Abstract
目的
探讨环状RNA(circRNA)在Luminal亚型乳腺癌细胞与正常乳腺细胞中表达谱的差异。
方法
分别提取Luminal亚型细胞MCF7和正常乳腺细胞MCF10A的总RNA;使用NanoDrop ND-1000对total RNA质量进行检测,用核糖核酸酶R分解total RNA以除去线性RNA并富集circRNA;利用随机引物法扩增富集的circRNA并转录成荧光cRNA;将荧光标记cRNA杂交到circRNA微阵列芯片上,扫描得到两种细胞的circRNA表达谱;在Agilent Feature Extraction软件中进行原始数据提取,并对采集到的阵列图像进行分位数归一化和后续数据处理,进行火山图和聚类热图分析;最后,选取差异表达倍数较高的3条circRNA进行实时荧光定量PCR(qPCR)鉴定。
结果
提取的MCF7和MCF10A细胞总RNA纯度高、完整度较好;扫描微阵列芯片上标记的荧光信号,发现两种细胞的circRNA表达谱明显不同;与MCF10A细胞相比,在MCF7细胞的全部12 910条circRNA表达归一化结果中,有5964条上调,81条表达水平一致,6865条下调;表达差异(Log fold change)在2倍以上的有343条,其中213条上调,130条下调;表达差异在5倍以上的有9条,其中8条上调,分别为:hsa_circRNA_061260(6.02倍)、hsa_circRNA_103933(5.96倍)、hsa_circRNA_005239(5.84倍)、hsa_circRNA_100689(5.69倍)、hsa_circRNA_004087(5.60倍)、hsa_circRNA_104420(5.25倍)、hsa_circRNA_104421(5.13倍)和hsa_circRNA_101222(5.03倍),1条下调:hsa_circRNA_ 104864(5.09倍);经qPCR鉴定hsa_circRNA_100689、hsa_circRNA_005239和hsa_circRNA_104864的差异表达趋势与芯片结果相符。
结论
Luminal亚型乳腺癌细胞与正常乳腺细胞的circRNA表达差异较大,其中表达上调或下调的circRNA有望成为Luminal亚型乳腺癌诊断的新靶标。
Keywords: 环状RNA, Luminal亚型, 乳腺癌, 表达谱
Abstract
Objective
To investigate the differences in the expression profiles of circular RNA (circRNA) between luminal breast cancer cells and normal breast cells.
Methods
Total RNA extracted from luminal breast cancer cells MCF7 and normal breast cells MCF10A was digested with Rnase R to remove linear RNAs and enrich circRNAs. The enriched circRNAs were amplified and transcribed into fluorescent cRNAs using a random priming method, and were hybridized onto the circRNA hybridization array. The circRNA expression profiles of MCF7 and MCF10A cells were analyzed using Agilent Feature Extraction software. Quantile normalization and subsequent data processing were performed, and volcano plot filtering and hierarchical clustering were utilized to analyze the circRNA expression patterns. The expressions of 3 circRNAs with significant log fold changes were validated using qPCR.
Results
The hybridization array data revealed significant differences in the circRNA expression profiles between MCF7 and MCF10A cells. Compared with those of MCF10A cells, the 12910 circRNAs expressed in MCF7 cells showed 5964 up-regulated, 81 consistently regulated, and 6865 down-regulated circRNAs; 343 circRNAs showed a log fold change by more than 2 folds, among which 213 circRNAs were up-regulated and 130 were down-regulated. Nine circRNAs showed differential expressions by more than 2 folds, including 8 up-regulated ones, namely hsa_circRNA_061260 (6.02 folds), hsa_circRNA_103933 (5.96 folds), hsa_circRNA_005239 (5.84 folds), hsa_circRNA_100689 (5.69 folds), hsa_circRNA_004087 (5.60 folds), hsa_circRNA_104420 (5.25 folds), hsa_circRNA_104421 (5.13 folds) and hsa_circRNA_101222 (5.03 folds); only one circRNA was down-regulated, namely hsa_circRNA_104864 (5.09 folds). The expressions of hsa_circRNA_100689, hsa_circRNA_005239 and hsa_circRNA_104864 were further validated by qPCR, which yielded consistent results with the microarray data.
Conclusion
The circRNA expression profiles differ significantly between luminal breast cancer cells and normal breast cells. These differentially expressed circRNAs may serve as potential novel targets for the diagnosis of luminal breast cancer.
