Abstract
Proteases are multifunctional, promiscuous enzymes that degrade proteins as well as peptides and drive important processes in health and disease. Current technology has enabled the construction of libraries of peptide substrates that detect protease activity, which provides valuable biological information. An ideal library would be orthogonal, such that each protease only hydrolyzes one unique substrate, however this is impractical due to off-target promiscuity (i.e., one protease targets multiple different substrates). Therefore, when a library of probes is exposed to a cocktail of proteases, each protease activates multiple probes, producing a convoluted signature. Computational methods for parsing these signatures to estimate individual protease activities primarily use an extensive collection of all possible protease-substrate combinations, which require impractical amounts of training data when expanding to search for more candidate substrates. Here we provide a computational method for estimating protease activities efficiently by reducing the number of substrates and clustering proteases with similar cleavage activities into families. We envision that this method will be used to extract meaningful diagnostic information from biological samples.
Author summary
The activity of enzymatic proteins, which are called proteases, drives numerous important processes in health and disease: including cancer, immunity, and infectious disease. Many labs have developed useful diagnostics by designing sensors that measure the activity of these proteases. However, if we want to detect multiple proteases at the same time, it becomes impractical to design sensors that only detect one protease. This is due to a phenomenon called protease promiscuity, which means that proteases will activate multiple different sensors. Computational methods have been created to solve this problem, but the challenge is that these often require large amounts of training data. Further, completely different proteases may be detected by the same subset of sensors. In this work, we design a computational method to overcome this problem by clustering similar proteases into "subfamilies", which increases estimation accuracy. Further, our method tests multiple combinations of sensors to maintain accuracy while minimizing the number of sensors used. Together, we envision that this work will increase the amount of useful information we can extract from biological samples, which may lead to better clinical diagnostics.
Introduction
Proteases are multifunctional enzymes that hydrolyze peptide bonds and are responsible for maintaining health in processes ranging from immunity to blood homeostasis, but are also drivers of diseases, including cancer and sepsis [1–10]. The ability to quantify the activity of proteases–of which there are >550 –in humans on a larger scale may provide valuable biological information, leading to improved diagnostic and therapeutic technologies. While Next Generation Sequencing technologies provide the ability to rapidly assess mRNA transcript levels of proteases, previous studies have shown a lack of correlation between expression and enzyme activity [11–13]. For this reason, countless platforms have been developed to sense and modulate protease activity both in vivo and in vitro, with the potential to extract useful physiological information [10, 14–29]. However, to completely resolve an individual's protease landscape (i.e., >550 proteases) with current technology would require a library of equal size, assuming all substrates are orthogonal (i.e., each protease hydrolyzes a unique substrate), which is impractical at this scale (i.e., on the order of 102). Further, current activity probes require experimental knowledge of protease-substrate specificity [30], which is difficult to completely map out because proteases are promiscuous [8], which means one protease is capable of hydrolyzing multiple different substrate sequences. Therefore, independent protease signatures become convolved when attempting to detect multiple proteases simultaneously, making it difficult to quantify the relative activity of each protease [31]. Previous studies have successfully developed computational algorithms to parse these signatures [32], but these methods may become complicated when applied to proteases with similar signatures in terms of their activities against substrates.
Here, to create a means for deconvolving protease signatures we develop a method, which requires limited prior knowledge of protease-substrate specificity. We demonstrate this method on a subset of blood proteases, including complement (e.g., C1r, MASP2, Factor D, etc.) and coagulation (e.g., Factor IIa, XIa, etc.) proteases, which display a high-degree of promiscuity and are involved in a range of hematological and immune disorders (i.e., clotting disorders, complement deficiencies, etc.) [33, 34]. To overcome the challenge of scaling to larger numbers of proteases, we use this method to improve experimental design by reducing the size of the substrate library. Furthermore, we cluster proteases with similar substrate activities into families, while maintaining high estimation accuracy. Under this framework, we lay the groundwork for understanding multiplexed protease-substrate signatures on a large scale, which may enable the future use of Massively Multiplexed Activity (MMA) libraries.
Methods
To improve experimental design for deconvolving protease composition of protease mixtures, we developed pipelines for estimating kinetic parameters from real experimental data, and simulating in silico experimental data. In this Method section, we introduce the individual components of the pipeline (Method "Cleavage dynamics approximation model" to "Quantifying estimation accuracy via Root-Mean-Square Error (RMSE)"). We then apply the pipeline to optimize the selection of substrates and cluster proteases into families (Method "Evaluating deconvolution performance and optimizing substrate selection").
The overall strategy for the deconvolution analysis consists of two optimization steps. The first step consists of learning the cleavage dynamics of every combination of one protease and one substrate by optimizing kinetic parameters for a modified Michaelis-Menten model [35, 36] (see details in Method "Modified Michaelis-Menten kinetics with saturation" and "Estimating kinetic parameters in the single-protease-single-substrate setting"). We then apply the kinetic parameters learned in the first step to estimate the mixing coefficients, which represent the individual concentrations of proteases in a mixture (see details in Method "Estimating mixing coefficients with multiple proteases "). In the case where a sufficient number of substrates are measured to deconvolve all individual proteases in a mixture, we screen for the optimal subset of substrates in order to reduce the required number of substrates. When highly correlated proteases exist in the mixture, which would require an impractically large number of substrates for deconvolution, we cluster the proteases into families via hierarchical clustering to enable deconvolution based on a reasonable number of substrates and achieve a higher accuracy at a lower resolution (Method "Evaluating deconvolution performance and optimizing substrate selection").
