Abstract
The genome of cells is constantly challenged by DNA damages from endogenous metabolism and environmental agents. These damages could potentially lead to genomic instability and thus to tumorigenesis. To cope with the threats, cells have evolved an intricate network, namely DNA damage response (DDR) system that senses and deals with the lesions of DNA. Although the DDR operates by relatively uniform principles, different tissues give rise to distinct types of DNA damages combined with high diversity of microenvironments across tissues. In this review, we discuss recent findings on specific DNA damage among different tissues as well as the main DNA repair way in corresponding microenvironments, highlighting tissue specificity of DDR and tumorigenesis. We hope the current review will provide further insights into molecular process of tumorigenesis and generate new strategies for cancer treatment.
Keywords: DNA damage response, tissue specificity, tumorigenesis, microenvironment, cancer treatment
Introduction
DNA damage is a serious threat to human health. It has been estimated that each cell in human body experiences approximately 100,000 DNA damage events per day1. These damages include occasional DNA mismatches during DNA replication in physiological processes, DNA strand breaks caused by aberrant topoisomerase (Topo) I and Topo II activity, and DNA lesions caused by reactive oxygen species (ROS)2,3. In addition, various stressors from the surrounding environment, such as ionizing radiation (IR) and ultraviolet (UV) light, as well as chemical factors including heavy metals, alkylating agents, nucleoside analogs and base analogs, can attack DNA, resulting in adduct formation, base deletion, DNA strand breakage, or crosslinking4-6. Some chemical agents are highly electrophilic and can react directly with DNA, while others must be metabolized before they can insult the nucleic acids7. DNA lesions can block DNA replication and transcription8. If not repaired timely and accurately, they may cause genomic instability or chromosomal abnormalities that threaten the integrity and functions of cells, tissues and organs9-11.
Living cells must constantly address genotoxic insults through a complex and coordinated process, known as DNA damage response (DDR) system12. The highly conserved DDR network consists of hierarchical pathways. The crucial components of the network can generally be classified as damage sensors, transducers, and effectors2. The DDR network can actively sense and signal problems in DNA to induce different cellular effects13. The fate of damaged cells is greatly influenced by multiple factors such as the extent of DNA lesions, the choice of DDR pathways, the rapidity and fidelity of DNA repair, and the status of the cells. Depending on these conditions, cells may temporarily fall into cell cycle arrest for DNA repair, or irreversibly undergo senescence or apoptosis6. Even failing to repair DNA lesions does not necessarily lead to fixation of potentially detrimental mutations, as such genetically aberrant cells are generally eliminated from the proliferative pool. This means that, DDR serves as a guardian of genomic integrity in a fairly broad sense.
While the association between defective DDR and predisposition to carcinoma has been recognized for decades, recent studies have revealed a novel role of DDR as a physiological barrier against tumorigenesis and tumor progression from early stages to advanced, invasive lesions14,15. Tumorigenesis is a multi-step process. An early step is the dysregulation of cell proliferation resulted from oncogenic activation or loss of tumor suppressors, which subsequently triggers DDR16. Such DDR activation achieves urgent disposal of precancerous cells, posing the cells for repair, senescence, or cell death. Thus, the process helps to delay or avoid uncontrolled cell growth and tumorigenesis. When progressing from pre-neoplastic abnormalities to cancers, this barrier has been observed to be diminished in most tissues through loss of certain DDR capabilities17. On the other hand, a cell with DDR deficiency indeed displays higher genomic instability and increased dependency on remaining DDR pathways, leading to a worse outcome during its DNA repair. This vicious circle overloads threats on the genome of the cell that may initiate its tumorigenesis.
General DNA damage and repair
In mammalian cells, major DNA repair types include nucleotide excision repair (NER), base excision repair (BER), mismatch repair (MMR), double-strand break (DSB) repair through homologous recombination (HR) or non-homologous end-joining (NHEJ), and Fanconi anemia (FA) DNA repair18. Single-strand breaks (SSBs) in which a 3′-hydroxyl adjoins a 5′-phosphate without missing nucleotides can be directly ligated19. Before discussing the features of DDR in different tissues, we briefly introduce the general functions of these DNA repair ways.
The NER pathway removes bulky adducts induced by UV light or platinum salts, and also the modified nucleotides that distort the structure of the double helix20,21. The two subpathways of NER, global genomic NER (GG-NER) and transcription-coupled NER (TC-NER), start differently22. The former is initiated by XPC and DDB1-DDB2 complex, while the latter relies on RNA polymerase II, CSA, CSB, and TFIIS to sense lesions23-25. Both of the subpathways lead to the recruitment of transcription factor II human (TFIIH) to unwind the DNA helix at the damaged site26. Xeroderma pigmentosum group A (XPA) and replication protein A (RPA) then orchestrate this open complex formation and stabilize the repair intermediate. The xeroderma pigmentosum group F (XPF) protein and the excision repair cross-complementation group 1 (ERCC1) protein form a designated complex, XPF-ERCC1, to cleave 5′ of the lesion, while xeroderma pigmentosum group G (XPG) executes the 3′ incision27. Replication factor C (RFC) then loads proliferating cell nuclear antigen (PCNA) to accommodate DNA polymerases for repair replication. And the final ligation step is carried out either by flap endonuclease 1 (FEN1) and DNA ligase I (LIG1) or by the ligase III-XRCC1 complex28. Different from NER, the BER pathway deals with small chemical alterations of DNA bases, abasic sites, and SSBs29. BER is initiated by one of at least 11 different damage-specific DNA glycosylases with or without apurinic/apyrimidinic endonuclease 1 (APE1)30. Glycosylases cleave the base-sugar bond, leaving an abasic site31. The resulting SSB can be filled in and ligated via short-patch or long-patch BER. The former recruits X-ray repair cross-complementing protein 1 (XRCC1), DNA ligase III (LIG3), poly ADP-ribose polymerase 1 (PARP-1), and DNA polymerase β (pol β) to fill the gap with a single nucleotide32. The latter loads polymerases that synthesize at least two nucleotides for long-patch gap filling, and this process requires PCNA and RFC33. Finally, the displaced strand is excised by FEN1, and the incision is ligated by LIG134. NER and BER are both carried out by multiprotein complexes comprised of approximately 20 components. Besides, both of the pathways require incision of the damaged DNA strand, with subsequent re-synthesis using the complementary strand as a template35. The size of repair patch differs, however, in that the NER patches are approximately 30 bases in length, whereas BER produces the patches of 1-6 bases in length36. Remarkably, NER and BER are carefully regulated by p53. p53 affects NER via the transcriptional activation of downstream effector genes including p48-XPE and Gadd45a37,38. As for BER, p53 might directly interact with BER proteins, including pol β and APE39-42. These effects are believed to be fundamental for p53 tumor suppressor to repair the vast majority of DNA damages incurred from various environmental stressors43.
MMR is another important DDR pathway for resisting tumorigenesis. This pathway mainly corrects replication errors, including nucleotide substitutions, deletions, and insertions. MMR is initiated when a heterodimer of the MutS homolog (MSH), MSH2-MSH6 (MutSα), recognizes a base-base mismatch or when MSH2-MSH3 (MutSβ) recognizes an insertion-deletion loop (IDL) longer than two base pairs44,45. These heterodimers recruit the postmeiotic segregation 2 (PMS2) protein bound to either MutL homolog 1 (MLH1) or MutL homolog 3 (MLH3), and the interactions activate PMS2 endonuclease activity46. Exonuclease 1 (EXO1) excises the DNA segment containing the mismatch, while RPA coats the single-stranded DNA (ssDNA). DNA polymerase works to fill the gap, and the DNA is then ligated. MMR pathway is usually robust in dividing cells, as mismatches occur frequently at the replication fork47. In accordance with this understanding, the genomes of MMR-deficient carcinomas are revealed to harbor considerable amount of somatic mutations48. Based on the indication that these MMR-deficient cancers may produce mutation-associated neoantigens recognized by the immune system, novel studies on immune checkpoint therapy were carried out, and further suggested that advanced solid tumors with MMR deficiency were sensitive to programmed death receptor-1 (PD-1) blockade48-50.
Among different types of DNA damage, DSBs are the most deleterious DNA lesions that may result in genome rearrangements, chromosomal translocations, and cell death. In mammalian cells, DSBs are repaired through either NHEJ or the HR process51,52. NHEJ, rapid but error-prone, is the main DSB repair option that may occur in each stage of the cell cycle53. The initiator of NHEJ, Ku70/80 heterodimer, has high affinity for the ends of DSBs54. DNA-bound Ku recruits DNA-protein kinase catalytic subunits (DNA-PKcs) to DSB ends to form a DNA-PK complex, activating the kinase activity of DNA-PKcs and facilitating ligation55. If the DSB ends are not compatible for ligation, end-processing enzymes are necessary for resecting the DNA ends, filling the gaps, and removing the blocking end groups. Crucial enzymes in this process include polynucleotide kinase 3′-phosphatase (PNKP), aprataxin and PNK-like factor (APLF), Artemis, polymerases μ and λ, and Werner (WRN)56. Finally, DNA ligase IV complex, consisting of DNA ligase IV (LIG4), X-ray repair cross-complementing protein 4 (XRCC4), and XRCC4-like factor (XLF), is responsible for ligation57. Despite its protective role in DSB lesions, NHEJ has been proposed to be involved in the formation of chromosomal translocations and act as a key source of genomic rearrangements and instability, especially when HR or other DDR pathways are defective58,59. Cells with mutations of genes in error-free repair pathways, such as XRCC2, FANCC, and BLM(RECQL3), may overload NHEJ for repair, resulting in a higher risk of cancer60-62.
Compared to NHEJ, HR-mediated DSB repair is more accurate. However, HR relies on the presence of sister chromatids, limiting the use of this pathway to cells in late G2 or S phase. In HR, DSBs are detected by Mre11-Rad50-Nbs1 (MRN) complex, which activates the ataxia telangiectasia mutated (ATM) checkpoint kinase for efficient phosphorylation of DNA repair factors. As MRN triggers 5′ to 3′ end resection of DSB ends, the range of resected DNA is extended, giving rise to single-stranded 3′ overhangs63,64. The ssDNA ends are rapidly bound and protected by RPA and are subsequently replaced by Rad51 recombinase for the generation of long nucleoprotein filaments65. Using the DNA strand of the undamaged chromatid as a template, Rad51 filaments assist in the homology search. After recombination repair, the DNA ends are joined by ligases. On the basis of HR mechanism, numerous new proteins and pathways with functions in HR have been identified in the past decades. For example, accumulated DNA damage due to inactivation or loss of BER, NER or translesion synthesis might be resolved or bypassed by HR during replication66-68. It is currently a consensus that overall DDR network, cell cycle checkpoints, and chromatin remodeling are critical for the onset, regulation, and efficiency of HR69-71. Due to its potent DSB execution, high repair fidelity, and broad crosstalk among pathways, HR can be viewed as the most important repair mechanism and a last resort for DNA repair. Tumors with mutated HR genes are usually characterized by gross gene rearrangements72.
Genes mutated in FA patients are found to interact with the DNA repair genes BRCA1 and FANCD1 (BRCA2) upon replication stalling and thus suppress tumorigenesis73. In particular, the FANCM-FAAP24-MHF1-MHF2 anchor complex detects lesions caused by interstrand crosslinks (ICLs), and recruits the core complex when activated74. The FA core complex is composed of FANCA, FANCB, FANCC, FANCE, FANCF, FANCG, FANCL, and the accessory proteins FAAP20 and FAAP10075. The complex monoubiquitylates each protein of the FANCI-FANCD2 (ID2) heterodimer. This process leads to the formation and loading of the ID2 complex at nuclear foci76,77. Ubiquitylated ID2 facilitates the DNA breakage at ICLs and translesion synthesis, and then signals downstream HR proteins such as FANCD1 (BRCA2), FANCJ (BRIP), FANCN (PALB2), and FANCO (RAD51C) to repair the collapsed replication fork induced by strand break78-83. Besides, FANCD2 mediates fork protection via RAD51 functions, which indicates a novel pathway connecting FA components to RAD51 and the BRCA1/2 tumor suppressors84. However, FA proteins are not classic HR factors, and cells from FA patients do not demonstrate severely defective HR repair of DSBs85. Therefore, the functional relationship between FA and HR proteins during replication stalling remains ambiguous and needs further exploration.
Tissue-specific DDR and tumorigenesis
Because of their surrounding environments, organs or tissues are faced with different insults which lead to various types of DNA damage86. Some tissues, such as the small intestine, are particularly sensitive to DNA damage, yet the colon is considerably more resistant87,88. While different pathways cooperate to accomplish DDR, cells with distinct origins differ in their capacities to sense, respond to, and resolve DNA lesions. For example, rapidly proliferating cells of the small intestine and colon have an efficient MMR pathway, while the NER, NHEJ, and HR pathways are more frequently initiated in skin cells for the repair of ICLs caused by UV radiation89-91. Clinical and experimental findings have provided extensive evidence for tissue-specific DNA repair. Mutations of DNA repair genes in different DDR pathways tend to have tissue-specific effects on patients, leading to tissue-specific phenotypes92-94. For example, variations in MMR pathway, particularly when involving MSH1, MSH2, MLH6, or PMS2, tend to increase the risk of colorectal cancers (CRCs), whereas mutations in SSB repair mechanism can have a higher chance to impair neurological functions94,95. People with mutations of XPA, XPC, XPE, and XPF in NER predispose xeroderma pigmentosum (XP) and subsequently cutaneous neoplasms, while hereditary or sporadic BRCA1, BRCA2, PALB2 and other HR mutations indicate greater incidence of breast and ovarian cancers93,96. It seems that these differences arise in accordance with the distinct characteristics of tissue cells. MMR plays an essential role in dividing cells of gut because of frequent mismatches at DNA replication fork. By contrast, neurons are more sensitive to SSBs and oxidative damage due to their postmitotic status and high metabolic rates, thus more dependent on corresponding repair. Sequencing of cancer genomes has in turn revealed a wide range of mutational signatures that vary in part due to their tissues of origin97.