Keywords: circRNA, luminal type, breast cancer, expression profiles
乳腺癌是女性最常见的恶性肿瘤,具有高度异质性,是典型的以分子分型指导治疗方案的恶性肿瘤,其中Luminal亚型乳腺癌约占乳腺癌发病总数的65%~ 70%[1-3]。发现新的早期诊断标志物对Luminal亚型乳腺癌的诊疗具有重要的指导作用。
环状RNA(circRNA)是一种共价闭合环状结构的非编码RNA,近年来逐渐成为分子生物学和临床诊断学研究的热点[4-5]。circRNA在不同的物种、组织、细胞中的表达不同,具有组织特异性[6-11],且circRNA对核酸酶不敏感,比线性RNA更为稳定,这使得circRNA在作为新型临床诊断标记物的开发应用上具有明显优势。circRNA还可作为miRNA的分子海绵而抑制miRNA的活性,并具有调控基因转录、结合RNA结合蛋白等功能[4]。circRNA在肿瘤的发生发展过程中发挥重要作用,其广泛性、保守性及组织特异性都预示着它有望有望成为新型肿瘤标志物和潜在靶点,为肿瘤的诊断和靶向治疗提供新的方向[6]。
目前,circRNA在乳腺癌中的研究报道较少,而且缺少从组学角度筛选差异调控circRNA的研究。Liang等[12]发现cycDENND4C是一种HIF1α相关的转录因子,在缺氧条件下可促进乳腺癌的增殖;对乳腺癌组织与癌旁组织的差异circRNA进行筛选,发现一些差异表达的circRNA,但该研究没有进行分型研究[13];let-7是报道较多的一种circRNA,其在乳腺癌、卵巢癌等多种肿瘤中表达下调,且let-7的下调与肿瘤的淋巴结转移和增殖水平相关。let-7能够下调致癌基因RAS和MYC的表达,并通过调控细胞周期相关基因影响乳腺癌增殖[14];目前circRNA在乳腺癌中的研究多集中于特定circRNA的作用机制或高通量的致病circRNA筛查等方面,而circRNA在特定亚型乳腺癌中的研究仅见于circGFRA1在三阴性乳腺癌中发挥内源竞争RNA (ceRNA)机制[15],circRNA在特定亚型乳腺癌中的表达谱及分子诊断价值尚未见报道。本文通过circRNA芯片对Luminal亚型乳腺癌细胞与正常乳腺细胞的circRNA表达谱进行差异比较,力求从整体水平上发现与Luminal亚型乳腺癌发生发展显著相关的circRNA,为具有致癌或抑癌作用的circRNA以及Luminal亚型乳腺癌的早期诊断提供依据。
1. 材料和方法
1.1. 细胞来源及培养
实验用的永生化乳腺正常上皮细胞MCF10A和Luminal亚型乳腺癌细胞MCF7均购自中国科学院上海细胞库。细胞使用DMEM培养基培养,在37 ℃,5% CO2的培养箱内生长。
1.2. 标本RNA质控
使用NanoDrop ND-1000测定MCF10A和MCF7细胞内总RNA浓度,RNA定量与质检结果见表 1,使用改良琼脂糖凝胶电泳评估标本RNA的完整性,结果见图 1。
1.
总RNA的定量与质检结果
Quantity and quality control of the total RNA extracted from the cells
| Sample ID | A260/280 Ratio | A260/230 Ratio | Conc (ng/pL) | Volume (pL) | Quantity (ng) | QC result (pass or fail) |
| The acceptable A260/280 ratio for pure total RNA is between 1.8 and 2.1. The acceptable A260/230 ratio for pure total RNA should be more than 1.8. | ||||||
| MCF10A-1 | 1.99 | 2.36 | 837.1 | 80 | 66968 | Pass |
| MCF10A-2 | 1.98 | 2.34 | 769.49 | 80 | 61559.2 | Pass |
| MCF10A-3 | 2.03 | 2.39 | 864.13 | 80 | 69130.4 | Pass |
| MCF7-1 | 2.05 | 2.29 | 893.75 | 60 | 53625 | Pass |
| MCF7-2 | 1.99 | 2.41 | 796.24 | 60 | 47774.4 | Pass |
| MCF7-3 | 2.01 | 2.4 | 918.99 | 60 | 55139.4 | Pass |
1.
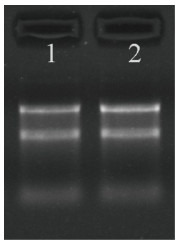
RNA琼脂糖凝胶电泳
RNA agarose gel electrophoresis. 1: Total RNA of MCF7 cells; 2: Total RNA sample of MCF1OA cells.
