Abstract
Objective
To explore the effect of dysregulation of epigenetic regulator EZH1 and EZH2 on the proliferation in MCL and the underlying mechanisms.
Methods
In this study, we elucidated the role of EZH1 and EZH2 overexpression by immunohistochemistry and correlated them to clinical outcome in 41 MCL patients. Quantitative real-time PCR and Western blot were applied to confirm the level of EZH1 and EZH2 in well-characterized MCL cell lines which were compared to those of naïve B cells. Then we manipulated the expression of EZH1 and EZH2 in MCL cells using CRISPR/Cas9 system to directly investigate their functional roles in MCL. We also evaluated the effect of two small molecule selective inhibitors, EPZ005687 and UNC1999, on MCL cell proliferation, cell cycle distribution and apoptosis in vitro. Finally, we performed RNA-sequencing (RNA-Seq) and Chromatin immunoprecipitation (ChIP) assay to further gain insight into the underlying molecular mechanisms.
Results
We found that EZH2 protein is overexpressed in approximately half of this cohort of MCL cases. More importantly, the overexpression of EZH2 is associated with poor OS in the patients. Nevertheless, simple EZH2 depletion in vitro has little impact on the viability of MCL cells, predominantly because of the consequent up-regulation of EZH1. Consistently, UNC1999, a dual EZH1/2 inhibitor, unlike the EZH2 selective inhibitor EPZ005687, exerts a potent inhibitory effect on MCL cells. Furthermore, we discover CDKN1C and TP53INP1 as the two important cell cycle regulators, the expression of which are repressed by EZH1/2 mediated epigenetic regulation and are restored by EZH1/2 dual inhibition.
Conclusions
Our study suggests that EZH2 participates in the pathogenesis of MCL which may serve as a potential biomarker for prognosis prediction. The dual inhibition of EZH1/2 is a promising therapeutic strategy for MCL.
Keywords: Mantle cell lymphoma, EZH1, EZH2, CRISPR/Cas9
Introduction
Mantle cell lymphoma (MCL) is a distinct subtype of B-cell non-Hodgkin’s lymphoma (NHL), which originates from naïve B cells1 and is different from other NHL in pathogenesis, histopathological features and clinical characteristics. The hallmark of MCL is chromosomal translocation t (11; 14) (q13; q32) which leads to aberrant cyclin D1 overexpression shown in vast majority of cases2. However, the overexpression of cyclin D1 alone was insufficient for the MCL lymphomagenesis in transgenic mice and simple knocking-down of cyclin D1 showed little effect on tumor cell survival and proliferation3-5. These data suggest that additional cyclin-independent mechanisms are involved in controlling the proliferation of MCL tumor cells, among which epigenetic dysregulation has drawn attention with therapeutic implications6.
Current treatment for MCL patients achieves a median overall survival (OS) of 3-6 years and novel agents are urgently needed7. Among the newly developed therapeutics, selected epigenetic agents, such as Polycomb Repressive Complex 2 (PRC2) inhibitor DZNep, have been demonstrated to be effective in suppressing MCL cells8. As one of the core subunits of PRC2, Enhancer of Zeste Homologous 2 (EZH2) is essential for B cell development, which makes it a promising therapeutic target. Normally, EZH2 represses Blimp1 and Irf4 expression through its histone lysine methyltransferase activity during the formation of germinal centers9. EZH2 overexpression and somatic mutations have been reported in hematopoietic malignancies10. Gain-of-function mutations in the SET domain (Y641) of EZH2 occur in 21.7% of germinal center B cell (GCB)-type diffused large B cell lymphoma (DLBCL) and 7.2% of follicular lymphoma (FL) patients11,12, but have not been demonstrated in MCL patients. Overexpression of mutated EZH2 induces germinal center hyperplasia and then drives it to become more susceptible to transformation to GCB-type DLBCL13. EZH1 is another mammalian homolog of Drosophila Ez which forms an additional non-canonical PRC2 complex (PRC2-EZH1) together with Suz12, Eed, and RbAP46/4814. Overexpression of EZH1 has been reported in 48.8% of grade 1 FL and almost 100% of maginal zone lymphoma of mucosa-associated lymphoid tissue15. Unlike EZH2, no somatic mutations have been demonstrated in EZH1 in hematopoietic malignancies16.
So far, the significance of EZH1 and EZH2 in MCL remains elusive. In this study, we aimed to examine the expression and functional role of EZH1 and EZH2 in MCL and their clinical significance.
Materials and methods
Clinical samples
A total of 41 confirmed primary MCL patients with formalin fixed paraffin-embedded (FFPE) tissues were included in this study. It consisted of 25 cases from the University of Nebraska Medical Center (Omaha, NE, USA) and 16 from the Tianjin Medical University Cancer Institute and Hospital (TMUCIH, Tianjin, China). Twenty lymph node specimens with reactive hyperplasia (RH) from TMUCIH were used as normal controls. All archived FFPE tissues were reviewed and classified by experienced hematopathologists according to the WHO Classification of Tumors of Haematopoietic and Lymphoid Tissue (Fourth Edition, 2017). This study was conducted under general consent from all patients and in accordance with the Declaration of Helsinki.
Cell culture, treatment, and reagents
Four MCL cell lines Granta-519, JVM-2, Mino, and Z138 were purchased from the ATCC (Manassas, VA, USA). Granta-519 was cultured with Dulbecco’s modified Eagle’s medium (DMEM) (Gibco, USA) and the rests were cultured with RPMI-1640 medium (Life Technologies, California, USA). Both media were supplemented with 10% fetal bovine serum (FBS) (Gibco, USA) and 1% penicillin/streptomycin. HEK293T cells were routinely maintained in the lab and grown in DMEM supplemented with 10% FBS. All cells were incubated at 37°C in a humidified atmosphere containing 5% CO2. Cells were plated at a density of 0.1 × 106 cells/flask in T25 flask for cell viability assay, at 0.5 × 106 cells/flask for apoptosis assays and cell cycle analysis, and at 1 × 106 cells/flask for Western blot. EPZ005687 and UNC1999 were purchased from Selleckchem (Houston, TX, USA). The compound was dissolved in anhydrous dimethyl sulfoxide (DMSO) to a stock solution of 10 mM, and then further diluted to the desired concentration for in vitro experiments.
Immunohistochemistry (IHC)
IHC was performed as previously described17. Briefly, 4 μm tissue sections of paraffin-embedded specimens were dewaxed in xylene and rehydrated in graded alcohol, peroxidase was quenched with 3% H2O2. The slides were then blocked with 2% bovine serum albumin (BSA) for 30 min and incubated with the primary rabbit anti-EZH2 polyclonal antibody (at 1:100 dilution, Invitrogen, Carlsbad, CA, USA) or rabbit anti-EZH1 polyclonal antibody (at 1:100 dilution, Invitrogen, Carlsbad, CA, USA) overnight at 4°C in a humid chamber. After washing thrice with phosphate-buffered saline (PBS), the slides were incubated with horseradish peroxidase-conjugated goat anti-rabbit IgG (Zhongshan, Beijing, China) for 1 h, followed by reaction with diaminobenzidine and counterstaining with Mayer’s hematoxylin. The negative control was performed by the omission of the primary antibody and substituting it with non-specific rabbit IgG. Antibody dilution and developing time were kept constant for all tissues.
