Abstract
Wild Saccharomyces cerevisiae strains are typically diploid. When faced with glucose and nitrogen limitation they can undergo meiosis and sporulate. Diploids can also enter a protective, nondividing cellular state or quiescence. The ability to enter quiescence is highly reproducible but shows broad natural variation. Some wild diploids can only enter cellular quiescence, which indicates that there are conditions in which sporulation is lost or selected against. Others only sporulate, but if sporulation is disabled by heterozygosity at the IME1 locus, those diploids can enter quiescence. W303 haploids can enter quiescence, but their diploid counterparts cannot. This is the result of diploidy, not mating type regulation. Introduction of SSD1 to W303 diploids switches fate, in that it rescues cellular quiescence and disrupts the ability to sporulate. Ssd1 and another RNA-binding protein, Mpt5 (Puf5), have parallel roles in quiescence in haploids. The ability of these mutants to enter quiescence, and their long-term survival in the quiescent state, can be rescued by exogenously added trehalose. The cell wall integrity pathway also promotes entry, maintenance, and recovery from quiescence through the Rlm1 transcription factor.
INTRODUCTION
Quiescence is a critical and conserved alternative to proliferation. Maintenance of this protective quiescent state and recovery from it promotes the long-term survival of both multi- and unicellular species. The advantage to studying quiescence in free-living, unicellular organisms is that there is no intervention or genetic manipulation required to follow the transition in or out of quiescence. In Saccharomyces cerevisiae, quiescence is typically induced by nutrient limitation, but it is not a starvation state. Faced with a waning nutrient supply, these cells down-regulate highly conserved signaling pathways that promote proliferation and redirect their gene expression and metabolism to stockpile nutrients and induce processes that promote long-term survival (Lillie and Pringle, 1980; Sillje et al., 1999; Francois and Parrou, 2001; De Virgilio, 2012; McKnight et al., 2015; Young et al., 2017).
Most of what we know about quiescence in budding yeast has come from studies of yeast cultures grown to saturation or stationary phase. However, stationary phase cultures contain at least three distinct cell types based on light scattering alone (Li et al., 2013). These three cell types arise in part from the highly asymmetric final cell divisions that cells undergo after they have taken up all the glucose from their medium (Johnston et al., 1977; Li et al., 2013) and the elaboration of highly fortified cell walls (Shimoi et al., 1998). One of these three cell types is quiescent (Q cells) and can be separated from the nonquiescent (non-Q) cells by density gradient sedimentation (Allen et al., 2006). These are unbudded cells with 1N DNA content that have accumulated high levels of glucose in the form of trehalose, which contributes to their high density (Shi et al., 2010). They are more thermotolerant and long-lived than their nonquiescent siblings (Allen et al., 2006; Li et al., 2009). They are primarily daughter cells and young mothers (Allen et al., 2006). This daughter enrichment is consistent with numerous studies indicating that daughter cells asymmetrically inherit higher quality proteins, mitochondria, and other organelles that promote their longevity (Lai et al., 2002; McFaline-Figueroa et al., 2011). The lighter nonquiescent cells are primarily old mothers that undergo reactive oxygen species accumulation and apoptosis. These cells lose the ability to reenter the cell cycle rapidly and those that can reenter do so much slower than Q cells (Lee et al., 2016).
In W303 haploid prototrophs, grown to saturation in rich medium, the Q cells make up about half the population. However, introduction of a full-length SSD1 allele to this strain enables 90% of the cells to fractionate as Q cells (Li et al., 2009). In this case the mother-daughter asymmetry persists in that the quiescent daughter cells have a longer life span than the quiescent mother cells (Li et al., 2013). There is also an Ssd1-mediated increase in stress tolerance and longevity in the nondividing state, but no significant increase in trehalose levels (Li et al., 2013). This indicates that trehalose is necessary but not sufficient to confer the properties of quiescent cells. Other factors, like Ssd1, contribute. Ssd1 levels increase as dividing cells age and this increase extends the number of times each cell can divide before senescence (Hu et al., 2018).
Ssd1 is an RNA-binding protein that binds to many mRNAs encoding cell wall proteins and delivers at least one of them to sites of polarized cell growth (Hogan et al., 2008; Jansen et al., 2009; Kurischko et al., 2011). Upon stress, or stationary phase, when cell division ceases, P bodies accumulate (Brengues et al., 2005; Teixeira et al., 2005) and Ssd1 delivers cell wall mRNAs to P bodies where they are translationally repressed, stored, or degraded (Holmes et al., 2004; Coller and Parker, 2005). Hence, a key function of Ssd1 is to regulate when and where cell wall growth and remodeling takes place. Lack of Ssd1 weakens cell walls and activates the cell wall integrity (CWI) checkpoint (Arias et al., 2011), which monitors cell wall defects and arrests division (Levin, 2011). Ssd1 and a second mRNA-binding protein (Mpt5/Puf5) function in parallel to maintain CWI and prolong life span in the dividing (Kaeberlein and Guarente, 2002) and nondividing states (Li et al., 2013). They also play overlapping, important roles in Q-cell formation in haploid cells (Li et al., 2013). Mpt5 activates the CWI pathway by translational repression of LRG1 mRNA (Duy et al., 2017), which encodes an inhibitor of the pathway (Stewart et al., 2007).
Ssd1 association with P bodies requires the low-complexity prion-like domain (PLD) in the N-terminus of Ssd1 (Kurischko and Broach, 2017) and is disrupted by Cbk1-dependent phosphorylation of the PLD. Failure to phosphorylate the Cbk1 sites in Ssd1 leads to its constitutive association with P bodies, acute translational repression of Ssd1-associated mRNAs, and cell lysis (Jorgensen et al., 2002; Kurischko et al., 2005; Jansen et al., 2009). Cbk1 is a Ndr/LATS family kinase that is localized to sites of polarized growth (Racki et al., 2000; Weiss et al., 2002), where it inhibits translational repression of cell wall mRNAs by Ssd1 and permits localized cell wall remodeling (Jansen et al., 2009). Cbk1 activity is modulated by Lre1 during the cell cycle (Mancini Lombardi et al., 2013) and in response to glucose availability (Versele and Thevelein, 2001).
We have quantified sporulation and quiescence entry in wild and laboratory yeast strains. Both are highly reproducible quantitative traits that show broad natural variation. Our previous work showed that Ssd1 and Mpt5 make overlapping contributions to quiescence entry in W303 haploids (Li et al., 2009, 2013). Our new data make it clear that Ssd1 is particularly important for the formation and longevity of quiescent cells in W303 diploids. We also find that the CWI pathway is required for surviving the entry into quiescence when Ssd1 function is compromised.
RESULTS
S. cerevisiae in the wild is typically diploid (Landry et al., 2006), and diploids can sporulate when glucose and nitrogen are scarce. This raises the possibility that S. cerevisiae has had little pressure to evolve an alternative to sporulation that would insure survival of diploid cells in the nondividing state. Alternatively, diploids may be equally capable of entering a quiescent cellular state when nutrients are limiting. To explore these possibilities, we assayed both laboratory and wild strains as haploids and diploids for their ability to enter a stress-tolerant and long-lived quiescent (Q) state and compared that to their ability to sporulate.
Quiescence is a quantitative trait accessible to both haploid and diploid S. cerevisiae strains
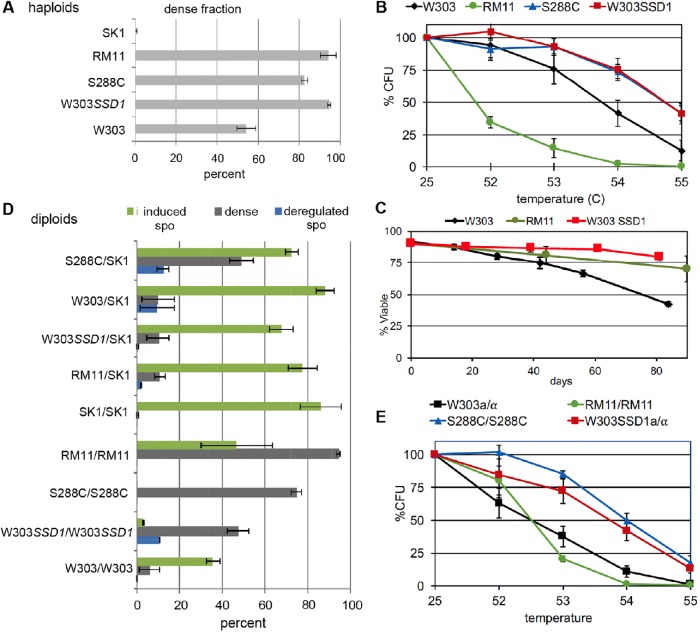
As noted above, haploid quiescent cells can be purified due to their high density compared with the nonquiescent cells within a saturated or stationary phase culture. We find that quiescent diploids can be reproducibly obtained by the same density gradient protocol (gray bars, Figures 1 and 2). This shift to high density is a characteristic of the transition to quiescence, but it does not singularly define the nondividing quiescent state. Hence, we also assayed the thermotolerance and longevity of these dense cells to identify Q cells. In parallel, we quantified sporulation in lab and wild diploids. We assayed what we will refer to as induced and deregulated sporulation. S. cerevisiae is induced to sporulate when shifted from rich medium to acetate (Esposito et al., 1969). This shift eliminates both nitrogen and glucose, which derepresses the master inducer of meiosis IME1, and triggers sporulation (green bars, Figures 1 and 2; Pinon, 1977; Kassir et al., 1988; Sagee et al., 1998; Weidberg et al., 2016). We use deregulated sporulation to refer to that which occurs without intervention, after 7 d of growth in rich medium, where the cells limit for glucose but not for nitrogen (Alvers et al., 2009). Under these conditions, two-spore asci can arise from a modified meiosis that produces two haploid nonsister spores with opposite mating type and maximal genetic diversity (Davidow et al., 1980; Neiman, 2011; Renicke et al., 2017). Asci sediment with the dense cell fraction, so we use microscopic examination to determine the percent of deregulated sporulation in the dense fraction (blue bars, Figures 1 and 2). Dense cells within this fraction are represented by the difference between the gray and blue bars. Budded cells are rare in all but one strain (red bars, Figure 2). It is worth noting that the conditions we use for generating Q cells and spores may not be ideal for all strains (Elrod et al., 2009). However, under these fixed conditions, we find Q-cell formation and both forms of sporulation to be highly reproducible, but quite variable among the strains we tested.