1.3. RNA标记与杂交
根据Arraystar公司提供的circRNA芯片(Human circular RNA Array V2.0)进行样本标记和序列杂交:用RNase R(Epeentre Technologies, USA)酶解总RNA以除去线性RNA并富集环状RNA,利用随机引物法(Arraystar Super RNA标记试剂盒,Arraystar)扩增circRNA并转录成荧光标记的cRNA;通过RNeasy Mini Kit(Qiagen)对标记的cRNA进行纯化;利用NanoDrop ND-1000测量cRNA(pmol Cy3/μg cRNA)的浓度和比活性;将标记的cRNA与circRNA芯片杂交:在1 μg标记的cRNA中加入5 μL 10×封闭剂和1 μL 25×裂解缓冲液进行片段化,60 ℃孵育30 min,加入25 μL 2×杂交缓冲液稀释标记物的cRNA;将50 μL杂交溶液滴加到circRNA表达微阵列载玻片上,在杂交炉中65 ℃孵育17 h;最后用Agilent Scanner G2505C洗涤玻片,固定并扫描。
1.4. 数据分析
1.4.1. 原始数据收集
用Agilent扫描仪扫描杂交阵列上的荧光信号,将扫描的图像导入Agilent Feature Extraction软件(11.0.1.1版)对采集到的阵列图像进行分析,提取原始数据。
1.4.2. 表达谱数据分析
使用R软件limma软件包进行原始数据的分位数归一化和数据处理,对低信号值的数据进行低强度过滤,保留标本中至少1个具有“P”或“M”标记(“所有目标值”)的circRNA以供进一步分析;通过折叠变化筛选鉴定不同表达的circRNA。采用层次聚类法显示不同样本间的circRNA表达差异。
1.4.3. CircRNA差异表达数据整理
通过火山图分析两组细胞circRNA表达水平的一致性。通过聚类热图归纳整理差异表达基因;CircRNA的表达倍数≥2且P < 0.05为显著差异表达,利用t检验进行差异显著性分析;根据差异倍数、P值等输出结果对差异表达的circRNA进行排序。
1.5. qPCR验证差异表达circRNA
使用Trizol法分别提取MCF7及MCF10A细胞的总RNA;逆转录试剂盒(Cat:AH341-01)及qPCR试剂盒(Cat:AQ131-01)选用北京全式金生物技术有限公司。逆转录反应体系:Total RNA 50 ng,5×SuperMix 4 μL,gDNA remover 1 μL,加RNase-free water至20 μL。轻轻混匀体系,50 ℃孵育15 min,85 ℃加热5 s失活逆转录酶及gDNA remover。qPCR反应体系:cDNA模板2 μL,上游引物0.2 μm,下游引物0.2 μm,2 × qPCR SuperMix 10 μL,加ddH2O至20 μL反应体系。qPCR反应条件:94 ℃ 30 s,94 ℃ 5 s,60 ℃ 30 s—40~45个循环,熔解曲线。本实验重复3次。
扩增引物:
hsa_circRNA_100689:上游引物:5'-ATGGCTCCGATGTGACCG
下游引物:5'-CGCCGACCTGTACTTCTTG
hsa_circRNA_005239:上游引物:5'-GCAGGGTCTGAGAATGAA
下游引物:5'-GGCAGTCAGCGTAGTTTT
hsa_circRNA_104864:上游引物:5'-TGTGGTAATGGTGGTTCT
下游引物:5'-AGGTACTCAGATAGGTGGAT
1.6. 统计学分析
采用SPSS 22.0统计软件,差异比较采用t检验,以P < 0.05为差异有统计学意义。
2. 结果
2.1. 标本总RNA质控
分别提取3组MCF10A和MCF7细胞的总RNA,利用紫外分光光度计测定总RNA的A260 nm/280 nm均在1.9~2.1之间,说明RNA纯度较好;RNA浓度在700~950 ng/μL。利用改良琼脂糖凝胶电泳检测总RNA提取质量。代表性的电泳结果如图 1所示,样本细胞提取的总RNA由28 s、18 s和5 s 3条清晰条带组成,RNA完整度较好。
2.2. circRNA芯片杂交结果
荧光标记的cRNA与circRNA探针杂交后,扫描得到circRNA杂交芯片阵列。如图 2所示。每个点代表一种circRNA,荧光亮度表示其相对表达水平,可见部分circRNA在MCF7和MCF10A细胞中的表达有较大差异。
2.
