Abstract
目的
研究长链非编码RNA linc00261在肝细胞癌(HCC)中的表达、功能及其临床意义。
方法
采用实时荧光定量PCR (qRT-PCR)法检测linc00261在74例肝癌、癌旁肝组织中的表达;采用卡方检验分析其表达量与临床病理指标的相关性;应用COX回归模型分析其表达水平与肝癌患者术后预后的关系;qRT-PCR检测linc00261在5株肝癌细胞株中的表达;在肝癌细胞株MHCC-LM3和SNU-449中采用siRNA(si-linc00261-1,si-linc00261-2,si-NC和空白对照组)和Lipofectamine3000进行转染敲低linc00261,活细胞计数(CCK8)、Transwell实验分别研究linc00261对肝癌细胞增殖,侵袭迁移能力的影响。
结果
HCC组织中linc00261的表达水平与肝癌患者术前血清AFP(P=0.032)、肿瘤大小(P=0.007)、有无微血管癌栓(P=0.01)、TNM分期(P= 0.02)显著相关;HCC组织中linc00261的低表达与患者的不良预后相关,其术后无瘤生存期明显缩短(P<0.05),是影响HCC患者术后无瘤生存期的一个独立危险因素;下调linc00261的表达可促进肝癌细胞株MHCC-LM3(P<0.01)和SNU-449(P<0.001)的迁移和侵袭能力,对细胞增殖无明显影响(P>0.05)。
结论
Linc00261有望成为肝癌切除术后的一种新的预后指标。
Keywords: linc00261, 肝细胞癌, 增殖, 迁移, 侵袭
Abstract
Objective
To investigate the expression of long noncoding RNA linc00261 in hepatocellular carcinoma (HCC) and its correlation with the clinicopathological features and postoperative outcomes of the patients.
Methods
Real-time fluorescence quantitative PCR (qRT-PCR) was used to detect the expression of linc00261 in surgical specimens of HCC and adjacent tissues from 74 patients. The correlation of the expression level of linc00261 in HCC with the clinicopathological parameters of the patients was analyzed using Chi-square test. The Cox's proportional hazards regression model was used to assess the value of linc00261 in predicting the prognosis of HCC patients after operation. The expression of linc00261 was also examined in 5 HCC cell lines using qRT-PCR. The HCC cell lines MHCC-LM3 and SNU-449 were transfected with small interfering RNAs targeting linc00261 for linc00261 knockdown, and the changes in the cell proliferation, migration and invasion abilities were observed using CCK-8 assay and Transwell assay.
Results
The expression level of linc00261 in HCC was significantly correlated with AFP (P=0.032), tumor size (P=0.007), microscopic vascular invasion (MVI; P=0.01), and TNM stage (P=002). The patients with lowered expressions of linc00261 in HCC tissues had a significantly shortened tumor-free survival time (P < 0.05), and a lowered expression of linc00261 was identified as an independent risk factor affecting postoperative recurrence-free survival time of the patients (P < 0.05). In HCC cell lines MHCC-LM3 and SNU-449 cells, linc00261 knockdown obviously promoted the cell migration and invasion (P < 0.01) but did not significantly affect cell proliferation (P > 0.05).
Conclusion
Linc00261 may serve as a new prognostic biomarker for predicting the postoperative outcomes of the patients with HCC.
Keywords: linc00261, hepatocellular carcinoma, proliferation, migration, invasion
原发性肝癌是世界上最为常见的肿瘤之一,在我国的恶性肿瘤中发病率和死亡率分别位居第4和第2位[1],严重危害人类健康,其中绝对大多数是肝细胞癌(HCC)[2]。HCC的发生发展是一多基因、多步骤的过程。乙型肝炎病毒感染是肝癌的主要原因之一,但其具体机制仍不清楚。在HCC的治疗方面,传统的放、化疗毒副作用较大,疗效欠佳;手术切除率低下,且复发转移率高[3]。因此,探究乙肝相关性肝癌的发生机制具有重要的价值。