FIGURE 1:
Haploid and diploid lab strains vary widely in their ability to form quiescent cells. (A) Dense cells were collected and quantified from the haploid strains indicated (n = 5, 2, 2, 8, 10 top to bottom). (B) Innate thermotolerance of the dense haploid cells (n = 7, 2, 2, 4 right to left). (C) Long-term viability of purified haploid Q cells (n = 2,2,3). (D) The dense fraction of the diploid strains indicated was quantified (gray bars, n = 5, 5, 5, 5, 10, 5, 6, 3, 9 top to bottom). This dense fraction includes quiescent cells and/or tetrad and dyad asci, which are the products of deregulated sporulation that occurs within the YEPD culture. The percent of asci within the dense fraction is shown in blue (n = 2). Each strains’ capacity to induce sporulation is shown in green (n = 3). (E) Dense fraction of diploid cells with no signs of sporulation were assayed for innate thermotolerance (n = 2). In these and all subsequent plots SSD1 refers to the SSD1 allele from S288c.
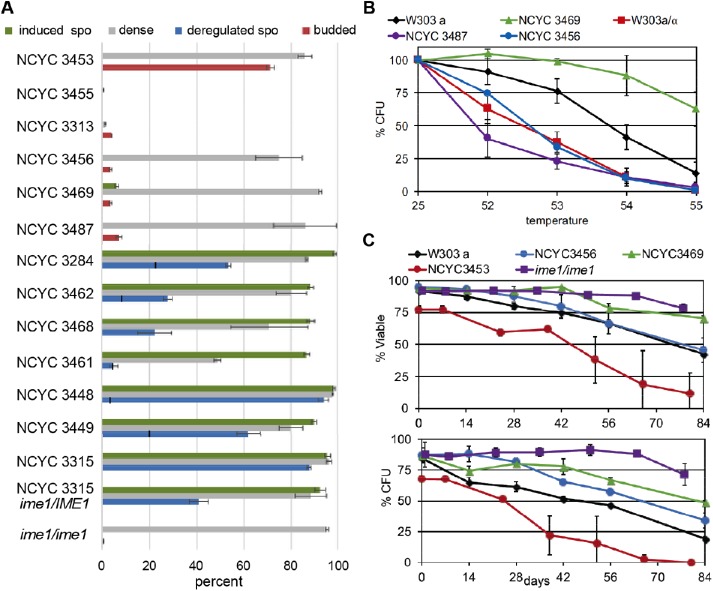
FIGURE 2:
Some wild diploids enter quiescence instead of sporulating. (A) As in Figure 1D, dense fraction was quantified (gray bars) and the percentage of asci resulting from deregulated sporulation within this fraction was counted microscopically (blue bars). The fraction left of the vertical line on the blue bars indicates the percentage of the deregulated sporulation that produced dyads. Efficiency of induced sporulation is shown by green bars. Red bars show the percent of budded cells within the dense fraction. Each strain was assayed in duplicate, except the bottom two ime1 mutant strains that were assayed in triplicate. NCYC 3315 ime1/IME1 is BY8014 and NCYC 3315 ime1/ime1 is BY8029. (B) Dense diploid cells with no signs of sporulation were assayed for innate thermotolerance (n = 7, 3, 4, 2, 2). (C) The dense cells produced by the strains indicated were assayed in duplicate for longevity in the nondividing state by monitoring viability and colony formation (CFU) over an 84-d time course in water.
Quiescence and sporulation are cell fate choices accessible to some but not all diploids
The wild haploid strain RM11 produces 96% dense cells, showing that in some genetic backgrounds nearly all cells are capable of obtaining the density of Q cells (Figure 1A). These dense RM11 cells are highly thermosensitive (Figure 1B), indicating that they lack at least one characteristic that has been associated with a protective quiescent state, but they have an extremely long life span in the nondividing state (Figure 1C), which is the cardinal feature of quiescent cells. The RM11 diploid can be induced to sporulate, but it shows no deregulated sporulation (Figure 1D). SK1, which is capable of highly efficient induced sporulation (Kane and Roth, 1974), shows no evidence of deregulated sporulation and completely fails to produce Q cells in the haploid or diploid state (Figure 1, A and D). S288c diploids are highly defective at sporulation under our conditions, but they are very efficient at entering quiescence (Figure 1, D and E; Allen et al., 2006). The W303 diploid can be induced to sporulate, but it produces very few dense cells (Figure 1D), and those that are produced are less thermotolerant than the W303 haploid Q cells (Figure 1, B and E). The same W303 strain carrying a wild-type SSD1 locus (from S288c) has the reverse fate, producing 10 times more dense cells with high thermotolerance and almost eliminating sporulation.
Interestingly, the very efficient induced sporulation of SK1 is dominant when SK1 is crossed to S288c, W303, W303SSD1, and RM11 (Figure 1D). Many genes have been identified that promote sporulation in SK1 (Deutschbauer and Davis, 2005; Ben-Ari et al., 2006) but the nature of this dominance has not been reported. The SK1 defect in Q-cell formation is also dominant in all but the S288c/SK1 diploids. These mixed background diploids display some deregulated sporulation (Figure 1D, blue bars), whereas the homozygous lab strains do not. It is likely that the lab strains were adopted in part because they do not sporulate in rich medium, which would result in mixtures of haploids and diploids. In contrast, deregulated sporulation is quite common in the wild strains we have tested (blue bars, Figure 2A).
The propensity to either sporulate or enter quiescence displayed by most of these strains suggests that there has been selective pressure to limit pathway choice to promote longevity in different environmental conditions. To see whether this restricted cell fate choice is observed in other wild diploids, we assayed Q-cell formation and sporulation in a set of wild diploids that had already been characterized for induced sporulation efficiency (Cubillos et al., 2011; Tomar et al., 2013). These authors found that one-third of the wild diploids they tested failed to induce sporulation when glucose and nitrogen was withdrawn. We assayed induced sporulation in their highest and lowest efficiency sporulators and our results are in complete agreement (Figure 2A). We also measured deregulated sporulation and the ability to enter quiescence.
The strains with the highest levels of induced sporulation varied in their ability to promote deregulated sporulation and form Q cells. NCYC 3448 (fruit), 3449 (cactus), and 3315 (oak) showed highly efficient deregulated sporulation. This indicates that these wild strains exclusively sporulate under conditions that trigger quiescence in other strains. This led us to wonder whether the path to quiescence is defective in these strains or whether it is just superceded by the drive to sporulate. To test this, we deleted the IME1 gene from one of the wild diploids showing efficient deregulated sporulation (NCYC 3315) and assayed the ime1/IME1 heterozygote and the ime1/ime1 homozygote (Figure 2A). Loss of one copy of IME1 had no impact on induced sporulation, but it clearly interfered with deregulated sporulation, indicating that Ime1 is haploinsufficient for this process (Figure 2A). The fraction of ime1/IME1 cells that did not sporulate in YEPD initiated cellular quiescence based on cell density increase. As expected, complete loss of Ime1 activity eliminated both induced and deregulated sporulation. Instead, all the ime1/ime1 cells developed the density of quiescent cells and these cells were extremely long lived in the nondividing state (Figure 2C). This indicates that this wild strain retains the capacity to enter quiescence, but it has evolved to exclusively sporulate under the conditions we tested.
The other four strains that achieved high levels of induced sporulation behaved like the ime1/IME1 heterozygote. With NCYC 3284 (soil), 3462 (palm), 3468 (palm), and 3461 (palm), the dense fraction was a mixture of cells and asci. This suggests that there are some environments in which it is beneficial to be capable of both cell fates. However, the dense cells could not be separated from the asci and hence, they could not be assayed to see whether they attained the longevity of quiescent cells.
Some wild diploids have evolved to enter quiescence instead of sporulating
The six poor sporulators also had a diverse response to glucose limitation. NCYC 3453 is a baker’s strain that had an unexpected phenotype. Ninety percent of these diploids attained the density of Q cells, but almost all of them were budded, which means either that they completely failed to arrest in G1 in response to glucose limitation or that mother and daughter cells fail to separate (Figure 2A). The cells in these 7-d cultures were 78 ± 3% viable with medium to large buds, but they had a short and variable life span in the nondividing state (Figure 2C). NCYC 3455 (clinical isolate) and NCYC 3313 (honey wine brewing) also produced neither Q cells nor spores. These three strains may lack the capacity to differentiate, or they may not recognize the same nutritional signals that trigger differentiation in most strains. The three other strains that were known to be defective in sporulation produced high levels of dense cells (NCYC 3456, 3469, and 3487). To see whether these dense cells had other properties of Q cells, we assayed thermotolerance and longevity. NCYC 3487 (baker’s strain) and NCYC 3456 (clinical isolate) were thermosensitive, but NCYC 3469 (fruit) had thermotolerance and longevity greater than that of the W303 haploid Q cells (Figure 2, B and C). These data show that wild diploids exist that have restricted pathway choice to enter a protective, quiescent cellular state instead of sporulating.
Quiescence can be regulated by ploidy
Although there are wild diploids that can form Q cells, there are others that cannot. W303 is an interesting example that forms Q cells efficiently as a haploid, but not as a diploid (Figure 1, A and D). Knowing that the truncated ssd1-2 allele in W303 (previously referred to as ssd1-d2) reduces Q-cell formation and longevity in haploids compared with the full-length SSD1 allele carried by S288c (Li et al., 2009, 2013), we also assayed W303SSD1 diploids. As noted above, W303 diploids can sporulate but they produce very few Q cells. W303SSD1 diploids have the reverse fate, producing 10 times more dense cells with high thermotolerance and almost eliminating sporulation. This indicates that, in W303, Ssd1 is an important regulator of these alternative cell fates, promoting quiescence in both haploids and diploids and interfering with sporulation in diploids. However, this is specific to the W303 background. Six of the strains we assayed carry the same S288c SSD1 allele. Three of these strains also primarily enter cellular quiescence (NCYC345, 3469, and 3487). However, RM11 can both sporulate and enter cellular quiescence, NCYC 3453 does neither, and the two others (NCYC3448 and 3449) have the opposite fate of only sporulating. All the other wild strains we analyzed have full-ength SSD1 alleles with one to seven amino acid differences from the SSD1 of S288c. The SSD1 alleles harbored by SK1 and NCYC3469 are identical, but SK1 only sporulates and NCYC3469 only enters cellular quiescence. Table 1 lists the SSD1 alleles residing in these strains and the phenotypic classes within which they reside. The S288c allele, referred to in the table as SSD1-1, is the most common and is represented in all four phenotypic classes. Clearly, there are other polymorphisms in these strains that are contributing to cell fate choice, but in W303, SSD1 is sufficient to cause a shift from sporulation to quiescence.
