Abstract
目的
探讨敲除长链非编码RNA MALAT-1基因对人喉鳞状细胞癌Hep-2细胞的影响。
方法
使用实时聚合酶链反应(RTPCR)以永生化鼻咽上皮(NPE)细胞株NP-69作为参照检测FaDu、Hep-2和鼻咽癌CNE-2Z细胞MALAT1的表达,使用shmalat1慢病毒转染Hep-2细胞,RT-PCR检测其MALAT1的表达。增殖速率分析,MTT法明确MALAT-1对细胞增殖的影响。流式细胞术分析细胞周期和细胞凋亡。采用Transwell小室检测Hep-2细胞的迁移。最后使用Matrigel侵袭实验评估shmalat1和shCtrl慢病毒转染的Hep-2细胞的侵袭能力。
结果
使用qPCR检测FaDu、Hep-2和鼻咽癌CNE-2Z细胞MALAT1的表达,结果发现与NP-69细胞相比,FaDu、Hep-2和鼻咽癌CNE-2Z细胞MALAT1的表达显著增加。敲除MALAT1明显抑制Hep-2细胞增殖。与MOCK和shCtrl细胞相比,MALAT1-shRNA慢病毒转染细胞S期细胞数量显著增加(P < 0.01)。此外,细胞在G2/M期的百分比下降(P < 0.01)。下调MALAT1时,Hep-2细胞的侵袭和迁移都受到抑制。
结论
MALAT1在喉鳞状细胞癌高表达。敲除长链非编码RNA MALAT-1基因对人喉鳞状细胞癌Hep-2细胞的增殖、凋亡、侵袭和迁移起抑制作用。
Keywords: 细胞凋亡, 长链非编码RNA, 喉肿瘤, MALAT1, 迁移和侵袭
Abstract
Objective
To investigate the effect of knocking down long chain non-coding RNA MALAT-1 gene on the biological behaviors of human laryngeal squamous cell carcinoma Hep-2 cells.
Method
With immortalized nasopharyngeal epithelial (NPE) cell line NP-69 as the reference, MALAT1 expression in FaDu, Hep-2 and nasopharyngeal carcinoma CNE-2Z cells were detected using real-time PCR. Hep-2 cells were transfected with shmalat1 lentivirus and the expression of MALAT1 was detected. MTT assay, flow cytometry, Transwell assay and M Atrigel invasiveness test were used to evaluate the effect of MALAT-1 knockdown on the proliferation, cell cycle, cell apoptosis, migration, and invasiveness of Hep-2 cells.
Results
Compared with NP-69 cells, Hep-2 cells, FaDu cells, and CNE-2Z cells all showed significantly increased MALAT-1 expression. In Hep-2 cells, knockdown of MALAT-1 significantly inhibited the cell proliferation, increased the cell percentage in S phase (P < 0.01), decreased the cell percentage in G2/M phase (P < 0.01), and attenuated the migration and invasiveness of the cells.
Conclusion
MALAT-1 is over-expressed in laryngeal squamous cell carcinoma, and knocking down MALAT-1 gene can significantly suppress the proliferation, invasion and migration and promotes apoptosis of the cancer cells.
Keywords: apoptosis, long chain non coded RNA, laryngeal neoplasms, MALAT1, migration and invasion
在世界范围内,头颈部肿瘤居所有肿瘤的第六位,每年全世界新发病人超过50万[1-2]。喉癌作为头颈部肿瘤的一种,在所有呼吸系统肿瘤中居第2位,大约2%~ 3%的呼吸系统恶性肿瘤为喉癌[3-4],其发病率具有显著的地理差异[5]。目前,以手术为主术后辅以放疗和化疗的综合治疗是治疗喉癌的基本方法[6]。然而,一项流行病学调查显示,在过去的20年间,喉癌的临床治疗效果尚不尽如人意[7]。因此,从分子水平开展喉癌的发生发展机制的研究将有助于提高喉癌的诊治水平。长链非编码RNA是一类转录本长度超过200核苷酸的RNA,本身并不编码或很少编码蛋白质[8-9]。长链非编码RNA在各种生物学过程和发病机制中起关键作用[10]。以往的研究结果表明,不同类型的肿瘤中lncRNAs表达具有差异性,提示lncRNAs可能成为癌症的潜在治疗靶点[11-12]。MALAT1是第1个癌症相关的lncRNAs。MALAT1广泛存在于人体组织,包括神经系统、肺、胰腺和其他器官[13-16]。亦有资料表明,MALAT1在肺癌、肝癌、乳腺癌、子宫内膜癌和胰腺癌中呈高表达[15, 17-19]。然而,与喉癌相关的MALAT1的数据有限。本研究的目的在于探明MALAT1在喉癌方面的作用。检测NP-69细胞中MALAT1的表达水平,并与采自头颈部肿瘤的3种鳞状细胞癌中MALAT1表达水平进行比较。另外,在Hep-2细胞中,用SiRNA有针对性的干扰MALAT1,并评价MALAT1对Hep-2细胞增殖、克隆形成、细胞凋亡和迁移的影响。