长链非编码RNA(lncRNA)是一类长度超过200个核苷酸的RNA分子,可定位于细胞核或细胞质中,曾一度被认为是基因组的“噪音”[4-5]。随着深入的研究发现,lncRNA可在转录、转录后以及表观遗传学水平上参与基因的调控[6]。LncRNA在肿瘤发生发展过程中,可起到促癌或抑癌的作用。Linc00261是位于20p11.21上的基因间lncRNA,目前实验研究表明linc00261在多种肿瘤中起到抑癌基因的作用。Yu等[7]研究发现linc00261在胃癌组织中的表达水平明显低于癌旁组织,并可通过抑制Slug蛋白表达水平,从而抑制胃癌的上皮细胞-间充质转化(EMT)进程。过表达linc00261可促进结肠癌细胞凋亡,增加对化疗药物顺铂的敏感性[8]。在人绒毛膜癌[9]和人类子宫内膜细胞系中[10],过表达linc00261可抑制细胞的侵袭和迁移能力。刘月[11]等通过RNA测序技术发现,linc00261在Barrett食管和食管腺癌组织中的表达水平明显高于正常食管磷状上皮组织,linc00261可负向调控其临位基因FOXA2的表达水平,从而促进Barrett食管的发生发展,但两者间的分子机制有待进一步研究。Linc00261在喉癌[12]和非小细胞肺癌[13]的癌组织中的表达水平明显低于癌旁组织,与临床病理参数进行相关性分析发现低表达linc00261提示患者的不良预后。而目前linc00261在HCC中的功能和作用未见详细报道。致力于研究探讨linc00261在HCC中的作用机制,可为HCC的诊疗提供一定的理论基础。
本研究拟采用实时荧光定量PCR检测linc00261在74例HCC及癌旁组织中的表达,并统计分析其表达与患者临床病理参数及术后无瘤生存时间的关系,以及检测沉默linc00261后对肝癌细胞株增殖,迁移和侵袭能力的变化,从而研究linc00261在HCC中的表达情况及其在HCC发生发展过程中所起的作用。
1. 材料和方法
1.1. 组织样本
选取2010年3月~2015年3月间在南方医科大学附属南方医院肝胆外科行根治性肝癌切除术并满足以下纳入标准的HCC患者74例。纳入标准为:(1)术前无其他治疗,如肝动脉栓塞化疗、射频消融治疗和免疫治疗等;(2)血清学检查提示乙肝表面抗原(+),但无丙肝感染;(3)术后病理学检查证实为原发性肝脏细胞癌;(4)切缘无癌残留;(5)临床资料完整。取肝癌组织及相应癌旁组织(距肿瘤边缘2 cm以上且经病理证实无癌细胞残余)于液氮中保存备用。
1.2. 随访
患者术后定期通过门诊复诊和电话随访的形式随访。复发定义为:术后经两种影像学检查证实存在肝癌特征性的占位性病变,或一种影像学检查提示存在肝癌特征性占位性病变,同时伴有甲胎蛋白水平(AFP)持续性增高(排除妊娠、生殖系胚胎源性肿瘤及活动性肝炎)[14]。术后无瘤生存时间定义为手术治疗日起至确诊复发或至末次随访时间所经历的时间,中位随访时间为30个月,随访内容包括:术后情况(进展、复发、死亡)、影像学检查变化、血清甲胎蛋白水平、肝功能等。
1.3. 主要试剂及仪器
RNAiso plus(9109)、逆转录试剂盒(RR047A)及qRT-PCR试剂盒(RR420)均购自Takara公司;Transwell小室、基质胶Matrigel (BD);ThermoND2000C超微量分光光度计(Thermo)、GeneAmp PCR system 9700扩增仪(PerkinElmer)、LightCycler®480 IISystem(Roche)。
1.4. 方法
1.4.1. 肝癌细胞的培养
人肝癌细胞株SNU-449和BEL-7402使用含10%胎牛血清和1%双抗的RPMI 1640培养基培养,人肝癌细胞株MHCC-LM3、SK-hep1和HepG2使用含10%胎牛血清和1%双抗的DMEM高糖培养基培养,于37 ℃、5%CO2温育箱中培养细胞,取生长状态良好的细胞用于试验。
1.4.2. Lipofectamine 3000法转染肝癌细胞株
取对数生长期的MHCC-LM3和SNU-449细胞接种于6孔板,细胞浓度为5×105/孔。当细胞生长至汇合度70%~80%时,按照Lipofectamine 3000说明书中的方法将各组SiRNA分别转染至MHCC-LM3和SNU-449细胞株的Si-linc00261-1、Si-linc00261-2和Si-NC组中,并设立空白对照组(未加siRNA及转染试剂的Mock组),进行下一步相关实验。
1.4.3. CCK-8法检测转染siRNA后MHCC-LM3和SNU-449细胞的增殖能力
将转染SiRNA 24 h后的各组MHCC-LM3和SNU-449细胞按照5×103/孔分别接种于96孔板中,每1组设置3个复孔。将96孔板置于细胞培养箱中培养,分别在0、24、48、72和96 h后加入CCK-8(10 μL/孔),然后避光培养2h,用酶联免疫检测仪检测450 nm波长处的吸光度,吸光度值为纵坐标,处理时间为横坐标绘制细胞生长曲线。
1.4.4. Transwell试验检测细胞的迁移和侵袭能力选取
肝癌细胞株MHCC-LM3和SNU-449进行细胞迁移和侵袭试验。迁移实验:SiRNA转染24 h后,胰酶消化细胞、离心弃上清,用无血清的DMEM培养基重悬细胞,以1×105/100 μL的细胞悬液接种到上室,下室加500 μL含10%FBS的DMEM培养液,48 h后取出小室,弃上室培养基,用棉签拭掉上室的细胞,4%多聚甲醛固定细胞15 min,结晶紫染色10 min,PBS漂洗3次,镜检穿出小室的细胞数并拍照。侵袭试验:1:8稀释50 mg/L的Matrigel后,加50 μL稀释后的Matrigel包被Transwell小室底部膜,待其风干,其余试验步骤同迁移实验。
1.4.5. 总RNA提取及逆转录反应
从液氮中取出组织,取适量至1 mL RNAiso plus中研磨充分,加200 μL氯仿后,剧烈震荡15 s,室温静置10 mins,4 ℃离心(12 000 r/min,15 min);取上清并按等体积加入异丙醇,上下颠倒混匀,4 ℃离心后收集沉淀;加1 mL 75%乙醇洗涤后晾干;加DEPC水溶解至适宜浓度,采用分光光度计测量总RNA浓度,A260/280在1.8~2.0之间为合格样品。按Takara公司逆转录试剂盒RR036A说明书所述加入试剂,使用SureCycler 8800 PCR仪(Agilent Technologie)进行逆转录反应,逆转录成的cDNA,-20 ℃保存备用。
1.4.6. 实时荧光定量PCR检测linc00261的表达
