Abstract
Background
Drug-resistant minority variants (DRMinVs) detected in patients who recently acquired human immunodeficiency virus type 1 (HIV-1) can be transmitted, generated de novo through virus replication, or technical errors. The first form is likely to persist and result in treatment failure, while the latter two could be stochastic and transient.
Methods
Ultradeep sequencing of plasma samples from 835 individuals with recent HIV-1 infection in the United Kingdom was performed to detect DRMinVs at a mutation frequency between 2% and 20%. Sequence alignments including >110 000 HIV-1 partial pol consensus sequences from the UK HIV Drug Resistance Database (UK-HDRD), linked to epidemiological and clinical data from the HIV and AIDS Reporting System, were used for transmission cluster analysis. Transmission clusters were identified using Cluster Picker with a clade support of >90% and maximum genetic distances of 4.5% or 1.5%, the latter to limit detection to likely direct transmission events.
Results
Drug-resistant majority variants (DRMajVs) were detected in 66 (7.9%) and DRMinVs in 84 (10.1%) of the recently infected individuals. High levels of clustering to sequences in UK-HDRD were observed for both DRMajV (n = 48; 72.7%) and DRMinV (n = 63; 75.0%) sequences. Of these, 43 (65.2%) with DRMajVs were in a transmission cluster with sequences that harbored the same DR mutation compared to only 3 (3.6%) sequences with DRMinVs (P < .00001, Fisher exact test). Evidence of likely direct transmission of DRMajVs was observed for 25/66 (37.9%), whereas none were observed for the DRMinVs (P < .00001).
Conclusions
Using a densely sampled HIV-infected population, we show no evidence of DRMinV transmission among recently infected individuals.
Keywords: HIV-1, Drug-resistant minority variants, Recent infection, Transmission cluster, Replication fitness
Phylogenetic reconstruction shows no evidence of drug-resistant minority variants (DRMinVs) in patients with recent human immunodeficiency virus type 1 infection resulting from a transmission event. Therefore, the majority of DRMinVs in people recently infected are most likely generated de novo.
The British HIV Association (BHIVA) human immunodeficiency virus (HIV) treatment guidelines recommend that genotyping be performed in order to rule out the presence of transmitted drug resistance and to guide the selection of first-line therapy regimens in individuals with diagnosed primary HIV infection [1]. Currently, this is performed by most diagnostic laboratories in the United Kingdom using Sanger capillary sequencing technology with a lower mutation frequency (MF) threshold of 20%. Ultradeep sequencing technologies capable of detecting variants present at a frequency as low as 1% of the virus population, for example, next-generation sequencing (NGS), are slowly being introduced into clinical microbiology diagnostic laboratories. However, the evidence for the clinical significance of drug-resistant minority variants (DRMinVs) is contradictory, and more data are required to inform their interpretation in clinical management of HIV-infected individuals using validated assays and large-scale clinical studies [2–10].
HIV exists as a population of multiple variants within an infected host [11, 12]. A majority of HIV-1 infections are thought to result from the transmission of a single virus clone, thus, multiple variants arise as a consequence of the error-prone nature of the HIV-1 replication mechanism [13–16]. The genetic diversity generated through this process is a harbinger of drug-resistant variants, which can lead to the failure of antiretroviral therapy (ART). Early detection of minority variants could therefore be of benefit to HIV-infected individuals undergoing ART.
Several factors may contribute to an association between DRMinVs and treatment failure. Mutational load, defined as the absolute copy number of drug-resistant variants per unit volume in an infected individual, is one factor [17]. This is a product of the MF and the patient’s viral load. For example, at 1% MF, the absolute copy number in an individual with a viral load of 103 copies/mL would be 10 copies/mL compared to 10 000 copies/mL in an individual with a viral load of 106 copies/mL. The genetic barrier to resistance is often linked to the association of non-nucleoside reverse transcriptase inhibitor (NNRTI) DRMinVs with virological failure [18]. This is the number of genetic changes required for a virus population to acquire robust resistance against a drug regimen. For most NNRTI regimens it takes only 1 or 2 genetic changes to cause high-level resistance [19, 20]. The genetic linkage of 2 or more drug-resistant mutations (DRMs) directed against different antiretroviral drug classes used in combination ART on a single genome is more likely to result in treatment failure than being present on separate genomes [21]. Other factors, such as adherence, pharmacodynamics, and pharmacokinetics that result in suboptimal drug concentrations, could also favor the outgrowth and subsequent dominance of DRMinVs.
However, it is also thought that the origin of a drug-resistant variant could influence whether it becomes clinically relevant. DRMinVs that arise under drug selection, either in treatment-experienced individuals or by transmission, are more likely to establish a persistent infection and impact treatment outcomes than those generated de novo in the absence of drug selection and as a consequence of viral replication [10, 22–25]. The transmission of DRMinVs, however, contradicts the current understanding that most HIV infections arise from a single virus clone, although it is possible that the drug-resistant variant could be transmitted as the sole virus or a majority population followed by reversion to wild type with residual persistence of the drug-resistant variant as a minority population. Several studies have investigated the origin or transmissibility of HIV-1 DRMinVs with inconsistent findings [10, 26–29].