1. 材料和方法
1.1. 材料
1.1.1. 细胞来源
FaDu、Hep-2和鼻咽癌CNE-2Z细胞。永生化鼻咽上皮(NPE)细胞株NP-69。所有细胞均购自中国科学院上海细胞库。
1.1.2. 主要试剂
DMEM培养基(生工生物技术)。10%胎牛血清(Gibco)。无血清的角质形成细胞(Gibco)。pgv248慢病毒载体(吉凯)。Matrigel基质胶(Corning)。Transwell小室(Corning)。Lipofectamin 2000(Invitrogen)。嘌呤霉素(生工生物)。TRIzol reagent(Invitrogen)。M-MLV逆转录酶(Promega)。SYBR premix Ex Taq酶(Takara)。MALAT-1引物(锐博生物)。DMSO(SigmaAldrich)。碘化丙啶(PI,50 μg/mL,Sigma-Aldrich)。核糖核酸酶A(100 μg/mL,生工生物科技)。膜联蛋白V凋亡检测试剂盒(eBioscience)。丝裂霉素C(Sigma Aldrich)。
1.2. 方法
1.2.1. 细胞培养和细胞转染
3种头颈部鳞状细胞株,包括两个喉肿瘤细胞株:FaDu、Hep-2和鼻咽癌CNE-2Z细胞。永生化鼻咽上皮(NPE)细胞株NP-69作为内参。所有细胞均购自中国科学院。头颈部鳞状细胞株存于DMEM培养基(生工生物),培养基内放入10%胎牛血清。NP-69细胞保存于无血清的角质形成细胞中(Gibco)。所有细胞系在37 ℃、5% CO2环境中培养。将专门针对MALAT1表达序列5'- GTA ACT GGC ATGTGA GCA A-3'的shRNA插入到pgv248慢病毒载体(吉凯)产生干扰shRNA(shMALAT1)。作为一个scrabmled控制(shCtrl)单元,将5'- TTC TCC GAA CGT GTC ACG TTT CAA GAG AAC GTG ACA CGTTCG GAG AA-3'克隆到慢病毒pgv248。根据制造商的说明,Lipofectamin 2000用于(Invitrogen)转染细胞与指定的构建。慢病毒感染的Hep-2细胞接种在6孔板上,接种密度为每孔5×104细胞,24 h后细胞感染shmalat1或shCtrl慢病毒的数量按感染复数(MOI)100计算。3 d后感染后,用荧光显微镜(Olympus xi-71)检测GFP表达来评价转染效率。选择嘌呤霉素感染后5 d,收获细胞,MALAT1敲除效率用逆转录聚合酶链反应评价。
1.2.2. 实时聚合酶链反应(RT-PCR)
从4株培养细胞中提取总RNA,TRIzol reagent(Invitrogen)。采用M-MLV逆转录酶(Promega)反转录成cDNA。使用SYBR premix Ex Taq酶表达进行检测(Takara)使用下面的引物:MALAT- 1上游引物,5'- CAG ACC ACC ACA GGT TTACAG-3';MALAT1下游引物,5'-AGA CCA TCC CAA AAT GCT TCA-3'。以GAPDH作为内参基因,GAPDH上游引物,5'-TGA CTT CAA CAG CGA CAC CCA-3';GAPDH下游引物,5'-CAC CCT GTT GCT GTA GCC AAA-3('锐博生物)。采用2-△△CT法计算MALAT-1的表达水平[20]。
1.2.3. 增殖速率分析
使用MTT法明确MALAT-1是否对细胞的增殖产生影响。将Hep-2细胞种植于96孔板,每孔2×103细胞数,24 h后进行转染。在转染后的第1、2、3、4、5天,细胞用最终浓度为0.2 mg/mL MTT试剂孵育4 h。除去上清液,每孔加入100 μL DMSO(SigmaAldrichA)培养3~5 min溶解甲晶体。使用微孔板读数器,指示细胞增殖的测量波长为490 nm。MTT检测在不同的时间点共记录3次。
1.2.4. 流式细胞术分析细胞周期
转染96 h后,细胞进行消化,用磷酸清洗一次缓冲盐水(PBS,pH 7.2-7.4)然后固定在4 ℃条件下于冰冷的70%乙醇1 h。碘化丙啶(PI,50 μg/mL,Sigma-Aldrich)和核糖核酸酶A(100 μg/mL,生工生物科技)添加到细胞培养液中,然后在37 ℃、暗箱中培养20 min。根据说明书,采用FACSCalibur(BD Biosciences)测定细胞周期时相。所有实验重复3次。
1.2.5. 流式细胞仪检测细胞凋亡分析
转染后24 h,用膜联蛋白V凋亡检测试剂盒对膜联蛋白V染色(eBioscience)观察细胞凋亡情况后流式细胞仪分析。每个实验重复3次。
1.2.6. 迁移实验
Hep-2细胞种植于6孔板上,每孔1×106细胞,37 ℃培养24 h。当达到80%~90%融合时,使用shmalat-1或shCtrl慢病毒转染细胞抑制其增殖。5 μm的丝裂霉素C(Sigma Aldrich)在转染后24 h加入细胞,培养2 h,然后用塑料无菌过滤头在单层表面进行划痕实验。单层细胞用PBS冲洗3次,用0.5%胎牛血清的DMEM置于37 ℃和5% CO2培养。倒置显微镜用于0、8、24 h后观察测量细胞迁移的平均迁移距离。计算如下:原来的差距距离–间隔24 h/初始间隙距离×100%。
