Abstract
目的
筛选并验证急性T淋巴细胞性白血病关键基因,并对其进行生物信息学分析。
方法
从公共数据库基因表达数据库(GEO)中下载T-ALL基因表达谱芯片GSE14317,应用R软件limma包筛选差异表达基因。利用DAVID数据库对差异基因进行GO功能富集分析和KEGG通路富集分析,同时利用STRING数据库及Cytoscape软件构建差异蛋白互作网络,应用JASPAR数据库构建转录因子靶基因网络,利用RT-PCR验证关键基因mRNA表达水平。
结果
GSE14317表达谱芯片中共有1443个差异基因,包括800个上调基因和643个下调基因,这些差异基因主要富集在细胞周期,造血细胞系,细胞因子间相互作用以及T细胞受体信号通路等。蛋白质相互作用网络图中显示节点最多的10个枢纽基因分别是CDK1,PIK3R1,CCNB1,CCNA2,CDC20,JUN,GNG11,PLK1,PCNA和CCNB1,这些基因在子网络中分别富集于趋化因子信号通路,泛素介导的蛋白降解以及细胞周期等。转录因子调节网络显示42个差异表达的转录因子,其中ELF5,HIC2和MEIS1与候选的9个关键基因启动子上游均有结合位点。RT-PCR结果显示除GNG11外其余9个关键基因表达情况与芯片结果一致,ELF5,HIC2和MEIS1表达均升高与芯片结果一致。
结论
CDK1,PIK3R1,CCNB1,CCNA2,CDC20,JUN,PLK1,PCNA,CCNB1,ELF5,HIC2和MEIS1在TALL发生中发挥重要作用,为T-ALL的治疗和诊断提供生物学靶标。
Keywords: 急性T淋巴细胞白血病, 生物信息学, 差异表达基因, 转录因子
Abstract
Objective
To explore the key genes in T-cell acute lymphoblastic leukemia (T-ALL) using bioinformatics method to better understand the pathogenic mechanisms of T-ALL.
Methods
The gene expression profiles of GSE14317 were obtained from Gene Expression Omnibus database. The differentially expressed genes (DEGs) in T-ALL were analyzed using R package Limma. The online analysis tool DAVID was used to perform the functional and pathway enrichment analysis. The proteinprotein interaction network was constructed by STRING and visualized by Cytoscape. Based on the JASPAR database, the transcription factors (TFs) of the hub genes were obtained. RT-PCR was used to test the mRNA expression level of the key genes.
Results
A total of 1443 DEGs were identified, including 800 up-regulated genes and 643 down-regulated genes. These DEGs were significantly enriched in the cell cycle, hematopoietic cell lineage, cytokine-cytokine receptor interaction and T cell receptor signaling pathway. The top 10 hub genes identified from the PPI networks included CDK1, PIK3R1, CCNB1, CCNA2, CDC20, JUN, GNG11, PLK1, PCNA and CCNB2, which were enriched in chemokine signaling pathway, ubiquition mediated proteolysis and cell cycle. In the TF-target gene network, 42 differentially expressed TFs were identified, among which ELF5, HIC2 and MEISI had binding sites with 9 of the candidate hub genes. RT-PCR showed that the mRNA expression level of all the candidate hub genes except for GNG11 were consistent with the gene expression profiles.
Conclusion
The hub genes CDK1, PIK3R1, CCNB1, CCNA2, CDC20, JUN, PLK1, PCNA, CCNB2, ELF5, HIC2 and MEISI participate in the occurrence of TALL. Our finding provides new insights into the pathogenesis of T-ALL.
