Abstract
In eukaryotic cells, organelle-specific protein quality control (PQC) is critical for maintaining cellular homeostasis. Despite the Golgi apparatus being the major protein processing and sorting site within the secretory pathway, how it contributes to PQC has remained largely unknown. Using different chemical biology-based protein unfolding systems, we reveal the segregation of unfolded proteins from folded proteins in the Golgi. Quality control (QC) substrates are subsequently exported in distinct carriers, which likely contain unfolded proteins as well as highly oligomerized cargo that mimic protein aggregates. At an additional sorting step, oligomerized proteins are committed to lysosomal degradation, while unfolded proteins localize to the endoplasmic reticulum (ER) and associate with chaperones. These results highlight the existence of checkpoints at which QC substrates are selected for Golgi export and lysosomal degradation. Our data also suggest that the steady-state ER localization of misfolded proteins, observed for several disease-causing mutants, may have different origins.
INTRODUCTION
Protein quality control (PQC) is highly important for cellular homoeostasis as well as for cell survival under stress (Klaips et al., 2018). In eukaryotic cells, organelle-specific PQC machineries carry out protein surveillance and funnel misfolded or mislocalized proteins into the appropriate quality control (QC) pathways (Richter et al., 2010; Araki and Nagata, 2011; Voos et al., 2016), which include attempts at protein refolding, sequestration (removal from the healthy proteome), and degradation (Wong and Cuervo, 2010; Kim et al., 2013; Sontag et al., 2017). One-third of all newly synthesized cellular proteins enter the secretory pathway at the endoplasmic reticulum (ER) (Braakman and Bulleid, 2011). Mechanisms that guard the ER proteome are crucial to ensure the proper folding of these newly synthesized proteins. ER-localized chaperones in conjunction with the ER-associated degradation (ERAD) machinery monitor the protein folding process and prevent the accumulation of misfolded proteins (Araki and Nagata, 2011): terminally misfolded proteins that undergo ERAD are retrotranslocated to the cytosol and are subsequently degraded by the proteasome (Wu and Rapoport, 2018). ER QC also targets proteins to the lysosome for degradation. This can occur directly via a route from the ER to the lysosome or it can involve an intermediate step of transient residency at the plasma membrane (PM) prior to degradation (Satpute-Krishnan et al., 2014; Fregno et al., 2018). The ER PQC machinery is also critical for post-ER compartments as it ensures that unfolded proteins, which are not (yet) subject to degradation, are retained in the ER (Geva and Schuldiner, 2014). Most ER chaperones contain a KDEL motif at their C-termini, which allows them to be retrieved from the ER-Golgi intermediate compartment (ERGIC) or the cis-Golgi (Hammond and Helenius, 1994; Yamamoto et al., 2001; Vavassori et al., 2013; Geva and Schuldiner, 2014). Via this route, escaped unfolded proteins can be recaptured. Rer1-mediated retrieval of escaped transmembrane proteins further restricts QC substrates and folding intermediates to the ER (Sato et al., 2004). Moreover, it has been suggested that by recycling through the ER, the folding state of some Golgi resident proteins and proteins targeted to the PM can be reevaluated (Cole et al., 1998; Storrie et al., 1998; Fossati et al., 2014). Collectively, these studies highlight the far-reaching implications of ER-associated QC with respect to post-ER compartments.
In contrast to the well-known role of the ER in monitoring proteins, the contribution of post-ER compartments to PQC is only beginning to be determined. A recent study suggests the ERGIC as an important QC checkpoint (Sirkis et al., 2017). It has been shown that a mutant form of the triggering receptor expressed on myeloid cells 2 protein, which is normally localized to the PM, is stopped at the ERGIC and transported back to the ER (Sirkis et al., 2017). Aberrant forms of other proteins that usually reside at the PM or are secreted have been found halted at the Golgi apparatus, highlighting that compartment as another post-ER QC checkpoint (Coughlan et al., 2004; Wang and Ng, 2010; Puig et al., 2016). Specifically, proteins carrying mutations in their transmembrane domains or unassembled subunits of multimeric transmembrane protein complexes are retained at the Golgi and degraded by the lysosome (Briant et al., 2017). Moreover, destabilized proteins such as genetic mutants of the prion protein and a mutant form of the α1-antitrypsin protein (ATZ) have been found in the Golgi to subsequently undergo lysosomal degradation (Stewart and Harris, 2005; Ashok and Hegde, 2009; Gelling et al., 2012). Other hints for a QC pathway targeting protein aggregates from the Golgi to lysosomes in mammalian cells come from early studies on the protease furin and from recent studies on highly oligomerized proteins (Wolins et al., 1997; Tewari et al., 2015). Induced oligomerization has been shown to promote the degradation of Golgi-localized proteins by the lysosome (Tewari et al., 2014, 2015).
Molecular mechanisms of PQC at the Golgi have been primarily studied in yeast. Using model substrates, these studies revealed that QC substrates can be targeted from the Golgi to the yeast vacuole (homologous to the mammalian lysosome) in a ubiquitin- or Vps10 (homologous to mammalian sortilin)-dependent manner (Hong et al., 1996; Reggiori and Pelham, 2002; Wang et al., 2011; Gelling et al., 2012). The ubiquitin E3 ligase Rsp5 is involved in targeting unfolded proteins from the Golgi for degradation (Wang et al., 2011). Moreover, a yeast-specific Golgi- and endosome-localized E3 ligase marks proteins carrying defects in their transmembrane domains with ubiquitin and thereby targets them to the vacuole (Reggiori and Pelham, 2002; Yang et al., 2018). This pathway becomes specifically important under stress conditions, such as starvation (Dobzinski et al., 2015). Another PQC mechanism important during stress that is induced by autophagy-deficiency is Golgi-membrane-associated degradation (Yamaguchi et al., 2016). This pathway compensates for loss of autophagy to ensure the degradation of cargo when anterograde trafficking is blocked (Yamaguchi et al., 2016). In addition to protein cargo from the secretory pathway, the Golgi also receives cargo from the endocytic pathway (Bonifacino and Rojas, 2006). For example, the infectious, aggregated form of the prion protein becomes a substrate for Golgi QC during early stages of infection (Goold et al., 2013). Specifically, the prion protein is internalized from the PM, targeted to the Golgi, and from there to the endo/lysosomal system for degradation (Goold et al., 2013). Collectively these studies show that, due to its central position at the secretory and endocytic pathways, the Golgi is exposed to a constant flux of different types of QC substrates and requires a QC system to mediate their identification, sorting, and export.
We have previously shown that induced protein unfolding in the Golgi elicits a transcriptional Golgi stress response (Serebrenik et al., 2018). Here we describe how the Golgi apparatus mitigates unfolded proteins and present a strategy for studying the post-ER secretory pathway QC machinery in mammalian cells. Our data identify the folding state of the lumenal domain of a transmembrane Golgi protein as a determinant for sorting within the Golgi. We show that unfolded trans-Golgi model proteins can exit the organelle in QC carriers targeted to the endo/lysosomal pathway. Our findings further point to a complex network of QC mechanisms in the secretory pathway that determine the fate of QC substrates. Unfolded proteins can abortively cycle between compartments (Golgi, ER, endosomes) without being degraded, suggesting that different commitment steps (i.e., Golgi export, lysosomal degradation, or ERAD) require different determinants. The unfolded trans-Golgi model protein eventually accumulates in the ER, demonstrating that several mechanisms can account for the steady-state ER localization of QC substrates.
RESULTS
Degradation of Golgi-localized proteins
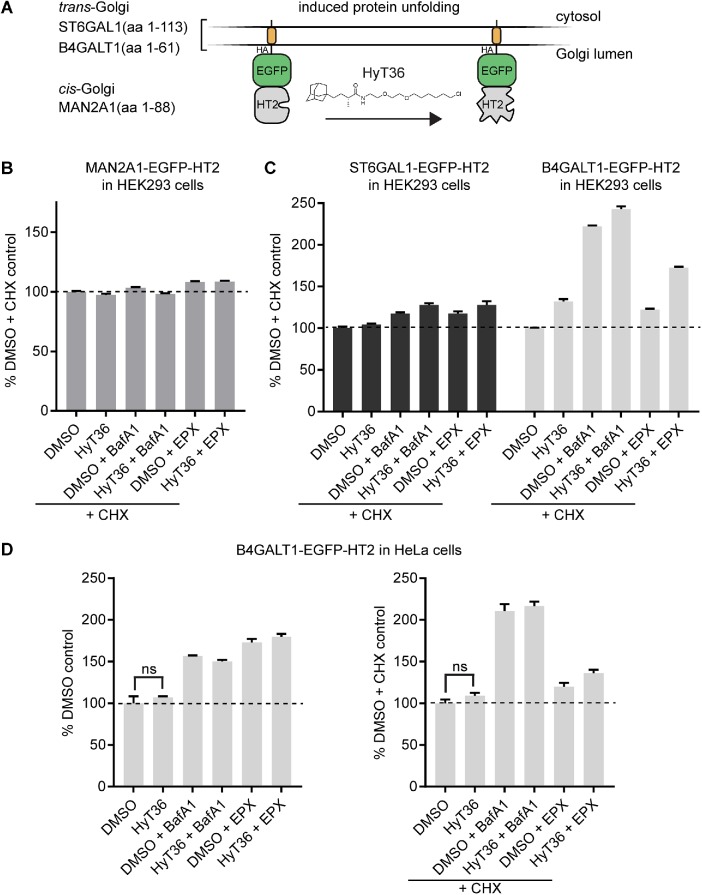
To study Golgi PQC, we generated a set of stable HEK293 cell lines expressing QC substrates in a doxycycline-inducible manner (Figure 1A). The Golgi-targeting sequences for these substrates were derived from the resident glycosyltransferases B4GALT1 and ST6GAL1 for the trans-Golgi and MAN2A1 for the cis-Golgi. To control the folding state of the lumenal domain, we incorporated the HaloTag2 (HT2) domain, which can be unfolded via hydrophobic tagging using the small molecule HT ligand, HyT36 (Neklesa et al., 2011; Tae et al., 2012). This chemical biology system has previously been shown to be amenable for investigation of ER and cytoplasmic QC systems (Neklesa et al., 2013; Raina et al., 2014). Moreover, we have demonstrated by limited proteolysis that B4GALT1-EGFP-HT2 (from now on referred to as B4GT-EGFP-HT2) is destabilized on HyT36 treatment (Serebrenik et al., 2018).