1.2.7. Transwell细胞迁移实验
采用Transwell小室检测Hep-2细胞的迁移。肿瘤细胞侵袭的膜是使用60 μL Matrigel基质胶(Corning)和120 μL DMEM制作,然后放置于Transwell小室的上室。在37 ℃孵育30 min,shmalat1或shCtrl转染细胞(1×106/mL)加入Transwell小室上室的Matrigel基质胶顶部。含30% FBS的趋化液600 μL加入Transwell小室下室,在37 ℃、5% CO2的无菌培养箱中孵育24 h。基质胶和迁移的细胞轻轻地从Transwell小室中取出,用Giemsa染色(Sigma-Aldrich, 3~5 min)。将小室置于光学显微镜下观察(Olympus XI-71)。随机选取5个视野,然后计数每个中倍视野中的细胞数,然后取平均数。
1.2.8. Transwell细胞侵袭试验
为了评估shmalat1和shCtrl慢病毒转染的Hep-2细胞的侵袭能力,使用Transwell细胞侵袭试验。首先合成Matrigel基质胶,500 μL的无血清培养基中加入24孔板的各孔37 ℃、孵育2 h。大约1×104 Hep-2细胞稀释于500 μL无血清培养基(Corning)后加入Matrigel侵袭小室,然后在下室中加入一个包含750 μL DMEM高糖培养基(含30%胎牛血清)的24孔板。在37 ℃、5% CO2环境培养24 h后,移去表面的顶细胞和Matrigel基质胶。100 μL的Giemsa染色液加入膜的底面,孵育3~5 min。将小室置于光学显微镜下观察(Olympus XI-71),随机选取5个视野,然后计数每个中倍视野中的细胞数,然后取平均数。
1.2.9. 统计分析
使用GraphPad PRISM(6.0.5)软件进行数据分析。正态分布的计量资料均以均数±标准差表示。采用t检验和单因素方差分析进行组间比较。以P < 0.05为差异有统计学意义。
2. 结果
2.1. FaDu、Hep-2和鼻咽癌CNE-2Z细胞MALAT1的表达以及敲除MALAT-1对Hep-2的影响
使用RT-PCR检测FaDu、Hep-2和鼻咽癌CNE-2Z细胞MALAT1的表达,结果发现与NP-69细胞相比,FaDu、Hep-2和鼻咽癌CNE-2Z细胞MALAT1的表达显著增加(P < 0.01,图 1A)。此外,在敲除MALAT1的细胞,其细胞生长与对照细胞相比没有受到影响(图 1B)。使用shmalat1慢病毒转染Hep-2细胞,其MALAT1表达减少91.5%(P < 0.01,图 1C)。因此shmalat1慢病毒有效地影响MALAT1的表达。敲除MALAT1抑制Hep-2细胞增殖(为了验证shmalat1敲除的抑制作用,使用MTT法评估Hep-2细胞的增殖)。与对照细胞相比,敲除MALAT1的Hep-2细胞接种后第4和第5天明显受到抑制(图 1D)。
1.
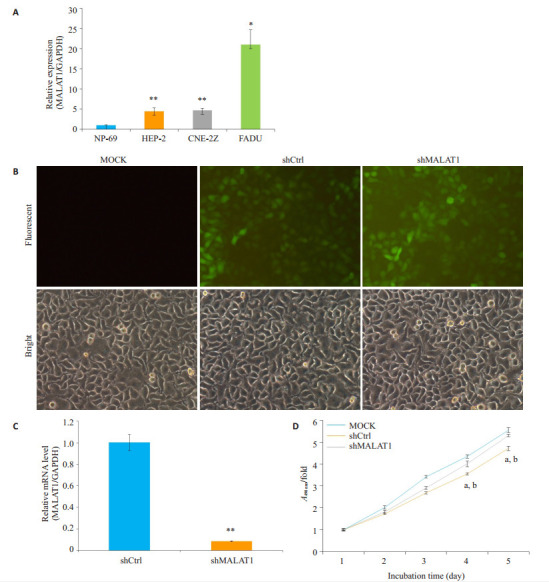
MALAT1基因在人喉鳞状细胞癌细胞系中的表达及其敲除对Hep-2细胞增殖的影响
Expression of MALAT1 in HNSCC cell lines and effect of MALAT1 knockdown on Hep- 2 cells. A: Expression of MALAT1 in human malignant HNSCC cell lines FaDu, Hep-2 and CNE-2Z, and NPE cell line NP-69. **P < 0.01 vs NP-69; *P < 0.05 vs NP-69; B: Efficiency of MALAT1 knockdown in Hep-2 cells; C: Representative images of Hep-2 cells transfected with ShMALAT1, MOCK or shCtrl lentivirus. **P < 0.01 vs shCtrl. D: Results of MTT assay. aP < 0.01 vs MOCK; bP < 0.01 vs shCtrl
2.2. Hep-2细胞MALAT-1下调对细胞周期进程的影响
Hep-2细胞MALAT-1下调导致细胞周期阻滞,为了确定MALAT1在细胞周期进程的重要性,我们通过流式细胞仪评估细胞周期各时相的Hep-2细胞(图 2A)。与MOCK和shCtrl细胞相比,MALAT1-shRNA慢病毒转染细胞S期细胞数量显著增加(P < 0.01,图 2B)。此外,细胞在G2/M期的百分比下降(P < 0.01)。
2.
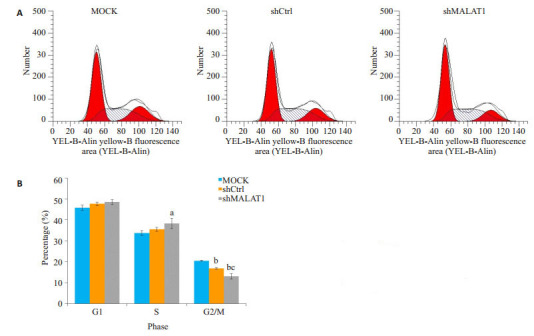
敲除MALAT1导致细胞周期阻滞于S期
MALAT1 knockdown causes cell cycle arrest in S phase. A: Flow cytometry analysis of cell cycle phases of Hep-2 cells; B: Proportions of cells in the G1, S and G2/M phases. aP < 0.05 vs MOCK, bP < 0.01 vs MOCK, cP < 0.01 vs shCtrl
2.3. 敲除MALAT1对Hep-2细胞克隆形成的抑制作用
经克隆形成实验测定,敲除MALAT1抑制Hep-2细胞克隆的能力。敲除MALAT1的Hep-2细胞克隆能力明显受损,下降约7.5%。shCtrl慢病毒处理过的细胞中观察到了平均124个,而在MALAT1敲除细胞观察到115个克隆细胞(图 3A、B)。
3.