Keywords: T-cell acute lymphoblastic leukemia, Bioinformatics analysis, differentially expressed gene, transcription factor
急性T淋巴细胞性白血病(T-ALL)起源于胸腺非成熟T淋巴细胞是一种血液系统恶性肿瘤[1-2],分别占儿童急性淋巴细胞白血病的15%和成人急性淋巴细胞白血病的25%左右[3],主要表现为循环白细胞增多,肝脾肿大,出血倾向以及中枢神经浸润等[4-5]。T-ALL的治疗主要有化疗,干细胞移植以及支持治疗,随着加强化疗的使用,T-ALL病人的预后有所改善,但5年无病生存率仍仅为60%~70%,许多T-ALL病人不能得到完全缓解最终导致复发[6]。目前针对T-ALL的治疗仍不容乐观,缺乏对T-ALL发生发展的分子机制的认知限制了对疾病的治疗能力。
T-ALL的发生发展是一个多基因多步骤协同过程, 存在多种分子及信号通路异常,仅关注单个或某几个基因的研究方法已经不能满足当前研究需要。基于高通量平台的基因表达谱芯片可以检测上万种基因mRNA水平变化,基因芯片技术联合生物信息学使得研究多个基因表达情况成为可能,现已用于分子诊断,癌症分型,预后预测以及新药研究等[7-9]。目前,针对T-ALL的基因芯片研究相对较少,在本文中,我们利用多个数据库对T-ALL病人基因表达谱芯片GSE14317进行全面综合分析,针对差异表达基因进行生物信息学分析并利用实时荧光定量PCR验证,旨在为临床筛选T-ALL的相关分子标志物及药物靶点提供理论基础。
1. 资料和方法
1.1. 表达谱芯片
本文自公共数据库基因表达数据库(GEO)中下载T-ALL基因表达谱芯片GSE14317[10],该芯片基于GPL571平台([HG-U133A_2]Affymetrix Human Genome U133A 2.0 Array),共有26例样本,包括19例T-ALL病人骨髓标本以及7例正常人胸腺CD4+T细胞对照标本。
1.2. 筛选差异表达基因
芯片数据进行数据过滤、标准化和对数化处理,将处理后的数据转化为基因表达矩阵,使用稳健多芯片平均标准化分析(RMA)方法对数据进行归一化处理。利用R语言limma包对T-ALL实验组与对照组比较分析,筛选差异表达基因。
1.3. 芯片数据生物信息学分析
利用DAVID(<a href="https://david.ncifcrf.gov/">https://david.ncifcrf.gov/</a>)在线数据库对差异基因进行GO功能富集分析以及KEGG通路富集分析。利用STRING等在线工具筛选交互作用评分大于0.9(高度可信)的蛋白质构建蛋白互作网络图,并用Cytoscape软件构建蛋白互作子网络,并对子网络基因进行通路富集分析。
1.4. RT-PCR验证基因表达
收集我院5例T-ALL病人骨髓标本以及3例正常人胸腺细胞作为对照,Trizol法提取细胞总RNA并逆转录为cDNA,利用SYBR荧光定量PCR法检测枢纽基因的mRNA表达水平。各基因的引物序列见表 1。最后根据2-△△Ct值计算基因相对表达量。
1.