FIGURE 1:
Degradation profiles of Golgi-localized model QC substrates. (A) Domain outline of Golgi QC substrates targeted to the cis- (MAN2A1-EGFP-HT2) and trans- (ST6GAL1- and B4GT-EGFP-HT2) Golgi. (B) Flow cytometry analysis quantifying the levels of MAN2A1-EGFP-HT2 in HEK293 cells on the indicated treatments for 6 h (EPX, epoxomicin; BafA1, bafilomycin A). CHX was added 1 h prior to the indicated treatments. (C) Same analysis as in B for HEK293 cells expressing B4GT-EGFP-HT2 or ST6GAL1-EGFP-HT2 (n = 2, data represent mean ± SEM). (D) Flow cytometry analysis quantifying the levels of B4GT-EGFP-HT2 in HeLa cells on the indicated treatments for 6 h in the absence (left) or presence (right) of CHX (n = 3, data represent mean ± SEM, results of t test are shown).
To identify Golgi PQC mechanisms, we first analyzed the degradation profiles of the fusion proteins using lysosome and proteasome inhibitors (bafilomycin A1 [BafA1] and epoxomicin, respectively) (Yoshimori et al., 1991; Meng et al., 1999). The experiments were performed in the presence and absence of the translation inhibitor cycloheximide (CHX) to distinguish between protein that is present at the Golgi and newly synthesized protein that could also be affected by the treatments when entering the secretory pathway at the ER. This analysis revealed that the cis-Golgi–localized MAN2A1-EGFP-HT2 is not turned over within the 6-h time frame of the experiment, given that its levels are not affected by any treatment in the presence of CHX (Figure 1B). Newly synthesized MAN2A1-EGFP-HT2 protein is, however, subject to turnover by the proteasome, and HyT36 treatment further sensitizes the newly synthesized protein to degradation, as shown by a decrease in levels in the absence of CHX (Supplemental Figure S1A).
As described previously for the B4GALT1-based construct expressed in HEK293 or HeLa cells, HyT36 treatment does not induce a decrease in levels of the trans-Golgi constructs (B4GT- and ST6GAL1-EGFP-HT2) in the presence or absence of CHX (Figure 1C and Supplemental Figure S1B) (Serebrenik et al., 2018). However, their homeostatic turnover is sensitive to lysosome inhibition by BafA1 and to proteasome inhibition by epoxomicin (Figure 1C and Supplemental Figure S1B), with the Golgi-localized B4GT-EGFP-HT2 construct showing the comparatively highest turnover, mediated by the lysosome (Figure 1C). The proteasome-mediated turnover largely affects newly synthesized protein as the effect of epoxomycin is strongly reduced in the presence of CHX. The same degradation profile is recapitulated in HeLa cells (Figure 1D). In summary, while hydrophobic tagging induces the degradation of cytosolic and ER-targeted fusion proteins (Neklesa et al., 2011; Raina et al., 2014), the tool does not lead to a reduction in the levels of any of the tested Golgi-localized proteins (Figure 1, B and C). This difference may be due to varying compartment-specific sensitivities to unfolded proteins and/or compartment-specific PQC mechanisms.
Unfolded proteins exit the Golgi in distinct carriers
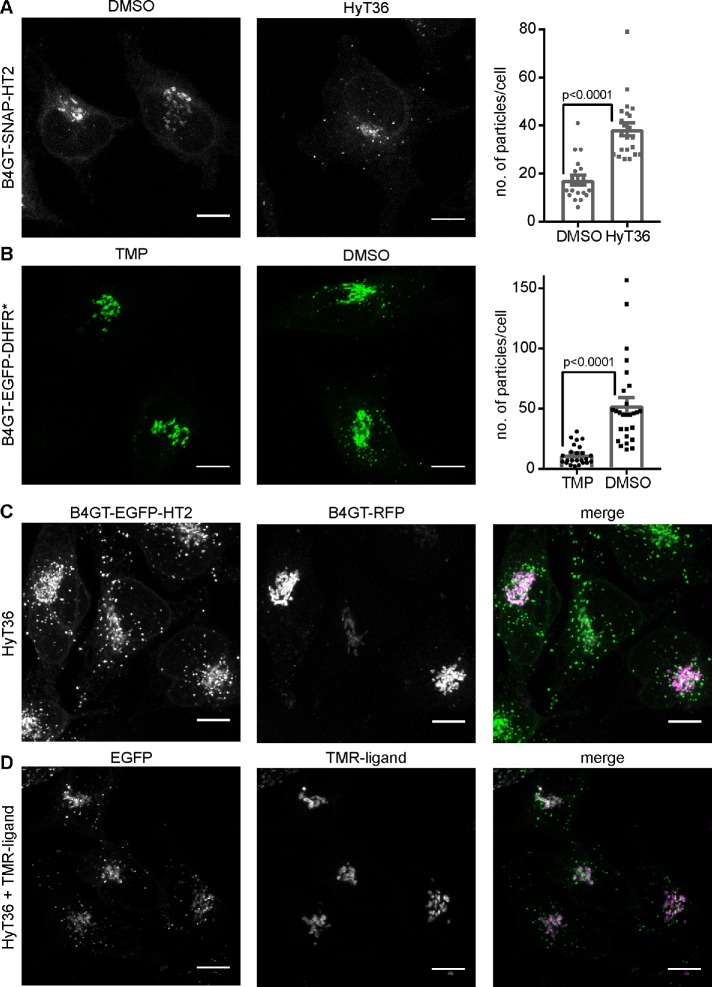
To gain detailed insight into the QC mechanisms operating at the Golgi, independent of protein degradation, we conducted a comprehensive imaging study in HeLa cells, which are well suited for microscopy studies of the Golgi. Given that the B4GT-EGFP-HT2 construct showed the lowest basal stability based on the degradation analysis, we chose to focus on this construct and investigate the additional effect of HyT36 on the metastable protein. To distinguish protein that comes from the Golgi from newly synthesized protein, we generated a B4GT-SNAP-HT2 construct that can be pulse-labeled with a fluorescent SNAP-tag ligand before treating cells with HyT36. In accordance with results obtained for the EGFP-based constructs (Figure 1D and Supplemental Figure S2A), levels of B4GT-SNAP-HT2 do not decrease on HyT36 treatment for 1 and 6 h (Supplemental Figure S2B). B4GT-SNAP-HT2 is localized to the Golgi under control conditions (dimethyl sulfoxide [DMSO]), yet 1 h after HyT36 treatment, carriers containing B4GT-SNAP-HT2 are clearly visible throughout the cell (Figure 2A). We quantified this phenotype by determining the number of particles per cell in FIJI and observed a significant increase on protein unfolding (Figure 2A).To synchronize the unfolding and export wave from the Golgi, we used a temperature block/release system when working with the EGFP-based constructs, where export from the Golgi is blocked by incubating cells at 20°C (Matlin and Simons, 1983). One hour after release of the temperature block, the unfolded B4GT-EGFP-HT2 protein is found in carriers distinct from the Golgi (Supplemental Figure S2C). This effect is also observed when a complementary protein unfolding system is used. The stability of DHFR*, a mutant protein domain (dihydrofolate reductase R12H, G67S, N18T, A19V), can be controlled with the small molecule stabilizer trimethoprim (TMP) (Iwamoto et al., 2010; Cho et al., 2013). We generated a B4GT-EGFP-DHFR* fusion construct and induced its expression in the presence of TMP. On TMP washout and release of the temperature block, B4GT-EGFP-DHFR* is found in carriers throughout the cell (Figure 2B). The formation of QC carriers with the HT2- and DHFR*-based unfolding systems corroborates the dependence of the observed phenotype on the folding state of the Golgi protein.
FIGURE 2:
Formation of Golgi QC carriers containing Golgi QC model substrates. (A) Left panel: representative confocal microscopy images of HeLa cells expressing B4GT-SNAP-HT2 labeled with a fluorescent SNAP ligand at time 0 and incubated with DMSO control or HyT36 for 1 h at 37°C. Right panel: particles per cell were quantified in FIJI (n = 19 for DMSO, n = 22 for HyT36). (B) Left panel: representative confocal microscopy images of HeLa cells expressing B4GT-EGFP-DHFR* incubated for 4 h at 20°C followed by 1 h at 37°C with the stabilizer TMP or TMP washed out and treated with DMSO. Right panel: Particles per cell were quantified in FIJI (n = 24 for TMP and n = 27 for DMSO). (C) B4GT-EGFP-HT2–expressing HeLa cells were transfected with B4GT-RFP and treated with HyT36 for 4 h at 20°C followed by 1 h at 37°C. (D) B4GT-EGFP-HT2–expressing HeLa cells were treated with HyT36 and a HT TMR ligand for 4 h at 20°C followed by 1 h at 37°C. All images are maximum projections of z-stacks. Scale bars correspond to 10 µm. Bar graphs represent mean ± SEM, p values (t test) are shown.