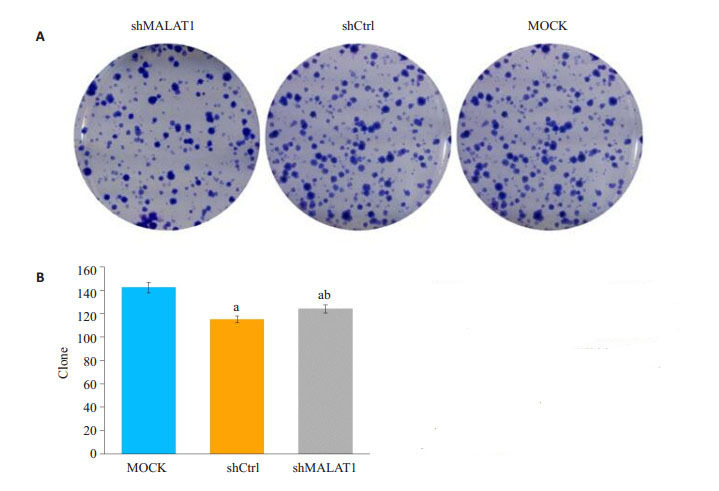
敲除MALAT1不影响人喉鳞状细胞癌Hep-2细胞集落形成
Knockdown of MALAT1 does not affect colony formation ability of human malignant LSCC Hep- 2 cells. A: Representative images of colony formation by Hep-2 cells; B: Colony formation by Hep-2 cells treated with empty lentivirus (MOCK), shCtrl and shMALAT1. aP < 0.01 vs MOCK, bP < 0.05 vs shCtrl
2.4. 流式细胞仪进行凋亡定量分析
MALAT1的低表达诱使Hep- 2细胞凋亡,经shmalat1转染的Hep-2细胞凋亡程度由Annexin v-apc染色和流式细胞仪检测(图 4)。如图 4A和4B所示,用流式细胞仪进行凋亡定量分析表明,与未转染的MOCK细胞或shCtrl细胞相比,敲除MALAT1的Hep-2细胞,其细胞凋亡明显加快(P < 0.01)。
4.
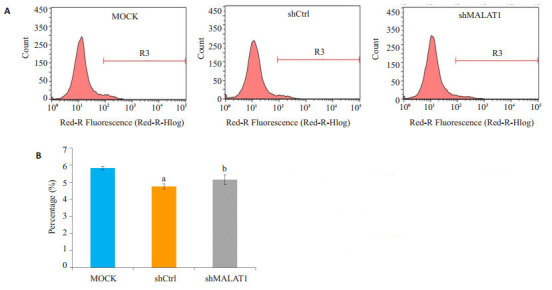
敲除MALAT1诱导人喉鳞状细胞癌Hep-2细胞凋亡
MALAT1 knockdown induces apoptosis in human malignant LSCC Hep-2 cells by Annexin-V staining and flow cytometric analysis. A: Representative images of cell apoptosis; B: Results of the flow cytometric analysis data, aP < 0.01 vs MOCK, bP < 0.05 vs MOCK
2.5. Transwell侵袭实验测定Hep-2细胞的侵袭迁移能力
MALAT1表达减少,抑制Hep-2细胞的迁移和侵袭,Hep-2细胞的侵袭迁移能力采用Transwell侵袭实验测定。Transwell迁移结果表明,迁移后24 h,与MOCK细胞和shCtrl细胞相比,细胞数量显著减少(P < 0.01,图 5A、C)。此外,在24 h的Matrigel侵袭实验中,与未转染的MOCK细胞和shCtrl转染细胞相比,敲除MALAT1侵袭Hep-2细胞数显著降低(图 5B、D)。
5.
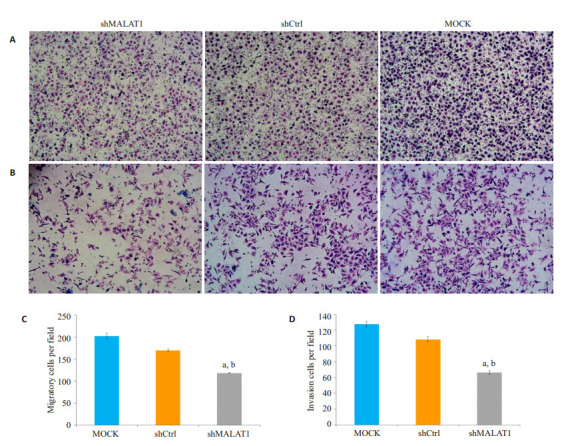
敲除MALAT1抑制Hep-2细胞的迁移和侵袭
MALAT1 knockdown inhibits migration and invasion of Hep-2 cells by Transwell and invasion assays. A: Representative images of a Transwell assay after 24 h of migration; B: Quantification of the number of migrated cells, aP < 0.01 vs MOCK, bP < 0.01 vs shCtrl; C: Representative images of an Invasion assay after 24h of migration; D: Quantification of the number of invaded cells, aP < 0.01 vs MOCK, bP < 0.01 vs shCtrl
2.6. 划痕实验评估经shmalat1或shCtrl慢病毒转染的Hep-2细胞的迁移力
为了验证以上结果,使用划痕实验评估经shMALAT1或shCtrl慢病毒转染的Hep-2细胞的迁移力,经过24 h迁移过程后。MOCK-treated细胞划痕修复了(29±3.0)%,shCtrl-treated细胞修复了(24±3.0)%。然而,在细胞中慢病毒转染shMALAT1划痕收窄只有(14±0.2)%(P < 0.01,图 6A、B)。
6.
