based, polymerization, constriction, and eventual fission cycle and that their structure and function deserve more in-depth study. Kalia et al. (2018) noted that rings with an ∼16-nm inner diameter might not be constricted enough to fission the double-membrane mitochondria (Figure 2e). Curiously, dynamin-2 may also play a role in mitochondrial division downstream of DRP1, which may explain how DRP1 could act as a “constrictase” by forming closed rings, while leaving the final fission step to a “modern” dynamin that is tuned to the dimensions of the DRP1-constricted ring (Lee et al., 2016). Alternatively, by destabilizing the inner membrane of the mitochondria, the rings themselves may induce membrane destabilization and fission. In support of this idea, a recent report suggests that DRP1 is sufficient for mitochondrial fission in murine embryonic fibroblasts that lack all dynamins-1, -2, and -3 (Kamerkar et al., 2018). This makes the rings on lipid membranes an important candidate for further biophysical and structural characterization. Finally, the mechanisms of membrane scission may be disparate or context-dependent for different dynamin family members, proceeding via helical oligomers for some, and via ring-shaped assemblies for others.
Together, new views of dynamins have illustrated ways in which DSPs bind nucleotides, undergo conformational changes, and remodel target membranes. Nucleotide binding in the G-domains opens up dynamins to assemble via stalks—and these movements are orchestrated by the conformational flexibility in the Stalk hinge and the GTPase hinge. As noted above, fascinating structural loops such as the L1NS and L2S also coordinate allosteric changes that enable dynamin fission-based functions. Further understanding of the structural changes within these elements in dynamins may enable us to understand membrane fission better and to develop targeted therapies for dynamin/DRP1-based pathogenesis. In disease systems such as encephalopathy, microcephaly, optic atrophy, and Charcot–Marie–Tooth disease, where dynamin function is compromised, targeted therapies could enable efficient fission (Zuchner et al., 2004; Waterham et al., 2007; Chang et al., 2010; Dhindsa et al., 2015; Vanstone et al., 2016). Conversely, systems where hyperfission enables diseases such as certain injuries related to hypoxia and reperfusion or specific malignancies, structure-based inhibition of dynamin/DRP1 assembly may be beneficial (Qian et al., 2013; Ding et al., 2017; Zhang et al., 2018). With new and future structures, we look toward understanding the full conformational repertoire and dynamics of DSPs with membranes and binding partners—as well as structure-inspired therapeutic interventions to tackle disease.
Acknowledgments
We thank Janet Shaw, Paul Thomas, Halil Aydin, Arthur Mello, and other members of the Frost lab for helpful discussions and comments on the manuscript. We also thank the anonymous reviewers for their insightful comments. This work was further supported by a Faculty Scholar grant from the HHMI (A.F.), the Searle Scholars Program (A.F.), National Institutes of Health (NIH) grant 1DP2GM110772-01 (A.F.), and NIH/National Institute of General Medical Sciences grant R01 GM127673-01 (A.F.). A.F. is a Chan Zuckerberg Biohub investigator.
Abbreviations used:
- BSE
bundle signaling element
- DRP1
dynamin-related protein 1
- DSP
dynamin superfamily protein
- GMPPCP
β,γ-methyleneguanosine 5′-triphosphate
- GTP
guanosine 5′-triphosphate
- Kcat
turnover number
- Km
Michaelis–Menten constant.