引物序列由上海生工生物工程股份有限公司合成。Linc00261上游引物为:GTCAGAAGGAAAGGCC GTGA;下游引物为:TGAGCCGAGATGAACA GGTG,退火温度为60 ℃。18sRNA的上游引物为:GTAACCCGTTGAACCCCATT;下游引物为:CCA TCCAATCGGTAGTAGCG,退火温度为60 ℃,18sRNA作为内参。实时荧光定量PCR反应,反应条件:预变性95 ℃ 30 s,之后40个循环(95 ℃ 5 s,65 ℃ 30 s,95 ℃ 5 s)。质量控制:每个样本均在35个循环前到达平台期,复孔间CT值相差小于0.5,溶解曲线为单峰,无非特异性产物信号。结果分析采用2-ΔΔCT法进行基因表达的相对定量[15]。
1.5. 统计学方法
应用IBM SPSS20.0软件进行统计学分析。组间比较采用单因素方差分析,P<0.05认为差异有统计学意义。以linc00261在肝癌组织中的表达量的中位数作为分界点,将所有病例分为高表达组(大于中位数)及低表达组(小于中位数)。采χ2检验分析linc00261在肝癌组织中表达量与临床病理指标的相关性。采用Kaplan-Meier法比较linc00261高表达及低表达患者的术后无瘤生存时间的差异,log-rank法进行假设检验;采用COX比例风险模型评价linc00261的表达对患者术后无瘤生存时间的预测作用。以α=0.05为检验水准,采用双侧检验。
2. 结果
2.1. Linc00261在肝癌组织中表达水平
实时荧光定量PCR发现,癌组织中linc00261表达低于癌旁组织的患者占约43.24%(32/74,图 1)。以linc00261在74例肝癌组织中的相对表达量(2-△△Ct)的中位数为界,将患者分为低表达组(n=37)、高表达(n= 37)两组,并与患者临床病理参数构建四格表做χ2分析。结果显示:HCC组织中linc00261的相对表达水平与患者血清AFP水平(P=0.032)、肿瘤大小(P=0.007)、微癌栓(P=0.01)、TNM分期(P=0.02)显著相关,与性别、年龄、有无肝硬化、组织分化程度、肿瘤数量、有无包膜等不相关(表 1)。
1.
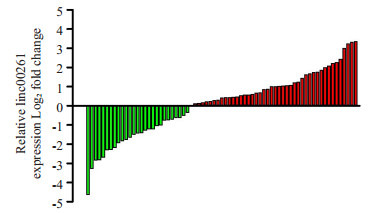
linc00261在肝细胞癌癌组织和非癌组织中的表达水平
Expression of linc00261 in surgical specimens of HCC and adjacent tissues.
1.
肝细胞癌中linc00261的相对表达水平与临床病理参数之间的关系
Correlation between linc00261 expression and clinicopathological characteristics of patients with hepatocellular carcinoma (n=74)
| Clinical parameter | Linc00261 | P | |
| High expression (n=37) | Low expression (n=37) | ||
| a: Portal vein tumor thrombosis; b: Micro-vascular invasion. | |||
| Gender | 0.207 | ||
| Male | 33 | 29 | |
| Female | 4 | 8 | |
| Age (year) | |||
| <60 | 30 | 30 | 1.000 |
| ≥60 | 7 | 7 | |
| AFP (μg/L) | 0.032 | ||
| <400 | 27 | 18 | |
| ≥400 | 10 | 19 | |
| Cirrhosis | 0.772 | ||
| Positive | 29 | 30 | |
| Negative | 8 | 7 | |
| Tumor number | 1.000 | ||
| =1 | 30 | 30 | |
| >1 | 7 | 7 | |
| Tumor size (cm) | 0.007 | ||
| ≤3 | 14 | 4 | |
| >3 | 23 | 33 | |
| Tumor capsule | 0.619 | ||
| Positive | 24 | 26 | |
| Negative | 13 | 11 | |
| Differentiation | 0.295 | ||
| Well | 7 | 5 | |
| Modrate | 26 | 23 | |
| Poor | 4 | 9 | |
| MVIa | 0.010 | ||
| Positive | 14 | 25 | |
| Negative | 23 | 12 | |
| PVTTb | 0.102 | ||
| Positive | 3 | 8 | |
| Negative | 34 | 29 | |
| TNM stage | 0.020 | ||
| Ⅰ+Ⅱ | 31 | 22 | |
| Ⅲ+Ⅳ | 6 | 15 | |
| BCLC stage | 0.155 | ||
| A | 26 | 18 | |
| B | 3 | 4 | |
| C | 8 | 15 | |
2.2. Linc00261在肝癌组织中的表达与术后无瘤生存期的关系
Kaplan-Meier生存分析发现,linc00261低表达组术后无瘤生存时间显著低于高表达组,其中位无瘤生存时间分别为7.66和70.37月(P<0.05,图 2)。
2.
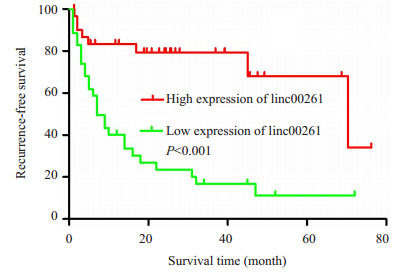
linc00261高表达组和低表达组术后无瘤生存时间的比较
Kaplan-Meier recurrence-free survival curves of patients with high and low expressions of linc00261.
2.3. 单因素、多因素分析
COX单因素回归分析评估各临床病理参数与患者术后无瘤生存时间的关系:肿瘤大小(P=0.004)、数目(P=0.013)、有无微癌栓(P<0.001)与门脉癌栓(P<0.001)、病理分化程度(P=0.001)及linc00261的相对表达水平(P<0.001)等与患者无瘤生存时间长短显著相关(表 2)。将前述单因素分析中有统计学意义的指标纳入COX多因素回归分析,结果显示:肿瘤数目(P=0.004)、有无微癌栓(P=0.007)、门脉癌栓(P=0.003)、病理分化程度(P= 0.047)及linc00261相对表达水平(P=0.004)为患者无瘤生存时间的独立预后因素(表 2)。
2.