TABLE 1:
SSD1 alleles used in this study.
| SSD1-1 | S288c, RM11, NCYC 3453,3487,3448, 3449 | Reference sequence (Saccharomyces Genome Database) |
| ssd1-2 | W303 | Terminates after 697 |
| SSD1-3 | NCYC 3456 | T827S |
| SSD1-4 | NCYC 3455, NCYC 3284 | T827S, S1190G, A1196P |
| SSD1-5 | NCYC 3315 | T827S, S1190G, A1196P, V1250A |
| SSD1-6 | SK1, NCYC 3469 | S377C, T693M, T827S, S1190G, A1196P |
| SSD1-7 | NCYC 3313 | G185D, S651F, T827S, S1190G, A1196P, V1250A |
| SSD1-8 | NCYC 3462, NCYC 3468, NCYC 3461 | S441G, E684K, T827S, R910K, N936S, S1190G, A1196P |
| SSD1 alleles present in different phenotypic classes of diploids | ||
| Spo- Q+ | SSD1-1 (S288c, 3487), SSD1-3 (3456), SSD1-6 (3469) | |
| Spo+ Q+ | SSD1-1(RM11), SSD1-4 (3284), SSD1-8 (3462, 3468, 3461) | |
| Spo+ Q− | SSD1-1 (3448,3449), ssd1-2 (W303), SSD1-5 (3315), SSD1-6 (SK1) | |
| Spo− Q− | SSD1-1 (3453), SSD1-4 (3455), SSD1-7 (3313) | |
Sequences were extracted from the Sanger Institute database. Allele numbers were arbitrarily set for clarity. Differences from the reference sequence (S288c) for each allele are listed at the top. At the bottom, the alleles and the strain numbers (in parentheses) for each phenotypic class are listed. The four-digit numbers refer to the NCYC strains.
To further investigate the diploid-specific defect in Q-cell formation in W303, we asked whether it was conferred by heterozygosity at the MAT locus. Haploids carry either the MATa or MATα mating type information, and when they mate, transcription factors encoded by the MATa and MATα loci act together to repress the mating program and promote sporulation (Strathern et al., 1981). As a result, mating is only accessible to haploids, and sporulation is accessible only to diploids. This led us to ask whether quiescence is also regulated by the MAT locus. W303a/α diploids produce 8 ± 6% dense cells (n = 8), which is sixfold fewer Q cells than the W303 MATa haploid produces, and those that are produced are thermosensitive (Figure 1). To see whether this failure to enter quiescence is regulated by heterozygosity at the MAT loci, we generated a MATα/matΔ diploid, which lacks MATa information (Strathern et al., 1981). This MATα/matΔ diploid produces only 0.7 ± 0.8% dense cells (n = 6), indicating that mating type heterozygosity is not required; diploidy per se is interfering with W303’s ability to enter quiescence.
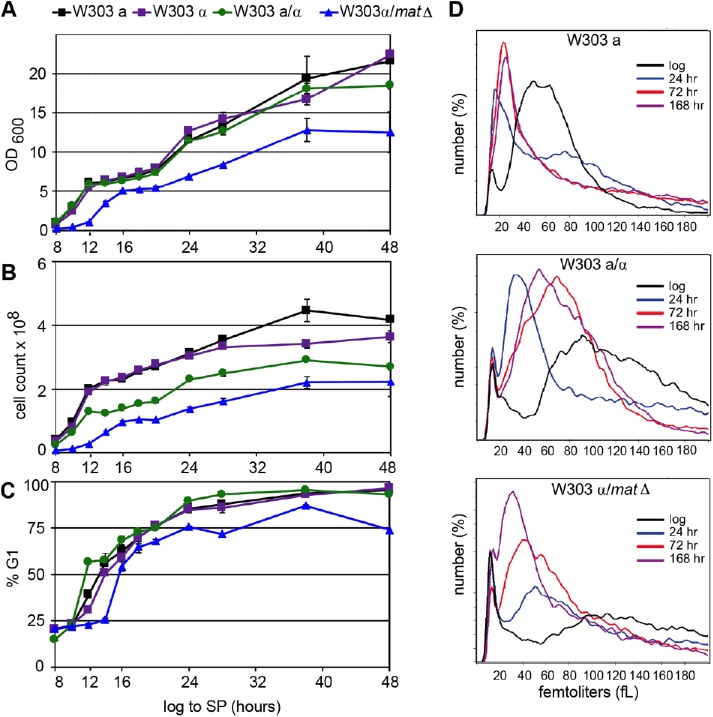
W303 diploids initially respond to glucose limitation like haploids
We have followed the W303 diploids and haploids as they transition from logarithmic growth to stationary phase (log to SP) to see where their behaviors diverge. The W303a and α haploids and the a/α diploids undergo the diauxic shift at the same time (12 h) and attain the same final optical density, but the larger diploids achieve that density at a lower cell number, as expected (Figure 3, A and B). The a/α diploid also shows the same G1 arrest profile as the haploids (Figure 3C). The MATα/matΔ grows more slowly, and never attains the cell density of the true a/α diploid. The MATα/matΔ is slower and somewhat less efficient at arresting in G1 (Figure 3C), but it initiates the G1 arrest before its diauxic shift (16 h) just like the true haploid and diploid strains. These data indicate that both diploid strains are responding to the waning glucose supply by prolonging G1, and they achieve a complete or nearly complete G1 arrest.
FIGURE 3:
W303 diploids display prolonged G1 phase and asymmetrical cell divisions as they enter stationary phase. Growth of W303 haploids and diploids from 8 to 48 h in YEPD was measured (n = 2) by optical density (A) and cell count by Coulter counter (B). The fraction of these cells in G1 was measured by flow cytometry (C). Cell volume distribution (D) for the haploid and diploids from log phase and three postdiauxic time points as indicated. A representative trace is shown for each (n = 4). Two independently constructed W303α/matΔ strains were assayed with identical results. Strain numbers are provided in Table 2. The SSD1 allele from S288c is referred to as SSD1-1 in the table to distinguish it from the other SSD1 alleles in the wild strains described in Table 1.
To see whether diploids also undergo asymmetrical cell divisions after the diauxic shift, we monitored cell size over this time course. During log phase growth, the diploids are more heterogeneous in size than the haploid and the MATα/matΔ culture contains a large fraction of very small particles (Figure 3D). These are most likely the inviable cells that comprise ∼50% of the population (Figure 4A). However, all three strains undergo asymmetrical cell divisions after the diauxic shift (Figure 3D). As previously observed (Li et al., 2013), the haploid shifts from a modal cell volume of ∼50 fl in log phase, to 16 fl postdiauxie (24 h), and then enlarges slightly to 26 fl after 7 d (168 h). In the case of the a/α diploid, cell size shifts from a very broad distribution with a peak at 91 fl to a remarkably tight distribution after 24 h, peaking at 34 fl, and then increasing in size to ∼60 fl at the later time points. This indicates that the true diploid undergoes asymmetrical cell divisions, just like the haploid, but it does not restrict growth to the same extent at later times. The MATα/matΔ cells also undergo an asymmetric cell division, shifting from >100 fl to 51 fl after 24 h, then, unlike either the haploid or the a/α diploid, these cells continue to shift to a smaller peak size, which reaches 32 fl after 168 h. We assayed two independently constructed MATα/matΔ strains and both behaved identically. On the basis of their G1 arrest and asymmetrical cell divisions, we conclude that diploids respond much like haploids as glucose becomes limiting, but some ploidy-sensitive phenomenon is preventing them from successfully transitioning to quiescence.
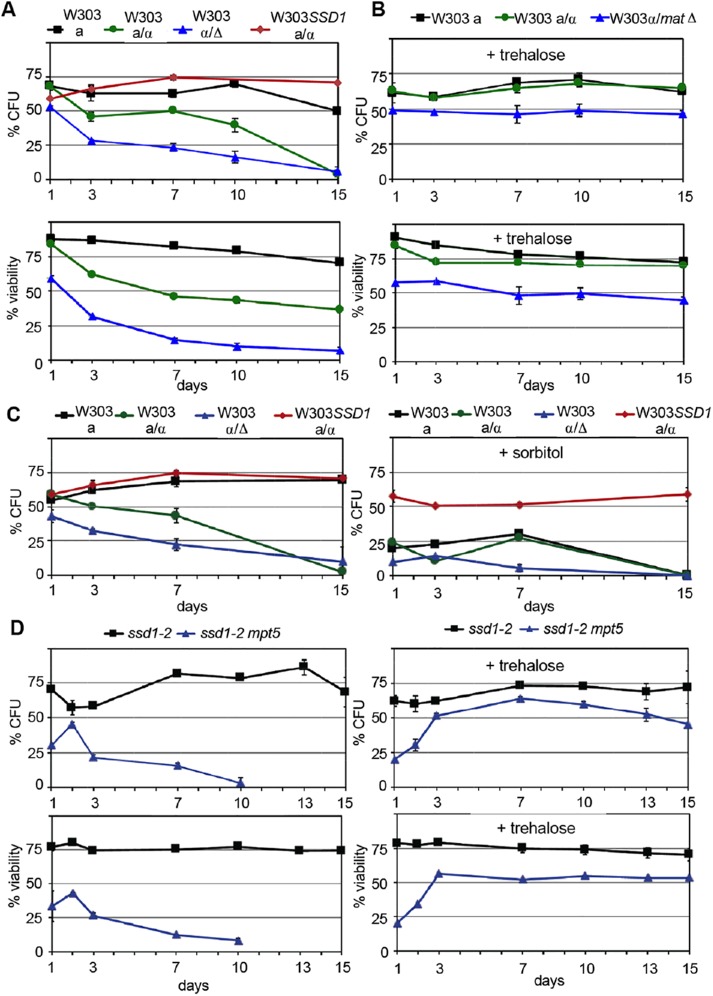
FIGURE 4:
Ssd1 and trehalose rescue the viability of W303 diploids as they enter quiescence. Strains listed in Table 2. (A) Percent of total cells that can form colonies (CFU) and remain viable of the strains indicated as they are cultured (in duplicate) in YEPD for 15 d. (B) As in A, but with the addition of 1% trehalose to the medium of the starting log phase culture, OD600 0.02 (n = 2). (C) Colony formation of the same strains with and without the addition of 1M sorbitol to the medium of the starting log phase cultures, OD600 0.02 (n = 2). (D) Colony formation and viability of two haploid strains (BY6500 W303 ssd1-2 and BY6672 W303 ssd1-2 mpt5) with (right) and without (left) the addition of 1% trehalose to the starting log phase culture, OD600 0.02 (n = 2). ssd1-2mpt5 cells grow slowly at 30°C and reach saturation a full day later than ssd1-2. This is likely why the double mutant continues to increase in cell number at early time points. Trehalose may also provide some thermoprotection to these cells.