Immunofluorescence staining of the Golgi marker Giantin shows that the structure of the Golgi remains intact while the unfolded protein is exported (Supplemental Figure S2D). The integrity of the Golgi on hydrophobic tagging of B4GT-EGFP-HT2 was also previously confirmed by electron microscopy (Serebrenik et al., 2018). We further confirmed that the QC carriers specifically contain unfolded protein by transfecting B4GT-EGFP-HT2–expressing cells with a construct comprising the identical B4GT-derived Golgi-targeting sequence fused to RFP. B4GT-RFP is present at the Golgi in HyT36-treated cells and not detected in the QC carriers (Figure 2C). Finally, we performed a competition experiment using a HT ligand with a fluorescent moiety (TMR) together with the hydrophobic tag HyT36 (Figure 2D). While EGFP-positive carriers are observed, the signal from the TMR ligand, which does not induce unfolding of HT2, is exclusively observed at the Golgi apparatus. These experiments clearly show that specifically unfolded proteins are present in the QC carriers.
Segregation of unfolded from folded proteins at the Golgi
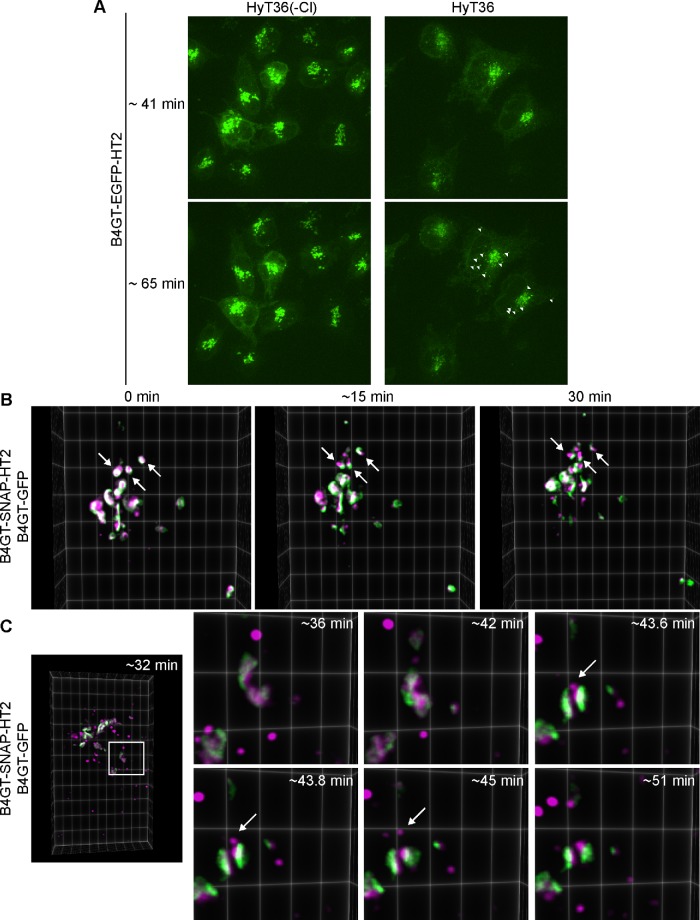
To monitor the exit of QC carriers from the Golgi, we performed live cell imaging. Spinning disk confocal imaging of B4GT-EGFP-HT2–expressing cells shows EGFP-positive QC carriers leaving the Golgi induced by HyT36 treatment (Supplemental Movie S1 and Figure 3A). Within a time frame of 1 h, carriers originating from the Golgi are found throughout the cell. In the control experiment performed with an inactive control compound (HyT36(-Cl)) (Serebrenik et al., 2018), the EGFP signal remains concentrated at the Golgi structure (Supplemental Movie S2 and Figure 3A). We further performed lattice light sheet microscopy (LLSM) imaging studies to visualize the export events in greater detail (Chen et al., 2014). To monitor the integrity of the Golgi during the process of QC carrier formation and identify sorting events, cells expressing B4GT-SNAP-HT2 were transfected with B4GT-GFP. The B4GT-SNAP-HT2 protein was labeled with a TMR-SNAP ligand and the cells were subsequently treated with HyT36. These studies clearly show that B4GT-SNAP-HT2 (the magenta signal from the TMR ligand) segregates from B4GT-GFP (the green signal) at the Golgi (Supplemental Movie S3 and Figure 3B). This phenotype is also apparent in fixed-cell imaging studies when comparing cells expressing B4GT-SNAP-HT2 and B4GT-GFP that have been treated with DMSO (control) or HyT36 for 45 min (Supplemental Figure S3). Moreover, carriers specifically containing B4GT-SNAP-HT2 are forming at the Golgi (Supplemental Movie S4 and Figure 3C). The B4GT-GFP protein is not incorporated into these carriers. The QC carriers containing unfolded protein then pinch off and are found throughout the cell. These data demonstrate a sorting process for unfolded proteins at the Golgi.
FIGURE 3:
Unfolded proteins are segregated from folded proteins at the Golgi. (A) Stills from spinning disk confocal imaging of HeLa cells expressing B4GT-EGFP-HT2 that were treated with HyT36 or the HyT36(-Cl) control. After incubation at 20°C, cells were transferred to the microscope and recording started 40 min thereafter for 70 min at 37°C. White arrowheads indicate individual carriers containing B4GT-EGFP-HT2. Images are maximum projections of z-stacks at 41 and 65 min at 37°C. (B) Stills from LLSM studies. After incubation at 20°C, cells were imaged at 37°C. Arrows indicate the separation of the B4GT-GFP and B4GT-SNAP-HT2 signal during a 30-min imaging period. (C) Zoom-in images of stills of LLSM covering the time frame of 30 to 60 min at 37°C. The white arrow indicates a carrier containing B4GT-SNAP-HT2 forming at the Golgi and pinching off. Images are three-dimensional projections rendered in ClearVolume.
Supplemental Movie 1.
B4GT-EGFP-HT2 expressing HeLa cells were treated with HyT36 for 4h at 20°C, transferred to the microscope heated to 37°C and after an additional 40 min of incubation, cells were imaged by spinning disk confocal microscopy for 70 min.
Supplemental Movie 2.
B4GT-EGFP-HT2 expressing HeLa cells were treated with the control compound HyT36(-Cl) for 4h at 20°C, transferred to the microscope heated to 37°C and after an additional 40 min of incubation, cells were imaged by spinning disk confocal microscopy for 70 min.
Supplemental Movie 3.
HeLa cells expressing B4GT-GFP and B4GT-SNAP-HT2labeled with a TMR-ligand were treated with HyT36 for 3h at 20°C, transferred to the microscope heated to 37°C and imaged by LLSM for 30 min.
Supplemental Movie 4.
HeLa cells expressing B4GT-GFP and B4GT-SNAP-HT2 labeled with a TMR-ligand were incubated with HyT36 for 3h at 20°C, for an additional 30 min at 37°C and then imaged by LLSM for 30 min.
Unfolded B4GT-HT2 proteins are targeted to the endo/lysosomal pathway
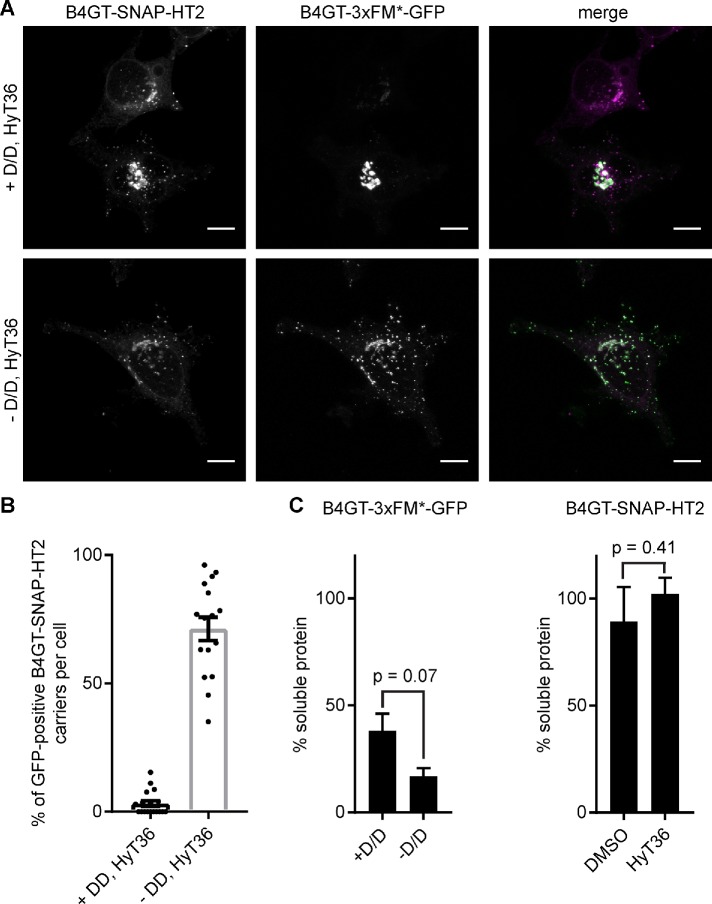
On the basis of the fact that the trans-Golgi model proteins are turned over by the lysosome (Figure 1, C and D), we presumed that the formation of the QC carriers may be the first step of targeting unfolded Golgi proteins to the endo/lysosomal system. Oligomerization has recently been shown to target proteins from the Golgi to lysosomes for degradation (Tewari et al., 2014, 2015). Small molecule-controlled oligomerization can be achieved by linking multiple FKBP F36M (FM*) domains that dimerize in the absence of the so-called D/D solubilizer (Rollins et al., 2000). Using a construct containing three consecutive FM* domains fused to Golgi-targeting sequences, Linstedt and colleagues showed that oligomerization induces the degradation of Golgi proteins in lysosomes (Tewari et al., 2015). We monitored the colocalization of the previously reported B4GT-3xFM*-GFP construct with a TMR-labeled B4GT-SNAP-HT2 version of the model protein sensitive to HyT36 treatment. In the presence of the D/D solubilizer, B4GT-3xFM*-GFP exists as a monomer and is exclusively present at the Golgi (Supplemental Figure S4A). When cells are treated with HyT36 and the D/D solubilizer is washed out, both model substrates are present in vesicular carriers (Figure 4A). Specifically, these experiments show that the B4GT-SNAP-HT2-containing carriers induced by HyT36 treatment also contain highly oligomerized B4GT-3xFM*-GFP proteins that are targeted to lysosomes for degradation (Figure 4A and Supplemental Figure S4B) (Tewari et al., 2015). To assess the aggregation propensity of the two different constructs, we lysed cells in a buffer with low detergent concentration and separated the soluble from the aggregated fraction by centrifugation. This analysis showed that B4GT-3xFM*-GFP is aggregation prone in the presence of the D/D solubilizer (∼38% apparent solubility) and even less soluble on 1-h washout of the compound (∼17% apparent solubility) (Figure 4C, left panel, and Supplemental Figure S4C). B4GT-SNAP-HT2 is nearly 100% soluble under control conditions (DMSO), as well as when cells are treated with HyT36 (Figure 4C, right panel, and Supplemental Figure S4D). These data highlight that aggregated and unfolded model QC substrates are present in the same carriers.