Furthermore, transgenic mouse models with reporter genes such as lacZ and lacI were used to quantify mutagenesis, and the results of the study have confirmed that mutation frequency varies considerably in a tissue-dependent manner98. Accordingly, the steady-state amount of oxidized DNA bases is proved to be higher in brain than in liver tissues, particularly in older mice99. By using oligonucleotide substrates to harbor SSBs in the nucleus and mitochondria, scientists have even found that testes contain higher glycosylase activity for BER than other tissues100. Though the precise underlying mechanism of tissue specificity in DDR and DNA repair has not yet been fully elucidated, many advanced findings have emerged and impelled our understanding in this field. Here, we will discuss the typical causes and types of DNA damage, the corresponding DDR or other DNA repair mechanisms, and the tumorigenesis in different tissues, hoping to provide further insights into DNA damage-induced tumors and new strategies of cancer treatments based on tissue specificity.
Skin
Skin is the largest organ of the human body, as it constitutes approximately 16% of the body mass101. Covering the body surface and serving as a protective barrier to isolate the internal environment of the body from the external environment, it plays a crucial role in resistance to daily stresses, such as harmful UV radiation from the sun, chemical agents, and infectious pathogens102. Remarkably, as many as 105 UV-induced photolesions caused by sunlight exposure are estimated to occur per day in every skin cell103. Ambient sunlight mainly contains UV-B (295–320 nm) and UV-A (320–400 nm), and each component can cause DNA damage through different mechanisms104. By interfering with nucleotide base pairing, UV radiation contributes to direct DNA photolesions, particularly (6,4)-photoproducts and cyclobutane pyrimidine dimers in skin cells105. These types of damage are mainly resolved through the NER pathway, indicating the significance of NER in skin disorder resistance106.
A large variety of skin maladies could be aroused if NER fails to repair DNA lesions in a timely manner107,108. The three most common diseases related to defective NER are XP, Cockayne syndrome (CS), and trichothiodystrophy (TTD). XP is a disease characterized by photosensitivity, ichthyosis (dry and scaly skin), and aberrant hyperpigmentation in skin areas exposed to sunlight. Notably, XP augments the susceptibility to UV-induced skin cancer (melanomas, basal and squamous cell carcinomas in particular) by 1000-fold109. Numerous mutations in genes, including XPA, ERCC3 (XPB), XPC, ERCC2 (XPD), DDB2 (XPE), ERCC4 (XPF), ERCC5 (XPG), and POLH (XPV), have been found in XP cells. These genes serve primarily to encode XP repair proteins in the NER pathway93,105. Depending on how much the mutations affect NER, different mutations can result in great heterogeneity in clinical phenotypes, and symptoms of XP can vary substantially in severity110. Although skin photosensitivity is presented in all three conditions, CS and TTD, unlike XP, have been found to be unrelated to increased skin cancer risk111. CS can be ascribed to mutations in ERCC8 (CSA), ERCC6 (CSB), XPB, XPD or XPG that induce defects in TC-NER. TTD is identical to CS, except for its additional cutaneous symptoms, including ichthyosis and brittle hair and nails. Mutations in TTDA, XPB, or XPD are the main genetic causes of TTD. Since these genes encode subunits of TFIIH, mutations in the genes give rise to destabilization or even dysfunction of the transcription factor112.
Cutaneous neoplasms are largely confined to patients with mutations in XPA, XPC, XPE and XPF93. Patients with CS due to defects in TC-NER, though photosensitive, are not prone to cancer, indicating the primary role of GG-NER in protecting DNA against UV-induced mutagenesis. Several studies on the effects of UV in NER-deficient mice have provided further evidence that Xpa-/-, Xpc-/- and Xpe-/- mice are prone to UV-induced skin cancer113-115. Moreover, Xpc-/- mice, defective merely in GG-NER, are not susceptible to edema or erythema, whereas Xpa-/- mice, which are defective in both GG-NER and TC-NER, are predisposed to both erythema and skin cancer induced by UV-B114,116. Thus, it can be concluded that, in GG-NER deficiency, mutations accumulate in areas of the skin exposed to UV radiation and prompt skin tumorigenesis117. By contrast, in most cases, TC-NER serves merely as an auxiliary mechanism against the cytotoxicity of DNA lesions118. Edema and erythema, rather than skin cancer, tend to be induced by UV when TC-NER is not intact. In addition, UV can cause mutations indirectly by generating ROS, such as hydrogen peroxide, hydroxyl radicals, and superoxide anions. ROS accumulation easily leads to oxidative lesions and SSBs119,120. The BER pathway is the main mechanism by which free radical lesions in DNA are repaired to prevent oxidative mutagenesis. Thus, defects in the BER pathway can also adversely affect skin121.
Liver
The liver is a highly specialized organ in vertebrates that mostly consists of hepatocytes122. Located in the right upper quadrant of the abdomen and inferior to the diaphragm in humans, the liver is believed to carry out as many as 500 different functions, typically in collaboration with other organs or systems123. This organ detoxifies various metabolites, synthesizes proteins, and produces bile, which aids in digestion by emulsifying lipids. The liver also plays a crucial role in metabolic processes, including the production of hormones, decomposition of red blood cells, and regulation of glycogen storage124-126.
Hepatic carcinoma is the sixth most prevalent cancer worldwide and the third leading cause of cancer death, of which hepatocellular carcinoma (HCC) is the principal histological subtype127. The development and progression of HCC is closely related to multiple risk factors, such as hepatitis B and C virus (HBV and HCV, respectively) infection, diabetes and obesity, aflatoxin exposure, alcohol consumption, diet contamination, and high levels of sex hormones128-130. These risk factors have been suggested to directly or indirectly foster the formation of DNA adducts and the production of ROS and/or reactive nitrogen species (RNS). For instance, hydroxyl radical has been shown to be the most damaging species that is responsible for numerous base modifications including thymine glycol, thymidine glycol, 5-(hydroxylmethyl)uracil, and 8-hydroxydeoxyguanosine131. In patients of chronic hepatitis C, higher level of 8-hydroxydeoxyguanosine in DNA extracted from liver tissue has been found132,133. These risk factors can potentially give rise to genome instability and thereby lead to neoplastic transformation. They may also target certain genes of DDR and/or DNA repair pathways. Aberrations in DNA repair proteins involved in HR, NHEJ, NER, BER, and MMR pathways, including p53, XRCC1, OGG1, ATM kinase, MRN complex, and PARP-1, have been reported to be associated with the development of HCC134-137.
Specifically, chronic HBV infection is one of the dominant risk factors for HCC development. It facilitates carcinogenesis via direct and/or indirect mechanisms. Direct effects of HBV include the interplay between HBV x protein (HBx) and host proteins. Viral DNA integration into the host genome can also arouse multitudinous mutations and chromosomal instability138. Indirect oncogenic effects of HBV involve oxidative stress and chronic inflammation. These events lead to accumulation of both genetic and epigenetic abnormalities in the liver, and result in chronic hepatitis, fibrosis, cirrhosis, tumorigenesis and tumor progression139. Chronic inflammation is characterized by the release of free radicals, the recruitment of immune cells to the liver, and the overexpression of cytokines such as interleukin 6 (IL-6), tumor necrosis factor α (TNF-α), and transforming growth factor β (TGF-β)140,141. In addition, according to cohort studies, diabetes and obesity increase the risk of HBV-associated HCC142,143. Aflatoxin B1 (AFB1), an established potent hepatocarcinogen, is metabolized to an active intermediate that binds to DNA. This process fosters a typical AGG to AGT transversion at the 249th codon of p53, and thus boosts clonal expansion of mutant cells144,145. A better understanding of relevant DDR factors in HCC can help us develop new strategies for HCC prevention and treatment.
Pancreas
The pancreas is a vital organ composed of exocrine and endocrine parts, and the exocrine part comprises more than 80% of the pancreatic mass146. The exocrine pancreas is constructed from a branching network of acinar and duct cells. To aid in protein regulation and carbohydrate digestion, the exocrine pancreas secretes digestive zymogens into the duodenum, and produces high concentrations (higher than 140 mM) of bicarbonate (HCO3–) in the pancreatic duct147,148. The endocrine components, on the other hand, include four types of specialized endocrine cells: α, β, δ, and pancreatic polypeptide (PP) cells. These cells gather into clusters called islets of Langerhans. Through secretion of different hormones into the bloodstream, the endocrine pancreas precisely regulates metabolism and glucose homeostasis146.
Pancreatic cancer is one of the most malignant and aggressive human cancers worldwide149. The average survival of pancreatic cancer patients is barely 6 months, and the five-year mortality is 97%–98%, commonly due to widespread metastasis150,151. In recent decades, studies on pancreatic cancer have detected certain types of DNA damage derived from exogenous carcinogen exposure and endogenous metabolic processes152. For example, aromatic DNA adducts and other types of DNA damage induced by smoking have been identified in the human pancreas153-155. Moreover, mutations in DDR factor genes, including BRCA2, PALB2, ATM, DNA-PK, CHK1, CHK2, FANCC, and FANCG, have been proven to be associated with chronic pancreatitis and pancreatic cancer149,156,157. In particular, mutations in BRCA2 and its partner PALB2 in HR are often involved in hereditary pancreatic cancer, indicating the key role of HR in suppressing hereditary pancreatic tumorigenesis158,159.
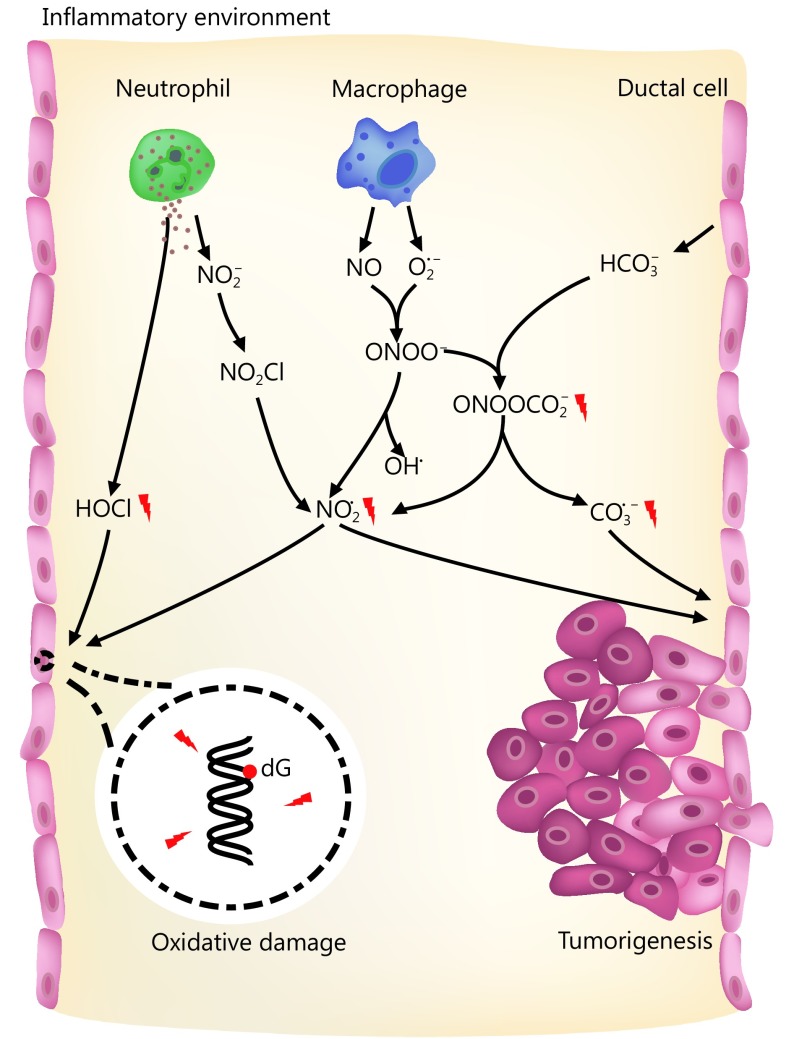
Established risk factors for pancreatic cancer include family history, aging, and smoking. Acute and chronic inflammation are also related to pancreatic cancer, especially hereditary pancreatic cancer160–162. During the development of pancreatitis, neutrophils and macrophages are recruited and activated163. These cells can generate large quantities of ROS and RNS to eradicate pathogens164,165. Many of the ROS and RNS are carcinogens that can potentially arouse structural alterations in DNA, interfere with signal transduction pathways, or affect the activity of stress-response genes166–168. For instance, the activity of myeloperoxidase (MPO) in neutrophils yields hypochlorous acid (HOCl), a potent oxidizing and halogenating agent169. In addition, MPO mediates the conversion of nitrite (NO2–) into the nitrogen dioxide radical (NO2·) through a nitryl chloride (NO2Cl) intermediate170. Activated macrophages can secrete the unstable substances nitric oxide (NO) and superoxide (O2·–), which then generate peroxynitrite (ONOO–) through interaction171. Though not a free radical itself, ONOO– homolyzes (t1/2 ~ 1 s) to form OH· and NO2·, thus causing 2-deoxyribose oxidation in DNA172. ONOO– also reacts with high concentrations of HCO3– to produce nitrosoperoxycarbonate (ONOOCO2–), a significant RNS in the pancreas. ONOOCO2– undergoes rapid homolysis (t1/2 ~ 50 ms) to generate the free radical NO2· and carbonate radicals (CO3·–), which may pose strong threats to genomic integrity and stability173. Deoxyguanosine (dG) has the least reduction potential among the four types of DNA deoxynucleosides, the produced radicals thereby preferentially cause nitration and oxidation of dG in DNA and probably of guanosine (G) in RNA164,174 (Figure 1). Low levels of oxidative lesions in DNA are quickly repaired via the BER pathway in most tissues175. However, many more RNS are formed in concentrated HCO3– in pancreatic ducts once inflammation occurs158. Excessive damage may overload the BER pathway and induce DNA SSBs. Accumulated SSB lesions can be duplicated during DNA replication and thus be converted into DSBs. Alternatively, DSBs may also be produced during BER if two SSBs in complementary strands are located close to each other3. When tissues bear gene mutations that impair HR, such as BRCA2 and PALB2 mutations, they are highly threated by these DNA lesions and may develop cancers.