We performed ultradeep sequencing of a partial HIV-1 pol gene from recently infected individuals in the United Kingdom sampled between 2011 and 2014 to detect sequences harboring DRMinVs. We used a phylogenetic approach to investigate the origin of DRMinVs and determine if they are a result of a transmission event. We performed transmission cluster analysis together with sequences from the UK HIV Drug Resistance Database (UK-HDRD), which contains the pol sequences from the vast majority of genotypic resistance test results performed in the United Kingdom since 1997.
METHODS
Samples and Data Collection
Between July 2011 and December 2014, 24 569 people were newly diagnosed with HIV-1 in England, Wales, and northern Ireland, of which 11 086 (45.1%) were tested using a recent infection testing algorithm (RITA) [30]. A total of 2043 (18.4%) were identified to have been infected within 5 months of sampling, of which 835 samples (i.e., 40.9% of recent infections) were subjected to NGS. This includes 442 samples from men who have sex with men sampled between July 2011 and December 2013 [30]. We also performed NGS on 186 samples from newly diagnosed individuals determined by RITA to be from long-standing infections. Linked clinical and demographic data were extracted from the HIV and AIDS Reporting System held at Public Health England.
A total of 111 807 partial pol sequences from 76 293 individuals generated by Sanger sequencing as part of routine clinical care in the United Kingdom from 1997 to 2014 collated by the UK-HDRD were also used.
Ultradeep Sequencing
A 1.3 kb region of the HIV pol gene (all of protease and the N-terminal 320 amino acids of reverse transcriptase) was amplified and sequenced using Illumina MiSeq, and the raw short-read data were analyzed using an in-house bioinformatics pipeline [30]. The drug resistance sequences generated in this study have been submitted to GenBank (accession numbers MH663717–MH663796).
Phylogenetic Analyses
The sequences from recently infected patients generated using NGS were compared to the UK-HDRD sequences. To minimize the number of sequences incorporated into any subsequent phylogenetic analysis, the NGS sequences and the UK-HDRD sequences were clustered on the basis of sequence similarity. Each NGS sequence was compared against a database of the UK-HDRD sequences using the Basic Local Alignment Search Tool (BLAST) [31]. For each NGS-generated sequence, an in-house Python script was used to identify all BLAST hits in the database with sequence identity >95%; the UK-HDRD sequences were put into clusters with the NGS-generated sequences. Clusters were merged when the same UK-HDRD sequence was identified as being greater than 95% similar to 2 or more NGS sequences. Duplicate sequences from the same patient were removed after matching using linked epidemiological data, resulting in a final dataset comprising 9182 sequences.
Sequences in the final dataset were aligned using MAFFT v7 [32] with minimal manual editing. Maximum likelihood phylogenetic reconstruction was performed using FastTree v2.1.9 [33] with a general time reversible (GTR) model of nucleotide substitution. Transmission clusters were identified from the phylogenetic trees using Cluster Picker v1.2.3 [34] at 4.5% or 1.5% maximum genetic distance threshold between all sequences in the cluster and a 90% minimum clade support threshold (Shimodaira-Hasegawa test).
RESULTS
Population Characteristics
The characteristics of the population of recently infected study participants are listed in Table 1. The majority were white men who acquired HIV through sex with other men. All participants were ART naive, and the majority were infected with HIV-1 subtype B with a median age of 32 years (interquartile range [IQR], 26–40).
Table 1.
Characteristics of the Cohort of People Recently Infected With Human Immunodeficiency Virus
| Characteristic | Category | Number of Sequences (% of total) |
|---|---|---|
| Gender | Male | 784 (93.9) |
| Female | 51 (6.1) | |
| Risk exposure | Men who have sex with men | 715 (85.6) |
| Heterosexual male | 50 (5.9) | |
| Heterosexual female | 46 (5.5) | |
| Intravenous drug users | 5 (0.6) | |
| Other/Unknown | 19 (2.3) | |
| Ethnicity | White | 600 (71.9) |
| Black (African/Caribbean/other) | 55 (6.6) | |
| Other/Unknown | 180 (21.6) | |
| Virus subtype | B | 560 (67.1) |
| Non-B (pure subtypes) | 134 (16.0) | |
| Circulating recombinant forms | 71 (8.5) | |
| Unique recombinant forms | 70 (8.4) | |
| Median age, years (interquartile range) | 32 (26–40) |
Prevalence of Drug-resistant Majority and Minority Variants in People Recently Infected
Ultradeep sequencing of the HIV-1 pol domain from the 835 recently infected individuals was performed to detect the presence of drug-resistant majority variants (DRMajVs) and DRMinVs. We define DRMajVs as those present at a frequency >20%, the equivalent of the limit of detection of Sanger-based sequencing, and DRMinVs as those present at a frequency between 2% and 20% [30]. Ninety-three DRMinVs were present in 84 of the 835 sequences (10.1%), whereas 80 DRMajVs were present in 66 sequences (7.9%), with a median MF of 99.2% (98.4%–99.5%) and 3.2% (2.5%–5.3%), respectively. The median depth of coverage at DRMinVs positions was 12 556 (IQR, 6769–24 890). Ten sequences had multiple DRMajVs, with 4 (n = 1), 3 (n = 1), and 2 (n = 8) variants, compared to 4 sequences that had multiple DRMinVs, with 3 (n = 1) and 2 (n = 3) variants. By drug class, the most common types of DRMajVs were L90M (n = 14; 63.6%) for protease inhibitors (PIs), T215Yrev (n = 19; 57.6%) for nucleoside reverse transcriptase inhibitors (NRTIs), and K103N (n = 18; 72.0%) for NNRTIs (Figure 1). In contrast, the most common DRMinVs were M46IL (n = 21; 42.9%) for PIs, D67NG (n = 11; 44.0%) for NRTIs, and G190E (n = 6; 31.6%) for NNRTIs. The majority of DRMinVs were against PIs (49/93; 52.7%), whereas DRMajVs were evenly distributed by drug class at 27.5% (n = 22) for PIs, 41.3% (n = 33) for NRTIs, and 31.3% (n = 25) for NNRTIs.