W303 diploids lose viability in stationary phase and SSD1 rescues viability and Q-cell formation
We also monitored colony-forming units (CFUs) and viability as the W303 diploids entered stationary phase and found that both diploids lose viability rapidly (Figure 4A). The W303 a/α diploid maintains viability longer, but it loses the ability to return to the cell cycle and form colonies almost as rapidly as do the W303 MATα/matΔ diploids. On the basis of the fact that W303 diploids carrying SSD1 form viable, long-lived Q cells (Figure 1, D and E), we expected that this diploid-specific phenotype would be rescued by SSD1, and that is indeed what we observe. Introduction of SSD1 to the W303 background rescues viability of the diploid (Figure 4A). Ssd1 is an RNA-binding protein and many of its target mRNAs encode cell wall proteins (Hogan et al., 2008; Jansen et al., 2009). As a result, some ssd1-2 phenotypes can be rescued by 1M sorbitol, an osmo-stabilizer (Kaeberlein and Guarente, 2002). Sorbitol does not rescue the diploid-specific lethality we observe as W303 diploids enter stationary phase. In fact, it causes more rapid death (Figure 4C).
Exogenously added trehalose rescues viability and the transition to quiescence of ssd1-2 diploids and ssd1-2 mpt5 haploids
In addition to being a glucose reserve, trehalose serves as a protectant from heat, oxidation, and other forms of stress (Elbein et al., 2003). This disaccharide of glucose accumulates to high levels as cells import the last glucose from their media (Lillie and Pringle, 1980). Several mutants have been identified that are defective in the accumulation or utilization of trehalose and these defects are associated with poor survival in stationary phase (Samokhvalov et al., 2004; Favre et al., 2008; Kyryakov et al., 2012; Ocampo et al., 2012; Cao et al., 2016). Some of these defects can also be rescued by addition of trehalose to the medium (Tapia et al., 2015; Cao et al., 2016). We found significantly less trehalose in the W303 diploid, and a further drop in the MATα/matΔ diploid (Table 2). This drop in trehalose is also rescued by SSD1. It could be that the loss of viability of W303 ssd1-2 diploids is preventing trehalose accumulation. Alternatively, failure to accumulate trehalose may cause cell death under glucose-limiting conditions. Whatever the cause, we found that addition of exogenous trehalose to 1% (26 mM) fully rescues the viability of these diploids (Figure 4B).
TABLE 2:
Trehalose levels are higher in W303 haploids than diploids.
| Strain | Genotype | Trehalose | SD | |
|---|---|---|---|---|
| BY6500 | MAT a | ssd1-2 | 596 | 60 (6) |
| BY6641 | MAT a | SSD1-1 | 641 | 52 (12) |
| BY6946 | MATa/α | ssd1-2/ssd1-2 | 309 | 83 (4) |
| BY6974 | MATα/mat∆ | ssd1-2/ssd1-2 | 142 | 39 (3) |
| BY6962 | MATa/α | SSD1-1/SSD1-1 | 551 | 26 (2) |
| BY6976 | MATa/α | SSD1-1/ssd1-2 | 615 | 41 (3) |
Trehalose was converted to glucose by trehalase and is reported as glucose equivalents (μg/ml). Samples were taken from YEPD cultures after 7 d of growth.
Number of measurements is in brackets.
In haploids, Ssd1 and Mpt5 play additive roles in CWI, Q-cell formation, and longevity (Li et al., 2013). Like ssd1-2 alone, the mpt5SSD1 haploid maintains high trehalose levels, but produces about half as many Q cells as wild type and these Q cells have reduced longevity in the nondividing state (Li et al., 2013). In contrast, the ssd1-2mpt5 haploid fails to produce Q cells, and these haploids lose 80% viability after 7 d of growth in YEPD (Li et al., 2013). They also show a sevenfold reduction in trehalose levels over that time course (Li et al., 2013). As above, we followed viability of the ssd1-2mpt5 haploid after addition of 1% trehalose (Figure 4D) and found that it fully rescued viability.
To see whether exogenous trehalose enables these cells to fully transition to the quiescent state, we grew these strains in YEPD plus trehalose for 7 d and fractionated them by density sedimentation. The added trehalose enabled nearly all of the cells to attain the density of quiescent cells (Figure 5A). We then transferred these dense cells to water and monitored their longevity in the nondividing state (Figure 5B). Not only were these dense cells greater than 75% viable at the start, but their ability to maintain viability in quiescence and reenter the cell cycle to form colonies rivaled that of most of the wild strains we assayed (Figure 2C). By these criteria, exogenously added trehalose rescues the ability to enter and maintain quiescence in these cells.
FIGURE 5:
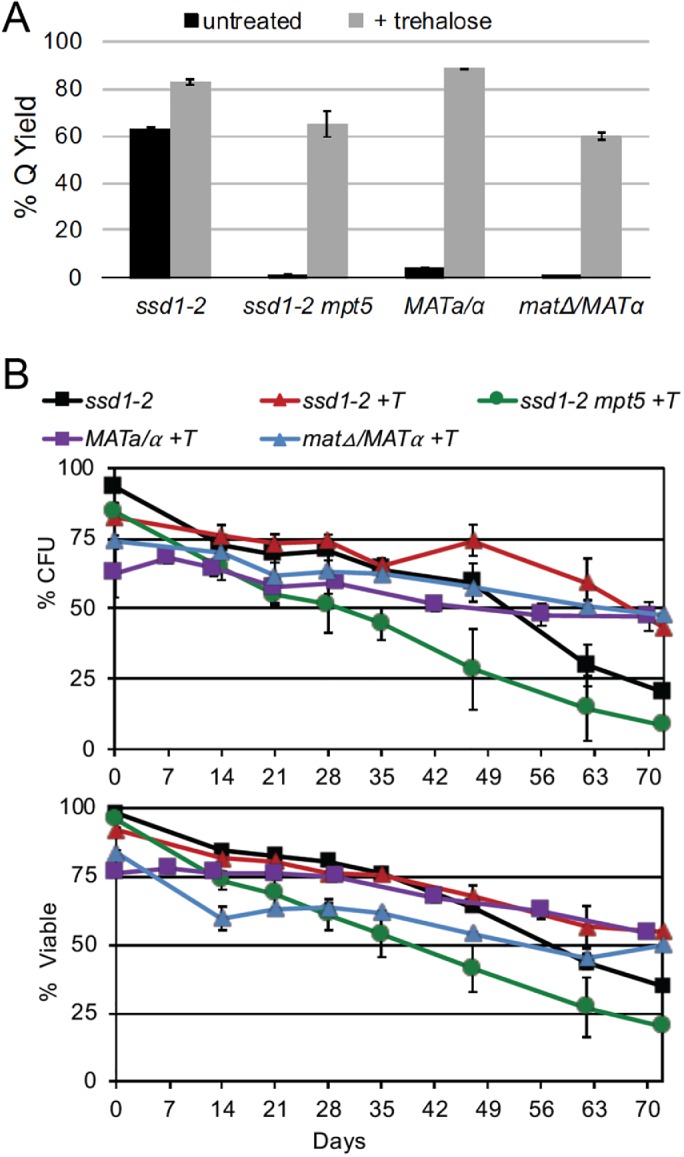
Trehalose supplementation rescues the ability to enter and maintain the quiescent state. (A) Yield of dense cells of W303-derived strains as indicated, with and without trehalose supplementation (n = 2). (B) Survival of those dense cells, incubated in water for 70 d with trehalose (+T) approaches or exceeds survival of ssd1-2 haploids (n = 2).
The CWI pathway plays a dual role in quiescence entry
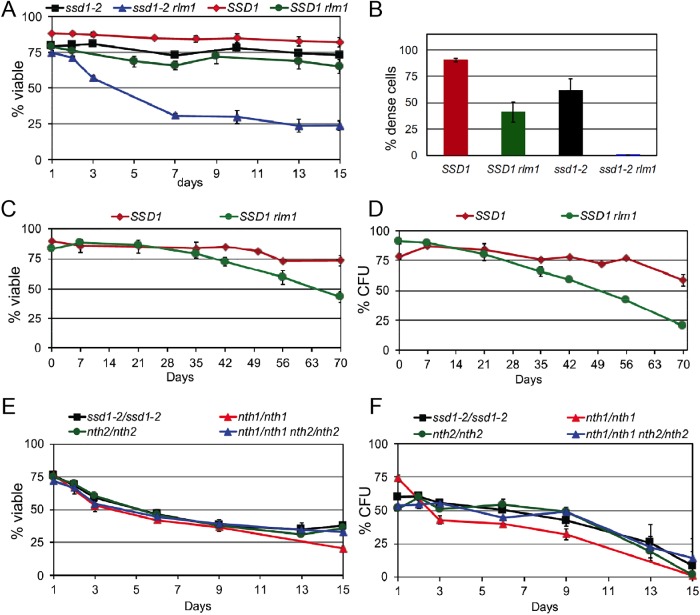
It has been shown that W303 ssd1-2 mpt5 cells lose viability rapidly in unbuffered minimal medium and this can be rescued by overproducing Pkc1, which activates the CWI pathway (Stewart et al., 2007). W303 ssd1-2 mpt5 cells are also thermosensitive (Kikuchi et al., 1994), and this thermosensitivity can be rescued by deleting Lrg1, which is a negative regulator of the CWI pathway (Stewart et al., 2007). Mpt5 binds (Gerber et al., 2004) and translationally represses Lrg1 (Stewart et al., 2007; Duy et al., 2017). These data suggest that activation of the CWI pathway is a critical function of Mpt5 under acid- (Burtner et al., 2009) and heat-stress conditions (Stewart et al., 2007). To see whether the CWI pathway plays an important role in quiescence entry, we deleted RLM1, which is the predominant downstream transcription factor activated by CWI (Watanabe et al., 1995; Levin, 2011). Figure 6A shows that rlm1 cells carrying a functional SSD1 gene survive growth to saturation in rich medium just like wild-type cells. The rlm1 ssd1-2 cells grow like wild type during logarithmic growth, but they lose viability rapidly as they reach saturation. We conclude that the CWI pathway plays a critical role during the transition to quiescence under conditions of cell wall stress. In contrast, when we purified dense cells, we found that rlm1 reduced yields in both SSD1 and ssd1-2 cells (Figure 6B) so the CWI pathway also plays a role in achieving the density of quiescent cells that is independent of Ssd1. We also assayed the longevity of the SSD1rlm1 dense cells in the nondividing state. Viability and to a greater extent the ability to reenter the cell cycle and form colonies is significantly reduced in SSD1rlm1 cells (Figure 6, C and D).