Linc00261与患者无瘤生存时间的单因素和多因素COX回归分析
Univariate and multivariate Cox regression analyses of linc00261 for predicting recurrence-free survival of the patients (n=74)
| Variables | HR | 95%CI | P |
| a: Portal vein tumor thrombosis; b: Micro-vascular invasion. | |||
| Univariate analysis | |||
| Age (<60 vs>60 years) | 1.433 | 0.721-2.847 | 0.305 |
| Gender (male vs female) | 0.744 | 0.335-1.653 | 0.468 |
| AFP (<400 vs ≥400) | 1.181 | 0.706-1.974 | 0.526 |
| Liver cirrhosis(Yes vs No) | 1.259 | 0.667-2.377 | 0.477 |
| Capsule(Yes vs No) | 0.897 | 0.509-1.580 | 0.706 |
| Tumor size (<3 vs ≥3 cm) | 3.913 | 1.564-9.794 | 0.004 |
| Tumor number | 2.155 | 1.176-3.984 | 0.013 |
| MVIa (Yes vs No) | 3.075 | 1.778-5.320 | 0.000 |
| PVTTb (Yes vs No) | 3.510 | 1.983-6.211 | 0.000 |
| Differentiation (well, moderate, poor) | 0.472 | 0.306-0.728 | 0.001 |
| Expression of Linc00261 (high vs low) | 0.215 | 0.098-0.475 | 0.000 |
| Multivariate analysis | |||
| Tumor size (<3 vs ≥3 cm) | 3.293 | 0.745-14.561 | 0.116 |
| Tumor number | 4.335 | 1.591-11.813 | 0.004 |
| MVI(Yes vs No) | 3.662 | 1.422-9.431 | 0.007 |
| PVTT(Yes vs No) | 3.762 | 1.549-9.136 | 0.003 |
| Differentiation (well, moderate, poor) | 0.549 | 0.304-0.991 | 0.047 |
| Linc00261 expression (high vs low) | 0.294 | 0.127-0.683 | 0.004 |
2.4. 检测Linc00261在肝癌细胞株中的表达及转染siRNA后linc00261的表达
采用qRT-PCR检测5株肝癌细胞株的linc00261表达水平(图 3A)。选择表达水平较高的MHCC-LM3和SNU-449进行小干扰沉默linc00261的表达,设Si1-linc00261组、Si2-linc00261组、Si-NC组和空白对照组(Mock)组,qRT-PCR检测转染siRNA 48 h后linc00261的表达水平,与空白对照组及Si-NC组的细胞相比,转染Si1-linc00261和Si2-linc00261的细胞的linc00261表达水平明显受到抑制(P<0.05,图 3B)。
3.
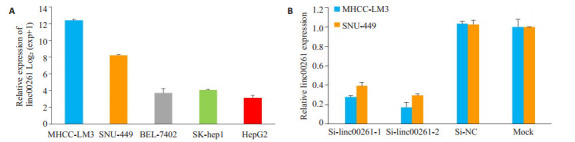
实时荧光定量PCR检测Linc00261表达水平
Expression of linc00261 detected by real-time PCR in different HCC cell lines (A) and in MHCC-LM3 and SNU-449 cells transfected with siRNAs (B).
2.5. 沉默linc00261后对肝癌细胞增殖的影响
采用CCK-8检测Si-linc00261-1组、Si-linc00261-2组,Si-NC组和空白对照组(Mock组)细胞的增殖情况,与Si-NC组和Mock组相比,小干扰敲低linc00261组在MHCC-LM3和SNU-449两株细胞中的细胞增殖能力未见明显变化(图 4A-B),无统计学差异(P>0.05)。
4.
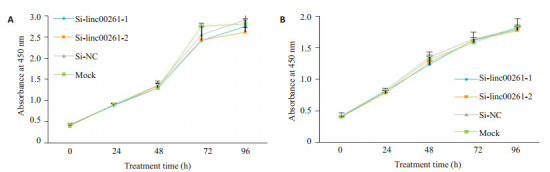
Linc00261的表达对肝癌细胞株MHCC-LM3和SNU-449增殖能力的影响
Effect of linc00261 knockdown on the proliferation of MHCC-LM3 and SNU-449 cells (P > 0.05).
2.6. 沉默linc00261后对肝癌细胞株迁移和侵袭能力的影响
与Si-NC组和Mock组相比,小干扰敲低linc00261组,在肝癌细胞系MHCC-LM3(P<0.01,图 5A-B)和SNU-449(P<0.001,图 6A-B)的迁移和侵袭能力明显增强。
5.