Recombinant protease activity assay
We tested a total of seven recombinant proteases in the activity assays. Complement proteases C1r (purity >90%), C1s (purity >95%), Complement Factor D (purity >90%), and Complement Factor I (purity >90%) were purchased from Sigma Aldrich. Complement protease MASP2 (purity >97%) was purchased from Biomatik. Coagulation proteases Factor IIa (purity >95%) and Factor XIa (purity >95%) were purchased from Haematologic Technologies. Initially, twenty peptide substrate sequences were curated from the literature, which represented discoveries from phage display screens as well as sequencing of physiological substrates [37–40]. We performed an initial screen with complement proteases which cleave after an arginine residue to identify the seven sequences used in these experiments (Fig A in S1 Text). To obtain the recombinant protease activity data, we first conjugated seven different c-terminus cysteine synthetic peptide substrates to amine functionalized 2 μm magnetic microparticles with SIA (i.e., succinimidyl iodoacetate), an amine-thiol crosslinker. The n-terminus of the peptides each contains one of seven unique glu-fib mass barcodes (Table 1). We then incubated a cocktail of these seven substrates (> 50 nM) with each of the seven recombinant proteases individually at 37°C on a spinner in PBS. At various time points between 0 and 400 minutes, we used a magnetic separator to remove the microparticles from the supernatant, which contained the hydrolyzed substrates plus mass barcodes. To provide a unique mass encoding for each of the seven substrates, we produced a family of mass reporters from Glu-fib with an isobaric mass-encoding strategy [41, 42]. By this method, all mass tags share the same parent mass so that peptides can be efficiently collected (i.e., MS-1) during tandem mass spectrometry (MS/MS), but can be differentiated after ion fragmentation (i.e., MS-2). Due to the fact that Glu-fib fragments into C-terminal y-type ions, we made mass codes centered on the y6 ion (i.e., GFFSAR). For each of the mass barcodes, we enriched the GFFSAR region with heavy amino acids, which resulted in sequences that varied by 1 Da each. To cancel out the resulting mass shifts, we balanced the remaining region (i.e., EGVNDNEE) by isotope enrichment. Mass spectrometry was performed by Syneos Health (Morrisville, NC) to quantify the amount of cleaved substrate at each time point. To summarize this method, 100 μL of sample volume and 25 μL of internal standard solution were UV-treated for 2 hours, using a UVP Analytik Jena UV Crosslinker CL-1000 oven. Sample cleanup was achieved using Mixed-mode anion exchange solid phase extraction. Chromatographic separation was achieved using a Waters XBridge C18 column, with the mobile phase composed of 0.1% formic acid in water and acetonitrile/trifluoroethanol. A gradient of 5% to 60% organic content at 0.6 mL/min over 3 minutes was employed. Analytes were analyzed using an AB Sciex 6500+ triple quadrupole mass spectrometer monitoring in MRM mode with an electrospray source set to positive ion mode. The total instrument run time was 5 minutes.
Table 1. Mass-barcoded peptide substrate sequences.
Table describing the sequences of the mass-barcoded peptide substrates along with their chemical modifications. ANP was used as a photocleavable linker to enable rapid detachment from the microparticles. 5-FAM was used for rapid quantification via fluorescence. Isotope enrichment modifications were used to distinguish mass barcodes for quantification with mass spectrometry.
| Substrate Name | Peptide sequence (N terminus on left)* | Modifications** |
|---|---|---|
| CC01 | e(*aa)(*aa)ndneeGFFsAr(ANP)K(5-FAM)GGLQRIYKC | 1st *aa = Gly(13C2); 2nd *aa = Val(U13C5,15N) |
| CC02 | eG(*aa)ndneeGF(*aa)s(*aa)r(ANP)K(5-FAM)GGKSVARTLLVKC | 1st *aa = Val(U13C5,15N); 2nd *aa = Phe(15N); 3rd *aa = Ala(15N) |
| CC03 | e(*aa)(*aa)ndneeGFFs(*aa)r(ANP)K(5-FAM)GGQRQRIIGGC | 1st *aa = Gly(U13C2,15N); 2nd *aa = Val(15N); 3rd *aa = Ala (U13C3,15N) |
| CC04 | e(*aa)Vndnee(*aa)FFs(*aa)r(ANP)K(5-FAM)GGKYLGRSYKVC | 1st *aa = Gly(13C2); 2nd *aa = Gly(13C2); 3rd *aa = Ala(U13C3,15N) |
| CC05 | eGVndnee(*aa)(*aa)Fs(*aa)r(ANP)K(5-FAM)GGGLQRALEIC | 1st *aa = Gly(U13C2,15N); 2nd *aa = Phe(15N); 3rd *aa = Ala(U13C3,15N) |
| CC06 | e(*aa)(*aa)ndnee(*aa)(*aa)(*aa)s(*aa)r(ANP)K(5-FAM)GGKTTGGRIYGGC | 1st *aa = Gly(13C2); 2nd *aa = Val(U13C5,15N); 3rd *aa = Gly(U13C2,15N); 4th *aa = Phe(15N); 5th *aa = Phe(15N); 6th *aa = Ala(15N); still include ANP and K5-FAM |
| CC07 | eG(*aa)ndnee(*aa)(*aa)Fs(*aa)r(ANP)K(5-FAM)GGQARGGSC | 1st *aa = Val(U13C5,15N); 2nd *aa = Gly(U13C2,15N); 3rd *aa = Phe(15N); 4th *aa = Ala(U13C3,15N) |
*ANP = Photocleavable linker 3-Amino-3-(2-nitrophenyl)propionic acid
*5-FAM = 5—Carboxyfluorescein
**Modifications represent heavy amino acids (i.e., isotope enrichment)
Cleavage dynamics approximation model
Michaelis-Menten kinetics as the approximation of cleavage dynamics
| (1) |
[S]: the remaining amount uncleaved substrate, [S] ∈ [0,1], [S]t = 0 = 1
V: the maximal rate of the reaction at the saturating substrate concentration
K: the substrate concentration when the reaction rate reaches half of V
n: the order of reaction
[α]: concentration of a protease
We use the Michaelis-Menten kinetics as the base model to approximate the cleavage dynamics [35, 36]. We add the mixing coefficient [α] (i.e., the concentration of a protease) is added to the original Michaelis-Menten model. On the left-hand side (LHS) of Eq (1), d[S]/dt represents the rate of change of the remaining uncleaved substrate. At t = 0, d[S]/dt is negative, which means that the substrate is being cleaved and [S] will decrease. As [S] decreases, the cleaving process slows down until [S] arrives at 0, where the reaction stops due to the depletion of the uncleaved substrates. However, in real experimental data, we noticed persistent non-zero saturation levels of uncleaved substrates, which motivated a modification of the model by adding a saturation term β (see details in Method "Modified Michaelis-Menten kinetics with saturation").
Modified Michaelis-Menten kinetics with saturation
Eq (2) is the modified Michaelis-Menten model for approximating the changing rate of one uncleaved substrate species when reacting with one protease. β is the saturation term representing the concentration of uncleaved substrate at which the reaction stops (when S = β, the RHS becomes 0). In subsequent discussions, we refer to V, K, n, and β as kinetic parameters, and [α] as the concentration mixing coefficient.
| (2) |
[S] ∈ [β,1], [S]t = 0 = 1.
β ∈ [0,1]
To generalize Eq (2) for modeling the dynamics of substrate cleavage by mixtures of proteases, subscripts i and j are introduced in Eq (3), which models the changing rate of the uncleaved substrates when reacting with multiple proteases.
| (3) |
[Si], i = 1, 2, 3, …, M
[αj], j = 1, 2, 3, …, N, ∀ j: [αj] > = 0
[Si] is the amount of uncleaved substrates of the ith substrate in the substrate library. [αj] is the concentration of jth protease in the mixture. Let M be the number of substrates and N be the number of proteases. Eq (3) assumes that no synergistic or antagonistic effect is involved among various proteases within the protease mixture when cleaving substrates.
Simulating in silico experiments
Simulating single-protease-single-substrate data
Given [S]t = 0 = 1, [α] = 1, and a set of specific kinetic parameter values (V, K, n, β), the amount of remaining uncleaved substrate [S]t = tz at a specific time t = tz can be calculated by numerically solving Eq (2). The kinetic parameter values are either randomly generated or estimated from real experimental data under single-protease-single-substrate setting (Method "Estimating kinetic parameters in the single-protease-single-substrate setting"). Let Q be the number of measurement time points, t = tz (z = 1, 2, …, Q). This simulation generates a Q×1 data vector representing the simulated amounts of the uncleaved substrate at Q time points for the single-protease-single-substrate scenario.
