Abstract
目的
探讨细胞分裂相关基因NUF2在乳腺癌的表达及临床意义。
方法
利用Oncomine数据库检测NUF2在乳腺癌组织中的表达情况,利用Kaplan-Meier Plotter数据库分析NUF2表达与乳腺癌患者总生存期和无复发生存期的关系,利用GEO数据库及GSEA分析NUF2对基因集富集的影响。利用String数据库分析与NUF2相关的蛋白并通过TIMER数据库验证NUF2表达情况与BUB1、MAD2L1、MYC表达情况的相关性。利用q-PCR验证在8对乳腺癌组织与配对的癌旁正常乳腺组织中,NUF2的mRNA表达情况。
结果
与正常乳腺组织相比,NUF2在乳腺癌中高表达(P < 0.001),且高表达组比低表达组具有较短的总生存期(HR = 1.52,P = 0.015)和无复发生存期(HR = 1.85,P < 0.001)。NUF2高表达组的乳腺癌样本主要富集在细胞周期、P53、G2/ M、DNA修复、MYC和PI3K-AKT-MTOR信号通路,这些通路主要与肿瘤的增殖、侵袭、转移和干性相关。结合String、基因集富集以及TIMER的结果,NUF2可能与BUB1、MAD2L1、MYC直接相互结合作用,促进乳腺癌的进展。q-PCR实验结果显示在8对配对乳腺癌组织中,6例癌组织中NUF2表达上调,2例癌组织中NUF2表达下调。
结论
NUF2基因在乳腺癌中高表达,其表达水平对预测乳腺癌患者的预后具有重要意义。
Keywords: 乳腺癌, NUF2, 数据库分析, 预后
Abstract
Objective
To investigate the expression of the cell division- associated gene NUF2 in breast cancer and its clinical significance.
Methods
The expression of NUF2 in breast cancer tissues was analyzed using Oncomine database. The relationship between the expression of NUF2 and the prognosis of breast cancer was analyzed using the Kaplan-Meier Plotter database. Gene set enrichment analysis (GSEA) and GEO database were used to investigate the effect of NUF2 on gene enrichment. The String database was utilized to analyze the proteins associated with NUF2. The TIMER database was analyzed to assess the correlations of NUF2 with BUB1, MAD2L1 and MYC. The expressions of NUF2 mRNA in 8 pairs of breast cancer tissues and adjacent tissues were verified by q-PCR.
Results
Compared with that in normal breast tissue, NUF2 was significantly overexpressed in breast cancer (P < 0.001). The overall survival time (HR = 1.52, P = 0.015) and the recurrence-free survival time (HR = 1.85, P = 3.2e-14) of the patients with high NUF2 expression were significantly shorter than those of patients with low NUF2 expression. In patients with high NUF2 expression, the enriched genes were involved mainly in cell cycle, P53, G2/M, DNA repair, MYC, and PI3K-AKT-MTOR signaling pathways, which were associated with tumor proliferation, invasion, metastasis and stemness. Combination of the results of String database, gene enrichment and TIMER database analyses suggested that NUF2 interacted directly with BUB1, MAD2L1, and MYC, which could promote the progression of breast cancer. The results of q-PCR showed that NUF2 expression was up-regulated in 6 cancer tissues and down-regulated in 2 cancer tissues.
Conclusion
NUF2 gene is overexpressed in breast cancer, and its expression level is important in predicting the prognosis of breast cancer.