1.
RNS and ROS in pancreas and the potential risk of tumorigenesis.
Colon
The colon is the terminal part of the digestive system. Unlike the small intestine, which functions primarily to absorb nutrients, the colon extracts water and salt from unabsorbed materials before they are discharged from the body. The colon is also the site of flora-aided fermentation. At the microscopic level, the colon is lined with a simple columnar epithelium with invaginations called colonic crypts. It has been estimated that each colonic crypt contains 1,500–4,900 cells, 5–6 of which are stem cells at the crypt base176. After cells are produced at the crypt base, they migrate upward along the axis of the crypt before detaching into the colonic lumen several days later177.
Due to its structural features and roles in the body, the colon is constantly challenged by tumorigenic factors, including dietary factors, gut microbiota-related factors, inflammation, and genetic mutations. As mismatches occur frequently at the replication fork in dividing cells, the MMR pathway is especially crucial in the colon47. Mutations in MMR genes typically lead to changes in the lengths of tandem nucleotide repeats located throughout the genome, producing a hypermutable phenotype known as microsatellite instability (MSI)178. MSI is found in approximately 15% of CRCs179. During replication, alterations in tandem nucleotide repeats give rise to temporary IDLs in DNA. If not corrected by MMR, these IDLs can generate frameshift or missense mutations in coding genes during subsequent rounds of DNA replication, ultimately resulting in the translation of aberrant proteins. In recent studies, MMR has also been indicated to be involved in multiple processes aside from DNA repair, such as miRNA processing and apoptosis induction180-182.
CRC is the third most prevalent cancer worldwide, leading to extensive morbidity and over 690,000 mortalities annually183. CRC progression is a gradual process in which normal colon epithelial cells (CECs) develop into polyps or colorectal adenoma and increasingly develop into colorectal adenocarcinoma184. Most CRC cases are sporadic, while approximately 30% of reported CRC cases are familial, with merely a small proportion having been well characterized185. Germline mutations in MMR genes, most commonly MLH1, MSH2, MSH6, and PMS2, have been proven to give rise to Lynch syndrome (also known as hereditary nonpolyposis colorectal cancer, HNPCC), the most prevalent form of hereditary CRC179. The majority of CRCs with high-frequency MSI (MSI-H), however, are sporadic non-Lynch syndrome cases186. These cases are caused by epigenetic inactivation of the MLH1 promoter via DNA hypermethylation. This inactivation often arises in tumors with a specific hypermethylation pathway, known as CpG island methylator phenotype (CIMP)187. Patients with MSI-H cancers, regardless of their germline or sporadic origins, have distinct pathological and clinical features, including increased numbers of tumor-infiltrating lymphocytes (TILs), frequent poor differentiation and mucinous histology, and proximal colon predominance188. Furthermore, patients with sporadic MSI-H tumors are of particular interest in epidemiological studies, especially with regard to the potential causative factors of cigarette smoking, female gender, and advanced age at diagnosis186. Thus, the defects of MMR gene in CRC could be synthetically lethal with the inhibition of genes controlling the response to oxidative DNA damage, such as DNA polymerase and PTEN-induced putative kinase 1 (PINK1)189. The synthetical lethality of MSH2 or MLH1 and DNA polymerase (POLB or POLG) causes an accumulation of 8-oxoG oxidative DNA lesions that kills cancer cells190. Likewise, inhibition of PINK1 in MMR-deficient colon cancer cell results in an elevation of ROS and the accumulation of both nuclear and mitochondrial oxidative DNA damage that limits cell proliferation191. These findings highlight targeted, mechanism-based therapeutic approaches against CRC.
There is growing evidence linking the gut microbiota, oxidative stress, and inflammation to the genetic and epigenetic alterations observed in CRC184. Advances in high-resolution next-generation sequencing (NGS) technologies have begun to facilitate investigation of the complex etiologic relationship between the microbiome and CRC. NGS analyses on human CRC tissues have enabled identification of microbial compositions related to subtypes of CRC. It is currently a consensus that the microbiota maintains a symbiotic relationship with its host and that it has various functions in both metabolism and immunity192,193. The gut microbiota and the byproducts or metabolites of the microbes therein can trigger inflammation in the colon, which accelerate colon tumorigenesis194. The microbes or products are recognized and bound by pattern recognition receptors (PRRs), such as Toll-like receptors (TLRs), on CECs and immune cells195. The binding of ligands to TLRs initiates downstream signaling cascades, leading to the activation of NF-κB. Active NF-κB regulates cell survival, proliferation, and inflammatory processes196. Cytokines produced by immune cells might also affect colon tumorigenesis through oncogene activation or gene silencing. For instance, TILs produce large amounts of IL-6, IL-17A, IL-17F, IL-21, IL-22, TNF-α, and other cytokines. These cytokines boost the proliferation of the HIT-29 and DLD-1 CRC cell lines by activating NF-κB and STAT3197. Clinically, MMR-deficient individuals have raised levels of peritumoral lymphocytes. Similar to the case for TILs, raised peritumoral lymphocyte levels are related to a higher risk of developing CRC in MMR-deficient patients198,199. Elevated levels of ROS produced by CECs and immune cells in response to TLR activation can also cause DNA damage and excessive proliferation of CECs200,201. Failure to repair ROS-mediated DNA damage could be an additional contributing mechanism to CRC progression.
Breast and ovary
Breasts and ovaries are crucial female organs closely related to reproduction and sexual characteristics. Each breast is composed of 15–25 secretory lobes embedded in subcutaneous adipose tissue. Each lobe is a compound tubular acinar gland. The acini and ducts are lined by an inner layer of cuboidal or columnar epithelial cells and an outer layer of myoepithelial cells. In females, the breasts contain mammary glands that produce and secrete milk to feed infants, while the ovaries are the female gonads. The ovary is composed of an outer cortex and an inner medulla, surrounded by a capsule. Ovaries play important roles in estrogen release, germ cell production, and hormone responses.
According to the Union for International Cancer Control World Cancer Congress in Paris in November 2016, breast cancer is the most commonly diagnosed cancer and the leading cause of cancer mortality among women worldwide, while ovarian cancer accounts for approximately 4% of global cancer cases and deaths among women183,202. Notably, functional mutations in BRCA1 confer lifetime risks of up to 90% for developing breast cancer and 40%–50% for ovarian cancer203-205. As a core DNA repair factor, BRCA1 plays crucial roles in DDR processes including cell cycle checkpoint activation and HR repair of DSBs97,206. However, it is unclear why BRCA1 defects tend to induce breast and ovarian cancers rather than other types of cancers. Knowledge of the underlying features of the breast and ovary would serve as a hotspot for understanding tumorigenesis in these two tissues.
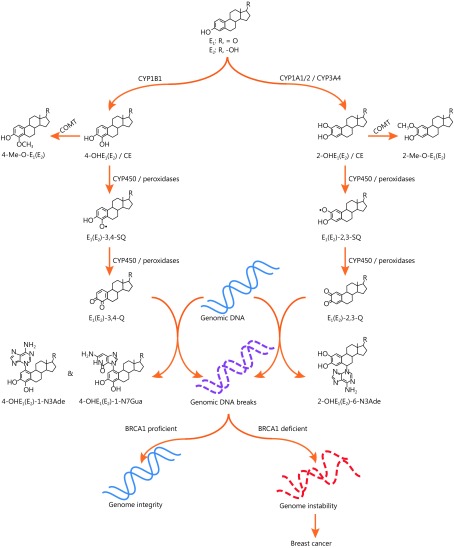
One of the most common metabolic materials in the breasts and ovaries is estrogen. Estrogen is produced and secreted by the ovaries, while the breasts are main target tissues207,208. As the major sex hormone of women, estrogen is essential for developing, regulating and maintaining the female reproductive system and secondary sex characteristics. In the human body, estrogen could be normally catabolized by catechol-o-methyltransferase (COMT) for maintaining normal cycling of the hormone209-212. However, when estrogen is present in excess, it can be oxidized into quinone radicals that induce oxidative damage in genomic DNA213-218, and repair of such lesions may require a BRCA1-dependent pathway219. An epidemiological study has also revealed that excessive exposure to estrogen is associated with a higher risk of breast cancer220. Thus, oxidative metabolites of estrogen may be the potential drivers of BRCA1-related breast cancer211 (Figure 2). Other mutations in the DNA repair genes ATM, CHEK2, PALB2, BARD1, BRIP1, RAD50, RAD51C, RAD51D, NBN, FANCM, and MRE11 may also increase the risk of breast and ovarian cancers96,221.
2.
Estrogen oxidation-induced DNA damage and breast cancer.
In parallel, estrogen acts on its two receptors, estrogen receptor α (ERα) and estrogen receptor β (ERβ). The binding of estrogen to its receptors regulates the expression of many genes in a well-organized way222. In particular, it activates the transcription of estrogen-responsive genes, such as genes of growth factors and cell cycle regulating cyclins, and proto-oncogenes, including c-MYC, c-FOS, and HER2/Neu223. The activated genes serve to stimulate cell proliferation and suppress apoptosis224. Estrogen also binds to membrane-bound G-protein-coupled estrogen receptor (GPER, GPR30) to rapidly exert nongenomic effects by activating second messenger systems225. When dysregulated, hyperactive estrogen signaling induces extortionate proliferation, giving rise to DNA damage accumulation and ultimately leading to malignant transformation17. Of note, estrogen signaling has been shown to interact with DDR and DNA repair pathways by regulating key effectors in DDR, including DNA-PKcs, ATM, ataxia telangiectasia and Rad3-related (ATR), BRCA1, BRCA2, and p53, and effector kinases in DNA repair mechanisms226. For instance, ERα may bind DNA-PKcs, stabilizing them and activating DDR through the NHEJ pathway227. ERα downregulates ATM by inhibiting ATM kinase expression via activation of microRNA-18a and microRNA-106a228. Moreover, the ATR protein is negatively regulated by ERα. ERα suppresses ATR activation, ATR-Chk1 signaling at the G2/M checkpoint, and the interaction between DNA Topo II binding protein 1 (TopBP1) and ATR at DNA damage sites. Thus, ER signaling affects not only estrogen-related carcinogenesis but also the processing of other genotoxic insults in estrogen-responsive tissues226.
In the past, DDR defects have been exploited to develop many commonly used chemo- and radio-therapies for breast and ovarian cancer. For example, platinum salts arouse DNA ICLs, the lesions recognized by the DDR and repaired by a combination of HR and NER. These agents can be effective in patients with ovarian carcinomas that commonly harbor defects in HR, especially high-grade serous ovarian cancer204. Today, an alternative approach to traditional therapy is to directly target specific components in DDR pathways through pharmacologic design. Recent years, PARP inhibitors are emerging as a promising new class of chemotherapeutic agents particularly effective against tumors bearing defective BRCA1 and BRCA2229,230. Although nonuniform explanations exist for anticancer mechanism of PARP inhibitors, all the studies highlight the importance of processes of DDR in tissues231-233. The generation of PARP inhibitors can be seen as a milestone of cancer treatment, which not only considerably improves the prognosis of breast and ovarian cancer, but also provides a novel idea for mechanism exploration and target therapy.
Hematopoietic system
The hematopoietic system has been well studied in terms of its development. All blood cell lineages derive from a limited number of hematopoietic stem cells (HSCs) through an actively amplifying progenitor compartment234. As it includes a pool of rapidly proliferating and regenerating cells, the hematopoietic system is also one of the most sensitive and vulnerable systems towards endogenous and exogenous genotoxic insults. HSCs originate from the aorta-gonad-mesonephros (AGM) region, travel to certain anatomic sites, including the placenta, spleen, liver, and lymph nodes of the developing fetus, and ultimately localize to the bone marrow cavity during late embryogenesis. After birth, fetal HSCs gradually develop into quiescent adult HSCs and remain in bone marrow throughout their lifetime.
It has been indicated that HSCs and other tissue-specific stem cells should have cellular and genomic protective mechanisms to maintain their functional potential throughout their lifespans. Indeed, increasing evidence suggests that HSCs are armed with multiple protective properties responsible for preserving their activity. For instance, the expression of telomerase and the dormant nature of HSCs help to prevent the uncapping of telomeres or the introduction of aberrations during replication235-237. ROS in HSCs are maintained at relatively low levels because of the low metabolic activity in these cells238. It is noteworthy that ATM-deficient HSCs contain increased level of ROS and display an overall function decline over time, resulting in eventual hematopoietic failure239. On clinic, isocitrate dehydrogenase-1 gene (IDH1) deficiency is a common driver of acute myeloid leukemia (AML). Mechanically, mutant IDH1 downregulates the expression of the DNA damage sensor ATM by altering its histone methylation. Decreased ATM results in impaired DNA repair and increased sensitivity to DNA damage, which reduces HSC self-renewal240.
Notably, the functions of prosurvival genes in Bcl-2 family and of p53-mediated DDR processes have been emphasized in HSCs for IR injury repair241,242. In quiescent HSCs, heightened expression of prosurvival genes, such as Bcl2, Bcl-Xl, Mcl1, and A1, inhibits cell death prompted by p53. In these cases, the proapoptotic genes Bax, Noxa, and Puma permit p21 to participate in a transient growth-arrest response for DNA repair243. Different from quiescent HSCs that employ the error-prone NHEJ pathway upon DNA damage, proliferating HSCs use the high-fidelity HR pathway to repair DSBs243. A study involving short-term culture of isolated quiescent HSCs revealed that γ-H2AX-marked DSBs are markedly reduced after HSCs enter the cell cycle, indicating that proliferating HSCs repair DSBs better than quiescent HSCs244. Also, this may be the reason why patients who undergo chemotherapy or radiotherapy tend to develop leukemias and lymphomas because the use of error-prone DNA repair in quiescent HSCs would potentially cause a large number of DNA lesions. The major outcome of the DDR in fetal HSCs is an increased level of apoptosis and cell elimination promoted by apoptosis-stimulating of p53 protein 1 (ASPP1). In contrast, adult HSCs demonstrate a very different response to IR, with DNA repair and overt cell survival being the principal outcomes243. This difference in response might be attributable to the different roles of HSCs during these stages. During embryogenesis, the vital goal of hematopoiesis is to amplify the stem cells and protect their genomic integrity to establish an HSC pool. This ensures blood homeostasis for the lifetime of the organism. In adults, however, the major function of the HSC compartment is to preserve blood homeostasis and quick response to hematopoietic needs associated with conditions such as infection and blood loss245. Moreover, HSCs are more resistant to IR than their downstream progeny, which prevents the exhaustion of the HSC pool246.