Footnotes
REFERENCES
- Adachi Y, Itoh K, Yamada T, Cerveny KL, Suzuki TL, Macdonald P, Frohman MA, Ramachandran R, Iijima M, Sesaki H. (2016). Coincident phosphatidic acid interaction restrains Drp1 in mitochondrial division. Mol Cell , 1034–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez FJD, He S, Perilla JR, Jang S, Schulten K, Engelman AN, Scheres SHW, Zhang P. (2017). CryoEM structure of MxB reveals a novel oligomerization interface critical for HIV restriction. Sci Adv , e1701264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand R, Eschenburg S, Reubold TF. (2016). Crystal structure of the GTPase domain and the bundle signalling element of dynamin in the GDP state. Biochem Biophys Res Commun , 76–80. [DOI] [PubMed] [Google Scholar]
- Bohuszewicz O, Liu J, Low HH. (2016). Membrane remodelling in bacteria. J Struct Biol , 3–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohuszewicz O, Low HH. (2018). Structure of a mitochondrial fission dynamin in the closed conformation. Nat Struct Mol Biol , 722–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui HT, Shaw JM. (2013). Dynamin assembly strategies and adaptor proteins in mitochondrial fission. Curr Biol , R891–R899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustillo-Zabalbeitia I, Montessuit S, Raemy E, Basanez G, Terrones O, Martinou JC. (2014). Specific interaction with cardiolipin triggers functional activation of dynamin-related protein 1. PLoS One , e102738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CR, Manlandro CM, Arnoult D, Stadler J, Posey AE, Hill RB, Blackstone C. (2010). A lethal de novo mutation in the middle domain of the dynamin-related GTPase Drp1 impairs higher order assembly and mitochondrial division. J Biol Chem , 32494–32503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappie JS, Acharya S, Leonard M, Schmid SL, Dyda F. (2010). G domain dimerization controls dynamin’s assembly-stimulated GTPase activity. Nature , 435–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappie JS, Dyda F. (2013). Building a fission machine—structural insights into dynamin assembly and activation. J Cell Sci , 2773–2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappie JS, Mears JA, Fang S, Leonard M, Schmid SL, Milligan RA, Hinshaw JE, Dyda F. (2011). A pseudoatomic model of the dynamin polymer identifies a hydrolysis-dependent powerstroke. Cell , 209–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinton RW, Francy CA, Ramachandran R, Qi X, Mears JA. (2016). Dynamin-related protein 1 oligomerization in solution impairs functional interactions with membrane-anchored mitochondrial fission factor. J Biol Chem , 478–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocucci E, Gaudin R, Kirchhausen T, Lippincott-Schwartz J. (2014). Dynamin recruitment and membrane scission at the neck of a clathrin-coated pit. Mol Biol Cell , 3595–3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhindsa RS, Bradrick SS, Yao X, Heinzen EL, Petrovski S, Krueger BJ, Johnson MR, Frankel WN, Petrou S, Boumil RM, et al (2015). Epileptic encephalopathy-causing mutations in DNM1 impair synaptic vesicle endocytosis. Neurol Genet , e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding M, Dong Q, Liu Z, Liu Z, Qu Y, Li X, Huo C, Jia X, Fu F, Wang X. (2017). Inhibition of dynamin-related protein 1 protects against myocardial ischemia-reperfusion injury in diabetic mice. Cardiovasc Diabetol , 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faelber K, Posor Y, Gao S, Held M, Roske Y, Schulze D, Haucke V, Noe F, Daumke O. (2011). Crystal structure of nucleotide-free dynamin. Nature , 556–560. [DOI] [PubMed] [Google Scholar]