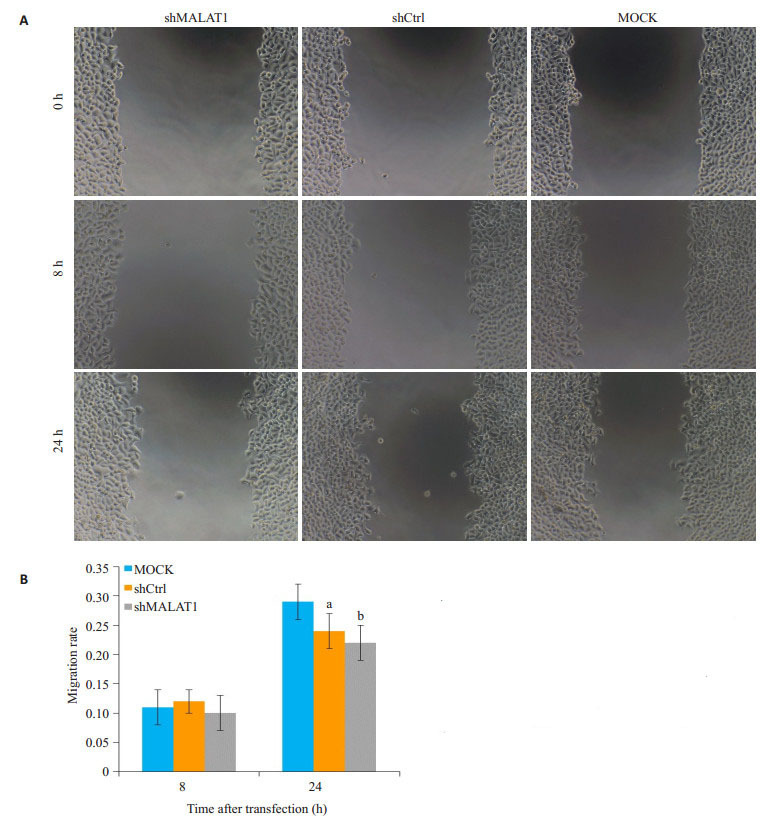
敲除MALAT1抑制Hep-2细胞的迁移
MALAT1 knockdown inhibits migration of Hep-2 cells by wound healing assay. A: Representative images of cell migration; B: Quantification of the migration rate in MOCK, shCtrl and shMALAT1-transfected cells based on the images shown in A, aP < 0.05 vs MOCK, bP < 0.01 vs MOCK
3. 讨论
随着学者对lncRNAs研究的不断深入,其在疾病发生发展中的作用被逐步揭示,但是其在细胞的各种功能还没有被广泛的研究[21-22]。异常表达的lncRNA与各种疾病,包括癌症的发生密切相关[23-25]。lncRNAMALAT1最初发现于肺转移性癌,之前文献报道,其他头颈部鳞癌细胞系,如鼻咽癌、Hone-1,5-8F和SUNE-1,其MALAT1表达上调[26],但其在喉鳞状细胞癌的作用仍然知之甚少[15]。为了探讨MALAT-1在头颈部鳞癌,尤其是喉癌的调节功能,我们比较了在头颈部鳞癌细胞MALAT1表达与NPE的细胞系MALAT1的表达。实验数据显示,相比于NP-69细胞,MALAT1在FaDu,Hep-2和CNE-2Z三种细胞系的表达明显上调。为了进一步证明,我们通过慢病毒转染细胞的方法沉默MALAT1,通过流式细胞仪评估细胞周期各时相的Hep-2细胞。与MOCK和shCtrl细胞相比,MALAT1- shRNA慢病毒转染细胞S期细胞数量显著增加,由于失去MALAT1,细胞阻滞在S期,并促进Hep-2细胞凋亡。我们的数据显示,MALAT1喉癌的发病机制中起着重要作用。
MALAT1最初在高转移性肺癌细胞发现[15]后来被证实在肿瘤细胞侵袭和转移中起关键作用,如胰腺癌[27]和黑色素瘤[28]。我们通过Transwell小室迁移实验、Transwell小室侵袭实验测定Hep-2细胞的侵袭迁移能力,结果显示敲除MALAT1的Hep-2细胞迁移和侵袭均受损。进一步验证,通过划痕实验评估经shmalat1或shCtrl慢病毒转染的Hep-2细胞的迁移力,发现MOCKtreated细胞划痕修复了(29±3.0)%,shCtrl-treated细胞修复了(24±3.0)%,shmalat1划痕收窄只有(14±0.2)%(P < 0.01)。表明,MALAT1的低表达干扰抑制Hep-2细胞的迁移。这与以往的研究结果是一致的,例如,Tano等[29]研究表明MALAT1沉默影响细胞迁移相关基因的表达情况,包括CTHRC1,CCT4,HMMR和ROD1。然而,MALAT1在细胞迁移和侵袭的分子机制知之甚少,有待进一步研究。
目前研究认为,MALAT1参与了细胞的调节增殖、细胞周期和凋亡[26, 30]。Yamada等[17]以前研究表明,与分泌期子宫内膜间质细胞比较,MALAT1在正常子宫内膜基质细胞和子宫内膜间质肉瘤表达水平均增高。这些结果表明,MALAT1可能在细胞增殖发挥重要作用,在本研究中,我们发现敲除MALAT-1降低Hep-2细胞增殖率。这表明,MALAT1可能在喉鳞状细胞癌细胞增殖中起重要作用。我们进一步的研究表明MALAT-1在表达下降诱导Hep-2细胞的细胞周期阻滞。Yang等[30]报道,与hnRNP C相互作用下调HeLa细胞和HepG2细胞MALAT1水平会诱导其阻滞在G2/M期。在细胞周期中,MALAT1的表达差异可以归因于不同的细胞类型的使用,在今后的研究中需要进一步研究和评估。我们发现,缺失MALAT1促进喉癌Hep-2细胞凋亡,这是与先前在各种癌细胞研究中发现的结果相一致[27, 31-32]。我们发现MALAT1在头颈部鳞癌细胞中的表达增高,而缺失MALAT1抑制增殖,诱导S期阻滞,促进细胞凋亡,并抑制Hep-2细胞的迁移和侵袭。总之,这些结果均显示MALAT1在喉鳞状细胞癌的增殖和迁移起关键作用,同时MALAT1表达增加,参与喉癌的发病机制。
Funding Statement
安徽省教育厅重点项目(KJ2015A284)
References
- 1.Jain P, Kumar P, Pai VR, et al. Neoadjuvant chemotherapy or chemoradiotherapy in head and neck cancer. Indian J Cancer. 2008;45(3):83–9. doi: 10.4103/0019-509X.44061. [Jain P, Kumar P, Pai VR, et al. Neoadjuvant chemotherapy or chemoradiotherapy in head and neck cancer[J]. Indian J Cancer, 2008, 45(3): 83-9.] [DOI] [PubMed] [Google Scholar]
- 2.Dwivedi R, Raturi D, Kandpal N, et al. Oxidative stress in patients with laryngeal carcinoma. Indian J Cancer. 2008;45(3):97–9. doi: 10.4103/0019-509X.44064. [Dwivedi R, Raturi D, Kandpal N, et al. Oxidative stress in patients with laryngeal carcinoma[J]. Indian J Cancer, 2008, 45(3): 97-9.] [DOI] [PubMed] [Google Scholar]
- 3.Yang M, Li W, Liu YY, et al. Promoter hypermethylation-induced transcriptional down-regulation of the gene MYCT1 in laryngeal squamous cell carcinoma. BMC Cancer. 2012;12(1):219. doi: 10.1186/1471-2407-12-219. [Yang M, Li W, Liu YY, et al. Promoter hypermethylation-induced transcriptional down-regulation of the gene MYCT1 in laryngeal squamous cell carcinoma[J]. BMC Cancer, 2012, 12(1): 219.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Micozkadioğlu D, Unal M, Pata YS, et al. Prognostic value of expression of p53, proliferating cell nuclear antigen, and c-erbB-2 in laryngeal carcinoma. http://www.ncbi.nlm.nih.gov/pubmed/%20%20%20%20%20%2018509272. Med Sci Monit. 2008;14(6):CR299–304. [Micozkadioğlu D, Unal M, Pata YS, et al. Prognostic value of expression of p53, proliferating cell nuclear antigen, and c-erbB-2 in laryngeal carcinoma[J]. Med Sci Monit, 2008, 14(6): CR299- 304.] [PubMed] [Google Scholar]