Due to its vigorous, sensitive, and amorphous properties, the hematopoietic system generally responses well to DDR inhibition treatments. Still, because of the significant diversity lying in blood cells of different stages and different types of hematopoietic cancers, DDR inhibitors vary in clinical indications. In a recent study, scientists find that a novel ATR kinase inhibitor AZD6738 leads to an accumulation of unrepaired DNA damage in p53- or ATM-defective chronic lymphocytic leukemia (CLL) cells. The synthetic lethality is achieved by mitotic catastrophe caused by defective cell cycle checkpoints, resulting in a selective cytotoxicity to both p53- and ATM-defective CLL cells247. Another interesting observation comes from AML, myelodysplastic syndrome, juvenile/chronic myelomonocytic leukemia, and T-cell acute lymphoblastic leukemia harboring KRAS mutations. The oncogenic KRAS causes a shift in DSB repair from the canonical-NHEJ pathway to a preferred use of the more error-prone alternative-NHEJ (alt-NHEJ) pathway248. Thus, targeting the components of the alt-NHEJ pathway could sensitize KRAS-mutated leukemic cells to standard chemotherapeutics and represents a promising approach for inducing synthetic lethality in cancer cells.
Conclusions
DNA damage and repair are inevitable and indispensable cellular processes that serve prominently to determine the fates of cells, organs, systems, and even the whole body. When DNA damage occurs, cells respond correspondingly. Though general mechanisms and DDR pathways are shared, the fates of cells greatly rely on tissue-specific processes. Aside from the tissue-specific expression of DDR conserved genes, epigenetic modulation, cytokines, hormones and cellular niches also precisely regulate cell fate in a tissue-specific manner. The combination of intrinsic properties and regulatory factors accounts for the specificity and precision of DDR.
DDR serves as not only a guardian of genomic integrity, but also an oncogene-inducible physiological barrier against tumorigenesis and early tumor progression. Mutations in different DDR pathways tend to arouse very different clinical events including tissue-specific tumorigenesis described above (Table 1). In the past decade, plenty of promising DDR inhibitors have been developed on clinic. The inhibition of DDR in a specific tissue or microenvironment preferentially kills cancer cells while spares normal cells, thus providing significant patient benefits over conventional chemotherapies249. Advances in NGS technologies would also facilitate the identification of patient subgroups or particular cancer types with DDR defects, and better instruct the personalized therapy250.
1.
Tissue specific DDR and tumorigenesis
| Tissue | DNA damaging agents | DNA damage | Major DDR | Common mutations in DDR and repair pathway | Tumorigenesis or related diseases |
| UV, ultraviolet; NER, nucleotide excision repair; XP, xeroderma pigmentosum; CS, Cockayne syndrome; TTD, trichothiodystrophy; ROS, reactive oxygen species; SSB, single strand break; BER, base excision repair; HBV, hepatitis B virus; HCV, hepatitis C virus; AFB1, Aflatoxin B1; RNS, reactive nitrogen species; HR, homologous recombination; NHEJ, non-homologous end joining; MMR, mismatch repair; HCC, hepatocellular carcinoma; IDL, insertion-deletion loop; MSI, microsatellite instability; CRC, colorectal cancer; CIMP, CpG island methylator phenotype; CE, catechol estrogen; SQ, semiquinone; Q, quinone; DSB, double strand break; IR, ionizing radiation; AML, acute myeloid leukemia; CLL, chronic lymphocytic leukemia. | |||||
| Skin | Sunlight (e.g., UV-B, UV-A) | Direct DNA photolesions (e.g., (6,4)-photoproducts, cyclobutane pyrimidine dimers) | NER | XPA, ERCC3 (XPB), XPC, ERCC2 (XPD), DDB2 (XPE), ERCC4 (XPF), ERCC5 (XPG), POLH (XPV) | XP |
| ERCC8 (CSA), ERCC6 (CSB), XPB, XPD, XPG | CS | ||||
| TTDA, XPB, XPD | TTD | ||||
| XPA, XPC, XPE, XPF | Cutaneous neoplasms | ||||
| ROS (e.g., hydrogen peroxide, hydroxyl radicals, superoxide anions) | Oxidative lesions, SSBs | BER | LIG1 | Skin maladies | |
| Liver | HBV and HCV infection, aflatoxins (e.g., AFB1), ROS, RNS | DNA adducts, oxidative lesions, genetic and epigenetic abnormalities | HR, NHEJ, NER, BER, MMR | p53, XRCC1, OGG1, ATM, MRE11A, RAD50, NBN, PARP1 | HCC |
| Pancreas | ROS (e.g., HOCl, O2·–, NO), RNS (e.g., NO2·, ONOOCO2–) | Aromatic DNA adducts, oxidative lesions (e.g., 2-deoxyribose oxidation, nitration and oxidation of dG), SSBs, DSBs | BER, HR | BRCA2, PALB2, ATM, DNA-PK, CHK1, CHK2, FANCC, FANCG | Chronic pancreatitis, pancreatic cancer |
| Colon | Replication errors, gut microbiota, inflammation, ROS | Mismatches, IDLs, frameshift or missense mutations | MMR | MLH1, MSH2, MSH6, PMS2 | MSI, CRCs (e.g., Lynch syndrome, CIMP) |
| Breast and ovary | Estrogens and their metabolites (CE-SQs, CE-Qs) | Oxidative lesions, depurinating adducts (e.g., 4-OHE-1-N3Ade, 4-OHE-1-N7Gua, 2-OHE-6-N3Ade), DSBs | HR | BRCA1, ATM, CHEK2, PALB2, BARD1, BRIP1, RAD50, RAD51C, RAD51D, NBN, FANCM, MRE11 | Breast cancer, ovarian cancer |
| Hematopoietic system | IR exposure, ROS, replication errors | DSBs, SSBs | HR, NHEJ, BER | IDH1, p53, Bcl2, Bcl-Xl, Mcl1, A1, Bax, Noxa, Puma, p21 | AML, CLL |
Still, the relationship between reported mutations and dysfunctions of the DDR proteins have yet to be defined. Comprehensive knowledge regarding the tissue-specific DNA damage and repair processes in different species, developmental stages, and cell cycle phases remains much limited. Prompt repair and cellular survival may not be the ultimate goal for each cell. Instead, tissues with distinct features and functions may prioritize different processes to maximize the overall wholesome state of the body. For instance, altruistic suicide of intestinal stem cells in response to DNA damage could prevent the transmission of precancerous mutations, which potentially explains the rarity of intestinal cancers compared to colonic neoplasms245. Why are certain pathways more sensitive and responsive towards DNA damage in some tissues than in others? What are the decisive elements among tissues? How do certain mutations or functional defects in DDR gradually develop into adverse consequences? The answers to these questions will provide profound insights into the mechanisms underlying the progression of tumorigenesis and novel clues for the development of effective anticancer treatments.
Acknowledgements
We are grateful to Dr. Ning Zhang for the insightful comments and suggestions on the manuscript. This work was supported by the National Natural Science Foundation of China (Grant No. 81622035, 81672610, and 81521002).
Conflict of interest statement
No potential conflicts of interest are disclosed.
References
- 1.Lindahl T Instability and decay of the primary structure of DNA. Nature. 1993;362:709–15. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 2.Sancar A, Lindsey-Boltz LA, Ünsal-Kaçmaz K, Linn S Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 3.Jackson SP, Bartek J The DNA-damage response in human biology and disease. Nature. 2009;461:1071–8. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borrego-Soto G, Ortiz-López R, Rojas-Martínez A Ionizing radiation-induced DNA injury and damage detection in patients with breast cancer. Genet Mol Biol. 2015;38:420–32. doi: 10.1590/S1415-475738420150019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sinha RP, Häder DP Uv-induced DNA damage and repair: a review. Photochem Photobiol Sci. 2002;1:225–36. doi: 10.1039/b201230h. [DOI] [PubMed] [Google Scholar]
- 6.Sun Y, Nelson PS Molecular pathways: involving microenvironment damage responses in cancer therapy resistance. Clin Cancer Res. 2012;18:4019–25. doi: 10.1158/1078-0432.CCR-11-0768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Straub KM, Burlingame AL Carcinogen binding to DNA. Biomed Mass Spectrom. 1981;8:431–5. doi: 10.1002/(ISSN)1096-9888. [DOI] [PubMed] [Google Scholar]
- 8.Polo SE, Jackson SP Dynamics of DNA damage response proteins at DNA breaks: a focus on protein modifications. Genes Dev. 2011;25:409–33. doi: 10.1101/gad.2021311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tidball AM, Neely MD, Chamberlin R, Aboud AA, Kumar KK, Han BY, et al Genomic instability associated with p53 knockdown in the generation of huntington’s disease human induced pluripotent stem cells. PLoS One. 2016;11:e0150372. doi: 10.1371/journal.pone.0150372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Musich PR, Zou Y Genomic instability and DNA damage responses in progeria arising from defective maturation of prelamin a. Aging. 2009;1:28–37. doi: 10.18632/aging.v1i1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lombardi S, Fuoco I, Di Fluri G, Costa F, Ricchiuti A, Biondi G, et al Genomic instability and cellular stress in organ biopsies and peripheral blood lymphocytes from patients with colorectal cancer and predisposing pathologies. Oncotarget. 2015;6:14852–64. doi: 10.18632/oncotarget.4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou BBS, Elledge SJ The DNA damage response: putting checkpoints in perspective. Nature. 2000;408:433–9. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- 13.Maréchal A, Zou L DNA damage sensing by the atm and atr kinases. Cold Spring Harb Perspect Biol. 2013;5:a012716. doi: 10.1101/cshperspect.a012716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shiloh Y ATM and related protein kinases: Safeguarding genome integrity. Nat Rev Cancer. 2003;3:155–68. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- 15.Gorgoulis VG, Vassiliou LVF, Karakaidos P, Zacharatos P, Kotsinas A, Liloglou T, et al Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434:907–13. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- 16.Bartkova J, Hamerlik P, Stockhausen MT, Ehrmann J, Hlobilkova A, Laursen H, et al Replication stress and oxidative damage contribute to aberrant constitutive activation of DNA damage signalling in human gliomas. Oncogene. 2010;29:5095–102. doi: 10.1038/onc.2010.249. [DOI] [PubMed] [Google Scholar]
- 17.Halazonetis TD, Gorgoulis VG, Bartek J An oncogene-induced DNA damage model for cancer development. Science. 2008;319:1352–5. doi: 10.1126/science.1140735. [DOI] [PubMed] [Google Scholar]
- 18.Walden H, Deans AJ The fanconi anemia DNA repair pathway: Structural and functional insights into a complex disorder. Annu Rev Biophys. 2014;43:257–78. doi: 10.1146/annurev-biophys-051013-022737. [DOI] [PubMed] [Google Scholar]
- 19.Lehnman IR DNA ligase: structure, mechanism, and function. Science. 1974;186:790–7. doi: 10.1126/science.186.4166.790. [DOI] [PubMed] [Google Scholar]
- 20.Gunz D, Hess MT, Naegeli H Recognition of DNA adducts by human nucleotide excision repair. Evidence for a thermodynamic probing mechanism. J Biol Chem. 1996;271:25089–98. doi: 10.1074/jbc.271.41.25089. [DOI] [PubMed] [Google Scholar]
- 21.Wood RD, Araujo SJ, Ariza RR, Batty DP, Biggerstaff M, Evans E, et al DNA damage recognition and nucleotide excision repair in mammalian cells. Cold Spring Harb Symp Quant Biol. 2000;65:173–82. doi: 10.1101/sqb.2000.65.173. [DOI] [PubMed] [Google Scholar]
- 22.Hanawalt PC, Spivak G Transcription-coupled DNA repair: two decades of progress and surprises. Nat Rev Mol Cell Biol. 2008;9:958–70. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- 23.Ford JM Regulation of DNA damage recognition and nucleotide excision repair: another role for p53. Mutat Res. 2005;577:195–202. doi: 10.1016/j.mrfmmm.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 24.Hanawalt PC Transcription-coupled repair and human disease. Science. 1994;266:1957–8. doi: 10.1126/science.7801121. [DOI] [PubMed] [Google Scholar]
- 25.Kuraoka I, Suzuki K, Ito S, Hayashida M, Kwei JSM, Ikegami T, et al RNA polymerase II bypasses 8-oxoguanine in the presence of transcription elongation factor TFIIS. DNA Repair (Amst) 2007;6:841–51. doi: 10.1016/j.dnarep.2007.01.014. [DOI] [PubMed] [Google Scholar]
- 26.Compe E, Egly JM TFIIH: when transcription met DNA repair. Nat Rev Mol Cell Biol. 2012;13:343–54. doi: 10.1038/nrm3350. [DOI] [PubMed] [Google Scholar]
- 27.Kirschner K, Melton DW Multiple roles of the ERCC1-XPF endonuclease in DNA repair and resistance to anticancer drugs. Anticancer Res. 2010;30:3223–32. [PubMed] [Google Scholar]
- 28.Mocquet V, Lainé JP, Riedl T, Zhou YJ, Lee MY, Egly JM Sequential recruitment of the repair factors during NER: the role of XPG in initiating the resynthesis step. EMBO J. 2008;27:155–67. doi: 10.1038/sj.emboj.7601948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Caldecott KW DNA single-strand break repair. Exp Cell Res. 2014;329:2–8. doi: 10.1016/j.yexcr.2014.08.027. [DOI] [PubMed] [Google Scholar]