Figure 1.
Drug-resistant majority variants (DRMajVs) and drug-resistant minority variants (DRMinVs) in people recently infected with human immunodeficiency virus type 1 (HIV-1). The number of sequences harboring DRMajVs (gray bars) and DRMinVs (black bars) by drug class in people recently infected with HIV-1. Abbreviations: DRMajV, drug-resistant majority variant; DRMinV, drug-resistant minority variant; NRTI, nucleoside reverse transcriptase inhibitor; NNRTI, nonnucleoside reverse transcriptase inhibitor; PI, protease inhibitor.
No Evidence for the Transmission of DRMinVs in People With Recent HIV-1 Infection
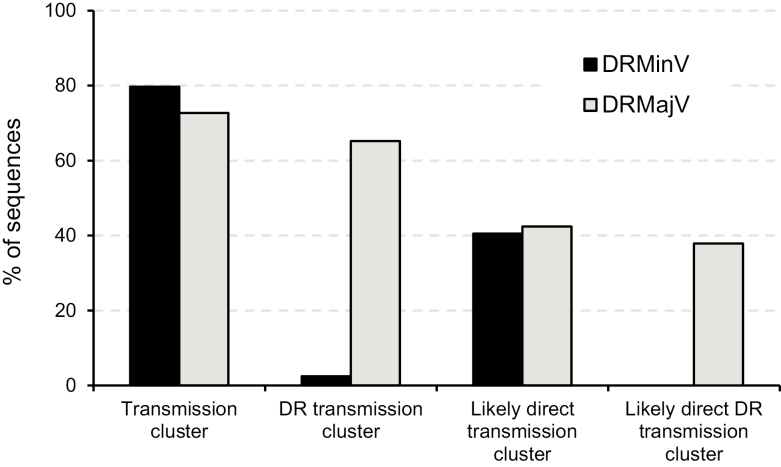
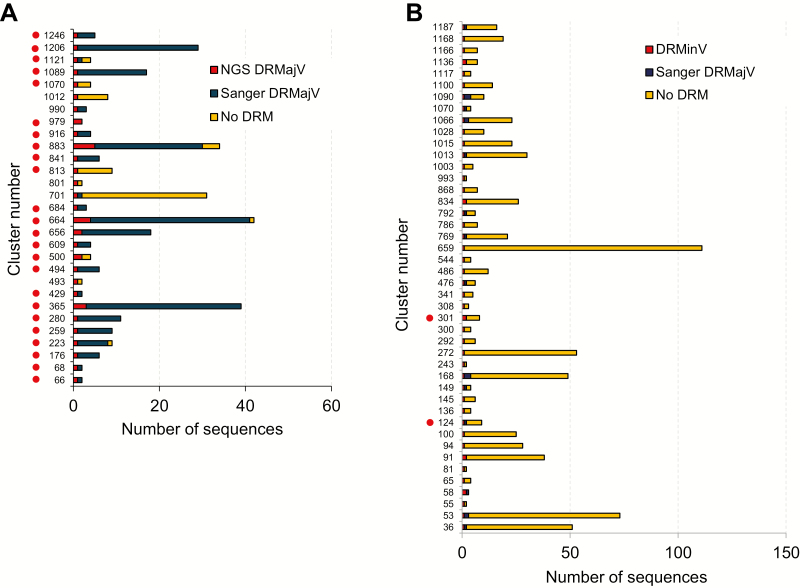
We performed transmission cluster analysis using the NGS-generated sequences from recently infected individuals and all the Sanger sequences in the UK-HDRD to investigate the possibility that DRMinVs in this population could be a result of a transmission event. A cluster of DRMajV (Sanger or NGS) and DRMinV sequences with the same drug resistance mutation (DRM) could suggest a potential DRMajV-DRMinV transmission, whereas a cluster of DRMinV sequences with the same DRM could suggest DRMinV-DRMinV transmission. The proportion of sequences present in a transmission cluster determined using a genetic distance of 4.5% and 90% bootstrap support was very high for both DRMajV (72.7%; 48/66) and DRMinV (75.0%; 63/84) NGS sequences (Figure 2). Of the DRMajV sequences, 43 (65.2%) were in a transmission cluster with sequences that harbored the same DRM compared to only 3 (3.6%) of DRMinV sequences (P < .00001, Fisher exact test). Of the 3 DRMinV sequences, 2 contained the D67G mutation in reverse transcriptase (RT) at 2.2% and 2.4% MF that were both present in the same transmission cluster together with 7 wild-type sequences. The third DRMinV sequence contained the K101E mutation in RT at 4.8% MF that was present in a transmission cluster that included 1 Sanger sequence with the K101E and M184I RT mutations and 7 wild-type sequences. Analysis of the distribution of DR-containing clusters showed that the majority of DRMajV NGS sequences (37/43; 86%) were present in transmission clusters where the majority of the sequences had the same DRM (Figure 3A). In contrast, all DRMinV sequences were present in clusters where the majority of the sequences did not contain any DRMs (Figure 3B).