Evaluation of immunostaining for EZH2 and EZH1 proteins
EZH2 and EZH1 expression were semi-quantitatively assessed based on the proportion and the staining intensity of the stained tumor nuclei cells as follows: staining proportion was classified as 1–4 (1 = 0%–25% positive cells, 2 = 26%–50% positive cells, 3 = 51%–75% and 4 ≥ 76%) and the intensity grade was scored as 0–3 (0 = negative staining, 1 = weak positive staining, 2 = moderate positive and 3 = strong positive). The final immunohistochemical score (IHS; ranging from 0 to 12) was obtained by multiplying the proportion and the intensity scores. The cutoff value for high and low expression was determined on the basis of a heterogeneity value measured through log-rank statistical analysis with respect to overall survival (OS). Only when the IHS ≥ 9 was defined as high expression. Two experienced pathologists independently evaluated both EZH1 and EZH2 staining and the consensus score is derived from the average of two independent IHS.
Western blot analysis
Cells were harvested and washed twice with ice-cold PBS and then centrifuged at 1000 rpm for 5 min, and lysed in 50–100 μl of ice-cold RIPA lysis buffer supplemented with 1 mM protease/phosphatase inhibitor cocktail. 10–40 μg of total protein (depending on different proteins) was electrophoresed by sodiumdodecyl sulfate-polyacrylamide (SDS-PAGE) gel. Separated proteins were then transferred to polyvinylidene difluoride (PVDF) membranes (Millipore, Billerica, MA, USA), which were then blocked in 5% BSA in Tris-buffer saline-Tween (TBST) for 2 h. Blots were probed by incubation with the primary antibodies at 4°C overnight. Primary antibodies employed in the study include EZH2 (07-689, at 1:1000 dilution, Millipore, Billerica, MA, USA), EZH1 (PA5-28710, at 1:100 dilution, Invitrogen, Carlsbad, CA, USA), H3K27me3 (07-449, at 1:2000 dilution, Millipore, Billerica, MA, USA), and Total H3 (04-928, at 1:5000 dilution, Millipore, Billerica, MA, USA). Blots were then washed and incubated with HRP-conjugated secondary antibody (Millipore, Billerica, MA, USA). Proteins were detected by enhanced chemiluminescence Plus reagents (Amersham Pharmacia, Piscataway, NJ) and visualized using the Chemiluminescence Imaging System. Signal quantification was obtained using Quantity One software (Bio-Rad Laboratories, USA) and normalized to Total H3. For each study, at least three independent experiments were performed.
Real-time quantitative reverse transcription-PCR
Total RNA was isolated from cultured cell lines using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) and subjected to reverse transcription using a cDNA synthesis kit (Vazyme, Piscataway, NJ, USA), with 1 μg of total RNA used as the template. Quantitative real-time PCR was performed using SYBR Green PCR Master Mix (Roche Diagnostics, Mannheim, Germany) with the 7500 Real-Time PCR System (Applied Biosystems, Inc. USA). The primers used for specific cDNA amplification were summarized in Supplementary Table S1. Before the experiments started, the standard curve was validated for calculations of template copy numbers in the indicated samples and melting curve analyses were employed to verify specific single product amplification. The relative quantity of the target mRNAs was estimated by the ∆∆CT value after normalizing to the expression level of β-actin. Experiments were repeated in triplicate independently.
S1.
The primer sequences list for qPCR and ChIP-qPCR
| Target | Sequences | |
| * The overhangs for ligation into the nickase sites were contained on the top and bottom of strand and in boldface. | ||
| qPCR target | Forward 5’ to 3’ | Reverse 5’ to 3’ |
| EZH2 | TACTTGTGGAGCCGCTGAC | TCTGCCACGTCAGATGGTG |
| EZH1 | AATATGGGAGCAAAGGCTCTGTATGTG | CACGAAGTTTCTTCCACTCTTCATTGAG |
| β-actin | CTGGACTTCGAGCAAGAGAT | GATGTCCACGTCACACTTCA |
| CDKN1C | GCGGTGAGCCAATTTAGAGC | TGCTACATGAACGGTCCCAG |
| TP53INP1 | GACTTCATAGATACTTGCACTGG | TTGTATCAGCCAAGCACTCAA |
| CRISPR target | Sequence of sgRNA | Sequence of Oligo 5’ to 3’* |
| EZH2 | GGTCGCGTCCGACACCCGGT | Oligo1 CACCGGTCGCGTCCGACACCCGGT |
| Oligo2 AAACACCGGGTGTCGGACGCGACC | ||
| EZH1 | TCGACAACTTAAACGGCTTC | Oligo1 CACCGGTCGACAACTTAAACGGCTTC |
| Oligo2 AAACGAAGCCGTTTAAGTTGTCGAC | ||
| ChIP-qPCR target | Forward 5’ to 3’ | Reverse 5’ to 3’ |
| CDKN1C promoter primer a | GCAGATAGCGGCTTCAGACT | GTCAGCTCCACTCTTTCCCC |
| CDKN1C promoter primer b | GTCCCATCCAGTTCACCCC | CTGTGAGCCAGCAGAGAGAC |
| TP53INP1 promoter primer a | GAGAGGTTGTCACCAACGCA | CAGCTCCCGGTTTTTGTTGC |
| TP53INP1 promoter primer b | GTAGAGGCCGCTCAATCACG | CTATAGGCAGGCGGAATCCAG |
Cell viability
Viable cell densities were determined by using the CellTiter 96 One Solution Cell Proliferation Assay (Promega, Madison, WI, USA) according to the manufacturer’s instructions. Briefly, cells were plated at a density of 10, 000/well in 96-well plates and incubated for 24–96 h with increasing concentrations of EPZ005687 or UNC1999. The cells were then incubated with MTS reagent (Sigma, M2128) for 3 h, and analyzed with 490 nm absorbance using microplate reader (Bio-Rad, Laboratories, USA). The data analysis was performed by GraphPad Prism (version 5.01, GraphPad Software, USA). The studies were independently conducted in at least three separate times and each experiment had six replicate wells on one plate.
Flow cytometry analysis
To determine cell cycle distribution, MCL cells were treated with increasing concentrations of EPZ005687 or UNC1999 for 48 h and then fixed by ice-cold 70% ethanol. Fixed cells were incubated overnight at –30°C and then washed with PBS and incubated with RNase and propidium iodide (PI) at room temperate in the dark for more than 30 min. DNA content was measured using a LSD Fortessa (BD Biosciences) and the cell cycle profile was analyzed by ModFit software (version 3.1). To detect apoptosis, FITC-Annexin V Apoptosis Detection Kit (BD Biosciences) was used. Briefly, treated cells were harvested and rinsed with PBS once and then resuspended in 500 μl of 1x Annexin V binding buffer following with addition of Annexin V-FITC and PI. After incubation at room temperate in the dark for more than 30 min, the samples were analyzed by a LSD Fortessa flow cytometer and the data were analyzed using FlowJo software (version 7.6).
Establishment of MCL cell line with down-regulation of EZH2 and EZH1/2
Crispr-Cas9 mediated EZH1/EZH2 knocking down was realized by transducing MCL cells with the LentiCRISPRv2 vector. The single guide RNA (sgRNA) sequences were listed in Supplementary Table S1. Briefly, Z138 cell line was stably transduced with lentivirus packaged with the lentiCRISPRv2 carrying the sgRNA, pMD2G envelope vector and psPAX2 packaging plasmid in HEK293T cells. Infectious lentivirus supernatants were harvested 48 h after transfection and again after 72 h, and filtered through 0.45 mm filter (Millipore, Billerica, MA, USA). Transduced cells were selected by 1 μg/mL puromycin (Sigma) for 2 days. Knocking down efficiency was determined by qRT-PCR and immunoblotting. For the double knocking-down, EZH1 sgRNAs were delivered by a lentiviral vector with Blasticidin resistance as previously described18.
