Abstract
目的
探讨长链非编码RNA-UFC1在肝癌侵袭和转移中的作用及机制。
方法
应用lincRNA-UFC1慢病毒载体转染人肝癌细胞株Huh7构建lincRNA-UFC1过表达组和对照组;应用lincRNA-UFC1干扰载体转染人肝癌细胞株BEL-7402构建lincRNA-UFC1干扰组和干扰对照组(scramble),通过实时定量PCR实验分析两种肝癌细胞中lincRNA-UFC1的表达水平。Transwell及划痕实验检测lincRNA-UFC1过表达组及干扰组相比对照组侵袭迁移能力的变化。Western blot检测lincRNAUFC1过表达组及干扰组相比对照组中GSK-3β/β-catenin蛋白的表达变化。利用GSK-3β/β-catenin信号通路抑制剂XAV939阻断后,观察Transwell及划痕实验中lincRNA-UFC1过表达对肝癌细胞侵袭迁移能力的影响。
结果
在肝细胞癌中,过表达lincRNA-UFC1相较于对照组,肝癌细胞的侵袭转移能力明显增加(P<0.001),而沉默lincRNA-UFC1的表达相较于scramble组,肝癌细胞侵袭转移能力明显减弱(P<0.001)。Western blot结果显示,过表达lincRNA-UFC1与对照组相比较,侵袭转移相关蛋白β-catenin和p-GSK-3β表达明显上调(P<0.001),而lincRNA-UFC1沉默表达后,结果相反。利用GSK-3β/β-catenin信号通路抑制剂XAV939处理肝癌细胞可以逆转lincRNA-UFC1促进肝癌细胞侵袭转移的功能(P<0.001)。
结论
在肝细胞癌中lincRNA-UFC1通过上调磷酸化GSK-3β及β-catenin促进肝癌细胞侵袭转移。
Keywords: lincRNA-UFC1, β-catenin, 磷酸化GSK3β, 肝细胞癌, 细胞侵袭转移
Abstract
Objective
To explore the role of Long noncoding RNA UFC1 (lincRNA-UFC1) in modulating the metastasis and invasion of hepatocellular carcinoma (HCC) cells and the underlying mechanism.
Methods
Human HCC cell line Huh7 was infected with the lentiviral vector carrying lincRNA-UFC1 to obtain a cell line with lincRNA-UFC1 overexpression. A short hairpin RNA (shRNA) targeting lincRNA-UFC1 was delivered in human HCC BEL-7402 cells via a lentiviral vector to obtain a cell line with lincRNA-UFC1 knockdown. Expression levels of lincRNA-UFC1 in the two HCC cell lines were detected using real-time PCR, and the changes in the cell invasion and migration in response to lincRNA-UFC1 overexpression or knockdown were analyzed using Transwell and wound-healing assays. The expressions of GSK-3β/β-catenin-related proteins in the cells were detected with Western blotting. XAV-939, a GSK-3β/β-catenin inhibitor, was used for assessing the impact of lincRNAUFC1 overexpression on the invasion and migration of the HCC cells through Transwell and wound-healing assays.
Results
Overexpression of lincRNA-UFC1 significantly promoted the invasion and migration of Huh7 cells as compared with the control cells (P < 0.001), while lincRNA-UFC1 knockdown obviously suppressed the invasion and migration of BEL-7402 cells (P < 0.001). The results of Western blotting showed that the expressions of proteins associated with the cell invasion and migration, namely β-catenin and P-GSK-3β, were significantly upregulated in response to lincRNA-UFC1 overexpression, and were obviously lowered after lincRNA-UFC1 knockdown. Treatment of the cells with XAV-939 significantly reversed the effect of lincRNA-UFC1 overexpression on the cell invasion and migration (P < 0.001).
Conclusion
lincRNA-UFC1 overexpresison promotes cell invasion and migration through the GSK-3β/β-catenin axis in HCC cells in vitro.