Figure 2.
Transmission cluster analysis of sequences harboring drug-resistant majority variants (DRMajVs) and drug-resistant minority variants (DRMinVs) from people with recent human immunodeficiency virus type 1 infections. Shown are the percentages of DRMajVs (gray bars) and DRMinVs (black bars) sequences that were present in a transmission cluster (genetic distance of 4.5%), a transmission cluster containing the same drug-resistance mutations (DRMs), a recent transmission cluster (genetic distance of 1.5%), and a recent transmission cluster containing the same DRM. Abbreviations: DRMajV, drug-resistant majority variant; DRMinV, drug-resistant minority variant; HIV, human immunodeficiency virus; UK HDRD, UK HIV Drug Resistance Database.
Figure 3.
Transmission cluster distribution of drug-resistant majority variant (DRMajV) and drug-resistant minority variant (DRMinV) sequences. A, The number of sequences in transmission clusters that contained DRMajVs from individuals recently infected with human immunodeficiency virus type 1 (HIV-1) and drug resistance mutations (DRMs) or no DRMs from the UK HIV Drug Resistance Database (UK HDRD). B, The number of sequences in transmission clusters that contained DRMinVs from individuals recently infected with HIV-1 and DRMs or no DRMs from the UK HDRD. Red dots represent clusters that contained recent DRMajV or DRMinV sequences together with DRMajV sequences from the UK HDRD harboring the same DRM. Abbreviations: DRMajV, drug-resistant majority variant; DRMinV, drug-resistant minority variant; UK HDRD, UK HIV Drug Resistance Database.
To limit detection to the most likely direct transmission events involving the NGS sequences from the acute infections, we used a genetic distance of 1.5% for transmission cluster analysis. This resulted in 28 (42.4%) DRMajV sequences compared to 32 (38.1%) DRMinV NGS sequences present in transmission clusters (Figure 2). Of the DRMajV sequences, 25/66 (37.9%) were present in a transmission cluster with other sequences with the same DRM compared to none of the DRMinV sequences (P < .00001). The median number of sequences per cluster was similar for both DRMajV and DRMinV transmission clusters (8 [IQR, 4–17] and 9 [IQR, 4–10], respectively).
Relationships Between MF, Mutational Load, and Transmission of DRMs
Further analysis of NGS data showed that a majority (46/50; 92.0%) of transmitted DRMs had a MF >90% compared to 1/50 (2.0%) with MF between 20% and 90% and 3/50 (8.0%) with MF <20% (Figure 4A). A positive correlation was observed between mutational load and MF (r = 0.51; P < .001, Spearman correlation) for the combined DRMajV and DRMinV dataset (Figure 4B). In contrast, no correlation was observed between mutational load and MF (r = 0.14; P = .321) for the DRMinV dataset alone (Figure 4C). Similar relationships were observed when the analysis was limited to sequences within transmission clusters (Supplementary Figure 1.
Figure 4.
Scatter graphs illustrating the distribution and relationships between mutation frequency, mutational load, and the transmission of drug resistance mutations (DRMs). A, Distribution of mutation frequency and its relationship to the transmission of DRMs. The mutation frequency of drug-resistant majority variant (DRMajV) and drug-resistant minority variant (DRMinV) is shown for the different drug classes: protease inhibitor (left-side, white area), nucleoside reverse transcriptase inhibitor (middle, gray area), and nonnucleoside reverse-transcriptase inhibitor (right-side, white area). Red dot indicates DRMajV or DRMinV sequence linked to sequence harboring the same DRM at 1.5% genetic distance threshold; orange dot indicates DRMajV or DRMinV sequence linked to sequence harboring the same DRM at 4.5% genetic distance threshold; green dot indicates DRMajV or DRMinV sequence present in a transmission cluster with wild-type sequences or sequences containing a different DRM; black dot indicates DRMajV or DRMinV sequence not present in a transmission cluster. B, Relationship between mutation frequency and mutational load of DRMajVs and DRMinVs. C, Relationship between mutation frequency and mutational load of DRMinVs only. Abbreviations: NRTI, nucleoside reverse transcriptase inhibitor; NNRTI, nonnucleoside reverse transcriptase inhibitor; PI, protease inhibitor.
DISCUSSION
In this study, we used a phylogenetic approach to determine the derivation of HIV-1 DRMinVs in people who were recently HIV infected in the United Kingdom. The presence of DRMs in this population is presumed to be a result of the transmission of a drug-resistant variant. It is thought that a transmitted drug resistance variant is more likely to persist because it was initially selected under drug pressure; however, the transmission of DRMinVs contradicts the current understanding that most sexually transmitted HIV-1 infections result from the transmission of a single clone. The relatively dense sampling of the UK HIV-1 epidemic enabled these analyses to be undertaken with high sensitivity. Unsurprisingly, the recent DRMajV and DRMinV NGS sequences were present in transmission clusters at high percentages of 72.7% and 75.0%, respectively, which is higher than the overall proportion of sequences in the UK HIV database that have previously been shown to be present in a transmission cluster at 52% [35].