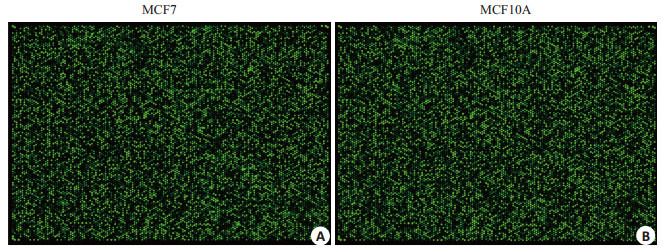
circRNA杂交芯片正列图
Results of circRNA hybridization chip. A: circRNA expression profile of MCF7 cells; B: circRNA expression profile of MCF10A profile.
2.3. 火山图分析
以标准化的MCF10A细胞circRNA表达水平作为横坐标,以标准化的MCF7细胞circRNA表达水平作为纵坐标,用ggplot2绘制火山图。大部分circRNA存在良好的线性关系,说明其表达水平在两种细胞间较为一致(图 3)。少数偏离线性范围的circRNA为差异表达circRNA(|Fold change|≥1.5,P < 0.05)。
3.
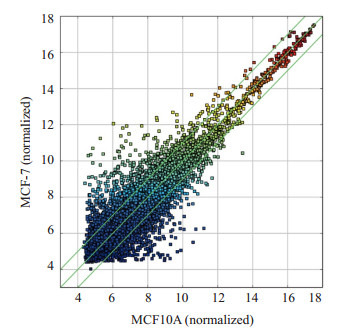
circRNA火山图
Volcano plot of circRNA expressions in the two cells.
2.4. 差异表达circRNA的聚类分析
将MCF10A与MCF7细胞中所有circRNA的表达数据绘制聚类热图,以颜色深浅表示circRNA表达水平的高低,红色代表高表达水平,绿色代表低表达水平。与正常乳腺细胞MCF10A相比,在Luminal亚型细胞MCF7中有5964条circRNA表达上调,81条表达一致,6865条下调(图 4)。差异表达2倍以上的circRNA有343条,其中213条上调,130条下调;差异表达5倍以上的有9条,其中8条上调,1条下调。
4.
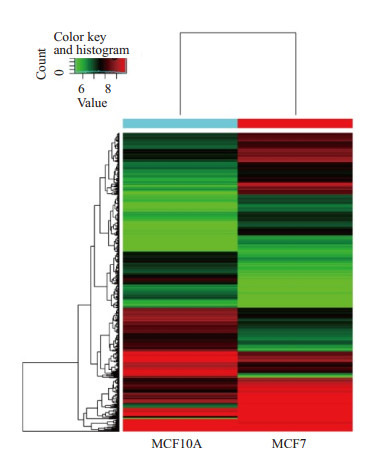
MCF10A与MCF7中circRNA表达水平聚类热图
Heat map of circRNA expressions in MCF10A and MCF7 cells.
从图 4中筛选出MCF7细胞中上调和下调最显著的20个circRNA,绘制成聚类热图(图 5)。其中差异表达变化5倍以上的有9条,其中8条上调分别是hsa_circRNA_061260(6.02倍)、hsa_circRNA_103933 (5.96倍)、hsa_circRNA_005239(5.84倍)、hsa_circRNA_ 100689(5.69倍)、hsa_circRNA_004087(5.60倍)、hsa_circRNA_104420(5.25倍)、hsa_circRNA_104421 (5.13倍)和hsa_circRNA_101222(5.03倍),1条下调:hsa_circRNA_104864(5.09倍)。
5.
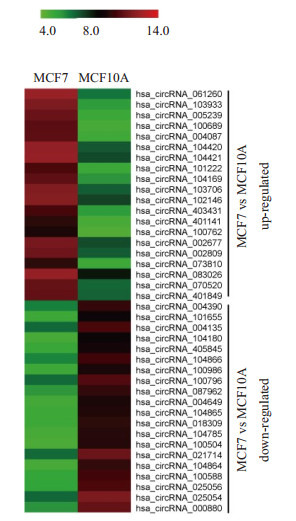
上调或下调倍数最高的20个circRNA的聚类热图分析
Heat map of the top 20 differentially expressed circRNAs in MCF7 cells.
2.5. 差异表达circRNA的验证
选择两个高表达且差异表达倍数在5倍以上的circRNA(hsa_circRNA_100689和hsa_circRNA_005239)以及1个低表达且差异表达倍数在5倍以上的circRNA (hsa_circRNA_104864)进行qPCR验证。3种circRNA在MCF7和MCF10A细胞中均有显著性差异表达,且差异表达趋势与芯片结果相符(图 6)。
6.
