Abstract
Objective
CSN6 is a vital subunit of the constitutive photomorphogenesis 9 (COP9) signalosome (CSN), which is responsible for development disorders and promotes ubiquitin-26S proteasome-dependent degradation in vitro and vivo. Its role in the tumor development of gastric cancer remains unclear. In this study, we investigated the role of CSN6 in gastric cancer progression.
Methods
Human gastric cancer samples were collected and immunohistochemistry was performed to identify the role of CSN6 in gastric cancer. The cell proliferation was measured by CCK-8 and the EdU incorporation method. Immunofluorescence localization and a co-immunoprecipitation study were used to show the interaction between the protein CSN6 and p16. Ubiquitination assay was performed to validate whether ubiquitination is involved in CSN6-mediated p16 degradation. BALB/c nude mice were used to produce a tumor model in order to test the effect of CSN6 on cancer growth in vivo.
Results
CSN6 expression was dramatically increased in gastric cancer tissues compared with paired adjacent non-tumor tissues and CSN6 was correlated with worse overall and disease-specific survival. Additionally, we also found that CSN6 downregulated p16 protein expression, thereby promoting gastric cancer cell growth and proliferation. Moreover, CSN6 interacted with p16 and a proteasome activator REGγ (PA28γ), thereby facilitating ubiquitin-independent degradation of p16.
Conclusions
CSN6 promoted the loss of p16-mediated tumor progression and played an important role in regulating ubiquitin-independent proteasomal degradation of p16.
Keywords: CSN6, gastric cancer, proliferation, p16, REGγ
Introduction
Gastric cancer (GC) is one of the most common types of malignancies and is associated with high mortality rates of cancer-related deaths worldwide1. There are nearly 1 million new GC cases each year making GC a particularly challenging malignancy2. Although surgical resection is considered to be the primary curative approach for this disease, the locoregional recurrence rate currently ranges from 24% to 54% after radical resection3. A multitude of causes have been identified that aberrant gene regulation may lead to GC4. Moreover, mutation of tumor suppressor genes and oncogenes is a primary mechanism underlying oncogenesis and cancer progression5. Molecular targeted therapies have been developed according to known oncogenic pathways. Thus, understanding the molecular mechanisms of GC will provide potential therapeutic targets for GC treatment.
The constitutive photomorphogenesis 9 (COP9) signalosome (CSN) was originally identified in Arabidopsis thaliana that mimic light-induced seedling development when grown in the dark6,7. The CSN is composed of eight canonical subunits and is involved in a number of developmental processes, for instance, signal transduction, DNA damage response, genome integrity, protein degradation, transcriptional activation and tumorigenesis8-10. Additionally, several studies suggested that CSN plays a key role in the regulation of cancer progression and tumor suppressive functions. Although a bulk of evidence has shown that the expression or localization of some CSN subunits was correlated to cancer progression or poor clinical outcome in many tumor types11, the detailed biological functions of the CSN’s subunits have not been well identified. Previous studies have illustrated the collaboration between CSN and the Ub-26S proteasome system in regulating the degradation of important cellular proteins such as cyclin-dependent kinase inhibitor p27kip12. Moreover, CSN is highly homologous to the lid sub-complex of the 26S proteasome and has developed similar functions with the ubiquitin-proteasome system13,14. The system consists of three key players: E1 ubiquitin-activating enzymes, E2 ubiquitin-conjugating enzymes and E3 ubiquitin ligases15. The CSN mediates a variety of cellular functions through its association with cullin-containing ubiquitin E3 ligases11,16. Previous studies have suggested that CSN6 associates with aberrant gene regulators such as MEK kinase 1 (MEKK1), E6-associated protein (E6AP) and epidermal growth factor receptor (EGFR) by facilitating ubiquitin-mediated degradation17-19. Moreover, CSN6 stabilizes COP1 through reducing COP1 self-ubiquitination and the CSN6-COP1 axis function as an E3 ubiquitin ligase involved in 14-3-3σ degradation to regulate cell growth and tumorigenicity10,20. It was identified that CSN could dedicate the protein to degradation by the Ub-26S proteasome system which demands ubiquitin21. However, the 20S proteasomes were shown to be capable of degrading proteins without the ubiquitin tagging or the presence of the 19S regulatory particle22. It was also identified that a proteasome activator REGγ (PA28γ) could present to proteasomes and increase the capacity of the 20S proteasome to degrade proteins23.
p16INK4a is a well known cyclin-dependent kinase inhibitor that arrests cells in early G1 as a specific inhibitor of D-type cyclin-dependent kinases24. p16 is encoded in the INK4a/ARF locus, which is situated on human chromosome 9p21. This locus contains the INK4a gene and ARF gene, which encode for the protein p16 and p14ARF, respectively, in humans25. The p16 protein plays an important role in inhibiting cell-cycle progression and is inactivated in diverse human cancers26. Consistently, inactivation of p16 has been implicated in the deregulation of cell cycle control27. Moreover, the loss of p16 attenuates cellular senescence and promotes carcinogenesis in human, which consequently may have prognostic implications28. Although the biochemical functions of p16INK4a with other cell cycle regulators have been studied expansively, little is known about its regulation. In the current study, we examined the mechanism underlying the CSN6-mediated p16 degradation.
To investigate the role of CSN6 in GC, we collected human GC samples and examined the clinicopathological and prognostic significance of CSN6. Immunoblotting analysis revealed that CSN6 was overexpressed in GC and was an important positive regulator of p16. Additionally, our data demonstrated that CSN6 promoted proliferation of GC by inhibiting the expression of p16 via an ubiquitination-independent degradation pathway. Taken together, our findings implied that the biological functions of CSN6 signaling regulation might become a viable avenue for inhibiting CSN6-induced cancer progression.
Materials and methods
Patients and samples
A GC tissue microarray (TMA) including 90 cases of cancer tissues and paired cancerous adjacent tissues was all enrolled from the Department of General Surgery, Affiliated Hospital of Xuzhou Medical University. The 180 dots were obtained from 90 patients who underwent radical gastrectomy. The array dot diameter was 1.5 mm, and each dot represented a tissue spot from one individual specimen that was selected and pathologically confirmed. The cases of TMA include 10 lost follow-up patients and 3 disease-free patients. Tumors were staged according to the revised 2010 Tumor Node Metastasis (TNM) staging system with 43 cases in stages II and 47 cases in stages III-IV. Follow-up information was obtained by reviewing patient medical records. The studies using human gastric tissue samples were approved by Xuzhou Human Subject Committee. Informed consent from the patients was obtained in all cases.
Ethical approval
This study was performed under a protocol approved by the Review Board of the Affiliated Hospital of Xuzhou Medical University, and all examinations were performed after obtaining written informed consents. The studies using human gastric tissue samples were approved by Xuzhou Human Subject Committee. Animal experiments were in conformance with the Institutional Animal Care and Use Committee of Xuzhou Medical University.
Immunohistochemistry
Immunohistochemistry was performed according to the streptavidin-peroxidase (Sp) method using a standard Sp Kit (Zhongshan biotech, Beijing, China). The TMA slides were dewaxed at 60°C for 20 minutes followed by two 10-minute washes with xylene and then rehydrated with graded ethanol and distilled water. Endogenous peroxidases were inhibited by 3% H2O2 for 30 minutes. Antigen retrieval was performed in a microwave oven with 10 mM citrate buffer (pH 6.0) at 95°C for 30 minutes. After 30 minutes blocking with 5% normal goat serum, the sections were incubated with anti-CSN6 antibody (1 : 50 dilution; Enzo Life Sciences, Switzerland) overnight at 4°C. The slides were then incubated for 1 hour with a biotin labeled secondary antibody, followed by diaminobenzidine (DAB; Zhongshan biotech, Beijing, China) substrate. The slides were sealed with cover slips after hematoxylin counterstain and Dehydration. The image was collected by light microscopy (Olympus BX-53 light microscope).