FIGURE 4:
Unfolded and aggregated Golgi QC substrates colocalize. (A) B4GT-SNAP-HT2 (labeled with TMR, magenta) expressing HeLa cells were transfected with B4GT-3xFM*-GFP (green). Cells were treated with HyT36 in the presence or absence of D/D solubilizer for 4 h at 20°C followed by 1 h 37°C. (B) Quantification of GFP-positive B4GT-SNAP-HT2 carriers per cell. (-D/D: n = 24, + D/D: n = 19). (C) Quantification of soluble B4GT-3xFM*-GFP (left panel) and B4GT-SNAP-HT2 (right panel) based on Western blot analysis. Raw data are shown in Supplemental Figure S4 (n = 2, data represent mean ± SD, results from t test are shown).
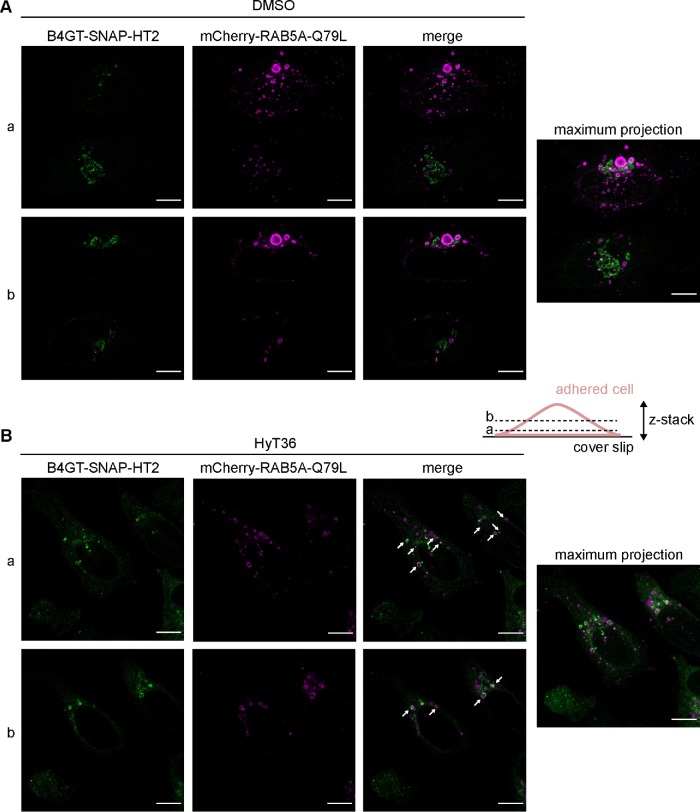
To identify the target compartment of the QC carriers, we transfected cells with an RFP-tagged version of RAB5, a marker for early endosomes. This experiment showed labeling of the B4GT-EGFP-HT2-containing carriers with Rab5 (Supplemental Figure S5, A and B). To better illustrate this effect, we used a mutant of the endosome-associated GTPase RAB5A, mCherry-RAB5A Q79L. Expression of this mutant leads to the formation of enlarged endosomes and multivesicular bodies (Stenmark et al., 1994; Wegner et al., 2010). Under control conditions (DMSO), a minor fraction of the B4GT-SNAP-HT2 protein can be observed in the mCherry-Rab5A Q79L-positive structures, likely representing protein that is tonically turned over by the lysosome (Figure 5A). On treatment with HyT36, mCherry-RAB5A Q79L-decorated structures are filled with the unfolded B4GT-SNAP-HT2 cargo (Figure 5B). In the larger mCherry-RAB5A Q79L structures, the signal from B4GT-SNAP-HT2 is also clearly observed as a ring-shaped structure pointing to the protein being part of the delimiting membrane of the multivesicular body (Figure 5B). Two slices of a z-stack covering the entire body of the cell are shown, as QC carriers and early endosomes are typically detected closer to the coverslip, while the signal from the Golgi apparatus is located closer to the center. Taken together, these data show that the B4GT-HT2 proteins are transported from the Golgi to the endo/lysosomal pathway on unfolding. The QC carriers are targeted to the lysosome; however, the unfolded HT2 protein is not degraded (Figure 1 and Supplemental Figure S2).
FIGURE 5:
QC carriers are targeted to the endo/lysosomal system. B4GT-SNAP-HT2 expressing HeLa cells were transfected with mCherry-RAB5A Q79L. Single slices and maximum projections of z-stacks for cells treated for 1 h at 37°C with DMSO (A) or HyT36 (B) are shown. Cartoon illustrates the individual slices of the z-stack (a and b). B4GT-SNAP-HT2 (labeled with Oregon Green), which is found in mCherry-RAB5A Q79L positive structures (magenta), is highlighted with white arrows. Cartoon illustrates the individual slices of the z-stack (a and b).
Unfolded Golgi proteins accumulate within the ER
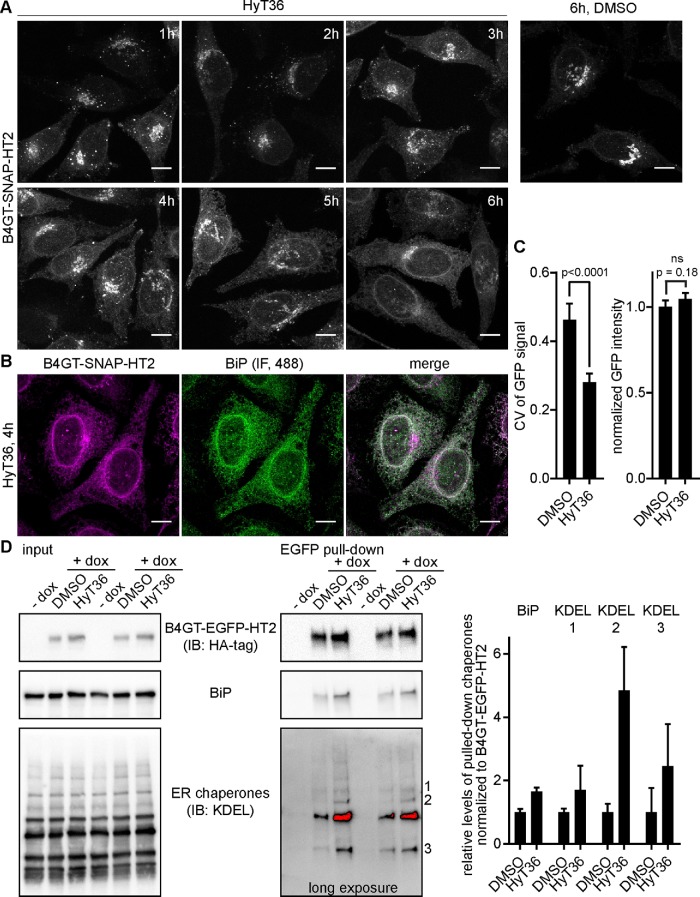
Even though the unfolded B4GT-HT2 fusion proteins are recognized for export from the Golgi and colocalize with oligomerized QC substrates targeted to lysosomes, they are not degraded. These data suggest that proteins can exit the pathway via the carriers before they are committed to lysosomal degradation. To identify the fate of the unfolded HT2 fusion protein, we performed an extended time course analysis of the B4GT-SNAP-HT2 construct. We pulse-labeled B4GT-SNAP-HT2 at time point 0 and followed its change in localization on HyT36 treatment. This pulse-chase analysis revealed that while the protein initially localizes to QC carriers (1–3 h posttreatment), it is detected in the ER at later time points after HyT36 treatment (starting at 3–6 h posttreatment) (Figure 6A). The reticular appearance of the B4GT-SNAP-HT2 signal and costaining of the ER marker BiP clearly demonstrate the ER localization of the unfolded reporter fusion (Figure 6B). We quantified this change in localization by determining the coefficient of variation (CV) of the EGFP signal per cell. Under control conditions (DMSO), the CV is comparatively high, reflecting the condensed EGFP signal at the Golgi apparatus (Figure 6C and Supplemental Figure S6A). On HyT36 treatment for 4 h, a more homogenous signal, which results in a significantly lower CV, is observed (Figure 6C and Supplemental Figure S6A). The image analysis also confirmed the results obtained by flow cytometry, showing no decrease of B4GT-EGFP-HT2 levels on HyT36 treatment (Figure 6C). The localization to the ER at later time points was also observed for the destabilized form of the B4GT-EGFP-DHFR* construct (Supplemental Figure S6B). Additionally, cotreatment of cells with HyT36 and the HT TMR ligand further confirmed that only unfolded B4GT-HT2 is localized to the ER (Supplemental Figure S6C). Once in the ER, unfolded B4GT-EGFP-HT2 associates with ER chaperones. In a pull-down experiment using lysates from HeLa cells expressing B4GT-EGFP-HT2, we find the ER chaperone BiP and other KDEL-containing chaperones associated with the Golgi model QC substrate on treatment with HyT36 (Figure 6D and Supplemental Figure S6D). Of note, the cells were treated with CHX before and during HyT36 treatment to exclude pull down of newly synthesized protein. Collectively these data show that B4GT-HT2 proteins are initially targeted to the endo/lysosomal system, yet are found in the ER where they engage protein chaperones at later time points. These data illustrate a clear difference between the distribution of aggregated and unfolded Golgi QC substrates among different pathways.