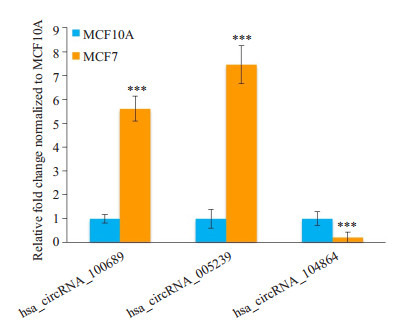
差异表达circRNA的qPCR验证
Validation of the differentially expressed circRNAs by qPCR. Compared with MCF10A, ***P < 0.001.
3. 讨论
circRNA与膀胱癌[16-17]、肝细胞癌[18-19]、胃癌[20]、大肠癌[21]、喉癌[22]、基底细胞癌[23]等多种恶性肿瘤的发生发展密切相关,但circRNA在乳腺癌中的报道却较少,并且尚未发现乳腺癌亚型特异性的circRNA。在缺氧条件下circDENND4C的表达水平升高,且能够促进乳腺癌细胞增殖[12]。Lu等[13]利用circRNA芯片筛选了乳腺癌组织与癌旁组织的差异表达circRNA,并预测了重要的circRNA-miRNA互作关系对,但该研究并没有对乳腺癌亚型进行细分。circRNA还与miRNA、lncRNA组成复杂的内源竞争性调控网络,从而影响Luminal亚型乳腺癌的发生发展[24-25]。研究表明miR- 34a与circRNA-LDHA组成的调控轴可促进乳腺癌增殖,而由雌激素受体调控的microRNA表达缺失可导致HER2信号通路过度激活,可能导致Luminal亚型乳腺癌患者预后较差[26-27]。然而,circRNA在Luminal亚型乳腺癌中的整体表达特征及其与正常乳腺上皮细胞的差异表达circRNA谱尚无人报道。本研究从整体水平研究circRNA在Luminal亚型乳腺癌细胞和正常乳腺细胞中的表达谱差异。研究采用Arraystar公司推出的全球首款circRNA芯片,其具有特异性剪接位点探针与外切酶预处理双重保障,能够准确检测样本中circRNA的表达,具有通量大、周期短、灵敏度高的优点。我们使用该circRNA芯片对Luminal亚型乳腺癌细胞MCF7和正常乳腺细胞MCF10A的circRNA进行分析,分别获得的两组细胞的circRNA表达谱和差异circRNA表达谱。与MCF10A相比,MCF7细胞中有5964条circRNA表达上调,6865条下调。差异表达2倍以上的有343条,其中213条上调,130条下调;差异表达5倍以上的有9条,其中8条上调,1条下调。
差异表达显著的circRNA可能参与了Luminal亚型乳腺癌的发生、发展与分子调控进程。hsa_circRNA_ 100689(2.24倍)和hsa_circRNA_005239(2.2倍)是差异表达倍数最高的两条circRNA,两条circRNA均命名为GFRA1。He等[15]发现circGFRA1表达上调与三阴性乳腺癌患者较差预后相关,并提出circGFRA1和GFRA1通过ceRNA机制调控miR-34a的生物学作用,提示circGFRA1可能是乳腺癌重要的致癌基因。本研究发现并验证的另外几种差异表达circRNA均属首次报道,尚没有文献报道其作用机理。在后续的研究中,我们将深入探索新发现的差异circRNA的生物学作用及作用机制,期望发现新的有望用于Luminal亚型乳腺癌临床诊断或治疗靶标的circRNA分子。
MCF7为雌激素受体阳性的乳腺癌细胞,该细胞株保留了多个分化乳腺上皮的特性,是研究Luminal亚型乳腺癌的最常用的细胞类型之一[28]。MCF10A是一种永生化的不具有致瘤性的乳腺正常上皮细胞,对胰岛素、糖皮质激素、表皮生长因子等外源刺激有响应,在电镜下观察,MCF10A细胞呈现典型的导管样细胞[29];而Hs 578bst是一种正常乳腺成纤维细胞,其来源于一个浸润性导管癌(Hs 578T的起源)旁的正常乳房组织,由于其具有成纤维细胞生长特性,并且没有ER受体[30],因此本研究选择MCF10A作为对照细胞。
综上所述,本研究绘制了Luminal亚型乳腺癌细胞与正常乳腺细胞的circRNA表达谱,发现了一些新的差异表达circRNA,为阐明Luminal亚型乳腺癌的致病机制和发现新的诊断靶标奠定了基础。
Biography
陈建新, 医学硕士, 主治医师, E-mail: 22068420@qq.com
Funding Statement
军队后勤科研项目(CWH17C017);广州市科技计划项目(201804010186)
Contributor Information
肖 斌 (Bin XIAO), Email: 22068420@qq.com.