引物信息
Sequences of the primers for RT-PCR
| Primer | Forward | Reversed |
| CDK1 | AACTACCTGGACCGCTTCCT | CCACTTGAGCTTGTT CACCA |
| PIK3R1 | AAGTGCCAGAGTGAAGTGGC | GTCCCGTCTGCTGTATCTCG |
| CCNB1 | TCTGCTGGGTGTAGGTCCTT | ACCAATGTCCCCAAGAGCTG |
| CCNA2 | CTGGTGGTCTGTGTTCTGTGA | TCTTGGATGCCAGTCTTACTC |
| CDC20 | CTTCCCTGCCAGACCGTATC | AGGATGTCACCAGAGCTTGC |
| JUN | CCAACTCATGCTAACGCAGC | TCTCTCCGTCGCAACTTGTC |
| GNG11 | TGCTTGGACCCAGTCTCAAA | GCGTCCCGAAACAACTGAAG |
| PLK1 | CCGCAATTACATGAGCGAGC | GGAGACTCAGGCGGTATGTG |
| PNCA | AGCCATATTGGAGATGCTGTTG | ATACTGAGTGTCACCGTTGACGA |
| CCNB2 | CACAGGATACACAGAGAATG | CTTGATGGCGATGAATTTAG |
| ELF5 | CAAGACTGTCACAGTCATAG | GTCAACCCGCTCCAAAA TTC |
| HIC2 | GCACTTCGTTTGCGGAGGC | CCGAAGAGCTTCAGGAAGC |
| MEIS1 | CTGTTGATGGTAGGCTGTAT | CTTCTTGGGACTGATGCTGGTG |
| GAPDH | CAGCGACACCCACTCCTCCACCTT | CATGAGGTCCACCACCCTGTTGCT |
1.5. 转录因子-靶基因网络构建
在UCSC数据库中下载核心枢纽基因启动子上游1000 bp序列,利用JASPAR数据库下载人类全基因转录因子序列,进行转录因子预测,将得到的阈值设为90%(高度可信)。筛选有差异表达的转录因子,将转录因子和枢纽基因导入Cytoscape软件构建转录因子-靶基因网络。
1.6. 统计学方法
实验结果采用SPSS19.0软件进行统计学分析,数据均以均数±标准差表示,采用单因素方差分析。P < 0.05为差异有统计学意义。
2. 结果
2.1. 基因表达谱芯片数据结果
样本均一化结果显示每个样本表达量的中位数均一致,表明样本已均一化处理(图 1)。本研究对样本进行贝叶斯检验,按照差异倍数 > 2,P < 0.05作为筛选条件,筛选T-ALL病人表达谱芯片中差异基因。结果发现共有1443个差异表达基因,其中上调的有800个,下调的有463个,选取最有差异的50个基因做热点图反应基因表达情况(图 2)。
1.
标本均一化处理
The expression spectrum data of GSE14317 profiles before or after normalization. A: Box plots of the expression spectrum data before normalization; B: Box plots of the expression spectrum data after normalization.
2.
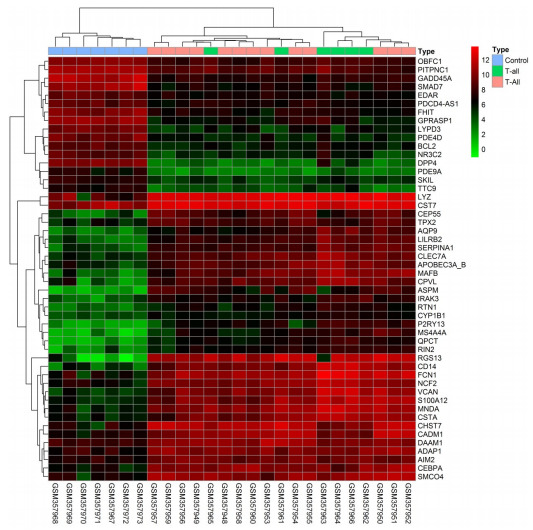
最有差异的50个基因热点图
Heat map of the top 50 differentially expressed genes. Red: up-regulation; green: down-regulation.
2.2. GO功能富集分析与KEGG通路分析
利用DAVID数据库对差异基因进行GO功能富集分析与KEGG通路分析。GO结果(P < 0.05和FDR < 0.05)显示上调的基因富集在细胞周期(GO:0007049)等,下调基因主要富集在蛋白刺激反应(GO:0051789)(表 2)。KEGG结果(P < 0.05和FDR < 0.05)显示上调基因主要富集于造血细胞系(hsa04640)和细胞周期(hsa04110)等,下调基因主要富集于细胞因子间相互作用(hsa04060)和T细胞受体信号通路(hsa04630)(表 3)。
2.