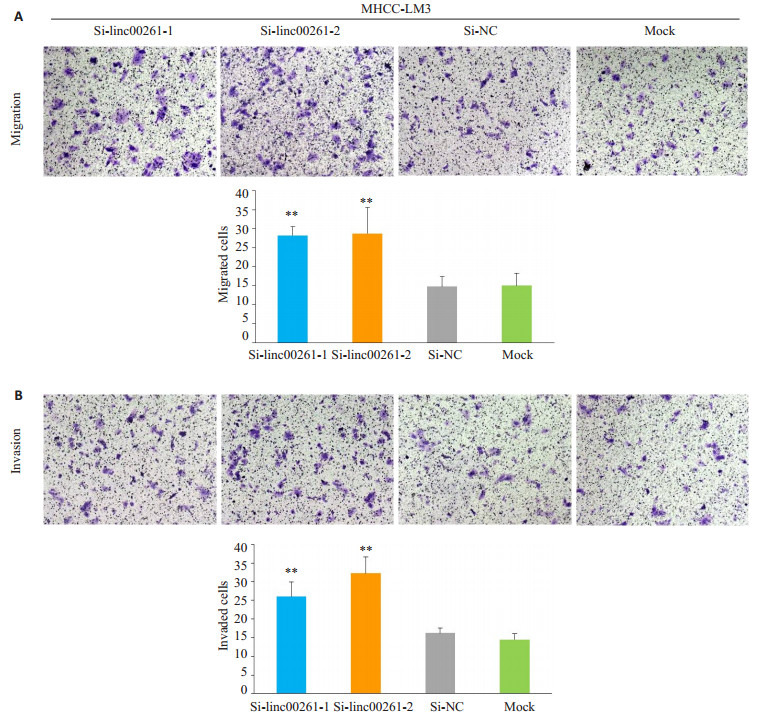
Linc00261的表达敲低后对肝癌细胞株MHCC-LM3迁移和侵袭能力的影响
Effect of linc00261 knockdown on cell migration (A) and invasion (B) of MHCC-LM3 cells (Original magnification:×200). (**P < 0.01 vs Si-NC and Mock).
6.
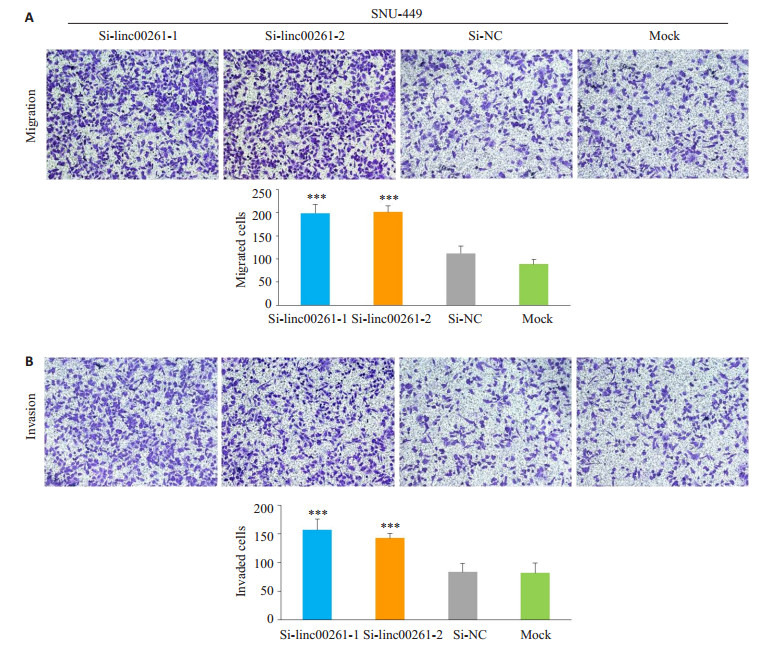
Linc00261的表达敲低后对肝癌细胞株SNU-449迁移和侵袭能力的影响
Effect of linc00261 knockdown on cell migration (A) and invasion (B) of SNU-449 cells (×200). (***P < 0.01 vs Si-NC and Mock).
3. 讨论
在本研究中,我们首次检测linc00261在HCC组织中的相对表达水平,同时统计分析发现linc00261的相对表达水平与患者术前血清AFP水平、肿瘤大小、微癌栓、TNM分期等显著相关;COX回归分析显示HCC组织中linc00261的低表达与患者的不良预后相关,其术后无瘤生存期明显缩短(P<0.05),是影响HCC患者术后无瘤生存期的一个独立危险因素;同时CCK-8细胞增殖实验发现,小干扰敲低linc00261后,对肝癌细胞的增殖无明显的影响,但敲低linc00261后可明显增强肝癌细胞的迁移和侵袭能力。
近年来,对于lncRNA在肿瘤中的研究主要探究其对肿瘤复发、转移和耐药方面的作用[16-17]。在HCC的发生发展过程中,一部分的lncRNA发生了突变或异常表达,可参与调节肝癌细胞的增殖[18-20]、侵袭及迁移能力[21-23],最终导致HCC的发生和进展。本实验证明linc00261在肝癌的发生发展过程中起到抑癌基因的作用,明确了沉默linc00261可促进肝癌细胞株的迁移侵袭能力的作用,但沉默linc00261在肝癌细胞株增殖能力上未见明显的影响。Linc00261对肝癌细胞株迁移和侵袭能力的影响,与目前敲低linc00261在胃癌、结肠癌及绒毛膜癌等肿瘤中起到促进肿瘤细胞迁移侵袭能力的作用相一致[7-9],对肝癌细胞增殖功能的影响未见明显差异,这可能与肿瘤特异性和肿瘤细胞特性相关。但linc00261的相对表达水平与患者肿瘤大小存在明显的相关性,在整个基因组中,基因间存在相互作用和联系,我们推测linc00261在低表达状态时未引发细胞增殖和凋亡相关的基因或信号通路的变化,但高表达linc00261或可能起到激活细胞凋亡或抑制细胞增殖的作用,有待进一步深入研究其具体机制。
上皮间充质转化是肿瘤细胞获得迁移和侵袭能力的基础,主要特征是上皮标志物E-cadherin表达下调、间质标志物N-cadherin和Vimentin等表达上调,致使细胞间链接丧失,黏附能力减弱,细胞迁移和侵袭能力增强[24-25]。研究表明,众多的lncRNAs参与调节癌症的EMT进程[26],可通过调控EMT相关转录因子的表达影响癌症的EMT进程。目前研究表明,Linc00261在胃癌中可抑制细胞的迁移和侵袭能力,抑制其EMT进程[7]。在本实验中,小干扰敲底linc00261后,肝癌细胞的迁移和侵袭能力明显增强,提示linc00261在HCC中可通过抑制EMT进程而抑制了肝癌细胞的迁移和侵袭能力,影响HCC的发生发展。癌栓的形成,是HCC预后不良的危险因素[27]。众多研究表明PVTT和MVI是HCC发生肝外转移的主要途径,两者与肿瘤的EMT进程及干细胞特性具有密切的关系[28-30]。Linc00261的表达水平与患者是否具有MVI显著相关,进一步说明linc00261可能不单影响HCC的EMT进程,还可能影响到肝癌细胞的干性特征,从而影响HCC的发生发展过程。而linc00261的表达水平与患者是否具有PVTT无明显相关性,我们认为可能的原因有:病例为随机入组,可能存在一定的样本差异性;可能与入组患者的样本量有关。本研究发现低表达linc00261患者术后无瘤生存时间明显短于高表达linc00261的患者,这为我们进一步研究揭示linc00261在HCC中的具体调控机制及作用打下基础。
综上所述,HCC组织中linc00261表达下调与肿瘤的恶性进展相关,linc00261可作为一个抑癌基因参与调控HCC迁移和侵袭能力的变化,可能参与调控HCC的EMT进程及干性特征,但其在HCC中的调控机制仍需深入研究。探究linc00261在HCC中的作用及分子机制,可为HCC的治疗提供新的思路。Linc00261可成为预测HCC患者术后预后的新指标。
Biography
陈占军,硕士,E-mail: 13265032757@163.com
Funding Statement
国家自然科学基金(8187111677)
Supported by National Natural Science Foundation of China (8187111677)
Contributor Information
陈 占军 (Zhanjun CHEN), Email: 13265032757@163.com.
杨 定华 (Dinghua YANG), Email: 13600039623@163.com.
References
- 1.Chen W, Zheng R, Zuo T, et al. National cancer incidence and mortality in China, 2012. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4779764/ Chin J Cancer Res. 2016;28(1):1–11. doi: 10.3978/j.issn.1000-9604.2016.02.08. [Chen W, Zheng R, Zuo T, et al. National cancer incidence and mortality in China, 2012[J]. Chin J Cancer Res, 2016, 28(1): 1-11.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schoenfeld I, Kraywinkel K. Epidemiology of hepatocellular carcinoma in germany. Der Onkologe. 2018;24(9):653–8. doi: 10.1007/s00761-018-0438-4. [Schoenfeld I, Kraywinkel K. Epidemiology of hepatocellular carcinoma in germany[J]. Der Onkologe, 2018, 24(9): 653-8.] [DOI] [Google Scholar]
- 3.Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet. 2012;379(9822):1245–55. doi: 10.1016/S0140-6736(11)61347-0. [Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma[J]. Lancet, 2012, 379(9822): 1245-55.] [DOI] [PubMed] [Google Scholar]