We observed a marked difference in the most common types of DRMajVs and DRMinVs in people recently infected, these being L90M and M46IL for PIs, T215rev and D67GN for NRTIs, and K103N and G190E for NNRTIs for DRMajVs and DRMinVs, respectively. Furthermore, there is discordance in the proportion of DRMinVs in people recently infected and DRMs detected in individuals experiencing treatment failure in the United Kingdom by drug class [30]. For example, PI mutations account for the majority of DRMinVs detected at 52%, whereas PI mutations are detected in less than 5% of people experiencing treatment failure in the UK [36]. Correspondingly, we found no significant evidence that DRMinVs in people recently infected are a result of a transmission event as only 3.6% (3/84) of DRMinVs were in a transmission cluster with other sequences with the same DRM compared to 65.2% (43/66) of DRMajVs. Most importantly, most of the observed DRMs were against old drugs and did not include DRMs such as K65R and M184V, which are also relevant in the era of preexposure prophylaxis.
It should be emphasized that these analyses do not rule out the possibility of DRMinV-to-DRMinV transmission. However, this is highly unlikely as it contradicts the current understanding that most HIV infections arise from the transmission of a single clone that would favor the transmission of the majority variant in sexual transmissions [13–16]. On the other hand, we did observe 2 DRMinVs containing the D67G RT mutation in the same transmission cluster, suggesting that DRMinV-to-DRMinV transmission could occur. This observation could either be a chance occurrence, as the 2 sequences were not in a likely direct transmission cluster, or the phylogenetic signal for direct relatedness could not reliably be discerned from the majority consensus sequences used in the phylogenetic reconstruction.
It is likely that DRMs are predominantly transmitted as a majority variant in recently infected individuals. The DRMs subsequently decay, albeit at different rates, and are thus more likely to be detected as minority variants in newly diagnosed, treatment-naive individuals with long-standing infections [37, 38]. There was a slight, but not statistically significant, increase in DRMinVs linked to the same DRM in a transmission cluster for those with long-standing infection (11.8%; 2/17) compared to those with recent infection (3.6%; 3/84; P = .196; Supplementary Figure 2. Of note, DRMinVs linked to transmitted resistance in both recent and long-standing infections were in RT and none were against protease even though the latter constituted the majority of DRMinVs detected in both populations. This suggests that DRMinVs in treatment-naive individuals may not be created equal and only a few mutation types could result from a transmission event. This could also explain the association of NNRTI DRMinVs with virological failure observed in some studies, whereas none have been reported to be associated with PIs [3, 8].
Alternatively, DRMinVs could be a result of technical errors introduced during sample processing or due to de novo virus replication errors [10, 28, 39]. We have previously shown that the effect from the former mostly contributes to variants detected below 2% in our assay and therefore are less likely to be the major source of the DRMinVs in this study as the variant threshold was set at ≥2% [30]. The replication process in HIV-1 is highly error prone and results in approximately 1 nucleotide mutation per genome per round of replication [40, 41]. Coupled with a high viral replication rate that can generate more than 1 billion virions in a single day in untreated individuals, it is postulated that a mutation occurs at every position in the HIV-1 genome every day. Interestingly, the most common DRMinVs were derived by G-A transition mutations, the most common replication error committed by HIV-1 and other lentiviral RTs [41], whereas the DRMajVs were a result of transversion mutations or more than 1 transition/transversion mutation. This suggests a different origin for DRMajVs and DRMinVs and that the latter could primarily be a result of viral and/or reverse-transcription polymerase chain reaction replication errors rather than drug-selection pressure. Of note, 87.5% of DRMajVs had a MF >90%, whereas 90% of DRMinVs had a MF <10%. In addition, there was no correlation between mutational load and MF for DRMinVs, also suggesting that most DRMinVs are likely generated de novo as a result of replication errors. These data indicate that a natural selection process may be at play that results in preservation of DRMs with reduced effect on viral fitness and a purge of DRMs that have a negative effect on viral fitness. This is in agreement with known effects on viral fitness of the common types of DRMajV and DRMinV described in this study [37, 42, 43] and the hypothesis that the frequency of minority variants is likely higher in recent infections because the effective population size is smaller and thus limits the effect of negative selection, which exponentially increases later during the infection [28].