上调基因功能和通路富集分析
Function and KEGG pathway analysis of the up-regulated genes.
| Term | Description | Count | P |
| up-regulated | |||
| GO: 0007049 | cell cycle | 89 | 9.94E-16 |
| GO: 0000278 | mitotic cell cycle | 57 | 1.48E-15 |
| GO: 0000280 | nuclear division | 43 | 1.70E-15 |
| GO: 0007067 | mitosis | 43 | 1.70E-15 |
| GO: 0000087 | M phase of mitotic cell cycle | 43 | 3.32E-15 |
| GO: 0022403 | cell cycle phase | 60 | 4.19E-15 |
| GO: 0006955 | immune response | 81 | 6.99E-15 |
| GO: 0048285 | organelle fission | 43 | 7.36E-15 |
| GO: 0000279 | M phase | 51 | 4.60E-14 |
| GO: 0022402 | cell cycle process | 70 | 4.79E-14 |
| KEGG: hsa04640 | Hematopoietic cell lineage | 18 | 5.94E-06 |
| KEGG: hsa04110 | Cell cycle | 19 | 2.61E-04 |
| KEGG: hsa05332 | Graft-versus-host disease | 9 | 0.001447 |
| KEGG: hsa04612 | Antigen processing and presentation | 13 | 0.002663 |
| KEGG: hsa04666 | Fc gamma R-mediated phagocytosis | 13 | 0.008088 |
| KEGG: hsa04662 | B cell receptor signaling pathway | 11 | 0.01045 |
| KEGG: hsa05322 | Systemic lupus erythematosus | 13 | 0.011134 |
| KEGG: hsa04620 | Toll-like receptor signaling pathway | 13 | 0.012956 |
| KEGG: hsa04060 | Cytokine-cytokine receptor interaction | 25 | 0.015748 |
| KEGG: hsa04650 | Natural killer cell mediated cytotoxicity | 15 | 0.020645 |
3.
上调基因功能和通路富集分析
Function and KEGG pathway analysis of the down-regulated genes
| Term | Description | Count | P |
| down-regulated | |||
| GO: 0051789 | Response to protein stimulus | 20 | 8.08E-09 |
| GO: 0006986 | Response to unfolded protein | 15 | 1.96E-07 |
| GO: 0001775 | Cell activation | 29 | 1.80E-06 |
| GO: 0046649 | Lymphocyte activation | 23 | 3.12E-06 |
| GO: 0010033 | Response to organic substance | 51 | 6.90E-06 |
| GO: 0042981 | Regulation of apoptosis | 55 | 7.44E-06 |
| GO: 0045321 | Leukocyte activation | 25 | 7.54E-06 |
| GO: 0043067 | Regulation of programmed cell death | 55 | 9.70E-06 |
| GO: 0010941 | Regulation of cell death | 55 | 1.09E-05 |
| GO: 0008284 | Positive regulation of cell proliferation | 34 | 1.79E-05 |
| KEGG: hsa04060 | Cytokine-cytokine receptor interaction | 28 | 1.79E-05 |
| KEGG: hsa04660 | T cell receptor signaling pathway | 16 | 5.25E-05 |
| KEGG: hsa04010 | MAPK signaling pathway | 26 | 1.81E-04 |
| KEGG: hsa04630 | Jak-STAT signaling pathway | 17 | 9.75E-04 |
| KEGG: hsa04062 | Chemokine signaling pathway | 18 | 0.002763 |
| KEGG: hsa04115 | p53 signaling pathway | 9 | 0.0085092 |
| KEGG: hsa05210 | Colorectal cancer | 10 | 0.0098568 |
| KEGG: hsa05220 | Chronic myeloid leukemia | 9 | 0.0150118 |
| KEGG: hsa05200 | Pathways in cancer | 23 | 0.0252524 |
| KEGG: hsa04012 | ErbB signaling pathway | 9 | 0.0335255 |
2.3. 蛋白质相互作用网络
对蛋白互作网络图分析发现,满足条件的共有694个节点,3348条关系,其中CDK1,PIK3R1,CCNB1,CCNA2,CDC20,JUN,GNG11,PLK1,PCNA和CCNB2基因具有最大互作关系。利用MCODE [11]构建高度蛋白互作子网络,以发现潜在的功能模块,筛选出最有意义的3个模块并进行信号通路富集分析(P < 0.05和FDR < 0.05),结果显示模块1主要富集于趋化因子信号通路,模块2主要富集于泛素介导的蛋白降解,模块3主要富集于细胞周期(图 3)。
3.