Evaluation of immunostaining
The evaluation of CSN6 staining was blindly and independently examined by two pathologists and a consensus was reached for each core. CSN6 staining intensity was scored 0 to 3 (0 = negative; 1 = weak; 2 = moderate; 3 = strong). The percentage of CSN6 positive stained cells was also scored into 4 categories: 1 (0%–25%), 2 (26%–50%), 3 (51%–75%), and 4 (76%–100%). The level of CSN6 staining was evaluated by immunoreactive score (IRS)29, which is calculated by multiplying the scores of staining intensity and the percentage of positive cells. Based on the IRS, CSN6 staining pattern was defined as low (IRS: 0–6) and high (IRS: 8–12).
Human tumor samples
Twelve pairs of primary gastric tumors with corresponding noncancerous gastric samples were obtained from the Affiliated Hospital of Xuzhou Medical University. Collection and usage of all patient materials and information were conducted according the institutional guidelines and the Declaration of Helsinki Principles. Primary gastric tumor samples were obtained from patients who had undergone operations to treat GC. Informed consent was obtained for use of these pathologic samples for research.
Cell culture and reagents
Human gastric cancer cell lines MGC-803, MKN-45 and AGS were obtained from the ATCC. MKN-45 and AGS cells were grown in RPMI1640 culture media (Hyclone) contain 10% fetal bovine serum (FBS, Invitrogen) supplemented with penicillin (100 U/ml) and streptomycin (100 mg/ml) (Life Technologies). The MGC-803 cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM)/10% FBS. Human gastric epithelial cell line GES was also obtained from the ATCC and cultured in RPMI1640 culture media (Hyclone)/10% FBS. All the cell lines were cytogenetically tested and authenticated before the cells were frozen. The frozen cells were thawed for a maximum of 2 months. We used the following antibodies: CSN6 (Enzo Life Sciences), p16 (Abcam), p21 (Abcam), p27 (Abcam), Flag (sigma), β-actin (Vicmed Life Sciences), Cyclin D (Cell Signaling), CDK4 (Cell Signaling), Ubiquitin (Santa Cruz), SKP2 (Absci), COP1 (Abcam), UBE1 (Proteintech), REGγ (Thermo Fisher).
Plasmid and stable cell line generation
Total RNA was extracted from U2OS cells by using the Qiagen RNeasy kit (Qiagen, Germantown, MD, USA), and first-strand cDNA was synthesized by the PrimeScript RT reagent kit (Takara) according to the manufacturer’s instructions. Then, the cDNA for CSN6 was amplified using Taq polymerase, and the following primers: 5′-AGCTAAGCTTGAAAATGGCGGCGGC-3′, forward, 5′-AGCTGAATTCCTTCAAGTACCCTCATCAG-3′, reverse. The cDNA was subcloned into the mammalian expression vector pcDNA3.1 at ECoRI and HindIII sites. The identity of the resulting clones was verified by sequencing.
The stable CSN6 overexpressed MKN45 and MGC803 cells were established by infected with lentiviruses, in which CSN6-control expression vectors and CSN6-overexpression vectors were respectively packed by Gene-Pharma (Soochow, China). Target cells were transfected with lentivirus for 48 h followed by selection with puromycin (Vicmed) for 30 days.
RNA interference
The siRNA (small interfering RNA) pool against CSN6-siRNA and siRNA NC were designed and synthesised by Gene-Pharma (Soochow, China). siRNA was transfected using siLentFect Lipid Reagent (Bio-Rad, Hercules, CA, USA) according to the manufacturer’s instructions. The sequences of the CSN6-siRNA are presented in Table 1:
1.
The sequences of the CSN6-siRNA
| Name | siRNA duplexes |
| si-CSN6#1 | 5′-CCGUGGAAGAGAAGAUUAUTT-3′ |
| 5′-AUAAUCUUCUCUUCCACGGTT-3′ | |
| si-CSN6#2 | 5′-GAGUCUGUCAUUGAUAUAATT-3′ |
| 5′-UUAUAUCAAUGACAGACUCTT-3′ | |
| si-CSN6#3 | 5′-GCCGAAAUAUCGAGGUGAUTT-3′ |
| 5′-AUCACCUCGAUAUUUCGGCTT-3′ |
Cells were grown to 50%~60% confluence before control/ CSN6 siRNA transfection. Six hours after transfection, the medium containing transfection reagents was removed and incubated in fresh medium.
Western blot analysis
Cells were harvested from the plates. The aliquots of cell extracts were separated on a 12% SDS-polyacrylamide gel. The proteins were then transferred to nitrocellulose membrane and incubated overnight at 4°C with appropriate primary antibodies. After incubation with peroxidase-coupled anti-mouse or anti-rabbit IgG at 37°C for 2 hours, membranes were then washed and scanned on the Chemiluminescence imaging analysis system (Tanon Biotechnology, Shanghai, China). Each western blot was repeated three times.
Co-immunoprecipitation
For co-immunoprecipitation (co-IP) assay, cell lysates for immunoprecipitation were incubated on a rocker with indicated antibodies at 4°C overnight: CSN6 (anti-rabbit, 1 : 50; Enzo Life Sciences), p16 (anti-mouse, 1 : 50; Abcam), Flag (anti-mouse, 1 : 50; sigma). Lysis buffer contained a cocktail of protease/phosphatase inhibitors (sigma). Then cell lysates were immunoprecipitated by Protein A/G beads (Santa Cruz). Beads were centrifuged at a low speed for 10 min and supernatant was discarded. Dried beads were mixed with 1x loading buffer and boiled for 5 min. Lysate samples were loaded onto gels as performed before.
Protein turnover assay
The cells were transfected with the indicated plasmids and incubated at 37°C with 5% CO2 for 24 h. Then CHX (cycloheximide) was added into the media at a final concentration of 100 μg/ml. The cells were harvested at the indicated times after CHX treatment. The protein levels were analyzed by immunoblotting. The density of protein was measured by densitometer and the integrated optical density was measured.
Ubiquitination assay
MKN-45 and MGC-803 cells were transfected or cotransfected with indicated plasmids. After 24 hours, cells were treated with 50 μg/mL of MG132 (Selleck Biotechnology, USA) for 6 h. The ubiquitinated proteins were immunoprecipitated with anti-p16. The protein complexes were then resolved by SDS-polyacrylamide gel and probed with anti-ubiquitin to visualize the level of ubiquitination.
Immunofluorescence localization
MKN-45 and MGC-803 cells were seeded on 12-mm coverslips in a 24-well plate (5 × 104 cells/well) and transfected with Flag-p16 plasmids. After 24 hours, cells were rinsed with PBS 15 min at room temperature. Then, cells were permeabilized with 0.1% Triton X-100 for 15 min and blocked using 10% FBS in PBS for 30 min. Cells were further incubated overnight with primary antibodies CSN6 (anti-rabbit, 1 : 200; Enzo Life Sciences), Flag (anti-mouse, 1 : 200; sigma). Next day, cells were incubated with secondary antibodies anti-mouse conjugated with Alexa Fluor 488 and anti-Rabbit conjugated with Alexa Fluor 555 (30 min) in 3% BSA prepared in PBS. Nuclei were visualized by DAPI. Images were acquired by immunofluorescence confocal laser scanning microscopy (Zeiss LSM 880).
CCK-8 assay
MKN-45 and MGC-803 cells were transfected with indicated plasmids. After 6h, cells (1 × 103 wells) were seeded in flat-bottomed 96-well plates for 4 days. Cell proliferation was evaluated by Cell Count Kit-8 (CCK-8, Vicmed, China) at various time points 24 h, 48 h, 72 h and 96 h according to the manufacturer’s instructions. Then absorbance was measured at 490 nm using an ELX-800 spectrometer reader (Bio-Tek Instruments, Winooski, USA). Each experiment was performed in triplicate.
