Abstract
目的
探讨不同浓度全反式维甲酸(ATRA)对小鼠肝癌细胞Hepa1-6成熟分化及细胞自噬水平的影响。
方法
以小鼠肝癌细胞Hepa1-6为研究对象,分别给予0.1、1、10 μmol/L浓度的ATRA处理,荧光定量PCR和Western blot检测肝细胞相关标志基因的表达,吲哚菁绿(ICG)及过碘酸-希夫(PAS)染色检测小鼠肝癌细胞Hepa1-6的成熟功能,透射电镜观测细胞连接及自噬小体,选择最适的ATRA作用浓度;Western blot检测自噬相关标志蛋白的表达,ptfLC3质粒转染细胞后共聚焦观察自噬流变化,检测自噬流是否通畅。
结果
相较于空白对照组,ATRA可呈浓度依赖性抑制肝前体标志蛋白AFP的表达,并促进肝细胞成熟标志ALB、CK18、TAT和ApoB的表达,ICG及PAS染色提示阳性细胞数明显增多;透射电镜结果显示ATRA诱导组的紧密连接和细胞骨架明显增多,可见高尔基复合体以及较多的自噬小体和自噬溶酶体。均以10 μmol/L组最为显著,作为最适终浓度。自噬相关标志蛋白LC3-Ⅱ、Beclin-1、RAB7、P62的表达不同程度增高,LC3-Ⅱ/LC3-Ⅰ的比值明显增加(P < 0.05),激光共聚焦可见10 μmol/LATRA处理组细胞胞浆内绿色亮点的自噬小体及红色亮点的自噬溶酶体均较对照组明显增多。
结论
ATRA可诱导小鼠肝癌细胞Hepa1-6的成熟分化,并促进细胞自噬水平增高。
Keywords: 原发性肝癌, 全反式维甲酸, 自噬, 细胞分化
Abstract
Objective
To investigate the effects of different concentrations of all-trans retinoic acid (ATRA) on the maturation, differentiation and autophagy of Hepa1-6 cells.Method Hepa1-6 cells were treated with 0.1, 1, and 10 μmol/L ATRA, and the changes in the expressions of hepatic specific markers were detected using real-time PCR and Western blotting.Indocyanine green (ICG) and periodic acid-schiff (PAS) staining was used to assess the functional maturation of Hepa1-6 cells, and the cellcell junction and autophagy were observed under transmission electron microscopy to determine the optimal concentration of ATRA for treatment.The expressions of autophagy- related markers in the cells were detected using Western blotting, and confocal microscopy was used to observe the autophagic flow in the cells transfected with ptfLC3 plasmid.
Results
Compared with the control cells, the hepatocytes treated with ATRA showed a concentration-dependent decrease in AFP expression and increase in the expressions of ALB, CK18, TAT and ApoB.ICG and PAS staining revealed significantly increased number of positive cells after ATRA treatment.Following ATRA treatment, the cells exhibited obviously increased tight junctions, cytoskeleton and number of autophagosomes under transmission electron microscopy.ATRA treatment resulted in significantly increased the expressions of autophagy-related markers LC3-Ⅱ, Beclin-1, RAB7 and P62 and also an increased ratio of LC3-Ⅱ/LC3-Ⅰ (P < 0.05).Confocal microscopy revealed obviously increased green and red spots in the cells after ATRA treatment.
Conclusion
ATRA can induce the maturation and differentiation and enhance the level of autophagy in Hepa1-6 cells.
Keywords: primary hepatic carcinoma, all-trans retinoic acid, autophagy, cell differentiation
原发性肝癌是临床上最常见的恶性肿瘤之一,位居恶性肿瘤死亡率第3位,我国为原发性肝癌的高发区。目前原发性肝癌临床仍然以手术治疗为主,辅以放疗和化疗的综合治疗,但整体效果仍不理想[1-3]。肝癌恶性程度高,复发转移率高,预后差,如何有效地提高肝癌的治疗效果,减轻广大肝癌患者的痛苦是目前急需解决的问题。全反式维甲酸(ATRA)是维生素A的活性代谢产物,通过其同源核受体对细胞生长,分化和凋亡具有强大的作用,体内外研究证实,ATRA可抑制肝癌细胞生长,诱导其凋亡,抑制肿瘤的侵袭和转移,提高化疗药物的敏感性[4-5]。本课题组前期研究也发现ATRA可逆转肝癌细胞上皮间质转化,诱导分化[6-8],但目前ATRA在肝癌临床治疗中还未被广泛应用,因此探讨ATRA作用于肝癌细胞的分子生物学机制很有必要。