FIGURE 6:
Golgi QC substrates localize to the ER. (A) Time course analysis B4GT-SNAP-HT2 protein localization in HeLa cells on labeling with a TMR-SNAP ligand at time 0 and treated with HyT36 for the indicated times. (B) Colocalization of B4GT-SNAP-HT2 labeled with a TMR-SNAP ligand and treated with HyT36 for 4 h at 37°C with the ER marker BiP visualized by immunofluorescence using an Alexa Fluor 488–conjugated secondary antibody. All images are maximum projections of z-stacks. Scale bars correspond to 10 µm. (C) B4GT-EGFP-HT2–expressing HeLa cells were treated with DMSO or HyT36 for 4 h. The CV and integrated intensity of the GFP signal per cell were quantified. GFP intensity was normalized to DMSO treatment (n = 16, results from t test are shown) (D) GFP-trap pull down of B4GT-EGFP-HT2 from HeLa cells on treatment with HyT36 or DMSO control for 4 h at 37°C. Cells were treated with CHX 1 h prior and during the indicated treatments. Western blot analysis and quantification (data represent mean ± SEM) of association with ER chaperones are shown.
DISCUSSION
The Golgi as a QC checkpoint
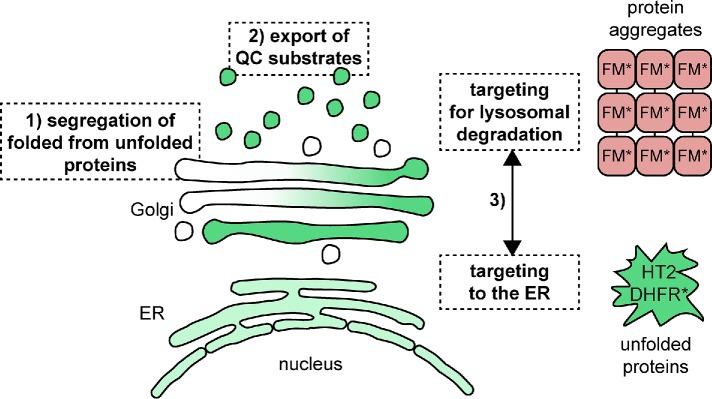
The Golgi apparatus is well known for its role in protein modification and processing (Moremen et al., 2012). In this study, we investigated how the Golgi deals with unfolded proteins to identify the contribution of the organelle to cellular PQC. Using chemical biology-based protein unfolding systems in combination with live cell imaging, we show that the folding state of the lumenal domain of a transmembrane protein in the Golgi can determine its transport route and ultimate localization. Protein unfolding induces export from the Golgi as visualized by live cell imaging. Specifically, unfolded Golgi proteins are first segregated from folded proteins and subsequently exported in QC carriers. We show that the carriers can contain both unfolded protein and highly oligomerized proteins, the latter mimicking protein aggregates (Tewari et al., 2015), demonstrating that different types of QC substrates may be part of the same carrier leaving the Golgi targeted to the endo/lysosomal system. The unfolded HT2 and DHFR* fusion proteins studied here are eventually sequestered in the ER, while the large oligomers formed by the FM* construct are degraded by the lysosome (Tewari et al., 2015). This suggests the existence of a QC checkpoint at the Golgi, where both types of QC substrates are selected for export and an additional sorting step, possibly on the QC carriers or on the endosome, which separates aggregated from unfolded QC substrates (Figure 7). At that additional checkpoint, aggregated or perhaps sufficiently misfolded proteins are selectively committed to lysosomal degradation, while redirecting the unfolded HT2 to the early secretory pathway may represent an attempt to refold the protein. This raises the question: which machinery decides the fate of unfolded and aggregated proteins? To this end, cytoplasmic proteins may be able to detect a difference in membrane curvature, which can be induced by aggregation of transmembrane proteins (McMahon and Boucrot, 2015), specifically selecting protein aggregates for lysosomal degradation. Alternatively, lumenal or transmembrane proteins, such as sorting receptors (Woodman and Futter, 2008; Progida and Bakke, 2016), may be able to specifically detect the aggregation state of the QC substrates. Individual unfolded proteins that are still freely diffusing in the membrane may be capable of leaving the carrier in complex with sorting receptors, while protein aggregates are not retrieved. This would allow selectively unfolded proteins to return to the Golgi and/or the ER. The association of the unfolded HT2 fusion protein with ER chaperones clearly demonstrates that it is recognized as unfolded in the ER and given that it is not targeted for ERAD, it may be regarded as a refoldable substrate protein. Based on these findings, it will be important to dissect the relationship between sequestration at the ER and abortive cycling to the endo/lysosomal system of the unfolded Golgi QC substrates. The same or similar characteristics of the QC substrate may spare it from both lysosomal degradation and ERAD. These data highlight that properties, such as aggregation status and possibly degree of unfolding of a Golgi QC substrate, determine its fate at different QC checkpoints after leaving the Golgi. A difference between the two pathways may be their kinetics. While the QC carriers form within 30–60 min on protein unfolding, possibly acting as the first line of defense, the ER localization is apparent only after 3–4 h. It should, however, be noted that this observation may be due to the fact that a larger amount of protein needs to be present at the ER to pass the detection limit of the microscope, given that the signal is less concentrated within the ER than in QC carriers and therefore the kinetic differential may not be as pronounced.
FIGURE 7:
Model of Golgi QC mechanisms: 1) unfolded proteins are separated from folded proteins at the Golgi; 2) subsequently, QC substrates are exported from the Golgi and targeted to the endo/lysosomal pathway; 3) at an additional QC checkpoint, aggregated proteins are committed to degradation at the lysosome. Unfolded Golgi proteins localize to the ER and associate with chaperones.
Given that proteins pass through the Golgi on their way to the PM, the endo/lysosomal-directed QC pathway may also be highly relevant to guard the proteome reaching the cell surface and the extracellular space. Other QC substrates following this pathway could include unassembled subunits of otherwise multimeric transmembrane protein complexes or proteins with mutations in their transmembrane domains. These types of QC substrates have been shown to escape the ER and are exported from the Golgi to the lysosome for degradation (Briant et al., 2017). Moreover, QC substrates that are halted at the Golgi on their way to the PM, such as select genetic prion protein mutants underlying Creutzfeldt-Jakob and Gerstmann-Sträussler disease (Ashok and Hegde, 2009) or select connexin32 mutants underlying X-linked Charcot-Marie-Tooth disease (VanSlyke et al., 2000), may be segregated from folded proteins but are not efficiently cleared from the organelle. At steady state, these mutants are found at the Golgi and similar to the trans-Golgi localized HT2-fusion proteins studied here, their turnover is mediated by the lysosome. The separation of unfolded from folded proteins may represent by itself a protective mechanism to maintain Golgi functionality in the presence of QC substrates. Given that the Golgi is in constant flux, it is important to provide a route through the organelle that is free from misfolded proteins which may interfere with important modification and sorting processes: cargo from the ER needs to be able to pass through the Golgi, acquire appropriate modifications, and reach downstream endomembrane compartments. The immediate isolation of unfolded proteins may support preserving Golgi functionality under protein folding stress, with the induction of the transcriptional Golgi stress response at a later stage further contributing to cellular adaptation to Golgi stress (Serebrenik et al., 2018).
The role of the ER in Golgi QC
The ER, which hosts a number of chaperones and degradation factors, plays a critical role in PQC for the secretory pathway (Araki and Nagata, 2011; Vincenz-Donnelly and Hipp, 2017). Several folding-deficient mutants and disease-associated misfolded proteins are sequestered at the ER (Hebert and Molinari, 2007). Notably, steady-state localization of QC substrates in the ER can have different origins. It may be due to primary retention of proteins after synthesis, or retrieval of unfolded proteins which escaped the ER. An additional checkpoint at the ERGIC can further limit the transport of QC substrates along the secretory pathway and ensure their availability for retrograde trafficking to the ER (Sirkis et al., 2017). Our study demonstrates that even when present at the trans-Golgi and/or exported in carriers targeted to the endo/lysosomal system, unfolded proteins can be transported back to the ER. Accordingly, retrograde trafficking to the ER from several downstream compartments of the secretory pathway is an important component of cellular PQC.
The ER has been shown to have a high capacity to counteract and buffer protein aggregation (Vincenz-Donnelly et al., 2018). It is conceivable that the spreading and dilution of QC substrates, which would otherwise be highly concentrated at the Golgi, within the large volume of the ER is by itself a QC strategy. Moreover, the presence of chaperones within the secretory pathway is largely restricted to the ER and their interaction with the Golgi QC substrates will further increase solubility and limit interaction with the healthy proteome (Braakman and Hebert, 2013). We have previously shown that the induced unfolding of B4GT-EGFP-HT2 is nontoxic (Serebrenik et al., 2018), suggesting that Golgi export and ER sequestration of the unfolded protein is an effective strategy to ensure cell survival under Golgi protein folding stress.