- 5.Che XH, Chen H, Xu ZM, et al. 14-3-3epsilon contributes to tumour suppression in laryngeal carcinoma by affecting apoptosis and invasion. BMC Cancer. 2010;10(1):306. doi: 10.1186/1471-2407-10-306. [Che XH, Chen H, Xu ZM, et al. 14-3-3epsilon contributes to tumour suppression in laryngeal carcinoma by affecting apoptosis and invasion[J]. BMC Cancer, 2010, 10(1): 306.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tu XP, Qiu QH, Chen LS, et al. Preoperative neutrophil-tolymphocyte ratio is an Independent prognostic marker in patients with laryngeal squamous cell carcinoma. BMC Cancer. 2015;15(1):743. doi: 10.1186/s12885-015-1727-6. [Tu XP, Qiu QH, Chen LS, et al. Preoperative neutrophil-tolymphocyte ratio is an Independent prognostic marker in patients with laryngeal squamous cell carcinoma[J]. BMC Cancer, 2015, 15 (1): 743.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yilmaz M, Karatas OF, Yuceturk B, et al. Alpha-B-crystallin expression in human laryngeal squamous cell carcinoma tissues. Head Neck. 2015;37(9):1344–8. doi: 10.1002/hed.v37.9. [Yilmaz M, Karatas OF, Yuceturk B, et al. Alpha-B-crystallin expression in human laryngeal squamous cell carcinoma tissues[J]. Head Neck, 2015, 37(9): 1344-8.] [DOI] [PubMed] [Google Scholar]
- 8.Bhan A, Mandal SS. Long noncoding RNAs: emerging stars in gene regulation, epigenetics and human disease. Chem Med Chem. 2014;9(9, SI):1932–56. doi: 10.1002/cmdc.201300534. [Bhan A, Mandal SS. Long noncoding RNAs: emerging stars in gene regulation, epigenetics and human disease[J]. Chem Med Chem, 2014, 9(9, SI): 1932-56.] [DOI] [PubMed] [Google Scholar]
- 9.Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21(6):354–61. doi: 10.1016/j.tcb.2011.04.001. [Wapinski O, Chang HY. Long noncoding RNAs and human disease [J]. Trends Cell Biol, 2011, 21(6): 354-61.] [DOI] [PubMed] [Google Scholar]
- 10.Liu MX, Chen X, Chen G, et al. A computational framework to infer human disease-associated long noncoding RNAs. PLoS One. 2014;9(1):e84408. doi: 10.1371/journal.pone.0084408. [Liu MX, Chen X, Chen G, et al. A computational framework to infer human disease-associated long noncoding RNAs[J]. PLoS One, 2014, 9(1): e84408.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diederichs S. The four dimensions of noncoding RNA conservation. Trends Genet. 2014;30(4):121–3. doi: 10.1016/j.tig.2014.01.004. [Diederichs S. The four dimensions of noncoding RNA conservation [J]. Trends Genet, 2014, 30(4): 121-3.] [DOI] [PubMed] [Google Scholar]
- 12.Xia H, Chen Q, Chen Y, et al. The lncRNA MALAT1 is a novel biomarker for gastric cancer metastasis. http://pubmedcentralcanada.ca/pmcc/articles/PMC5302908/ Oncotarget. 2016;7(35):56209–18. doi: 10.18632/oncotarget.10941. [Xia H, Chen Q, Chen Y, et al. The lncRNA MALAT1 is a novel biomarker for gastric cancer metastasis[J]. Oncotarget, 2016, 7 (35): 56209-18.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gutschner T, Hämmerle M, Diederichs S. MALAT1 -- a paradigm for long noncoding RNA function in cancer. J Mol Med (Berl) 2013;91(7):791–801. doi: 10.1007/s00109-013-1028-y. [Gutschner T, Hämmerle M, Diederichs S. MALAT1 -- a paradigm for long noncoding RNA function in cancer[J]. J Mol Med (Berl), 2013, 91(7): 791-801.] [DOI] [PubMed] [Google Scholar]