差异基因蛋白互作网络
Top 3 modules screened from the protein-protein network. A: Module 1; B: Module 2; C: Module 3.
2.4. RT-PCR验证结果
利用RT-PCR验证CDK1,PIK3R1,CCNB1,CCNA2,CDC20,JUN,GNG11,PLK1,PCNA和CCNB2在T-ALL病人中的mRNA表达情况。结果显示CDK1,CCNB2,CDC20,PLK1,CCNB2的mRNA表达量较正常对照组增加(P < 0.05),CNB1,CCNA2,PCNA的mRNA表达量较正常对照组明显增加(P < 0.01)。PIK3R1的mRNA表达量较正常对照组降低(P < 0.05),JUN的mRNA表达量较正常对照组明显降低(P < 0.01),与芯片结果数据分析一致。但GNG11的mRNA表达量与正常对照组相比无统计学意义(P > 0.05,图 4)。
4.
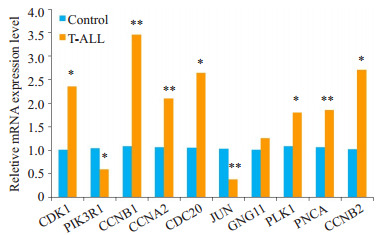
各基因在T-ALL病人中mRNA表达水平的变化
Changes in mRNA expressions of the identified DEGs in T-ALL patients. *P < 0.05 vs normal control group, **P < 0.01 vs normal control group.
2.5. 转录因子-靶基因调节网络
利用JASPAR数据库预测9个关键基因启动子上游转录因子结合位点,筛选出42个差异表达转录因子,其中ELF5,HIC2和MEIS1与候选的9个关键基因启动子上游均有结合位点。将转录因子和关键基因导入Cytoscape软件构建转录因子-靶基因网络(图 5)。
5.
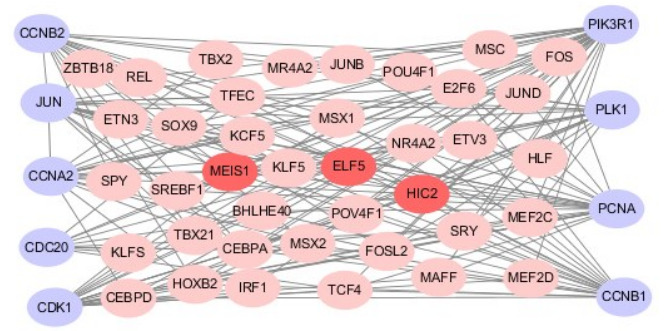
转录因子-靶基因网络
The TFs-target genes regulatory network of T-ALL; the grey nodes represent hub genes, the pink nodes represent transcription factors and the croci nodes represent the three most significant TFs.
2.6. 转录因子mRNA表达水平
用RT-PCR验证ELF5,HIC2和MEIS1在T-ALL病人中的mRNA表达情况。结果显示ELF5和HIC2 MEIS1的mRNA表达量较正常对照组增加(P < 0.05)MEIS1的mRNA表达量较正常对照组明显增加(P < 0.01,图 6),与芯片结果一致。
6.