Gene expression analysis using RNA-Sequencing
Total RNA was extracted from UNC1999 treated cells (coupled with DMSO treated cells in both the Z138 and Mino cells). RNA quality was then assessed using an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, Calif). An equal amount of total RNA with a 28S/18S ratio ≥ 1.0 and RNA Integrity Number (RIN) ≥ 7.0 from two individuals in each group (one sample to sample compared and 4 pairs of samples were tested totally) was pooled together to construct cDNA libraries and then whole transcriptome RNA-Sequencing (RNA-Seq) was carried out for differential gene expression analysis through a BGISEQ-500 platform. We selected gene sets that were upregulated or downregulated more than two-fold in both cell lines. Gene ontology analysis was performed using the DAVID web-based software (https://david.ncifcrf.gov/).
Chromatin immunoprecipitation (ChIP) assay
To prepare chromatin from cells, Z138 and Mino cells were harvested after UNC1999 treatment and washed twice with ice-cold PBS. Formaldehyde was added to 1% final concentration and cells were fixed for 10 min at RT before addition of glycine at 0.125 M final concentration and incubated for 5 min. Crosslinked cells were washed with PBS and transferred to a SDS buffer containing protease inhibitors. The cells were then re-suspend in ice-cold IP Buffer (1 volume SDS buffer: 0.5 volume Triton dilution buffer) for sonication. Chromatin was sonicated 30 × 1 min in a bioruptor (Diagenode) using a 30 sec on/off program at high power. Approximate 200-500 base pairs (bp) DNA were generated and confirmed by agarose gel electrophoresis. For Histone-ChIP assays, 100 μg of chromatin was used as the template for the specific IP and diluted at 1:100 for the input. Samples were incubated with 5 μg of specific antibody anti trimethyl-histone H3K27 (07-449, Millipore) or with normal mouse IgG antibody (12-371, Millipore) overnight at 4°C with gentle rotation and then incubated with 40 μl of magnetic Dynabeads Protein G (Invitrogen, NY, USA) on a rotating wheel for 4 h at 4°C to form immune complexes. Each sample was washed twice with low salt buffer (0.1% SDS, Triton-X-100, 2mM EDTA, 20 mM Tris-HCl pH = 8.1, 150 mM NaCl) and with high salt buffer (0.1% SDS, Triton-X-100, 2 mM EDTA, 20 mM Tris-HCl pH = 8.1, 500 mM NaCl). After that, the immune complexes were reversed to free DNA and then purified with DNA purification kit (Qiagen, Hilden, Germany) and re-suspended in 50 μl of pre-heated distilled water. After immunoprecipitation, the DNA quantity of each sample was measured. Two different regions were amplified using the same DNA amount and the primers were summarized in Supplementary Table S1.
Statistical analysis
Data are represented as the mean ± s.e.m. Chi-square test or Fisher’s exact test was used to determine statistical significance. The Kaplan-Meier method was used to estimate the OS distributions, and the log-rank test was performed to compare the survival difference. Person’s rank correlation coefficient was used to evaluate the relationship between the variables. The difference was considered significant when the P-value was < 0.05 and all reported P values were two-sided. These statistical analyses were performed with the Statistical Package for the Social Sciences, version 17.0 (SPSS Inc., Chicago, IL, USA) statistical software.
Results
Different expression of EZH1/2 in MCL cases and cell lines
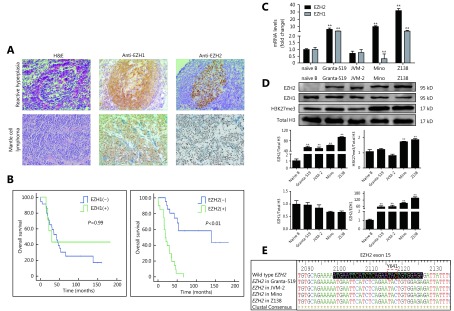
We first studied the EZH1 and EZH2 proteins expression in 41 primary MCL FFPE samples by IHC (Supplementary Table S2 and S3) and compared the results with those in benign lymph nodes with follicular reactive hyperplasia (RH) (n = 20). In the RH setting, EZH1-positive cells were seen within the mantle zone whereas EZH2-positive cells were mainly located in the germinal centers (Figure 1A). In MCL cases, strong nuclear EZH1 staining of tumor cells was seen in 7/41 cases, while strong nuclear EZH2 expression was seen in 20/41 cases (Figure 1A and Supplementary Table S4). Further analysis in this cohort of patients revealed no significant correlation between EZH1 positivity and clinical outcome, whereas EZH2 overexpression was strongly correlated with poorer OS (P < 0.01, Figure 1B). The median survival of those patients with EZH2 overexpression was only 2 years, compared to 9.6 years of those negative cases (Figure 1B). These results suggest that EZH2 is likely to contribute to the pathogenesis of MCL and may serve as a biomarker to predict the clinical course and prognosis. We next examined the mRNA and protein levels of EZH1 and EZH2 in four MCL cell lines, Granta-519, JVM-2, Mino, and Z138. Compared to the naïve B-cells, all four MCL cell lines, except JVM-2, showed at least 9-fold increase in the EZH2 mRNA expression, while Granta-519 and Z138 also showed a high level of EZH1 mRNA (Figure 1C). The protein levels of EZH1 in MCL cell lines were comparable with naïve B-cells, whereas EZH2 protein was remarkably overexpressed in all of the four cell lines compared with naïve B cells in which EZH2 protein was barely detected. Notably, the level of H3K27me3 which reflects the function of the PRC2 complex was significantly increased in Z138 and Mino cells, the two bearing the highest EZH2 expression (Figure 1D and Supplementary Figure S1). We also probed the gene status of EZH2 by DNA sequencing and found that all four MCL cell lines harbored wild-type EZH2 (Figure 1E).
S2.