Keywords: breast cancer, NUF2, database analysis, prognosis
乳腺癌发病率和死亡率均占女性恶性肿瘤的首位,其发生发展过程中受到多种癌基因、抑癌基因、细胞因子、非编码RNA等调控,随着对乳腺癌研究的深入,寻找诊断标志物和靶向治疗成为研究的热点[1-2]。细胞分裂相关基因NUF2在细胞有丝分裂过程中起到稳定着丝粒和使染色体正确分离的作用[3]。据报道,NUF2在多种恶性肿瘤中均有较高表达,并且对肿瘤的形成和发展起着重要作用[4]。为了保持基因组的完整性,细胞周期中必须进行适当和准确的染色体分离,这需要染色体、动粒和纺锤体之间进行适当的协调,而NUF2复合体作为有丝分裂中着丝粒的相互作用和纺锤体装配的重要检查点,能够稳定动粒-微管和保障染色体的正常分离[5]。当肿瘤细胞内高表达的NUF2被干扰后,虽然纺锤体形成正常,但运动蛋白不能形成纺锤体微管的附件,也不能激活纺锤体装配检查点,导致染色体的异常分离,诱导有丝分裂细胞发生细胞死亡[6]。而NUF2在乳腺癌中尚未见报道,其在乳腺癌中的作用仍不清楚。基因芯片数据库可通过融合多芯片以及大样本进行分析,得出较客观结果,为NUF2基因在乳腺癌中的深入研究提供理论依据。本研究使用Oncomine和Kaplan-Meier Plotter数据库分析NUF2在乳腺癌中的表达及其与预后的关系,利用GEO数据库、GSEA分析、String以及TIMER数据库探索NUF2在乳腺癌的发生发展中的作用机制,并通过q-PCR验证NUF2在乳腺癌组织中的表达情况。
1. 资料和方法
1.1. Oncomine数据库分析
Oncomine数据库是癌症基因芯片数据库和整合数据的挖掘平台,旨在挖掘癌症基因信息,可用于比较癌症和正常组织的基因差异表达分析。本研究设置的检索条件:(1)Gene:NUF2;(2)Analysis Type:cancer vs normal analysis;(3)Cancer Type:breast cancer;(4)Data Type:mRNA;(5)设定条件:P-value:1E-4,Fold Change:2,Gene Rank:Top 10%。可得到NUF2在不同癌症的表达情况与在乳腺癌组织中mRNA表达水平升高。
1.2. Kaplan-Meier Plotter数据库分析
在数据库中设置条件:(1)Gene:NUF2(223381_at);(2)Split patients by:Auto select best cutoff。其中收录了626例乳腺癌患者总生存期和目的基因的表达信息,分析数据库内NUF2表达与乳腺癌患者总生存期的关系。收录了1764例乳腺癌患者复发情况和目的基因的表达信息,分析NUF2表达与乳腺癌无复发生存期的关系。
1.3. 基因集富集分析
将GEO数据库中GDS5027数据集中的乳腺癌样本根据NUF2(223381_at)表达值的中位数分为NUF2高表达组和NUF2低表达组。采用GSEA2.24版本进行基因集富集分析(GSEA),以hallmark基因集和KEGG基因集作为参考基因集,按照default weighted enrichment statistic的方法,以分析置换次数为1000次,FDR < 0.25,P < 0.05为条件,分析NUF2的表达水平对各种生物通路可能的作用机制。
1.4. String数据库分析
String数据库是分析基因或蛋白相互作用的在线检索工具,其中包含了已证实的和预测的蛋白质与蛋白质相互作用的生物数据库。该数据库可与本研究中的基因集富集分析结果结合,提供乳腺癌中NUF2相关蛋白的相互作用分析。
1.5. TIMER数据库分析
利用TIMER数据库,在乳腺癌组织中对NUF2的表达与BUB1、MAD2L1、MYC的表达进行Spearman相关性分析。本研究设置条件:(1)cancer type:breast invasive carcinoma,共1093例;(2)gene symbols(Yaxis):NUF2;(3)gene symbols(X-axis):BUB1、MAD2L1、MYC;(4)correlation adjusted by:tumor purity;可得到在乳腺癌组织中NUF2的表达情况与BUB1、MAD2L1、MYC的表达情况的相关性。
1.6. q-PCR检测
使用Trizol-氯仿-异丙醇方法提取RNA,逆转录为cDNA,按照Takara试剂盒的说明进行PCR扩增,以GAPDH为对照,比较8对乳腺癌配对组织中NUF2的mRNA的相对表达情况。引物序列如下:NUF2上游引物为5'-TACCATTCACCAATTTAGTTACT-3',下游引物为5'-TAGAATATCAGCAGTCTCAAAG-3';GAPDH上游引物为5'-ACAGCCCGCAGGATCAGGAAA-3';下游引物为5'-AACACGCTTCACGGGCACTC-3'。实验重复3次。
2. 结果
2.1. Oncomine数据库分析在mRNA水平上,NUF2基因在乳腺癌中的表达
Oncomine数据库显示NUF2基因在乳腺癌中表达显著高于正常乳腺组织,共5个子数据库,有612个样本。与正常乳腺组织相比,NUF2基因在乳腺癌中高表达(图 1)。在TCGA Breast芯片中的4个子数据库中,NUF2基因在乳腺癌中的表达是正常乳腺组织的6.091、9.245、4.857、9.678倍(P < 0.05);在Richardson Breast 2芯片中,NUF2在乳腺癌中的表达是正常乳腺组织的18.906倍(P < 0.05,表 1、图 2)。
1.
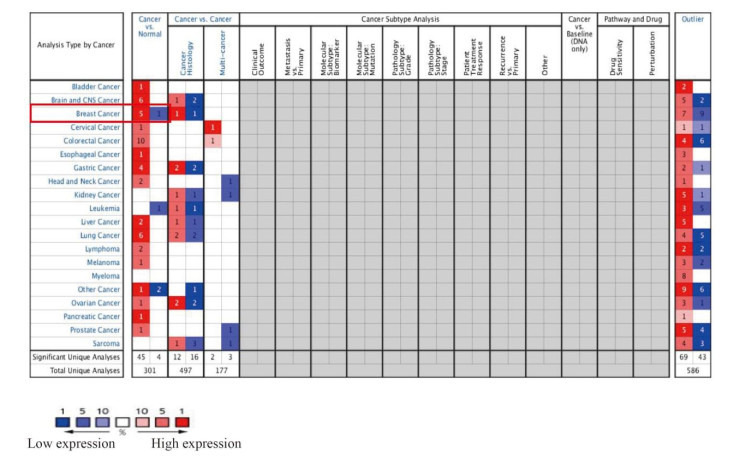
Oncomine中NUF2在不同肿瘤中的表达情况
Expression of NUF2 in different tumors in Oncomine
1.