李 林海 (Linhai1 LI), Email: mature303@126.com.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. doi: 10.3322/caac.21387. [Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017[J]. CA Cancer J Clin, 2017, 67(1): 7-30. ] [DOI] [PubMed] [Google Scholar]
- 2.Russnes HG, Lingjaerde OC, Borresen-Dale AL, et al. Breast cancer molecular stratification: from intrinsic subtypes to integrative clusters. Am J Pathol. 2017;187(10):2152–62. doi: 10.1016/j.ajpath.2017.04.022. [Russnes HG, Lingjaerde OC, Borresen-Dale AL, et al. Breast cancer molecular stratification: from intrinsic subtypes to integrative clusters [J]. Am J Pathol, 2017, 187(10): 2152-62. ] [DOI] [PubMed] [Google Scholar]
- 3.Taherian-Fard A, Srihari S, Ragan MA. Breast cancer classification: linking molecular mechanisms to disease prognosis. Brief Bioinform. 2015;16(3):461–74. doi: 10.1093/bib/bbu020. [Taherian-Fard A, Srihari S, Ragan MA. Breast cancer classification: linking molecular mechanisms to disease prognosis[J]. Brief Bioinform, 2015, 16(3): 461-74. ] [DOI] [PubMed] [Google Scholar]
- 4.Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–8. doi: 10.1038/nature11928. [Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency[J]. Nature, 2013, 495 (7441): 333-8. ] [DOI] [PubMed] [Google Scholar]
- 5.Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–8. doi: 10.1038/nature11993. [Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges[J]. Nature, 2013, 495 (7441): 384-8. ] [DOI] [PubMed] [Google Scholar]
- 6.Zheng QP, Bao CY, Guo WJ, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. http://busqueda.bvsalud.org/portal/resource/pt/mdl-27050392. Nat Commun. 2016;7(3):11215. doi: 10.1038/ncomms11215. [Zheng QP, Bao CY, Guo WJ, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs [J]. Nat Commun, 2016, 7(3): 11215. ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lyu D, Huang S. The emerging role and clinical implication of human exonic circular RNA. RNA Biol. 2017;14(8):1000–6. doi: 10.1080/15476286.2016.1227904. [Lyu D, Huang S. The emerging role and clinical implication of human exonic circular RNA[J]. RNA Biol, 2017, 14(8): 1000-6. ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19(2):141–57. doi: 10.1261/rna.035667.112. [Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats[J]. RNA, 2013, 19(2): 141-57. ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bachmayr-Heyda A, Reiner AT, Auer K, et al. Correlation of circular RNA abundance with proliferation--exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues. http://europepmc.org/abstract/MED/25624062. Sci Rep. 2015;5(2):8057. doi: 10.1038/srep08057. [Bachmayr-Heyda A, Reiner AT, Auer K, et al. Correlation of circular RNA abundance with proliferation--exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues [J]. Sci Rep, 2015, 5(2): 8057. ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li F, Zhang L, Li W, et al. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/β-catenin pathway. http://europepmc.org/abstract/MED/25749389. Oncotarget. 2015;6(8):6001–13. doi: 10.18632/oncotarget.3469. [Li F, Zhang L, Li W, et al. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/β-catenin pathway [J]. Oncotarget, 2015, 6(8): 6001-13. ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qian Y, Lu Y, Rui C, et al. Potential significance of circular RNA in human placental tissue for patients with preeclampsia. Cell Physiol Biochem. 2016;39(4):1380–90. doi: 10.1159/000447842. [Qian Y, Lu Y, Rui C, et al. Potential significance of circular RNA in human placental tissue for patients with preeclampsia[J]. Cell Physiol Biochem, 2016, 39(4): 1380-90. ] [DOI] [PubMed] [Google Scholar]
- 12.Liang G, Liu Z, Tan L, et al. HIF1α-associated circDENND4C promotes proliferation of breast cancer cells in hypoxic environment. http://orca.cf.ac.uk/103038/ Anticancer Res. 2017;37(8):4337–43. doi: 10.21873/anticanres.11827. [Liang G, Liu Z, Tan L, et al. HIF1α-associated circDENND4C promotes proliferation of breast cancer cells in hypoxic environment [J]. Anticancer Res, 2017, 37(8): 4337-43. ] [DOI] [PubMed] [Google Scholar]