Simulating data with multiple proteases
Given a library of M -substrates and a mixture of N -proteases, coefficient [αj] as the concentration of jth protease in the mixture, and (Vij, Kij, nij, βij) as kinetic parameters for the reaction between the ith substrate and the jth protease, the amount of remaining uncleaved ith substrate [Si]t = tz at a specific time t = tz can be calculated by numerically integrating Eq (3). The values of the kinetic parameters and mixing coefficients are either randomly generated or estimated from real experimental data (Method "Estimating mixing coefficients with multiple proteases"). This simulation generates an M×Q matrix as the simulated data.
Estimating kinetic parameters and mixing coefficients
Estimating kinetic parameters in the single-protease-single-substrate setting
For each single-protease-single-substrate combination, we measure/simulate reaction products at Q time points after reaction starts, resulting in a data vector Y with dimension Q×1. In addition to simulated in silico experiments (Method "Simulating single-protease-single-substrate data"), Y can also be collected from real experiments under the single-protease-single-substrate setting. The problem of estimating the kinetic parameters can be formulated as the following optimization problem:
| (4) |
Using the “active-set” algorithm [43–45], this optimization problem leads to one set of kinetic parameters that can best fit the data Y of the specific protease-substrate combination (details see S2 Text). For a collection of single-protease-single-substrate settings of M substrates and N proteases, optimization is performed for each of the M×N combinations, resulting in kinetic parameter matrices (V, K, n, β), each of which has a dimension of M×N.
Estimating mixing coefficients with multiple proteases
Once kinetic parameters for all single-protease-single-substrate combinations have been estimated, we move to estimate the mixing coefficients of proteases in a protease mixture. Let M be the number of substrates, N be the number of proteases in the mixture, and Q be the number of measurement times for the reaction between each substrate and the mixture. The problem of estimating the mixing coefficients can be formulated into an optimization problem as follows:
| (5) |
[Si], i = 1, 2, 3, …, M
[αj], j = 1, 2, 3, …, N, ∀ j: [αj] > = 0
Y is an M×Q data matrix either generated from in silico simulation (Method "Simulating data with multiple proteases") or collected from real experiment under the multi-proteases setting. This optimization problem, solved by “active-set” algorithm [43–45], generates estimations of the mixing coefficients of proteases in the mixture (details see S2 Text).
Quantifying estimation accuracy via Root-Mean-Square Error (RMSE)
Once estimated mixing coefficients of a protease mixture have been obtained, the estimation accuracy is evaluated by the root-mean-square error (RMSE) metric. This metric is commonly used in machine learning to quantify accuracy for regression analysis [46]. An example of quantifying estimation error using RMSE is in Table 2.
Table 2. The first row is true α in P mixtures, of which each has N proteases.
The second row is estimated α. The RMSEs for individual proteases (R1, …, RN) are calculated in the third row, and the overall RMSE will be the average of all individual RMSEs. In the simulation setting, P is the number of repetitions we applied. The repetition time is P = 200.
| Protease 1 | Protease 2 | … | Protease N | |
|---|---|---|---|---|
| True α | … | |||
| Estimated | … | |||
| RMSE (Protease) | … | |||
| RMSE (overall) | ||||
Evaluating deconvolution performance and optimizing substrate selection
Given the kinetic parameter values of the reactions between a set of proteases and a set of substrates, we would like to evaluate whether we can accurately deconvolve mixtures of the proteases by measuring their cleaving activities against the substrates. We first simulate the in silico experimental data corresponding to the single-protease-single-substrate scenario (Eq 2) and simulate the in silico experimental data for protease mixtures reacting with multiple substrates (Eq 3). We then estimate the kinetic parameter values based on the simulated single-protease-single-substrate data (Eq 4). Finally, we estimate the mixing coefficients based on the estimated kinetic parameters and the simulated experimental data for protease mixtures reacting with multiple substrates (Eq 5), and evaluate the deconvolution accuracy using RMSE. In this analysis pipeline, we choose to estimate the single-protease-single-substrate kinetic parameters because the true kinetic parameter values are often unavailable in practice. Using this pipeline, we can evaluate the expected deconvolution performance for a given set of proteases using a given set of substrates and then derive optimal experimental designs for choosing the most suitable substrates for deconvolving the protease mixtures.
Results
Recombinant protease substrate specificity
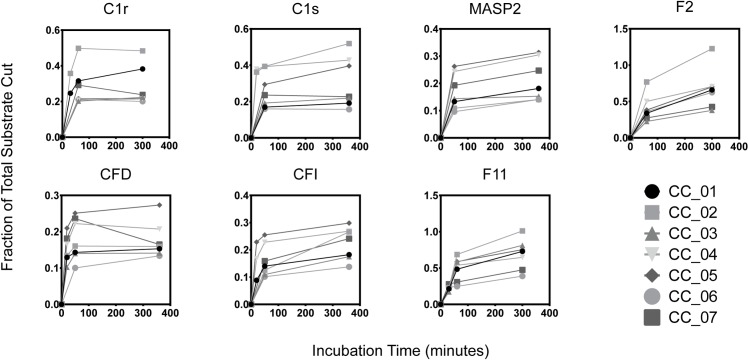
To obtain kinetic protease activity data we incubated 7 serum proteases from the complement and coagulation cascades with 7 protease substrates (Fig 1). These results showed that while each protease hydrolyzed the library of probes with different velocities, each signature was not necessarily linearly independent. Interestingly, certain proteases that showed similar activity toward the panel of substrates are involved in different physiological processes. For example, MASP2 and CFI showed similar activity signatures against this panel of substrates, although each are involved in different pathways of the complement system (e.g., MASP2 is in the lectin pathway, CFI is in the alternative pathway). In other words, this demonstrates that proteases may be related at the activity level, but may be involved in different physiological processes. Additionally, each protease showed unique early saturation levels, which we characterized with the parameter β.
Fig 1. Recombinant protease cleavage assays of seven complement and coagulation cascade proteins.
From left to right, top to bottom, abbreviations are: Complement proteins C1r and C1s, MASP2, Coagulation Factor IIa, Complement Factor D, Complement Factor I, and Coagulation Factor XIa. Each trace represents a different peptide substrate (CC01–07).