Keywords: lincRNA-UFC1, β-catenin, p-GSK3, hepatocellular carcinoma, tumor progression
肝细胞癌(HCC)在肿瘤相关死亡中居第3位[1]。近年来在肝癌的治疗上取得了一定的进展[2],但由于肝癌容易发生远处转移导致肝癌患者的预后较差[3-4]。因此,研究肝癌侵袭转移的分子机制对提高肝癌患者的预后有着重要的意义。
长链非编码RNA(lncRNAs)是在一类哺乳动物细胞中大量存在的,不编码蛋白质的RNA,其长度大于200个核苷酸[5-8]。越来越多的证据证明lncRNAs在癌症中发挥着重要的作用,如乳腺癌,结直肠癌,前列腺癌,肝细胞癌等[9-12]。在乳腺癌中,NF-κB相互作用lncRNA(NKILA)通过与NF-κB/IκB复合物结合,负向调控NF-κB信号活性,从而抑制乳腺癌转移和疾病进展,是乳腺癌患者预后良好的分子指标[11]。在肝癌中,TGF-β激活的lncRNA(lncRNA-ATB)通过诱导上皮间质转换(EMT)促进肝癌细胞的侵袭、转移,并且与患者预后不良密切相关[10]。因此,lncRNAs也许可以成为预示癌症预后的标志物和治疗的靶点[13-14]。在既往的研究中,我们课题组发现lincRNA-UFC1在肝癌中高表达,其高表达预示着肝癌患者的不良预后,进一步机制探讨发现lincRNA-UFC1在miRNA-34a的调控下,通过直接与HuR结合而增强β-catenin mRNA的稳定性,并诱导β-catenin的核转位而激活β-catenin下游信号通路,促进肝癌细胞的增殖[15]。然而,lincRNA-UFC1对肝癌细胞的侵袭转移的作用尚未明确。因此,本研究通过探讨lincRNA-UFC1对肝细胞癌侵袭、转移能力的影响,揭示了lincRNA-UFC1在肝癌发生发展过程中促肿瘤的作用及其基本机制,为肝癌精准治疗提供了新的潜在靶点。
1. 材料和方法
1.1. 材料
肝癌细胞系Huh7和BEL-7402(中国科学院上海细胞库);胎牛血清、高糖DMEM培养基、PRMI 1640培养基(GIBCO);胰蛋白酶消化液(吉诺);Lipofectamin 2000、TRIzol(Invitrogen);RIPA蛋白裂解液(罗氏);慢病毒(吉凯基因);β-catenin抑制剂XAV939(GLPBIO);BCA蛋白浓度测定试剂盒(碧云天);PVDF膜(Bio-red);SYBR®PrimeScript®RT- PCR Kit(Perfect Real Time)(TAKARA);内参β-actin引物由上海英潍捷基公司合成;β-actin mRNA引物(5'-TCTTCGCTTTGTCCTTTCGT-3';5'-GCTGGAAGGTGGACAACGA-3');β-catenin、GSK3β、P-GSK3β、β-actin单克隆抗体(Santa Cruz);脱脂奶粉(CST)、羊抗鼠/兔二抗(北京中杉金桥),Western blot电泳系统(Bio-Rad)。
1.2. 细胞培养
人肝癌细胞株Huh7、BEL-7402采用10%胎牛血清的高糖DMEM培养基培养,BEL-7402采用含10%胎牛血清的PRMI 1640培养基培养,置于37 ℃、5% CO2培养箱内培养,倒置显微镜下观察细胞生长情况,每2~3 d更换培养基,3~4 d用含EDTA的胰酶消化、传代培养,取生长状态良好的细胞用于实验。
1.3. 慢病毒载体构建和肝癌细胞转染