Clonogenic assay
The CSN6 transfectants and control cells were seeded in six-well culture plates at 200 cells per well in triplicate. Medium was changed every 3 days over 10 days of foci formation. At the end of the period, cell monolayer was stained in crystal violet solution (0.5% crystal violet, 20% methanol) and then destained by wash with water. Foci were then counted and photographed.
EdU incorporation immunofluorescence
The EdU staining was performed following keyFlour488 Click-iT EdU imaging detection kit (KeyGEN Biotech, Nanjing, China) instrument. MKN-45 and MGC-803 cells were transfected with indicated plasmid or siRNA and seeded on coverslips in 24-well plates. Then cells were immobilized by 3.7% neutral methanol for 15min and permeabilized with 0.1% Triton X-100 for 15min. Next, cells were incubated with 10 μM EdU for 30 min. After treating with 3% BSA in PBS, Hoechst33342 was used for nuclear staining. Cell numbers of EdU-staining were counted per field. Data are shown from a typical experiment performed in triplicate.
Cell-cycle analysis
Cells were harvested and washed with PBS three times after transfected with control siRNA/siCSN6 or pcDNA3.1-control/pcDNA3.1-CSN6 expression. Then, the cells were fixed with 70% cold ethanol at 4°C overnight and resuspended in 500μl DNA staining solution and incubated for 30 min at room temperature. Samples were then analyzed using FACScan flow cytometer (Becton-Dicskinson Biosciences, San Jose, CA, USA). Data on cell cycle distribution were analyzed using ModFit LT 3.0 software. Each experiment was performed in triplicate.
Tumorigenesis in nude mice
20 female BALB/c nude mice were purchased from HFK Biotechnology (Beijing, China). Animal studies have been allowed by the ethics association. Mice were randomly divided into experimental groups. Vector control/CSN6-expressing MKN-45 or MGC-803 (1 × 106) cells were harvested in 150 μl liquid medium. Then per injection were injected subcutaneously into the dorsal flanks of mice. Tumor volumes were measured and recorded per 5 days for up to 35 days. Then the tumors were removed for data analysis. The protocols for animal studies were approved by the Institutional Animal Care and Use Committee of Xuzhou Medical University.
Statistical analysis
All experiments were performed at least three times unless otherwise indicated. Data are shown as means ± SD. Two-tailed Student’s t-test was performed to calculate significance in an interval of 95% confidence level. Statistical differences between the means for the different groups were evaluated with Instat 5.0 (GraphPAD software, San Diego, CA) using one-way analysis of variance (ANOVA). For TMA, the association between CSN6 staining and the clinicopathologic parameters of the GC patients were evaluated by χ2 test. Difference between each patient’s tumors with its normal counterpart was evaluated by paired χ2 test. Kaplan-Meier method and log-rank test were used to evaluate the correlation between CSN6 expression and patient survival. A value of P < 0.05 was considered statistically significant.
Results
CSN6 protein level is increased in human GC and high CSN6 expression is correlated with poor patient prognosis
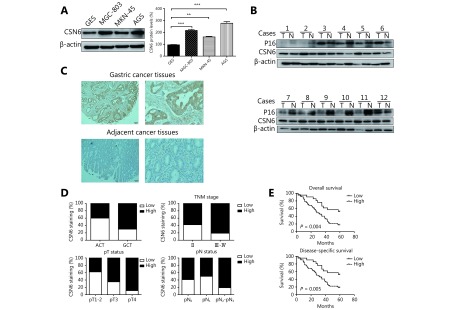
In order to investigate CSN6 expression in GC cells and tissues, Western blot and immunohistochemistry staining were carried out in GC tissues and non-tumor tissues. Western blot analysis demonstrated the higher expression of CSN6 protein in 3 human GC cell lines compared to normal gastric epithelium cell line (Figure 1A). Dysregulated CSN6 may be associated with various gene expressions in human tumors. Then we examined the p16 expression paralleling the levels of CSN6 protein of 12 patients in matched normal and cancerous gastric tissues. We found that 10 of 12 cancer samples had higher levels of CSN6 correlated with lower p16 expression as compared to matched normal tissues (Figure 1B). To identify the correlation of CSN6 expression with clinicopathological parameters, immunohistochemistry staining was performed in TMA slide containing GC tissues and paired adjacent tumor tissues (Figure 1C). We found that higher expression of CSN6 was observed in carcinoma tissues compared with adjacent tumor tissues (Figure 1D). The clinicopathological characteristics of CSN6 expression in GC patients was also investigated by microarray analysis and is summarized in Table 2. We observed that high expression of CSN6 protein was significantly related to some clinicopathological features, such as TNM stage (P = 0.019), depth of invasion-pT status (P = 0.000) and lymph node metastasis-pN status (P = 0.028) (Figure 1D). For the reason that CSN6 expression is remarkably increased in GC, we sought to determine whether CSN6 expression is associated with the prognosis. Survival analysis revealed that high level of CSN6 was associated with overall survival in 80 cases (P = 0.004, log-rank test, Figure 1E top panel). Furthermore, we observed the relationship between CSN6 status and GC specific survival. It was observed that CSN6 was also correlated with disease-specific survival (P = 0.005, log-rank test, Figure 1E bottom panel). Thus, CSN6 may serve as a novel molecular prognostic indicator for GC.
1.
CSN6 is increased in GC tissues and is associated with clinicopathologic parameters in GC. (A) Western blot analysis was used to evaluate the protein levels of CSN6 in various GC cell lines and a normal gastric epithelium cell line. β-actin served as a loading control. (B) Expression status of CSN6 and p16 in matched cancerous (T) and normal (N) regions isolated from GC patients. Cell lysates from 12 primary human GC samples were immunoblotted with the indicated antibodies. (C) Representative photos of CSN6 expression patterns in gastric tumors. The top panel shows the GC tissues, whereas the bottom panel depicts matched tumor adjacent tissues (original magnification, 100×). The magnifying detail of the immunohistochemical analysis for each case can be shown on the right side (original magnification, 400×). (D) Compared with the tumor adjacent cancer tissues (ACT), the overall expression level of CSN6 in the gastric cancer tissues (GCT) was significantly higher, P < 0.01, χ2 test. Increased CSN6 expression was correlated with TNM stage, P < 0.05, χ2 test. Increased CSN6 expression was correlated with pT status, P < 0.05, χ2 test. Increased CSN6 expression was correlated with pN status, P < 0.05, χ2 test. (E) Kaplan–Meier estimates of the probability of 7-year overall survival according to low and high CSN6 expression of 80 patients with GC, P = 0.004, log-rank test. High CSN6 expression correlated with a poorer 7-year disease-specific cumulative survival for 77 GC patients, P = 0.005, log-rank test.
2.