- 30.Krokan HE, Bjørås M Base excision repair. Cold Spring Harb Perspect Biol. 2013;5:a012583. doi: 10.1101/cshperspect.a012583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mol CD, Izumi T, Mitra S, Tainer JA DNA-bound structures and mutants reveal abasic DNA binding by APE1 DNA repair and coordination. Nature. 2000;403:451–6. doi: 10.1038/35000249. [DOI] [PubMed] [Google Scholar]
- 32.Caldecott KW DNA damage responses and neurological disease. Preface. DNA Repair (Amst) 2008;7:1009. doi: 10.1016/j.dnarep.2008.04.011. [DOI] [PubMed] [Google Scholar]
- 33.Kim YJ, Wilson III DM Overview of base excision repair biochemistry. Curr Mol Pharmacol. 2012;5:3–13. doi: 10.2174/1874467211205010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lindahl T, Barnes DE Repair of endogenous DNA damage. Cold Spring Harb Symp Quant Biol. 2000;65:127–33. doi: 10.1101/sqb.2000.65.127. [DOI] [PubMed] [Google Scholar]
- 35.Matsumoto Y, Kim K, Hurwitz J, Gary R, Levin DS, Tomkinson AE, et al Reconstitution of proliferating cell nuclear antigen-dependent repair of apurinic/apyrimidinic sites with purified human proteins. J Biol Chem. 1999;274:33703–8. doi: 10.1074/jbc.274.47.33703. [DOI] [PubMed] [Google Scholar]
- 36.Friedberg EC, Bardwell AJ, Bardwell L, Feaver WJ, Kornberg RD, Svejstrup JQ, et al Nucleotide excision repair in the yeast Saccharomyces cerevisiae: its relationship to specialized mitotic recombination and RNA polymerase II basal transcription . Philos Trans R Soc Lond B Biol Sci. 1995;347:63–8. doi: 10.1098/rstb.1995.0010. [DOI] [PubMed] [Google Scholar]
- 37.Smith ML, Ford JM, Hollander MC, Bortnick RA, Amundson SA, Seo YR, et al p53-mediated DNA repair responses to UV radiation: studies of mouse cells lacking p53, p21, and/or gadd45 genes . Mol Cell Biol. 2000;20:3705–14. doi: 10.1128/MCB.20.10.3705-3714.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hwang BJ, Ford JM, Hanawalt PC, Chu G Expression of the p48 xeroderma pigmentosum gene is p53-dependent and is involved in global genomic repair . Proc Natl Acad Sci U S A. 1999;96:424–8. doi: 10.1073/pnas.96.2.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Offer H, Zurer I, Banfalvi G, Reha'k M, Falcovitz A, Milyavsky M, et al p53 modulates base excision repair activity in a cell cycle-specific manner after genotoxic stress. Cancer Res. 2001;61:88–96. [PubMed] [Google Scholar]
- 40.Zhou JM, Ahn J, Wilson SH, Prives C A role for p53 in base excision repair. EMBO J. 2001;20:914–23. doi: 10.1093/emboj/20.4.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seo YR, Fishel ML, Amundson S, Kelley MR, Smith ML Implication of p53 in base excision DNA repair: in vivo evidence . Oncogene. 2002;21:731–7. doi: 10.1038/sj.onc.1205129. [DOI] [PubMed] [Google Scholar]
- 42.Jayaraman L, Murthy KG, Zhu C, Curran T, Xanthoudakis S, Prives C Identification of redox/repair protein ref-1 as a potent activator of p53. Genes Dev. 1997;11:558–70. doi: 10.1101/gad.11.5.558. [DOI] [PubMed] [Google Scholar]
- 43.Seo YR, Jung HJ The potential roles of p53 tumor suppressor in nucleotide excision repair (NER) and base excision repair (BER) Exp Mol Med. 2004;36:505–9. doi: 10.1038/emm.2004.64. [DOI] [PubMed] [Google Scholar]
- 44.Jiricny J Postreplicative mismatch repair. Cold Spring Harb Perspect Biol. 2013;5:a012633. doi: 10.1101/cshperspect.a012633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Genschel J, Littman SJ, Drummond JT, Modrich P Isolation of mutSβ from human cells and comparison of the mismatch repair specificities of mutSβ and mutSα. J Biol Chem. 1998;273:19895–901. doi: 10.1074/jbc.273.31.19895. [DOI] [PubMed] [Google Scholar]
- 46.Masuda K, Banno K, Yanokura M, Kobayashi Y, Kisu I, Ueki A, et al Relationship between DNA mismatch repair deficiency and endometrial cancer. Mol Biol Int. 2011;2011:256063. doi: 10.4061/2011/256063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dion V Tissue specificity in DNA repair: lessons from trinucleotide repeat instability. Trends Genet. 2014;30:220–9. doi: 10.1016/j.tig.2014.04.005. [DOI] [PubMed] [Google Scholar]
- 48.Le DT, Durham JN, Smith KN, Wang H, Bartlett BR, Aulakh LK, et al Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science. 2017;357:409–13. doi: 10.1126/science.aan6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lengauer C, Kinzler KW, Vogelstein B Genetic instabilities in human cancers. Nature. 1998;396:643–9. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 50.Smyrk TC, Watson P, Kaul K, Lynch HT Tumor-infiltrating lymphocytes are a marker for microsatellite instability in colorectal carcinoma. Cancer. 2001;91:2417–22. doi: 10.1002/(ISSN)1097-0142. [DOI] [PubMed] [Google Scholar]
- 51.Moore JK, Haber JE Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in saccharomyces cerevisiae. Mol Cell Biol. 1996;16:2164–73. doi: 10.1128/MCB.16.5.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ohle C, Tesorero R, Schermann G, Dobrev N, Sinning I, Fischer T Transient RNA-DNA hybrids are required for efficient double-strand break repair. Cell. 2016;167:1001–13. doi: 10.1016/j.cell.2016.10.001. [DOI] [PubMed] [Google Scholar]
- 53.Lieber MR The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Blier PR, Griffith AJ, Craft J, Hardin JA Binding of Ku protein to DNA. Measurement of affinity for ends and demonstration of binding to nicks. J Biol Chem. 1993;268:7594–601. [PubMed] [Google Scholar]
- 55.Gottlieb TM, Jackson SP The DNA-dependent protein kinase: requirement for DNA ends and association with Ku antigen. Cell. 1993;72:131–42. doi: 10.1016/0092-8674(93)90057-W. [DOI] [PubMed] [Google Scholar]
- 56.Davis AJ, Chen BPC, Chen DJ DNA-PK: a dynamic enzyme in a versatile DSB repair pathway. DNA Repair (Amst) 2014;17:21–9. doi: 10.1016/j.dnarep.2014.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cottarel J, Frit P, Bombarde O, Salles B, Negrel A, Bernard S, et al A noncatalytic function of the ligation complex during nonhomologous end joining. J Cell Biol. 2013;200:173–86. doi: 10.1083/jcb.201203128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hastings PJ, Lupski JR, Rosenberg SM, Ira G Mechanisms of change in gene copy number. Nat Rev Genet. 2009;10:551–64. doi: 10.1038/nrg2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bunting SF, Nussenzweig A End-joining, translocations and cancer. Nat Rev Cancer. 2013;13:443–54. doi: 10.1038/nrc3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Park DJ, Lesueur F, Nguyen-Dumont T, Pertesi M, Odefrey F, Hammet F, et al Rare mutations in XRCC2 increase the risk of breast cancer . Am J Hum Genet. 2012;90:734–9. doi: 10.1016/j.ajhg.2012.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stephens PJ, Tarpey PS, Davies H, Van Loo P, Greenman C, Wedge DC, et al The landscape of cancer genes and mutational processes in breast cancer. Nature. 2012;486:400–4. doi: 10.1038/nature11017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thompson ER, Doyle MA, Ryland GL, Rowley SM, Choong DYH, Tothill RW, et al Exome sequencing identifies rare deleterious mutations in DNA repair genes FANCC and BLM as potential breast cancer susceptibility alleles . PLoS Genet. 2012;8:e1002894. doi: 10.1371/journal.pgen.1002894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ohsawa R, Seol JH, Tyler JK At the intersection of non-coding transcription, DNA repair, chromatin structure, and cellular senescence. Front Genet. 2013;4:136. doi: 10.3389/fgene.2013.00136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mimitou EP, Symington LS Nucleases and helicases take center stage in homologous recombination. Trends Biochem Sci. 2009;34:264–72. doi: 10.1016/j.tibs.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 65.Pellegrini L, Yu DS, Lo T, Anand S, Lee M, Blundell TL, et al Insights into DNA recombination from the structure of a RAD51-BRCA2 complex. Nature. 2002;420:287–93. doi: 10.1038/nature01230. [DOI] [PubMed] [Google Scholar]
- 66.Swanson RL, Morey NJ, Doetsch PW, Jinks-Robertson S Overlapping specificities of base excision repair, nucleotide excision repair, recombination, and translesion synthesis pathways for DNA base damage in Saccharomyces cerevisiae . Mol Cell Biol. 1999;19:2929–35. doi: 10.1128/MCB.19.4.2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hoeijmakers JH Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–74. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 68.Smirnova M, Klein HL Role of the error-free damage bypass postreplication repair pathway in the maintenance of genomic stability. Mutat Res. 2003;532:117–35. doi: 10.1016/j.mrfmmm.2003.08.026. [DOI] [PubMed] [Google Scholar]
- 69.Huertas P, Cortés-Ledesma F, Sartori AA, Aguilera A, Jackson SP CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature. 2008;455:689–92. doi: 10.1038/nature07215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stante M, Minopoli G, Passaro F, Raia M, Vecchio LD, Russo T Fe65 is required for Tip60-directed histone H4 acetylation at DNA strand breaks. Proc Natl Acad Sci USA. 2009;106:5093–8. doi: 10.1073/pnas.0810869106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Esashi F, Christ N, Gannon J, Liu YL, Hunt T, Jasin M, et al CDK-dependent phosphorylation of BRCA2 as a regulatory mechanism for recombinational repair. Nature. 2005;434:598–604. doi: 10.1038/nature03404. [DOI] [PubMed] [Google Scholar]
- 72.Patel KJ, Yu VPCC, Lee H, Corcoran A, Thistlethwaite FC, Evans MJ, et al Involvement of brca2 in DNA repair. Mol Cell. 1998;1:347–57. doi: 10.1016/S1097-2765(00)80035-0. [DOI] [PubMed] [Google Scholar]
- 73.Moldovan GL, D'Andrea AD How the fanconi anemia pathway guards the genome. Annu Rev Genet. 2009;43:223–49. doi: 10.1146/annurev-genet-102108-134222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ali AM, Singh TR, Meetei AR FANCM-FAAP24 and FANCJ: FA proteins that metabolize DNA. Mutat Res. 2009;668:20–6. doi: 10.1016/j.mrfmmm.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Alpi AF, Pace PE, Babu MM, Patel KJ Mechanistic insight into site-restricted monoubiquitination of FANCD2 by Ube2t, FANCL, and FANCI. Mol Cell. 2008;32:767–77. doi: 10.1016/j.molcel.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 76.Dorsman JC, Levitus M, Rockx D, Rooimans MA, Oostra AB, Haitjema A, et al Identification of the fanconi anemia complementation group I gene, FANCI. Cell Oncol. 2017;29:211–8. doi: 10.1155/2007/151968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sims AE, Spiteri E, Sims III RJ, Arita AG, Lach FP, Landers T, et al FANCI is a second monoubiquitinated member of the Fanconi anemia pathway. Nat Struct Mol Biol. 2007;14:564–7. doi: 10.1038/nsmb1252. [DOI] [PubMed] [Google Scholar]
- 78.Long DT, Räschle M, Joukov V, Walter JC Mechanism of RAD51-dependent DNA interstrand cross-link repair. Science. 2011;333:84–7. doi: 10.1126/science.1204258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ciccia A, Elledge SJ The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Godthelp BC, Van Buul PP, Jaspers NGJ, Elghalbzouri-Maghrani E, Van Duijn-Goedhart A, Arwert F, et al Cellular characterization of cells from the fanconi anemia complementation group, FA-D1/BRCA2. Mutat Res. 2006;601:191–201. doi: 10.1016/j.mrfmmm.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 81.Schwab RA, Nieminuszczy J, Shin-ya K, Niedzwiedz W FANCJ couples replication past natural fork barriers with maintenance of chromatin structure. J Cell Biol. 2013;201:33–48. doi: 10.1083/jcb.201208009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Xia B, Dorsman JC, Ameziane N, De Vries Y, Rooimans MA, Sheng Q, et al Fanconi anemia is associated with a defect in the BRCA2 partner PALB2. Nat Genet. 2007;39:159–61. doi: 10.1038/ng1942. [DOI] [PubMed] [Google Scholar]
- 83.Vaz F, Hanenberg H, Schuster B, Barker K, Wiek C, Erven V, et al Mutation of the RAD51C gene in a fanconi anemia-like disorder . Nat Genet. 2010;42:406–9. doi: 10.1038/ng.570. [DOI] [PubMed] [Google Scholar]
- 84.Schlacher K, Wu H, Jasin M A distinct replication fork protection pathway connects fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer cell. 2012;22:106–16. doi: 10.1016/j.ccr.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nakanishi K, Cavallo F, Perrouault L, Giovannangeli C, Moynahan ME, Barchi M, et al Homology-directed fanconi anemia pathway cross-link repair is dependent on DNA replication. Nat Struct Mol Biol. 2011;18:500–3. doi: 10.1038/nsmb.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Englander EW DNA damage response in peripheral nervous system: coping with cancer therapy-induced DNA lesions. DNA Repair (Amst) 2013;12:685–90. doi: 10.1016/j.dnarep.2013.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gudkov AV, Komarova EA The role of p53 in determining sensitivity to radiotherapy. Nat Rev Cancer. 2003;3:117–29. doi: 10.1038/nrc992. [DOI] [PubMed] [Google Scholar]