各子数据库的情况
The situation of each sub-database
| Database | P | Fold change | Total sample (n) | Normal (n) | Breast cancer (n) |
| TCGA Breast | 1.5e-29 | 6.091 | 64 | 61 | 3 |
| 4.46e-37 | 9.245 | 137 | 61 | 76 | |
| 3.97e-18 | 4.857 | 97 | 61 | 36 | |
| 4.74e-44 | 9.678 | 450 | 61 | 389 | |
| Richardson Breast 2 | 1.48e-13 | 18.906 | 47 | 7 | 40 |
2.
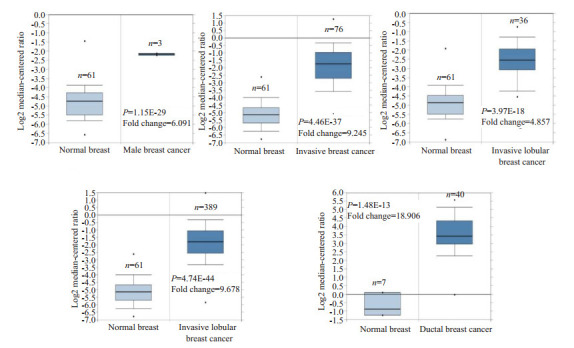
Oncomine各子数据库中NUF2在正常乳腺组织和乳腺癌组织中mRNA的表达
mRNA expression of NUF2 in normal breast tissues and breast cancer tissues in each sub-database of Oncomine
2.2. NUF2基因表达水平与乳腺癌患者预后的相关性
为明确NUF2基因是否能作为判断乳腺癌患者预后的分子标志物,利用Kaplan-Meier Plotter数据库分析NUF2基因表达与乳腺癌患者生存期的关系。NUF2基因高表达组(n = 406)总体生存期低于低表达组(n = 220),差异具有统计学意义(HR = 1.52,P = 0.015,图 3A)。NUF2基因高表达组(n = 936)无复发生存期低于低表达组(n = 828),差异具有统计学意义(HR = 1.85,P < 0.05,图 3B)。经生存分析发现,乳腺癌NUF2基因的表达与预后相关。
3.
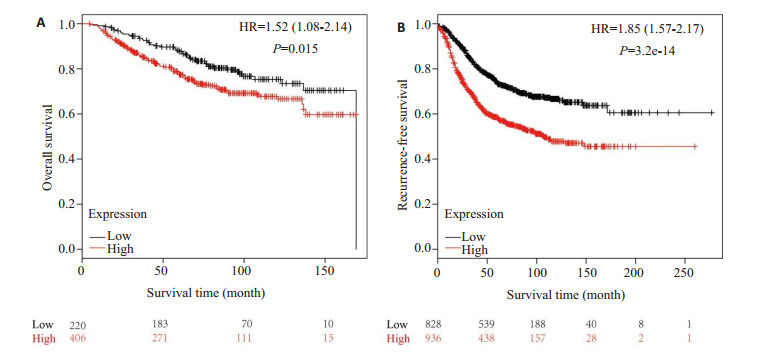
NUF2基因表达水平与乳腺癌患者生存期的关系
Relationship between the expression level of NUF2 and survival of breast cancer patients. A: Correlation between the expression level of NUF2 and overall survival of breast cancer patients; B: Correlation between the expression level of NUF2 and recurrence-free survival of breast cancer
2.3. NUF2基因在乳腺癌中高表达的基因集富集分析
明确NUF2基因与乳腺癌的预后相关性后,进一步分析NUF2基因促进乳腺癌发展的可能机制。将GDS5027数据集中的156个乳腺癌样本按照NUF2(223381_at)表达值的中位数分为NUF2高表达组(n = 78)和NUF2低表达组(n = 78),以hallmark基因集和KEGG基因集作为参考数据集进行GSEA分析。NUF2基因高表达组乳腺癌样本主要富集在细胞周期、P53、G2/M、DNA修复、MYC和PI3K-AKT-MTOR信号通路(图 4),NUF2可能通过以上生物学过程促进乳腺癌的发生发展。
4.
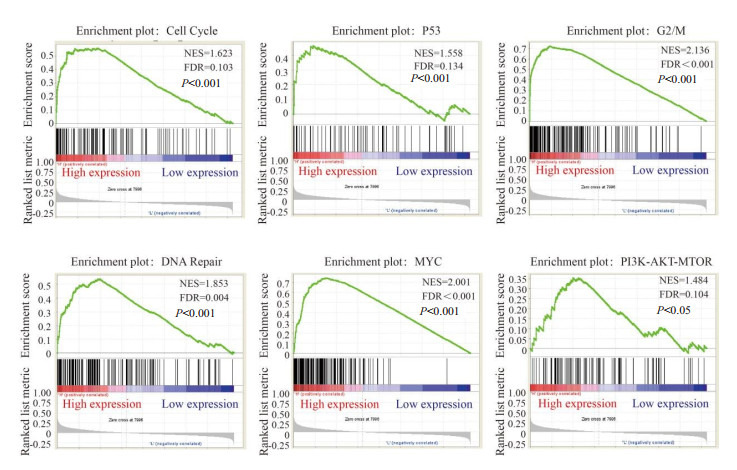
NUF2的基因集富集分析
Gene set enrichment analysis of NUF2
2.4. 与NUF2相关蛋白的分析
使用String数据库分析NUF2蛋白的相互作用,发现多个蛋白与NUF2相互作用,包括BUB1、MAD2L1、ZWINT、CENPE、CASC5、SPC25、MIS12、NDC80、SPC24、CENPF(图 5A)。GSEA分析发现在细胞周期生物学过程中,与上调的NUF2呈正相关的蛋白有BUB1、MAD2L1、MYC,同时MYC信号通路也随着NUF2的表达升高而富集(图 5B)。
5.