自噬即自体吞噬,是一种在真核细胞中保守的细胞自耗途径,细胞在应激状态可激活这一分解代谢过程,通过消化自身内部受损、变性及衰老的蛋白质和细胞器来维持内环境稳定,实现物质循环利用,保持细胞活力[9]。研究证实自噬可参与多种干细胞的分化及多种化疗药物抗肿瘤作用[10-11],而ATRA及其维甲酸信号也可参与调节细胞的自噬水平,目前已有较多研究发现ATRA可通过诱导自噬缓解髓系白血病[12-14],但其对肝脏疾病自噬的影响尚无报道,本研究以小鼠肝癌细胞Hepa1-6为对象,探讨ATRA诱导肝癌细胞分化过程中细胞的自噬水平及自噬流是否通畅,有助于阐明ATRA诱导肝癌细胞良性转归的分子生物学机制,同时为ATRA应用于临床肝癌治疗提供重要的理论基础和实验依据。
1. 材料和方法
1.1. 主要材料与试剂
小鼠肝癌细胞Hepa1-6;DMEM完全培养液(Dulbecco modified Eagle medium)、胰蛋白酶及胎牛血清(Gibco);β-actin、ALB、AFP、CK18抗体(Santa Cruz),LC3、Beclin-1、RAB7、P62抗体(CST),蛋白裂解液及除抗体外的Western blot所用试剂(碧云天);RNA提取试剂盒及Real-time PCR试剂盒(Bioteck),逆转录试剂盒(Promega);PCR引物合成(华大基因),ATRA及吲哚菁绿(ICG)(Sigma);过碘酸-希夫(PAS)染色试剂盒(索莱宝);ptfLC3双荧光质粒(addgene)。
1.2. 细胞培养及分组
将小鼠肝癌细胞Hepa1-6接种于含10%胎牛血清,100 U/mL青霉素和100 µg/mL链霉素的DMEM完全培养液中,置于含5% CO2的37 ℃恒温箱中孵育,待贴壁生长至80%~90%的汇合率时,用胰蛋白酶进行消化、传代。取对数生长期的细胞进行试验,待细胞贴壁后分别加入0.1、1、10 μmol/L终浓度的ATRA,同时设置空白对照组。以下实验均重复至少3次。
1.3. Real-time PCR
ATRA处理7 d后检测肝细胞相关标志蛋白基因水平的表达情况。试剂盒提取出各组细胞的总RNA,并逆转录成cDNA。使用Primer3程序设计相关基因的引物(表 1)。cDNA样品稀释10倍后进行Real-time PCR扩增:72 ℃ 3 min;94 ℃ 3 min;94 ℃ 10 s,55 ℃ 20 s,72 ℃ 20 s,共40个循环;溶解曲线65 ℃~95 ℃,每5 s增加0.5 ℃,读板。所有样本通过β-actin的表达水平进行标准化。
1.
Real-time PCR引物序列
Sequences of primers for real-time PCR
| Name of genes | Sequences of primers (5'-3') | |
| Forward | Reverse | |
| AFP | ACGAGGAAAGCCCCTCAG | GCCATTCCCTCACCACAG |
| ALB | CCAGACATTCCCCAATGC | CAAGTTCCGCCCTGTCAT |
| CK18 | GCCATTCCCTCACCACAG | ACAGAGCCACCCCAGACA |
| TAT | ACCTTCAATCCCATCCGA | TCCCGACTGGATAGGTAG |
| ApoB | CATGTGATCCCCACAGCA | TCCCAGGACCATGGAAAA |
| β-actin | AGGGAAATCGTGCGTGAC | CGCTCGTTGCCAATAGTGA |
1.4. PAS染色
ATRA诱导小鼠肝癌细胞Hepa1-6 9 d后,室温下PBS洗3次,4%多聚甲醛固定10 min,PBS再洗3次,0.5%高碘酸液处理5 min,流水冲洗5 min,滴加Schiff染液染色15 min,流水清洗5 min,置于显微镜下观察,每组选取大于10个非重复视野拍照。
1.5. ICG摄取释放实验
如上ATRA处理细胞9 d,移除培养基后PBS洗3次,加入1 mg/mL ICG染液200 μL,放回含5% CO2的37 ℃恒温箱1 h,PBS洗3次,显微镜下观察,每组选取大于10个非重复视野拍照。随后移除孔内PBS,换回原来的培养基,置于含5% CO2的37 ℃恒温箱孵育6 h,显微镜下观察细胞ICG释放情况。
1.6. 透射电镜观察细胞自噬水平
ATRA处理3 d后收集贴壁细胞。PBS洗3次,加入1 mL PBS将细胞刮下,1200 r/min离心10 min,吸弃上清后加入4%戊二醛固定,送至重庆医科大学生命科学研究院电镜室进行电镜样品固定、漂洗、脱水、浸透、包埋,超薄切片后进行透射电镜观察。
1.7. 激光共聚焦观察细胞自噬情况
细胞贴壁后即用ptfLC3质粒转染细胞,转染6 h后诱导组加入10 μmol/L终浓度的ATRA。诱导后48 h和72 h激光共聚焦显微镜下观察细胞内红色及绿色亮点,每组随机选取大于10个非重复视野拍照。
1.8. Western blot检测肝细胞标志蛋白及自噬相关标志蛋白的表达
ATRA处理后提取细胞总蛋白,BCA法测定蛋白浓度。12% SDS-PAGE 50 V电泳30 min+100 V电泳1 h,PVDF膜100 V转膜,5%脱脂牛奶室温封闭1 h,一抗β-actin(1:200)、ALB(1:200)、AFP(1:200)、CK18(1: 200)、LC3(1:1000)、Beclin-1(1:1000)、RAB7(1:1000)、P62(1:1000)置于4 ℃孵育过夜,对应二抗室温孵育1 h。ECL发光显影,Image-J软件分析Western blot条带灰度值。
肝细胞标志物的检测时间点是ATRA处理后7 d,自噬标志物的检测时间点是ATRA处理后3 d,在结果部分描述。
1.9. 统计学分析
所有数据以均数±标准差表达,SPSS13.0软件进行统计学检验,组间比较采用单因素方差分析,两两比较采用SNKq检验,P < 0.05表示差异有统计学意义。
2. 结果
2.1. ATRA诱导小鼠肝癌细胞的成熟分化
Real-time PCR及Western blot结果显示,ATRA诱导后7 d,肝前体标志蛋白AFP基因水平及蛋白水平的表达均明显下调,而成熟肝细胞标志蛋白ALB、CK18、TAT、ApoB表达显著增强,且呈浓度依赖性,以10 μmol/L组最为明显(图 1,2,P < 0.05)。
1.