1.3.1. 建立稳定高表达lincRNA-UFC1基因的肝癌细胞株
以肝癌细胞cDNA为模板,PCR扩增(引物含交换配对碱基、Age I酶切位点)之后1%琼脂糖凝胶电泳,胶回收试剂盒回收目的基因片段与pGC-LV(吉凯公司)同时进行AgeⅠ酶切,DNA连接酶将目的片段与酶切的pGC-LV连接,构建载体pGC-LV-lincRNA-UFC1,测序验证。病毒包装和感染:建立稳定高表达lincRNAUFC1基因的肝癌细胞株:采用磷酸钙转染法,将pGCLV-lincRNA-UFC1(主体质粒)、pHelper1.0(gag/pol元件)、pHelper2.0(VSVG元件)共转染293 Z毒质粒,梯度稀释法测定慢病毒滴度。于24孔板内分别接种4×104细胞,用慢病毒悬液感染肝癌细胞Huh7,48 h后加入puromycin筛选抗性克隆。
1.3.2. 建立lincRNA-UFC1干扰的肝癌细胞株
合成lincRNA-UFC1的shRNA正义链(带有MluI酶切位点)和反义链(带有ClaI酶切位点)的DNA Oligo序列,应用DNA退火buffer梯度退火(95 ℃ 2 min,之后每90 sec下降1 ℃~25 ℃),形成DNA双链后,行2%的琼脂糖凝胶电泳,应用胶回收试剂盒回收双链DNA,同时对PLVTHM载体进行MluI和ClaI双酶切,应用DNA连接酶将shRNA与酶切的PLVTHM连接,以构建载体pLVTHM-shRNA-lincRNA-UFC1,测序验证。病毒包装和感染:采用磷酸钙转染法,将pLVTHM-shRNAlincRNA- UFC1(主体质粒)、包装载体(psPAX2和pMD2.G)共转染293FT细胞,培养48~72 h后收集病毒上清,4 ℃ 50 000×g离心2.5 h沉淀病毒质粒,梯度稀释法测定慢病毒滴度。于24孔板内接种4×104个细胞,用慢病毒悬液感染肝癌细胞BEL-7402,48 h后荧光显微镜下观察转染效率,利用流式细胞仪进行细胞分选获得GFP阳性细胞,建立稳定细胞株
1.4. 实时定量RT-PCR(qRT-PCR)
按说明书用Trizol提取收集细胞中的总RNA,A260 nm/A280 nm在1.8~2.0之间的RNA样本进行逆转录。我们用PrimeScript逆转录PCR(RT-PCR)试剂盒按照说明书特异性逆转录lincRNA-UFC1,U6作内参。我们用SYBR Premix PCR试剂盒按照说明书在LC480仪器上进行实时定量PCR实验。所有实验重复3次。我们用delta-CT求基因相当表达量方法分析lincRNAUFC1的表达水平。
1.5. Western blot实验
取转染48 h后的细胞,弃培养基,用0.01 mol/L的PBS冲洗3次后每孔加入80 μL RIPA裂解液充分裂解,10 min震荡1次,共震荡3次,15 000 r/min离心15 min后取上清,BCA法测定总蛋白浓度。将蛋白样品加入适量6×loading buffer,煮沸10 min后即可用10%聚丙烯酰胺凝胶电泳。90 V电压转膜2 h。5% BSA(TBST溶解)室温封闭1 h后加入相应比例稀释的β-catenin一抗(1:1000)、GSK-3β一抗(1:500)、p-GSK-3β一抗(1:500)及β-actin一抗(1:1000),4 ℃孵育过夜,TBST漂洗8 min×4次后加入适当(1:10 000)浓度的二抗,室温孵育1 h。TBST漂洗5 min×6次。暗室内浸入ECL液显影定影,并使胶片曝光,洗片。
1.6. 肿瘤体外侵袭实验