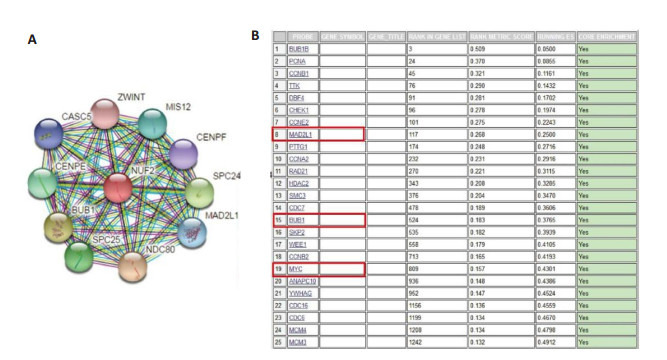
NUF2的相互作用蛋白
The interacting proteins of NUF2. A: The interacting protein of NUF2 in String database; B: Enriched genes in cell cycle signaling pathway in gene set enrichment analysis
TIMER数据库Spearman相关性分析显示,乳腺癌组织中NUF2与BUB1具有强相关性(r = 0.801,P = 2.76e-223,图 6A)、NUF2与MAD2L1具有强相关性(r = 0.743,P = 3.15e-175,图 6B)、NUF2与MYC具有弱相关性(r = 0.183,P = 6.09e-09,图 6C)。
6.
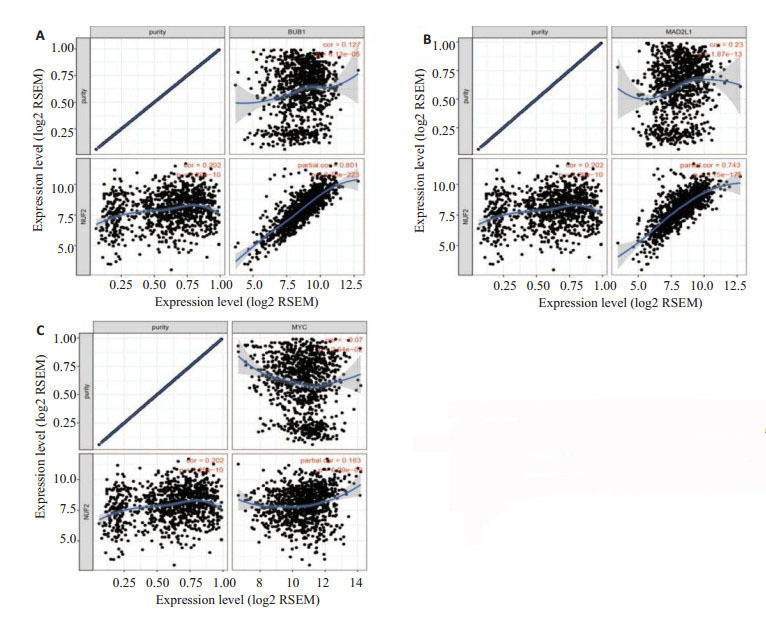
通过TIMER数据库进行Spearman相关性分析
Spearman correlation analysis in TIMER database. A: Correlation analysis between NUF2 and BUB1; B: Correlation analysis between NUF2 and MAD2L1; C: Correlation analysis between NUF2 and MYC
2.5. 通过q-PCR验证在mRNA水平上,NUF2基因在乳腺癌中的表达
通过q-PCR验证在8对乳腺癌配对组织中NUF2的mRNA表达情况,结果显示在8对乳腺癌配对组织中,6例癌组织中NUF2的表达显著高于配对的正常乳腺组织,2例癌组织中NUF2表达低于配对的正常乳腺组织(图 7)。
7.