Patients’ characteristics and CSN6 expression
| Variables | CSN6 staining | Total | P* | |
| Low (%) | High (%) | |||
| *P values are obtained from χ2 test. | ||||
| All points | 27 (30) | 63 (70) | 90 | |
| Age (years) | ||||
| ≤ 57 | 7 (23.3) | 23 (76.7) | 30 | 0.329 |
| > 57 | 20 (33.3) | 40 (66.7) | 60 | |
| Gender | ||||
| Male | 22 (31.4) | 48 (68.6) | 70 | 0.580 |
| Female | 5 (25) | 15 (75) | 20 | |
| Tumor size (cm) | ||||
| ≤ 7 | 17 (27.9) | 44 (72.1) | 61 | 0.522 |
| > 7 | 10 (34.5) | 19 (65.5) | 29 | |
| pT status | ||||
| pT1-2 | 16 (61.5) | 10 (38.5) | 26 | 0.000 |
| pT3 | 6 (35.3) | 11 (64.7) | 17 | |
| pT4 | 5(10.6) | 42 (89.4) | 47 | |
| pN status | ||||
| pN0 | 9 (40.9) | 13 (59.1) | 22 | 0.028 |
| pN1 | 8 (50) | 8 (50) | 16 | |
| pN2-pN3 | 10 (19.2) | 42 (80.8) | 52 | |
| TNM stage | ||||
| II | 18 (41.9) | 25 (58.1) | 43 | 0.019 |
| III-IV | 9 (19.1) | 38 (80.9) | 47 | |
CSN6 promotes cell proliferation, colony formation and cell cycle progression
Results from the CCK-8 assays indicated that CSN6 was a positive regulator of cell growth (Figure 2A). Furthermore, clonogenic analysis showed that CSN6 transfectants evidently grew faster and formed larger colonies than control cells. Meanwhile, the colonies were fewer in CSN6-knockdown cells compared with the control groups (Figure 2B). Consistently, CSN6-overexpression in GC cells increased EdU incorporation and CSN6-knockdown reduced EdU incorporation compared with the control groups (Figure 2C). These results suggested that CSN6 may promote the cell proliferation and colony formation of GC cells.
2.
Effects of CSN6 expression on proliferation in GC cells. (A) CCK-8 assay was performed in MKN-45 cells and MGC-803 cells after transfection of pCDNA3.1-CSN6/pCDNA3.1 empty vector expression plasmids and control siRNA/CSN6 siRNA for 24, 48, 72 and 96h. (B) The effects of CSN6 on the colony formation ability of CSN6-knockdown and CSN6-overexpression in MKN-45 and MGC-803 cell lines. (C) CSN6 expression promotes cell proliferation of EdU incorporation. MKN-45 and MGC-803 cells were transfected with CSN6 and fixed for anti-EdU staining (original magnification, 100×). EdU-positive cells were counted and are presented as a bar graph. (D) The percentage of G1 and S population cells was measured by flow cytometry after CSN6 overexpression in MKN-45 and MGC-803 cells. After the knockdown of CSN6 in GC cells, the percentage of cells at G1 and S stage was calculated using ModFit LT 3.0 software. The data represent the mean ± SD (*P ≤ 0.05, **P ≤ 0.01). All experiments were performed in triplicate.
To validate whether CSN6 has an effect on cell cycle, the flow cytometric analysis was performed to examine the cell cycle distribution in GC cells. Flow cytometric analyses in Figure 2D (top panel) revealed that CSN6 significantly decreased the cells in the G1 phase from 65.28% to 49.54% on average in MKN-45 cells and from 68.76% to 51.93% in MGC-803 cells. The cell population in the S phase was increased from 22.33% to 34.68% in MKN-45 cells and from 21.38% to 32.47% in MGC-803 cells. Increasing expression of CSN6 significantly decreased the G1 cell population and increased the S phase cells, showing that overexpression of CSN6 markedly enhanced the G1/S transition. Consistently, the attenuation of CSN6 expression significantly increased the G1 cell population and decreased the S phase cells, showing that the attenuation of CSN6 expression in GC cells markedly arrested the G1/S transition (Figure 2D bottom panel). Taken together, these data demonstrated that CSN6 could promote cell cycle progression in GC cells.
CSN6 downregulates p16 in a dose-dependent manner
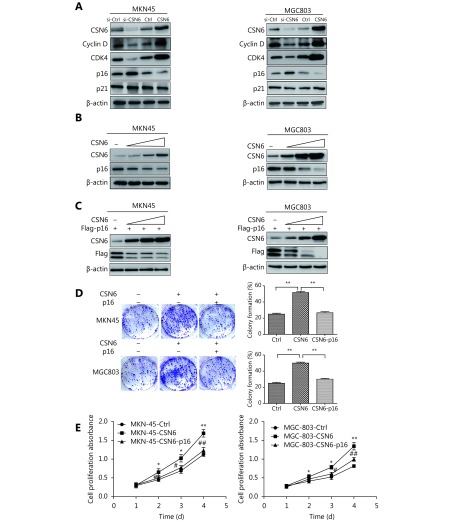
To explore the molecular mechanism underlying CSN6-induced cell cycle progression, we measured the expression of some G1 cell cycle regulatory proteins. We found that CSN6 greatly affected the protein expression of cyclin D, CDK4 and p16 (Figure 3A). Previous studies have found that the expression of CDK4 and cyclin D was reduced in the CSN6-knockdown cells19. It is well known that the p16 and p21 proteins are cyclin-dependent kinase inhibitors. To investigate the mechanisms of CSN6 regulating cancer progression in GC cells, we performed western blot to detect the relationship between CSN6 and cyclin-dependent kinase inhibitor protein levels in GC cells. Although the results showed that there was no significant change in the p21 in both cell lines (Figure 3A), it was shown that CSN6 could downregulate the steady-state expression of endogenous p16 in a dose-dependent manner in GC cells (Figure 3B). Similar results were obtained for levels of exogenous p16 (Figure 3C). Together, these results suggested that CSN6 was able to negatively regulate p16 in a dose-dependent manner. Furthermore, we investigated if CSN6 promoted cell proliferation via reducing p16 expression. As we invalidated before, CSN6 promoted the cell proliferation and cell clone formation, and the restoration of p16 rescued the phenotypes of CSN6 overexpression (Figure 3D-3E).
3.
CSN6 negatively regulates the expression of p16. (A) Protein expression levels of CSN6, cyclin D, CDK4, p16 and p21 were analyzed by Western blot in MKN-45 and MGC-803 cells after knockdown or overexpression of CSN6. Lysates were immunoblotted with the indicated antibodies. (B) CSN6 reduced the protein level of p16 in a dose-dependent manner in MKN-45 and MGC-803 cell lines. Equal amounts of cell lysates were immunoblotted with the indicated antibodies. (C) MKN-45 and MGC-803 cells were co-transfected with p16 plasmid and increasing amounts of CSN6. CSN6 negatively regulates the steady-state expression of p16 in a dose-dependent manner. Equal amounts of cell lysates were immunoblotted with the indicated antibodies. (D) Overexpression of CSN6 promoted the cell clone formation. Restoration of p16 rescued the augmention of cell clone formation. (E) Overexpression of CSN6 promoted cell proliferation. Restoration of p16 rescued the promotion of cell proliferation. The data represent the mean ± SD (* or #P ≤ 0.05, ** or ##P ≤ 0.01).
CSN6 downregulates p16 via proteasomal degradation
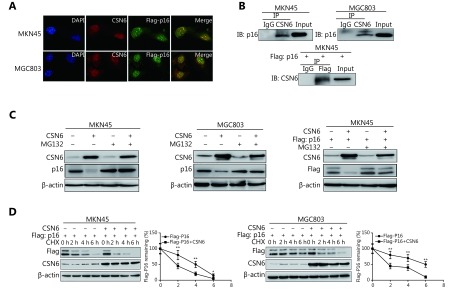
The above observations prompted us to investigate whether CSN6 has a direct effect on p16. The immunofluorescence localization study showed that the protein CSN6 colocalized with p16 (Figure 4A). In addition, co-IP showed association between endogenous CSN6 and p16 in MKN-45 and MGC-803 cells. Additionally, exogenous interaction of the two proteins in cells was also shown by co-IP experiments (Figure 4B). We next investigated how CSN6 interacted with p16 to regulate the protein level of p16. GC cells were transfected with the indicated plasmids and treated with the proteasome inhibitor MG132. The data showed that CSN6-mediated downregulation of p16 was rescued by MG132. Consistently, CSN6-mediated downregulation of exogenous p16 was also prevented by proteasome inhibitor MG132 (Figure 4C). Accordingly, in MKN-45 cells, the p16 turnover rate was more rapid in CSN6-overexpression than in control cells in the presence of the de novo protein synthesis inhibitor cycloheximide (CHX). CSN6 also reduced the p16 half-life period in MGC-803 cells (Figure 4D). The above results suggested that CSN6 downregulated p16 protein at the post-transcriptional level. Thus, CSN6 may downregulate p16 by proteasomal degradation.
4.
The 26S proteasome is involved in CSN6-mediated p16 degradation. (A) MKN-45 or MGC-803 cells were transfected with Flag-p16. The CSN6 colocalized with Flag-p16 (original magnification, 400×). The photos of CSN6 and Flag-p16 immunofluorescence staining are merged, and are labeled as Merge. (B) The MKN-45 or MGC-803 cell lysates were analyzed by immunoprecipitation with CSN6 and were immunoblotted with anti-p16. CSN6 interacted with endogenous p16 (top panel). The indicated plasmids were transfected into MKN-45 cells. Lysates were analyzed by immunoprecipitation with Flag and were immunoblotted with anti-CSN6. CSN6 interacted with extraneous p16 (bottom panel). (C) Proteasome inhibitor rescued CSN6-mediated degradation of p16. MKN-45 or MGC-803 cells were transfected with the indicated plasmids and treated with or without proteasome inhibitor MG132 (50 μg/mL, 6 h). Cell lysates were immunoblotted with the indicated antibodies. (D) CSN6 increased p16 turnover. GC cells were transfected with the indicated plasmids and were then treated with cycloheximide (CHX; 100 μg/mL) for the indicated times. The immunoblot of Flag-p16 at each time point was measured using a densitometer. The turnover of p16 is indicated graphically.
The 20S proteasome ubiquitin-independent degradation pathway is involved in CSN6-mediated downregulation of p16
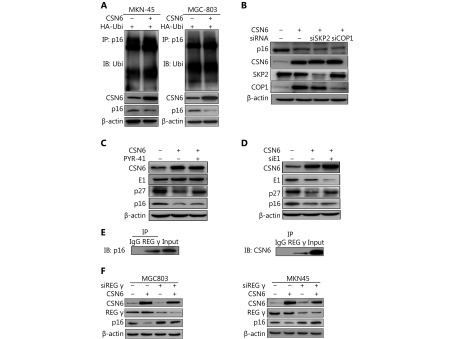
In many cases, modulation of protein levels largely relies on ubiquitination-mediated degradation by the 26S proteasome. We then validated whether ubiquitination is involved in CSN6-mediated p16 degradation. Thus, we performed ubiquitination assay and found that CSN6 overexpression did not increase the endogenous ubiquitination level of p16 (Figure 5A). The COP9 signalosome usually collaborates with an E3 ligase such as SKP2 or COP1 to degrade target proteins through ubiquitination30,31. We then examined whether CSN6 reduced the expression of p16 through E3 ubiquitin ligases. As shown in Figure 5B, the knockdown of SKP2 or COP1 did not rescue the degradation of p16 with the enhancement of CSN6. In the ubiquitination pathway, ubiquitin is activated in a two-step reaction by an E1 ubiquitin-activating enzyme. Then GC cells were treated with PYR-41 (an ubiquitin E1 inhibitor), and the results showed that p27 which is identified to be degraded by ubiquitination31, was upregulated with the inhibition of E1 activity. However, p16 did not change when the E1 activity was inhibited (Figure 5C). Furthermore, we detected the expression of p27 and p16 proteins in E1-knockdown cells and we found that the protein level of p27 increased and the p16 expression did not change (Figure 5D). Thus, we may reach a conclusion that CSN6 promotes p16 degradation through an ubiquitin-independent pathway.
5.
CSN6 regulates p16 in a ubiquitin-independent proteasomal manner. (A) MKN-45 and MGC803 cells were transfected with control vector or CSN6 and HA-Ubi. The cell lysates were immunoprecipitated with anti-p16 and immunoblotted with anti-ubiquitin antibody. Equal amounts of the cell lysates were analyzed by immunoblotting with the indicated antibodies. (B) MKN-45 cells were transfected with siRNA SKP2 or siRNA COP1 plus the indicated plasmids. Cell lysates were immunoblotted with the indicated antibodies. (C) MKN-45 cells were treated with PYR-41 for 30 min plus the indicated plasmids. Cells were lysed and analyzed by Western blot for E1, p27 and p16. (D) MKN-45 cells were transfected with E1 siRNA and CSN6 plasmid. Western blot assay was performed to detect the expression of p27 and p16 proteins. (E) Interaction of endogenous REGγ with endogenous p16 and CSN6. Equal amounts of MKN-45 cell lysates were immunoprecipitated with REGγ antibody and immunoblotted with anti-p16 and anti-CSN6 antibody. (F) MKN-45 or MGC803 cells were transfected with siRNA REGγ and the indicated plasmids. Cell lysates were immunoblotted with the indicated antibodies.
It has been identified that proteins can be targeted for ubiquitin-independent degradation by the 20S proteasome, which requires proteasome activators such as REGγ to facilitate degradation32. To explore whether the 20S proteasome activator REGγ was involved in CSN6 induced p16 degradation, we performed co-IP to examine the interaction of the proteins. Our results showed that REGγ bound with CSN6 and p16 (Figure 5E). Moreover, CSN6-mediated downregulation of p16 was rescued by knockdown of REGγ (Figure 5F). Taken together, the above data demonstrated that CSN6 downregulated p16 stability through regulating the 20S proteasome ubiquitin-independent degradation pathway.
CSN6 promotes cancer growth via regulating p16 protein level in xenograft model
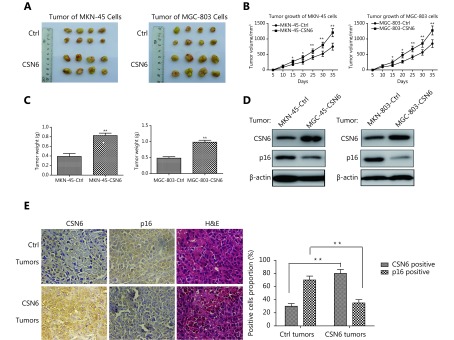
To further investigate the role of CSN6 in the promotion of tumorigenesis, we established xenograft cancer model by subcutaneously inoculating stably transfected CSN6 in MKN-45 and MGC-803 cells. To avoid individual variations in mice, the corresponding control cells were subcutaneously injected into the left side of the same mice. CSN6 increased the tumor growth rate and tumor weight compared with vector control-transfected cells (Figure 6A-C). Western blot showed that treatment with CSN6 overexpression GC cells mediated tumor growth and reduced p16 level in tumors (Figure 6D). We then examined the protein expression of CSN6 and p16 in tumors isolated from nude mice by immunohistochemistry. More CSN6 positive signals and fewer p16 signals were observed in CSN6-overexpressing tumors than in the control group (Figure 6E). Clearly, our study showed that overexpression of CSN6 bestowed survival advantages to MKN-45 and MGC-803 cells for the tumorigenesis in mice. Collectively, CSN6 could promote tumorigenesis through regulating the p16 protein level during the development of tumors.
6.
Involvement of CSN6 and p16 in tumorigenesis. (A-C) Overexpression of CSN6 promoted tumorigenicity. CSN6-overexpressing MKN-45 or MGC-803 stable transfectants were subcutaneously injected into the right side of the dorsal flanks of eight nude mice. Simultaneously, their corresponding control cells were injected into the left side of the same mice. The tumor volumes of the individual mice were measured every 5 days. Tumors were collected at the end of the assay, and the tumor weight of each group was measured. The data represent the mean±SD (*P ≤ 0.05, **P ≤ 0.01). (D) CSN6 expression in xenograft mouse models regulated protein levels of p16. MKN-45 or MGC-803 cells with CSN6 overexpression or control vector were subcutaneously injected into the nude mice. The indicated tumor samples were isolated and analyzed by immunoblotting with the indicated antibodies. (E) Overexpression of CSN6 diminished p16 expression in tumors. Tumors were collected and representative tumor sections are shown (original magnification, 400×). The statistical graph indicates the proportion of positive cells (**P ≤ 0.01).
Discussion
In the COP9 signalosome, both CSN6 and CSN5 own the MPN domain33, which is involved in controlling Cullin deneddylation activity34. The MPN domain bears a resemblance to the active site residues of metalloproteases that are involved in proteasome associated deneddylation activity35,36. CSN6 is a critical ubiquitination regulator involved in cell cycle regulation10. However, its role in GC remains to be determined. Here, we discovered that CSN6 has a critical role in controlling the protein level of p16INK4a. p16 is a tumor suppressor and performs multiple biological functions such as the induction of senescence, cell apoptosis, DNA repair and the inhibition of cell cycle progression24,37-39. Here we report for the first time that CSN6 plays an important role in GC and is a negative regulator of p16 that promotes tumorigenesis.
Recent studies have progressively focused on tumor-associated genes that are presumed to be responsible for cancer development40,41. In the present study, we investigated the role of CSN6 and p16 proteins in GC. It is worth noting that in our collection of GC examples, CSN6 expression was increased in GC tissues compared with normal gastric tissues. Furthermore, we examined the association between CSN6 expression and clinicopathological characteristics, and our data showed that high expression of CSN6 was significantly correlated with TNM stage. In addition, Kaplan-Meier analysis showed that high expression of CSN6 was associated with poor overall survival in GC patients. In general, these observations suggested that CSN6 is an important prognostic factor in GC and could play a potential role in GC proliferation. Because tumor cell proliferation and growth are essential steps in the process of carcinogenesis, we then investigated the effects of CSN6 on cell proliferation and cell cycle regulation in GC. Our results demonstrated that CSN6 promoted the growth and proliferation capacities of cancer cells by G1 arrest. On the basis of our in vitro studies, we found that CSN6 overexpression significantly promoted the formation of tumors in nude mice. Moreover, the expression trends of CSN6 and p16 in CSN6-treated tumor groups were consistent with the results of in vitro studies, which demonstrated that CSN6 promoted cancer cell growth by reducing p16 expression. Taken together, the in vivo study about CSN6 provides more evidence to support the contributions of CSN6 in tumor development.
Cell cycle progression is regulated by a family of protein kinases named cyclin-dependent kinases (CDKs), which are regulated by different mechanisms including interaction with two families of inhibitors (CKIs), the Cip-Kip family and the INK4 family. The negative correlation between CSN6 and p16 in both cancer cell lines and tumor tissues is particularly interesting, as the p16 protein level is associated with the prognosis of many types of human cancer. The cell cycle inhibitors of the INK4-class include p15INK4b, p16INK4a, p18INK4c, and p19INK4a 42. Our mechanistic studies indicated that CSN6 interacted with p16, and that CSN6-mediated p16 downregulation was suppressed by a proteasome inhibitor. In addition, the turnover rate of p16 in the presence of the de novo protein synthesis inhibitor was upregulated by CSN6. These results demonstrated that the reduction of p16 depended on the proteasomal degradation, and that CSN6 may function as an accelerator during p16 degradation.
It was identified that CSN subunits may promote Cullin activity in three distinct ways: promoting deneddylation, indirectly counteracting spurious ubiquitination and stimulating Cullin-4 function43. Cullins are a family of hydrophobic proteins that provide a scaffold for ubiquitin ligases (E3). They combine with RING proteins to form Cullin-RING ubiquitin ligases (CRLs) that are highly diverse and play a role in myriad cellular processes. Cullin-dependent ubiquitin ligases regulate a variety of cellular and developmental processes by recruiting specific proteins for ubiquitin-mediated degradation44. In addition, CSN has also been proposed to serve as a regulator of the proteasome45,46. CSN6, as a subunit of CSN, is involved in regulating cullin-based E3 ligases and works together with E3 ligase COP1 to regulate the ubiquitination process of p2730. There is a mutual regulatory relationship between CSN6 and E3 ligase E6AP, which are involved in ubiquitination regulation to target protein degradation47. The ubiquitin-dependent degradation process is the major system that is involved in proteasomal degradation. Thus, it raises the question of whether CSN6 targets human p16 through a ubiquitin-dependent degradation pathway. It was identified that the ubiquitin-mediated proteolysis system mediates proteasomal degradation by attaching a polyubiquitin chain to lysine residues in the target protein. However, the primary sequence of the p16 protein does not contain any lysine residues. It is our goal to confirm whether CSN6 promotes the degradation of p16 via an ubiquitination pathway. However, our study showed that CSN6 did not promote the p16 ubiquitination during the degradation process. Thus, these observations present a paradox: How does CSN6 reduce p16 expression? Is there another manner of regulation for CSN6 to decrease p16 protein other than CSN6 controlling ubiquitination? The mechanism underlying CSN6-mediated p16 degradation in GC remains to be explored.
Although the ubiquitin-dependent proteasomal degradation process is considered the major route that mediates proteasomal degradation23, proteins can also be targeted for degradation by the 20S proteasome, which does not require ubiquitylation or the presence of the 19S regulatory particle31. Our study showed that the E3 ligases were not involved in CSN6-mediated regulation of p16 protein degradation (Figure 5B). It was identified that the E1 ubiquitin-activating enzyme initiates ubiquitination by modifying ubiquitin and is essential to the ubiquitination pathway. Then, GC cells were treated with ubiquitin E1 inhibitor PYR-41, and the result showed that p16 did not change with the inhibition of E1 activity. Furthermore, the GC cells were transfected with E1 siRNA to attenuate E1 expression and we found that the protein level of p27 increased and p16 expression did not change. Thus, the E1 ubiquitin-activating enzyme was not involved in p16 protein degradation. Thus, we may reach a conclusion that p16 degradation is ubiquitin independent. On the basis of our mechanism studies, we proposed that p16 may be regulated by CSN6 via 20S proteasome ubiquitin-independent degradation. The 20S proteasome is a cylindrically shaped complex with a heterodimeric structure (α7β7β7α7 subunits)48. It is associated with some proteasome activators such as REGγ and PA200 regulators that activate the proteolytic capacity of the complex49. We subsequently investigated whether REGγ could present cell cycle regulator p16 to proteasomes for degradation. It was validated that REGγ could interact with both CSN6 and p16 proteins. Furthermore, the decrease of REGγ could rescue CSN6-mediated downregulation of p16. These observations demonstrated that CSN6 might promote the 20S proteasomal ubiquitin-independent degradation of p16.
Conclusions
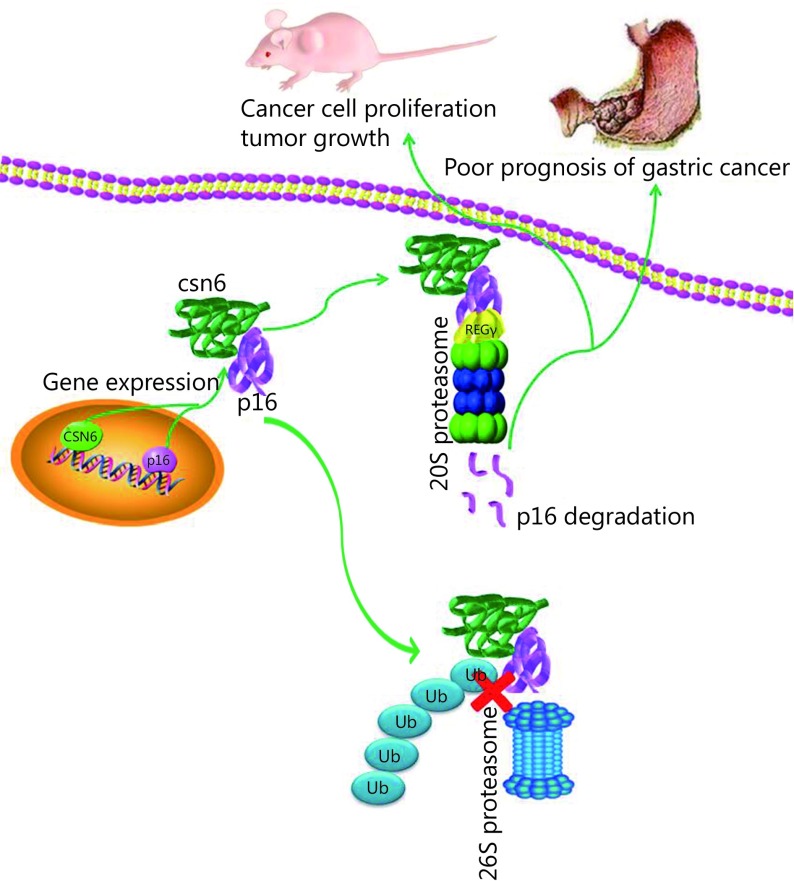
Taken together, our study provides evidence that increased CSN6 expression was significantly correlated with GC progression and was a novel prognostic factor of worse outcome in GC patients. Furthermore, CSN6 decreased the stability of tumor suppressor p16 in an ubiquitin-independent proteasomal degradation pathway and thereby promoted tumorigenicity (Figure 7). Our study provides the in vitro and in vivo evidence that CSN6-p16 serves as an important target for GC therapy. Targeting the CSN6 may be a useful therapeutic strategy for cancer intervention in p16-deficient cancer.
7.
Model of the effect of CSN6 on modulating p16. CSN6 decreased the stability of tumor suppressor p16 in a ubiquitin-independent proteasomal degradation pathway and thereby promoted cancer cell proliferation, tumor growth, and poor prognosis of GC.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant No. 81572349, 81872080), Jiangsu Provincial Medical Talent (Grant No. ZDRCA2016055) and the Science and Technology Department of Jiangsu Province (Grant No. BK20181148).
Conflict of interest statement
No potential conflicts of interest are disclosed.
Contributor Information
Jun Song, Email: songjunwk@126.com.
Dongsheng Pei, Email: dspei@xzhmu.edu.cn.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.v61:2. [DOI] [PubMed] [Google Scholar]
- 2.Chen WQ, Zheng RS, Zeng HM, Zhang SW The updated incidences and mortalities of major cancers in China, 2011. Chin J Cancer. 2015;34:53. doi: 10.1186/s40880-015-0042-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lowy AM, Feig BW, Janjan N, Rich TA, Pisters PWT, Ajani JA, et al A pilot study of preoperative chemoradiotherapy for resectable gastric cancer. Ann Surg Oncol. 2001;8:519–24. doi: 10.1007/s10434-001-0519-1. [DOI] [PubMed] [Google Scholar]
- 4.Stock M, Otto F Gene deregulation in gastric cancer. Gene. 2005;360:1–19. doi: 10.1016/j.gene.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 5.Xue Z, Vis DJ, Bruna A, Sustic T, van Wageningen S, Batra AS, et al MAP3K1 and MAP2K4 mutations are associated with sensitivity to MEK inhibitors in multiple cancer models . Cell Res. 2018;28:719–29. doi: 10.1038/s41422-018-0044-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wei N, Chamovitz DA, Deng XW Arabidopsis COP9 is a component of a novel signaling complex mediating light control of development. Cell. 1994;78:117–24. doi: 10.1016/0092-8674(94)90578-9. [DOI] [PubMed] [Google Scholar]
- 7.Chamovitz DA, Wei N, Osterlund MT, von Arnim AG, Staub JM, Matsui M, et al The COP9 complex, a novel multisubunit nuclear regulator involved in light control of a plant developmental switch. Cell. 1996;86:115–21. doi: 10.1016/S0092-8674(00)80082-3. [DOI] [PubMed] [Google Scholar]
- 8.Chen B, Zhao RY, Su CH, Linan M, Tseng C, Phan L, et al CDK inhibitor p57Kip2 is negatively regulated by COP9 signalosome subunit 6 . Cell Cycle. 2012;11:4633–41. doi: 10.4161/cc.22887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xue YW, Chen J, Choi HH, Phan L, Chou PC, Zhao RY, et al HER2-Akt signaling in regulating COP9 signalsome subunit 6 and p53. Cell Cycle. 2012;11:4181–90. doi: 10.4161/cc.22413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choi HH, Gully C, Su CH, Velazquez-Torres G, Chou PC, Tseng C, et al COP9 signalosome subunit 6 stabilizes COP1, which functions as an E3 ubiquitin ligase for 14-3-3σ. Oncogene. 2011;30:4791–801. doi: 10.1038/onc.2011.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Richardson KS, Zundel W The emerging role of the COP9 signalosome in cancer. Mol Cancer Res. 2005;3:645–53. doi: 10.1158/1541-7786.MCR-05-0233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tomoda K, Kubota Y, Kato JY Degradation of the cyclin-dependent-kinase inhibitor p27Kip1 is instigated by Jab1 . Nature. 1999;398:160–5. doi: 10.1038/18230. [DOI] [PubMed] [Google Scholar]
- 13.Wei N, Deng XW The COP9 signalosome. Annu Rev Cell Dev Biol. 2003;19:261–86. doi: 10.1146/annurev.cellbio.19.111301.112449. [DOI] [PubMed] [Google Scholar]
- 14.Scheel H, Hofmann K Prediction of a common structural scaffold for proteasome lid, COP9-signalosome and eIF3 complexes. BMC Bioinformatics. 2005;6:71. doi: 10.1186/1471-2105-6-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang GW, Cai HC Ubiquitin-proteasome system and sperm DNA repair: an update. Nat J Androl. 2016;22:834–7. [PubMed] [Google Scholar]
- 16.Wolf DA, Zhou CS, Wee S The COP9 signalosome: an assembly and maintenance platform for cullin ubiquitin ligases? Nat Cell Biol. 2003;5:1029–33. doi: 10.1038/ncb1203-1029. [DOI] [PubMed] [Google Scholar]
- 17.Shin J, Phan L, Chen J, Lu ZM, Lee MH CSN6 positively regulates c-Jun in a MEKK1-dependent manner. Cell Cycle. 2015;14:3079–87. doi: 10.1080/15384101.2015.1078030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao RY, Yeung SCJ, Chen J, Iwakuma T, Su CH, Chen B, et al Subunit 6 of the COP9 signalosome promotes tumorigenesis in mice through stabilization of MDM2 and is upregulated in human cancers. J Clin Invest. 2011;121:851–65. doi: 10.1172/JCI44111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hou J, Deng Q, Zhou J, Zou J, Zhang Y, Tan P, et al CSN6 controls the proliferation and metastasis of glioblastoma by CHIP-mediated degradation of EGFR. Oncogene. 2017;36:1134–44. doi: 10.1038/onc.2016.280. [DOI] [PubMed] [Google Scholar]
- 20.Choi HH, Lee MH CSN6-COP1 axis in cancer. Aging. 2015;7:461–2. doi: 10.18632/aging.v7i7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bech-Otschir D, Kraft R, Huang XH, Henklein P, Kapelari B, Pollmann C, et al COP9 signalosome-specific phosphorylation targets p53 to degradation by the ubiquitin system. EMBO J. 2001;20:1630–9. doi: 10.1093/emboj/20.7.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hwang J, Winkler L, Kalejta RF Ubiquitin-independent proteasomal degradation during oncogenic viral infections. Biochim Biophys Acta. 2011;1816:147–57. doi: 10.1016/j.bbcan.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ben-Nissan G, Sharon M Regulating the 20S proteasome ubiquitin-independent degradation pathway. Biomolecules. 2014;4:862–84. doi: 10.3390/biom4030862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Serrano M, Hannon GJ, Beach D A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature. 1993;366:704–7. doi: 10.1038/366704a0. [DOI] [PubMed] [Google Scholar]
- 25.Sharpless NE INK4a/ARF: a multifunctional tumor suppressor locus . Mutat Res. 2005;576:22–38. doi: 10.1016/j.mrfmmm.2004.08.021. [DOI] [PubMed] [Google Scholar]
- 26.Sandhu C, Peehl DM, Slingerland J p16INK4A mediates cyclin dependent kinase 4 and 6 inhibition in senescent prostatic epithelial cells . Cancer Res. 2000;60:2616–22. [PubMed] [Google Scholar]
- 27.Ben-Saadon R, Fajerman I, Ziv T, Hellman U, Schwartz AL, Ciechanover A The tumor suppressor protein p16INK4a and the human papillomavirus oncoprotein-58 E7 are naturally occurring lysine-less proteins that are degraded by the ubiquitin system: direct evidence for ubiquitination at the N-terminal residue . J Biol Chem. 2004;279:41414–21. doi: 10.1074/jbc.M407201200. [DOI] [PubMed] [Google Scholar]
- 28.Remo A, Pancione M, Zanella C, Manfrin E p16 expression in prostate cancer and nonmalignant lesions: novel findings and review of the literature. Appl Immunohistochem Mol Morphol. 2016;24:201–6. doi: 10.1097/PAI.0000000000000171. [DOI] [PubMed] [Google Scholar]
- 29.Remmele W, Stegner HE Recommendation for uniform definition of an immunoreactive score (IRS) for immunohistochemical estrogen receptor detection (ER-ICA) in breast cancer tissue. Pathologe. 1987;8:138–40. [PubMed] [Google Scholar]
- 30.Denti S, Fernandez-Sanchez ME, Rogge L, Bianchi E The COP9 signalosome regulates Skp2 levels and proliferation of human cells. J Biol Chem. 2006;281:32188–96. doi: 10.1074/jbc.M604746200. [DOI] [PubMed] [Google Scholar]
- 31.Choi HH, Guma S, Fang LK, Phan L, Ivan C, Baggerly K, et al Regulating the stability and localization of CDK inhibitor p27Kip1 via CSN6-COP1 axis . Cell Cycle. 2015;14:2265–73. doi: 10.1080/15384101.2015.1046655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sánchez-Lanzas R, Castaño JG Proteins directly interacting with mammalian 20S proteasomal subunits and ubiquitin-independent proteasomal degradation. Biomolecules. 2014;4:1140–54. doi: 10.3390/biom4041140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Birol M, Enchev RI, Padilla A, Stengel F, Aebersold R, Betzi S, et al Structural and biochemical characterization of the Cop9 signalosome CSN5/CSN6 heterodimer. PLoS One. 2014;9:e105688. doi: 10.1371/journal.pone.0105688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang XH, Ordemann J, Pratschke J, Dubiel W Overexpression of COP9 signalosome subunits, CSN7A and CSN7B, exerts different effects on adipogenic differentiation. FEBS Open Bio. 2016;6:1102–12. doi: 10.1002/feb4.2016.6.issue-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maytal-Kivity V, Reis N, Hofmann K, Glickman MH MPN+, a putative catalytic motif found in a subset of MPN domain proteins from eukaryotes and prokaryotes, is critical for Rpn11 function. BMC Biochem. 2002;3:28. doi: 10.1186/1471-2091-3-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lyapina S, Cope G, Shevchenko A, Serino G, Tsuge T, Zhou CS, et al Promotion of NEDD8-CUL1 conjugate cleavage by COP9 signalosome. Science. 2001;292:1382–5. doi: 10.1126/science.1059780. [DOI] [PubMed] [Google Scholar]
- 37.Hara E, Smith R, Parry D, Tahara H, Stone S, Peters G Regulation of p16CDKN2 expression and its implications for cell immortalization and senescence. Mol Cell Biol. 1996;16:859–67. doi: 10.1128/MCB.16.3.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kataoka M, Wiehle S, Spitz F, Schumacher G, Roth JA, Cristiano RJ Down-regulation of bcl-2 is associated with p16INK4-mediated apoptosis in non-small cell lung cancer cells . Oncogene. 2000;19:1589–95. doi: 10.1038/sj.onc.1203466. [DOI] [PubMed] [Google Scholar]
- 39.Shapiro GI, Edwards CD, Ewen ME, Rollins BJ p16INK4A participates in a G1 arrest checkpoint in response to DNA damage . Mol Cell Biol. 1998;18:378–87. doi: 10.1128/MCB.18.1.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ahmat Amin MKB, Shimizu A, Zankov DP, Sato A, Kurita S, Ito M, et al Epithelial membrane protein 1 promotes tumor metastasis by enhancing cell migration via copine-III and Rac1 . Oncogene. 2018;37:5416–34. doi: 10.1038/s41388-018-0286-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang KX, Ji WX, Yu YF, Li ZM, Niu XM, Xia WL, et al FGFR1-ERK1/2-SOX2 axis promotes cell proliferation, epithelial-mesenchymal transition, and metastasis in FGFR1-amplified lung cancer. Oncogene. 2018;37:5340–54. doi: 10.1038/s41388-018-0311-3. [DOI] [PubMed] [Google Scholar]
- 42.Souza-Rodrígues E, Estanyol JM, Friedrich-Heineken E, Olmedo E, Vera J, Canela N, et al Proteomic analysis of p16ink4a-binding proteins . Proteomics. 2007;7:4102–11. doi: 10.1002/(ISSN)1615-9861. [DOI] [PubMed] [Google Scholar]
- 43.Cope GA, Deshaies RJ COP9 signalosome: a multifunctional regulator of SCF and other cullin-based ubiquitin ligases. Cell. 2003;114:663–71. doi: 10.1016/S0092-8674(03)00722-0. [DOI] [PubMed] [Google Scholar]
- 44.Donaldson TD, Noureddine MA, Reynolds PJ, Bradford W, Duronio RJ Targeted disruption of Drosophila Roc1b reveals functional differences in the Roc subunit of Cullin-dependent E3 ubiquitin ligases . Mol Biol Cell. 2004;15:4892–903. doi: 10.1091/mbc.e04-03-0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun Y, Wilson MP, Majerus PW Inositol 1,3,4-trisphosphate 5/6-kinase associates with the COP9 signalosome by binding to CSN1. J Biol Chem. 2002;277:45759–64. doi: 10.1074/jbc.M208709200. [DOI] [PubMed] [Google Scholar]
- 46.Wilson MP, Sun Y, Cao L, Majerus PW Inositol 1,3,4-trisphosphate 5/6-kinase is a protein kinase that phosphorylates the transcription factors c-Jun and ATF-2. J Biol Chem. 2001;276:40998–1004. doi: 10.1074/jbc.M106605200. [DOI] [PubMed] [Google Scholar]
- 47.Gao SJ, Fang LK, Phan LM, Qdaisat A, Yeung SCJ, Lee MH COP9 signalosome subunit 6 (CSN6) regulates E6AP/UBE3A in cervical cancer. Oncotarget. 2015;6:28026–41. doi: 10.18632/oncotarget.4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tomko RJ Jr, Hochstrasser M Molecular architecture and assembly of the eukaryotic proteasome. Annu Rev Biochem. 2013;82:415–45. doi: 10.1146/annurev-biochem-060410-150257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ma CP, Slaughter CA, DeMartino GN Identification, purification, and characterization of a protein activator (PA28) of the 20 S proteasome (macropain) J Biol Chem. 1992;267:10515–23. [PubMed] [Google Scholar]