In conclusion, we find no evidence that DRMinVs detected in people recently infected with HIV are a result of a transmission event, suggesting that their detection to inform first-line treatment is unlikely to be of clinical benefit and might unnecessarily limit treatment options. Most importantly, future longitudinal studies should focus on determining the treatment outcomes in the people identified to harbor the DRMinVs, which would better inform the clinical significance of DRMinVs in this population. This finding does not extend to the detection of DRMinVs in treatment-experienced individuals where the variants would have emerged under drug-selection pressure and could therefore adversely affect treatment outcomes.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
UK HIV Drug Resistance Database. Steering committee: David Asboe, Anton Pozniak (Chelsea & Westminster Hospital, London); Patricia Cane (Public Health England [PHE], Porton Down); David Chadwick (South Tees Hospitals NHS Trust, Middlesbrough); Duncan Churchill (Brighton and Sussex University Hospitals National Health Service [NHS] Trust); Duncan Clark (Barts Health NHS Trust, London); Simon Collins (HIV i-Base, London); Valerie Delpech (National Infection Service, PHE); Samuel Douthwaite (Guy’s and St. Thomas’ NHS Foundation Trust, London); David Dunn, Esther Fearnhill, Kholoud Porter, Anna Tostevin, Oliver Stirrup (Institute for Global Health, University College London [UCL]); Christophe Fraser (University of Oxford); Anna Maria Geretti (Institute of Infection and Global Health, University of Liverpool); Rory Gunson (Gartnavel General Hospital, Glasgow); Antony Hale (Leeds Teaching Hospitals NHS Trust); Stéphane Hué (London School of Hygiene and Tropical Medicine); Linda Lazarus (Expert Advisory Group on AIDS Secretariat, PHE); Andrew Leigh-Brown (University of Edinburgh); Tamyo Mbisa (National Infection Service, PHE); Nicola Mackie (Imperial NHS Trust, London); Chloe Orkin (Barts Health NHS Trust, London); Eleni Nastouli, Deenan Pillay, Andrew Phillips, Caroline Sabin (University College London, London); Erasmus Smit (PHE, Birmingham Heartlands Hospital); Kate Templeton (Royal Infirmary of Edinburgh); Peter Tilston (Manchester Royal Infirmary); Erik Volz (Imperial College London, London); Ian Williams (Mortimer Market Centre, London); Hongyi Zhang (Addenbrooke’s Hospital, Cambridge). Coordinating center: Institute for Global Health, UCL (David Dunn, Keith Fairbrother, Esther Fearnhill, Kholoud Porter, Anna Tostevin, Oliver Stirrup). Centers contributing data: Clinical Microbiology and Public Health Laboratory, Addenbrooke’s Hospital, Cambridge (Justine Dawkins); Guy’s and St Thomas’ NHS Foundation Trust, London (Siobhan O’Shea, Jane Mullen); PHE—Public Health Laboratory, Birmingham Heartlands Hospital, Birmingham (Erasmus Smit); Antiviral Unit, National Infection Service, PHE, London (Tamyo Mbisa); Imperial College Health NHS Trust, London (Alison Cox); King’s College Hospital, London (Richard Tandy); Medical Microbiology Laboratory, Leeds Teaching Hospitals NHS Trust (Tracy Fawcett); Specialist Virology Centre, Liverpool (Mark Hopkins); Department of Clinical Virology, Manchester Royal Infirmary, Manchester (Peter Tilston); Department of Virology, Royal Free Hospital, London (Clare Booth, Ana Garcia-Diaz); Edinburgh Specialist Virology Centre, Royal Infirmary of Edinburgh (Lynne Renwick); Department of Infection & Tropical Medicine, Royal Victoria Infirmary, Newcastle (Matthias L Schmid, Brendan Payne); South Tees Hospitals NHS Trust, Middlesbrough (David Chadwick); Department of Virology, Barts Health NHS Trust, London (Jonathan Hubb); Molecular Diagnostic Unit, Imperial College, London (Simon Dustan); University College London Hospitals (Stuart Kirk); West of Scotland Specialist Virology Laboratory, Gartnavel, Glasgow (Rory Gunson, Amanda Bradley-Stewart).
Author contributions. J. L. M. conceived the idea for the study. All authors were involved in collecting the data. G. M. coordinated recent infection testing algorithm testing. J. L. performed the next-generation sequencing experiments and initial sequence data analysis. J. L. M., D. F. B., and R. M. performed the bioinformatics analyses. A. S. H. performed mutational load data analysis. J. L. M. wrote the first draft of the manuscript. All authors reviewed the manuscript and agreed to publish it.
Acknowledgments. The authors thank the staff of Antiviral Unit and Clinical Service Unit in the Virus Reference Department, PHE for providing laboratory support and Kieren Lythgow for additional bioinformatics support. They also thank the National Institute for Health Research Health Protection Research Unit (NIHR HPRU) Blood Borne and Sexually Transmitted Infections Steering Committee: Caroline Sabin (director), Anthony Nardone (PHE lead), Catherine Mercer, Gwenda Hughes, Jackie Cassell, Greta Rait, Samreen Ijaz, Tim Rhodes, Sema Mandal, Kholoud Porter, and William Rosenberg for reviewing the manuscript.
Disclaimer. The views expressed in this publication are those of the author(s) and not necessarily those of the NHS, the National Institute for Health Research, the Department of Health, or PHE.
Financial support. This research was funded by PHE and by the NIHR and undertaken by the NIHR HPRU in Blood Borne and Sexually Transmitted Infections at UCL in partnership with PHE and in collaboration with the London School of Hygiene and Tropical Medicine. The UK HIV Drug Resistance Database is supported by the MRC (award number 164587).