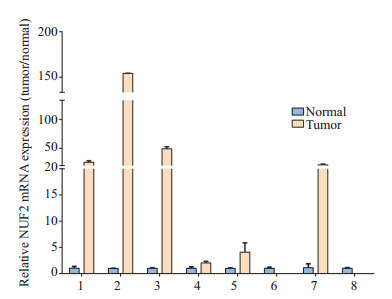
q-PCR验证NUF2在8对乳腺癌配对组织中mRNA的表达
NUF2 mRNA expression in 8 pairs of breast cancer and adjacent tissues by q-PCR
3. 讨论
多数肿瘤细胞都表现出染色体的不稳定[7]。NUF2的异常表达,能够引起有丝分裂失调[5,8]。NUF2的表达情况与肿瘤的发展关系密切,可作为不良预后的生物标志物,在胰腺癌、神经胶质瘤、肝癌中通过siRNA抑制NUF2的表达,能够减缓肿瘤的生长[9-11]。但是,目前NUF2在乳腺癌中的研究却尚未有报道。本研究首先利用Oncomine数据库验证NUF2在不同恶性肿瘤中的表达,发现与正常乳腺组织相比,乳腺癌中NUF2的表达显著升高。进一步通过q-PCR对乳腺癌配对组织进行验证,发现在乳腺癌组织中,NUF2的mRNA水平显著上调。本研究结果显示NUF2高表达组的总生存期和无复发生存期均显著低于NUF2低表达组。表明NUF2高表达的乳腺癌患者预后较差,更容易发生复发。
本研究进一步利用GEO数据库中GDS5027中156例乳腺癌样本进行基因富集分析,发现NUF2与细胞周期、P53、G2/M、DNA修复、MYC和PI3K-AKT-MTOR信号通路关系密切。与正常细胞相比,肿瘤细胞多具有染色体的改变,包括染色体的丢失、增加,多倍体以及其他改变[12-13]。近年来的研究表明,在细胞增殖中,遗传物质复制的精确性,是由细胞周期驱动机制、监测点的机制以及DNA修复机制之间互相协调共同维持的,而肿瘤的发生具有一个明显的共同特征:细胞周期调控机制紊乱[14-15]。P53在人类细胞周期G1期监测点起着关键性作用,超过50%的肿瘤都存在着P53基因的突变,尤其在乳腺癌、结肠癌、肺癌、胰腺癌中突变更为常见[16-19]。同样地,在结直肠癌和胃癌中,通过敲低NUF2的表达,肿瘤细胞生长受到显着抑制,并且细胞周期中sub-G1占比明显增加[20]。在卵巢癌和非小细胞癌中,降低NUF2的表达能促进肿瘤细胞的凋亡[21-22]。而DNA修复基因的突变,引起细胞DNA的修复异常,可导致原癌基因的激活,也可导致抑癌基因的失活,是引起正常细胞发生恶变的重要机制[23]。DNA修复功能异常可能便于休眠中肿瘤细胞从免疫系统中逃逸,肿瘤细胞从而积累突变导致肿瘤复发[24]。C-MYC基因是MYC基因家族的重要成员之一,在乳腺癌中C-MYC的异常激活会激活其致癌转化作用,使细胞具有高度增生潜能,开始向恶性表型转化,提高肿瘤细胞的干性和增殖能力[25-26]。PI3K/Akt/mTOR信号转导通路具有抑制细胞凋亡、促进细胞增殖,促进细胞周期的运行等作用,在乳腺癌的发生、发展以及治疗的过程中发挥着重要作用[27]。同时在曲妥珠单抗的治疗的乳腺癌患者中,MYC基因的复制增多、不稳定的着丝粒状态以及PI3K的激活会影响患者的预后[28-29],而NUF2的高表达会引起着丝粒的不稳定,并且与MYC、PI3K通路相关。
肿瘤干细胞被认为是造成治疗抵抗和复发的重要原因,在乳腺癌中BUB1是有丝分裂检查点中的丝氨酸/苏氨酸激酶,在染色体分离中起重要作用,能维持乳腺癌干细胞的干性[30]。MAD2L1在浸润性导管癌中显著高表达,调控肿瘤细胞的增殖能力,并与患者的总体生存期相关[31]。
综上所述,NUF2可能为乳腺癌患者预后的不利因素,其通过多种途径来促进肿瘤的增殖以及肿瘤细胞的侵袭能力,进而影响乳腺癌患者的临床预后,有望成为乳腺癌复发、不良预后的重要生物标志物之一。然而,NUF2是如何调控这些生物信号通路以及可能结合的下游蛋白来发挥促进肿瘤进展的作用,仍需要进一步的研究。
Biography
孙景波,硕士,E-mail: sunjingbo@i.smu.edu.cn
Funding Statement
广州市天河区科技计划(201504KW038)
Contributor Information
孙 景波 (Jingbo SUN), Email: sunjingbo@i.smu.edu.cn.
刘 晓珑 (Xiaolong LIU), Email: g_xlliu@126.com.