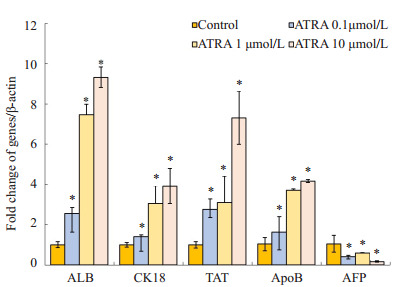
Real-time PCR检测不同浓度ATRA诱导后小鼠肝癌细胞Hepa1-6肝细胞标志mRNA水平的表达
mRNA expressions of hepatic specific markers in ATRA-induced Hepa1-6 cells detected by real-time PCR (*P < 0.05 vs control group).
2.
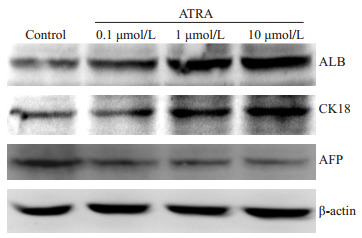
Western blot检测不同浓度ATRA诱导后小鼠肝癌细胞Hepa1-6肝细胞标志蛋白水平的表达
Protein expression of hepatic specific markers in ATRA-induced Hepa1-6 cells detected by Western blotting.
2.2. ATRA促进小鼠肝癌细胞功能成熟
ICG摄取实验及PAS染色可用于检测肝细胞的代谢解毒及糖原合成功能[15-16]。对照组仅有极少数细胞摄取吲哚菁绿,而ATRA诱导后10 d摄取吲哚菁绿的细胞数明显增多,且呈剂量依赖性增高。与ICG结果相似,对照组胞浆内几乎没有糖原合成,而ATRA处理组的细胞胞浆内的紫红色明显增加且呈剂量依赖性(图 3)。
3.
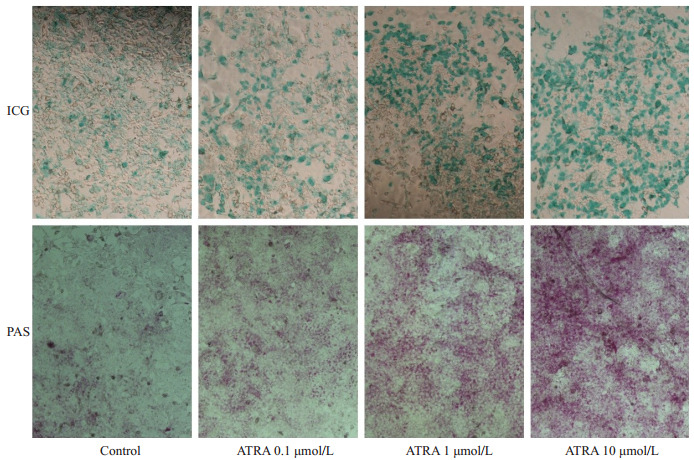
不同浓度ATRA诱导小鼠肝癌细胞Hepa1-6的ICG摄取和糖原合成能力
ICG uptake and glycogen storage function ofATRA-induced Hepa1-6 cells (Original magnification: ×100).
2.3. ATRA促进小鼠肝癌细胞内的自噬活性
透射电镜是目前检测细胞自噬水平的金标准[17],可直观检测细胞内自噬小体及自噬溶酶体的结构形态和数量。Control组细胞较幼稚,细胞连接较少,没有自噬小体及自噬溶酶体的形成,而ATRA诱导组发现有吞噬了细胞器的双层膜结构的自噬溶酶体,成熟细胞器增多,出现桥粒,紧密连接等细胞间连接,且随ATRA浓度增高越多,10 μmol/L的处理组还发现了具有分泌蛋白功能的高尔基复合体(图 4)。
4.
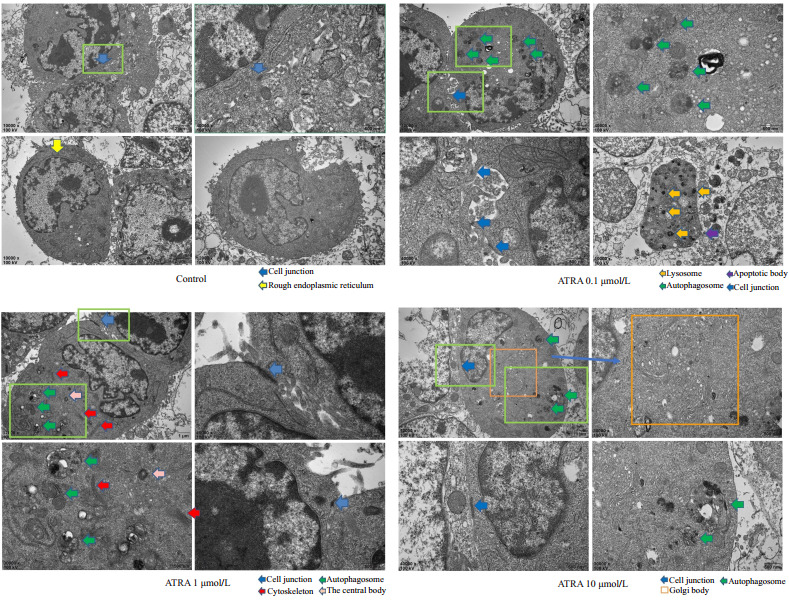
透射电镜观察小鼠肝癌细胞Hepa1-6内自噬小体及细胞连接
Autophagosome and cell junction in the cytoplasm of Hepa1-6 cells observed by transmission electron microscopy.