- 88.Barker N, Van Es JH, Kuipers J, Kujala P, Van Den Born M, Cozijnsen M, et al Identification of stem cells in small intestine and colon by marker gene lgr5 . Nature. 2007;449:1003–7. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 89.Chao EC, Lipkin SM Molecular models for the tissue specificity of DNA mismatch repair-deficient carcinogenesis. Nucleic Acids Res. 2006;34:840–52. doi: 10.1093/nar/gkj489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mayor S Researcher from study alleging link between MMR and autism warns of measles epidemic. BMJ. 2003;327:1069. doi: 10.1136/bmj.327.7423.1069. [DOI] [Google Scholar]
- 91.D'Errico M, Lemma T, Calcagnile A, De Santis LP, Dogliotti E Cell type and DNA damage specific response of human skin cells to environmental agents. Mutat Res. 2007;614:37–47. doi: 10.1016/j.mrfmmm.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 92.Al-Tassan N, Chmiel NH, Maynard J, Fleming N, Livingston AL, Williams GT, et al Inherited variants of MYH associated with somatic G:C→T:A mutations in colorectal tumors . Nat Genet. 2002;30:227–32. doi: 10.1038/ng828. [DOI] [PubMed] [Google Scholar]
- 93.DiGiovanna JJ, Kraemer KH Shining a light on xeroderma pigmentosum. J Invest Dermatol. 2012;132:785–96. doi: 10.1038/jid.2011.426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lynch HT, De La Chapelle A Genetic susceptibility to non-polyposis colorectal cancer. J Med Genet. 1999;36:801–18. [PMC free article] [PubMed] [Google Scholar]
- 95.Takashima H, Boerkoel CF, John J, Saifi GM, Salih MAM, Armstrong D, et al Mutation of TDP1, encoding a topoisomerase I-dependent DNA damage repair enzyme, in spinocerebellar ataxia with axonal neuropathy . Nat Genet. 2002;32:267–72. doi: 10.1038/ng987. [DOI] [PubMed] [Google Scholar]
- 96.Economopoulou P, Dimitriadis G, Psyrri A Beyond BRCA: new hereditary breast cancer susceptibility genes . Cancer Treat Rev. 2015;41:1–8. doi: 10.1016/j.ctrv.2014.10.008. [DOI] [PubMed] [Google Scholar]
- 97.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SAJR, Behjati S, Biankin AV, et al Signatures of mutational processes in human cancer. Nature. 2013;500:415–21. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kohler SW, Provost GS, Fieck A, Kretz PL, Bullock WO, Putman DL, et al Analysis of spontaneous and induced mutations in transgenic mice using a lambda ZAP/lacl shuttle vector . Environ Mol Mutagen. 1991;18:316–21. doi: 10.1002/(ISSN)1098-2280. [DOI] [PubMed] [Google Scholar]
- 99.Kovtun IV, Liu Y, Bjoras M, Klungland A, Wilson SH, McMurray CT OGG1 initiates AGE-dependent CAG trinucleotide expansion in somatic cells. Nature. 2007;447:447–52. doi: 10.1038/nature05778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Karahalil B, Hogue BA, De Souza-Pinto NC, Bohr VA Base excision repair capacity in mitochondria and nuclei: tissue-specific variations. FASEB J. 2002;16:1895–902. doi: 10.1096/fj.02-0463com. [DOI] [PubMed] [Google Scholar]
- 101.D'Orazio J, Jarrett S, Amaro-Ortiz A, Scott T UV radiation and the skin. Int J Mol Sci. 2013;14:12222–48. doi: 10.3390/ijms140612222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Fuchs E Scratching the surface of skin development. Nature. 2007;445:834–42. doi: 10.1038/nature05659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hoeijmakers JHJ DNA damage, aging, and cancer. N Engl J Med. 2009;361:1475–85. doi: 10.1056/NEJMra0804615. [DOI] [PubMed] [Google Scholar]
- 104.Sklar LR, Almutawa F, Lim HW, Hamzavi I Effects of ultraviolet radiation, visible light, and infrared radiation on erythema and pigmentation: a review. Photochem Photobiol Sci. 2012;12:54–64. doi: 10.1039/c2pp25152c. [DOI] [PubMed] [Google Scholar]
- 105.Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA repair and mutagenesis. 2nd ed. Washington, DC, USA: ASM Press; 2005.
- 106.Singh B, Schneider M, Knyazev P, Ullrich A UV-induced EGFR signal transactivation is dependent on proligand shedding by activated metalloproteases in skin cancer cell lines. Int J Cancer. 2009;124:531–9. doi: 10.1002/ijc.v124:3. [DOI] [PubMed] [Google Scholar]
- 107.Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP, et al A landscape of driver mutations in melanoma. Cell. 2012;150:251–63. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sato M, Nishigori C, Zghal M, Yagi T, Takebe H Ultraviolet-specific mutations in p53 gene in skin tumors in xeroderma pigmentosum patients. Cancer Res. 1993;53:2944–6. [PubMed] [Google Scholar]
- 109.Van Steeg H, Kraemer KH Xeroderma pigmentosum and the role of UV-induced DNA damage in skin cancer. Mol Med Today. 1999;5:86–94. doi: 10.1016/s1357-4310(98)01394-x. [DOI] [PubMed] [Google Scholar]
- 110.Kraemer KH, Patronas NJ, Schiffmann R, Brooks BP, Tamura D, Digiovanna JJ Xeroderma pigmentosum, trichothiodystrophy and cockayne syndrome: a complex genotype–phenotype relationship. Neuroscience. 2007;145:1388–96. doi: 10.1016/j.neuroscience.2006.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Nance MA, Berry SA Cockayne syndrome: review of 140 cases. Am J Med Genet. 1992;42:68–84. doi: 10.1002/(ISSN)1096-8628. [DOI] [PubMed] [Google Scholar]
- 112.Giglia-Mari G, Coin F, Ranish JA, Hoogstraten D, Theil A, Wijgers N, et al A new, tenth subunit of TFIIH is responsible for the DNA repair syndrome trichothiodystrophy group A. Nat Genet. 2004;36:714–9. doi: 10.1038/ng1387. [DOI] [PubMed] [Google Scholar]
- 113.Itoh T, Cado D, Kamide R, Linn S DDB2 gene disruption leads to skin tumors and resistance to apoptosis after exposure to ultraviolet light but not a chemical carcinogen. Proc Natl Acad Sci USA. 2004;101:2052–7. doi: 10.1073/pnas.0306551101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Berg RJW, Ruven HJT, Sands AT, De Gruijl FR, Mullenders LHF Defective global genome repair in XPC mice is associated with skin cancer susceptibility but not with sensitivity to UVB induced erythema and edema. J Invest Dermatol. 1998;110:405–9. doi: 10.1111/j.1523-1747.1998.00173.x. [DOI] [PubMed] [Google Scholar]
- 115.Nakane H, Takeuchi S, Yuba S, Saijo M, Nakatsu Y, Murai H, et al High incidence of ultraviolet-B-or chemical-carcinogen-induced skin tumours in mice lacking the xeroderma pigmentosum group A gene. Nature. 1995;377:165–8. doi: 10.1038/377165a0. [DOI] [PubMed] [Google Scholar]
- 116.De Vries A, Van Oostrom CTM, Hofhuis FMA, Dortant PM, Berg RJW, De Gruijl FR, et al Increased susceptibility to ultraviolet-B and carcinogens of mice lacking the DNA excision repair gene XPA. Nature. 1995;377:169–73. doi: 10.1038/377169a0. [DOI] [PubMed] [Google Scholar]
- 117.Melis JPM, Luijten M, Mullenders LHF, Van Steeg H The role of XPC: implications in cancer and oxidative DNA damage. Mutat Res. 2011;728:107–17. doi: 10.1016/j.mrrev.2011.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Schumacher B, Garinis GA, Hoeijmakers JHJ Age to survive: DNA damage and aging. Trends Genet. 2008;24:77–85. doi: 10.1016/j.tig.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 119.Hanson KM, Gratton E, Bardeen CJ Sunscreen enhancement of UV-induced reactive oxygen species in the skin. Free Radic Biol Med. 2006;41:1205–12. doi: 10.1016/j.freeradbiomed.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 120.Trzeciak AR, Mohanty JG, Jacob KD, Barnes J, Ejiogu N, Lohani A, et al Oxidative damage to DNA and single strand break repair capacity: relationship to other measures of oxidative stress in a population cohort. Mutat Res. 2012;736:93–103. doi: 10.1016/j.mrfmmm.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Webster ADB, Barnes DE, Arlett CF, Lehmann AR, Lindahl T Growth retardation and immunodeficiency in a patient with mutations in the DNA ligase I gene. Lancet. 1992;339:1508–9. doi: 10.1016/0140-6736(92)91266-B. [DOI] [PubMed] [Google Scholar]
- 122.Elias H, Bengelsdorf H The structure of the liver of vertebrates. Acta Anat. 1952;14:297–337. doi: 10.1159/000140715. [DOI] [PubMed] [Google Scholar]
- 123.Adams DH. Hepatology: a textbook of liver disease, 4th edition. Edited by Zakim, Boyer. Philadelphia: Saunders, 2003, £230.00, pp 1765. ISBN 0-7216-9051-3. Gut. 2003; 52: 1230-1.
- 124.Abdel-Misih SRZ, Bloomston M Liver anatomy. Surg Clin North Am. 2010;90:643–53. doi: 10.1016/j.suc.2010.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Maher JJ Exploring alcohol's effects on liver function. Alcohol Health Res World. 1997;21:5–12. [PMC free article] [PubMed] [Google Scholar]
- 126.Knell AJ Liver function and failure: the evolution of liver physiology. J Roy Coll Phys Lond. 1980;14:205–8. [PMC free article] [PubMed] [Google Scholar]
- 127.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.v61:2. [DOI] [PubMed] [Google Scholar]
- 128.Arzumanyan A, Reis HMGPV, Feitelson MA Pathogenic mechanisms in hbv- and hcv-associated hepatocellular carcinoma. Nat Rev Cancer. 2013;13:123–35. doi: 10.1038/nrc3449. [DOI] [PubMed] [Google Scholar]
- 129.Nordenstedt H, White DL, El-Serag HB The changing pattern of epidemiology in hepatocellular carcinoma. Dig Liver Dis. 2010;42:S206–14. doi: 10.1016/S1590-8658(10)60507-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Yang SF, Chang CW, Wei RJ, Shiue YL, Wang SN, Yeh YT Involvement of DNA damage response pathways in hepatocellular carcinoma. Biomed Res Int. 2014;2014:153867. doi: 10.1155/2014/153867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Tanaka H, Fujita N, Sugimoto R, Urawa N, Horiike S, Kobayashi Y, et al Hepatic oxidative DNA damage is associated with increased risk for hepatocellular carcinoma in chronic hepatitis C. Br J Cancer. 2008;98:580–6. doi: 10.1038/sj.bjc.6604204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Mahmood S, Kawanaka M, Kamei A, Izumi A, Nakata K, Niiyama G, et al Immunohistochemical evaluation of oxidative stress markers in chronic hepatitis C. Antioxid Redox Signal. 2004;6:19–24. doi: 10.1089/152308604771978318. [DOI] [PubMed] [Google Scholar]
- 133.Fujita N, Horiike S, Sugimoto R, Tanaka H, Iwasa M, Kobayashi Y, et al Hepatic oxidative DNA damage correlates with iron overload in chronic hepatitis C patients. Free Radic Biol Med. 2007;42:353–62. doi: 10.1016/j.freeradbiomed.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 134.Puisieux A, Galvin K, Troalen F, Bressac B, Marcais C, Galun E, et al Retinoblastoma and p53 tumor suppressor genes in human hepatoma cell lines. FASEB J. 1993;7:1407–13. doi: 10.1096/fasebj.7.14.8224613. [DOI] [PubMed] [Google Scholar]
- 135.Yuan T, Wei JY, Luo J, Liu MG, Deng SL, Chen P Polymorphisms of base-excision repair genes hOGG1 326cys and XRCC1 280His increase hepatocellular carcinoma risk. Dig Dis Sci. 2012;57:2451–7. doi: 10.1007/s10620-012-2192-6. [DOI] [PubMed] [Google Scholar]
- 136.Machida K, McNamara G, Cheng KTH, Huang J, Wang CH, Comai L, et al Hepatitis C virus inhibits DNA damage repair through reactive oxygen and nitrogen species and by interfering with the ATM-NBS1/Mre11/Rad50 DNA repair pathway in monocytes and hepatocytes. J Immunol. 2010;185:6985–98. doi: 10.4049/jimmunol.1000618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Cun YP, Dai N, Xiong CJ, Li MX, Sui JD, Qian CY, et al Silencing of APE1 enhances sensitivity of human hepatocellular carcinoma cells to radiotherapy in vitro and in a xenograft model . PLoS One. 2013;8:e55313. doi: 10.1371/journal.pone.0055313. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 138.Aoki H, Kajino K, Arakawa Y, Hino O Molecular cloning of a rat chromosome putative recombinogenic sequence homologous to the hepatitis B virus encapsidation signal. Proc Natl Acad Sci USA. 1996;93:7300–4. doi: 10.1073/pnas.93.14.7300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zemel R, Issachar A, Tur-Kaspa R The role of oncogenic viruses in the pathogenesis of hepatocellular carcinoma. Clin Liver Dis. 2011;15:261–79. doi: 10.1016/j.cld.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 140.Hussain SP, Hofseth LJ, Harris CC Radical causes of cancer. Nat Rev Cancer. 2003;3:276–85. doi: 10.1038/nrc1046. [DOI] [PubMed] [Google Scholar]
- 141.Takamatsu S, Noguchi N, Kudoh A, Nakamura N, Kawamura T, Teramoto K, et al Influence of risk factors for metabolic syndrome and non-alcoholic fatty liver disease on the progression and prognosis of hepatocellular carcinoma. Hepatogastroenterology. 2008;55:609–14. [PubMed] [Google Scholar]