Validating the RMSE for evaluating protease deconvolution
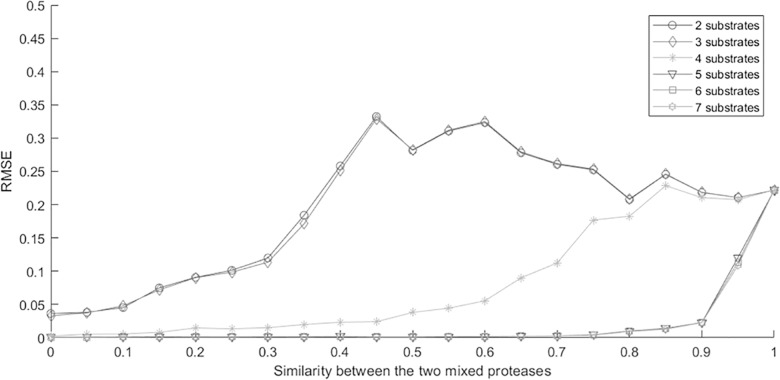
To verify the efficacy of using root mean squared error (RMSE) to approximate estimation accuracy, we simulated a series of 2-protease mixtures with increasing levels of similarity between the two proteases, which represented deconvolution problems with an increasing level of difficulty. In the simulations, the number of observed time points Q was 2, which matched our experimental time points shown in Fig 1. More specifically, we first simulated two proteases (p1,p2) by randomly generating their kinetic parameters against multiple substrates. Since the kinetic parameters were randomly generated, these two proteases were independent of each other. We then generated a series of intermediate proteases by linearly combining the two sets of kinetic parameters: p3 = λp1+(1−λ)p2, λ = 0,0.05,0.1,0.15,…,1. After that, we (1) simulated substrate cleavage data of protein mixtures of p1 and p3 defined by varying values for λ, (2) performed optimizations to estimate the mixing coefficients of the mixtures, and (3) applied RMSE to evaluate the estimation accuracy. Intuitively, the estimation problem is more difficult for cases where the mixed proteases are highly correlated (λ close to 1). In addition, we simulated cases with varying numbers of substrates (i.e., 2–7 substrates) and, in general, the more substrates that were measured, the easier it was to deconvolve the protease mixtures. In these simulations, the RMSE is expected to be larger for more difficult cases, and smaller for relatively easier cases.
In Fig 2, the horizontal axis represented the λ value for generating the protease p3, which meant that simulation cases from left to right had an increasing level of similarity between proteases p1 and p3, and thus had an increasing level of difficulty for deconvolving protease mixtures of the two proteases. Each curve represented a different series of simulations with a particular number of substrates. In Fig 2, the simulation series with a larger number of substrates led to smaller RMSEs. Note that the 2- and 3-substrates curves largely overlapped, and the 5-, 6-, and 7-substrates curves also largely overlapped. In each simulated series with a specific number of substrates, the RMSE increased in general with respect to the horizontal axis that represented an increasing level of difficulty. In the 2- and 3-substrates curves, the changes of RMSE were not monotonic. This was mainly because, with a limited number of substrates in the simulation, λ around 0.5 already represented quite difficult situations that led to very large RMSE with high variance. For the subsequent larger λ values representing even more difficult situations, the slight decrease of the subsequent RMSEs was due to the high variance when the RMSE was large. Overall, the observed RMSEs showed expected trends with respect to the level of difficulty of the simulated cases, validating that the RMSE is a useful evaluation metric.
Fig 2. RMSE reflected the level of difficulties in deconvolution of the simulated protease mixtures.
The x-axis represents the similarity between the two proteases in the mixture. A higher similarity between proteases led to a higher correlation between their cleavage dynamics, and thus higher difficulty in deconvolution. The y-axis was the RMSE of the estimated mixing coefficients. For each substrates-proteases setting, repetition P = 200 was used to calculate the corresponding reported RMSE. Each curve represented a simulation series with a different number of substrates. A smaller number of substrates corresponded to more difficult situations for deconvolution analysis.
Optimizing choices of substrates
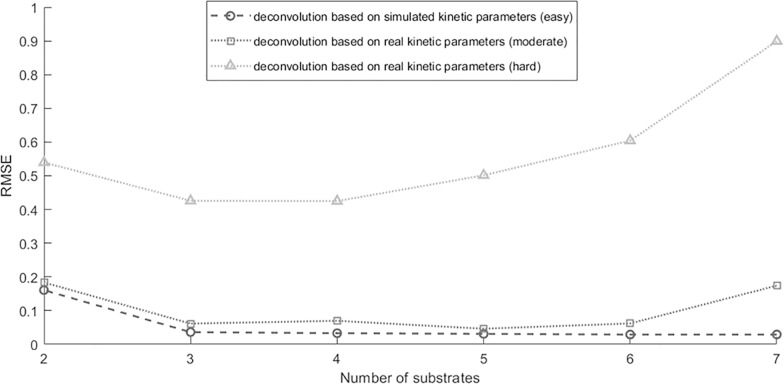
To demonstrate the feasibility of optimizing choices of substrates, we considered 3 proteases and 7 substrates, with their single-protease-single-substrate kinetic parameters randomly generated. We first evaluated the accuracy for deconvolving mixtures of the 3 proteases using all 7 substrates, which resulted in low RMSE as shown by the right-most point on the dashed-circle line in Fig 3. We then removed one substrate and evaluated the RMSE for deconvolution with 6 substrates. All 7 possibilities were evaluated, and the best RMSE was reported as the second right-most point on the dashed line, which was virtually the same as the 7-substrate scenario. We iterated this analysis, removing one substrate that had the least impact on RMSE in each iteration, until only 2 substrates remained. As shown in Fig 3, the RMSE remained low until the number of substrates reduced from 3 to 2. This was because the single-protease-single-substrate kinetic parameters were randomly generated, which represented 3 proteases that had independent substrate cleavage activities. In other words, at least three substrates were needed to estimate the activity of 3 independent proteases.
Fig 3. Influence of substrate library’s size on the deconvolution accuracy.
The x-axis represented the number of substrates applied for deconvolution and the y-axis represented the resulting RMSE. The dashed curve with circles represented RMSE for deconvolving mixtures of three simulated proteases that are independent, and showed increased RMSE as the number of substrates decreased from 7 down to 2. The dotted curves with squares and with triangles represented RMSE for deconvolving mixtures of three real proteases that are slightly correlated and highly correlated accordingly, where the RMSEs were relatively high even when the number of substrates was 7. Repetition time P = 200 was applied for each substrates-proteases setting. Discussion regarding the decrease of RMSE when reducing the number of substrates were discussed in Supplementary Figure B—G.
We performed two sets of similar analyses using 3 of the 7 proteases and the 7 substrates in our real experimental data in section "Recombinant Protease Substrate Specificity". One set of analyses was based on proteases MASP2, C1r, and F2, which were from 3 different proteases families, and the other set of analyses was based on proteases MASP2, CFI, and CFD, which were highly correlated in terms of their substrate cleavage dynamics. The single-protease-single-substrate kinetic parameters were estimated from real experimental data. All subsequent analyses were the same as the above where kinetic parameters were randomly generated. As shown by the dotted-triangle curve in Fig 3, deconvolving the three highly correlated proteases was quite difficult with large the RMSE regardless of how many substrates were used. The dotted-square curve in Fig 3 was similar to the analysis where kinetic parameters were randomly generated, indicating that the three proteases from different protease families had relative independent cleavage dynamics against the substrates. Interestingly, the performance actually improved in both dotted curves when the number of substrates reduced from 7 to 5 (or 4). This was because the first few substrates being removed had extremely similar cleavage dynamics against all the proteases (details in Figs B-G in S3 Text). Those substrates were not only uninformative but also sources of confusion for the deconvolution analysis. Therefore, effective deconvolution of protease mixtures required a decent number of substrates with uncorrelated cleavage dynamics against the proteases. However, correlated substrate cleavage dynamics is ubiquitous, especially among proteases in the same physiological family. When deconvolving mixtures containing highly correlated proteases, even a large number of substrates may not lead to satisfactory deconvolution performances. This motivated us to investigate a less ambitious goal of deconvolving protease families, instead of deconvolving individual proteases.