Studying Golgi QC with chemical biology tools
In this study, we used different chemical biology tools to investigate PQC in the secretory pathway. To this end, we targeted properly folded proteins to the Golgi apparatus, thereby avoiding initial retention in the ER. Applying hydrophobic tagging in the Golgi clearly demonstrated the difference among cytoplasmic, ER, and Golgi QC. While the addition of HyT36 leads to a reduction in the levels of cytoplasmic and ER-localized EGFP-HT2 (∼75% and ∼40%, respectively), none of the Golgi-localized constructs tested here are sensitized to degradation on hydrophobic tagging (Neklesa et al., 2011; Raina et al., 2014). This suggests that the threshold for committing a protein to a degradation pathway from the Golgi is higher than for other compartments. Moreover, we find differences in turnover among the different Golgi model substrates showing that they exhibit different basal stabilities that may be related to their targeting sequence. Based on our degradation analysis, the B4GT-based model substrates are well suited for studies on Golgi QC. Induced unfolding of the B4GT-based model substrates allowed us to uncover QC mechanisms that specifically operate at the Golgi. Hydrophobic tagging of the HT2 domain and destabilization of DHFR* reveal the export of unfolded protein from the Golgi. Being able to control the folding state with a small molecule allowed the following of a “wave” of unfolded protein, which further enabled live cell microscopy studies of Golgi export events in great detail. In our study, it was particularly valuable that we were able to successfully combine different chemical biology tools inducing the unfolding or aggregation of proteins to permit concurrent examination of different aberrantly formed reporter proteins. Our study shows that when an HT2-based construct and an FM*-based construct are present in the same cell, they do not interfere with each other: the constructs are responsive only to their corresponding small molecule (de)stabilizer and are not advanced into QC pathways along with the other system. On the basis of the imaging studies, no cross-talk between the two tools was observed. The combination of different chemical biology tools enables parallel monitoring of aggregates being targeted to lysosomes and unfolded proteins eventually reaching the ER. The complementarity of the HT2-, DHFR*-, and FM*-based systems further allows designing experiments involving the concomitant induction of QC pathways at different sites of the cell and monitoring of the interplay of organelle-specific PQC. Our study highlights the potential of chemical biology tools that allow controlling protein folding state for unraveling post-ER QC mechanisms. On the basis of our findings, future work will address the molecular mechanisms underlying Golgi QC in mammalian cells. Moreover, it will be important to extend the work to endosome- and PM-related PQC mechanisms to fully understand the collective contribution of post-ER endomembrane compartments to cellular protein homeostasis.
MATERIALS AND METHODS
Reagents and antibodies
Reagents and antibodies were purchased from the following suppliers: anti-Giantin (Abcam; ab24586, 1:1000), anti-BiP for immunofluorescence (Abcam ab21685, 1:500), anti-BiP for Western blot (Cell signaling, 3183S, 1:1000), anti-KDEL (Abcam ab176333, 1:1000), anti-GFP (Santa Cruz sc-9996, 1:1000), anti-HA (Cell signaling 3724, 1:1000), anti-GAPDH (Cell signaling 2118, 1:2000), TMP (Sigma), CHX (Sigma), doxycycline (Sigma), fibronectin (EMD Millipore), Hoechst (Thermo Fisher Scientific; 10 mg/ml in H2O, 1:2000) D/D solubilizer (Clonetech), BafA1 (Alfa Aesar), epoxomicin (EMD Millipore), SNAP-tag ligands (SNAP Cell TMR Star and Oregon Green [New England Biolabs]), HT TMR ligand (Promega), and DSP Crosslinker (Thermo Fisher Scientific). HyT36 and HyT36(-Cl) were previously described (Tae et al., 2012; Serebrenik et al., 2018).
DNA constructs
Cloning of B4GT-EGFP-HT2 was previously described (Serebrenik et al., 2018). B4GT-SNAP-HT2 was cloned by replacing the EGFP domain of B4GT-EGFP-HT2 using Gibson assembly (Gibson et al., 2009). ST6GAL1-EGFP-HT2 and MAN2A1-EGFP-HT2 were cloned by replacing the transmembrane and cytoplasmic domain of B4GT-EGFP-HT2 with the first 113 and 88 amino acids of ST6GAL1 (Homo sapiens) and MAN2A1 (Mus musculus), respectively, via USER cloning (Nour-Eldin et al., 2010). B4GT-EGFP-DHFR* was created by replacing HT2 from B4GT-EGFP-HT2 with the DHFR domain from pBMN YFP-DHFR(DD) (a gift from Thomas Wandless [Stanford University], Addgene plasmid #29326). mCherry-Rab5CA(Q79L) was a gift from Sergio Grinstein (Addgene plasmid #35138). RFP-Rab5a and B4GT-GFP were gifts from James Rothman (Yale University), from which B4GT-RFP was derived by replacing the GFP domain with RFP via USER cloning. B4GT-3xFM*-GFP was a gift from Adam Linstedt (Carnegie Mellon University).
Cell culture methods and cell treatments
HeLa Flp-in Trex cells (a gift from Stephen Taylor, University of Manchester) and HEK293 Flp-in Trex cells (Thermo Fisher Scientific) were grown in 5% CO2 at 37°C in DMEM (Life Technologies) containing 1% Pen/Strep (Life Technologies) and 10% fetal bovine serum (FBS) (Biological Industries). Cells were regularly tested for mycoplasma infection using the MycoAltert kit (Lonza). Drug treatments at 20°C and following 37°C were performed in L-15 (Life Technologies) containing 1% Pen/Strep and 10% FBS.
For labeling SNAP-tagged proteins, the fluorescent SNAP-tag ligands were diluted to a final concentration of 2 µM in culture medium. Cells were incubated in the solution for 30 min followed by three washes in culture medium and an additional incubation for 30 min before subsequent drug treatment. All doxycycline treatments were performed at 100 ng/ml, CHX treatments at 20 µg/ml, BafA1 treatments at 100 nM, epoxomicin treatments at 1 µM, HyT36 treatments at 10 µM, TMP treatments at 10 µM, and D/D solubilizer at 1 µM. For the competition experiment in Figure 2 and Supplemental Figure S6, HyT36 and the TMR HT ligand were mixed at a final concentration of 10 µM and 5 nM, respectively, in culture medium.
DNA transfections were performed using Lipofectamine2000 (Thermo Fisher Scientific) according to the manufacturer’s instructions. For experiments in Figure 4A and Supplemental Figure S4, stable B4GT-SNAP-HT2 Flp-in Trex HeLa cells were transfected with B4GT-3xFM-GFP. After 16 h, cells were trypsinized and transferred onto coverslips in medium containing doxycycline and D/D solubilizer. Cells were allowed to adhere to the coverslip for 24 h. On labeling with the TMR-SNAP ligand, cells were treated with HyT36 or DMSO in the presence or absence of D/D solubilizer. Treatments were performed in L-15 + 1% Pen/Strep + 10% FBS for 4 h at 20°C followed by 1 h at 37°C.
Flow cytometry
Cultured cells were trypsinized and resuspended in DMEM + 1% Pen/Strep + 10% FBS. GFP fluorescence was measured on a BD FACSCalibur (BD Biosciences) for HEK293 cells and on a BD LSRFortessa X-20 (BD Bioscience) for HeLa cells. Data were analyzed using FlowingSoftware (provided by the Turku Centre for Biotechnology, University of Turku and Åbo Akademi University, Turku, Finland) and plotted in Prism (GraphPad).
Laser scanning confocal microscopy
Flp-In Trex HeLa cells were plated on coverslips coated with fibronectin. The cells were grown in DMEM + 1% Pen/Strep + 10% FBS containing doxycycline and were allowed to adhere to the coverslips for 20–24 h. On the indicated treatment, cells were fixed using 4% paraformaldehyde in PBS (phosphate-buffered saline) for 20 min at RT. For immunofluorescence, cells were permeabilized and blocked in 10% FBS in PBS + 0.01% Triton X-100 and subsequently incubated with primary antibody diluted in 1% bovine serum albumin (BSA) in PBS + 0.01% Triton X-100 o/n at 4°C and α-rabbit secondary antibody conjugated to Alexa Fluor 488 or 547 (Thermo Fisher Scientific) diluted 1:500 in 1% BSA in PBS + 0.01% Triton X-100 for 1 h at room temperature. Finally, coverslips were mounted onto microscopy slides using Vectashield mounting medium (Vector Laboratories). Images were taken on a Zeiss LSM 800. z-Stacks covering the entire body of the cells were recorded with 1-µm spacing using a 40× or 63× Plan-Apochromat lens, 1.3 and 1.4 NA, respectively.
Live cell spinning disk confocal microscopy
B4GT-EGFP-HT2 expressing Flp-in Trex HeLa cells were grown in µ-slide 4-well dishes (ibidi) for 24 h in DMEM + 1% Pen/Strep + 10% FBS containing doxycycline. The medium was exchanged to L-15 + 1% Pen/Strep + 10% FBS containing HyT36 or HyT36(-Cl) and cells were incubated for 4 h at 20°C. The dishes were transferred to the microscope equipped with an incubator set to 37°C. Cells were imaged on an inverted microscope (Olympus IX81) using UltraVIEW VoX spinning disk confocal fluorescence illumination (PerkinElmer) and equipped with a 60× oil 1.4 NA oil phase objective. z-Stacks (1.5-μm slices) spanning the entire volume of the cells were acquired on a 1Kx1K EMCCD camera (Hamamatsu Photonics) using Volocity Software (Improvision, Coventry, UK).