- 14.He X, He Y, Xi B, et al. LncRNAs expression in preeclampsia placenta reveals the potential role of LncRNAs contributing to preeclampsia pathogenesis. PLoS One. 2013;8(11):e81437. doi: 10.1371/journal.pone.0081437. [He X, He Y, Xi B, et al. LncRNAs expression in preeclampsia placenta reveals the potential role of LncRNAs contributing to preeclampsia pathogenesis[J]. PLoS One, 2013, 8(11): e81437.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ji P, Diederichs S, Wang WB, et al. MALAT-1, a novel noncoding RNA, and thymosin beta 4 predict metastasis and survival in earlystage non-small cell lung cancer. Oncogene. 2003;22(39):8031–41. doi: 10.1038/sj.onc.1206928. [Ji P, Diederichs S, Wang WB, et al. MALAT-1, a novel noncoding RNA, and thymosin beta 4 predict metastasis and survival in earlystage non-small cell lung cancer[J]. Oncogene, 2003, 22(39): 8031- 41.] [DOI] [PubMed] [Google Scholar]
- 16.Bernard D, Prasanth KV, Tripathi V, et al. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J. 2010;29(18):3082–93. doi: 10.1038/emboj.2010.199. [Bernard D, Prasanth KV, Tripathi V, et al. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression[J]. EMBO J, 2010, 29(18): 3082-93.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamada K, Kano J, Tsunoda H, et al. Phenotypic characterization of endometrial stromal sarcoma of the uterus. Cancer Sci. 2006;97(2):106–12. doi: 10.1111/cas.2006.97.issue-2. [Yamada K, Kano J, Tsunoda H, et al. Phenotypic characterization of endometrial stromal sarcoma of the uterus[J]. Cancer Sci, 2006, 97 (2): 106-12.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin R, Maeda S, Liu C, et al. A large noncoding RNA is a marker for murine hepatocellular carcinomas and a spectrum of human carcinomas. Oncogene. 2007;26(6):851–8. doi: 10.1038/sj.onc.1209846. [Lin R, Maeda S, Liu C, et al. A large noncoding RNA is a marker for murine hepatocellular carcinomas and a spectrum of human carcinomas[J]. Oncogene, 2007, 26(6): 851-8.] [DOI] [PubMed] [Google Scholar]
- 19.Yoshimoto R, Mayeda A, Yoshida M, et al. MALAT1 long noncoding RNA in cancer. Biochim Biophys Acta. 2016;1859(1):192–9. doi: 10.1016/j.bbagrm.2015.09.012. [Yoshimoto R, Mayeda A, Yoshida M, et al. MALAT1 long noncoding RNA in cancer[J]. Biochim Biophys Acta, 2016, 1859(1): 192-9.] [DOI] [PubMed] [Google Scholar]
- 20.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29(9):e45. doi: 10.1093/nar/29.9.e45. [Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR[J]. Nucleic Acids Res, 2001, 29(9): e45.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bonasio R, Shiekhattar R. Regulation of transcription by long noncoding RNAs. http://d.old.wanfangdata.com.cn/OAPaper/oai_pubmedcentral.nih.gov_3858397. Annual review of genetics. 1994;48:51–2. doi: 10.1146/annurev-genet-120213-092323. [Bonasio R and Shiekhattar R.Regulation of transcription by long noncoding RNAs. Annual review of genetics, 1994, 48: 51-2.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Quinodoz S, Guttman M. Long noncoding RNAs: an emerging Link between gene regulation and nuclear organization. Trends Cell Biol. 2014;24(11):651–63. doi: 10.1016/j.tcb.2014.08.009. [Quinodoz S, Guttman M. Long noncoding RNAs: an emerging Link between gene regulation and nuclear organization[J]. Trends Cell Biol, 2014, 24(11): 651-63.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen LL, Zhao JC. Functional analysis of long noncoding RNAs in development and disease. Adv Exp Med Biol. 2014;825:129–58. doi: 10.1007/978-1-4939-1221-6. [Chen LL, Zhao JC. Functional analysis of long noncoding RNAs in development and disease[J]. Adv Exp Med Biol, 2014, 825: 129- 58.] [DOI] [PubMed] [Google Scholar]
- 24.Hajjari M, Khoshnevisan A, Shin YK. Molecular function and regulation of long non-coding RNAs: paradigms with potential roles in cancer. Tumour Biol. 2014;35(11):10645–63. doi: 10.1007/s13277-014-2636-z. [Hajjari M, Khoshnevisan A, Shin YK. Molecular function and regulation of long non-coding RNAs: paradigms with potential roles in cancer[J]. Tumour Biol, 2014, 35(11): 10645-63.] [DOI] [PubMed] [Google Scholar]