Deconvolving protease families
As mentioned above, deconvolving protease mixtures can be challenging and may require an impractically large number of substrates when proteases with highly correlated substrate cleavage dynamics exist in the mixture. Therefore, we proposed to verify the efficacy of clustering highly correlated proteases into families, and deconvolving the activity signatures by estimating the mixing concentration of the protease families, rather than individual proteases.
Deconvolving simulated protease families
We first simulated a scenario with 9 proteases and 7 substrates, in which the 9 proteases formed three families. Each family contained 3 highly correlated proteases, but the families were independent of each other. To generate the 3 highly correlated proteases in one family in silico, we took a similar strategy as described in Section "Validating the RMSE for evaluating protease deconvolution". For each protease family, we randomly generated the kinetic parameters of 3 proteases (p1, p2, and p3) cleaving 7 substrates, and then generated 2 proteases p4 and p5 that correlated p1 using the following combinations p4 = λp1+(1−λ)p2, and p5 = λp1+(1−λ)p3. Proteases p1, p4 and p5 form the family. Here, λ was either 0.9 or 0.6, representing a proteases family containing highly correlated proteases or moderately correlated proteases. We repeated the above three times to generate the kinetic parameters for the three families of proteases.
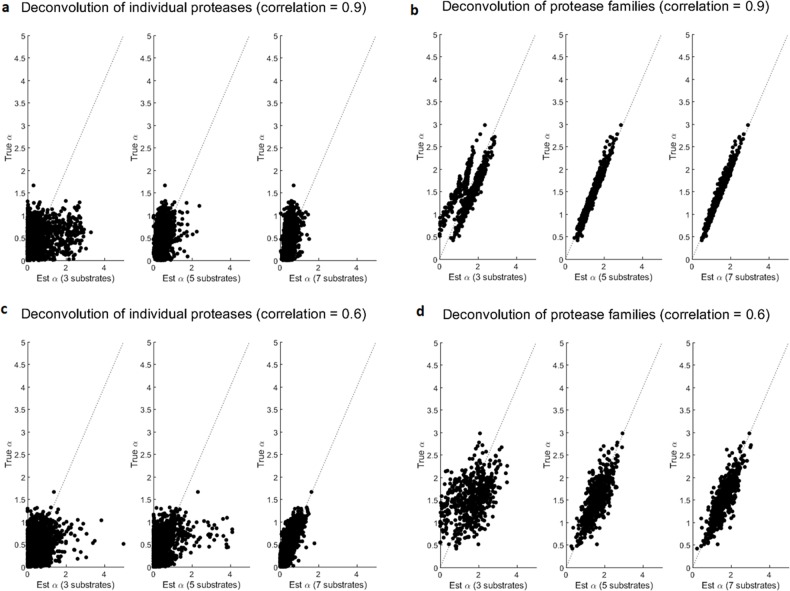
After generating the single-protease-single-substrate kinetic parameters with family structures, we simulated data for the single-protease-single-substrate setting and the multi-proteases-multi-substrates setting. We then evaluated the performance for deconvolving the 9 individual proteases using 3, 5, or 7 substrates. Fig 4A and 4C showed scatter plots of the true simulated mixing coefficients versus the estimated mixing coefficients, where the estimation performance was poor. The only exception was the case in the third plot of Fig 4C, where the protease was moderately correlated (λ = 0.6) and the number of substrates was 7. This was the least challenging case simulated here for deconvolving individual proteases, where the estimated mixing coefficients roughly tracked the true mixing coefficients. Overall, with the presence of correlated proteases, although the protease families were independent, it was difficult to deconvolve the mixing coefficients of the individual proteases.
Fig 4. Comparison between deconvolution of individual proteases and deconvolution of protease families.
The x axis represented the estimated mixing coefficients. The y axis represented the true simulated mixing coefficients. Consider mixtures of 9 proteases from 3 independent families that contain highly correlated proteases within each family. (a) When deconvolving the 9 individual proteases, the estimated mixing coefficients for the individual proteases showed poor agreements with their true simulated values, regardless of whether 3, 5, or 7 substrates were used. (b) When deconvolving the 3 protease families, the estimated mixing coefficients for the protease families showed high agreement with the sum of the family members’ true mixing coefficients. (c-d) Simulation of protease families that contain moderately correlated proteases showed similar results, where deconvolution of individual proteases was difficult but deconvolution of protease families was accurate.
Using the same simulated data as above, we evaluated the possibility for deconvolving protease families. In order to perform deconvolution at the protease family level, we used Eq (4) to estimate one set of kinetic parameters for each family, by treating the simulated single-protease-single-substrate data for protease members in the same family as replicates of a "representative" protease for the family. After estimating the kinetic parameters for the three protease families, we then optimized Eq (5) to estimate the mixing coefficient of the protease families. Ideally, the estimated mixing coefficient for one protease family should approximate the sum of the true mixing coefficients of members in the family, which was indeed what we observed in the results shown in Fig 4B and 4D. Meanwhile, the performance difference between Fig 4B and 4D indicated that successful deconvolution of protease families required members within each family to be decently correlated. This analysis demonstrated the feasibility of accurately deconvolving protease families, while the deconvolution of individual proteases was difficult.