- Fahrner JA, Liu R, Perry MS, Klein J, Chan DC. (2016). A novel de novo dominant negative mutation in DNM1L impairs mitochondrial fission and presents as childhood epileptic encephalopathy. Am J Med Genet A , 2002–2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson KM, Lemmon MA, Schlessinger J, Sigler PB. (1994). Crystal structure at 2.2 Å resolution of the pleckstrin homology domain from human dynamin. Cell , 199–209. [DOI] [PubMed] [Google Scholar]
- Ferguson SM, De Camilli P. (2012). Dynamin, a membrane-remodelling GTPase. Nat Rev Mol Cell Biol , 75–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson SM, Raimondi A, Paradise S, Shen H, Mesaki K, Ferguson A, Destaing O, Ko G, Takasaki J, Cremona O, et al (2009). Coordinated actions of actin and BAR proteins upstream of dynamin at endocytic clathrin-coated pits. Dev Cell , 811–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford MG, Jenni S, Nunnari J. (2011). The crystal structure of dynamin. Nature , 561–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francy CA, Clinton RW, Fröhlich C, Murphy C, Mears JA. (2017). Cryo-EM studies of Drp1 reveal cardiolipin interactions that activate the helical oligomer. Sci Rep , 10744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fröhlich C, Grabiger S, Schwefel D, Faelber K, Rosenbaum E, Mears J, Rocks O, Daumke O. (2013). Structural insights into oligomerization and mitochondrial remodelling of dynamin 1-like protein. EMBO J , 1280–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S, von der Malsburg A, Dick A, Faelber K, Schroder GF, Haller O, Kochs G, Daumke O. (2011). Structure of myxovirus resistance protein a reveals intra- and intermolecular domain interactions required for the antiviral function. Immunity , 514–525. [DOI] [PubMed] [Google Scholar]
- Gao S, von der Malsburg A, Paeschke S, Behlke J, Haller O, Kochs G, Daumke O. (2010). Structural basis of oligomerization in the stalk region of dynamin-like MxA. Nature , 502–506. [DOI] [PubMed] [Google Scholar]
- Haller O, Gao S, von der Malsburg A, Daumke O, Kochs G. (2010). Dynamin-like MxA GTPase: structural insights into oligomerization and implications for antiviral activity. J Biol Chem , 28419–28424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haller O, Staeheli P, Schwemmle M, Kochs G. (2015). Mx GTPases: dynamin-like antiviral machines of innate immunity. Trends Microbiol , 154–163. [DOI] [PubMed] [Google Scholar]
- Hatch AL, Gurel PS, Higgs HN. (2014). Novel roles for actin in mitochondrial fission. J Cell Sci , 4549–4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimah JR, Hinshaw JE. (2019). Structural insights into the mechanism of dynamin superfamily proteins. Trends Cell Biol , 257–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalia R, Talledge N, Frost A. (2015). Structural and functional studies of membrane remodeling machines. Methods Cell Biol , 165–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalia R, Wang RY, Yusuf A, Thomas PV, Agard DA, Shaw JM, Frost A. (2018). Structural basis of mitochondrial receptor binding and constriction by DRP1. Nature , 401–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamerkar SC, Kraus F, Sharpe AJ, Pucadyil TJ, Ryan MT. (2018). Dynamin-related protein 1 has membrane constricting and severing abilities sufficient for mitochondrial and peroxisomal fission. Nat Commun , 5239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochs G, Reichelt M, Danino D, Hinshaw JE, Haller O. (2005). Assay and functional analysis of dynamin-like Mx proteins. Methods Enzymol , 632–643. [DOI] [PubMed] [Google Scholar]
- Koenig JH, Ikeda K. (1989). Disappearance and reformation of synaptic vesicle membrane upon transmitter release observed under reversible blockage of membrane retrieval. J Neurosci , 3844–3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L, Sochacki KA, Wang H, Fang S, Canagarajah B, Kehr AD, Rice WJ, Strub M-P, Taraska JW, Hinshaw JE. (2018). Cryo-EM of the dynamin polymer assembled on lipid membrane. Nature , 258–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labrousse AM, Zappaterra MD, Rube DA, Bliek AMVD. (1999). C. elegans dynamin-related protein DRP-1 controls severing of the mitochondrial outer membrane. Mol Cell , 815–826. [DOI] [PubMed] [Google Scholar]