References
- 1.Manders K, van de Poll-Franse LV, Creemers GJ, et al. Clinical management of women with metastatic breast cancer: a descriptive study according to age group. http://d.old.wanfangdata.com.cn/OAPaper/oai_doaj-articles_382315c55609c9ead9e1932912882c85. BMC Cancer. 2006;18(6):179–92. doi: 10.1186/1471-2407-6-179. [Manders K, van de Poll-Franse LV, Creemers GJ, et al. Clinical management of women with metastatic breast cancer: a descriptive study according to age group[J]. BMC Cancer, 2006, 18(6): 179-92.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Newman G, Gonzalez-Perez RR. Leptin-cytokine crosstalk in breast cancer. Mol Cell Endocrinol. 2014;382(1):570–82. doi: 10.1016/j.mce.2013.03.025. [Newman G, Gonzalez-Perez RR. Leptin-cytokine crosstalk in breast cancer[J]. Mol Cell Endocrinol, 2014,382(1): 570-82.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu D, Ding X, Du J, et al. Human NUF2 interacts with centromereassociated protein E and is essential for a stable spindle microtubulekinetochore attachment. J Biol Chem. 2007;282(29):21415–24. doi: 10.1074/jbc.M609026200. [Liu D, Ding X, Du J, et al. Human NUF2 interacts with centromereassociated protein E and is essential for a stable spindle microtubulekinetochore attachment[J]. J Biol Chem, 2007,282(29): 21415-24.] [DOI] [PubMed] [Google Scholar]
- 4.Rieder CL, Salmon ED. The vertebrate cell kinetochore and its roles during mitosis. Trends Cell Biol. 1998;8(8):310–8. doi: 10.1016/S0962-8924(98)01299-9. [Rieder CL, Salmon ED. The vertebrate cell kinetochore and its roles during mitosis[J]. Trends Cell Biol, 1998, 8(8): 310-8.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Obara W, Sato F, Takeda K, et al. Phase I clinical trial of cell division associated 1 (CDCA1) peptide vaccination for castration resistant prostate cancer. Cancer Sci. 2017;108(7):1452–7. doi: 10.1111/cas.2017.108.issue-7. [Obara W, Sato F, Takeda K, et al. Phase I clinical trial of cell division associated 1 (CDCA1) peptide vaccination for castration resistant prostate cancer[J]. Cancer Sci, 2017,108(7): 1452-7.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang T, Zhou Y, Qi ST, et al. Nuf2 is required for chromosome segregation during mouse oocyte meiotic maturation. Cell Cycle. 2015;14(16):2701–10. doi: 10.1080/15384101.2015.1058677. [Zhang T, Zhou Y, Qi ST, et al. Nuf2 is required for chromosome segregation during mouse oocyte meiotic maturation[J]. Cell Cycle, 2015, 14(16): 2701-10.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sansregret L, Vanhaesebroeck B, Swanton C. Determinants and clinical implications of chromosomal instability in cancer. Nat Rev Clin Oncol. 2018;15(3):139–51. doi: 10.1038/nrclinonc.2017.198. [Sansregret L, Vanhaesebroeck B, Swanton C. Determinants and clinical implications of chromosomal instability in cancer[J]. Nat Rev Clin Oncol, 2018, 15(3): 139-51.] [DOI] [PubMed] [Google Scholar]
- 8.Sugimasa H, Taniue K, Kurimoto A, et al. Heterogeneous nuclear ribonucleoprotein K upregulates the kinetochore complex component NUF2 and promotes the tumorigenicity of colon cancer cells. Biochem Biophys Res Commun. 2015;459(1):29–35. doi: 10.1016/j.bbrc.2015.02.043. [Sugimasa H, Taniue K, Kurimoto A, et al. Heterogeneous nuclear ribonucleoprotein K upregulates the kinetochore complex component NUF2 and promotes the tumorigenicity of colon cancer cells [J]. Biochem Biophys Res Commun, 2015,459(1): 29-35.] [DOI] [PubMed] [Google Scholar]
- 9.Hu P, Chen X, Sun J, et al. siRNA-mediated knockdown against NUF2 suppresses pancreatic cancer proliferation in vitro and in vivo. http://www.wanfangdata.com.cn/details/detail.do?_type=perio&id=Doaj000003819057. Biosci Rep. 2015;35(1):79–86. doi: 10.1042/BSR20140124. [Hu P, Chen X, Sun J, et al. siRNA-mediated knockdown against NUF2 suppresses pancreatic cancer proliferation in vitro and in vivo [J]. Biosci Rep, 2015, 35(1): 79-86.] [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10.Huang SK, Qian JX, Yuan BQ, et al. SiRNA-mediated knockdown against NUF2 suppresses tumor growth and induces cell apoptosis in human glioma cells. http://www.wanfangdata.com.cn/details/detail.do?_type=perio&id=5a7ffc8b91ea1c7871e9e54c6cb2e2c6. Cell Mol Biol. 2014;60(4):30–6. [Huang SK, Qian JX, Yuan BQ, et al. SiRNA-mediated knockdown against NUF2 suppresses tumor growth and induces cell apoptosis in human glioma cells[J]. Cell Mol Biol, 2014, 60(4): 30-6.] [PubMed] [Google Scholar]