Deconvolving protease families derived from real data
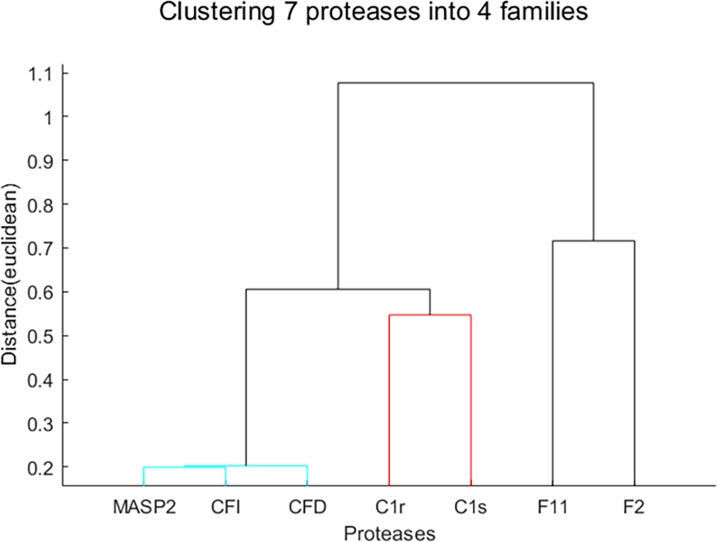
To further validate the idea of deconvolving protease families, we used the real single-protease-single-substrate data in Section "Recombinant Protease Substrate Specificity". We represented each protease by a vector containing the collection of all data points for this protease in the single-protease-single-substrate assays in Fig 1. We then clustered proteases by agglomerative hierarchical clustering using single linkage and Euclidean Distance. To group the proteases into families, we chose a cut-off value for distance to be 0.6. This allowed us to cluster proteases with correlated hydrolytic activities into the same family. After the clustering analysis, we grouped the 7 individual proteases into 4 computationally-derived families: (1) Complement protein MASP2 (lectin pathway) and Complement Factor I and D (alternative pathway), (2) Complement proteins C1r and C1s (classical pathway), (3) Coagulation factor IIa, and (4) Coagulation factor XIa. (Fig 5). Our results showed that the computationally-derived clusters reflect the physiological pathways they are involved in (i.e., group 1 and 2 are complement pathways and group 3 and 4 are coagulation pathways). This is also reflected at the sequence level, where clustered proteases C1r and C1s share 38.5% sequence identity while C1s shares only 27.9% and 31.2% identity with CFD and F11, respectively [47].
Fig 5. Hierarchical clustering of individual proteases led to 4 families.
MASP2+CFI+CFD formed the most strongly correlated family, C1r + C1s formed the moderately correlated family, and F11 and F2 each served as its own family.
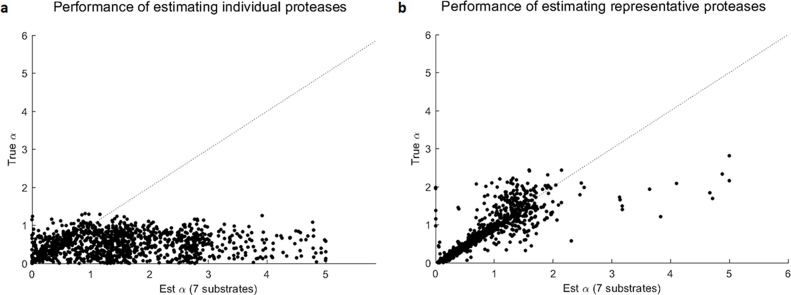
After that, we performed the same analysis as above, including simulation of the multi-protease-multi-substrate data, estimation of the kinetic parameters for the protease families, and estimation of the mixing coefficients for the protease families. We also performed deconvolution for the individual proteases. As shown in Fig 6, the deconvolution of the individual proteases performed poorly, while the estimated mixing coefficients of the protease families decently tracked the simulated true mixing coefficients. The deconvolution accuracy of these protease families was lower than the above analysis of randomly generated kinetic parameters. This was because the randomly generated kinetic parameters led to simulated protease families that were independent of each other, whereas the protease families derived from real data were not as independent. Since human proteases are known to organize in a nested hierarchy of protease families and subfamilies, the correlation among them makes deconvolution of individual protease cumbersome and impractical. This analysis provides a practical alternative for sensing and characterizing protease activities at the level of protease families.
Fig 6. Performances of estimating representative families and individual proteases using the same 7 proteases.
(a) Deconvolution of 7 individual proteases was no better than a random guess since the scatter dots spread out widely on the horizontal direction. (b) After clustering 7 individual proteases into 4 protease families as shown in Fig 5, deconvolution accuracy increased, with mixing coefficients of protease families being close to the sum of individual mixing coefficients within according protease families.
Discussion
While significant advances in measuring large amounts of biological information have been made by using genome-wide sequencing techniques, the lack of correlation between expression and activity is a major limitation [13]. Further, the primary drivers of physiological processes in health and disease are enzymes (e.g., proteases, kinases), meaning valuable information is stored in the real-time activity of these proteins. One of the major challenges in scaling up to multiplexed libraries for protease activity analysis is substrate design, due to the difficulty of screening for specific substrates. Due to the promiscuity of proteases, the goal of designing substrates with both high sensitivity and high specificity for all human proteases is experimentally challenging. To date, the major advancements in this substrate design include multiplex substrate profiling by mass spectrometry (MSP-MS) [48–50] and randomized substrate screens with phage display [51], as well as computational approaches for receptor-ligand prediction [52, 53]. Here, we present a framework for evaluating the activity contributions of individual proteases within a complex mixture with minimal substrate design. However, the MSP-MS and phage display methods provide the ability to develop peptide substrates for proteases ad hoc, which we envision could be combined with our algorithm to enable more strategic control over the foundational protease signatures. Further, these methods provide the ability to identify the dominant proteases in a biological sample, which could also be used to pre-emptively narrow our list of target proteases.
Fundamentally, however, human proteases are organized in a hierarchy of protease families that consist of proteases with highly correlated activities against many substrates. Deconvolution of highly correlated proteases would require an impractically large number of substrates. Therefore, we propose to cluster highly correlated proteases into families, and estimate the relative activity contributions of these families. We demonstrated the feasibility of accurately deconvolving protease families when the deconvolution of individual proteases was difficult. We envision that this may allow for the rapid characterization and investigation of physiologically relevant protease families, which can effectively screen the entire protease landscape before homing in on specific protease targets.