- 4.Han BW, Chen YQ. Potential pathological and functional links between long noncoding RNAs and hematopoiesis. http://europepmc.org/abstract/med/23962981. Sci Signal. 2013;6(289):e5. doi: 10.1126/scisignal.2004099. [Han BW, Chen YQ. Potential pathological and functional links between long noncoding RNAs and hematopoiesis[J]. Sci Signal, 2013, 6(289): e5.] [DOI] [PubMed] [Google Scholar]
- 5.Necsulea A, Soumillon M, Warnefors MA, et al. The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature. 2014;505(7485):635. doi: 10.1038/nature12943. [Necsulea A, Soumillon M, Warnefors MA, et al. The evolution of lncRNA repertoires and expression patterns in tetrapods[J]. Nature, 2014, 505(7485): 635.] [DOI] [PubMed] [Google Scholar]
- 6.Emst C, Morton CC. Identification and function of long non-coding RNA. http://d.old.wanfangdata.com.cn/Periodical/shykdxxb201803004. Front Cell Neurosci. 2013;7:168. doi: 10.3389/fncel.2013.00168. [Emst C, Morton CC. Identification and function of long non-coding RNA[J]. Front Cell Neurosci, 2013, 7: 168.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu YC, Li LJ, Zheng ZQ, et al. Long non-coding RNA linc00261 suppresses gastric cancer progression via promoting Slug degradation. J Cell Mol Med. 2017;21(5):955–67. doi: 10.1111/jcmm.2017.21.issue-5. [Yu YC, Li LJ, Zheng ZQ, et al. Long non-coding RNA linc00261 suppresses gastric cancer progression via promoting Slug degradation[J]. J Cell Mol Med, 2017, 21(5): 955-67.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang ZK, Yang L, Wu LL, et al. Long non-coding RNA LINC00261 sensitizes human colon cancer cells to cisplatin therapy. http://www.ncbi.nlm.nih.gov/pubmed/29267503. Braz J Med Biol Res. 2018;51(2):e6793. doi: 10.1590/1414-431X20176793. [Wang ZK, Yang L, Wu LL, et al. Long non-coding RNA LINC00261 sensitizes human colon cancer cells to cisplatin therapy [J]. Braz J Med Biol Res, 2018, 51(2): e6793.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang YA, Xue K, Guan YH, et al. Long noncoding RNA LINC00261 suppresses cell proliferation and invasion and promotes cell apoptosis in human choriocarcinoma. Oncol Res. 2017;25(5):733–42. doi: 10.3727/096504016X14772362173376. [Wang YA, Xue K, Guan YH, et al. Long noncoding RNA LINC00261 suppresses cell proliferation and invasion and promotes cell apoptosis in human choriocarcinoma[J]. Oncol Res, 2017, 25(5): 733-42.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sha LX, Huang LX, Luo XS, et al. Long non-coding RNA LINC00261 inhibits cell growth and migration in endometriosis. J Obstet Gynaecol Res. 2017;43(10):1563–9. doi: 10.1111/jog.2017.43.issue-10. [Sha LX, Huang LX, Luo XS, et al. Long non-coding RNA LINC00261 inhibits cell growth and migration in endometriosis[J]. J Obstet Gynaecol Res, 2017, 43(10): 1563-9.] [DOI] [PubMed] [Google Scholar]
- 11.刘 月, 陈 虹羽, 邵 青, et al. 长链非编码RNA LINC00261在Barrett食管, 食管腺癌组织中的表达及其作用. http://d.old.wanfangdata.com.cn/Periodical/dsjydxxb201701011. 第三军医大学学报. 2017;39(01):67–71. [刘月, 陈虹羽, 邵青, 等.长链非编码RNA LINC00261在Barrett食管, 食管腺癌组织中的表达及其作用[J].第三军医大学学报, 2017, 39 (01): 67-71.] [Google Scholar]
- 12.张 春明, 高 伟, 吴 勇延, et al. 长链非编码RNA LINC00261在喉鳞状细胞癌中的表达及意义. http://www.wanfangdata.com.cn/details/detail.do?_type=perio&id=QKC20172017021700089385. 临床耳鼻咽喉头颈外科杂志. 2017;31(1):68–71. doi: 10.13201/j.issn.1001-1781.2017.01.018. [张春明, 高伟, 吴勇延, 等.长链非编码RNA LINC00261在喉鳞状细胞癌中的表达及意义[J].临床耳鼻咽喉头颈外科杂志, 2017, 31(1): 68-71.] [DOI] [PubMed] [Google Scholar]