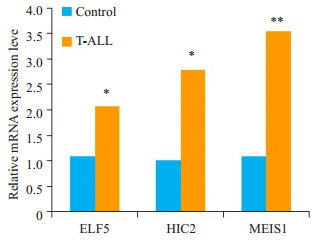
转录因子在T-ALL病人中mRNA表达水平的变化
Changes in mRNA expressions of the identified TFs in T-ALL patients. *P < 0.05 vs normal control group, **P < 0.01 vs normal control group.
3. 讨论
急性T淋巴细胞性白血病(T-ALL)是一种血液系统急进性恶性肿瘤,占急性淋巴细胞白血病(ALL)的15%~25%左右。目前T-ALL的治疗主要有化疗,干细胞移植以及支持治疗,近年来T-ALL的治疗取得了较大改善,但仍有25%的患者对T-ALL的治疗耐受[12]或出现严重副作用,探讨研究T-ALL发生发展的具体机制仍是当务之急。基因芯片技术联合生物信息学方法突破传统的小规模研究方式为全面综合分析多基因调控事件在T-ALL发生发展的作用提供了可能。通过对差异表达基因构建蛋白互作网络分析显示节点最多的10个基因:CDK1,PIK3R1,CCNB1,CCNA2,CDC20,JUN,GNG11,PLK1,PCNA和CCNB1。利用RT-PCR验证基因表达情况发现除GNG11外,其余9个关键基因表达情况与芯片结果一致。
CDK1是丝氨酸/苏氨酸蛋白激酶家族的一员,通过催化不同底物磷酸化实现对细胞周期不同时相的推进和转化作用。此外Lu等[13]研究发现,在人类黑色素瘤中,CDK1可以使P53凋亡刺激蛋白抑制因子(IASPP)磷酸化,促进IASPP单体进入核,并暴露其P53结合位点,导致P53抑制增加。在T-ALL中上调的CDK1可能通过促进细胞周期并抑制P53途径加重疾病进程。细胞周期蛋白CCNB1,CCNA2和CCNB2与细胞周期密切相关, 在控制细胞周期G1/S向G1/M转换中起着必不可少的作用,它们参与多种肿瘤细胞周期调控,如结直肠癌,乳腺癌,肺癌等。细胞周期相关蛋白CDC20是纺锤体组装检查点的靶向物和有丝分裂后期促进复合体的正调控因子,参与细胞周期中某些蛋白质的泛素化降解,使细胞周期以单向的方式推进[14]。Bie等[15]发现在胶质母细胞瘤中CDC20的表达受BRCA1调控与疾病的分级有关。BRCA1是多种肿瘤的易感基因,在本文中BRCA1与CDC20表达均升高,据此可以推测BRCA1与T-ALL的分级有关,提示研究抗BRCA1的药物可以为T-ALL的治疗提供靶向。PLK1属于PLKs家族成员,是一种广泛存在于真核生物中的丝氨酸/苏氨酸蛋白激酶。PLK1在多种肿瘤中高表达并与预后有关,Shao等[16]研究发现PLK1可以介导Numb的磷酸化,抑制P53的转录活性,P53失去抑癌功能最终导致肿瘤的发生。PCNA与细胞DNA合成密切相关,在细胞增殖的启动上起重要作用,目前PCNA作为诊断肿瘤增殖的指标,随疾病的进程逐渐增多[17]。PIK3R1在多种肿瘤中有较高的突变率[18],参与PI3K-AKT信号通路。Remke [19]发现异常的PI3K-AKT信号通路与白血病的生成以及T-ALL早期治疗效果差有关。JUN属于转录因子AP-1家族成员之一,JUN和FOS共同组成异源二聚体复合物AP-1,AP-1在肿瘤中扮演重要的作用参与多种细胞生物过程,如细胞增殖,细胞死亡,细胞周期以及细胞分化等。Scupoli等[20]报道在T-ALL中CXCL12可以激活JNK/AP-1信号通路导致IL-8增多影响T-ALL进程。
4.