Supporting information
(DOCX)
(DOCX)
(DOCX)
(ZIP)
Acknowledgments
The authors would like to thank Quoc Mac (Georgia Tech) and Dr. David Myers (Georgia Tech & Emory) for helpful discussions. The authors would like to thank Anthony Mancino (Syneos) for technical expertise in processing mass spectrometry samples.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This work was funded by an NIH Director’s New Innovator Award (Award No. DP2HD091793). B.A.H is supported by the NSF GRFP, National Institutes of Health GT BioMAT Training Grant under Award Number 5T32EB006343 and the Georgia Tech President's Fellowship. This material is based upon work supported by the National Science Foundation Graduate Research Fellowship under Grant No. DGE-1650044 (B.A.H.). G.A.K. holds a Career Award at the Scientific Interface from the Burroughs Welcome Fund. This work was performed in part at the Georgia Tech Institute for Electronics and Nanotechnology, a member of the National Nanotechnology Coordinated Infrastructure (NNCI), which is supported by the National Science Foundation (Grant ECCS-1542174). This work was partially supported by funding from the National Science Foundation (CCF1552784). P.Q. is an ISAC Marylou Ingram Scholar and a Carol Ann and David D. Flanagan Faculty Fellow. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Bird PI, Trapani JA, Villadangos JA. Endolysosomal proteases and their inhibitors in immunity. Nature Reviews Immunology. 2009;9:871 10.1038/nri2671 [DOI] [PubMed] [Google Scholar]
- 2.de Souza GA, de Godoy LMF, Mann M. Identification of 491 proteins in the tear fluid proteome reveals a large number of proteases and protease inhibitors. Genome Biology. 2006;7(8):R72 10.1186/gb-2006-7-8-r72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frees D, Brondsted L, Ingmer H. Bacterial proteases and virulence. Sub-cellular biochemistry. 2013;66:161–92. Epub 2013/03/13. 10.1007/978-94-007-5940-4_7 . [DOI] [PubMed] [Google Scholar]
- 4.Heutinck KM, ten Berge IJ, Hack CE, Hamann J, Rowshani AT. Serine proteases of the human immune system in health and disease. Molecular immunology. 2010;47(11–12):1943–55. Epub 2010/06/12. 10.1016/j.molimm.2010.04.020 . [DOI] [PubMed] [Google Scholar]
- 5.Hua Y, Nair S. Proteases in cardiometabolic diseases: Pathophysiology, molecular mechanisms and clinical applications. Biochimica et biophysica acta. 2015;1852(2):195–208. 10.1016/j.bbadis.2014.04.032 PMC4225191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ingmer H, Brondsted L. Proteases in bacterial pathogenesis. Research in microbiology. 2009;160(9):704–10. Epub 2009/09/26. 10.1016/j.resmic.2009.08.017 . [DOI] [PubMed] [Google Scholar]
- 7.Koblinski JE, Ahram M, Sloane BF. Unraveling the role of proteases in cancer. Clinica chimica acta; international journal of clinical chemistry. 2000;291(2):113–35. Epub 2000/02/17. 10675719. 10.1016/s0009-8981(99)00224-7 [DOI] [PubMed] [Google Scholar]
- 8.Lopez-Otin C, Bond JS. Proteases: multifunctional enzymes in life and disease. The Journal of biological chemistry. 2008;283(45):30433–7. Epub 2008/07/25. 10.1074/jbc.R800035200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pérez-Silva JG, Español Y, Velasco G, Quesada V. The Degradome database: expanding roles of mammalian proteases in life and disease. Nucleic Acids Research. 2016;44(D1):D351–D5. 10.1093/nar/gkv1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sanman LE, Bogyo M. Activity-based profiling of proteases. Annual review of biochemistry. 2014;83:249–73. Epub 2014/06/07. 10.1146/annurev-biochem-060713-035352 . [DOI] [PubMed] [Google Scholar]
- 11.Miyamoto S, Ochiai A, Boku N, Ohtsu A, Tahara M, Yoshida S, et al. Discrepancies between the gene expression, protein expression, and enzymatic activity of thymidylate synthase and dihydropyrimidine dehydrogenase in human gastrointestinal cancers and adjacent normal mucosa. International journal of oncology. 2001;18(4):705–13. Epub 2001/03/17. 10.3892/ijo.18.4.705 . [DOI] [PubMed] [Google Scholar]
- 12.Takumi O, Michiharu S, Kazuhiko N, Hajime N, Tsuyoshi O, Sumito S, et al. [Correlation between enzymatic activity and gene expression of orotate phosphoribosyl transferase (OPRT) in colorectal cancer]. Gan to kagaku ryoho Cancer & chemotherapy. 2002;29(13):2515–9. Epub 2003/01/01. . [PubMed] [Google Scholar]
- 13.Yin J, Zhang D, Zhuang J, Huang Y, Mu Y, Lv S. Study on the Correlation between Gene Expression and Enzyme Activity of Seven Key Enzymes and Ginsenoside Content in Ginseng in Over Time in Ji’an, China. International Journal of Molecular Sciences. 2017;18(12):2682 10.3390/ijms18122682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dudani JS, Jain PK, Kwong GA, Stevens KR, Bhatia SN. Photoactivated Spatiotemporally-Responsive Nanosensors of in Vivo Protease Activity. ACS Nano. 2015;9(12):11708–17. 10.1021/acsnano.5b05946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edgington LE, Verdoes M, Bogyo M. Functional imaging of proteases: recent advances in the design and application of substrate-based and activity-based probes. Current opinion in chemical biology. 2011;15(6):798–805. Epub 2011/11/22. 10.1016/j.cbpa.2011.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fonovic M, Bogyo M. Activity based probes for proteases: applications to biomarker discovery, molecular imaging and drug screening. Current pharmaceutical design. 2007;13(3):253–61. Epub 2007/02/23. 10.2174/138161207779313623 . [DOI] [PubMed] [Google Scholar]
- 17.Holt BA, Bellavia MC, Potter D, White D, Stowell SR, Sulchek T. Fc microparticles can modulate the physical extent and magnitude of complement activity. Biomaterials science. 2017;5(3):463–74. Epub 2017/01/10. 10.1039/c6bm00608f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holt BA, Mac QD, Kwong GA. Nanosensors to Detect Protease Activity In Vivo for Noninvasive Diagnostics. JoVE. 2018;(137):e57937 10.3791/57937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kwon EJ, Dudani JS, Bhatia SN. Ultrasensitive tumour-penetrating nanosensors of protease activity. Nature biomedical engineering. 2017;1 Epub 2017/10/04. 10.1038/s41551-017-0054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kwong GA, Dudani JS, Carrodeguas E, Mazumdar EV, Zekavat SM, Bhatia SN. Mathematical framework for activity-based cancer biomarkers. Proceedings of the National Academy of Sciences. 2015;112(41):12627–32. 10.1073/pnas.1506925112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nomura DK, Dix MM, Cravatt BF. Activity-based protein profiling for biochemical pathway discovery in cancer. Nature reviews Cancer. 2010;10(9):630–8. 10.1038/nrc2901 PMC3021511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schuerle S, Dudani JS, Christiansen MG, Anikeeva P, Bhatia SN. Magnetically Actuated Protease Sensors for in Vivo Tumor Profiling. Nano Letters. 2016;16(10):6303–10. 10.1021/acs.nanolett.6b02670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stein V, Alexandrov K. Protease-based synthetic sensing and signal amplification. Proceedings of the National Academy of Sciences. 2014;111(45):15934–9. 