- 13.Liu Y, Xiao N, Xu SF. Decreased expression of long non-coding RNA LINC00261 is a prognostic marker for patients with non-small cell lung cancer: a preliminary study. http://www.ncbi.nlm.nih.gov/pubmed/29272004. Eur Rev Med Pharmacol Sci. 2017;21(24):5691–5. doi: 10.26355/eurrev_201712_14014. [Liu Y, Xiao N, Xu SF. Decreased expression of long non-coding RNA LINC00261 is a prognostic marker for patients with non-small cell lung cancer: a preliminary study[J]. Eur Rev Med Pharmacol Sci, 2017, 21(24): 5691-5.] [DOI] [PubMed] [Google Scholar]
- 14.中华人民共和国卫生和计划生育委员会医政医管局 原发性肝癌诊疗规范(2017年版) 中华消化外科杂志. 2017;16(7):635–47. doi: 10.3760/cma.j.issn.1673-9752.2017.07.001. [中华人民共和国卫生和计划生育委员会医政医管局.原发性肝癌诊疗规范(2017年版) [J].中华消化外科杂志, 2017, 16(7): 635-47.] [DOI] [Google Scholar]
- 15.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–8. doi: 10.1006/meth.2001.1262. [Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method[J]. Methods, 2001, 25(4): 402-8.] [DOI] [PubMed] [Google Scholar]
- 16.Heery R, Finn SP, Cuffe S, et al. Long Non-Coding RNAs: key regulators of epithelial-mesenchymal transition, tumour drug resistance and cancer stem cells. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC5406713/ Cancers (Basel) 2017;9(4):E38. doi: 10.3390/cancers9040038. [Heery R, Finn SP, Cuffe S, et al. Long Non-Coding RNAs: key regulators of epithelial-mesenchymal transition, tumour drug resistance and cancer stem cells[J]. Cancers (Basel), 2017, 9(4): E38.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li D, Liu XE, Zhou J, et al. Long noncoding RNA HULC modulates the phosphorylation of YB-1 through serving as a scaffold of extracellular signal-regulated kinase and YB-1 to enhance hepatocarcinogenesis. Hepatology. 2017;65(5):1612–27. doi: 10.1002/hep.29010. [Li D, Liu XE, Zhou J, et al. Long noncoding RNA HULC modulates the phosphorylation of YB-1 through serving as a scaffold of extracellular signal-regulated kinase and YB-1 to enhance hepatocarcinogenesis[J]. Hepatology, 2017, 65(5): 1612-27.] [DOI] [PubMed] [Google Scholar]
- 18.Du YM, Kong GY, You XN, et al. Elevation of highly up-regulated in liver cancer (HULC) by hepatitis B virus X protein promotes hepatoma cell proliferation via down-regulating p18. J Biol Chem. 2012;287(31):26302–11. doi: 10.1074/jbc.M112.342113. [Du YM, Kong GY, You XN, et al. Elevation of highly up-regulated in liver cancer (HULC) by hepatitis B virus X protein promotes hepatoma cell proliferation via down-regulating p18[J]. J Biol Chem, 2012, 287(31): 26302-11.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cao CH, Sun JY, Zhang DY, et al. The long intergenic noncoding RNA UFC1, a target of MicroRNA 34a, interacts with the mRNA stabilizing protein HuR to increase levels of beta-Catenin in HCC cells. Gastroenterology. 2015;148(2):415–U249. doi: 10.1053/j.gastro.2014.10.012. [Cao CH, Sun JY, Zhang DY, et al. The long intergenic noncoding RNA UFC1, a target of MicroRNA 34a, interacts with the mRNA stabilizing protein HuR to increase levels of beta-Catenin in HCC cells[J]. Gastroenterology, 2015, 148(2): 415-U249.] [DOI] [PubMed] [Google Scholar]
- 20.Deng L, Yang SB, Xu FF, et al. Long noncoding RNA CCAT1 promotes hepatocellular carcinoma progression by functioning as let-7 sponge. J Exp Clin Cancer Res. 2015;34:18. doi: 10.1186/s13046-015-0136-7. [Deng L, Yang SB, Xu FF, et al. Long noncoding RNA CCAT1 promotes hepatocellular carcinoma progression by functioning as let-7 sponge[J]. J Exp Clin Cancer Res, 2015, 34: 18.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang L, Yang F, Yuan JH, et al. Epigenetic activation of the MiR-200 family contributes to H19-mediated metastasis suppression in hepatocellular carcinoma. Carcinogenesis. 2013;34(3):577–86. doi: 10.1093/carcin/bgs381. [Zhang L, Yang F, Yuan JH, et al. Epigenetic activation of the MiR-200 family contributes to H19-mediated metastasis suppression in hepatocellular carcinoma[J]. Carcinogenesis, 2013, 34(3): 577-86.] [DOI] [PubMed] [Google Scholar]