- 142.Chiang CH, Huang KC Association between metabolic factors and chronic hepatitis B virus infection. World J Gastroenterol. 2014;20:7213–6. doi: 10.3748/wjg.v20.i23.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.El-Serag HB, Tran T, Everhart JE Diabetes increases the risk of chronic liver disease and hepatocellular carcinoma. Gastroenterology. 2004;126:460–8. doi: 10.1053/j.gastro.2003.10.065. [DOI] [PubMed] [Google Scholar]
- 144.Johnson WW, Guengerich FP Reaction of aflatoxin B1 exo-8,9-epoxide with DNA: kinetic analysis of covalent binding and DNA-induced hydrolysis . Proc Natl Acad Sci USA. 1997;94:6121–5. doi: 10.1073/pnas.94.12.6121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Bressac B, Kew M, Wands J, Ozturk M Selective G to T mutations of p53 gene in hepatocellular carcinoma from southern Africa. Nature. 1991;350:429–31. doi: 10.1038/350429a0. [DOI] [PubMed] [Google Scholar]
- 146.Dunne RF, Hezel AF Genetics and biology of pancreatic ductal adenocarcinoma. Hematol Oncol Clin North Am. 2015;29:595–608. doi: 10.1016/j.hoc.2015.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Domschke S, Domschke W, Rösch W, Konturek SJ, Sprügel W, Mitznegg P, et al Inhibition by somatostatin of secretin-stimulated pancreatic secretion in man: a study with pure pancreatic juice. Scand J Gastroenterol. 1977;12:59–63. [PubMed] [Google Scholar]
- 148.Lee MG, Ohana E, Park HW, Yang D, Muallem S Molecular mechanism of pancreatic and salivary gland fluid and HCO3– secretion . Physiol Rev. 2012;92:39–74. doi: 10.1152/physrev.00011.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Osterman M, Kathawa D, Liu DG, Guo H, Zhang C, Li M, et al Elevated DNA damage response in pancreatic cancer. Histochem Cell Biol. 2014;142:713–20. doi: 10.1007/s00418-014-1245-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Hidalgo M Pancreatic cancer. N Engl J Med. 2010;362:1605–17. doi: 10.1056/NEJMra0901557. [DOI] [PubMed] [Google Scholar]
- 151.Campbell PJ, Yachida S, Mudie LJ, Stephens PJ, Pleasance ED, Stebbings LA, et al The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature. 2010;467:1109–13. doi: 10.1038/nature09460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Li DH, Xie KP, Wolff R, Abbruzzese JL Pancreatic cancer. Lancet. 2004;363:1049–57. doi: 10.1016/S0140-6736(04)15841-8. [DOI] [PubMed] [Google Scholar]
- 153.Li DH, Firozi PF, Zhang WQ, Shen JJ, Digiovanni J, Lau S, et al DNA adducts, genetic polymorphisms, and K-ras mutation in human pancreatic cancer . Mutat Res. 2002;513:37–48. doi: 10.1016/S1383-5718(01)00291-1. [DOI] [PubMed] [Google Scholar]
- 154.Thompson PA, Seyedi F, Lang NP, Macleod SL, Wogan GN, Anderson KE, et al Comparison of DNA adduct levels associated with exogenous and endogenous exposures in human pancreas in relation to metabolic genotype. Mutat Res. 1999;424:263–74. doi: 10.1016/S0027-5107(99)00024-X. [DOI] [PubMed] [Google Scholar]
- 155.Kadlubar FF, Anderson KE, Häussermann S, Lang NP, Barone GW, Thompson PA, et al Comparison of DNA adduct levels associated with oxidative stress in human pancreas. Mutat Res. 1998;405:125–33. doi: 10.1016/S0027-5107(98)00129-8. [DOI] [PubMed] [Google Scholar]
- 156.Van Der Heijden MS, Yeo CJ, Hruban RH, Kern SE Fanconi anemia gene mutations in young-onset pancreatic cancer. Cancer Res. 2003;63:2585–8. [PubMed] [Google Scholar]
- 157.Rustgi AK Familial pancreatic cancer: genetic advances. Genes Dev. 2014;28:1–7. doi: 10.1101/gad.228452.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Li M, Chen Q, Ma T, Yu XC Targeting reactive nitrogen species suppresses hereditary pancreatic cancer. Proc Natl Acad Sci USA. 2017;114:7106–11. doi: 10.1073/pnas.1702156114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Tischkowitz MD, Sabbaghian N, Hamel N, Borgida A, Rosner C, Taherian N, et al Analysis of the gene coding for the BRCA2-interacting protein PALB2 in familial and sporadic pancreatic cancer . Gastroenterology. 2009;137:1183–6. doi: 10.1053/j.gastro.2009.06.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Guerra C, Schuhmacher AJ, Cañamero M, Grippo PJ, Verdaguer L, Pérez-Gallego L, et al Chronic pancreatitis is essential for induction of pancreatic ductal adenocarcinoma by K-Ras oncogenes in adult mice. Cancer Cell. 2007;11:291–302. doi: 10.1016/j.ccr.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 161.Mujica VR, Barkin JS, Go VLW, Study Group Participants Acute pancreatitis secondary to pancreatic carcinoma. Pancreas. 2000;21:329–32. doi: 10.1097/00006676-200011000-00001. [DOI] [PubMed] [Google Scholar]
- 162.Ekbom A, McLaughlin JK, Nyrén O Pancreatitis and the risk of pancreatic cancer. N Engl J Med. 1993;329:1502–3. doi: 10.1056/NEJM199311113292016. [DOI] [PubMed] [Google Scholar]
- 163.Bogdan C Nitric oxide and the immune response. Nat Immunol. 2001;2:907–16. doi: 10.1038/ni1001-907. [DOI] [PubMed] [Google Scholar]
- 164.Dedon PC, Tannenbaum SR Reactive nitrogen species in the chemical biology of inflammation. Arch Biochem Biophys. 2004;423:12–22. doi: 10.1016/j.abb.2003.12.017. [DOI] [PubMed] [Google Scholar]
- 165.Coussens LM, Werb Z Inflammation and cancer. Nature. 2002;420:860–7. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Halliwell B, Aruoma OI DNA damage by oxygen-derived species its mechanism and measurement in mammalian systems. FEBS Lett. 1991;281:9–19. doi: 10.1016/0014-5793(91)80347-6. [DOI] [PubMed] [Google Scholar]
- 167.Schreck R, Albermann K, Baeuerle PA Nuclear factor κB: an oxidative stress-responsive transcription factor of eukaryotic cells (a review) Free Radic Res Commun. 1992;17:221–37. doi: 10.3109/10715769209079515. [DOI] [PubMed] [Google Scholar]
- 168.Horváthová M, Jahnová E, Gazdík F Detection of intracellular cytokines during antioxidant supplementation in corticoid-dependent asthmatics and modulation of adhesion molecule expression on cultured endothelial cells. Biol Trace Elem Res. 2001;83:17–30. doi: 10.1385/BTER:83:1. [DOI] [PubMed] [Google Scholar]
- 169.Van Der Vliet A, Eiserich JP, Halliwell B, Cross CE Formation of reactive nitrogen species during peroxidase-catalyzed oxidation of nitrite. A potential additional mechanism of nitric oxide-dependent toxicity. J Biol Chem. 1997;272:7617–25. doi: 10.1074/jbc.272.12.7617. [DOI] [PubMed] [Google Scholar]
- 170.Eiserich JP, Hristova M, Cross CE, Jones AD, Freeman BA, Halliwell B, et al Formation of nitric oxide-derived inflammatory oxidants by myeloperoxidase in neutrophils. Nature. 1998;391:393–7. doi: 10.1038/34923. [DOI] [PubMed] [Google Scholar]
- 171.Sawa T, Ohshima H Nitrative DNA damage in inflammation and its possible role in carcinogenesis. Nitric Oxide. 2006;14:91–100. doi: 10.1016/j.niox.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 172.Tretyakova NY, Burney S, Pamir B, Wishnok JS, Dedon PC, Wogan GN, et al Peroxynitrite-induced DNA damage in the supF gene: correlation with the mutational spectrum . Mutat Res. 2000;447:287–303. doi: 10.1016/S0027-5107(99)00221-3. [DOI] [PubMed] [Google Scholar]
- 173.Hoon LC, Dedon PC, Deen WM Kinetic analysis of intracellular concentrations of reactive nitrogen species. Chem Res Toxicol. 2008;21:2134–47. doi: 10.1021/tx800213b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Yermilov V, Yoshie Y, Rubio J, Ohshima H Effects of carbon dioxide/bicarbonate on induction of DNA single-strand breaks and formation of 8-nitroguanine, 8-oxoguanine and base-propenal mediated by peroxynitrite. FEBS Lett. 1996;399:67–70. doi: 10.1016/S0014-5793(96)01288-4. [DOI] [PubMed] [Google Scholar]
- 175.Maynard S, Schurman SH, Harboe C, De Souza-Pinto NC, Bohr VA Base excision repair of oxidative DNA damage and association with cancer and aging. Carcinogenesis. 2009;30:2–10. doi: 10.1093/carcin/bgn250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Nooteboom M, Johnson R, Taylor RW, Wright NA, Lightowlers RN, Kirkwood TBL, et al Age-associated mitochondrial DNA mutations lead to small but significant changes in cell proliferation and apoptosis in human colonic crypts. Aging Cell. 2010;9:96–9. doi: 10.1111/ace.2010.9.issue-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Baker AM, Cereser B, Melton S, Fletcher AG, Rodriguez-Justo M, Tadrous PJ, et al Quantification of crypt and stem cell evolution in the normal and neoplastic human colon. Cell Rep. 2014;8:940–7. doi: 10.1016/j.celrep.2014.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Lynch HT, Snyder CL, Shaw TG, Heinen CD, Hitchins MP Milestones of lynch syndrome: 1895-2015. Nat Rev Cancer. 2015;15:181–94. doi: 10.1038/nrc3878. [DOI] [PubMed] [Google Scholar]
- 179.Boland CR, Goel A Microsatellite instability in colorectal cancer. Gastroenterology. 2010;138:2073–87. doi: 10.1053/j.gastro.2009.12.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Mao GG, Lee S, Ortega J, Gu LY, Li GM Modulation of microRNA processing by mismatch repair protein mutLα. Cell Res. 2012;22:973–85. doi: 10.1038/cr.2012.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Pabla N, Ma ZW, McIlhatton MA, Fishel R, Dong Z hMSH2 recruits ATR to DNA damage sites for activation during DNA damage-induced apoptosis. J Biol Chem. 2011;286:10411–8. doi: 10.1074/jbc.M110.210989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Luo YH, Lin FT, Lin WC ATM-mediated stabilization of hMutL DNA mismatch repair proteins augments p53 activation during DNA damage. Mol Cell Biol. 2004;24:6430–44. doi: 10.1128/MCB.24.14.6430-6444.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–86. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 184.Li SKH, Martin A Mismatch repair and colon cancer: mechanisms and therapies explored. Trends Mol Med. 2016;22:274–89. doi: 10.1016/j.molmed.2016.02.003. [DOI] [PubMed] [Google Scholar]
- 185.Jasperson KW, Tuohy TM, Neklason DW, Burt RW Hereditary and familial colon cancer. Gastroenterology. 2010;138:2044–58. doi: 10.1053/j.gastro.2010.01.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Poynter JN, Siegmund KD, Weisenberger DJ, Long TI, Thibodeau SN, Lindor N, et al Molecular characterization of MSI-H colorectal cancer by MLHI promoter methylation, immunohistochemistry, and mismatch repair germline mutation screening . Cancer Epidemiol Biomarkers Prev. 2008;17:3208–15. doi: 10.1158/1055-9965.EPI-08-0512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JPJ CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci USA. 1999;96:8681–6. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Jass JR, Do KA, Simms LA, Iino H, Wynter C, Pillay SP, et al Morphology of sporadic colorectal cancer with DNA replication errors. Gut. 1998;42:673–9. doi: 10.1136/gut.42.5.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Lord CJ, Ashworth A The DNA damage response and cancer therapy. Nature. 2012;481:287–94. doi: 10.1038/nature10760. [DOI] [PubMed] [Google Scholar]
- 190.Martin SA, McCabe N, Mullarkey M, Cummins R, Burgess DJ, Nakabeppu Y, et al DNA polymerases as potential therapeutic targets for cancers deficient in the DNA mismatch repair proteins MSH2 or MLH1. Cancer Cell. 2010;17:235–48. doi: 10.1016/j.ccr.2009.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Martin SA, Hewish M, Sims D, Lord CJ, Ashworth A Parallel high-throughput RNA interference screens identify PINK1 as a potential therapeutic target for the treatment of DNA mismatch repair-deficient cancers. Cancer Res. 2011;71:1836–48. doi: 10.1158/0008-5472.CAN-10-2836. [DOI] [PubMed] [Google Scholar]
- 192.Tremaroli V, Bäckhed F Functional interactions between the gut microbiota and host metabolism. Nature. 2012;489:242–9. doi: 10.1038/nature11552. [DOI] [PubMed] [Google Scholar]
- 193.Sommer F, Bäckhed F The gut microbiota — masters of host development and physiology. Nat Rev Microbiol. 2013;11:227–38. doi: 10.1038/nrmicro2974. [DOI] [PubMed] [Google Scholar]
- 194.Louis P, Hold GL, Flint HJ The gut microbiota, bacterial metabolites and colorectal cancer. Nat Rev Microbiol. 2014;12:661–72. doi: 10.1038/nrmicro3344. [DOI] [PubMed] [Google Scholar]
- 195.Abreu MT Toll-like receptor signalling in the intestinal epithelium: how bacterial recognition shapes intestinal function. Nat Rev Immunol. 2010;10:131–44. doi: 10.1038/nri2707. [DOI] [PubMed] [Google Scholar]