Live cell LLSM
B4GT-SNAP-HT2 expressing Flp-in Trex HeLa cells were transfected with B4GT-GFP. After 24 h cells were trypsinized and transferred to a 3.5-cm dish lined with 5-mm coverslips in DMEM + 1% Pen/Strep + 10% FBS containing doxycycline. After 24 h, cells were labeled with a TMR-SNAP ligand and following treated with HyT36 in L-15 + 1% Pen/Strep + 10% FBS at 20°C until transferred to the LLSM microscope equipped with an incubation chamber set to 37°C in live cell imaging solution (Thermo Fisher Scientific A14291DJ). For generating the light-sheet pattern, inner and outer NAs of 0.325 and 0.4, respectively, were used. Images were taken by exciting with 488- and 560-nm lasers with an exposure time of 5 ms and an interval of ∼5 s between time points.
Image analysis
All microscopy images were processed and analyzed with FIJI (Schindelin et al., 2012). Quantification of carriers in Figure 2 and Supplemental Figure S2 was performed from maximum-intensity projections of z-stacks. Individual cells were defined as regions of interest (ROIs). After background subtraction, images were equally thresholded and the number of particles (10–150 pixels, circularity: 0.2–1.0) per cell (i.e., per ROI) was quantified with the AnalyzeParticle plugin. Data were plotted in Prism (GraphPad). For Figure 4A, the quantification of GFP-positive B4GT-SNAP-HT2 containing carriers was performed from maximum-intensity projections of z-stacks. B4GT-SNAP-HT2 particles were selected with the AnalyzeParticle plugin and the mean GFP fluorescence was determined in the selected area. Particles ≥80 GFP mean fluorescence were considered positive. LLSM imaging data were analyzed with the ClearVolume plugin (Royer et al., 2015).
Automated imaging and image analysis
To determine the change in localization of the B4GT-EGFP-HT2 construct from the Golgi to the ER, HeLa cells were plated in 384-well plates in culture medium containing doxycycline, lipofectamine2000 and a RISC-free siRNA (Dharmacon). After 24 h, culture medium was exchanged and after an additional 24 h incubation, cells were treated for 4 h with HyT36 or DMSO. Cells were fixed with 4% PFA in PBS, washed 3× with PBS, stained with Hoechst, and imaged with a GE IN Cell analyzer 2200, with five fields of view per well. Images were analyzed with Cell Profiler (Carpenter et al., 2006). Cells were identified first by the nuclear stain and following by the GFP signal. The integrated intensity and the CV of the GFP signal were determined. The mean per well was calculated and used to generate the mean and SD over 16 wells for each condition. Data were plotted in Prism (GraphPad).
Pull-down studies and Western blot analysis
Stable B4GT-EGFP-HT2 Flp-in Trex HeLa cells were grown in 15-cm dishes. The cells were treated with doxycycline for 20 h, followed by CHX treatment started 1 h prior to HyT36 and DMSO (control) treatments for an additional 4 h. The cells were washed with PBS and incubated with 1 mM DSP Crosslinker for 10 min at RT. The cross-linking reaction was quenched by the addition of Tris, pH 7.5, to a final concentration of 25 mM. Cells were lysed in 50 mM Tris, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% NP-40 supplemented with cOmplete protease inhibitor cocktail (Roche). Cell lysates were clarified by centrifugation at 20,000 × g for 10 min at 4°C. The supernatant after centrifugation (SOL) was incubated with magnetic agarose GFP-trap beads (ChromoTek) for 90 min at 4°C. The beads were then washed 3× with 50 mM Tris, pH 7.5, 150 mM NaCl. Proteins bound to the beads were eluted in SDS–PAGE sample buffer.
Samples were applied to SDS–PAGE gels and transferred to nitrocellulose membranes. After blocking in milk, membranes were incubated with primary and secondary antibodies and finally developed at a Bio-Rad ChemiDoc Imager. Quantification of Western blots was performed in ImageLab (Bio-Rad).
To analyze the aggregation propensity of B4GT-3xFM*-GFP and B4GT-SNAP-HT2, HeLa cells were lysed in 50 mM Tris, pH 7.5, 150 mM NaCl, 1 mM EDTA, 0.1% NP-40 supplemented with cOmplete protease inhibitor cocktail. A sample for SDS–PAGE was taken from the whole cell lysate (WCL) and the SOL at 20,000 × g for 10 min at 4°C. For quantification of the percentage of soluble protein, bands on GFP- or HA-Western blots were first normalized to GAPDH and then to the WCL lane.
Supplementary Material
Acknowledgments
We thank James Rothman and Francesca Bottanelli for helpful discussions and the Yale Center for Molecular Discovery for recording images on the GE IN Cell analyzer 2200. This work was supported by National Institutes of Health Grant no. R35CA197589 and EMBO Long-Term Fellowship Grant no. 1102-2014 to D.H. Y.V.S. was supported in part by National Institutes of Health Grant no. 5T32GM06754 3-12.
Abbreviations used:
- ATZ
α1-antitrypsin protein
- BafA1
bafilomycin A1
- B4GT-EGFP-HT2
B4GALT1-EGFP-HT2
- BSA
bovine serum albumin
- CHX
cycloheximide
- CV
coefficient of variation
- DHFR
dihydrofolate reductase
- ER
endoplasmic reticulum
- ERAD
ER-associated degradation
- ERGIC
ER-Golgi intermediate compartment
- FBS
fetal bovine serum
- HT
HaloTag
- LLSM
lattice light sheet microscopy
- PBS
phosphate-buffered saline
- PM
plasma membrane
- PQC
protein quality control
- QC
quality control
- ROI
regions of interest
- SOL
supernatant after centrifugation
- TMP
trimethoprim
- WCL
whole cell lysate.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E19-01-0069) on June 5, 2019.
REFERENCES
- Araki K, Nagata K. (2011). Protein folding and quality control in the ER. Cold Spring Harb Perspect Biol , a007526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashok A, Hegde RS. (2009). Selective processing and metabolism of disease-causing mutant prion proteins. PLoS Pathog , e1000479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino JS, Rojas R. (2006). Retrograde transport from endosomes to the trans-Golgi network. Nat Rev Mol Cell Biol , 568–579. [DOI] [PubMed] [Google Scholar]
- Braakman I, Bulleid NJ. (2011). Protein folding and modification in the mammalian endoplasmic reticulum. Annu Rev Biochem , 71–99. [DOI] [PubMed] [Google Scholar]
- Braakman I, Hebert DN. (2013). Protein folding in the endoplasmic reticulum. Cold Spring Harb Perspect Biol , a013201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briant K, Johnson N, Swanton E. (2017). Transmembrane domain quality control systems operate at the endoplasmic reticulum and Golgi apparatus. PLoS One , e0173924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J, et al. (2006). CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol , R100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen BC, Legant WR, Wang K, Shao L, Milkie DE, Davidson MW, Janetopoulos C, Wu XS, Hammer JA, 3rd, Liu Z, et al. (2014). Lattice light-sheet microscopy: imaging molecules to embryos at high spatiotemporal resolution. Science , 1257998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho U, Zimmerman SM, Chen LC, Owen E, Kim JV, Kim SK, Wandless TJ. (2013). Rapid and tunable control of protein stability in Caenorhabditis elegans using a small molecule. PLoS One , e72393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole NB, Ellenberg J, Song J, DiEuliis D, Lippincott-Schwartz J. (1998). Retrograde transport of Golgi-localized proteins to the ER. J Cell Biol , 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coughlan CM, Walker JL, Cochran JC, Wittrup KD, Brodsky JL. (2004). Degradation of mutated bovine pancreatic trypsin inhibitor in the yeast vacuole suggests post-endoplasmic reticulum protein quality control. J Biol Chem , 15289–15297. [DOI] [PubMed] [Google Scholar]
- Dobzinski N, Chuartzman SG, Kama R, Schuldiner M, Gerst JE. (2015). Starvation-dependent regulation of Golgi quality control links the TOR signaling and vacuolar protein sorting pathways. Cell Rep , 1876–1886. [DOI] [PubMed] [Google Scholar]
- Fossati M, Colombo SF, Borgese N. (2014). A positive signal prevents secretory membrane cargo from recycling between the Golgi and the ER. EMBO J , 2080–2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fregno I, Fasana E, Bergmann TJ, Raimondi A, Loi M, Solda T, Galli C, D’Antuono R, Morone D, Danieli A, et al. (2018). ER-to-lysosome-associated degradation of proteasome-resistant ATZ polymers occurs via receptor-mediated vesicular transport. EMBO J 37, e99259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelling CL, Dawes IW, Perlmutter DH, Fisher EA, Brodsky JL. (2012). The endosomal protein-sorting receptor sortilin has a role in trafficking alpha-1 antitrypsin. Genetics , 889–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geva Y, Schuldiner M. (2014). The back and forth of cargo exit from the endoplasmic reticulum. Curr Biol , R130–R136. [DOI] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, 3rd, Smith HO. (2009). Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods , 343–345. [DOI] [PubMed] [Google Scholar]
- Goold R, McKinnon C, Rabbanian S, Collinge J, Schiavo G, Tabrizi SJ. (2013). Alternative fates of newly formed PrPSc upon prion conversion on the plasma membrane. J Cell Sci , 3552–3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond C, Helenius A. (1994). Quality control in the secretory pathway: retention of a misfolded viral membrane glycoprotein involves cycling between the ER, intermediate compartment, and Golgi apparatus. J Cell Biol , 41–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert DN, Molinari M. (2007). In and out of the ER: protein folding, quality control, degradation, and related human diseases. Physiol Rev , 1377–1408. [DOI] [PubMed] [Google Scholar]