FIGURE 6:
Rlm1 promotes entry into quiescence and becomes essential when Ssd1 activity is compromised. (A) Viabililty of W303-derived strains as indicated as they transition from logarithmic growth to stationary phase. Strains used are BY6500 W303 ssd1-2, BY7442 W303 ssd1-2 rlm1, By6641 W303 SSD1, and BY8197 W303 SSD1 rlm1. (B) Yield of dense cells after 7 d of growth to saturation. (C, D) Percent viability and colony formation of dense cells incubated in water of BY6641 W303 SSD1 and BY8197 W303 SSD1 rlm1. (E, F) Percent viability and colony formation of W303-derived diploids as they transition from logarithmic growth to stationary phase. Strains used are W303 ssd1-2/ssd1-2 diploid (BY6946) and isogenic diploids: nth1::KAN/nth1::NAT (BY7967), nth2::HYG/nth2::HYG (BY8152), or nth1:KAN/nth1::NAT nth2::HYG/nth2::HYG (BY8161). Experiments were carried out in duplicate.
Among the genes activated by Rlm1 is the NTH1 trehalase gene, which encodes the primary enzyme that degrades intracellular trehalose (Eleutherio et al., 2015). Trehalose is a critical energy source in cells exiting quiescence (Shi et al., 2010) and this may explain why rlm1 cells are defective in cell cycle reentry. The prospect that Ssd1 might translationally repress trehalase mRNA to promote entry and maintenance of quiescence led us to ask whether deletion of the two intracellular trehalase genes (NTH1 and NTH2; Nwaka et al., 1995) could rescue the ssd1-2 diploid. Figure 6, E and F, shows that loss of either or both of these intracellular trehalase activities does not rescue the ssd1-2 diploid.
DISCUSSION
S. cerevisiae in the wild is typically diploid (Landry et al., 2006) and its ability to sporulate is a quantitative trait that has been exploited to help define the mechanics and regulation of sporulation (Neiman, 2011). We have shown that the ability to enter quiescence is another highly variable but reproducible quantitative trait of both wild and laboratory S. cerevisiae strains. In lab strains, sporulation is triggered by glucose and nitrogen deprivation, but several of the wild strains have lost the requirement for nitrogen starvation, which we refer to as deregulated sporulation. Disrupting sporulation by deleting both copies of IME1 enabled one of these diploids to enter quiescence, so it is capable of entering quiescence, but there is a hierarchy of decision making that favors sporulation under these conditions. The polymorphism(s) responsible for deregulated sporulation have not been identified, but it is a common trait among the wild strains we assayed and might be relevant in the context of fungal pathogens (Huang and Hull, 2017).
Some wild diploids readily form Q cells and have lost the ability to sporulate, so there must be environmental conditions under which sporulation cannot occur or is disadvantageous. For example, Q cells are typically highly thermotolerant (Allen et al., 2006; Li et al., 2013), but sporulation fails at high temperature (Esposito and Esposito, 1969). Entering and recovering from quiescence is more rapid and less energy-consuming than spore production and germination. Cellular quiescence also enables cells to maintain a diploid state. These differences suggest that the ability to sporulate could be selected against or lost in environments where quiescence is more advantageous.
In the W303 lab strain, the pathway to quiescence is largely restricted to haploids due to the presence of the truncated and defective ssd1-2 gene. Ssd1 is an RNA-binding protein (Uesono et al., 1997) that is highly pleiotropic, interacting with more than 800 genes (S. cerevisiae Genome Database). When the truncated ssd1-2 allele is replaced with the full-length SSD1 allele from S288c, it doubles Q-cell formation in haploids and they are longer lived in the nondividing state (Li et al., 2009). W303 diploids produce very few Q cells, but introduction of SSD1 increases Q-ell formation and reduces sporulation by about 10-fold. This indicates that introducing SSD1 into W303 is sufficient to shift its diploid fate from primarily sporulation to primarily cellular quiescence. The mechanism of this switch is unknown.
The fact that Ssd1 function is much more critical in diploids than in haploids for entry into quiescence is likely to be due to its role in cell wall remodeling. Cell wall mRNAs are Ssd1’s most common targets (Hogan et al., 2008; Jansen et al., 2009; Wanless et al., 2014), and loss of Ssd1 function confers sensitivity to many cell wall disrupting agents (Lopez-Garcia et al., 2010). Cell wall fortification is a key step in the transition to quiescence (Li et al., 2015) and ssd1-2 interferes with this process (Li et al., 2013). Interestingly, 3% of yeast proteins show more than twofold differences in level between haploids and diploids and 20% of the proteins that are overexpressed in haploids are involved in cell wall organization (de Godoy et al., 2008). At least eight of the Ssd1-bound cell wall mRNAs encode proteins that are present at up to fivefold higher levels in haploids than diploids (Tos1, Scw4, Uth1, Cts1, Scw10, Cis3, Sim1, and Bgl2; de Godoy et al., 2008). Three of these (Scw4, Scw10, and Cts1) are haploinsufficient for competitive growth in diploids (Pir et al., 2012), so they are clearly limiting in the larger diploids. Mpt5 also binds several of the same cell wall mRNAs that Ssd1 binds (Lapointe et al., 2017), including four that are underrepresented in diploids (Tos1, Uth1, Cis3, and Bgl2). These findings support the possibility that, in diploids, which are larger and relatively limited for cell wall proteins, Ssd1 is critical for the cell wall remodeling required to survive the transition to quiescence. In haploids, where many cell wall proteins are in great excess, Ssd1 function is less critical and/or Mpt5 can compensate for its loss.
After 7 d of growth, ssd1-2 mpt5 haploids and ssd1-2 diploids are largely inviable and they have very low levels of trehalose (Table 2; Li et al., 2013). The addition of exogenous trehalose restores their viability and enables them to enter a long-lived quiescence state. It is possible that trehalose levels are low because these cells are dying and the rescue by trehalose is due to its role as a stress protectant. Trehalose protects cells by binding to the inner and outer layers of the plasma membrane (Eleutherio et al., 2015). Secreted trehalose protects yeast from dehydration and reactive oxygen species accumulation (Eleutherio et al., 1993; da Costa Morato Nery et al., 2008). Recently it has been shown that intracellular trehalose plays a direct role in preventing desiccation-induced protein aggregation in budding yeast (Kim et al., 2018). P bodies (Brengues et al., 2005) and other protein aggregates are induced by glucose depletion and there are several examples in which they promote cellular adaptation to environmental change (Liu et al., 2012; Holmes et al., 2013; Simpson-Lavy et al., 2017; Kim et al., 2018). The lethality we observe could be due to toxic aggregations and could be rescued by the disaggregation properties of trehalose.
It is also possible that trehalose accumulation is promoted by Ssd1 and Mpt5 directly. Regulation of Ace2 and morphogenesis (RAM) signaling and its downstream kinase Cbk1 regulate cell wall synthesis and remodeling during the cell cycle (Weiss, 2012). It is essential that Cbk1 kinase phosphorylate Ssd1 and inhibit its translational repression of cell wall mRNAs during periods of cell wall expansion (Figure 7). This is an ancient pathway, remarkably conserved in Schizosaccharomyces pombe (Nunez et al., 2016). Cbk1 may also bind (Gogl et al., 2015) and inhibit (Bourens et al., 2009) Mpt5. Cbk1 activity is inhibited by Lre1 during the cell cycle and in response to glucose limitation (Versele and Thevelein, 2001; Mancini Lombardi et al., 2013). Lre1 overproduction or loss of Cbk1, both conditions that increase translational repression by Ssd1, increase trehalose levels, and loss of Lre1 activity, which reduces Ssd1 activity, also reduces trehalose levels (Versele and Thevelein, 2001). These effects do not involve obvious changes in the mRNA levels of trehalose metabolism genes or depend on their known regulators (Versele and Thevelein, 2001). These findings raise the possibility that there is a novel form of trehalose regulation during the transition to quiescence that involves Lre1-dependent inactivation of Cbk1. That novel regulation could be translational repression of mRNAs responsible for trehalose utilization or secretion by Ssd1. However, these Lre1-mediated effects on trehalose levels were observed in W303 ssd1-2 haploids (Versele and Thevelein, 2001). So, if there is such a pathway, Mpt5 or something else would have to substitute for Ssd1. In addition, no obvious trehalose-associated mRNAs have been identified that bind Ssd1 (Hogan et al., 2008; Jansen et al., 2009). However, Lre1 mRNA is an Ssd1 target. It is bound and destabilized by Ssd1 (Hogan et al., 2008; Jansen et al., 2009), so it is both an upstream activator of Ssd1 and a downstream target of Ssd1-mediated repression. This negative feedback, eliminating Lre1 activity, may be important to restore Cbk1 activity and enable Cbk1 to perform its other functions, including activation of Rlm1 (Martin-Yken et al., 2003; Kuravi et al., 2011), which initiates most of the transcriptional program of CWI signaling (Figure 7; Watanabe et al., 1995; Levin, 2011).