- 196.Wang JQ, Jeelall YS, Ferguson LL, Horikawa K Toll-like receptors and cancer: MYD88 mutation and inflammation. Front Immunol. 2014;5:367. doi: 10.3389/fimmu.2014.00367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.De Simone V, Franzè E, Ronchetti G, Colantoni A, Fantini MC, Di Fusco D, et al Th17-type cytokines, IL-6 and TNF-α synergistically activate STAT3 and NF-kB to promote colorectal cancer cell growth. Oncogene. 2015;34:3493–503. doi: 10.1038/onc.2014.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, et al PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;372:2509–20. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Win AK, Buchanan DD, Rosty C, MacInnis RJ, Dowty JG, Dite GS, et al Role of tumour molecular and pathology features to estimate colorectal cancer risk for first-degree relatives. Gut. 2015;64:101–10. doi: 10.1136/gutjnl-2013-306567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.West AP, Brodsky IE, Rahner C, Woo DK, Erdjument-Bromage H, Tempst P, et al TLR signalling augments macrophage bactericidal activity through mitochondrial ROS. Nature. 2011;472:476–80. doi: 10.1038/nature09973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Myant KB, Cammareri P, McGhee EJ, Ridgway RA, Huels DJ, Cordero JB, et al ROS production and NF-κB activation triggered by RAC1 facilitate WNT-driven intestinal stem cell proliferation and colorectal cancer initiation. Cell Stem Cell. 2013;12:761–73. doi: 10.1016/j.stem.2013.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Torre LA, Islami F, Siegel RL, Ward EM, Jemal A Global cancer in women: burden and trends. Cancer Epidemiol Biomarkers Prev. 2017;26:444–57. doi: 10.1158/1055-9965.EPI-16-0858. [DOI] [PubMed] [Google Scholar]
- 203.Rahman N, Stratton MR The genetics of breast cancer susceptibility. Annu Rev Genet. 1998;32:95–121. doi: 10.1146/annurev.genet.32.1.95. [DOI] [PubMed] [Google Scholar]
- 204.Turner N, Tutt A, Ashworth A Hallmarks of 'BRCAness' in sporadic cancers. Nat Rev Cancer. 2004;4:814–9. doi: 10.1038/nrc1457. [DOI] [PubMed] [Google Scholar]
- 205.Drost R, Bouwman P, Rottenberg S, Boon U, Schut E, Klarenbeek S, et al BRCA1 RING function is essential for tumor suppression but dispensable for therapy resistance. Cancer Cell. 2011;20:797–809. doi: 10.1016/j.ccr.2011.11.014. [DOI] [PubMed] [Google Scholar]
- 206.Huen MSY, Sy SMH, Chen JJ BRCA1 and its toolbox for the maintenance of genome integrity. Nat Rev Mol Cell Biol. 2010;11:138–48. doi: 10.1038/nrm2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Cui J, Shen Y, Li RN Estrogen synthesis and signaling pathways during aging: from periphery to brain. Trends Mol Med. 2013;19:197–209. doi: 10.1016/j.molmed.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Yaghjyan L, Colditz GA Estrogens in the breast tissue: a systematic review. Cancer Causes Control. 2011;22:529–40. doi: 10.1007/s10552-011-9729-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Clegg DJ Minireview: the year in review of estrogen regulation of metabolism. Mol Endocrinol. 2012;26:1957–60. doi: 10.1210/me.2012-1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Raftogianis R, Creveling C, Weinshilboum R, Weisz J Estrogen metabolism by conjugation. J Natl Cancer Inst Monogr. 2000;2000:113–24. doi: 10.1093/oxfordjournals.jncimonographs.a024234. [DOI] [PubMed] [Google Scholar]
- 211.Li M, Chen Q, Yu XC Chemopreventive effects of ROS targeting in a murine model of BRCA1-deficient breast cancer. Cancer Res. 2017;77:448–58. doi: 10.1158/0008-5472.CAN-16-2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Salama SA, Kamel M, Awad M, Nasser AHB, Al-Hendy A, Botting S, et al Catecholestrogens induce oxidative stress and malignant transformation in human endometrial glandular cells: protective effect of catechol-O-methyltransferase . Int J Cancer. 2008;123:1246–54. doi: 10.1002/ijc.v123:6. [DOI] [PubMed] [Google Scholar]
- 213.Li KM, Todorovic R, Devanesan P, Higginbotham S, Köfeler H, Ramanathan R, et al Metabolism and DNA binding studies of 4-hydroxyestradiol and estradiol-3,4-quinone in vitro and in female ACI rat mammary gland in vivo . Carcinogenesis. 2004;25:289–97. doi: 10.1093/carcin/bgg191. [DOI] [PubMed] [Google Scholar]
- 214.Yager JD Catechol-O-methyltransferase: characteristics, polymorphisms and role in breast cancer . Drug Discov Today Dis Mech. 2012;9:e41–6. doi: 10.1016/j.ddmec.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Tsuchiya Y, Nakajima M, Yokoi T Cytochrome P450-mediated metabolism of estrogens and its regulation in human. Cancer Lett. 2005;227:115–24. doi: 10.1016/j.canlet.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 216.Zahid M, Kohli E, Saeed M, Rogan E, Cavalieri E The greater reactivity of estradiol-3,4-quinone vs estradiol-2,3-quinone with DNA in the formation of depurinating adducts: implications for tumor-initiating activity. Chem Res Toxicol. 2006;19:164–72. doi: 10.1021/tx050229y. [DOI] [PubMed] [Google Scholar]
- 217.Mannistö PT, Kaakkola S Catechol-O-methyltransferase (COMT): biochemistry, molecular biology, pharmacology, and clinical efficacy of the new selective COMT inhibitors . Pharmacol Rev. 1999;51:593–628. [PubMed] [Google Scholar]
- 218.Bolton JL, Shen L p-quinone methides are the major decomposition products of catechol estrogen o-quinones . Carcinogenesis. 1996;17:925–9. doi: 10.1093/carcin/17.5.925. [DOI] [PubMed] [Google Scholar]
- 219.Savage KI, Matchett KB, Barros EM, Cooper KM, Irwin GW, Gorski JJ, et al BRCA1 deficiency exacerbates estrogen-induced DNA damage and genomic instability. Cancer Res. 2014;74:2773–84. doi: 10.1158/0008-5472.CAN-13-2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Fuhrman BJ, Schairer C, Gail MH, Boyd-Morin J, Xu X, Sue LY, et al Estrogen metabolism and risk of breast cancer in postmenopausal women. J Natl Cancer Inst. 2012;104:326–39. doi: 10.1093/jnci/djr531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Swift M, Reitnauer PJ, Morrell D, Chase CL Breast and other cancers in families with ataxia-telangiectasia. N Engl J Med. 1987;316:1289–94. doi: 10.1056/NEJM198705213162101. [DOI] [PubMed] [Google Scholar]
- 222.Wang L, Di LJ BRCA1 and estrogen/estrogen receptor in breast cancer: where they interact? Int J Biol Sci. 2014;10:566–75. doi: 10.7150/ijbs.8579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Chang CY, McDonnell DP Molecular pathways: the metabolic regulator estrogen-related receptor α as a therapeutic target in cancer. Clin Cancer Res. 2012;18:6089–95. doi: 10.1158/1078-0432.CCR-11-3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Musgrove EA, Sutherland RL Biological determinants of endocrine resistance in breast cancer. Nat Rev Cancer. 2009;9:631–43. doi: 10.1038/nrc2713. [DOI] [PubMed] [Google Scholar]
- 225.Filardo EJ, Quinn JA, Bland KI, Frackelton Jr AR Estrogen-induced activation of erk-1 and erk-2 requires the G protein-coupled receptor homolog, GPR30, and occurs via trans-activation of the epidermal growth factor receptor through release of HB-EGF . Mol Endocrinol. 2000;14:1649–60. doi: 10.1210/mend.14.10.0532. [DOI] [PubMed] [Google Scholar]
- 226.Caldon CE Estrogen signaling and the DNA damage response in hormone dependent breast cancers. Front Oncol. 2014;4:106. doi: 10.3389/fonc.2014.00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Pedram A, Razandi M, Evinger AJ, Lee E, Levin ER Estrogen inhibits ATR signaling to cell cycle checkpoints and DNA repair. Mol Biol Cell. 2009;20:3374–89. doi: 10.1091/mbc.e09-01-0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Castellano L, Giamas G, Jacob J, Coombes RC, Lucchesi W, Thiruchelvam P, et al The estrogen receptor-α-induced microRNA signature regulates itself and its transcriptional response. Proc Natl Acad Sci USA. 2009;106:15732–7. doi: 10.1073/pnas.0906947106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, et al Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–7. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 230.Audeh MW, Carmichael J, Penson RT, Friedlander M, Powell B, Bell-McGuinn KM, et al Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and recurrent ovarian cancer: a proof-of-concept trial . Lancet. 2010;376:245–51. doi: 10.1016/S0140-6736(10)60893-8. [DOI] [PubMed] [Google Scholar]
- 231.Xu GT, Chapman JR, Brandsma I, Yuan JS, Mistrik M, Bouwman P, et al REV7 counteracts DNA double-strand break resection and affects PARP inhibition. Nature. 2015;521:541–4. doi: 10.1038/nature14328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Farmer H, McCabe N, Lord CJ, Tutt ANJ, Johnson DA, Richardson TB, et al Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy . Nature. 2005;434:917–21. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 233.Lord CJ, Ashworth A PARP inhibitors: synthetic lethality in the clinic. Science. 2017;355:1152–8. doi: 10.1126/science.aam7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Orkin SH, Zon LI Hematopoiesis: an evolving paradigm for stem cell biology. Cell. 2008;132:631–44. doi: 10.1016/j.cell.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Morrison SJ, Prowse KR, Ho P, Weissman IL Telomerase activity in hematopoietic cells is associated with self-renewal potential. Immunity. 1996;5:207–16. doi: 10.1016/S1074-7613(00)80316-7. [DOI] [PubMed] [Google Scholar]
- 236.Foudi A, Hochedlinger K, Van Buren D, Schindler JW, Jaenisch R, Carey V, et al Analysis of histone 2B-GFP retention reveals slowly cycling hematopoietic stem cells. Nat Biotechnol. 2009;27:84–90. doi: 10.1038/nbt.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Allsopp RC, Morin GR, DePinho R, Harley CB, Weissman IL Telomerase is required to slow telomere shortening and extend replicative lifespan of HSCS during serial transplantation. Blood. 2003;102:517–20. doi: 10.1182/blood-2002-07-2334. [DOI] [PubMed] [Google Scholar]
- 238.Nombela-Arrieta C, Pivarnik G, Winkel B, Canty KJ, Harley B, Mahoney JE, et al Quantitative imaging of haematopoietic stem and progenitor cell localization and hypoxic status in the bone marrow microenvironment. Nat Cell Biol. 2013;15:533–43. doi: 10.1038/ncb2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Ito K, Hirao A, Arai F, Matsuoka S, Takubo K, Hamaguchi I, et al Regulation of oxidative stress by ATM is required for self-renewal of haematopoietic stem cells. Nature. 2004;431:997–1002. doi: 10.1038/nature02989. [DOI] [PubMed] [Google Scholar]
- 240.Inoue S, Li WY, Tseng A, Beerman I, Elia AJ, Bendall SC, et al Mutant IDH1 downregulates ATM and alters DNA repair and sensitivity to DNA damage independent of TET2. Cancer Cell. 2016;30:337–48. doi: 10.1016/j.ccell.2016.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Seita J, Rossi DJ, Weissman IL Differential DNA damage response in stem and progenitor cells. Cell Stem Cell. 2010;7:145–7. doi: 10.1016/j.stem.2010.07.006. [DOI] [PubMed] [Google Scholar]
- 242.Asai T, Liu Y, Bae N, Nimer SD The p53 tumor suppressor protein regulates hematopoietic stem cell fate. J Cell Physiol. 2011;226:2215–21. doi: 10.1002/jcp.22561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Mohrin M, Bourke E, Alexander D, Warr MR, Barry-Holson K, Le Beau MM, et al Hematopoietic stem cell quiescence promotes error-prone DNA repair and mutagenesis. Cell Stem Cell. 2010;7:174–85. doi: 10.1016/j.stem.2010.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Beerman I, Seita J, Inlay MA, Weissman IL, Rossi DJ Quiescent hematopoietic stem cells accumulate DNA damage during aging that is repaired upon entry into cell cycle. Cell Stem Cell. 2014;15:37–50. doi: 10.1016/j.stem.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Blanpain C, Mohrin M, Sotiropoulou PA, Passegué E DNA-damage response in tissue-specific and cancer stem cells. Cell Stem Cell. 2011;8:16–29. doi: 10.1016/j.stem.2010.12.012. [DOI] [PubMed] [Google Scholar]
- 246.Meijne EI, Van Der Winden-van Groenewegen RJ, Ploemacher RE, Vos O, David JA, Huiskamp R The effects of x-irradiation on hematopoietic stem cell compartments in the mouse. Exp Hematol. 1991;19:617–23. [PubMed] [Google Scholar]
- 247.Kwok M, Davies N, Agathanggelou A, Smith E, Oldreive C, Petermann E, et al ATR inhibition induces synthetic lethality and overcomes chemoresistance in TP53- or ATM-defective chronic lymphocytic leukemia cells . Blood. 2016;127:582–95. doi: 10.1182/blood-2015-05-644872. [DOI] [PubMed] [Google Scholar]
- 248.Hähnel PS, Enders B, Sasca D, Roos WP, Kaina B, Bullinger L, et al Targeting components of the alternative NHEJ pathway sensitizes KRAS mutant leukemic cells to chemotherapy. Blood. 2014;123:2355–66. doi: 10.1182/blood-2013-01-477620. [DOI] [PubMed] [Google Scholar]
- 249.Curtin NJ DNA repair dysregulation from cancer driver to therapeutic target. Nat Rev Cancer. 2012;12:801–17. doi: 10.1038/nrc3399. [DOI] [PubMed] [Google Scholar]
- 250.Kandoth C, McLellan MD, Vandin F, Ye K, Niu BF, Lu C, et al Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333–9. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]