The clinical treatment history of investigated patients
| PATHNO | Gender | DXAGE | STAGE | CURRX | OS | ALIVE | EZH1 | EZH2 | PREVRX |
| PATHNO = pathology ID number; SEX = gender: 2=female, 1=male; DXAGE = age at date of diagnosis; STAGE = Ann Arbor stage; CURRX = initial therapy code: 50=W&W; 53=CAPBOP3; 63=CAPBOP4; 73=CNOP; 82=Rituxan, Fludara, Adria or Mitoxantrone, Cytoxan & VCR; 83=Fludara/R-CHOP; 84=R-CHOP->R-HyperCVAD; 91= HyperCVAD; 92=R-HyperCVAD; 93=CHOP; 94=R-CHOP; 97=R-CHOP & Bortezomib; 98=Leukeran & Prednisone; OS = overall survival in months; ALIVE = patient status, alive=0, dead=1; EZH1 = EZH1 result: low expression=0, high expression=1; EZH2 = EZH2 result: low expression=0, high expression=1; PREVRX = previous therapy given (‘-’ indicates this is ade novo case) | |||||||||
| NEB_S89-6719 | 1 | 64 | 4 | 63 | 53 | 1 | 0 | 0 | – |
| NEB_S91-1644 | 1 | 60 | 4 | 63 | 24 | 1 | 0 | 1 | – |
| NEB_S96-747 | 1 | 63 | 4 | 73 | 5 | 1 | 0 | 1 | – |
| NEB_S97-6596 | 1 | 65 | 4 | 73 | 22 | 1 | 1 | 0 | – |
| NEB_S97-9354 | 2 | 80 | 3 | 73 | 24 | 1 | 0 | 0 | – |
| NEB_S97-10825 | 2 | 59 | 4 | 73 | 20 | 1 | 0 | 1 | – |
| NEB_S97-11946 | 1 | 55 | 4 | 73 | 47 | 1 | 0 | 1 | – |
| NEB_SC01-1791 | 1 | 85 | 4 | 82 | 42 | 1 | 0 | 1 | – |
| NEB_SC04-1638 | 2 | 65 | 2 | 94 | 45 | 1 | 0 | 1 | – |
| NEB_S03-21240 | 1 | 70 | 4 | 94 | 150 | 0 | 0 | 0 | – |
| NEB_S03-19827 | 2 | 57 | 4 | 92 | 35 | 1 | 0 | 1 | – |
| NEB_S02-4321 | 1 | 46 | 4 | 92 | 161 | 0 | 0 | 0 | – |
| NEB_S02-21874 | 1 | 59 | 4 | 94 | 66 | 1 | 0 | 1 | – |
| NEB_SC05-3291 | 1 | 37 | 4 | 92 | 19 | 1 | 0 | 1 | – |
| NEB_SC03-305 | 1 | 76 | 2 | 50 | 18 | 1 | 0 | 1 | – |
| NEB_S98-12406 | 1 | 75 | 4 | 94 | 19 | 1 | 1 | 0 | – |
| NEB_SC05-3291 | 1 | 37 | 4 | 92 | 31 | 1 | 0 | 0 | – |
| NEB_SC06-760 | 1 | 73 | 4 | 94 | 16 | 1 | 0 | 1 | – |
| NEB_SC06-3687 | 1 | 88 | 3 | 98 | 0.3 | 1 | 0 | 1 | – |
| NEB_SC06-1183 | 1 | 78 | 4 | 94 | 43 | 1 | 0 | 0 | – |
| NEB_S07-463 | 1 | 71 | 4 | 97 | 35 | 1 | 0 | 1 | – |
| NEB_S07-5289 | 1 | 60 | 4 | 92 | 105 | 0 | 0 | 0 | – |
| NEB_S95-1283 | 1 | 58 | 3 | 73 | 51 | 1 | 0 | 0 | – |
| NEB_S89-3218 | 2 | 77 | 4 | 63 | 139 | 1 | 0 | 0 | – |
| NEB_SP01-5878 | 2 | 49 | 4 | 91 | 183 | 0 | 1 | 0 | – |
| TMU_344322 | 1 | 54 | 4 | 93 | 30 | 1 | 0 | 1 | – |
| TMU_362565 | 2 | 69 | 4 | 93 | 30 | 1 | 1 | 1 | – |
| TMU_363595 | 1 | 57 | 4 | 93 | 16 | 1 | 0 | 1 | – |
| TMU_364180 | 2 | 46 | 2 | 84 | 70 | 0 | 0 | 0 | – |
| TMU_395043 | 1 | 53 | 3 | 93 | 115 | 0 | 0 | 0 | – |
| TMU_395456 | 1 | 64 | 2 | 91 | 128 | 0 | 0 | 0 | – |
| TMU_398817 | 1 | 67 | 4 | 92 | 131 | 0 | 0 | 0 | – |
| TMU_411072 | 2 | 71 | 4 | 94 | 15 | 1 | 0 | 1 | – |
| TMU_412975 | 1 | 56 | 4 | 94 | 15 | 1 | 1 | 1 | – |
| TMU_418367 | 1 | 62 | 3 | 84 | 118 | 0 | 0 | 0 | – |
| TMU_425675 | 1 | 64 | 4 | 92 | 121 | 0 | 1 | 0 | – |
| TMU_435280 | 2 | 66 | 2 | 94 | 131 | 0 | 0 | 0 | – |
| TMU_435293 | 2 | 62 | 4 | 94 | 16 | 1 | 0 | 1 | – |
| TMU_438376 | 2 | 63 | 4 | 91 | 137 | 0 | 1 | 0 | – |
| TMU_444479 | 1 | 59 | 3 | 94 | 28 | 0 | 0 | 0 | – |
| TMU_446227 | 1 | 61 | 4 | 94 | 1 | 1 | 0 | 1 | – |
S3.
Clinicopathologic characteristics of 41 patients with MCL
| Features | n (%) | EZH2 | χ2 | P | EZH1 | χ2 | P | ||
| Positive | Negative | Positive | Negative | ||||||
| Gender | 0.01 | 0.92 | 0.753 | 0.386 | |||||
| Male | 29 (70.7) | 14 | 15 | 4 | 25 | ||||
| Female | 12 (29.3) | 6 | 6 | 3 | 9 | ||||
| Age (years) | 0.196 | 0.658 | 0.234 | 0.629 | |||||
| < 60 | 15 (36.6) | 8 | 7 | 2 | 13 | ||||
| ≥ 60 | 26 (63.4) | 12 | 14 | 5 | 21 | ||||
| Ann Arbor stage | 2.221 | 0.136 | 2.114 | 0.146 | |||||
| Ⅰ~Ⅱ | 5 (12.2) | 4 | 1 | 2 | 3 | ||||
| Ⅲ~Ⅳ | 36 (87.8) | 16 | 20 | 5 | 31 | ||||
| LDH value | 1.177 | 0.278 | 0.041 | 0.839 | |||||
| Normal | 22 (53.7) | 9 | 13 | 4 | 18 | ||||
| High | 19 (46.3) | 11 | 8 | 3 | 16 | ||||
| B-symptoms | 0.028 | 0.867 | 0.041 | 0.839 | |||||
| Negative | 19 (46.3) | 9 | 10 | 3 | 16 | ||||
| Positive | 22 (53.7) | 11 | 11 | 4 | 18 | ||||
1.
EZH1 and EZH2 are differently expressed in naïve B cells and MCL. (A) Expression of EZH1 and EZH2 in reactive germinal centers and MCL cases were determined by immunohistochemistry (20 x). (B) Overall survival of MCL cases was analyzed by Kaplan-Meier plot based on the expression of EZH1 and EZH2. (C) The mRNA and (D) protein levels of EZH1 and EZH2 as well as H3K27me3 in naïve B cells and 4 MCL cell lines were determined by qRT-PCR and Western blot. (E) Gene sequence alignment showing the representative Y641 status of EZH2 in four MCL cell lines. ** P < 0.01.
S4.
IHC results of EZH2 and EZH1 in primary MCL patients and benign lymph nodes follicular reactive hyperplasia
| n | EZH2 | χ2 | P | EZH1 | χ2 | P | |||
| Positive | Negative | Positive | Negative | ||||||
| **P < 0.01 | |||||||||
| Primary MCL | 41 | 20 | 21 | 11.414 | 0.001** | 7 | 34 | 0.078 | 0.780 |
| Reactive hyperplasia | 20 | 1 | 19 | 4 | 16 | ||||
S1.