- 11.Liu Q, Dai SJ, Li H, et al. Silencing of NUF2 inhibits tumor growth and induces apoptosis in human hepatocellular carcinomas. Asian Pac J Cancer Prev. 2014;15(20):8623–9. doi: 10.7314/APJCP.2014.15.20.8623. [Liu Q, Dai SJ, Li H, et al. Silencing of NUF2 inhibits tumor growth and induces apoptosis in human hepatocellular carcinomas[J]. Asian Pac J Cancer Prev, 2014, 15(20): 8623-9.] [DOI] [PubMed] [Google Scholar]
- 12.Lomonosov M, Anand S, Sangrithi M, et al. Stabilization of stalled DNA replication forks by the BRCA2 breast cancer susceptibility protein. Genes Dev. 2003;17(24):3017–22. doi: 10.1101/gad.279003. [Lomonosov M, Anand S, Sangrithi M, et al. Stabilization of stalled DNA replication forks by the BRCA2 breast cancer susceptibility protein[J]. Genes Dev, 2003, 17(24): 3017-22.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Venkitaraman AR. Tumour suppressor mechanisms in the control of chromosome stability: insights from BRCA2. Mol Cells. 2014;37(2):95–9. doi: 10.14348/molcells.2014.2346. [Venkitaraman AR. Tumour suppressor mechanisms in the control of chromosome stability: insights from BRCA2[J]. Mol Cells, 2014, 37 (2): 95-9.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu DZ, Li CF, Zhang X, et al. Skp2-MacroH2A1-CDK8 axis orchestrates G2/M transition and tumorigenesis. http://www.wanfangdata.com.cn/details/detail.do?_type=perio&id=76a36ce2bcc2625e6d1b29a84f772903. Nat Commun. 2015;28(6):641–57. doi: 10.1038/ncomms7641. [Xu DZ, Li CF, Zhang X, et al. Skp2-MacroH2A1-CDK8 axis orchestrates G2/M transition and tumorigenesis[J]. Nat Commun, 2015, 28(6): 641-57.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Umbreit NT, Gestaut DR, Tien JF, et al. The Ndc80 kinetochore complex directly modulates microtubule dynamics. http://d.old.wanfangdata.com.cn/OAPaper/oai_pubmedcentral.nih.gov_3479545. Mol Biol Cell. 2012;23(40):16113–8. doi: 10.1073/pnas.1209615109. [Umbreit NT, Gestaut DR, Tien JF, et al. The Ndc80 kinetochore complex directly modulates microtubule dynamics[J]. Mol Biol Cell, 2012, 23(40): 16113-8.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramalingam V, Varunkumar KA, Rajaram R. p53 mediated transcriptional regulation of long non-coding RNA by 1-hydroxy-1- norresistomycin triggers intrinsic apoptosis in adenocarcinoma lung cancer. Chem Biol Interact. 2018;287(2):1–12. doi: 10.1016/j.cbi.2018.03.016. [Ramalingam V, Varunkumar KA, Rajaram R. p53 mediated transcriptional regulation of long non-coding RNA by 1-hydroxy-1- norresistomycin triggers intrinsic apoptosis in adenocarcinoma lung cancer[J]. Chem Biol Interact, 2018,287(2): 1-12.] [DOI] [PubMed] [Google Scholar]
- 17.Chen G, Zhou T, Li Y, et al. p53 target miR-29c-3p suppresses colon cancer cell invasion and migration through inhibition of PHLDB2. Biochem Biophys Res Commun. 2017;487(1):90–5. doi: 10.1016/j.bbrc.2017.04.023. [Chen G, Zhou T, Li Y, et al. p53 target miR-29c-3p suppresses colon cancer cell invasion and migration through inhibition of PHLDB2 [J]. Biochem Biophys Res Commun, 2017,487(1): 90-5.] [DOI] [PubMed] [Google Scholar]
- 18.Surget S, Khoury MP, Bourdon JC. Uncovering the role of p53 splice variants in human malignancy: a clinical perspective. http://d.old.wanfangdata.com.cn/OAPaper/oai_pubmedcentral.nih.gov_3872270. Onco Targets Ther. 2014;7(1):57–67. doi: 10.2147/OTT.S53876. [Surget S, Khoury MP, Bourdon JC. Uncovering the role of p53 splice variants in human malignancy: a clinical perspective[J]. Onco Targets Ther, 2014, 7(1): 57-67.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.You D, Jung SP, Jeong Y, et al. Wild-type p53 controls the level of fibronectin expression in breast cancer cells. Oncol Rep. 2017;38(4):2551–7. doi: 10.3892/or.2017.5860. [You D, Jung SP, Jeong Y, et al. Wild-type p53 controls the level of fibronectin expression in breast cancer cells[J]. Oncol Rep, 2017, 38 (4): 2551-7.] [DOI] [PubMed] [Google Scholar]
- 20.Kaneko N, Miura K, Gu ZD, et al. siRNA-mediated knockdown against CDCA1 and KNTC2, both frequently overexpressed in colorectal and gastric cancers, suppresses cell proliferation and induces apoptosis. Biochem Biophys Res Commun. 2009;390(4):1235–40. doi: 10.1016/j.bbrc.2009.10.127. [Kaneko N, Miura K, Gu ZD, et al. siRNA-mediated knockdown against CDCA1 and KNTC2, both frequently overexpressed in colorectal and gastric cancers, suppresses cell proliferation and induces apoptosis[J]. Biochem Biophys Res Commun, 2009,390 (4): 1235-40.] [DOI] [PubMed] [Google Scholar]