- Lackner LL, Horner JS, Nunnari J. (2009). Mechanistic analysis of a dynamin effector. Science , 874–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JE, Westrate LM, Wu H, Page C, Voeltz GK. (2016). Multiple dynamin family members collaborate to drive mitochondrial division. Nature , 139–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legesse-Miller A, Massol RH, Kirchhausen T. (2003). Constriction and Dnm1p recruitment are distinct processes in mitochondrial fission. Mol Biol Cell , 1953–1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low HH, Sachse C, Amos LA, Lowe J. (2009). Structure of a bacterial dynamin-like protein lipid tube provides a mechanism for assembly and membrane curving. Cell , 1342–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks B, Stowell MHB, Vallis Y, Mills IG, Gibson A, Hopkins CR, McMahon HT. (2001). GTPase activity of dynamin and resulting conformation change are essential for endocytosis. Nature , 231–235. [DOI] [PubMed] [Google Scholar]
- Mears JA, Lackner LL, Fang S, Ingerman E, Nunnari J, Hinshaw JE. (2011). Conformational changes in Dnm1 support a contractile mechanism for mitochondrial fission. Nat Struct Mol Biol , 20–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra P, Chan DC. (2014). Mitochondrial dynamics and inheritance during cell division, development and disease. Nat Rev Mol Cell Biol , 634–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morlot S, Galli V, Klein M, Chiaruttini N, Manzi J, Humbert F, Dinis L, Lenz M, Cappello G, Roux A. (2012). Membrane shape at the edge of the dynamin helix sets location and duration of the fission reaction. Cell , 619–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuga D, Keegan BR, Brisch E, Thatcher JW, Hermann GJ, Bleazard W, Shaw JM. (1998). The dynamin-related GTPase, Dnm1p, controls mitochondrial morphology in yeast. J Cell Biol , 333–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannuzzo M, McDargh ZA, Deserno M. (2018). The role of scaffold reshaping and disassembly in dynamin driven membrane fission. Elife , e39441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park RJ, Shen H, Liu L, Liu X, Ferguson SM, De Camilli P. (2013). Dynamin triple knockout cells reveal off target effects of commonly used dynamin inhibitors. J Cell Sci , 5305–5312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pucadyil TJ, Schmid SL. (2008). Real-time visualization of dynamin-catalyzed membrane fission and vesicle release. Cell , 1263–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian W, Wang J, Van Houten B. (2013). The role of dynamin-related protein 1 in cancer growth: a promising therapeutic target? Expert Opin Ther Targets , 997–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran R, Schmid SL. (2018). The dynamin superfamily. Curr Biol , R411–R416. [DOI] [PubMed] [Google Scholar]
- Reubold TF, Faelber K, Plattner N, Posor Y, Ketel K, Curth U, Schlegel J, Anand R, Manstein DJ, Noe F, et al (2015). Crystal structure of the dynamin tetramer. Nature , 404–408. [DOI] [PubMed] [Google Scholar]
- Roux A, Uyhazi K, Frost A, De Camilli P. (2006). GTP-dependent twisting of dynamin implicates constriction and tension in membrane fission. Nature , 528–531. [DOI] [PubMed] [Google Scholar]
- Sheffer R, Douiev L, Edvardson S, Shaag A, Tamimi K, Soiferman D, Meiner V, Saada A. (2016). Postnatal microcephaly and pain insensitivity due to a de novo heterozygous DNM1L mutation causing impaired mitochondrial fission and function. Am J Med Genet A , 1603–1607. [DOI] [PubMed] [Google Scholar]