FIGURE 7:
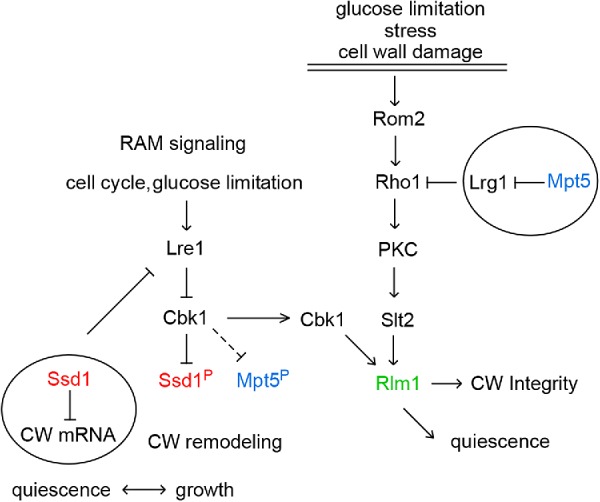
Model for how the RAM (regulation of Ace2 and morphogenesis) and the CWI (cell wall integrity) pathways regulate the transition between growth and quiescence. In response to glucose limitation, Lre1 inhibits the conserved NDR/LATS kinase Cbk1 in the RAM pathway (left) and prevents it from phosphorylating and inhibiting Ssd1. This enables Ssd1 to sequester and inhibit cell wall (CW) mRNAs in P bodies (depicted by circles) and prevent budding and cell wall expansion. Ssd1 and Mpt5 may play overlapping roles in this process. Mpt5 may also be a Cbk1 target. In addition to CW mRNAs, Ssd1 also binds and represses Lre1 mRNA. This negative feedback releases Cbk1 to activate Rlm1. The CWI pathway (right) is activated by stress, nutrient limitation, and cell wall damage. This involves Mpt5-mediated destabilization of the LRG1 mRNA. Lrg1 is a Rho1 GTPase activating protein (GAP) that must be neutralized for full CWI pathway activation. The downstream Slt2 kinase and Cbk1 are both required to activate the Rlm1 transcription factor. Rlm1 activation initiates the transcriptional program that protects cells from stress and promotes the entry, maintenance, and recovery from quiescence.
The RAM and CWI signaling pathways have compensatory roles in cell survival. CWI is constitutively activated in the absence of Ssd1 (Arias et al., 2011). This reinforces the importance of Ssd1 in cell wall remodeling and the role of the CWI pathway in protecting against cell wall defects. Hyperactivation of CWI rescues the stress sensitivity of mpt5 ssd1-2 cells (Stewart et al., 2007), and overproduction of Ssd1 rescues CWI pathway defects (Costigan et al., 1992; Lee et al., 1993). We have deleted Rlm1, the predominant transcription factor activated by CWI signaling, and shown that it is required for the viability of ssd1-2 cells during the transition to quiescence. In addition, Rlm1 is required for producing the dense cells that are characteristic of the transition to quiescence, and for the ability of these dense cells to maintain this protective nondividing state and reenter the cell cycle when nutrients are restored.
Taken together, our data indicate that both the RAM and CWI pathways are important during the transition from growth to quiescence. Cbk1 plays a role in both pathways (Figure 7). In the RAM pathway, Cbk1 must be inhibited to allow Ssd1 to bind and translationally repress cell wall mRNAs (Jansen et al., 2009). In the CWI pathway, Cbk1 is required for the activation of Rlm1 (Martin-Yken et al., 2003; Kuravi et al., 2011; Mancini Lombardi et al., 2013). We speculate that these may be sequential steps. When glucose is limiting, inhibition of Cbk1 by Lre1 enables Ssd1 to repress cell wall (CW) mRNAs and prevent budding and cell wall expansion. Subsequent sequestration of Lre1 mRNA by Ssd1 would restore Cbk1 activity and enable it to activate the Rlm1-dependent transcriptional program that further promotes the transition to quiescence. Sequential targeting of Cbk1 first to Ssd1 and then to Rlm1 would be consistent with the observed delay in Rlm1 activation (Kuravi et al., 2011). Maximum Rlm1 activity occurs 60 min after heat shock and 120 min after caffeine treatment.
These data reveal the complex and polymorphic nature of the budding yeast response to the transition from growth to quiescence. Many polymorphisms that influence sporulation have been identified, but those that promote cellular quiescence are much less well understood. We have shown that Ssd1 is sufficient to shift W303 diploids from predominantly sporulating, to predominantly entering cellular quiescence. However, the same SSD1 allele in other strains does not confer the same fate shift. We have also demonstrated the importance of Mpt5, the CWI pathway, and trehalose in maintaining viability during this transition, especially when Ssd1 is defective. Understanding the way cells enter, maintain, and reverse the quiescent state is key to understanding growth control. These processes are likely to be conserved, just as the fundamentals of the cell duplication process are conserved. Budding yeast is an ideal model for such studies because we can quantify and monitor these processes in a natural setting. We can also take advantage of the natural variation in these processes to identify key regulators that are fungi-specific. Most anti-fungal drugs target growing cells and there is considerable evidence that cells can resist these agents by persisting in a quiescent state (Bojsen et al., 2017). Targeted inactivation of fungi-specific gene products that promote or maintain quiescence could lead to a new class of anti-fungal treatments.
MATERIALs AND METHODS
Strains
Strains are all prototrophic wild and laboratory strains cultured in rich medium (YEPD [yeast extract 10 g/l, Bactopeptone 20 g/l, adenine hemisulfate 55 mg/l, dextrose 20 g/l]). BY6500 and BY6501 are prototrophic W303a and α haploids (Li et al., 2009) and the homozygous diploid is BY6946. These strains are rad5 can1-100 and carry a C-terminal truncation allele of SSD1, referred to historically as ssd1-d2. We will refer to this allele as ssd1-2. The prototrophic W303 MATa (BY6641) and MATα (BY6564) are isogenic except they carry the full-length SSD1 allele that is also found in S288c and RM11 (Li et al., 2009). This allele is historically referred to as SSD1-V, but we will refer to it as SSD1. These haploids were crossed to produce the homozygous diploid (BY6962). The RM11 MATα haploid and diploid are ho AMN1 versions of the wild yeast (BY6941 and BY6944, respectively). SK1 MATa haploid and diploid strains are ho::hisG prototrophs (BY7018 and BY7043). The S288c MATa haploid and diploid are BY6942 and BY6945. Prototrophic diploids were isolated as zygotes (Amberg et al., 2006) and confirmed to sporulate and to have the size and DNA content of diploids on the Coulter counter and flow cytometer, respectively. NCYC strains are from the National Collection of Yeast Cultures. The NCYC 3315 diploid heterozygous for ime1 (BY8014) was made by replacing one copy of IME1 with the hygromycin B resistance gene hphMX4 from pAG32 (Goldstein and McCusker, 1999). The ime1/ime1 homozygote (BY8029) was made by replacing the other IME1 copy with a nourseothricin resistance gene natMX3 from pAG35 (Goldstein and McCusker, 1999). The mpt5 and rlm1 null mutants were made by replacing the coding sequence with the G418 resistance genes KanMX from pFA6a-KanMX (Longtine et al., 1998) in the W303 ssd1-2 or SSD1 background. Using the same drug marker replacement strategy and the vectors described above, we constructed the nth1, nth2, and nth1nth2 homozygous diploids. Strain numbers and specific markers used are provided in the relevant figure legends. When diploids cannot be selected directly, we mate the strains for at least 4 h, streak for single colonies and pick a dozen large colonies from each mating. These potential diploids are purified, sized on a Beckman Coulter counter Z2, and sporulated and assayed for ploidy by flow cytometry.
Induced and deregulated sporulation assays
The efficiency of induced sporulation was assayed by patching cells onto a YEPD plate and growing them for 4 h at 30°C before transfer to sporulation plates. Sporulation plates roughly follow the SPO recipe in Elrod et al. (2009). They contain 1.5% potassium acetate, 0.1% glucose, and 0.25% yeast extract, supplemented with 10 μg/ml adenine and uracil; 5 μg/ml methionine, arginine, histidine, leucine, lysine, and tryptophan; 2 μg/ml tyrosine; 1 μg/ml proline; and 25 μg/ml phenylalanine from a filter sterilized 40× stock solution. Sporulation plates were incubated for 4 d at 25°C; then a sample was resuspended in water. At least 200 cells, dyads, and/or tetrads were counted in triplicate by phase contrast microscopy. Deregulated sporulation was observed within the dense fraction of cells purified after 7 d of culturing in YEPD and quantified as above.
Quiescence and viability assays
Dense cells were purified through a percoll gradient as previously reported (Allen et al., 2006) using a 25 ml gradient and loading 200 OD600 units of cells. Q-cell yield is calculated as the fraction of cells that sediment to the bottom 9 ml of the gradient. Longevity of purified dense cells was assayed in water as described (Li et al., 2009). Survival was assayed by CFUs and by viability using the FungaLight (Invitrogen L34952) flow cytometer assay. All assays are plotted with error bars and the number of biological replicates is provided in the figure legends.
Innate thermotolerance was assayed by incubation at the designated temperatures for 10 min in a PCR machine. Purified Q cells were resuspended at one OD600 and 50 μl was transferred to a 0.5-ml PCR tube and placed in a prewarmed PCR machine. After 10 min, 0.45 ml cold water was added to each tube and then they were diluted and plated for survivors.
Trehalose assay
The trehalose assay is based on that described by Parrou et al. (1997) and modified by Shi et al. (2010). Cells were grown for 7 d in rich YEPD medium. Five OD600 units of cells were washed and transferred to 0.125 ml 0.25M Na2CO3 in a screw-capped tube and stored at −80°C. Cells were broken, digested with trehalase (T8778; Sigma-Aldrich) to release glucose, and assayed for glucose in a 96-well format as described (Shi et al., 2010).
Acknowledgments
We thank Sarah Zanders for stimulating discussions and yeast strains. Thanks also to Wenying Shou for the RM11 derivatives and S288c. This work was supported by National Institute of Aging Grant no. R21 AG048595 and National Institute of General Medical Sciences Grant no. R01 GM120318-01 to L.L.B.
Abbreviations used:
- CWI
cell wall integrity
- DS
diauxic shift
- NDR/Lats
nuclear Dbf2-related/large tumor suppressor
- Q
quiescent
- RAM
regulation of Ace2 and morphogenesis
- SP
stationary phase
- Spo
sporulation.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E19-04-0190) on May 29, 2019.