模块信号通路分析
KEGG pathway analysis of top 3 modules
| Modules | GeneSet | P | FDR | Nodes |
| Module 1 | Chemokine signaling pathway | 6.49E-17 | 8.88E-14 | ADCY1, CCR1, PF4, CXCR2, GNG11, CXCR3, CXCL6, CCL5, CXCL10, CCR9, CCR7, CCR6, PPBP, CXCR5, CXCR4, CCR4 |
| Cytokine-cytokine receptor interaction | 9.71E-12 | 7.79E-09 | CCR1, PF4, CXCR2, CXCR3, CXCL6, CCL5, CXCL10, CCR9, CCR7, CCR6, CXCR5, PPBP, CXCR4, CCR4 | |
| Module 2 | Ubiquitin mediated proteolysis | 3.73E-12 | 3.62E-09 | UBE2E3, CBLB, SOCS3, SOCS1, RBX1, CDC20, UBE2C, SMURF2, NEDD4L, FBXO2, UBA3, KEAP1 |
| Module 3 | Cell cycle | 4.09E-06 | 0.003418 | CCNB1, CDK1, CCNB2, PLK1, PCNA, BUB1B, CHEK1 |
转录因子通过与靶基因启动子上游结合调控基因表达。因此基于JASPAR数据库,我们对筛选出的9个关键基因进行转录因子预测,共有42个差异表达转录因子,其中ELF5,HIC2和MEIS1与9个关键基因启动子上游均有结合位点。ELF5参与细胞增殖和肿瘤形成,在急性髓性白血病中ELF5抑制P53/P21信号通路导致白血病的发生[21]。MEIS1属于TALE转录因子家族,在正常的胚胎造血中发挥重要作用,在儿童急性白血病中MEIS1与HOXA9共表达加速白血病进程[22]。肿瘤高甲基化基因2(HIC2)是HIC1家族一员,研究发现启动子高甲基化与白血病的发生密切相关,去甲基化治疗是治疗白血病的又一重要途径。目前针对HIC2在T-ALL中的具体作用机制报道较少仍需进一步研究。
综上所述,应用生物信息学方法综合系统的分析了T-ALL差异表达基因,筛选并验证11个关键基因,从分子水平为探讨T-ALL发生发展机制提供了理论依据。筛选出的关键基因为T-ALL的早期诊断及治疗靶点提供了新的方向,为下一步研究T-ALL的发生机制提供了有意义的信息。
Biography
蒋光洁, 硕士研究生, E-mail: 306118416@qq.com
Funding Statement
国家自然科学基金(81373444,81570142)
Supported by National Natural Science Foundation of China (81373444, 81570142)
Contributor Information
蒋 光洁 (Guangjie JIANG), Email: 306118416@qq.com.
邹 琳 (Lin ZOU), Email: cmmc_sm@163.com.