- 13.Lu LS, Sun J, Shi PY, et al. Identification of circular RNAs as a promising new class of diagnostic biomarkers for human breast cancer. http://pubmed.cn/28484086. Oncotarget. 2017;8(27):44096–107. doi: 10.18632/oncotarget.17307. [Lu LS, Sun J, Shi PY, et al. Identification of circular RNAs as a promising new class of diagnostic biomarkers for human breast cancer [J]. Oncotarget, 2017, 8(27): 44096-107. ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson SM, Grosshans H, Shingara J, et al. RAS is regulated by the let-7 MicroRNA family. Cell. 2005;120(5):635–47. doi: 10.1016/j.cell.2005.01.014. [Johnson SM, Grosshans H, Shingara J, et al. RAS is regulated by the let-7 MicroRNA family[J]. Cell, 2005, 120(5): 635-47. ] [DOI] [PubMed] [Google Scholar]
- 15.He R, Liu P, Xie X, et al. circGFRA1 and GFRA1 act as ceRNAs in triple negative breast cancer by regulating miR-34a. J Exp Clin Cancer Res. 2017;36(1):145. doi: 10.1186/s13046-017-0614-1. [He R, Liu P, Xie X, et al. circGFRA1 and GFRA1 act as ceRNAs in triple negative breast cancer by regulating miR-34a[J]. J Exp Clin Cancer Res, 2017, 36(1): 145. ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhong Z, Huang M, Lv M, et al. Circular RNA MYLK as a competing endogenous RNA promotes bladder cancer progression through modulating VEGFA/VEGFR2 signaling pathway. Cancer Lett. 2017;403:305–17. doi: 10.1016/j.canlet.2017.06.027. [Zhong Z, Huang M, Lv M, et al. Circular RNA MYLK as a competing endogenous RNA promotes bladder cancer progression through modulating VEGFA/VEGFR2 signaling pathway [J]. Cancer Lett, 2017, 403: 305-17. ] [DOI] [PubMed] [Google Scholar]
- 17.Huang M, Zhong Z, Lv M, et al. Comprehensive analysis of differentially expressed profiles of lncRNAs and circRNAs with associated co-expression and ceRNA networks in bladder carcinoma. http://www.oncotarget.com/index.php?journal=oncotarget&page=article&op=view&path%5B%5D=9706. Oncotarget. 2016;7(30):47186–200. doi: 10.18632/oncotarget.9706. [Huang M, Zhong Z, Lv M, et al. Comprehensive analysis of differentially expressed profiles of lncRNAs and circRNAs with associated co-expression and ceRNA networks in bladder carcinoma [J]. Oncotarget, 2016, 7(30): 47186-200. ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qin M, Liu G, Huo X, et al. Hsa_circ_0001649: A circular RNA and potential novel biomarker for hepatocellular carcinoma. Cancer Biomark. 2016;16(1):161–9. doi: 10.3233/CBM-150552. [Qin M, Liu G, Huo X, et al. Hsa_circ_0001649: A circular RNA and potential novel biomarker for hepatocellular carcinoma[J]. Cancer Biomark, 2016, 16(1): 161-9. ] [DOI] [PubMed] [Google Scholar]
- 19.Shang X, Li G, Liu H, et al. Comprehensive circular RNA profiling reveals that hsa_circ_0005075, a new circular RNA biomarker, is involved in hepatocellular crcinoma development. Medicine (Baltimore) 2016;95(22):e3811. doi: 10.1097/MD.0000000000003811. [Shang X, Li G, Liu H, et al. Comprehensive circular RNA profiling reveals that hsa_circ_0005075, a new circular RNA biomarker, is involved in hepatocellular crcinoma development[J]. Medicine (Baltimore), 2016, 95(22): e3811. ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li P, Chen S, Chen H, et al. Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clin Chim Acta. 2015;444:132–6. doi: 10.1016/j.cca.2015.02.018. [Li P, Chen S, Chen H, et al. Using circular RNA as a novel type of biomarker in the screening of gastric cancer[J]. Clin Chim Acta, 2015, 444: 132-6. ] [DOI] [PubMed] [Google Scholar]