REFERENCES
- Allen C, Buttner S, Aragon AD, Thomas JA, Meirelles O, Jaetao JE, Benn D, Ruby SW, Veenhuis M, Madeo F, Werner-Washburne M. (2006). Isolation of quiescent and nonquiescent cells from yeast stationary-phase cultures. J Cell Biol , 89–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvers AL, Fishwick LK, Wood MS, Hu D, Chung HS, Dunn WA, Jr, Aris JP. (2009). Autophagy and amino acid homeostasis are required for chronological longevity in Saccharomyces cerevisiae. Aging Cell , 353–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amberg DC, Burke DJ, Strathern JN. (2006). Picking zygotes. CSH Protoc , pdb.prot4185. [DOI] [PubMed] [Google Scholar]
- Arias P, Diez-Muniz S, Garcia R, Nombela C, Rodriguez-Pena JM, Arroyo J. (2011). Genome-wide survey of yeast mutations leading to activation of the yeast cell integrity MAPK pathway: novel insights into diverse MAPK outcomes. BMC Genomics , 390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Ari G, Zenvirth D, Sherman A, David L, Klutstein M, Lavi U, Hillel J, Simchen G. (2006). Four linked genes participate in controlling sporulation efficiency in budding yeast. PLoS Genet , e195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bojsen R, Regenberg B, Folkesson A. (2017). Persistence and drug tolerance in pathogenic yeast. Curr Genet , 19–22. [DOI] [PubMed] [Google Scholar]
- Bourens M, Panozzo C, Nowacka A, Imbeaud S, Mucchielli MH, Herbert CJ. (2009). Mutations in the Saccharomyces cerevisiae kinase Cbk1p lead to a fertility defect that can be suppressed by the absence of Brr1p or Mpt5p (Puf5p), proteins involved in RNA metabolism. Genetics , 161–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brengues M, Teixeira D, Parker R. (2005). Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science , 486–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burtner CR, Murakami CJ, Kennedy BK, Kaeberlein M. (2009). A molecular mechanism of chronological aging in yeast. Cell Cycle , 1256–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L, Tang Y, Quan Z, Zhang Z, Oliver SG, Zhang N. (2016). Chronological lifespan in yeast is dependent on the accumulation of storage carbohydrates mediated by Yak1, Mck1, and Rim15 kinases. PLoS Genet , e1006458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller J, Parker R. (2005). General translational repression by activators of mRNA decapping. Cell , 875–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costigan C, Gehrung S, Snyder M. (1992). A synthetic lethal screen identifies SLK1, a novel protein kinase homolog implicated in yeast cell morphogenesis and cell growth. Mol Cell Biol , 1162–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubillos FA, Billi E, Zorgo E, Parts L, Fargier P, Omholt S, Blomberg A, Warringer J, Louis EJ, Liti G. (2011). Assessing the complex architecture of polygenic traits in diverged yeast populations. Mol Ecol , 1401–1413. [DOI] [PubMed] [Google Scholar]
- da Costa Morato Nery D, da Silva CG, Mariani D, Fernandes PN, Pereira MD, Panek AD, Eleutherio EC. (2008). The role of trehalose and its transporter in protection against reactive oxygen species. Biochim Biophys Acta , 1408–1411. [DOI] [PubMed] [Google Scholar]
- Davidow LS, Goetsch L, Byers B. (1980). Preferential occurrence of nonsister spores in two-spored asci of Saccharomycescerevisiae: evidence for regulation of spore-wall formation by the spindle pole body. Genetics , 581–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Godoy LM, Olsen JV, Cox J, Nielsen ML, Hubner NC, Frohlich F, Walther TC, Mann M. (2008). Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature , 1251–1254. [DOI] [PubMed] [Google Scholar]
- Deutschbauer AM, Davis RW. (2005). Quantitative trait loci mapped to single-nucleotide resolution in yeast. Nat Genet , 1333–1340. [DOI] [PubMed] [Google Scholar]
- De Virgilio C. (2012). The essence of yeast quiescence. FEMS Microbiol Rev , 306–339. [DOI] [PubMed] [Google Scholar]
- Duy DL, Suda Y, Irie K. (2017). Cytoplasmic deadenylase Ccr4 is required for translational repression of LRG1 mRNA in the stationary phase. PLoS One , e0172476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbein AD, Pan YT, Pastuszak I, Carroll D. (2003). New insights on trehalose: a multifunctional molecule. Glycobiology , 17R–27R. [DOI] [PubMed] [Google Scholar]
- Eleutherio EC, Araujo PS, Panek AD. (1993). Role of the trehalose carrier in dehydration resistance of Saccharomyces cerevisiae. Biochim Biophys Acta , 263–266. [DOI] [PubMed] [Google Scholar]
- Eleutherio E, Panek A, De Mesquita JF, Trevisol E, Magalhaes R. (2015). Revisiting yeast trehalose metabolism. Curr Genet , 263–274. [DOI] [PubMed] [Google Scholar]
- Elrod SL, Chen SM, Schwartz K, Shuster EO. (2009). Optimizing sporulation conditions for different Saccharomyces cerevisiae strain backgrounds. Methods Mol Biol , 21–26. [DOI] [PubMed] [Google Scholar]
- Esposito MS, Esposito RE. (1969). The genetic control of sporulation in Saccharomyces. I. The isolation of temperature-sensitive sporulation-deficient mutants. Genetics , 79–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito MS, Esposito RE, Arnaud M, Halvorson HO. (1969). Acetate utilization and macromolecular synthesis during sporulation of yeast. J Bacteriol , 180–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favre C, Aguilar PS, Carrillo MC. (2008). Oxidative stress and chronological aging in glycogen-phosphorylase-deleted yeast. Free Radic Biol Med , 1446–1456. [DOI] [PubMed] [Google Scholar]
- Francois J, Parrou JL. (2001). Reserve carbohydrates metabolism in the yeast Saccharomyces cerevisiae. FEMS Microbiol Rev , 125–145. [DOI] [PubMed] [Google Scholar]
- Gerber AP, Herschlag D, Brown PO. (2004). Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PLoS Biol , E79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogl G, Schneider KD, Yeh BJ, Alam N, Nguyen Ba AN, Moses AM, Hetenyi C, Remenyi A, Weiss EL. (2015). The structure of an NDR/LATS kinase-mob complex reveals a novel kinase-coactivator system and substrate docking mechanism. PLoS Biol , e1002146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein AL, McCusker JH. (1999). Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast , 1541–1553. [DOI] [PubMed] [Google Scholar]
- Hogan DJ, Riordan DP, Gerber AP, Herschlag D, Brown PO. (2008). Diverse RNA-binding proteins interact with functionally related sets of RNAs, suggesting an extensive regulatory system. PLoS Biol , e255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes LE, Campbell SG, De Long SK, Sachs AB, Ashe MP. (2004). Loss of translational control in yeast compromised for the major mRNA decay pathway. Mol Cell Biol , 2998–3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes DL, Lancaster AK, Lindquist S, Halfmann R. (2013). Heritable remodeling of yeast multicellularity by an environmentally responsive prion. Cell , 153–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Xia B, Postnikoff SD, Shen ZJ, Tomoiaga AS, Harkness TA, Seol JH, Li W, Chen K, Tyler JK. (2018). Ssd1 and Gcn2 suppress global translation efficiency in replicatively aged yeast while their activation extends lifespan. Elife , e35551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M, Hull CM. (2017). Sporulation: how to survive on planet Earth (and beyond). Curr Genet , 831–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen JM, Wanless AG, Seidel CW, Weiss EL. (2009). Cbk1 regulation of the RNA-binding protein Ssd1 integrates cell fate with translational control. Curr Biol , 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston GC, Pringle JR, Hartwell LH. (1977). Coordination of growth with cell division in the yeast Saccharomyces cerevisiae. Exp Cell Res , 79–98. [DOI] [PubMed] [Google Scholar]
- Jorgensen P, Nelson B, Robinson MD, Chen Y, Andrews B, Tyers M, Boone C. (2002). High-resolution genetic mapping with ordered arrays of Saccharomyces cerevisiae deletion mutants. Genetics , 1091–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, Guarente L. (2002). Saccharomyces cerevisiae MPT5 and SSD1 function in parallel pathways to promote cell wall integrity. Genetics , 83–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane SM, Roth R. (1974). Carbohydrate metabolism during ascospore development in yeast. J Bacteriol , 8–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassir Y, Granot D, Simchen G. (1988). IME1, a positive regulator gene of meiosis in S. cerevisiae. Cell , 853–862. [DOI] [PubMed] [Google Scholar]
- Kikuchi Y, Oka Y, Kobayashi M, Uesono Y, Toh-e A, Kikuchi A. (1994). A new yeast gene, HTR1, required for growth at high temperature, is needed for recovery from mating pheromone-induced G1 arrest. Mol Gen Genet , 107–116. [DOI] [PubMed] [Google Scholar]
- Kim SX, Camdere G, Hu X, Koshland D, Tapia H. (2018). Synergy between the small intrinsically disordered protein Hsp12 and trehalose sustain viability after severe desiccation. Elife , e38337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuravi VK, Kurischko C, Puri M, Luca FC. (2011). Cbk1 kinase and Bck2 control MAP kinase activation and inactivation during heat shock. Mol Biol Cell , 4892–4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurischko C, Broach JR. (2017). Phosphorylation and nuclear transit modulate the balance between normal function and terminal aggregation of the yeast RNA-binding protein Ssd1. Mol Biol Cell , 3057–3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurischko C, Kim HK, Kuravi VK, Pratzka J, Luca FC. (2011). The yeast Cbk1 kinase regulates mRNA localization via the mRNA-binding protein Ssd1. J Cell Biol , 583–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurischko C, Weiss G, Ottey M, Luca FC. (2005). A role for the Saccharomyces cerevisiae regulation of Ace2 and polarized morphogenesis signaling network in cell integrity. Genetics , 443–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyryakov P, Beach A, Richard VR, Burstein MT, Leonov A, Levy S, Titorenko VI. (2012). Caloric restriction extends yeast chronological lifespan by altering a pattern of age-related changes in trehalose concentration. Front Physiol , 256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai CY, Jaruga E, Borghouts C, Jazwinski SM. (2002). A mutation in the ATP2 gene abrogates the age asymmetry between mother and daughter cells of the yeast Saccharomyces cerevisiae. Genetics , 73–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry CR, Townsend JP, Hartl DL, Cavalieri D. (2006). Ecological and evolutionary genomics of Saccharomyces cerevisiae. Mol Ecol , 575–591. [DOI] [PubMed] [Google Scholar]