- Hong E, Davidson AR, Kaiser CA. (1996). A pathway for targeting soluble misfolded proteins to the yeast vacuole. J Cell Biol , 623–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamoto M, Bjorklund T, Lundberg C, Kirik D, Wandless TJ. (2010). A general chemical method to regulate protein stability in the mammalian central nervous system. Chem Biol , 981–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YE, Hipp MS, Bracher A, Hayer-Hartl M, Hartl FU. (2013). Molecular chaperone functions in protein folding and proteostasis. Annu Rev Biochem , 323–355. [DOI] [PubMed] [Google Scholar]
- Klaips CL, Jayaraj GG, Hartl FU. (2018). Pathways of cellular proteostasis in aging and disease. J Cell Biol 217, 51–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matlin KS, Simons K. (1983). Reduced temperature prevents transfer of a membrane glycoprotein to the cell surface but does not prevent terminal glycosylation. Cell , 233–243. [DOI] [PubMed] [Google Scholar]
- McMahon HT, Boucrot E. (2015). Membrane curvature at a glance. J Cell Sci , 1065–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng L, Mohan R, Kwok BH, Elofsson M, Sin N, Crews CM. (1999). Epoxomicin, a potent and selective proteasome inhibitor, exhibits in vivo antiinflammatory activity. Proc Natl Acad Sci USA , 10403–10408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moremen KW, Tiemeyer M, Nairn AV. (2012). Vertebrate protein glycosylation: diversity, synthesis and function. Nat Rev Mol Cell Biol , 448–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neklesa TK, Noblin DJ, Kuzin A, Lew S, Seetharaman J, Acton TB, Kornhaber G, Xiao R, Montelione GT, Tong L, et al. (2013). A bidirectional system for the dynamic small molecule control of intracellular fusion proteins. ACS Chem Biol , 2293–2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neklesa TK, Tae HS, Schneekloth AR, Stulberg MJ, Corson TW, Sundberg TB, Raina K, Holley SA, Crews CM. (2011). Small-molecule hydrophobic tagging-induced degradation of HaloTag fusion proteins. Nat Chem Biol , 538–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nour-Eldin HH, Geu-Flores F, Halkier BA. (2010). USER cloning and USER fusion: the ideal cloning techniques for small and big laboratories. Methods Mol Biol , 185–200. [DOI] [PubMed] [Google Scholar]
- Progida C, Bakke O. (2016). Bidirectional traffic between the Golgi and the endosomes - machineries and regulation. J Cell Sci , 3971–3982. [DOI] [PubMed] [Google Scholar]
- Puig B, Altmeppen HC, Ulbrich S, Linsenmeier L, Krasemann S, Chakroun K, Acevedo-Morantes CY, Wille H, Tatzelt J, Glatzel M. (2016). Secretory pathway retention of mutant prion protein induces p38-MAPK activation and lethal disease in mice. Sci Rep , 24970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raina K, Noblin DJ, Serebrenik YV, Adams A, Zhao C, Crews CM. (2014). Targeted protein destabilization reveals an estrogen-mediated ER stress response. Nat Chem Biol , 957–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F, Pelham HR. (2002). A transmembrane ubiquitin ligase required to sort membrane proteins into multivesicular bodies. Nat Cell Biol , 117–123. [DOI] [PubMed] [Google Scholar]
- Richter K, Haslbeck M, Buchner J. (2010). The heat shock response: life on the verge of death. Mol Cell , 253–266. [DOI] [PubMed] [Google Scholar]
- Rollins CT, Rivera VM, Woolfson DN, Keenan T, Hatada M, Adams SE, Andrade LJ, Yaeger D, van Schravendijk MR, Holt DA, et al. (2000). A ligand-reversible dimerization system for controlling protein-protein interactions. Proc Natl Acad Sci USA , 7096–7101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royer LA, Weigert M, Gunther U, Maghelli N, Jug F, Sbalzarini IF, Myers EW. (2015). ClearVolume: open-source live 3D visualization for light-sheet microscopy. Nat Methods , 480–481. [DOI] [PubMed] [Google Scholar]
- Sato M, Sato K, Nakano A. (2004). Endoplasmic reticulum quality control of unassembled iron transporter depends on Rer1p-mediated retrieval from the golgi. Mol Biol Cell , 1417–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satpute-Krishnan P, Ajinkya M, Bhat S, Itakura E, Hegde RS, Lippincott-Schwartz J. (2014). ER stress-induced clearance of misfolded GPI-anchored proteins via the secretory pathway. Cell , 522–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods , 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serebrenik YV, Hellerschmied D, Toure M, Lopez-Giraldez F, Brookner D, Crews CM. (2018). Targeted protein unfolding uncovers a Golgi-specific transcriptional stress response. Mol Biol Cell , 1284–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirkis DW, Aparicio RE, Schekman R. (2017). Neurodegeneration-associated mutant TREM2 proteins abortively cycle between the ER and ER-Golgi intermediate compartment. Mol Biol Cell , 2723–2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sontag EM, Samant RS, Frydman J. (2017). Mechanisms and functions of spatial protein quality control. Annu Rev Biochem , 97–122. [DOI] [PubMed] [Google Scholar]
- Stenmark H, Parton RG, Steele-Mortimer O, Lutcke A, Gruenberg J, Zerial M. (1994). Inhibition of rab5 GTPase activity stimulates membrane fusion in endocytosis. EMBO J , 1287–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart RS, Harris DA. (2005). A transmembrane form of the prion protein is localized in the Golgi apparatus of neurons. J Biol Chem , 15855–15864. [DOI] [PubMed] [Google Scholar]
- Storrie B, White J, Rottger S, Stelzer EH, Suganuma T, Nilsson T. (1998). Recycling of golgi-resident glycosyltransferases through the ER reveals a novel pathway and provides an explanation for nocodazole-induced Golgi scattering. J Cell Biol , 1505–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tae HS, Sundberg TB, Neklesa TK, Noblin DJ, Gustafson JL, Roth AG, Raina K, Crews CM. (2012). Identification of hydrophobic tags for the degradation of stabilized proteins. Chembiochem , 538–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tewari R, Bachert C, Linstedt AD. (2015). Induced oligomerization targets Golgi proteins for degradation in lysosomes. Mol Biol Cell , 4427–4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tewari R, Jarvela T, Linstedt AD. (2014). Manganese induces oligomerization to promote down-regulation of the intracellular trafficking receptor used by Shiga toxin. Mol Biol Cell , 3049–3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanSlyke JK, Deschenes SM, Musil LS. (2000). Intracellular transport, assembly, and degradation of wild-type and disease-linked mutant gap junction proteins. Mol Biol Cell , 1933–1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vavassori S, Cortini M, Masui S, Sannino S, Anelli T, Caserta IR, Fagioli C, Mossuto MF, Fornili A, van Anken E, et al. (2013). A pH-regulated quality control cycle for surveillance of secretory protein assembly. Mol Cell , 783–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincenz-Donnelly L, Hipp MS. (2017). The endoplasmic reticulum: A hub of protein quality control in health and disease. Free Radic Biol Med , 383–393. [DOI] [PubMed] [Google Scholar]
- Vincenz-Donnelly L, Holthusen H, Korner R, Hansen EC, Presto J, Johansson J, Sawarkar R, Hartl FU, Hipp MS. (2018). High capacity of the endoplasmic reticulum to prevent secretion and aggregation of amyloidogenic proteins. EMBO J , 337–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voos W, Jaworek W, Wilkening A, Bruderek M. (2016). Protein quality control at the mitochondrion. Essays Biochem , 213–225. [DOI] [PubMed] [Google Scholar]
- Wang S, Ng DT. (2010). Evasion of endoplasmic reticulum surveillance makes Wsc1p an obligate substrate of Golgi quality control. Mol Biol Cell , 1153–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Thibault G, Ng DT. (2011). Routing misfolded proteins through the multivesicular body (MVB) pathway protects against proteotoxicity. J Biol Chem , 29376–29387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegner CS, Malerod L, Pedersen NM, Progida C, Bakke O, Stenmark H, Brech A. (2010). Ultrastructural characterization of giant endosomes induced by GTPase-deficient Rab5. Histochem Cell Biol , 41–55. [DOI] [PubMed] [Google Scholar]
- Wolins N, Bosshart H, Kuster H, Bonifacino JS. (1997). Aggregation as a determinant of protein fate in post-Golgi compartments: role of the luminal domain of furin in lysosomal targeting. J Cell Biol , 1735–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong E, Cuervo AM. (2010). Integration of clearance mechanisms: the proteasome and autophagy. Cold Spring Harb Perspect Biol , a006734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodman PG, Futter CE. (2008). Multivesicular bodies: co-ordinated progression to maturity. Curr Opin Cell Biol , 408–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Rapoport TA. (2018). Mechanistic insights into ER-associated protein degradation. Curr Opin Cell Biol , 22–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi H, Arakawa S, Kanaseki T, Miyatsuka T, Fujitani Y, Watada H, Tsujimoto Y, Shimizu S. (2016). Golgi membrane-associated degradation pathway in yeast and mammals. EMBO J , 1991–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K, Fujii R, Toyofuku Y, Saito T, Koseki H, Hsu VW, Aoe T. (2001). The KDEL receptor mediates a retrieval mechanism that contributes to quality control at the endoplasmic reticulum. EMBO J , 3082–3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Arines FM, Zhang W, Li M. (2018). Sorting of a multi-subunit ubiquitin ligase complex in the endolysosome system. eLife 7, e33116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimori T, Yamamoto A, Moriyama Y, Futai M, Tashiro Y. (1991). Bafilomycin A1, a specific inhibitor of vacuolar-type H(+)-ATPase, inhibits acidification and protein degradation in lysosomes of cultured cells. J Biol Chem , 17707–17712. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.