- 21.Xie H, Ren X, Xin S, et al. Emerging roles of circRNA_001569 targeting miR-145 in the proliferation and invasion of colorectal cancer. http://pubmed.cn/27058418. Oncotarget. 2016;7(18):26680–91. doi: 10.18632/oncotarget.8589. [Xie H, Ren X, Xin S, et al. Emerging roles of circRNA_001569 targeting miR-145 in the proliferation and invasion of colorectal cancer [J]. Oncotarget, 2016, 7(18): 26680-91. ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xuan LJ, Qu LM, Zhou H, et al. Circular RNA: a novel biomarker for progressive laryngeal cancer. http://europepmc.org/abstract/MED/27158380. Am J Transl Res. 2016;8(2):932–9. [Xuan LJ, Qu LM, Zhou H, et al. Circular RNA: a novel biomarker for progressive laryngeal cancer[J]. Am J Transl Res, 2016, 8(2): 932-9. ] [PMC free article] [PubMed] [Google Scholar]
- 23.Sand M, Bechara FG, Sand D, et al. Circular RNA expression in basal cell carcinoma. Epigenomics. 2016;8(5):619–32. doi: 10.2217/epi-2015-0019. [Sand M, Bechara FG, Sand D, et al. Circular RNA expression in basal cell carcinoma[J]. Epigenomics, 2016, 8(5): 619-32. ] [DOI] [PubMed] [Google Scholar]
- 24.Chen J, Xu J, Li Y, et al. Competing endogenous RNA network analysis identifies critical genes among the different breast cancer subtypes. http://www.oncotarget.com/index.php?journal=oncotarget&page=article&op=view&path%5B%5D=14361. Oncotarget. 2017;8(6):10171–84. doi: 10.18632/oncotarget.14361. [Chen J, Xu J, Li Y, et al. Competing endogenous RNA network analysis identifies critical genes among the different breast cancer subtypes [J]. Oncotarget, 2017, 8(6): 10171-84. ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou S, Wang L, Yang Q, et al. Systematical analysis of lncRNAmRNA competing endogenous RNA network in breast cancer subtypes. Breast Cancer Res Treat. 2018;169(2):267–75. doi: 10.1007/s10549-018-4678-1. [Zhou S, Wang L, Yang Q, et al. Systematical analysis of lncRNAmRNA competing endogenous RNA network in breast cancer subtypes [J]. Breast Cancer Res Treat, 2018, 169(2): 267-75. ] [DOI] [PubMed] [Google Scholar]
- 26.Xiao X, Huang X, Ye F, et al. The miR-34a-LDHA axis regulates glucose metabolism and tumor growth in breast cancer. Sci Rep. 2016;6:21735. doi: 10.1038/srep21735. [Xiao X, Huang X, Ye F, et al. The miR-34a-LDHA axis regulates glucose metabolism and tumor growth in breast cancer [J]. Sci Rep, 2016, 6: 21735. ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bailey ST, Westerling T, Brown M. Loss of estrogen-regulated microRNA expression increases HER2 signaling and is prognostic of poor outcome in luminal breast cancer. Cancer Res. 2015;75(2):436–45. doi: 10.1158/0008-5472.CAN-14-1041. [Bailey ST, Westerling T, Brown M. Loss of estrogen-regulated microRNA expression increases HER2 signaling and is prognostic of poor outcome in luminal breast cancer [J]. Cancer Res, 2015, 75 (2): 436-45. ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Comşa Ş, Cîmpean AM, Raica M. The story of MCF-7 breast cancer cell line: 40 years of experience in research. http://ar.iiarjournals.org/content/35/6/3147. Anticancer Res. 2015;35(6):3147–54. [Comşa Ş, Cîmpean AM, Raica M. The story of MCF-7 breast cancer cell line: 40 years of experience in research[J]. Anticancer Res, 2015, 35(6): 3147-54. ] [PubMed] [Google Scholar]
- 29.Santner SJ, Dawson PJ, Tait L, et al. Malignant MCF10CA1 cell lines derived from premalignant human breast epithelial MCF10AT cells. Breast Cancer Res Treat. 2001;65(2):101–10. doi: 10.1023/A:1006461422273. [Santner SJ, Dawson PJ, Tait L, et al. Malignant MCF10CA1 cell lines derived from premalignant human breast epithelial MCF10AT cells [J]. Breast Cancer Res Treat, 2001, 65(2): 101-10. ] [DOI] [PubMed] [Google Scholar]
- 30.Wang X, Sang X, Diorio C, et al. In vitro interactions between mammary fibroblasts (Hs 578Bst) and cancer epithelial cells (MCF- 7) modulate aromatase, steroid sulfatase and 17β-hydroxysteroid dehydrogenases. Mol Cell Endocrinol. 2015;412:339–48. doi: 10.1016/j.mce.2015.05.032. [Wang X, Sang X, Diorio C, et al. In vitro interactions between mammary fibroblasts (Hs 578Bst) and cancer epithelial cells (MCF- 7) modulate aromatase, steroid sulfatase and 17β-hydroxysteroid dehydrogenases [J]. Mol Cell Endocrinol, 2015, 412: 339-48.] [DOI] [PubMed] [Google Scholar]