2.4. ATRA上调小鼠肝癌细胞内的自噬蛋白表达,自噬流通畅
综上所述,ATRA可有效诱导小鼠肝癌细胞Hepa1-6的成熟分化并促进其成熟肝细胞的代谢解毒和糖原合成储存功能,且以10 μmol/L作用最为显著,因此选择10 μmol/L作为最适终浓度。
LC3、Beclin-1、RAB7、P62是细胞自噬的标志蛋白。Western blot结果显示,10 μmol/LATRA诱导小鼠肝癌细胞Hepa1-6 3 d后,LC3-Ⅱ、Beclin-1以及RAB7的蛋白表达均有明显上调,LC3-Ⅱ/LC3-Ⅰ比值显著增高(图 5),而P62蛋白水平稍有增高。
5.
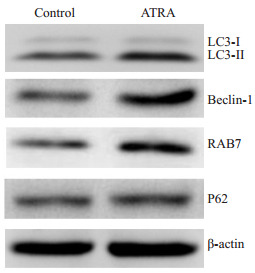
Western blot检测自噬相关标志蛋白的表达
Protein expressions of autophagy-related markers detected by Western blotting.
用ptfLC3双荧光质粒转染细胞,标记及追踪自噬小体以及自噬流的变化,10 μmol/L ATRA诱导48 h及72 h后,激光共聚焦扫描显微镜成像后的结果显示如图 6。对照组细胞内呈现部分绿色亮点(代表自噬小体),而红色亮点(代表自噬溶酶体)较少,提示对照组有部分自噬小体形成;ATRA诱导48 h后,绿色及红色亮点均有明显增多,诱导72 h后,部分细胞不见绿色荧光及亮点,红色亮点仍有明显增多。
6.
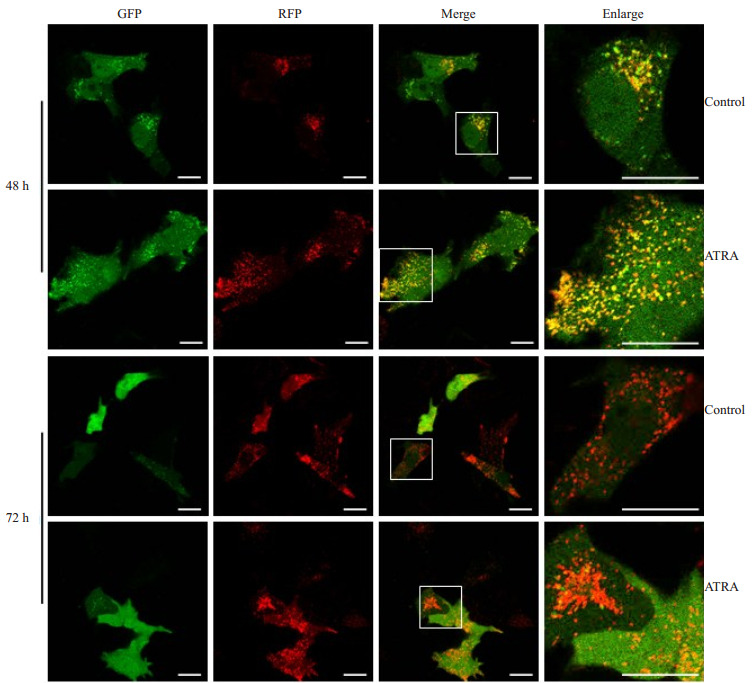
ptfLC3双荧光质粒转染48 h和72 h后追踪自噬活性
Autophagy activity in the cells detected 48 and 72 h after ptfLC3 transfection.Scare bar=20 μm.
3. 讨论
全反式维甲酸(ATRA)是目前公认的能有效诱导干细胞和肿瘤细胞分化的重要因子[18-19]。我们的前期研究发现,ATRA可促进小鼠肝癌细胞Hepa1-6的分化[8],Orfali等发现,ATRA及其信号通路可提高细胞内自噬的活性[11, 20],但ATRA促进小鼠肝癌细胞Hepa1-6分化的作用与自噬的关系目前尚无报道。
本研究结果显示,ATRA诱导后,肝前体标志蛋白AFP在基因水平和蛋白水平的表达均有明显下调,而成熟肝细胞的标志蛋白ALB、CK18、TAT、ApoB表达显著增强,ICG摄取实验及PAS糖原染色发现相较于对照组,ATRA诱导后细胞分化能力增强,透射电镜下ATRA诱导组细胞更加成熟,且上述结果均随ATRA作用浓度的增大越明显。因此,我们证实了ATRA可有效诱导小鼠肝癌细胞hepa1-6的成熟分化,使其表达一定的成熟肝细胞功能,10 μmol/L为最适作用浓度。
自噬是一种高度保守的进化过程,它通过降解细胞内受损的蛋白质及细胞器,将代谢产物从溶酶体再次释放并循环到代谢及生物合成中,防止细胞废物产生的毒性积累,以维持细胞内稳态[21-25]。自噬与恶性肿瘤、心脏疾病和神经退行性变等有着密切关系[26-27],自噬相关基因ATG6/Beclin-1在40%~75%的人类前列腺癌,乳腺癌和卵巢癌中有丢失,在某些基于细胞的测定中,抑制自噬可促进癌细胞生长,并且Beclin-1杂合突变的小鼠有使肝肺肿瘤和淋巴瘤发展的可能[21, 23-24]。在一些模型中,自噬可抑制癌症起始阶段的组织损伤,慢性炎症,DNA损伤反应和基因组不稳定性,从而抑制了癌症的发生[21, 24-25]。