以聚碳酸酯微孔膜(孔径8 μm)分隔侵袭小室的上下室,滤膜上包被人工基底膜胶(ECMatrix)。实验前在侵袭小室上室加入无血清的培养液2 h,以使ECMatrix水化。细胞用胰酶消化制成单细胞悬液,无血清的培养液洗3遍后,细胞计数。小室的上室接种肿瘤细胞200 μL(含1×105细胞);用300 μL 10%胎牛血清完全培养液作为趋化因子置于下室。37 ℃、5% CO2湿化培养箱内孵育24 h,取出小室,用棉签擦去上室内未穿过滤膜的细胞,取出滤膜,中性甲醛固定,苏木精染色。在200倍光镜下随机取5个视野计数,取其均值代表浸润力量值。
1.7. 细胞划痕实验
先用marker笔在6孔板背后,用直尺比着,均匀平行得划横线,每隔1 cm1道,横穿过孔。每孔至少穿过5条线。加入约5×105细胞。第2天用枪头比着直尺,垂至于背后的横线划痕。用PBS洗细胞3次,去处划下的细胞,加入无血清培养基。放入37 ℃ 5% CO2培养箱湿化培养箱内孵育。按0、24、36 h取样,拍照。
1.8. 统计分析
所有计量数据采用均数±标准差表示,采用SPSS 20.0统计软件进行分析,样本均数的比较采用OneWay ANOVA检验,双侧P<0.05为差异有统计学意义。
2. 结果
2.1. LincRNA-UFC1促进肝癌细胞的侵袭
沉默或过表达lincRNA-UFC1后,在Huh7细胞和BEL-7402细胞中,lincRNA-UFC1的水平分别具有明显的明显升高或降低,差异具有统计学意义(图 1A,P<0.001)。Huh7细胞沉默lincRNA-UFC1表达后,体外侵袭小室检测结果显示细胞侵袭能力明显下降(图 1B,P<0.001)。BEL-7402细胞过表达lincRNA-UFC1后,体外侵袭小室检测结果显示BEL-7402细胞侵袭能力明显增加(图 1C,P<0.001)。
1.
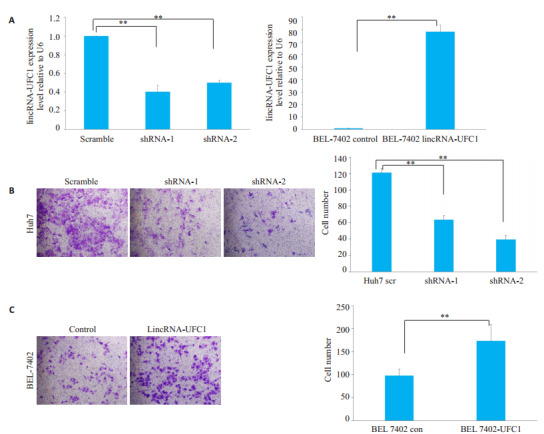
LincRNA-UFC1促进肝癌细胞侵袭
LincRNA-UFC1 promotes HCC cell invasion. A: lincRNA-UFC1 expression detected by qRT-PCR in Huh7 and BEL-7402 cells infected by lentiviruses carrying lincRNA-UFC1 short hairpin RNA (shRNA)-1 or shRNA-2 or a scrambled shRNA, or by lentivirus expressing the full-length human lincRNA-UFC1 sequence or the empty vector; B-C: Transwell invasion assay of Huh7 and BEL-7402 cells with lincRNA-UFC1 overexpression or knockdown(Original magnification: ×200). Scr: Scramble; con: Control; sh: shRNA; **P < 0.001.
2.2. LincRNA促进肝癌细胞迁移能力
转染lincRNA-UFC1的干扰慢病毒后,Huh7细胞的迁移能力明显下降(图 2A,P<0.001),而转染lincRNA-UFC1过表达慢病毒后,BEL-7402细胞迁移能力明显上升(图 2B,P<0.001)。
2.
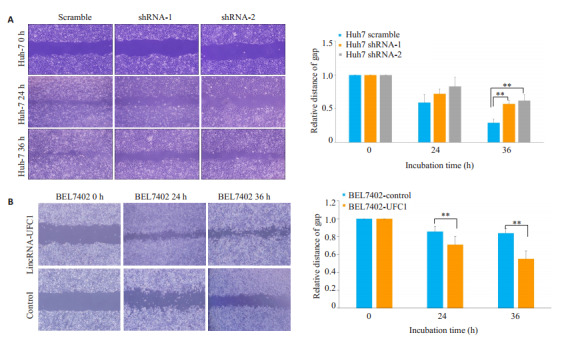
LincRNA-UFC1促进肝癌细胞迁移
LincRNA-UFC1 promotes HCC cell migration (×200). A: Migration of Huh7 cells analyzed by wound-healing assay after infection with lentiviruses carrying lincRNA-UFC1 short hairpin RNA (shRNA)-1 or shRNA-2 or a scrambled shRNA; B: Wound-healing assays for assessing migration of BEL-7402 cells afte infection by a lentivirus expressing full-length human lincRNA-UFC1 sequence or by a empty vector. **P < 0.001.
2.3. LincRNA-UFC1通过激活GSK-3β/β-catenin信号轴促进肝癌细胞侵袭转移
低表达lincRNA-UFC1后,β-catenin及p-GSK-3β的蛋白水平都有显著的下调,而过表达lincRNA-UFC1后,β-catenin及p-GSK-3β的蛋白水平明显上调(图 3A)。利用GSK- 3β/β- catenin信号通路抑制剂XAV939(0.75 μmol/L)处理肝癌细胞后,Transwell结果提示:lincRNA-UFC1过表达肝癌细胞侵袭能力显著减弱(图 3B,P<0.001);同时,划痕实验结果提示:lincRNAUFC1过表达肝癌细胞迁移能力显著减弱(图 3C,P<0.001)。
3.
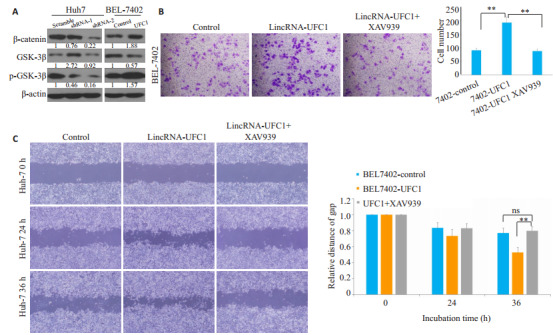
LincRNA-UFC1可以激活GSK-3β/β-catenin信号轴
LincRNA-UFC1 activates GSK-3β/β-catenin axis. A: β-catenin, GSK-3β, and p-GSK-3β expression levels detected by Western blotting after lincRNA- UFC1 overexpression or silencing; B: Transwell assays showing suppression of cell invasion in BEL-7402 cells with lincRNA-UFC1 overexpression following treatment with GSK-3β/β-catenin inhibitor XAV-939; C: Wound-healing assays showing suppressed cell invasion in BEL-7402 cells with lincRNA-UFC1 overexpression following XAV-939 treatment. **P < 0.001.
3. 讨论
近年的研究表明,lncRNA直接参与肿瘤细胞增殖、凋亡、侵袭及转移等过程[16-18],其能够通过调控SRA,PCAT-1等关键蛋白的表达发挥其致癌作用[19-20],其异常表达与肿瘤的发生、发展密切相关[19-22]。既往研究发现,绝大多数类型的lncRNA定位于胞核[23],很小部分类型富集于胞浆,而胞浆内的lncRNA在蛋白定位、mRNA翻译及维持mRNA稳定性的过程中起到重要调控作用[24-26]。我们课题组前期研究发现,lincRNA-UFC1不仅在肝癌组织中高表达,在癌栓中表达也呈明显的强阳性,其表达水平与肝癌的大小和BCLC分期呈正相关,提示lincRNA-UFC1可能与肝癌细胞的侵袭转移密切相关[15, 27]。与此同时,其他研究者[28]发现癌组织中lincRNA-UFC1高表达的宫颈癌患者预后较差,并且在宫颈癌细胞株Hela和Sila中沉默lincRNA-UFC1后,肿瘤细胞的侵袭、迁移和增殖能力受到抑制,进一步的研究中发现lincRNA-UFC1的癌基因作用是通过E2F1- linc-UFC1/miR-34a/FOXP3通路激活和加强的。Yu[29]发现结直肠癌患者的TNM分期也与lincRNA-UFC1的表达水平呈正相关,结直肠癌细胞株中沉默lincRNAUFC1能够使肿瘤细胞发生细胞周期G1阻滞,从而抑制肿瘤细胞的增殖,以及诱导细胞凋亡。上述研究也进一步支持lincRNA-UFC1可能参与多种人类肿瘤的发生发展,针对lincRNA-UFC1的干预治疗有可能阻断其致癌和促癌作用。
本研究也通过从机制上进一步探讨了lincRNAUFC1参与调节肿瘤增殖和迁移的分子机制。我们发现了lincRNA-UFC1/GSK-3β/β-catenin信号通路对于维持肝癌细胞侵袭和迁移能力的重要性,并通过通路抑制剂进一步验证该通路参与肝癌侵袭转移的机制。根据既往研究不同肿瘤中lincRNA-UFC1作用通路各有不同,提示lincRNA-UFC1的促癌作用是通过复杂而高效的网络而发挥出来的。既往研究还表明lincRNA发挥生物学作用主要通过影响特异性靶基因或者与microRNA相互作用在细胞转录和翻译过程中发挥着动态调节作用。如miR-21靶向作用肿瘤抑制基因lncRNA GAS5并抑制GAS5表达从而发挥促癌作用[30]。这些发现也进一步提示我们可以从不同分子机制(如lincRNA-UFC1影响的关键靶基因或与lincRNA-UFC1相互作用的microRNA)上探讨lincRNA-UFC1参与调节肝癌发生发展的过程。这些研究对于深入了解lincRNA-UFC1生物学过程至关重要。
lincRNA-UFC1除了参与恶性肿瘤的发生发展外,其在良性病中也发挥重要致病作用。而在良性疾病骨关节炎里,lincRNA-UFC1能够通过与miR-34a结合而促进软骨细胞生成,抑制细胞凋亡[31]。其他lincRNA,如心脏肥大相关因子(CHRF)可以作为miR-489的内源性吸附“海绵”参与调节Myd88表达并促使心肌肥大[32]。所以,lincRNA-UFC1在肿瘤的发生过程中,总体起到促进肿瘤发展的癌基因作用,而在良性疾病中,lincRNA-UFC1起到的是促进细胞增殖的作用。因此lincRNA-UFC1可能是肿瘤发生发展过程中的一个重要基因节点。
本研究仍存在一些缺陷,尽管我们通过体外实验的方法,展示了lincRNA-UFC1在肝癌发生、发展中的作用及其可能的机制,揭示了其潜在的临床应用价值,但是由于缺乏体内试验的结果和数据,调节lincRNAUFC1表达水平能否为肝癌的预防和治疗提供新的手段和方法,尚需要进一步的深入研究。
本研究中,我们发现了lincRNA-UFC1在肝细胞癌的侵袭转移中起着重要的作用,并且其通过调控GSK- 3β磷酸化水平对Wnt/β-catenin通路起调节作用,进而调节肝癌细胞侵袭转移能力,提示lincRNA-UFC1可能成为潜在的肝癌治疗的分子靶标。
Biography
王剑,硕士,主治医师,E-mail: wangj_gz1981@163.com
Funding Statement
国家自然科学基金(81472711,81702390);广东省自然科学基金(2018030310285,2017A030310105)
Supported by National Natural Science Foundation of China (81472711, 81702390)
Contributor Information
王 剑 (Jian WANG), Email: wangj_gz1981@163.com.
董 忠谊 (Zhongyi DONG), Email: dongzy1317@foxmail.com.
References
- 1.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.v68.6. [DOI] [PubMed] [Google Scholar]
- 2.Young S, Craig P, Golzarian J. Current trends in the treatment of hepatocellular carcinoma with transarterial embolization: a crosssectional survey of techniques. https://link.springer.com/article/10.1007/s00330-018-5782-7. Eur Radiol. 2018;29(6):3287–95. doi: 10.1007/s00330-018-5782-7. [DOI] [PubMed] [Google Scholar]
- 3.Li JH, Ma WJ, Wang GG, et al. Clinicopathologic significance and prognostic value of programmed cell death ligand 1 (PD-L1) in patients with hepatocellular carcinoma: a Meta-Analysis. Front Immunol. 2018;9:2077. doi: 10.3389/fimmu.2018.02077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu GY, Wu J, Wang BH, et al. Importance of tumor size at diagnosis as a prognostic factor for hepatocellular carcinoma survival: a population-based study. Cancer Manag Res. 2018;10:4401–10. doi: 10.2147/CMAR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang F, Zhang L, Zhang CG. Long noncoding RNAs and tumorigenesis: genetic associations, molecular mechanisms, and therapeutic strategies. Tumor Biol. 2016;37(1):163–75. doi: 10.1007/s13277-015-4445-4. [DOI] [PubMed] [Google Scholar]
- 6.Tay Y, Rinn J. Pandolfi PP.the multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505(7483):344–52. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43(6, SI):904–14. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cesana M, Cacchiarelli D, Legnini I, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147(2):358–69. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yuan SX, Yang F, Yang Y, et al. Long noncoding RNA associated with microvascular invasion in hepatocellular carcinoma promotes angiogenesis and serves as a predictor for hepatocellular carcinoma patients' poor recurrence-free survival after hepatectomy. Hepatology. 2012;56(6):2231–41. doi: 10.1002/hep.25895. [DOI] [PubMed] [Google Scholar]
- 10.Huang J, Zhou N, Watabe K, et al. Long non-coding RNA UCA1 promotes breast tumor growth by suppression of p27 (Kip1) J] Cell Death Dis. 2014;5:e1008. doi: 10.1038/cddis.2013.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takahashi Y, Sawada G, Kurashige J, et al. Amplification of PVT-1 is involved in poor prognosis via apoptosis inhibition in colorectal cancers. http://www.wanfangdata.com.cn/details/detail.do?_type=perio&id=41a726c494c20e8ce47807bb7387a892. Br J Cancer. 2014;110(1):164–71. doi: 10.1038/bjc.2013.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang L, Zhao HY, Xu YQ, et al. Systematic identification of lincRNA-based prognostic biomarkers by integrating lincRNA expression and copy number variation in lung adenocarcinoma. https://onlinelibrary.wiley.com/doi/abs/10.1002/ijc.31865. Int J Cancer. 2019;144(7):1723–34. doi: 10.1002/ijc.31865. [DOI] [PubMed] [Google Scholar]
- 13.Yan YM, Yu J, Liu HC, et al. Construction of a long non-coding RNA-associated ceRNA network reveals potential prognostic lncRNA biomarkers in hepatocellular carcinoma. Pathol Res Pract. 2018;214(12):2031–8. doi: 10.1016/j.prp.2018.09.022. [DOI] [PubMed] [Google Scholar]
- 14.Cao CH, Sun JY, Zhang DY, et al. The long intergenic noncoding RNA UFC1, a target of MicroRNA 34a, interacts with the mRNA stabilizing protein HuR to increase levels of beta-Catenin in HCC cells. Gastroenterology. 2015;148(2):415–U249. doi: 10.1053/j.gastro.2014.10.012. [DOI] [PubMed] [Google Scholar]
- 15.Liu Z, Zhao P, Han Y, et al. LincRNA FEZF1-AS1 is associated with prognosis in lung adenocarcinoma and promotes cell proliferation, migration and invasion. Oncol Res. 2018;27(1):39–45. doi: 10.3727/096504018X15199482824130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma JL, Yang YE, Huo DS, et al. LincRNA-RoR/miR-145 promote invasion and metastasis in triple-negative breast cancer via targeting MUC1. Biochem Biophys Res Commun. 2018;500(3):614–20. doi: 10.1016/j.bbrc.2018.04.119. [DOI] [PubMed] [Google Scholar]
- 17.Jia M, Jiang L, Wang YD, et al. lincRNA-p21 inhibits invasion and metastasis of hepatocellular carcinoma through notch signalinginduced epithelial-mesenchymal transition. Hepatol Res. 2016;46(11):1137–44. doi: 10.1111/hepr.v46.11. [DOI] [PubMed] [Google Scholar]
- 18.Prensner JR, Iyer MK, Balbin OA, et al. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat Biotechnol. 2011;29(8):742–9. doi: 10.1038/nbt.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cooper C, Guo J, Yan Y, et al. Increasing the relative expression of endogenous non-coding Steroid Receptor RNA Activator (SRA) in human breast cancer cells using modified oligonucleotides. Nucleic Acids Res. 2009;37(13):4518–31. doi: 10.1093/nar/gkp441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Louro R, Smirnova AS, Verjovski-Almeida S. Long intronic noncoding RNA transcription: expression noise or expression choice? https://www.sciencedirect.com/science/article/pii/S0888754308003042. Genomics. 2009;93(4):291–8. doi: 10.1016/j.ygeno.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 21.Okamura D, Maeda I, Taniguchi H, et al. Cell cycle gene-specific control of transcription has a critical role in proliferation of primordial germ cells. Genes Dev. 2012;26(22):2477–82. doi: 10.1101/gad.202242.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tuck AC, Tollervey D. A transcriptome-wide Atlas of RNP composition reveals diverse classes of mRNAs and lncRNAs. Cell. 2013;154(5):996–1009. doi: 10.1016/j.cell.2013.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang JF, Guo YJ, Zhao CX, et al. Hepatitis B virus X protein (HBx)-Related long noncoding RNA (lncRNA) Down-Regulated expression by HBx (Dreh) inhibits hepatocellular carcinoma metastasis by targeting the intermediate filament protein vimentin. Hepatology. 2013;57(5):1882–92. doi: 10.1002/hep.26195. [DOI] [PubMed] [Google Scholar]
- 24.Wilusz Cj WJ, Translation--The ML. RNA. Mol Cell. 2012;47(4):495–6. doi: 10.1016/j.molcel.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 25.Yoon JH, Abdelmohsen K, Srikantan S, et al. LincRNA-p21 suppresses target mRNA translation. Mol Cell. 2012;47(4):648–55. doi: 10.1016/j.molcel.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu L, Dong Z, Liang J, et al. As an Independent prognostic factor, FAT10 promotes hepatitis B virus-related hepatocellular carcinoma progression via Akt/GSK3 beta pathway. Oncogene. 2014;33(7):909–20. doi: 10.1038/onc.2013.236. [DOI] [PubMed] [Google Scholar]
- 27.Xi J, Feng J, Zeng ST, et al. Long noncoding RNA UFC1 is activated by E2F1 and exerts oncogenic properties by functioning as a ceRNA of FOXP3. Cancer Med. 2018;7(7):3301–10. doi: 10.1002/cam4.2018.7.issue-7. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Zhang Z, Zhu Z, Watabe K, et al. Negative regulation of lncRNA GAS5 by miR-21. Cell Death Differ. 2013;20(11):1558–68. doi: 10.1038/cdd.2013.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu T, Shan TD, Li JY, et al. Knockdown of linc-UFC1 suppresses proliferation and induces apoptosis of colorectal cancer. Cell Death Dis. 2016;7:e2228. doi: 10.1038/cddis.2016.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang G, Wu YD, Xu D, et al. Long noncoding RNA UFC1 promotes proliferation of chondrocyte in osteoarthritis by acting as a sponge for miR-34a. DNA Cell Biol. 2016;35(11):691–5. doi: 10.1089/dna.2016.3397. [DOI] [PubMed] [Google Scholar]
- 31.Wang K, Liu F, Zhou LY, et al. The long noncoding RNA CHRF regulates cardiac hypertrophy by targeting miR-489. Circ Res. 2014;114(9):1377–88. doi: 10.1161/CIRCRESAHA.114.302476. [DOI] [PubMed] [Google Scholar]