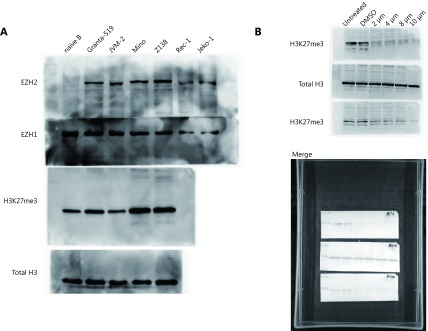
Full gel images of Western blot depicted in Figure 1D and Figure 3G. (A) Protein levels of EZH1 and EZH2 as well as H3K27me3 in naïve B cells and 4 MCL cell lines were determined by western blot. (B) Z138 and Mino cells were treated with increasing dose of UNC1999 for 48 h and determined with the level of EZH2, EZH1 and H3K27me3.
EZH2 depletion has little impact on MCL cells
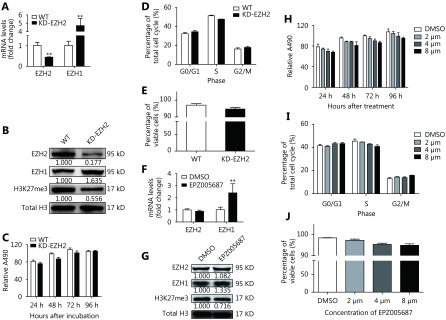
To determine the significance of EZH2 overexpression in MCL, we performed a loss-of-function study using CRISPR/Cas9 system to knock down (KD) EZH2 in Z138 cells. KD efficiency was determined on both mRNA and protein level. We observed an approximate 50% reduction of EZH2 at mRNA level and more than 80% of protein loss after lentivirus mediated CRISPR/Cas9 KD (Figure 2A and 2B). Furthermore, we found that EZH2 KD in Z138 cells downregulated H3K27me3 level by approximate 50% (Figure 2B), in accordance with the crucial role of EZH2 in H3K27me3. Interestingly, despite the substantial depletion of both EZH2 and H3K27me3, cell viability was barely affected as determined by both proliferation and apoptosis analysis (Figure 2C, 2D and 2E), suggesting that either EZH2 had little functional role in MCL or its function loss was compensated through a feedback mechanism. To this end, we examined EZH1 and surprisingly found the expression was remarkably increased upon EZH2 KD as showed on both mRNA and protein level (Figure 2A and 2B). To confirm the finding, we treated Z138 cells with a selective EZH2 inhibitor, EPZ005687 and found the inhibition of EZH2 also significantly elevated the mRNA and protein level of EZH1 (Figure 2F and 2G). Consistent with CRISPR/Cas9 KD, EPZ005687 treatment only induced slight repression on the cell viability (Figure 2H) and had basically no impact on the cell cycle progression and apoptosis (Figure 2I and 2J). Taken together, these data suggest that inhibition of EZH2 alone is insufficient to inhibit MCL cell proliferation, which is likely because of the feedback upregulation and functional compensation from EZH1.
2.
EZH2 depletion has little impact on MCL cells. (A, B) Z138 cells were introduced with CRISPR/Cas9 system targeting EZH2. After 4 weeks of puromycin selection, EZH1 and EZH2 expression were determined on mRNA and protein levels respectively. (C-E) Upon EZH2 KD, cell proliferation was determined by MTS assay; cell cycle progression and apoptosis were determined by flow cytometry. (F-J) Z138 cells were treated with EPZ005687 and determined with EZH1/EZH2 expression, cell proliferation, cell cycle progression, and apoptosis. Data are represented as the mean ± SD. All experiments were repeated at least three times. ** P < 0.01.
Dual inhibition of EZH1/2 exhibits a potent inhibitory effect on MCL cells
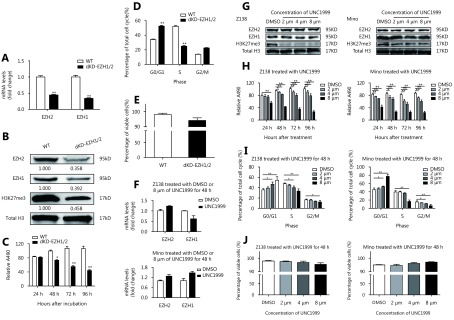
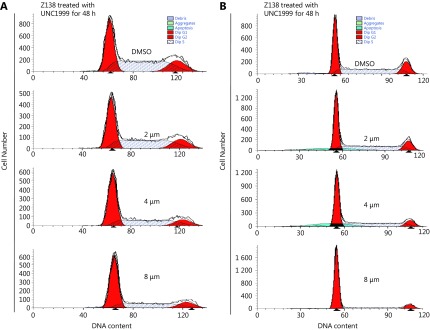
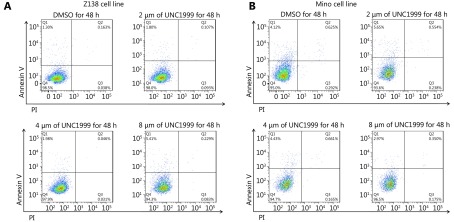
To verify the compensatory effect of EZH1, we further knocked down EZH1 in EZH2-KD Z138 cells using the CRISPR/Cas9 system. The EZH1 mRNA and protein levels were decreased by 60% and 80% respectively (Figure 3A and 3B). Notably, double KD of EZH1 and EZH2 remarkably repressed cell viability (Figure 3C). Moreover, we found that the double-KD significantly induced G0/G1 phase arrest whereas had an only minor impact on apoptosis (Figure 3D and 3E), suggesting that the inhibitory effect of double-KD predominantly resulted from inhibition of cell proliferation. In addition, EZH1/EZH2 double-KD further decreased the H3K27me3 level compared with EZH2 single-KD, supporting that both enzymes are responsible for promoting H3K27me3 in MCL. To further confirm the results, we applied an EZH1/EZH2 dual inhibitor, UNC1999 which targets the SAM binding pocket and showed only marginal effect on EZH1/EZH2 expression (Figure 3F and 3G) in MCL cells. Consistent with CRISPR KD, UNC1999 treatment significantly decreased H3K27me3 levels and inhibited the cell viability in both Z138 and Mino cells (Figure 3G and 3H). Cell cycle and apoptosis analysis also confirmed that the proliferation suppression upon EZH1/EZH2 inhibition resulted from G0/G1 arrest while apoptosis was barely affected (Figure 3I and 3J, and Supplementary Figure S2 and S3). Taken together, these data suggest that EZH1 and EZH2 are functionally overlapped, and EZH1 can compensate the functional loss of EZH2, thus dual inhibition may be required to achieve efficient proliferation suppression in MCL cells.
3.
Double inhibition of EZH1/2 represses the proliferation of MCL cells. (A, B) Z138-EZH2 KD cells were introduced with CRISPR/cas9 system targeting EZH1. After 4 weeks of puromycin and Blasticidin double selection, the EZH1/EZH2 expression was determined on mRNA and protein levels respectively. (C-E) Upon EZH1/EZH2 double-KD, cell proliferation was determined by MTS assay, cell cycle progression and apoptosis were determined by flow cytometry. (F) Z138 and Mino cells were treated with 8μm of UNC1999 for 48 h, and measured with EZH1/EZH2 mRNA expression by qRT-PCR. (G) Z138 and Mino cells were treated with increasing dose of UNC1999 for 48 h and determined with the EZH2, EZH1 and H3K27me3 level. (H-J) Z138 and Mino cells were treated with increasing dose of UNC1999 and determined with cell proliferation, cell cycle progression and apoptosis. Data are represented as the mean ± SD. All experiments were repeated at least three times. * P < 0.05 and ** P < 0.01.