ATRA诱导后电镜下观察发现细胞内自噬小体增多,且随ATRA浓度增高越多。ptfLC3双荧光质粒可以同时直观地观察细胞的自噬水平及自噬流变化情况,LC3-Ⅰ主要在胞质内散在分布,共聚焦扫描显微镜下呈现片状分布,LC3-Ⅱ则主要定位于各阶段自噬囊泡的内外膜上,共聚焦扫描显微镜下呈现出聚集的绿色亮点,因此绿色亮点即代表细胞内的自噬小体。由于绿色荧光在自噬小体与溶酶体结合形成自噬溶酶体时会淬灭,而红色荧光对酸性环境的耐受力较强不易发生降解,因此红色亮点代表晚期的自噬溶酶体[28-29]。结果显示,对照组细胞内有部分绿色亮点,而红色亮点较少,提示对照组细胞有基础的自噬小体形成,处于自噬的早期阶段;ATRA诱导后48~72 h细胞内红色亮点明显增多,提示自噬溶酶体形成增多,细胞自噬水平增高,且自噬流通畅。自噬小体和自噬溶酶体可及时清除细胞内损坏的细胞器及各种代谢产物,防止有害物质堆积,利于细胞生存,我们认为ATRA对细胞成熟分化的作用也可能与自噬小体和自噬溶酶体的这种清除作用有关。
LC3是目前被最广泛应用的自噬标志蛋白,它参与了自噬小体膜的形成,包括LC3-Ⅰ和LC3-Ⅱ两种蛋白形式,两者之间可相互转化。其中LC3-Ⅰ广泛分布在胞质内,经泛素化修饰后与自噬小体膜表面的磷脂酰乙醇胺结合形成LC3-Ⅱ,LC3-Ⅱ/Ⅰ的比值与自噬小体的数量成正相关,在一定程度上可反映细胞的自噬活性[30]。Beclin-1主要定位于高尔基体、内质网及线粒体,通过Beclin-1与液泡分选蛋白34(Vps34)的复合体募集其他自噬相关蛋白在自噬小体形成早期发挥了重要作用[31]。RAB7(Ras相关蛋白7)是自噬小体与溶酶体融合过程中的一个重要分子,它参与自噬小体的成熟,运输以及最终与溶酶体的融合,代表自噬的通量[32]。ATRA诱导后,LC3-Ⅱ、Beclin-1和RAB7的表达均明显增高,LC3-Ⅱ/I比值增高。P62(也被称为SQSTM1)是自噬流下游的标志蛋白,它可连接LC3和泛素化的底物,当自噬小体和溶酶体融合时被降解[33]。ATRA作用后P62的表达稍有增高。有报道称P62与LC3的变化并不一定相反,在短期饥饿时,诱发细胞自噬,这会使作为选择性底物的P62降低,但在长时间(~4~8 h)饥饿后,P62 mRNA转录上调,自噬产生足够的细胞内氨基酸提供原料,因此细胞通过从头蛋白质合成将P62蛋白水平恢复到基础水平[34]。Chu等也发现,在急性脊髓损伤并发肺损伤后24~72 h肺部自噬水平增高,但P62的水平也增高[35]。
王卓等研究在髓系白血病中ATRA通过抑制mTOR通路并激活Atg1-PI3KC3-Atg5自噬通路来促进自噬体的形成[36];Zhong C等则发现,ATRA通过促进细胞自噬保护肝细胞免受缺血再灌注损伤,其可能分子机制是依赖于Foxo3/p-Akt/Foxo1信号通路调控[37];Shi L等认为全反式维甲酸通过针对keap1/nrf2的信号通路增强了人体神经胶质瘤细胞的自噬作用[38]。然而对于ATRA是如何上调肝癌细胞的自噬水平还需进一步研究。
因此,我们初步证实了ATRA诱导后可促进小鼠肝癌细胞Hepa1-6的成熟分化,并表达一定的成熟肝细胞功能,并且在此过程中细胞自噬水平明显增加,自噬可能参与了ATRA诱导小鼠肝癌细胞Hepa1-6成熟分化的过程。本研究为进一步探讨ATRA诱导肝癌细胞成熟分化的分子生物学机制奠定了坚实的基础,也为ATRA用于肝癌的临床治疗提供了重要思路。
Biography
方姝煜,硕士研究生,E-mail: 347064066@qq.com
Funding Statement
重庆市自然科学基金(csct2016jcyjA0228)
Contributor Information
方 姝煜 (Shuyu FANG), Email: 347064066@qq.com.
毕 杨 (Yang BI), Email: yang_bi@hospital.cqmu.edu.cn.
References
- 1.Cillo U, Vitale A, Polacco M, et al. Liver transplantation for hepatocellular carcinoma through the lens of transplant benefit. Hepatology. 2017;65(5):1741–8. doi: 10.1002/hep.28998. [Cillo U, Vitale A, Polacco M, et al.Liver transplantation for hepatocellular carcinoma through the lens of transplant benefit[J].Hepatology, 2017, 65(5): 1741-8.] [DOI] [PubMed] [Google Scholar]
- 2.Saberi B, Dadabhai AS, Durand CM, et al. Challenges in treatment of hepatitis C among patients with hepatocellular carcinoma. Hepatology. 2017;66(2):661–3. doi: 10.1002/hep.v66.2. [Saberi B, Dadabhai AS, Durand CM, et al.Challenges in treatment of hepatitis C among patients with hepatocellular carcinoma[J].Hepatology, 2017, 66(2): 661-3.] [DOI] [PubMed] [Google Scholar]
- 3.Zhang S, Yue M, Shu R, et al. Recent advances in the management of hepatocellular carcinoma. https://www.ncbi.nlm.nih.gov/pubmed/27273938. J BUON. 2016;21(2):307–11. [Zhang S, Yue M, Shu R, et al.Recent advances in the management of hepatocellular carcinoma[J].J BUON, 2016, 21(2): 307-11.] [PubMed] [Google Scholar]
- 4.Siddikuzzaman, Guruvayoorappan C, Berlin GV. All trans retinoic acid and cancer. Immunopharmacol Immunotoxicol. 2011;33(2):241–9. doi: 10.3109/08923973.2010.521507. [Siddikuzzaman, Guruvayoorappan C, Berlin GV.All trans retinoic acid and cancer[J].Immunopharmacol Immunotoxicol, 2011, 33(2): 241-9.] [DOI] [PubMed] [Google Scholar]
- 5.Qian XJ, He JS, Zhao Y, et al. Inhibition of p38 MAPK phosphorylation is critical for bestatin to enhance ATRA-Induced cell differentiation in acute promyelocytic leukemia NB4 cells. Am J Ther. 2016;23(3):e680–9. doi: 10.1097/01.mjt.0000433950.01406.b3. [Qian XJ, He JS, Zhao Y, et al.Inhibition of p38 MAPK phosphorylation is critical for bestatin to enhance ATRA-Induced cell differentiation in acute promyelocytic leukemia NB4 cells[J].Am J Ther, 2016, 23(3): e680-9.] [DOI] [PubMed] [Google Scholar]
- 6.Huang JY, Bi Y, Zhu GH, et al. Retinoic acid signalling induces the differentiation of mouse fetal liver-derived hepatic progenitor cells. Liver Int. 2009;29(10):1569–81. doi: 10.1111/liv.2009.29.issue-10. [Huang JY, Bi Y, Zhu GH, et al.Retinoic acid signalling induces the differentiation of mouse fetal liver-derived hepatic progenitor cells [J].Liver Int, 2009, 29(10): 1569-81.] [DOI] [PubMed] [Google Scholar]
- 7.周 建武, 何 昀, 龚 梦嘉, et al. 重组siRARγ腺病毒构建及其对小鼠肝脏祖细胞分化的影响. http://www.wanfangdata.com.cn/details/detail.do?_type=perio&id=jcyxylc201401005. 基础医学与临床. 2014;34(1):22–8. [周建武, 何昀, 龚梦嘉, 等.重组siRARγ腺病毒构建及其对小鼠肝脏祖细胞分化的影响[J].基础医学与临床, 2014, 34(1): 22-8.] [Google Scholar]
- 8.Cui JJ, Gong MJ, He Y, et al. All- trans retinoic acid inhibits proliferation, migration, invasion and induces differentiation of hepa1-6 cells through reversing EMT in vitro. Int J Oncol. 2016;48(1):349–57. doi: 10.3892/ijo.2015.3235. [Cui JJ, Gong MJ, He Y, et al.All- trans retinoic acid inhibits proliferation, migration, invasion and induces differentiation of hepa1-6 cells through reversing EMT in vitro[J].Int J Oncol, 2016, 48(1): 349-57.] [DOI] [PubMed] [Google Scholar]
- 9.Hurley JH, Schulman BA. Atomistic autophagy: the structures of cellular self-digestion. Cell. 2014;157(2):300–11. doi: 10.1016/j.cell.2014.01.070. [Hurley JH, Schulman BA.Atomistic autophagy: the structures of cellular self-digestion[J].Cell, 2014, 157(2): 300-11.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Humbert M, Federzoni EA, Tschan MP. Distinct TP73-DAPK2- ATG5 pathway involvement in ATO-mediated cell death versus ATRA-mediated autophagy responses in APL. J Leukoc Biol. 2017;102(6):1357–70. doi: 10.1189/jlb.1A0317-132R. [Humbert M, Federzoni EA, Tschan MP.Distinct TP73-DAPK2- ATG5 pathway involvement in ATO-mediated cell death versus ATRA-mediated autophagy responses in APL[J].J Leukoc Biol, 2017, 102(6): 1357-70.] [DOI] [PubMed] [Google Scholar]
- 11.Yang Z, Klionsky DJ. Eaten alive: a history of macroautophagy. Nat Cell Biol. 2010;12(9):814–22. doi: 10.1038/ncb0910-814. [Yang Z, Klionsky DJ.Eaten alive: a history of macroautophagy[J]Nat Cell Biol, 2010, 12(9): 814-22.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Orfali N, Mckenna SL, Cahill MR, et al. Retinoid receptor signaling and autophagy in acute promyelocytic leukemia. Exp Cell Res. 2014;324(1):1–12. doi: 10.1016/j.yexcr.2014.03.018. [Orfali N, Mckenna SL, Cahill MR, et al.Retinoid receptor signaling and autophagy in acute promyelocytic leukemia[J].Exp Cell Res, 2014, 324(1): 1-12.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Orfali N, O'donovan TR, Nyhan MJ, et al. Induction of autophagy is a key component of all-trans-retinoic acid-induced differentiation in leukemia cells and a potential target for pharmacologic modulation. Exp Hematol. 2015;43(9):781–93. doi: 10.1016/j.exphem.2015.04.012. [Orfali N, O'donovan TR, Nyhan MJ, et al.Induction of autophagy is a key component of all-trans-retinoic acid-induced differentiation in leukemia cells and a potential target for pharmacologic modulation [J].Exp Hematol, 2015, 43(9): 781-93.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cao HL, Xie J, Guo L, et al. All-trans retinoic acid induces autophagic degradation of ubiquitin-like modifier activating enzyme 3 in acute promyelocytic leukemia cells. Leuk Lymphoma. 2018;59(5):1222–30. doi: 10.1080/10428194.2017.1365850. [Cao HL, Xie J, Guo L, et al.All-trans retinoic acid induces autophagic degradation of ubiquitin-like modifier activating enzyme 3 in acute promyelocytic leukemia cells[J].Leuk Lymphoma, 2018, 59 (5): 1222-30.] [DOI] [PubMed] [Google Scholar]
- 15.Yamada T, Yoshikawa M, Kanda S, et al. In vitro differentiation of embryonic stem cells into hepatocyte-like cells identified by cellular uptake of indocyanine green. Stem Cells. 2002;20(2):146–54. doi: 10.1634/stemcells.20-2-146. [Yamada T, Yoshikawa M, Kanda S, et al.In vitro differentiation of embryonic stem cells into hepatocyte-like cells identified by cellular uptake of indocyanine green[J].Stem Cells, 2002, 20(2): 146-54.] [DOI] [PubMed] [Google Scholar]
- 16.Kamo N, Yasuchika K, Fujii H, et al. Two populations of Thy1- positive mesenchymal cells regulate in vitro maturation of hepatic progenitor cells. Am J Physiol Gastrointest Liver Physiol. 2007;292(2):G526–34. doi: 10.1152/ajpgi.00241.2006. [Kamo N, Yasuchika K, Fujii H, et al.Two populations of Thy1- positive mesenchymal cells regulate in vitro maturation of hepatic progenitor cells[J].Am J Physiol Gastrointest Liver Physiol, 2007, 292(2): G526-34.] [DOI] [PubMed] [Google Scholar]
- 17.Klionsky DJ, Abdelmohsen K, Abe A, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition) Autophagy. 2016;12(1):1–222. doi: 10.1080/15548627.2015.1100356. [Klionsky DJ, Abdelmohsen K, Abe A, et al.Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition) [J].Autophagy, 2016, 12(1): 1-222.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arisi MF, Starker RA, Addya S, et al. All trans-retinoic acid (ATRA) induces re-differentiation of early transformed breast epithelial cells. Int J Oncol. 2014;44(6):1831–42. doi: 10.3892/ijo.2014.2354. [Arisi MF, Starker RA, Addya S, et al.All trans-retinoic acid (ATRA) induces re-differentiation of early transformed breast epithelial cells [J].Int J Oncol, 2014, 44(6): 1831-42.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu WJ, Jiang NJ, Guo QL, et al. ATRA and as2O3 regulate differentiation of human hematopoietic stem cells into granulocyte progenitor via alteration of HoxB8 expression. https://es.scribd.com/document/336456025/Targeted-Cancer-Therapy. Eur Rev Med Pharmacol Sci. 2015;19(6):1055–62. [Liu WJ, Jiang NJ, Guo QL, et al.ATRA and as2O3 regulate differentiation of human hematopoietic stem cells into granulocyte progenitor via alteration of HoxB8 expression[J].Eur Rev Med Pharmacol Sci, 2015, 19(6): 1055-62.] [PubMed] [Google Scholar]
- 20.Eriksen AB, Torgersen ML, Holm KL, et al. Retinoic acid-induced IgG production in TLR-activated human primary B cells involves ULK1-mediated autophagy. Autophagy. 2015;11(3):460–71. doi: 10.1080/15548627.2015.1009797. [Eriksen AB, Torgersen ML, Holm KL, et al.Retinoic acid-induced IgG production in TLR-activated human primary B cells involves ULK1-mediated autophagy[J].Autophagy, 2015, 11(3): 460-71.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.White E, Mehnert JM, Chan CS. Autophagy, metabolism, and cancer. Clin Cancer Res. 2015;21(22):5037–46. doi: 10.1158/1078-0432.CCR-15-0490. [White E, Mehnert JM, Chan CS.Autophagy, metabolism, and cancer [J].Clin Cancer Res, 2015, 21(22): 5037-46.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Farrow JM, Yang JC, Evans CP. Autophagy as a modulator and target in prostate cancer. Nat Rev Urol. 2014;11(9):508–16. doi: 10.1038/nrurol.2014.196. [Farrow JM, Yang JC, Evans CP.Autophagy as a modulator and target in prostate cancer[J].Nat Rev Urol, 2014, 11(9): 508-16.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.White E. The role for autophagy in cancer. J Clin Invest. 2015;125(1):42–6. doi: 10.1172/JCI73941. [White E.The role for autophagy in cancer[J].J Clin Invest, 2015, 125(1): 42-6.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Villar VH, Merhi F, Djavaheri-Mergny MA. Glutaminolysis and autophagy in cancer. Autophagy. 2015;11(8):1198–208. doi: 10.1080/15548627.2015.1053680. [Villar VH, Merhi F, Djavaheri-Mergny MA.Glutaminolysis and autophagy in cancer[J].Autophagy, 2015, 11(8): 1198-208.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hönscheid P, Datta K, Muders MH. Autophagy: detection, regulation and its role in cancer and therapy response. Int J Radiat Biol. 2014;90(8):628–35. doi: 10.3109/09553002.2014.907932. [Hönscheid P, Datta K, Muders MH.Autophagy: detection, regulation and its role in cancer and therapy response[J].Int J Radiat Biol, 2014, 90(8): 628-35.] [DOI] [PubMed] [Google Scholar]
- 26.Lucin KM, Wyss- Coray T. Targeting autophagy for diseasetherapy. Nat Biotechnol. 2013;31(4):322–3. doi: 10.1038/nbt.2554. [Lucin KM, Wyss- Coray T.Targeting autophagy for diseasetherapy [J].Nat Biotechnol, 2013, 31(4): 322-3.] [DOI] [PubMed] [Google Scholar]
- 27.Nixon RA. The role of autophagy in neurodegenerative disease. Nat Med. 2013;19(8):983–97. doi: 10.1038/nm.3232. [Nixon RA.The role of autophagy in neurodegenerative disease[J].Nat Med, 2013, 19(8): 983-97.] [DOI] [PubMed] [Google Scholar]
- 28.Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescenttagged LC3. Autophagy. 2007;3(5):452–60. doi: 10.4161/auto.4451. [Kimura S, Noda T, Yoshimori T.Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescenttagged LC3[J].Autophagy, 2007, 3(5): 452-60.] [DOI] [PubMed] [Google Scholar]
- 29.Shanware NP, Bray K, Eng CH, et al. Glutamine deprivation stimulates mTOR-JNK-dependent chemokine secretion. Nat Commun. 2014;5:4900. doi: 10.1038/ncomms5900. [Shanware NP, Bray K, Eng CH, et al.Glutamine deprivation stimulates mTOR-JNK-dependent chemokine secretion[J].Nat Commun, 2014, 5: 4900.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dancourt J, Melia TJ. Lipidation of the autophagy proteins LC3 and GABARAP is a membrane-curvature dependent process. Autophagy. 2014;10(8):1470–1. doi: 10.4161/auto.29468. [Dancourt J, Melia TJ.Lipidation of the autophagy proteins LC3 and GABARAP is a membrane-curvature dependent process[J].Autophagy, 2014, 10(8): 1470-1.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Backer JM. The regulation and function of Class Ⅲ PI3Ks: novel roles for Vps34. https://www.ncbi.nlm.nih.gov/pubmed/18215151. Biochem J. 2008;410(1):1–17. doi: 10.1042/BJ20071427. [Backer JM.The regulation and function of Class Ⅲ PI3Ks: novel roles for Vps34[J].Biochem J, 2008, 410(1): 1-17.] [DOI] [PubMed] [Google Scholar]
- 32.Hyttinen JM, Niittykoski M, Salminen AA. Maturation of autophagosomes and endosomes: a key role for Rab7. https://www.ncbi.nlm.nih.gov/pubmed/23220125. Biochim BiophysActa. 2013;1833(3):503–10. doi: 10.1016/j.bbamcr.2012.11.018. [Hyttinen JM, Niittykoski M, Salminen AA.Maturation of autophagosomes and endosomes: a key role for Rab7[J].Biochim BiophysActa, 2013, 1833(3): 503-10.] [DOI] [PubMed] [Google Scholar]
- 33.Pankiv S, Clausen TH, Lamark T, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282(33):24131–45. doi: 10.1074/jbc.M702824200. [Pankiv S, Clausen TH, Lamark T, et al.p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy[J].J Biol Chem, 2007, 282(33): 24131-45.] [DOI] [PubMed] [Google Scholar]
- 34.Sahani MH, Itakura E, Mizushima N. Expression of the autophagy substrate SQSTM1/p62 is restored during prolonged starvation depending on transcriptional upregulation and autophagy- derived amino acids. Autophagy. 2014;10(3):431–41. doi: 10.4161/auto.27344. [Sahani MH, Itakura E, Mizushima N.Expression of the autophagy substrate SQSTM1/p62 is restored during prolonged starvation depending on transcriptional upregulation and autophagy- derived amino acids[J].Autophagy, 2014, 10(3): 431-41.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chu R, Wang J, Bi Y, et al.The kinetics of autophagy in the lung following acute spinal cord injury in rats[J].Spine, 2018, [Epub ahead of print].
- 36.王卓.自噬调节全反式维甲酸诱导髓系白血病细胞分化的机制研究[D].长沙: 中南大学, 2011.
- 37.Zhong C, Pu L, Fang M, et al. Retinoic acid receptor alpha promotes autophagy to alleviate liver ischemia and reperfusion injury. World J Gastroenterol. 2015;21(43):12381–91. doi: 10.3748/wjg.v21.i43.12381. [Zhong C, Pu L, Fang M, et al.Retinoic acid receptor alpha promotes autophagy to alleviate liver ischemia and reperfusion injury[J].World J Gastroenterol, 2015, 21(43): 12381-91.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shi L, Li H, Zhan Y. All-trans retinoic acid enhances temozolomideinduced autophagy in human glioma cells U251 via targeting Keap1/ Nrf2/ARE signaling pathway. Oncol Lett. 2017;14(3):2709–14. doi: 10.3892/ol.2017.6482. [Shi L, Li H, Zhan Y.All-trans retinoic acid enhances temozolomideinduced autophagy in human glioma cells U251 via targeting Keap1/ Nrf2/ARE signaling pathway[J].Oncol Lett, 2017, 14(3): 2709-14.] [DOI] [PMC free article] [PubMed] [Google Scholar]