- 21.Hayama S, Daigo Y, Kato T, et al. Activation of CDCA1-KNTC2, members of centromere protein complex, involved in pulmonary carcinogenesis. Cancer Res. 2006;66(21):10339–48. doi: 10.1158/0008-5472.CAN-06-2137. [Hayama S, Daigo Y, Kato T, et al. Activation of CDCA1-KNTC2, members of centromere protein complex, involved in pulmonary carcinogenesis[J]. Cancer Res, 2006, 66(21): 10339-48.] [DOI] [PubMed] [Google Scholar]
- 22.Numnum TM, Makhija S, Lu B, et al. Improved anti-tumor therapy based upon infectivity-enhanced adenoviral delivery of rna interference in ovarian carcinoma cell lines. Gynecol Oncol. 2008;108(1):34–41. doi: 10.1016/j.ygyno.2007.08.096. [Numnum TM, Makhija S, Lu B, et al. Improved anti-tumor therapy based upon infectivity-enhanced adenoviral delivery of rna interference in ovarian carcinoma cell lines[J]. Gynecol Oncol, 2008,108(1): 34-41.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Foster SS, De S, Johnson LK, et al. Cell cycle- and DNA repair pathway-specific effects of apoptosis on tumor suppression. Proc NatlAcad Sci USA. 2012;109(25):9953–8. doi: 10.1073/pnas.1120476109. [Foster SS, De S, Johnson LK, et al. Cell cycle- and DNA repair pathway-specific effects of apoptosis on tumor suppression[J]. Proc NatlAcad Sci USA, 2012,109(25): 9953-8.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Evans EB, Lin SY. New insights into tumor dormancy: targeting DNArepair pathways. World J Clin Oncol. 2015;6(5):80–8. doi: 10.5306/wjco.v6.i5.80. [Evans EB, Lin SY. New insights into tumor dormancy: targeting DNArepair pathways[J]. World J Clin Oncol, 2015, 6(5): 80-8.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Janghorban M, Farrell AS, Allen-Petersen BL, et al. Targeting cMYC by antagonizing PP2A inhibitors in breast cancer. Proc Natl Acad Sci USA. 2014;111(25):9157–62. doi: 10.1073/pnas.1317630111. [Janghorban M, Farrell AS, Allen-Petersen BL, et al. Targeting cMYC by antagonizing PP2A inhibitors in breast cancer[J]. Proc Natl Acad Sci USA, 2014,111(25): 9157-62.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shachaf CM, Felsher DW. Tumor dormancy and MYC inactivation: pushing cancer to the brink of normalcy. Cancer Res. 2005;65(11):4471–4. doi: 10.1158/0008-5472.CAN-05-1172. [Shachaf CM, Felsher DW. Tumor dormancy and MYC inactivation: pushing cancer to the brink of normalcy[J]. Cancer Res, 2005, 65 (11): 4471-4.] [DOI] [PubMed] [Google Scholar]
- 27.Paplomata E, O'regan R. The PI3K/AKT/mTOR pathway in breast cancer: targets, trials and biomarkers. Ther Adv Med Oncol. 2014;6(4):154–66. doi: 10.1177/1758834014530023. [Paplomata E, O'regan R. The PI3K/AKT/mTOR pathway in breast cancer: targets, trials and biomarkers[J]. Ther Adv Med Oncol, 2014, 6(4): 154-66.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hennessy BT, Smith DL, Ram PT, et al. Exploiting the PI3K/AKT pathway for cancer drug discovery. Nat Rev Drug Discov. 2005;4(12):988–1004. doi: 10.1038/nrd1902. [Hennessy BT, Smith DL, Ram PT, et al. Exploiting the PI3K/AKT pathway for cancer drug discovery[J]. Nat Rev Drug Discov, 2005, 4 (12): 988-1004.] [DOI] [PubMed] [Google Scholar]
- 29.Gogas H, Kotoula V, Alexopoulou Z, et al. MYC copy gain, chromosomal instability and PI3K activation as potential markers of unfavourable outcome in trastuzumab-treated patients with metastatic breast cancer. J Transl Med. 2016;14(1):136–45. doi: 10.1186/s12967-016-0883-z. [Gogas H, Kotoula V, Alexopoulou Z, et al. MYC copy gain, chromosomal instability and PI3K activation as potential markers of unfavourable outcome in trastuzumab-treated patients with metastatic breast cancer[J]. J Transl Med, 2016, 14(1): 136-45.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Han JY, Han YK, Park GY, et al. Bub1 is required for maintaining cancer stem cells in breast cancer cell lines. Sci Rep. 2015;42(5):993–106. doi: 10.1038/srep15993. [Han JY, Han YK, Park GY, et al. Bub1 is required for maintaining cancer stem cells in breast cancer cell lines[J]. Sci Rep, 2015, 42(5): 993-106.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li C, Luo L, Wei S, et al. Identification of the potential crucial genes in invasive ductal carcinoma using bioinformatics analysis. Oncotarget. 2018;9(6):6800–13. doi: 10.18632/oncotarget.23239. [Li C, Luo L, Wei S, et al. Identification of the potential crucial genes in invasive ductal carcinoma using bioinformatics analysis[J]. Oncotarget, 2018, 9(6): 6800-13.] [DOI] [PMC free article] [PubMed] [Google Scholar]