S2.
UNC1999 induced G0/G1 arrest in MCL cells. DNA content histograms showed cell cycle alteration after 48 h treatment of UNC1999 in Z138 (A) and Mino (B) cells.
S3.
UNC1999 has little impact on apoptosis in MCL cells. Z138 (A) and Mino (B) cells were treated with increasing dose of UNC1999 for 48 h and determined with apoptosis by Annexin V and PI staining followed by flow cytometry quantification.
Dual inhibition of EZH1/2 induces cell cycle arrest by reactivating CDKN1C and TP53INP1 in MCL cells.
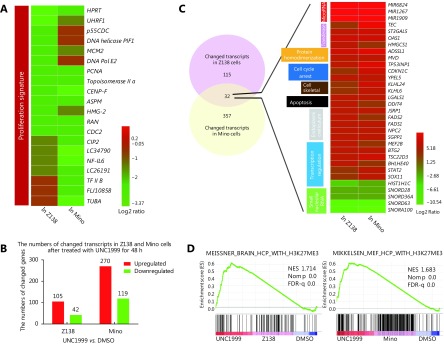
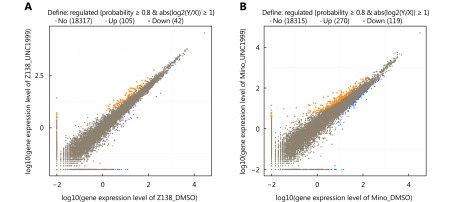
Next, we sought to determine the molecular mechanism of EZH1/EZH2 double-inhibition induced proliferation arrest in MCL. We performed RNA-seq based gene expression profiling analysis in Z138 and Mino cells treated with UNC1999. First, we examined the proliferation signatures established in our previous study19, and found that the majority of the signature genes were substantially down-regulated by UNC1999 treatment in both cells (Figure 4A, P = 0.066 in Z138, P = 0.068 in Mino cells), in accordance with the inhibitory effect of EZH1/2 depletion. In general, there were 105 transcripts in Z138 cells and 270 transcripts in Mino cells significantly up-regulated; 42 and 119 transcripts significantly down-regulated, respectively, upon UNC1999 treatment (Figure 4B and Supplementary Figure S4). Among the downregulated genes, five were commonly identified in both cell lines with four of them being small nucleolar RNA; among the upregulated genes, twenty-seven were commonly identified in both cell lines. Functionally, these commonly altered genes fell into categories of cell cycle regulation, apoptosis, transcription regulation and microRNAs (Figure 4C). GSEA analysis demonstrated that the altered genes were highly enriched in the H3K27me3 signatures (Figure 4D), further confirming that the repression of H3K27me3 plays a major role in the cytostatic property of EZH1/EZH2 depletion.
4.
Gene expression alterations upon EZH1/EZH2 double inhibition. Z138 and Mino cells were treated with UNC1999 for 48 h and then performed with RNA-Seq assay. (A) The heat-map shows the FPKM fold change of the proliferation signature genes. (B) The overall numbers of altered transcripts upon UNC1999 treatment. (C) Venn diagram and heat-map show that the altered genes overlapped between Z138 and Mino involve multiple cell functions. (D) GSEA plots demonstrate that the altered genes are highly enriched in the H3K27me3 signature. NES, NOM P value, and FDR are indicated.
S4.
Gene expression profiling of MCL cells treated with UNC1999. Scatter diagrams shows gene expression level determined by RNA-Seq in Z138 (A) and Mino (B) cells. The genes with more than two-fold change are highlighted.
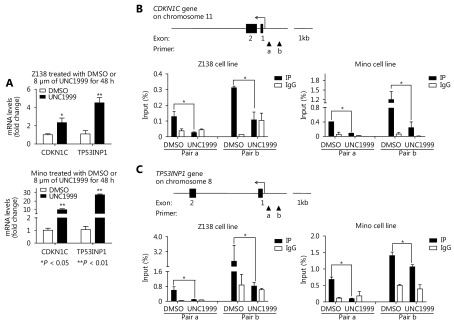
Particularly, in the up-regulated gene panel, we noticed two important cell cycle regulators CDKN1C and TP53INP1, both of which are tumor suppressors and have anti-proliferative activity. Increased expression of these two genes, to a large extent, explains the proliferation suppression by EZH1/EZH2 depletion. To confirm these findings, we performed qRT-PCR analysis and observed CDKN1C was up-regulated 2.3 and 9.8 fold in Z138 and Mino cells respectively whereas TP53INP1 was up-regulated 4.5 and 27.5 fold respectively upon UNC1999 treatment (Figure 5A). Furthermore, we performed ChIP assay to pull down H3K27me3 and then analyzed the promoter segment abundance of both CDKN1C and TP53INP1. Notably, H3K27me3 was significantly depleted from the two promoters in both Z138 and Mino cells with UNC1999 treatment (Figure 5B and 5C). These data strongly suggest that the expression of CDKN1C and TP53INP1 are repressed by H3K27me3 and the dual inhibition of EZH1/EZH2 abolishes the repression, inducing proliferation arrest and thus representing a novel therapeutic strategy for MCL.
5.
CDKN1C and TP53INP1 are suppressed by H3K27me3 and restored by UNC1999 treatment in MCL cells. (A) Z138 and Mino cells were treated with UNC1999 for 48 h and determined with CDKN1C and TP53INP1 expression by qRT-PCR. (B, C) H3K27me3 abundance on the promoters of CDKN1C and TP53INP1 were determined by ChIP-qPCR in Z138 and Mino cells respectively with UNC1999 treatment for 48 h. Data are represented as the mean ± SD. All experiments were repeated at least three times. * P < 0.05 and ** P < 0.01.
Discussion
Mounting evidence suggests that epigenetic regulation involves in the program of B lymphocytes development and differentiation20,21. During the course of normal B-cell development, the expression pattern of EZH2 and EZH1 are different. EZH2 decreases gradually after the pro-B stage, reaching the trough in naïve B cells. The expression of EZH1, on the contrary, gradually increased along with B cell development22. Remarkably, EZH2 is significantly upregulated when naïve B cells receive antigen stimulation and immigrate into the germinal center. It is thus reasonable to assume that the overexpression of EZH2 in naïve B-cells represents an “active” state that may contribute to the malignant transformation. In the current study, we found that EZH2 is overexpressed in about half of the MCL cases, and the overexpression is closely related to the inferior clinical outcome, supporting the notion that EZH2 is involved in the pathogenesis of MCL and its overexpression may serve as a novel biomarker for prognosis prediction.
Unexpectedly, we found that inhibition of EZH2 alone is unable to induce functional alteration on tumor cells. Our further investigation revealed that the concomitant increase of EZH1 compensated the functional loss of EZH2, which was validated by the substantial proliferation suppression resulted from EZH1/EZH2 dual inhibition. Although EZH2 and EZH1 are homologous and share high sequence identity, up to 76% overall and 96% of the SET domains, their functions are not redundant23. It has been demonstrated that PRC2-EZH2 mainly exhibits the PKMT activity while PRC2-EZH1 compacts the chromatin directly14. Besides, EZH2 has other functions than epigenetic transcriptional silencing as evidenced by the activation of CCND1 expression in natural killer/T-cell lymphoma cells and activation of STAT3 in glioblastoma multiforme24,25. Notwithstanding, our results suggest that EZH2 and EZH1 are, at least partially, functional overlapped, which is consistent with previous observation that PRC2-EZH1 coexists with PRC2-EZH2 in catalyzing H3K27me3 at target genes26,27. A recent study also demonstrated that EZH1 plays an essential role in myelodysplastic disorders upon EZH2 loss28. These findings indicate that EZH1 and EZH2 share common tumorigenic properties and may cooperatively participate in the cancer development. Importantly, we found that higher level of EZH2 is associated with lower level of EZH1 which was markedly upregulated after EZH2 inhibition. Future studies need to address whether EZH1 and EZH2 are directly repressed by each other and the significance of such regulation in physiological and pathological conditions.