Potential conflicts of interest. A. P. reports grants and personal fees from ViiV, Gilead, Janssen, and Merck outside the submitted work; was a principal investigator on a test and treat program in Tanzania; and was part of a research group investigating new antiretroviral regimens in South Africa. P. K. reports grants from Gilead Sciences outside the submitted work. C. S. reports grants from Medical Research Council and NIHR during the conduct of the study and personal fees from Gilead Sciences, ViiV, and Janssen-Cilag outside the submitted work. All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Contributor Information
UK HIV Drug Resistance Database:
David Asboe, Anton Pozniak, Patricia Cane, David Chadwick, Duncan Churchill, Duncan Clark, Simon Collins, Valerie Delpech, Samuel Douthwaite, David Dunn, Esther Fearnhill, Kholoud Porter, Anna Tostevin, Oliver Stirrup, Christophe Fraser, Anna Maria Geretti, Rory Gunson, Antony Hale, Stéphane Hué, Linda Lazarus, Andrew Leigh-Brown, Tamyo Mbisa, Nicola Mackie, Chloe Orkin, Eleni Nastouli, Deenan Pillay, Andrew Phillips, Caroline Sabin, Erasmus Smit, Kate Templeton, Peter Tilston, Erik Volz, Ian Williams, Hongyi Zhang, David Dunn, Keith Fairbrother, Esther Fearnhill, Kholoud Porter, Anna Tostevin, Oliver Stirrup, Justine Dawkins, Siobhan O’Shea, Jane Mullen, Erasmus Smit, Tamyo Mbisa, Alison Cox, Richard Tandy, Tracy Fawcett, Mark Hopkins, Peter Tilston, Clare Booth, Ana Garcia-Diaz, Lynne Renwick, Matthias L Schmid, Brendan Payne, David Chadwick, Jonathan Hubb, Simon Dustan, Stuart Kirk, Rory Gunson, and Amanda Bradley-Stewart
References
- 1. Churchill D, Waters L, Ahmed N, et al. . British HIV Association guidelines for the treatment of HIV-1-positive adults with antiretroviral therapy 2015. HIV Med 2016; 17(Suppl 4):s2–104. [DOI] [PubMed] [Google Scholar]
- 2. Johnson JA, Li JF, Wei X, et al. . Minority HIV-1 drug resistance mutations are present in antiretroviral treatment-naïve populations and associate with reduced treatment efficacy. PLoS Med 2008; 5:e158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Li JZ, Paredes R, Ribaudo HJ, et al. . Low-frequency HIV-1 drug resistance mutations and risk of NNRTI-based antiretroviral treatment failure: a systematic review and pooled analysis. JAMA 2011; 305:1327–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lataillade M, Chiarella J, Yang R, et al. . Prevalence and clinical significance of HIV drug resistance mutations by ultra-deep sequencing in antiretroviral-naïve subjects in the CASTLE study. PLoS One 2010; 5:e10952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Simen BB, Simons JF, Hullsiek KH, et al. ; Terry Beirn Community Programs for Clinical Research on AIDS . Low-abundance drug-resistant viral variants in chronically HIV-infected, antiretroviral treatment-naive patients significantly impact treatment outcomes. J Infect Dis 2009; 199:693–701. [DOI] [PubMed] [Google Scholar]
- 6. Cozzi-Lepri A, Noguera-Julian M, Di Giallonardo F, et al. ; CHAIN Minority HIV-1 Variants Working Group . Low-frequency drug-resistant HIV-1 and risk of virological failure to first-line NNRTI-based ART: a multicohort European case-control study using centralized ultrasensitive 454 pyrosequencing. J Antimicrob Chemother 2015; 70:930–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Charpentier C, Lee GQ, Rodriguez C, et al. . Highly frequent HIV-1 minority resistant variants at baseline of the ANRS 139 TRIO trial had a limited impact on virological response. J Antimicrob Chemother 2015; 70:2090–6. [DOI] [PubMed] [Google Scholar]
- 8. Perrier M, Visseaux B, Landman R, et al. . No impact of HIV-1 protease minority resistant variants on the virological response to a first-line PI-based regimen containing darunavir or atazanavir. J Antimicrob Chemother 2018; 73:173–6. [DOI] [PubMed] [Google Scholar]
- 9. Raymond S, Nicot F, Pallier C, et al. ; French National Agency for Research on AIDS and Viral Hepatitis (ANRS) AC11 Resistance Study Group . Impact of human immunodeficiency virus type 1 minority variants on the virus response to a rilpivirine-based first-line regimen. Clin Infect Dis 2018; 66:1588–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gianella S, Delport W, Pacold ME, et al. . Detection of minority resistance during early HIV-1 infection: natural variation and spurious detection rather than transmission and evolution of multiple viral variants. J Virol 2011; 85:8359–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Perelson AS, Neumann AU, Markowitz M, Leonard JM, Ho DD. HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science 1996; 271:1582–6. [DOI] [PubMed] [Google Scholar]
- 12. Coffin JM. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science 1995; 267:483–9. [DOI] [PubMed] [Google Scholar]