- Lapointe CP, Preston MA, Wilinski D, Saunders HAJ, Campbell ZT, Wickens M. (2017). Architecture and dynamics of overlapped RNA regulatory networks. RNA , 1636–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HY, Cheng KY, Chao JC, Leu JY. (2016). Differentiated cytoplasmic granule formation in quiescent and non-quiescent cells upon chronological aging. Microb Cell , 109–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KS, Irie K, Gotoh Y, Watanabe Y, Araki H, Nishida E, Matsumoto K, Levin DE. (1993). A yeast mitogen-activated protein kinase homolog (Mpk1p) mediates signalling by protein kinase C. Mol Cell Biol , 3067–3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin DE. (2011). Regulation of cell wall biogenesis in Saccharomyces cerevisiae: the cell wall integrity signaling pathway. Genetics , 1145–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Lu Y, Qin LX, Bar-Joseph Z, Werner-Washburne M, Breeden LL. (2009). Budding yeast SSD1-V regulates transcript levels of many longevity genes and extends chronological life span in purified quiescent cells. Mol Biol Cell , 3851–3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Miles S, Breeden LL. (2015). A genetic screen for Saccharomyces cerevisiae mutants that fail to enter quiescence. G3 (Bethesda) , 1783–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Miles S, Melville Z, Prasad A, Bradley G, Breeden LL. (2013). Key events during the transition from rapid growth to quiescence in budding yeast require posttranscriptional regulators. Mol Biol Cell , 3697–3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillie SH, Pringle JR. (1980). Reserve carbohydrate metabolism in Saccharomyces cerevisiae: responses to nutrient limitation. J Bacteriol , 1384–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu IC, Chiu SW, Lee HY, Leu JY. (2012). The histone deacetylase Hos2 forms an Hsp42-dependent cytoplasmic granule in quiescent yeast cells. Mol Biol Cell , 1231–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. (1998). Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cervisiae. Yeast , 953–961. [DOI] [PubMed] [Google Scholar]
- Lopez-Garcia B, Gandia M, Munoz A, Carmona L, Marcos JF. (2010). A genomic approach highlights common and diverse effects and determinants of susceptibility on the yeast Saccharomyces cerevisiae exposed to distinct antimicrobial peptides. BMC Microbiol , 289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancini Lombardi I, Palani S, Meitinger F, Darieva Z, Hofmann A, Sharrocks AD, Pereira G. (2013). Lre1 directly inhibits the NDR/Lats kinase Cbk1 at the cell division site in a phosphorylation-dependent manner. Curr Biol , 1736–1745. [DOI] [PubMed] [Google Scholar]
- Martin-Yken H, Dagkessamanskaia A, Basmaji F, Lagorce A, Francois J. (2003). The interaction of Slt2 MAP kinase with Knr4 is necessary for signalling through the cell wall integrity pathway in Saccharomyces cerevisiae. Mol Microbiol , 23–35. [DOI] [PubMed] [Google Scholar]
- McFaline-Figueroa JR, Vevea J, Swayne TC, Zhou C, Liu C, Leung G, Boldogh IR, Pon LA. (2011). Mitochondrial quality control during inheritance is associated with lifespan and mother-daughter age asymmetry in budding yeast. Aging Cell , 885–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnight JN, Boerma JW, Breeden LL, Tsukiyama T. (2015). Global promoter targeting of a conserved lysine deacetylase for transcriptional shutoff during quiescence entry. Mol Cell , 732–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiman AM. (2011). Sporulation in the budding yeast Saccharomyces cerevisiae. Genetics , 737–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunez I, Rodriguez Pino M, Wiley DJ, Das ME, Chen C, Goshima T, Kume K, Hirata D, Toda T, Verde F. (2016). Spatial control of translation repression and polarized growth by conserved NDR kinase Orb6 and RNA-binding protein Sts5. Elife , e14216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nwaka S, Kopp M, Holzer H. (1995). Expression and function of the trehalase genes NTH1 and YBR0106 in Saccharomyces cerevisiae. J Biol Chem , 10193–10198. [DOI] [PubMed] [Google Scholar]
- Ocampo A, Liu J, Schroeder EA, Shadel GS, Barrientos A. (2012). Mitochondrial respiratory thresholds regulate yeast chronological life span and its extension by caloric restriction. Cell Metab , 55–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrou JL, Teste MA, Francois J. (1997). Effects of various types of stress on the metabolism of reserve carbohydrates in Saccharomyces cerevisiae: genetic evidence for a stress-induced recycling of glycogen and trehalose. Microbiology (Pt 6), 1891–1900. [DOI] [PubMed] [Google Scholar]
- Pinon R. (1977). Effects of ammonium ions on sporulation of Saccharomyces cerevisiae. Exp Cell Res , 367–378. [DOI] [PubMed] [Google Scholar]
- Pir P, Gutteridge A, Wu J, Rash B, Kell DB, Zhang N, Oliver SG. (2012). The genetic control of growth rate: a systems biology study in yeast. BMC Syst Biol , 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racki WJ, Becam AM, Nasr F, Herbert CJ. (2000). Cbk1p, a protein similar to the human myotonic dystrophy kinase, is essential for normal morphogenesis in Saccharomyces cerevisiae. EMBO J , 4524–4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renicke C, Allmann AK, Lutz AP, Heimerl T, Taxis C. (2017). The mitotic exit network regulates spindle pole body selection during sporulation of Saccharomyces cerevisiae. Genetics , 919–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagee S, Sherman A, Shenhar G, Robzyk K, Ben-Doy N, Simchen G, Kassir Y. (1998). Multiple and distinct activation and repression sequences mediate the regulated transcription of IME1, a transcriptional activator of meiosis-specific genes in Saccharomyces cerevisiae. Mol Cell Biol , 1985–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samokhvalov V, Ignatov V, Kondrashova M. (2004). Reserve carbohydrates maintain the viability of Saccharomyces cerevisiae cells during chronological aging. Mech Ageing Dev , 229–235. [DOI] [PubMed] [Google Scholar]
- Shi L, Sutter BM, Ye X, Tu BP. (2010). Trehalose is a key determinant of the quiescent metabolic state that fuels cell cycle progression upon return to growth. Mol Biol Cell , 1982–1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimoi H, Kitagaki H, Ohmori H, Iimura Y, Ito K. (1998). Sed1p is a major cell wall protein of Saccharomyces cerevisiae in the stationary phase and is involved in lytic enzyme resistance. J Bacteriol , 3381–3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sillje HH, Paalman JW, ter Schure EG, Olsthoorn SQ, Verkleij AJ, Boonstra J, Verrips CT. (1999). Function of trehalose and glycogen in cell cycle progression and cell viability in Saccharomyces cerevisiae. J Bacteriol , 396–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson-Lavy K, Xu T, Johnston M, Kupiec M. (2017). The Std1 activator of the Snf1/AMPK kinase controls glucose response in yeast by a regulated protein aggregation. Mol Cell , 1120–1133.e1123. [DOI] [PubMed] [Google Scholar]
- Stewart MS, Krause SA, McGhie J, Gray JV. (2007). Mpt5p, a stress tolerance- and lifespan-promoting PUF protein in Saccharomyces cerevisiae, acts upstream of the cell wall integrity pathway. Eukaryot Cell , 262–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strathern JN, Hicks J, Herskowitz I. (1981). Control of cell type in yeast by the mating type locus. The α1-α2 hypothesis. J Mol Biol , 357–372. [DOI] [PubMed] [Google Scholar]
- Tapia H, Young L, Fox D, Bertozzi CR, Koshland D. (2015). Increasing intracellular trehalose is sufficient to confer desiccation tolerance to Saccharomyces cerevisiae. Proc Natl Acad Sci USA , 6122–6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira D, Sheth U, Valencia-Sanchez MA, Brengues M, Parker R. (2005). Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA , 371–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomar P, Bhatia A, Ramdas S, Diao L, Bhanot G, Sinha H. (2013). Sporulation genes associated with sporulation efficiency in natural isolates of yeast. PLoS One , e69765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uesono Y, Toh-e A, Kikuchi Y. (1997). Ssd1p of Saccharomyces cerevisiae associates with RNA. J Biol Chem , 16103–16109. [DOI] [PubMed] [Google Scholar]
- Versele M, Thevelein JM. (2001). Lre1 affects chitinase expression, trehalose accumulation and heat resistance through inhibition of the Cbk1 protein kinase in Saccharomyces cerevisiae. Mol Microbiol , 1311–1326. [DOI] [PubMed] [Google Scholar]
- Wanless AG, Lin Y, Weiss EL. (2014). Cell morphogenesis proteins are translationally controlled through UTRs by the Ndr/LATS target Ssd1. PLoS One , e85212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Irie K, Matsumoto K. (1995). Yeast RLM1 encodes a serum response factor-like protein that may function downstream of the Mpk1 (Slt2) mitogen-activated protein kinase pathway. Mol Cell Biol , 5740–5749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidberg H, Moretto F, Spedale G, Amon A, van Werven FJ. (2016). Nutrient control of yeast gametogenesis is mediated by TORC1, PKA and energy availability. PLoS Genet , e1006075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss EL. (2012). Mitotic exit and separation of mother and daughter cells. Genetics , 1165–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss EL, Kurischko C, Zhang C, Shokat K, Drubin DG, Luca FC. (2002). The Saccharomyces cerevisiae Mob2p–Cbk1p kinase complex promotes polarized growth and acts with the mitotic exit network to facilitate daughter cell–specific localization of Ace2p transcription factor. J Cell Biol , 885–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young CP, Hillyer C, Hokamp K, Fitzpatrick DJ, Konstantinov NK, Welty JS, Ness SA, Werner-Washburne M, Fleming AB, Osley MA. (2017). Distinct histone methylation and transcription profiles are established during the development of cellular quiescence in yeast. BMC Genomics , 107. [DOI] [PMC free article] [PubMed] [Google Scholar]