References
- 1.Girardi T, Vicente C, Cools J, et al. The genetics and molecular biology of T-ALL. Blood. 2017;129(9):1113–23. doi: 10.1182/blood-2016-10-706465. [Girardi T, Vicente C, Cools J, et al. The genetics and molecular biology of T-ALL[J]. Blood, 2017, 129(9): 1113-23.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Karrman K, Johansson B. Pediatric T-cell acute lymphoblastic leukemia. Genes Chromosomes Cancer. 2017;56(2):89–116. doi: 10.1002/gcc.v56.2. [Karrman K, Johansson B. Pediatric T-cell acute lymphoblastic leukemia[J]. Genes Chromosomes Cancer, 2017, 56(2): 89-116.] [DOI] [PubMed] [Google Scholar]
- 3.Kramer AC, Kothari A, Wilson WC, et al. Dnmt3a regulates T-cell development and suppresses T-ALL transformation. Leukemia. 2017;31(11):2479–90. doi: 10.1038/leu.2017.89. [Kramer AC, Kothari A, Wilson WC, et al. Dnmt3a regulates T-cell development and suppresses T-ALL transformation[J]. Leukemia, 2017, 31(11): 2479-90.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Donnellan W, Mineishi S, Wicker J, et al. Gamma-Delta T cell acute lymphoblastic leukemia: a Single-Center experience. Glob J Cancer Ther. 2016;2(1):26–9. doi: 10.17352/gjct. [Donnellan W, Mineishi S, Wicker J, et al. Gamma-Delta T cell acute lymphoblastic leukemia: a Single-Center experience[J]. Glob J Cancer Ther, 2016, 2(1): 26-9.] [DOI] [Google Scholar]
- 5.Jost TR, Borga C, Radaelli EA, et al. Role of CXCR4-mediated bone marrow colonization in CNS infiltration by T cell acute lymphoblastic leukemia. J Leukoc Biol. 2016;99(6):1077–87. doi: 10.1189/jlb.5MA0915-394R. [Jost TR, Borga C, Radaelli EA, et al. Role of CXCR4-mediated bone marrow colonization in CNS infiltration by T cell acute lymphoblastic leukemia[J]. J Leukoc Biol, 2016, 99(6): 1077-87.] [DOI] [PubMed] [Google Scholar]
- 6.Sutton R, Shaw PJ, Venn NC, et al. Persistent MRD before and after allogeneic BMT predicts relapse in children with acute lymphoblastic leukaemia. Br J Haematol. 2015;168(3):395–404. doi: 10.1111/bjh.13142. [Sutton R, Shaw PJ, Venn NC, et al. Persistent MRD before and after allogeneic BMT predicts relapse in children with acute lymphoblastic leukaemia[J]. Br J Haematol, 2015, 168(3): 395-404.] [DOI] [PubMed] [Google Scholar]
- 7.Hoelzer D, Thiel E, Arnold R, et al. Successful subtype oriented treatment strategies in adult T-All; results of 744 patients treated in three consecutive GMALL studies. http://news.medlive.cn/hema/info-progress/show-125886_112.html. Blood. 2009;114(22):137. [Hoelzer D, Thiel E, Arnold R, et al. Successful subtype oriented treatment strategies in adult T-All; results of 744 patients treated in three consecutive GMALL studies[J]. Blood, 2009, 114(22): 137.] [Google Scholar]
- 8.Damavandi MD, Braconi C, Cascione LA, et al. Global gene expression profiling of mice tumor-derived organoids identifies key microRNAs and metabolic genes involved in CRC progression. https://www.researchgate.net/profile/Michele_Ghidini/publication/277632237_Abstract_3083_Global_gene_expression_profiling_of_mice_tumor-derived_organoids_identifies_key_microRNAs_and_metabolic_genes_involved_in_CRC_progression/links/56046eb008ae8e08c08a9b4c.pdf. Cancer Res. 2015;75(15):3038. [Damavandi MD, Braconi C, Cascione LA, et al. Global gene expression profiling of mice tumor-derived organoids identifies key microRNAs and metabolic genes involved in CRC progression[J]. Cancer Res, 2015, 75(15): 3038.] [Google Scholar]
- 9.Kulasingam V, Diamandis EP. Strategies for discovering novel cancer biomarkers through utilization of emerging technologies. Nat Clin Pract Oncol. 2008;5(10):588–99. doi: 10.1038/ncponc1187. [Kulasingam V, Diamandis EP. Strategies for discovering novel cancer biomarkers through utilization of emerging technologies[J]. Nat Clin Pract Oncol, 2008, 5(10): 588-99.] [DOI] [PubMed] [Google Scholar]
- 10.Pise-Masison CA, Radonovich M, Dohoney K, et al. Gene expression profiling of ATL patients: compilation of disease-related genes and evidence for TCF4 involvement in BIRC5 gene expression and cell viability. Blood. 2009;113(17):4016–26. doi: 10.1182/blood-2008-08-175901. [Pise-Masison CA, