Abstract
目的
应用生物信息学技术探索胃癌发病机制,为胃癌的防治提供生物信息学依据。
方法
用GEO2R在线工具分析GSE79973中胃癌组织和正常胃黏膜组织的差异表达基因(Differentially expressed genes, DEGs),通过DAVID数据库对DEGs进行GO分析和KEGG通路富集分析,然后通过STRING数据库构建蛋白质相互作用网络,用Cytoscape软件进行关键基因(Hub基因)筛选和功能模块分析,并在GEPIA数据库对Hub基因进行验证,用Target Scan数据库预测调控靶基因的microRNAs,并用OncomiR分析microRNAs在胃癌组织中的表达及其与生存预后的关系。
结果
共筛选出181个在胃癌中差异表达的基因。蛋白质互作网络筛选出10个Hub基因。DEGs功能分析主要涉及蛋白质消化吸收、PI3K-Akt信号通路、ECM-受体相互作用、血小板激活信号通路。GEPIA数据库验证显示COL1A1在胃癌组织中高表达,并和胃癌患者的不良预后有关。miR-129-5p与COL1A1 mRNA的3'UTR结合。与正常组织相比,miR-129-5p在胃癌组织中表达明显下调,且与胃癌患者预后具有一定相关性。
结论
miR-129-5p调控的COL1A1是胃癌潜在的治疗靶点。
Keywords: 胃癌, 差异表达基因, COL1A1, miR-129-5p, 生物信息学分析
Abstract
Objective
To explore the pathogenesis of gastric cancer through a bioinformatic approach to provide evidence for the prevention and treatment of gastric cancer.
Methods
The differentially expressed genes (DEGs) in gastric cancer and normal gastric mucosa in GSE79973 dataset were analyzed using GEO2R online tool. GO analysis and KEGG pathway enrichment analysis of the DEGs in DAVID database were performed. The protein interaction network was constructed using STRING database, and the key genes (Hub genes) were screened and their functional modules were analyzed using Cytoscape software. The GEPIA database was used to validate the Hub genes, and the Target Scan database was used to predict the microRNAs that regulate the target genes; OncomiR was used to analyze the expressions of the microRNAs in gastric cancer tissues and their relationship with the survival outcomes of the patients.
Results
A total of 181 DEGs were identified in gastric cancer, and 10 hub genes were screened by the protein- protein interaction network. Functional analysis showed that these DEGs were involved mainly in protein digestion and absorption, PI3K-Akt signaling pathway, ECM-receptor interaction and platelet activation signal pathway. GEPIA database validation showed that COL1A1 was highly expressed in gastric cancer tissues and was associated with a poor prognosis of patients with gastric cancer. MiR-129-5p was found to bind to the 3'UTR of COL1A1 mRNA, and compared with that in normal tissues, miR-129-5p expression was obviously down-regulated in gastric cancer tissues, and was correlated with the prognosis of the patients.
Conclusion
COL1A1 under regulation by MiR-129-5p is a potential therapeutic target for gastric cancer.
Keywords: gastric cancer, differentially expressed genes, COL1A1, miR-129-5p, bioinformatic analysis
胃癌是我国第2大肿瘤[1],5年生存率全球仅为10%[2],其高发病率和高死亡率严重威胁着人类健康,随着诊疗技术的发展,胃癌的发病率和死亡率在一些发达国家有稳步下降的趋势[3],然而,亚洲仍有很高的发病率[4],因此,深入探究胃癌的发病机制及新的治疗方法就显得尤为重要。近年来,大量生物标志物已应用于胃癌的早期诊断[5-6],然而,这些生物标志物并没有被很好的整合,并且,这些生物标志物可在多种肿瘤中被检测到[7-8],因此,对胃癌诊断和治疗的特异性靶点还需要进行深入的研究。
COL1A1是胶原家族的重要成员,被认为与癌症发生有关,COL1A1的异常表达在多种癌症中均有报道[9-11]。此外,Zang等[12]发现COL1A1在胃癌中存在差异表达,但COL1A1在胃癌中的临床意义仍不清楚。基因表达谱(GEO)数据库为癌症相关基因表达谱的生物信息学挖掘提供了可能[13]。本研究通过生物信息学方法筛选出胃癌芯片数据GSE79973中胃癌组织和正常胃黏膜组织的DEGs,对DEGs进行GO分析和KEGG通路富集分析,然后通过构建蛋白质-蛋白质相互作用(PPI)网络,筛选出Hub基因并验证,同时预测调控COL1A1的miRNAs,旨在为胃癌分子机制的进一步研究提供生物信息学依据,也为我们进行基因个体化治疗提供新的途径。
1. 材料和方法
1.1. 芯片数据来源
本研究从GEO (<a href="https://www.ncbi.nlm.nih.gov/geo/">https://www.ncbi.nlm.nih.gov/geo/</a>)数据库下载基因芯片数据集GSE79973,芯片总共包含20例样本,其中10例正常胃黏膜组织和10例胃腺癌组织样本,其芯片平台是GPL570[HG-U133_Plus_ 2] Affymetrix Human Genome U133 Plus 2.0 Array,表达数据为expression profiling by array,种属为Homo sapiens。
1.2. 数据处理
用GEO2R(<a href="https://www.ncbi.nlm.nih.gov/geo/geo2r/">https://www.ncbi.nlm.nih.gov/geo/geo2r/</a>)<sup>[<xref ref-type="bibr" rid="b14">14</xref>]</sup>在线工具分析胃癌样本与正常样本基因数据。将胃癌组织芯片GSE79973矩阵数据的探针名转化为基因名,对原始数据进行去重等处理后,以|logFC|>2且<italic>P</italic> < 0.01的标准筛选出DEGs,用SangerBox软件绘制火山图。
1.3. DEGs的富集分析
为深入了解这些DEGs,我们用DAVID(the Database for Annotation, Visualization and Integrated Discovery,<a href="http://david.abcc.ncifcrf.gov/">http://david.abcc.ncifcrf.gov/</a>)在线分析数据库<sup>[<xref ref-type="bibr" rid="b15">15</xref>]</sup>对DEGs进行GO和KEGG通路富集分析<sup>[<xref ref-type="bibr" rid="b16">16</xref>-<xref ref-type="bibr" rid="b17">17</xref>]</sup>,以<italic>P</italic> < 0.05为差异有统计学意义。
1.4. PPI网络构建和关键基因筛选
通过在线分析网站STRING(Search Tool for the Rtrieval of Interacting Genes, <a href="https://string-db.org/">https://string-db.org/</a>)<sup>[<xref ref-type="bibr" rid="b18">18</xref>]</sup>得到DEGs的蛋白互作网络,以TSV格式导出,将所得源文件导入Cytoscape<sup>[<xref ref-type="bibr" rid="b19">19</xref>]</sup>进行可视化分析,用插件cytoHubba进行Hub基因分析,选用MCC算法,选取前10个Hub基因。
1.5. PPI功能模块分析
为进一步明确胃癌可能的信号通路,我们在进行PPI网络构建后,用Cytoscape软件中MCODE插件对PPI网络进行聚类分析后得到PPI功能模块,然后用DAVID数据库将功能模块中的基因进行KEGG pathway分析。
1.6. 关键基因验证分析
为进一步验证Hub基因,我们利用GEPIA(Gene Expression Profiling Interactive Analysis, <a href="http://gepia.cancer-pku.cn">http://gepia.cancer-pku.cn</a>)数据库<sup>[<xref ref-type="bibr" rid="b20">20</xref>]</sup>分析Hub基因在胃癌组织和正常组织中的表达水平,并绘制Hub基因的KaplanMeiter生存曲线。
1.7. COL1A1和microRNAs关系预测
为了解COL1A1参与胃癌的发生发展机制,我们通过在线数据库Target Scan 7.2(<a href="http://www.targetscan.org/">http://www.targetscan.org/</a>)预测与COL1A1相互作用的microRNAs。
1.8. microRNAs在胃癌组织的表达及其与生存预后的关系
基于OncomiR数据库(<a href="http://www.oncomir.org">http://www.oncomir.org</a>)分析miRNA在胃癌组织和正常组织中的表达,并对其进行预后分析。
2. 结果
2.1. 胃癌和正常组织的DEGs
通过对基因芯片GSE79973进行数据分析,结果显示有181个DEGs(胃癌组/正常对照组),其中上调基因和下调基因分别为57个和124个(图 1)。
1.
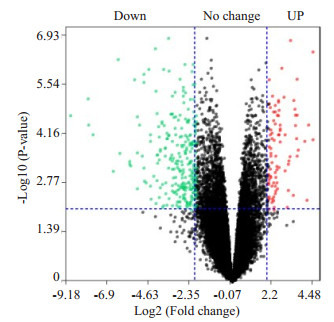
差异表达基因火山图
Volcano plot of the differential expressed genes in gastric cancer
2.2. GO和KEGG通路富集分析
GO可分为生物过程(biological process, BP)、细胞组成(cellular component, CC)和分子功能(molecular function, MF)。采用DAVID对181个DEGs进行GO和KEGG通路富集分析,结果如表 1所示。DEGs主要涉及细胞黏附、细胞外基质组织、氧化还原过程、胶原蛋白分解代谢、异物的代谢等生物过程,细胞学组成分析显示这些基因大多参与细胞外泌体、细胞外基质、细胞外区等的组成。分子功能的变化主要集中在锌、铁离子结合、相同的蛋白结合、细胞外基质结构组成、肝素结合、氧化还原酶活性、血红素结合、氧气结合等。KEGG通路富集分析表明,差异基因主要涉及PI3K-Akt信号通路、ECM-受体相互作用、蛋白质消化吸收、化学致癌作用、视黄醇的新陈代谢、细胞色素P450代谢通路、矿物质的吸收、胃酸分泌等。
1.
胃癌相关差异表达基因的GO和KEGG通路富集分析
Enrichment analysis of GO and KEGG pathway of the differentially expressed genes in gastric cancer
| Category | ID | Term | Count | P |
| BP | GO:0007155 | Cell adhesion | 20 | 1.00E-08 |
| BP | GO:0030198 | Extracellular matrix organization | 17 | 8.62E-12 |
| BP | GO:0055114 | Oxidation-reduction process | 13 | 0.004225507 |
| BP | GO:0030574 | Collagen catabolic process | 10 | 2.99E-09 |
| BP | GO:0006805 | Xenobiotic metabolic process | 8 | 4.07E-06 |
| BP | GO:0001501 | Skeletal system development | 8 | 1.57E-04 |
| BP | GO:0030199 | Collagen fibril organization | 7 | 8.18E-07 |
| BP | GO:0008202 | Steroid metabolic process | 7 | 1.49E-06 |
| BP | GO:0007586 | Digestion | 7 | 1.44E-05 |
| BP | GO:0034220 | Ion transmembrane transport | 7 | 0.008647397 |
| BP | GO:0001525 | Angiogenesis | 7 | 0.011406595 |
| BP | GO:0051216 | Cartilage development | 6 | 1.35E-04 |
| CC | GO:0005576 | Extracellular region | 45 | 3.21E-13 |
| CC | GO:0005615 | Extracellular space | 37 | 1.51E-10 |
| CC | GO:0070062 | Extracellular exosome | 32 | 0.047465783 |
| CC | GO:0005887 | Integral component of plasma membrane | 19 | 0.040478934 |
| CC | GO:0005578 | Proteinaceous extracellular matrix | 15 | 4.36E-08 |
| CC | GO:0031012 | Extracellular matrix | 14 | 9.79E-07 |
| CC | GO:0005788 | Endoplasmic reticulum lumen | 13 | 5.98E-08 |
| CC | GO:0005581 | Collagen trimer | 11 | 4.00E-09 |
| CC | GO:0016324 | Apical plasma membrane | 11 | 1.43E-04 |
| CC | GO:0009986 | Cell surface | 11 | 0.013726185 |
| CC | GO:0005604 | Basement membrane | 6 | 4.73E-04 |
| CC | GO:0031090 | Organelle membrane | 6 | 7.37E-04 |
| MF | GO:0008270 | Zinc ion binding | 16 | 0.049404886 |
| MF | GO:0042802 | Identical protein binding | 13 | 0.017914572 |
| MF | GO:0005201 | Extracellular matrix structural constituent | 9 | 6.31E-08 |
| MF | GO:0008201 | Heparin binding | 8 | 3.16E-04 |
| MF | GO:0016491 | Oxidoreductase activity | 8 | 0.001188406 |
| MF | GO:0020037 | Heme binding | 7 | 8.49E-04 |
| MF | GO:0019825 | Oxygen binding | 6 | 3.69E-05 |
| MF | GO:0016705 | Oxidoreductase activity, acting on paired donors, with Incorporation or reduction of molecular oxygen | 6 | 9.43E-05 |
| MF | GO:0004497 | Monooxygenase activity | 6 | 1.03E-04 |
| MF | GO:0005178 | Integrin binding | 6 | 0.001598275 |
| MF | GO:0005506 | Iron ion binding | 6 | 0.007975728 |
| MF | GO:0008392 | Arachidonic acid epoxygenase activity | 5 | 5.14E-06 |
| KEGG pathway | hsa04151 | PI3K-Akt signaling pathway | 12 | 0.001311204 |
| KEGG pathway | hsa04512 | ECM-receptor interaction | 10 | 3.88E-07 |
| KEGG pathway | hsa04974 | Protein digestion and absorption | 10 | 4.29E-07 |
| KEGG pathway | hsa04510 | Focal adhesion | 10 | 4.07E-04 |
| KEGG pathway | hsa05204 | Chemical carcinogenesis | 9 | 2.34E-06 |
| KEGG pathway | hsa00830 | Retinol metabolism | 7 | 6.72E-05 |
| KEGG pathway | hsa00982 | Drug metabolism-cytochrome P450 | 7 | 9.47E-05 |
| KEGG pathway | hsa00980 | Metabolism of xenobiotics by cytochrome P450 | 7 | 1.52E-04 |
| KEGG pathway | hsa04978 | Mineral absorption | 6 | 1.12E-04 |
| KEGG pathway | hsa04971 | Gastric acid secretion | 6 | 0.001205941 |
| KEGG pathway | hsa05146 | Amoebiasis | 6 | 0.006138929 |
| KEGG pathway | hsa00010 | Glycolysis/Gluconeogenesis | 4 | 0.03749949 |
2.3. 差异表达基因的PPI网络分析
将181个显著差异基因输入STRING数据库中,然后将所得数据导入Cytoscape中,利用插件cytoHubba找出前10个Hub基因,分别为COL1A1、COL1A2、COL4A1、COL2A1、SERPINH1、COL6A3、COL11A1、COL10A1、COL12A1、COL8A1(图 2)。
2.
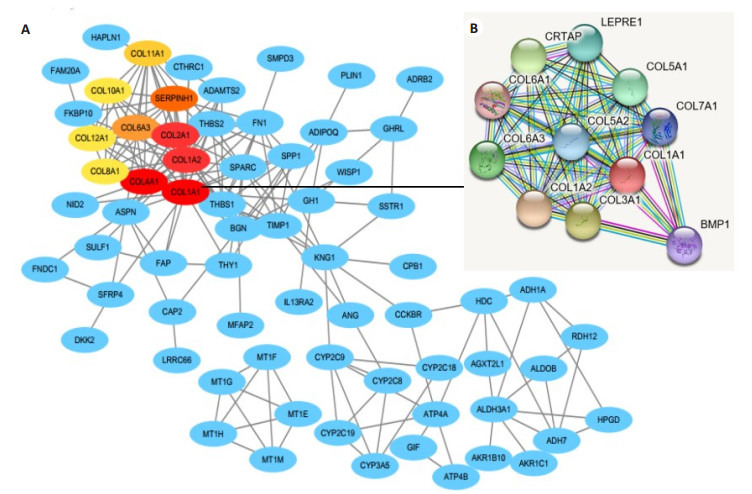
差异基因编码蛋白质的PPI分析图和关键基因筛选
PPI analysis of the proteins encoded by the differential genes and screening of the key genes. A: PPI network for the DEGs; B: Amplification of the network for PPI associated with COL1A1.
2.4. PPI功能模块分析
我们用Cytoscape软件中MCODE插件对PPI网络进行聚类分析后得到不同的PPI功能模块,Score得分最高的模块如图 3所示。然后我们通过DAVID在线分析工具对模块中包含的基因进行KEGG pathway分析,主要涉及蛋白质消化吸收、PI3K-Akt信号通路、ECM-受体相互作用、血小板激活信号通路(表 2)。
3.
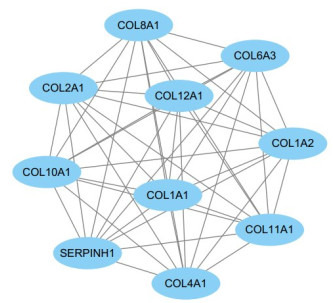
功能模块图
Functional module diagram
2.
功能模块内基因的KEGG Pathway分析
KEGG pathway analysis of the genes in the functional modules
| Category | ID | Term | Count | P |
| KEGG pathway | hsa04974 | Protein digestion and absorption | 7 | 3.69E-12 |
| KEGG pathway | hsa04512 | ECM-receptor interaction | 5 | 3.51E-07 |
| KEGG pathway | hsa05146 | Amoebiasis | 5 | 7.80E-07 |
| KEGG pathway | hsa04510 | Focal adhesion | 5 | 1.12E-05 |
| KEGG pathway | hsa04151 | PI3K-Akt signaling pathway | 5 | 8.61E-05 |
| KEGG pathway | hsa04611 | Platelet activation | 4 | 1.27E-04 |
2.5. 关键基因验证
用GEPIA数据库进一步验证分析了10个Hub基因在胃癌组织(408例)和正常组织(211例)的表达水平中的表达情况,发现除了COL2A1在胃癌组织中低表达外,其他9个Hub基因均在胃癌组织中高表达,差异有统计学意义(P < 0.05,图 4)。最后我们用GEPIA数据库绘制了Hub基因高表达胃癌组织和低表达胃癌组织的Kaplan-Meiter生存曲线,结果显示COL1A1、COL4A1、COL12A1高表达的胃癌组织的生存率低于低表达组织,差异具有统计学意义(P < 0.05),与患者不良预后密切相关(图 5)。COL1A1的高表达与不良预后的相关性更加显著。
4.
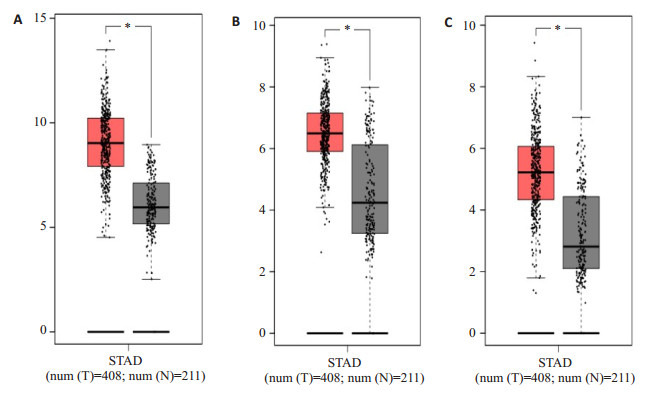
胃癌关键基因在肿瘤组织及正常组织中的表达水平
Expression levels of the key genes in gastric cancer and normal tissues. A: COL1A1 expression level; B: COL4A1expression level; C: COL12A1expression level. *P < 0.05 vs normal tissue. The X axis represents tissue type, T the tumor, and N the normal tissue. The Y axis represents log2(TPM +1). TPM: Number of transcripts per million reads.
5.
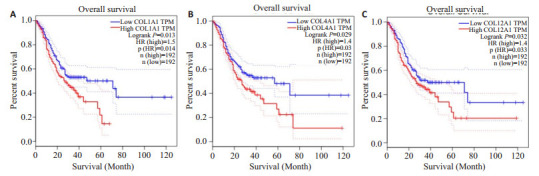
关键基因对胃癌患者生存分析的验证结果
Validation of the key genes in survival analysis of the patients with gastric cancer. A: COL1A1 validation result; B: COL4A1 validation result; C: COL12A1validation result. The red line represents the high expression group, and the blue line represents the low expression group. HR: Risk ratio.
6.
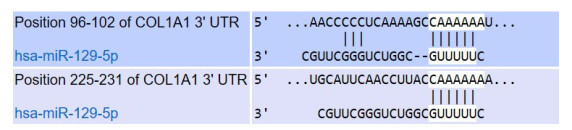
COL1A1 mRNA 3'UTR中miR-129-5p结合位点的预测结果
Prediction of miR-129-5p binding sites in COL1A1 mRNA3'UTR.
2.6. COL1A1与miRNA相互作用预测结果
用Target Scan数据库预测到miR-129-5p直接与COL1A1 mRNA的3'UTR结合,是COL1A1转录后调节因子(图 5)。
2.7. miR-129-5p在胃癌中的表达水平与生存预后分析
经OncomiR数据库检索发现,miR-129-5p在胃癌组织中的表达显著低于正常组织(P=3.32e-05,图 7A)。为分析miR-129-5p与胃癌生存预后之间的关系,我们使用此数据库进一步分析了miR-129-5p在胃癌组织中的表达水平与生存期的关系,结果发现,低表达组生存期时间短于正常组织,但差异不具有统计学意义(P=0.1182,图 7B)。
7.
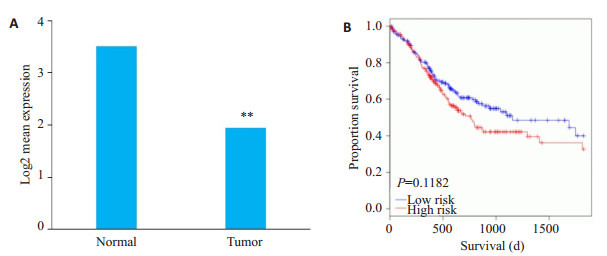
miR-129-5p在胃癌中的表达与其生存预后分析
Expression of miR-129-5p in gastric cancer and analysis of the survival outcomes of the patients. A: Expression of miR-129-5p in gastric cancer (**P < 0.05 vs normal); B: Relationship between miR-129-5p expression level and the survival outcomes.
3. 讨论
胃癌早期诊断具有一定难度,大多数胃癌患者确诊时已是晚期[21],已失去最佳治疗时机,死亡率一直居高不下。因此,探究新的早期肿瘤生物标志物对胃癌的防治具有一定价值。本研究采用生物信息学方法对GEO数据库中的胃腺癌组织和正常胃黏膜组织的基因芯片数据进行分析。首先比较胃癌组织和正常胃黏膜组织中的基因表达情况,共筛选出181个DEGs(胃癌组/正常对照组),其中上调基因和下调基因分别为57个和124个。为进一步了解DEGs,我们进行了GO和KEGG通路富集分析,DEGs的生物过程主要涉及细胞黏附、氧化还原过程、胶原蛋白分解代谢等,细胞学组成分析显示这些基因大多参与细胞外泌体、细胞外基质、细胞外区等的组成。分子功能的变化主要集中在锌、铁离子结合、相同的蛋白结合、细胞外基质结构组成、肝素结合、氧化还原酶活性、血红素结合、氧气结合等。正常情况下,机体的氧化还原过程处于动态平衡状态,而细胞氧化还原环境持续遭到破坏,则可能导致肿瘤的发生[22]。功能模块分析显示:KEGG通路主要涉及蛋白质消化吸收、PI3K-Akt信号通路、ECM-受体相互作用、血小板激活信号通路。这与一项胃癌关键基因的生物信息学分析的研究结果相似[23]。PI3K-Akt通路在许多肿瘤中都具有较高的易感性[24]。PI3K-Akt通路通过促进细胞增殖,在肿瘤细胞侵袭、转移中起着重要的作用[25]。
PPI网络筛选出10个Hub基因,由GEPIA验证得知COL1A1(Collagen, type Ⅰ, alpha 1)的高表达与不良预后显著相关,有研究已证实此结果[26]。最近有研究[27]提出了COL1A1可作为胃癌早期筛查的标志。Ⅰ型胶原是纤维胶原家族的主要成分,主要参与细胞外基质结构的组成,被认为是一种肿瘤相关基因[28],可能参与了肿瘤的侵袭和进展[29],有研究表明[30],COL1A1的上调有助于卵巢癌细胞对顺铂耐药。为进一步了解COL1A1参与胃癌发生发展的分子机制,我们预测了调控COL1A1的转录后调节因子miRNAs,miRNA是内源性小型非编码RNA分子,其长度为18-24个核苷酸,可通过诱导mRNA降解或通过与mRNA的3'-UTR的互补结合而抑制mRNA[31]。预测结果显示miR-129-5p可直接与COL1A1 mRNA的3'UTR结合。miR-129-5p是一种有效的肿瘤抑制因子[32-33],为验证胃癌中miR- 129-5p与COL1A1的关系,我们通过OncomiR数据库检索了miR-129-5p在胃癌中的表达与生存预后,结果显示miR-129-5p在胃癌组织中的表达显著低于正常组织(P=3.32e-05),生存期也短于正常组织。由此得出miR-129-5p调控的COL1A1是胃癌潜在的治疗靶点。这与最近的一项miR-129-5p通过抑制COL1A1来抑制胃癌细胞的侵袭和增殖[34]的研究结果一致。
综上所述,我们通过生物信息学分析确定了差异表达的基因,由富集分析和蛋白互作可知,COL1A1在胃癌中是一种高表达分子。此外,在胃癌中预测到miR- 129-5p可下调COL1A1的表达。COL1A1应该是miR- 129-5p调控胃癌治疗的靶点。为了得到更准确的相关性结果,还需要进行一系列的实验来验证预测结果。
Biography
杨万霞,在读硕士研究生,E-mail: lnyangwx@163.com
Funding Statement
甘肃省重点研发计划项目(18YF1FA108);兰大二院萃英计划面上项目(CY2018-MS10);兰大二院萃英计划临床拔尖技术项目(CY2018-BJ04)
Contributor Information
杨 万霞 (Wanxia YANG), Email: lnyangwx@163.com.
尤 崇革 (Chongge YOU), Email: youchg@lzu.edu.cn.
References
- 1.Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CACancer J Clin. 2016;66(2):115–32. doi: 10.3322/caac.21338. [Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015 [J]. CACancer J Clin, 2016, 66(2): 115-32.] [DOI] [PubMed] [Google Scholar]
- 2.Orditura M, Galizia G, Sforza V, et al. Treatment of gastric cancer. World J Gastroenterol. 2014;20(7):1635–49. doi: 10.3748/wjg.v20.i7.1635. [Orditura M, Galizia G, Sforza V, et al. Treatment of gastric cancer[J]. World J Gastroenterol, 2014, 20(7): 1635-49.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Malvezzi M, Bonifazi M, Bertuccio P, et al. An age-period-cohort analysis of gastric cancer mortality from 1950 to 2007 in Europe. Ann Epidemiol. 2010;20(12):898–905. doi: 10.1016/j.annepidem.2010.08.013. [Malvezzi M, Bonifazi M, Bertuccio P, et al. An age-period-cohort analysis of gastric cancer mortality from 1950 to 2007 in Europe[J]. Ann Epidemiol, 2010, 20(12): 898-905.] [DOI] [PubMed] [Google Scholar]
- 4.Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CACancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262. [Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012[J]. CACancer J Clin, 2015, 65(2): 87-108.] [DOI] [PubMed] [Google Scholar]
- 5.Moghbeli M, Makhdoumi Y, Soltani Delgosha M, et al. ErbB1 and ErbB3 co-over expression as a prognostic factor in gastric cancer. Biol Res. 2019;52(1):2. doi: 10.1186/s40659-018-0208-1. [Moghbeli M, Makhdoumi Y, Soltani Delgosha M, et al. ErbB1 and ErbB3 co-over expression as a prognostic factor in gastric cancer[J]. Biol Res, 2019, 52(1): 2.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.吴 永伟, 赵 刚. CEA、CA724、CA199与PGⅠ、PGⅡ、PGR联合检测在胃癌早期诊断中的价值分析. 川北医学院学报. 2018;33(6):836–9. doi: 10.3969/j.issn.1005-3697.2018.06.008. [吴永伟, 赵刚. CEA、CA724、CA199与PGⅠ、PGⅡ、PGR联合检测在胃癌早期诊断中的价值分析[J].川北医学院学报, 2018, 33(6): 836-9.] [DOI] [Google Scholar]
- 7.Hasholzner U, Baumgartner L, Stieber P, et al. Clinical signifcance of the tumour markers CA 125 Ⅱ and CA 72-4 in ovarian carcinoma. Int J Cancer. 1996;69(4):329–34. doi: 10.1002/(ISSN)1097-0215. [Hasholzner U, Baumgartner L, Stieber P, et al. Clinical signifcance of the tumour markers CA 125 Ⅱ and CA 72-4 in ovarian carcinoma [J]. Int J Cancer, 1996, 69(4): 329-34.] [DOI] [PubMed] [Google Scholar]
- 8.Luo G, Liu C, Guo M, et al. Potential Biomarkers in Lewis Negative Patients With Pancreatic Cancer. Ann Surg. 2017;265(4):800–05. doi: 10.1097/SLA.0000000000001741. [Luo G, Liu C, Guo M, et al. Potential Biomarkers in Lewis Negative Patients With Pancreatic Cancer[J].Ann Surg, 2017, 265(4): 800-05.] [DOI] [PubMed] [Google Scholar]
- 9.Barcus CE, O'Leary KA, Brockman JL, et al. Elevated collagen- Ⅰ augments tumor progressive signals, intravasation and metastasis of prolactin-induced estrogen receptor alpha positive mammary tumor cells. Breast Cancer Res. 2017;19(1):9. doi: 10.1186/s13058-017-0801-1. [Barcus CE, O'Leary KA, Brockman JL, et al. Elevated collagen- Ⅰ augments tumor progressive signals, intravasation and metastasis of prolactin-induced estrogen receptor alpha positive mammary tumor cells[J]. Breast Cancer Res, 2017, 19(1): 9.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ibanez de Caceres I, Dulaimi E, Hoffman AM, et al. Identification of novel target genes by an epigenetic reactivation screen of renal cancer. Cancer Res. 2006;66(10):5021–8. doi: 10.1158/0008-5472.CAN-05-3365. [Ibanez de Caceres I, Dulaimi E, Hoffman AM, et al. Identification of novel target genes by an epigenetic reactivation screen of renal cancer[J]. Cancer Res, 2006, 66(10): 5021-8.] [DOI] [PubMed] [Google Scholar]
- 11.Lv J, Guo L, Wang JH, et al. Biomarker identification and transregulatory network analyses in esophageal adenocarcinoma and Barrett's esophagus. World J Gastroenterol. 2019;25(2):233–44. doi: 10.3748/wjg.v25.i2.233. [Lv J, Guo L, Wang JH, et al. Biomarker identification and transregulatory network analyses in esophageal adenocarcinoma and Barrett's esophagus[J]. World J Gastroenterol, 2019, 25(2): 233-44.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zang S, Guo R, Xing R, et al. Identification of differentiallyexpressed genes in intestinal gastric cancer by microarray analysis. Genom Proteom Bioinformat. 2014;12(6):276–83. doi: 10.1016/j.gpb.2014.09.004. [Zang S, Guo R, Xing R, et al. Identification of differentiallyexpressed genes in intestinal gastric cancer by microarray analysis [J]. Genom Proteom Bioinformat, 2014, 12(6): 276-83.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tinker AV, Boussioutas A, Bowtell DD. The challenges of gene expression microarrays for the study of human cancer. Cancer Cell. 2006;9(5):333–9. doi: 10.1016/j.ccr.2006.05.001. [Tinker AV, Boussioutas A, Bowtell DD. The challenges of gene expression microarrays for the study of human cancer[J]. Cancer Cell, 2006, 9(5): 333-9.] [DOI] [PubMed] [Google Scholar]
- 14.Davis S, Meltzer PS. GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics. 2007;23(14):1846–7. doi: 10.1093/bioinformatics/btm254. [Davis S, Meltzer PS. GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor[J]. Bioinformatics, 2007, 23(14): 1846-7.] [DOI] [PubMed] [Google Scholar]
- 15.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources [J]. Nat Protoc, 2009, 4(1): 44-57.] [DOI] [PubMed] [Google Scholar]
- 16.Sherman BT, Huang DW, Tan Q, et al. DAVID Knowledgebase: a gene- centered database integrating heterogeneous gene annotation resources to facilitate high-throughput gene functional analysis. BMC Bioinformatics. 2007;8:426. doi: 10.1186/1471-2105-8-426. [Sherman BT, Huang DW, Tan Q, et al. DAVID Knowledgebase: a gene- centered database integrating heterogeneous gene annotation resources to facilitate high-throughput gene functional analysis[J]. BMC Bioinformatics, 2007, 8: 426.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiao X, Sherman BT, Huang D W, et al. DAVID-WS: a stateful web service to facilitate gene/protein list analysis. Bioinformatics. 2012;28(13):1805–6. doi: 10.1093/bioinformatics/bts251. [Jiao X, Sherman BT, Huang D W, et al. DAVID-WS: a stateful web service to facilitate gene/protein list analysis[J]. Bioinformatics, 2012, 28(13): 1805-6.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Szklarczyk D, Franceschini A, Wyder S, et al. STRING v10: proteinprotein interaction networks, integrated over the tree of life. NucleicAcids Res. 2015;43(Database issue):D447–52. doi: 10.1093/nar/gku1003. [Szklarczyk D, Franceschini A, Wyder S, et al. STRING v10: proteinprotein interaction networks, integrated over the tree of life[J]. NucleicAcids Res, 2015, 43(Database issue): D447-52.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–504. doi: 10.1101/gr.1239303. [Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks[J]. Genome Res, 2003, 13(11): 2498-504.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. NucleicAcids Res. 2017;45(W1):W98–102. doi: 10.1093/nar/gkx247. [Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses[J]. NucleicAcids Res, 2017, 45(W1): W98-102.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zong L, Abe M, Seto Y, et al. The challenge of screening for early gastric cancer in China. Lancet. 2016;388(10060):2606. doi: 10.1016/S0140-6736(16)32226-7. [Zong L, Abe M, Seto Y, et al. The challenge of screening for early gastric cancer in China[J]. Lancet, 2016, 388(10060): 2606.] [DOI] [PubMed] [Google Scholar]
- 22.杨 梦祺, 刘 盼盼, 黄 蓬. 肿瘤氧化还原代谢与干预. http://d.old.wanfangdata.com.cn/Periodical/zgshywzz201609004. 中国生化药物杂志. 2016;36(9):16–23. [杨梦祺, 刘盼盼, 黄蓬.肿瘤氧化还原代谢与干预[J].中国生化药物杂志, 2016, 36(9): 16-23.] [Google Scholar]
- 23.Sun C, Yuan Q, Wu D, et al. Identification of core genes and outcome in gastric cancer using bioinformatics analysis. Oncotarget. 2017;8(41):70271–80. doi: 10.18632/oncotarget.20082. [Sun C, Yuan Q, Wu D, et al. Identification of core genes and outcome in gastric cancer using bioinformatics analysis[J]. Oncotarget, 2017, 8(41): 70271-80.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brugge J, Hung MC, Mills GB. A new mutational AKTivation in the PI3K pathway. Cancer Cell. 2007;12(2):104–7. doi: 10.1016/j.ccr.2007.07.014. [Brugge J, Hung MC, Mills GB. A new mutational AKTivation in the PI3K pathway[J]. Cancer Cell, 2007, 12(2): 104-7.] [DOI] [PubMed] [Google Scholar]
- 25.Hao NB, Tang B, Wang GZ, et al. Hepatocyte growth factor (HGF) upregulates heparanase expression via the PI3K/Akt/NF-kappaB signaling pathway for gastric cancer metastasis. Cancer Lett. 2015;361(1):57–66. doi: 10.1016/j.canlet.2015.02.043. [Hao NB, Tang B, Wang GZ, et al. Hepatocyte growth factor (HGF) upregulates heparanase expression via the PI3K/Akt/NF-kappaB signaling pathway for gastric cancer metastasis[J]. Cancer Lett, 2015, 361(1): 57-66.] [DOI] [PubMed] [Google Scholar]
- 26.Zhuo C, Li X, Zhuang H, et al. Elevated THBS2, COL1A2, and SPP1 expression levels as predictors of gastric cancer prognosis. Cell Physiol Biochem. 2016;40(6):1316–24. doi: 10.1159/000453184. [Zhuo C, Li X, Zhuang H, et al. Elevated THBS2, COL1A2, and SPP1 expression levels as predictors of gastric cancer prognosis[J]. Cell Physiol Biochem, 2016, 40(6): 1316-24.] [DOI] [PubMed] [Google Scholar]
- 27.Li J, Ding Y, Li A. Identification of COL1A1 and COL1A2 as candidate prognostic factors in gastric cancer. World J Surg Oncol. 2016;14(1):297. doi: 10.1186/s12957-016-1056-5. [Li J, Ding Y, Li A. Identification of COL1A1 and COL1A2 as candidate prognostic factors in gastric cancer[J]. World J Surg Oncol, 2016, 14(1): 297.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hayashi M, Nomoto S, Hishida M, et al. Identification of the collagen type 1 α 1 gene (COL1A1) as a candidate survival-related factor associated with hepatocellular carcinoma. BMC Cancer. 2014;14(1):108. doi: 10.1186/1471-2407-14-108. [Hayashi M, Nomoto S, Hishida M, et al. Identification of the collagen type 1 α 1 gene (COL1A1) as a candidate survival-related factor associated with hepatocellular carcinoma[J]. BMC Cancer, 2014, 14(1): 108.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wolf K, Alexander S, Schacht V, et al. Collagen-based cell migration models in vitro and in vivo. Semin Cell Dev Biol. 2009;20(8):931–41. doi: 10.1016/j.semcdb.2009.08.005. [Wolf K, Alexander S, Schacht V, et al. Collagen-based cell migration models in vitro and in vivo[J]. Semin Cell Dev Biol, 2009, 20(8): 931-41.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu PN, Yan MD, Lai HC, et al. Downregulation of miR-29 contributes to cisplatin resistance of ovarian cancer cells. http://www.wanfangdata.com.cn/details/detail.do?_type=perio&id=58cdacbcf2ba92aaa4e728269e7e0f2a. Int J Cancer. 2013;134(3):542–51. doi: 10.1002/ijc.28399. [Yu PN, Yan MD, Lai HC, et al. Downregulation of miR-29 contributes to cisplatin resistance of ovarian cancer cells[J]. Int J Cancer, 2013, 134(3): 542-51.] [DOI] [PubMed] [Google Scholar]
- 31.Sionov RV. MicroRNAs and glucocorticoid-induced apoptosis in lymphoid malignancies. http://d.old.wanfangdata.com.cn/OAPaper/oai_pubmedcentral.nih.gov_3569899. ISRN Hematol. 2013;2013(13):348212. doi: 10.1155/2013/348212. [Sionov RV. MicroRNAs and glucocorticoid-induced apoptosis in lymphoid malignancies[J]. ISRN Hematol, 2013, 2013(13): 348212.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Han H, Li W, Shen H, et al. microRNA-129-5p, a c-Myc negative target, affects hepatocellular carcinoma progression by blocking the Warburg effect. J Mol Cell Biol. 2016;8(5):400–10. doi: 10.1093/jmcb/mjw010. [Han H, Li W, Shen H, et al. microRNA-129-5p, a c-Myc negative target, affects hepatocellular carcinoma progression by blocking the Warburg effect[J]. J Mol Cell Biol, 2016, 8(5): 400-10.] [DOI] [PubMed] [Google Scholar]
- 33.王 球玉, 唐 珺, 周 佽想, et al. miR-129在乳腺癌中表达下调及其对乳腺癌细胞迁移运动的影响. http://www.wanfangdata.com.cn/details/detail.do?_type=perio&id=shenglxb201204006. 生理学报. 2012;64(4):403–11. [王球玉, 唐珺, 周佽想, 等. miR-129在乳腺癌中表达下调及其对乳腺癌细胞迁移运动的影响[J].生理学报, 2012, 64(4): 403-11.] [PubMed] [Google Scholar]
- 34.Wang Q, Yu J. MiR-129-5p suppresses gastric cancer cell invasion and proliferation by inhibiting COL1A1. Biochem Cell Biol. 2018;96(1):19–25. doi: 10.1139/bcb-2016-0254. [Wang Q, Yu J. MiR-129-5p suppresses gastric cancer cell invasion and proliferation by inhibiting COL1A1[J]. Biochem Cell Biol, 2018, 96(1): 19-25.] [DOI] [PubMed] [Google Scholar]