- 13. Abrahams MR, Anderson JA, Giorgi EE, et al. ; CAPRISA Acute Infection Study Team; Center for HIV-AIDS Vaccine Immunology Consortium . Quantitating the multiplicity of infection with human immunodeficiency virus type 1 subtype C reveals a non-Poisson distribution of transmitted variants. J Virol 2009; 83:3556–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Haaland RE, Hawkins PA, Salazar-Gonzalez J, et al. . Inflammatory genital infections mitigate a severe genetic bottleneck in heterosexual transmission of subtype A and C HIV-1. PLoS Pathog 2009; 5:e1000274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Keele BF, Giorgi EE, Salazar-Gonzalez JF, et al. . Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci U S A 2008; 105:7552–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li H, Bar KJ, Wang S, et al. . High multiplicity infection by HIV-1 in men who have sex with men. PLoS Pathog 2010; 6:e1000890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gupta S, Lataillade M, Kyriakides TC, et al. . Low-frequency NNRTI-resistant HIV-1 variants and relationship to mutational load in antiretroviral-naïve subjects. Viruses 2014; 6:3428–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tang MW, Shafer RW. HIV-1 antiretroviral resistance: scientific principles and clinical applications. Drugs 2012; 72:e1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sluis-Cremer N, Tachedjian G. Mechanisms of inhibition of HIV replication by non-nucleoside reverse transcriptase inhibitors. Virus Res 2008; 134:147–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vingerhoets J, Tambuyzer L, Azijn H, et al. . Resistance profile of etravirine: combined analysis of baseline genotypic and phenotypic data from the randomized, controlled phase III clinical studies. AIDS 2010; 24:503–14. [DOI] [PubMed] [Google Scholar]
- 21. Gega A, Kozal MJ. New technology to detect low-level drug-resistant HIV variants. Future Virol 2011; 6:17–26. [Google Scholar]
- 22. Parisi SG, Mazzi R, Boldrin C, et al. . Drug-resistance mutations can be archived very early in HIV primary infection. AIDS 2006; 20:1337–8. [DOI] [PubMed] [Google Scholar]
- 23. Pao D, Andrady U, Clarke J, et al. . Long-term persistence of primary genotypic resistance after HIV-1 seroconversion. J Acquir Immune Defic Syndr 2004; 37:1570–3. [DOI] [PubMed] [Google Scholar]
- 24. Metzner KJ, Leemann C, DiGF, et al. . Reappearance of minority K10 nority K103N HIV-1 variants after interruption of ART initiated during primary HIV-1 infection. PLoS One 2011; 6:e21734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Metzner KJ, Rauch P, Braun P, et al. . Prevalence of key resistance mutations K65R, K103N, and M184V as minority HIV-1 variants in chronically HIV-1 infected, treatment-naïve patients. J Clin Virol 2011; 50:156–61. [DOI] [PubMed] [Google Scholar]
- 26. Metzner KJ, Scherrer AU, Preiswerk B, et al. . Origin of minority drug-resistant HIV-1 variants in primary HIV-1 infection. J Infect Dis 2013; 208:1102–12. [DOI] [PubMed] [Google Scholar]
- 27. Lipscomb JT, Switzer WM, Li JF, Masciotra S, Owen SM, Johnson JA. HIV reverse-transcriptase drug resistance mutations during early infection reveal greater transmission diversity than in envelope sequences. J Infect Dis 2014; 210:1827–37. [DOI] [PubMed] [Google Scholar]
- 28. Chaillon A, Nakazawa M, Wertheim JO, et al. . No substantial evidence for sexual transmission of minority HIV drug resistance mutations in men who have sex with Men. J Virol 2017; 91:e00769-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li JZ. HIV-1 drug-resistant minority variants: sweating the small stuff. J Infect Dis 2014; 209:639–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cunningham E, Chan YT, Aghaizu A, et al. . Enhanced surveillance of HIV-1 drug resistance in recently infected MSM in the UK. J Antimicrob Chemother 2017; 72:227–34. [DOI] [PubMed] [Google Scholar]
- 31. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol 1990; 215:403–10. [DOI] [PubMed] [Google Scholar]
- 32. Katoh K, Rozewicki J, Yamada KD. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform 2017 Sep 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Price MN, Dehal PS, Arkin AP. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 2010; 5(3):e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ragonnet-Cronin M, Hodcroft E, Hue S, et al. . Automated analysis of phylogenetic clusters. BMC Bioinf 2013; 14:317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Leigh Brown AJ, Lycett SJ, Weinert L, Hughes GJ, Fearnhill E, Dunn DT. Transmission network parameters estimated from HIV sequences for a nationwide epidemic. J Infect Dis 2011; 204:1463–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. HIV drug resistance in the UK. 2019. Available at: http://www.hivrdb.org.uk/hiv-drug-resistance-uk . [Google Scholar]
- 37. Castro H, Pillay D, Cane P, et al. . Persistence of HIV-1 transmitted drug resistance mutations. J Infect Dis 2013; 208:1459–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jain V, Sucupira MC, Bacchetti P, et al. . Differential persistence of transmitted HIV-1 drug resistance mutation classes. J Infect Dis 2011; 203:1174–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Orton RJ, Wright CF, Morelli MJ, et al. . Distinguishing low frequency mutations from RT-PCR and sequence errors in viral deep sequencing data. BMC Genomics 2015; 16:229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mansky LM, Pearl DK, Gajary LC. Combination of drugs and drug-resistant reverse transcriptase results in a multiplicative increase of human immunodeficiency virus type 1 mutant frequencies. J Virol 2002; 76:9253–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Abram ME, Ferris AL, Shao W, Alvord WG, Hughes SH. Nature, position, and frequency of mutations made in a single cycle of HIV-1 replication. J Virol 2010; 84:9864–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wertheim JO, Oster AM, Johnson JA, et al. . Transmission fitness of drug-resistant HIV revealed in a surveillance system transmission network. Virus Evol 2017; 3:vex008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kuhnert D, Kouyos R, Shirreff G, et al. . Quantifying the fitness cost of HIV-1 drug resistance mutations through phylodynamics. PLoS Pathog 2018; 14:e1006895. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.