- 25.Gupta RA, Shah N, Wang KC, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464(7291):1071–6. doi: 10.1038/nature08975. [Gupta RA, Shah N, Wang KC, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis [J]. Nature, 2010, 464(7291): 1071-6.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin C, Yan BC, Lu Q, et al. The role of MALAT1/miR-1/slug axis on radioresistance in nasopharyngeal carcinoma. Tumor Biol. 2016;37(3):4025–33. doi: 10.1007/s13277-015-4227-z. [Jin C, Yan BC, Lu Q, et al. The role of MALAT1/miR-1/slug axis on radioresistance in nasopharyngeal carcinoma[J]. Tumor Biol, 2016, 37(3): 4025-33.] [DOI] [PubMed] [Google Scholar]
- 27.Jiao F, Hu H, Yuan CC, et al. Elevated expression level of long noncoding RNA MALAT-1 facilitates cell growth, migration and invasion in pancreatic cancer. Oncol Rep. 2014;32(6):2485–92. doi: 10.3892/or.2014.3518. [Jiao F, Hu H, Yuan CC, et al. Elevated expression level of long noncoding RNA MALAT-1 facilitates cell growth, migration and invasion in pancreatic cancer[J]. Oncol Rep, 2014, 32(6): 2485-92.] [DOI] [PubMed] [Google Scholar]
- 28.Tian YJ, Zhang XY, Hao YH, et al. Potential roles of abnormally expressed long noncoding RNA UCA1 and Malat-1 in metastasis of melanoma. Melanoma Res. 2014;24(4):335–41. doi: 10.1097/CMR.0000000000000080. [Tian YJ, Zhang XY, Hao YH, et al. Potential roles of abnormally expressed long noncoding RNA UCA1 and Malat-1 in metastasis of melanoma[J]. Melanoma Res, 2014, 24(4): 335-41.] [DOI] [PubMed] [Google Scholar]
- 29.Tano K, Mizuno R, Okada T, et al. MALAT-1 enhances cell motility of lung adenocarcinoma cells by influencing the expression of motility-related genes. FEBS Lett. 2010;584(22):4575–80. doi: 10.1016/j.febslet.2010.10.008. [Tano K, Mizuno R, Okada T, et al. MALAT-1 enhances cell motility of lung adenocarcinoma cells by influencing the expression of motility-related genes[J]. FEBS Lett, 2010, 584(22): 4575-80.] [DOI] [PubMed] [Google Scholar]
- 30.Yang F, Yi F, Han X, et al. MALAT-1 interacts with hnRNP C in cell cycle regulation. FEBS Lett. 2013;587(19):3175–81. doi: 10.1016/j.febslet.2013.07.048. [Yang F, Yi F, Han X, et al. MALAT-1 interacts with hnRNP C in cell cycle regulation[J]. FEBS Lett, 2013, 587(19): 3175-81.] [DOI] [PubMed] [Google Scholar]
- 31.Lai MC, Yang Z, Zhou L, et al. Long non-coding RNA MALAT-1 overexpression predicts tumor recurrence of hepatocellular carcinoma after liver transplantation. Med Oncol. 2012;29(3):1810–6. doi: 10.1007/s12032-011-0004-z. [Lai MC, Yang Z, Zhou L, et al. Long non-coding RNA MALAT-1 overexpression predicts tumor recurrence of hepatocellular carcinoma after liver transplantation[J]. Med Oncol, 2012, 29(3): 1810-6.] [DOI] [PubMed] [Google Scholar]
- 32.Fang D, Yang H, Lin J, et al. 17β-estradiol regulates cell proliferation, colony formation, migration, invasion and promotes apoptosis by upregulating miR-9 and thus degrades MALAT-1 in osteosarcoma cell MG-63 in an estrogen receptor-independent manner. Biochem Biophys Res Commun. 2015;457(4):500–6. doi: 10.1016/j.bbrc.2014.12.114. [Fang D, Yang H, Lin J, et al. 17β-estradiol regulates cell proliferation, colony formation, migration, invasion and promotes apoptosis by upregulating miR-9 and thus degrades MALAT-1 in osteosarcoma cell MG-63 in an estrogen receptor-independent manner[J]. Biochem Biophys Res Commun, 2015, 457(4): 500-6.] [DOI] [PubMed] [Google Scholar]