- Shpetner HS, Vallee RB. (1989). Identification of dynamin, a novel mechanochemical enzyme that mediates interactions between microtubules. Cell , 421–432. [DOI] [PubMed] [Google Scholar]
- Srinivasan S, Dharmarajan V, Reed DK, Griffin PR, Schmid SL. (2016). Identification and function of conformational dynamics in the multidomain GTPase dynamin. EMBO J , 443–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowell MH, Marks B, Wigge P, McMahon HT. (1999). Nucleotide-dependent conformational changes in dynamin: evidence for a mechanochemical molecular spring. Nat Cell Biol , 27–32. [DOI] [PubMed] [Google Scholar]
- Strack S, Wilson TJ, Cribbs JT. (2013). Cyclin-dependent kinases regulate splice-specific targeting of dynamin-related protein 1 to microtubules. J Cell Biol , 1037–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundborger AC, Fang S, Heymann JA, Ray P, Chappie JS, Hinshaw JE. (2014). A dynamin mutant defines a superconstricted prefission state. Cell Rep , 734–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweitzer SM, Hinshaw JE. (1998). Dynamin undergoes a GTP-dependent conformational change causing vesiculation. Cell , 1021–1029. [DOI] [PubMed] [Google Scholar]
- Takei K, Haucke V, Slepnev V, Farsad K, Salazar M, Chen H, De Camilli P. (1998). Generation of coated intermediates of clathrin-mediated endocytosis on protein-free liposomes. Cell , 131–141. [DOI] [PubMed] [Google Scholar]
- Takei K, McPherson PS, Schmid S, De Camilli P. (1995). Tubular membrane invaginations coated by dynamin rings are induced by GTP-γS in nerve terminals. Nature , 186–190. [DOI] [PubMed] [Google Scholar]
- Vanstone JR, Smith AM, McBride S, Naas T, Holcik M, Antoun G, Harper ME, Michaud J, Sell E, Chakraborty P, et al (2016). DNM1L-related mitochondrial fission defect presenting as refractory epilepsy. Eur J Hum Genet , 1084–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakabayashi J, Zhang Z, Wakabayashi N, Tamura Y, Fukaya M, Kensler TW, Iijima M, Sesaki H. (2009). The dynamin-related GTPase Drp1 is required for embryonic and brain development in mice. J Cell Biol , 805–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterham HR, Koster J, van Roermund CWT, Mooyer PAW, Wanders RJA, Leonard JV. (2007). A lethal defect of mitochondrial and peroxisomal fission. N Engl J Med , 1736–1741. [DOI] [PubMed] [Google Scholar]
- Xu S, Cherok E, Das S, Li S, Roelofs BA, Ge SX, Polster BM, Boyman L, Lederer WJ, Wang C, et al (2016). Mitochondrial E3 ubiquitin ligase MARCH5 controls mitochondrial fission and cell sensitivity to stress-induced apoptosis through regulation of MiD49 protein. Mol Biol Cell , 349–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Liu Y, Tang Y, Wang X, Li Z, Li R, Ti Z, Gao W, Bai J, Lv Y. (2018). Increased mitochondrial fission is critical for hypoxia-induced pancreatic beta cell death. PLoS One , e0197266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Gerassimov N, Lee DM, Jimah JR, Kim S, Luvsanjav D, Winkelman J, Mettlen M, Abrams ME, Kalia R, et al (2019). The mechanisms of dynamin-actin interaction. BioRxiv, .org/10.1101/586461. [Google Scholar]
- Zhao J, Liu T, Jin S, Wang X, Qu M, Uhlen P, Tomilin N, Shupliakov O, Lendahl U, Nister M. (2011). Human MIEF1 recruits Drp1 to mitochondrial outer membranes and promotes mitochondrial fusion rather than fission. EMBO J , 2762–2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, Cahill SM, Lemmon MA, Fushman D, Schlessinger J, Cowburn D. (1996). Identification of the binding site for acidic phospholipids on the PH domain of dynamin: implications for stimulation of GTPase activity. J Mol Biol , 14–21. [DOI] [PubMed] [Google Scholar]
- Zuchner S, Mersiyanova IV, Muglia M, Bissar-Tadmouri N, Rochelle J, Dadali EL, Zappia M, Nelis E, Patitucci A, Senderek J, et al (2004). Mutations in the mitochondrial GTPase mitofusin 2 cause Charcot–Marie–Tooth neuropathy type 2A. Nat Genet , 449–451. [DOI] [PubMed] [Google Scholar]