10.1073/pnas.1405220111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.To T-L, Piggott BJ, Makhijani K, Yu D, Jan YN, Shu X. Rationally designed fluorogenic protease reporter visualizes spatiotemporal dynamics of apoptosis in vivo. Proceedings of the National Academy of Sciences. 2015;112(11):3338–43. 10.1073/pnas.1502857112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Villanueva J, Nazarian A, Lawlor K, Tempst P. Monitoring Peptidase Activities in Complex Proteomes by MALDI-TOF Mass Spectrometry. Nature protocols. 2009;4(8):1167–83. 10.1038/nprot.2009.88 PMC2760592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Villanueva J, Shaffer DR, Philip J, Chaparro CA, Erdjument-Bromage H, Olshen AB, et al. Differential exoprotease activities confer tumor-specific serum peptidome patterns. The Journal of Clinical Investigation. 2006;116(1):271–84. 10.1172/JCI26022 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Warren AD, Gaylord ST, Ngan KC, Dumont Milutinovic M, Kwong GA, Bhatia SN, et al. Disease detection by ultrasensitive quantification of microdosed synthetic urinary biomarkers. Journal of the American Chemical Society. 2014;136(39):13709–14. Epub 2014/09/10. 10.1021/ja505676h [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mac QD, Mathews DV, Kahla JA, Stoffers CM, Delmas OM, Holt BA, et al. Non-invasive early detection of acute transplant rejection via nanosensors of granzyme B activity. Nature biomedical engineering. 2019. 10.1038/s41551-019-0358-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holt BA, Kwong GA. Bacterial defiance as a form of prodrug failure. bioRxiv. 2019:556951 10.1101/556951 [DOI] [Google Scholar]
- 30.Overall CM, Blobel CP. In search of partners: linking extracellular proteases to substrates. Nature Reviews Molecular Cell Biology. 2007;8:245 10.1038/nrm2120 [DOI] [PubMed] [Google Scholar]
- 31.Kwong GA, von Maltzahn G, Murugappan G, Abudayyeh O, Mo S, Papayannopoulos IA, et al. Mass-encoded synthetic biomarkers for multiplexed urinary monitoring of disease. Nat Biotech. 2013;31(1):63–70. http://www.nature.com/nbt/journal/v31/n1/abs/nbt.2464.html#supplementary-information. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller MA, Barkal L, Jeng K, Herrlich A, Moss M, Griffith LG, et al. Proteolytic Activity Matrix Analysis (PrAMA) for simultaneous determination of multiple protease activities. Integr Biol (Camb). 2011;3(4):422–38. Epub 2010/12/25. 10.1039/c0ib00083c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Triplett DA. Coagulation and bleeding disorders: review and update. Clinical chemistry. 2000;46(8 Pt 2):1260–9. Epub 2000/08/06. . [PubMed] [Google Scholar]
- 34.Pettigrew HD, Teuber SS, Gershwin ME. Clinical significance of complement deficiencies. Annals of the New York Academy of Sciences. 2009;1173:108–23. Epub 2009/09/18. 10.1111/j.1749-6632.2009.04633.x . [DOI] [PubMed] [Google Scholar]
- 35.Menten L, Michaelis M. Die kinetik der invertinwirkung. Biochem Z. 1913;49:333–69. [Google Scholar]
- 36.Keener JP, Sneyd J. Mathematical physiology: Springer; 1998. [Google Scholar]
- 37.Kam CM, McRae BJ, Harper JW, Niemann MA, Volanakis JE, Powers JC. Human complement proteins D, C2, and B. Active site mapping with peptide thioester substrates. The Journal of biological chemistry. 1987;262(8):3444–51. Epub 1987/03/15. . [PubMed] [Google Scholar]
- 38.Tsiftsoglou SA, Sim RB. Human Complement Factor I Does Not Require Cofactors for Cleavage of Synthetic Substrates. The Journal of Immunology. 2004;173(1):367–75. 10.4049/jimmunol.173.1.367 [DOI] [PubMed] [Google Scholar]
- 39.Gosalia DN, Salisbury CM, Ellman JA, Diamond SL. High Throughput Substrate Specificity Profiling of Serine and Cysteine Proteases Using Solution-phase Fluorogenic Peptide Microarrays. Molecular & Cellular Proteomics. 2005;4(5):626–36. 10.1074/mcp.M500004-MCP200 [DOI] [PubMed] [Google Scholar]
- 40.Budayova-Spano M, Grabarse W, Thielens NM, Hillen H, Lacroix M, Schmidt M, et al. Monomeric Structures of the Zymogen and Active Catalytic Domain of Complement Protease C1r: Further Insights into the C1 Activation Mechanism. Structure. 2002;10(11):1509–19. 10.1016/S0969-2126(02)00881-X. [DOI] [PubMed] [Google Scholar]
- 41.Thompson A, Schafer J, Kuhn K, Kienle S, Schwarz J, Schmidt G, et al. Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal Chem. 2003;75(8):1895–904. Epub 2003/04/26. 10.1021/ac0262560 . [DOI] [PubMed] [Google Scholar]
- 42.Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Molecular & cellular proteomics: MCP. 2004;3(12):1154–69. Epub 2004/09/24. 10.1074/mcp.M400129-MCP200 . [DOI] [PubMed] [Google Scholar]
- 43.Han S-P. A globally convergent method for nonlinear programming. Journal of optimization theory and applications. 1977;22(3):297–309. [Google Scholar]
- 44.Powell MJ. A fast algorithm for nonlinearly constrained optimization calculations Numerical analysis: Springer; 1978. p. 144–57. [Google Scholar]
- 45.Powell MJ. The convergence of variable metric methods for nonlinearly constrained optimization calculations Nonlinear programming 3: Elsevier; 1978. p. 27–63. [Google Scholar]
- 46.Hastie T, Tibshirani R, Friedman J. The Elements of Statistical Learning 2009. [Google Scholar]
- 47.O'Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016;44(D1):D733–45. Epub 2015/11/11. 10.1093/nar/gkv1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.O'Donoghue AJ, Eroy-Reveles AA, Knudsen GM, Ingram J, Zhou M, Statnekov JB, et al. Global identification of peptidase specificity by multiplex substrate profiling. Nat Methods. 2012;9(11):1095–100. Epub 2012/10/02. 10.1038/nmeth.2182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ivry SL, Sharib JM, Dominguez DA, Roy N, Hatcher SE, Yip-Schneider M, et al. Global protease activity profiling provides differential diagnosis of pancreatic cysts. Clinical Cancer Research. 2017:clincanres.2987.016. 10.1158/1078-0432.ccr-16-2987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Winter MB, La Greca F, Arastu-Kapur S, Caiazza F, Cimermancic P, Buchholz TJ, et al. Immunoproteasome functions explained by divergence in cleavage specificity and regulation. eLife. 2017;6:e27364 10.7554/eLife.27364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kretz CA, Tomberg K, Van Esbroeck A, Yee A, Ginsburg D. High throughput protease profiling comprehensively defines active site specificity for thrombin and ADAMTS13. Scientific Reports. 2018;8(1):2788 10.1038/s41598-018-21021-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Goncalves-Almeida VM, Pires DE, de Melo-Minardi RC, da Silveira CH, Meira W, Santoro MM. HydroPaCe: understanding and predicting cross-inhibition in serine proteases through hydrophobic patch centroids. Bioinformatics (Oxford, England). 2012;28(3):342–9. Epub 2011/12/16. 10.1093/bioinformatics/btr680 . [DOI] [PubMed] [Google Scholar]
- 53.Pires DE, de Melo-Minardi RC, da Silveira CH, Campos FF, Meira W Jr. aCSM: noise-free graph-based signatures to large-scale receptor-based ligand prediction. Bioinformatics (Oxford, England). 2013;29(7):855–61. Epub 2013/02/12. 10.1093/bioinformatics/btt058 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(DOCX)
(ZIP)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.