In the cell cycle regulation, cyclin-dependent kinases (CDKs) form complexes with distinctive types of cyclin proteins, promoting the cycle progression; whereas cyclin-dependent kinase inhibitors (CDKNs) inhibit the kinase activities of Cyclin/CDK complexes at appropriate time points, halting cycle progression29. Known to date, the family of CDKNs includes CDKN1A (P21), Kip1/CDKN1B (P27), Kip2/CDKN1C (p57), CDKN2A (p16), CDKN2B (p15), CDKN2C (p18), CDKN2D (p19) and CDKN3. CDKN1C is a recently identified CDKN and was found to inhibit Cyclin D-CDK4, Cyclin D-CDK6, Cyclin E-CDK2 and Cyclin A-CDK2 complexes inducing G0/G1 arrest without requirement of either pRb or p5330. In breast cancer cells, CDKN1C was demonstrated to be a direct target of EZH231. In acute lymphoblastic leukemia cells, CDKN1C is restored by Rapamycin treatment and participates in Rapamycin mediated cell cycle arrest32. However, the role of CDKN1C in MCL are poorly studied. In this study, we showed that CDKN1C as well as TP53INP1, another important cell cycle regulator, are directly sequestered by H3K27me3, and for the first time, we demonstrated that the inhibition of EZH1/EZH2 up-regulated the expression of CDKN1C and TP53INP1. These results strongly support that epigenetic modification plays an important role in cell cycle progression and contributes to the pathogenesis of MCL.
Given the hallmark cyclin D1 dysregulation in MCL, cell cycle targeting is of great value in the clinical administration. UNC1999 is a new generation of SAM-competitive small molecular inhibitor which directly docks into the SAM pocket of both EZH2 and EZH1 SET domain inhibiting their enzymatic activities without altering EZH protein levels33. It is more potent than most of the kinase inhibitors used in pre-clinical studies and in clinical trials on cell cycle inhibition. The CDK4/6 inhibitor PD0332991, although has already been used for MCL in phase I/II clinical trials, achieved only 18% overall response rate34. UNC1999 is also more effective than DZNep which indirectly inhibits the function of EZH2 by promoting degradation of the PRC2 complex35. The biochemical character of UNC1999 ensures the integrity of the PRC2 complex and maintains the protein-protein interactions among the PRC2 subunits. This is totally different from DZNep which lacks specificity and reduces the methylation state at multiple histone residues targeted not only by EZH236. This is an important concern, because PRC2 is a multimeric protein complex in which absence of any component may cause secondary effect, including recruitment of new members to maintain its biological function and activation of other pathways, such as STAT5 signaling to compensate its function37. Therefore, UNC1999 is prone to be more effective and has fewer tendencies with resistance. Further clinical studies are warranted to demonstrate its in vivo efficacy.
Conclusions
Taken together, in this study we demonstrated that epigenetic dysregulation characterized by EZH2 overexpression plays an important role in the MCL development. The dual inhibition of EZH1/EZH2 exhibited potent anti-cancer effect and may serve as a promising strategy for the management of MCL patients.
Acknowledgments
This study was supported by a grant from the National Natural Science Foundation of China (Grant No. 81372539).
Conflict of interest statement
No potential conflicts of interest are disclosed.
References
- 1.Weisenburger DD, Armitage JO Mantle cell lymphoma--an entity comes of age. Blood. 1996;87:4483–94. [PubMed] [Google Scholar]
- 2.Fu K, Weisenburger DD, Greiner TC, Dave S, Wright G, Rosenwald A, et al Cyclin D1-negative mantle cell lymphoma: a clinicopathologic study based on gene expression profiling. Blood. 2005;106:4315–21. doi: 10.1182/blood-2005-04-1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bodrug SE, Warner BJ, Bath ML, Lindeman GJ, Harris AW, Adams JM Cyclin D1 transgene impedes lymphocyte maturation and collaborates in lymphomagenesis with the myc gene. EMBO J. 1994;13:2124–30. doi: 10.1002/embj.1994.13.issue-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klier M, Anastasov N, Hermann A, Meindl T, Angermeier D, Raffeld M, et al Specific lentiviral shRNA-mediated knockdown of cyclin D1 in mantle cell lymphoma has minimal effects on cell survival and reveals a regulatory circuit with cyclin D2. Leukemia. 2008;22:2097–105. doi: 10.1038/leu.2008.213. [DOI] [PubMed] [Google Scholar]
- 5.Lovec H, Grzeschiczek A, Kowalski MB, Möröy T Cyclin D1/bcl-1 cooperates with myc genes in the generation of B-cell lymphoma in transgenic mice. EMBO J. 1994;13:3487–95. doi: 10.1002/embj.1994.13.issue-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang J, Jima D, Moffitt AB, Liu QQ, Czader M, Hsi ED, et al The genomic landscape of mantle cell lymphoma is related to the epigenetically determined chromatin state of normal b cells. Blood. 2014;123:2988–96. doi: 10.1182/blood-2013-07-517177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vose JM Mantle cell lymphoma: 2015 update on diagnosis, risk-stratification, and clinical management. Am J Hematol. 2015;90:739–45. doi: 10.1002/ajh.v90.8. [DOI] [PubMed] [Google Scholar]
- 8.Fiskus W, Rao R, Balusu R, Ganguly S, Tao JG, Sotomayor E, et al Superior efficacy of a combined epigenetic therapy against human mantle cell lymphoma cells. Clin Cancer Res. 2012;18:6227–38. doi: 10.1158/1078-0432.CCR-12-0873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caganova M, Carrisi C, Varano G, Mainoldi F, Zanardi F, Germain PL, et al Germinal center dysregulation by histone methyltransferase EZH2 promotes lymphomagenesis. J Clin Invest. 2013;123:5009–22. doi: 10.1172/JCI70626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bracken AP, Pasini D, Capra M, Prosperini E, Colli E, Helin K EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer . EMBO J. 2003;22:5323–35. doi: 10.1093/emboj/cdg542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morin RD, Johnson NA, Severson TM, Mungall AJ, An JH, Goya R, et al Somatic mutations altering EZH2(Tyr641) in follicular and diffuse large b-cell lymphomas of germinal-center origin. Nat Genet. 2010;42:181–5. doi: 10.1038/ng.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McCabe MT, Ott HM, Ganji G, Korenchuk S, Thompson C, Van Aller GS, et al EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012;492:108–12. doi: 10.1038/nature11606. [DOI] [PubMed] [Google Scholar]