- 22.Cai B, Song XQ, Cai JP, et al. HOTAIR: a cancer-related long noncoding RNA. Neoplasma. 2014;61(4):379–91. doi: 10.4149/neo_2014_075. [Cai B, Song XQ, Cai JP, et al. HOTAIR: a cancer-related long noncoding RNA[J]. Neoplasma, 2014, 61(4): 379-91.] [DOI] [PubMed] [Google Scholar]
- 23.Yuan JH, Yang F, Wang F, et al. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25(5):666–81. doi: 10.1016/j.ccr.2014.03.010. [Yuan JH, Yang F, Wang F, et al. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma[J]. Cancer Cell, 2014, 25(5): 666-81.] [DOI] [PubMed] [Google Scholar]
- 24.Nieto MA, Huang RY, Jackson RA, et al. EMT: 2016. Cell. 2016;166(1):21–45. doi: 10.1016/j.cell.2016.06.028. [Nieto MA, Huang RY, Jackson RA, et al. EMT: 2016[J]. Cell, 2016, 166(1): 21-45.] [DOI] [PubMed] [Google Scholar]
- 25.Brabletz T, Kalluri R, Nieto MA, et al. EMT in cancer. Nat Rev Cancer. 2018;18(2):128–34. doi: 10.1038/nrc.2017.118. [Brabletz T, Kalluri R, Nieto MA, et al. EMT in cancer[J]. Nat Rev Cancer, 2018, 18(2): 128-34.] [DOI] [PubMed] [Google Scholar]
- 26.Dhamija S, Diederichs S. From junk to master regulators of invasion: lncRNA functions in migration, EMT and metastasis. Int J Cancer. 2016;139(2):269–80. doi: 10.1002/ijc.30039. [Dhamija S, Diederichs S. From junk to master regulators of invasion: lncRNA functions in migration, EMT and metastasis[J]. Int J Cancer, 2016, 139(2): 269-80.] [DOI] [PubMed] [Google Scholar]
- 27.Du M, Chen LL, Zhao J, et al. Microvascular invasion (MVI) is a poorer prognostic predictor for small hepatocellular carcinoma. BMC Cancer. 2014;14:38. doi: 10.1186/1471-2407-14-38. [Du M, Chen LL, Zhao J, et al. Microvascular invasion (MVI) is a poorer prognostic predictor for small hepatocellular carcinoma[J]. BMC Cancer, 2014, 14: 38.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li T, Xie J, Shen C, et al. Upregulation of long noncoding RNA ZEB1-AS1 promotes tumor metastasis and predicts poor prognosis in hepatocellular carcinoma. Oncogene. 2016;35(12):1575–84. doi: 10.1038/onc.2015.223. [Li T, Xie J, Shen C, et al. Upregulation of long noncoding RNA ZEB1-AS1 promotes tumor metastasis and predicts poor prognosis in hepatocellular carcinoma[J]. Oncogene, 2016, 35(12): 1575-84.] [DOI] [PubMed] [Google Scholar]
- 29.Li T, Xie J, Shen C, et al. Amplification of long noncoding RNA ZFAS1 promotes metastasis in hepatocellular carcinoma. Cancer Res. 2015;75(15):3181–91. doi: 10.1158/0008-5472.CAN-14-3721. [Li T, Xie J, Shen C, et al. Amplification of long noncoding RNA ZFAS1 promotes metastasis in hepatocellular carcinoma[J]. Cancer Res, 2015, 75(15): 3181-91.] [DOI] [PubMed] [Google Scholar]
- 30.Takizawa D, Kakizaki S, Sohara N, et al. Hepatocellular carcinoma with portal vein tumor thrombosis: clinical characteristics, prognosis, and patient survival analysis. Dig Dis Sci. 2007;52(11):3290–5. doi: 10.1007/s10620-007-9808-2. [Takizawa D, Kakizaki S, Sohara N, et al. Hepatocellular carcinoma with portal vein tumor thrombosis: clinical characteristics, prognosis, and patient survival analysis[J]. Dig Dis Sci, 2007, 52 (11): 3290-5.] [DOI] [PubMed] [Google Scholar]