- 13.Béguelin W, Popovic R, Teater M, Jiang YW, Bunting KL, Rosen M, et al EZH2 is required for germinal center formation and somatic EZH2 mutations promote lymphoid transformation. Cancer Cell. 2013;23:677–92. doi: 10.1016/j.ccr.2013.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Margueron R, Li GH, Sarma K, Blais A, Zavadil J, Woodcock CL, et al Ezh1 and ezh2 maintain repressive chromatin through different mechanisms. Mol Cell. 2008;32:503–18. doi: 10.1016/j.molcel.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abdalkader L, Oka T, Takata K, Sato H, Murakami I, Otte AP, et al Aberrant differential expression of EZH1 and EZH2 in polycomb repressive complex 2 among B- and T/NK-cell neoplasms. Pathology. 2016;48:467–82. doi: 10.1016/j.pathol.2016.05.002. [DOI] [PubMed] [Google Scholar]
- 16.Shih AH, Abdel-Wahab O, Patel JP, Levine RL The role of mutations in epigenetic regulators in myeloid malignancies. Nat Rev Cancer. 2012;12:599–612. doi: 10.1038/nrc3343. [DOI] [PubMed] [Google Scholar]
- 17.Dong L, Lv HJ, Li W, Song Z, Li LF, Zhou SY, et al Co-expression of PD-L1 and p-AKT is associated with poor prognosis in diffuse large B-cell lymphoma via PD-1/PD-L1 axis activating intracellular AKT/mTOR pathway in tumor cells . Oncotarget. 2016;7:33350–62. doi: 10.18632/oncotarget.9061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bi CF, Zhang X, Lu T, Zhang XY, Wang XH, Meng BL, et al Inhibition of 4EBP phosphorylation mediates the cytotoxic effect of mechanistic target of rapamycin kinase inhibitors in aggressive B-cell lymphomas. Haematologica. 2017;102:755–64. doi: 10.3324/haematol.2016.159160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosenwald A, Wright G, Wiestner A, Chan WC, Connors JM, Campo E, et al The proliferation gene expression signature is a quantitative integrator of oncogenic events that predicts survival in mantle cell lymphoma. Cancer Cell. 2003;3:185–97. doi: 10.1016/S1535-6108(03)00028-X. [DOI] [PubMed] [Google Scholar]
- 20.Velichutina I, Shaknovich R, Geng H, Johnson NA, Gascoyne RD, Melnick AM, et al EZH2-mediated epigenetic silencing in germinal center B cells contributes to proliferation and lymphomagenesis. Blood. 2010;116:5247–55. doi: 10.1182/blood-2010-04-280149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wesemann DR, Portuguese AJ, Magee JM, Gallagher MP, Zhou XL, Panchakshari RA, et al Reprogramming IgH isotype-switched B cells to functional-grade induced pluripotent stem cells. Proc Natl Acad Sci USA. 2012;109:13745–50. doi: 10.1073/pnas.1210286109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Su IH, Basavaraj A, Krutchinsky AN, Hobert O, Ullrich A, Chait BT, et al Ezh2 controls B cell development through histone H3 methylation and Igh rearrangement . Nat Immunol. 2003;4:124–31. doi: 10.1038/ni876. [DOI] [PubMed] [Google Scholar]
- 23.Margueron R, Reinberg D The polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–9. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yan J, Ng SB, Tay JL, Lin BH, Koh TL, Tan J, et al EZH2 overexpression in natural killer/T-cell lymphoma confers growth advantage independently of histone methyltransferase activity. Blood. 2013;121:4512–20. doi: 10.1182/blood-2012-08-450494. [DOI] [PubMed] [Google Scholar]
- 25.Kim E, Kim M, Woo DH, Shin Y, Shin J, Chang N, et al Phosphorylation of EZH2 activates STAT3 signaling via STAT3 methylation and promotes tumorigenicity of glioblastoma stem-like cells . Cancer Cell. 2013;23:839–52. doi: 10.1016/j.ccr.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hidalgo I, Herrera-Merchan A, Ligos JM, Carramolino L, Nuñez J, Martinez F, et al Ezh1 is required for hematopoietic stem cell maintenance and prevents senescence-like cell cycle arrest. Cell Stem Cell. 2012;11:649–62. doi: 10.1016/j.stem.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 27.Shen XH, Liu YC, Hsu YJ, Fujiwara Y, Kim J, Mao XH, et al EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol Cell. 2008;32:491–502. doi: 10.1016/j.molcel.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mochizuki-Kashio M, Aoyama K, Sashida G, Oshima M, Tomioka T, Muto T, et al Ezh2 loss in hematopoietic stem cells predisposes mice to develop heterogeneous malignancies in an ezh1-dependent manner. Blood. 2015;126:1172–83. doi: 10.1182/blood-2015-03-634428. [DOI] [PubMed] [Google Scholar]
- 29.Hirama T, Koeffler HP Role of the cyclin-dependent kinase inhibitors in the development of cancer. Blood. 1995;86:841–54. [PubMed] [Google Scholar]
- 30.Matsuoka S, Edwards MC, Bai C, Parker S, Zhang P, Baldini A, et al p57KIP2, a structurally distinct member of the p21CIP1 Cdk inhibitor family, is a candidate tumor suppressor gene. Genes Dev. 1995;9:650–62. doi: 10.1101/gad.9.6.650. [DOI] [PubMed] [Google Scholar]
- 31.Yang XJ, Karuturi RKM, Sun F, Aau M, Yu K, Shao RG, et al CDKN1C (p57KIP2) is a direct target of EZH2 and suppressed by multiple epigenetic mechanisms in breast cancer cells . PLoS One. 2009;4:e5011. doi: 10.1371/journal.pone.0005011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li HB, Kong XL, Cui G, Ren CC, Fan SJ, Sun LL, et al Rapamycin restores p14, p15 and p57 expression and inhibits the mTOR/p70S6K pathway in acute lymphoblastic leukemia cells. Int J Hematol. 2015;102:558–68. doi: 10.1007/s12185-015-1858-1. [DOI] [PubMed] [Google Scholar]
- 33.Konze KD, Ma AQ, Li FL, Barsyte-Lovejoy D, Parton T, MacNevin CJ, et al An orally bioavailable chemical probe of the lysine methyltransferases EZH2 and EZH1. ACS Chem Biol. 2013;8:1324–34. doi: 10.1021/cb400133j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leonard JP, LaCasce AS, Smith MR, Noy A, Chirieac LR, Rodig SJ, et al Selective CDK4/6 inhibition with tumor responses by PD0332991 in patients with mantle cell lymphoma. Blood. 2012;119:4597–607. doi: 10.1182/blood-2011-10-388298. [DOI] [PubMed] [Google Scholar]
- 35.Miranda TB, Cortez CC, Yoo CB, Liang G, Abe M, Kelly TK, et al DZNep is a global histone methylation inhibitor that reactivates developmental genes not silenced by DNA methylation. Mol Cancer Ther. 2009;8:1579–88. doi: 10.1158/1535-7163.MCT-09-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee JK, Kim KC DZNep, inhibitor of S-adenosylhomocysteine hydrolase, down-regulates expression of SETDB1 H3K9me3 HMTase in human lung cancer cells. Biochem Biophys Res Commun. 2013;438:647–52. doi: 10.1016/j.bbrc.2013.07.128. [DOI] [PubMed] [Google Scholar]
- 37.Yoo KH, Oh S, Kang K, Hensel T, Robinson GW, Hennighausen L Loss of EZH2 results in precocious mammary gland development and activation of STAT5-dependent genes. Nucleic Acids Res. 2015;43:8774–89. doi: 10.1093/nar/gkv776. [DOI] [PMC free article] [PubMed] [Google Scholar]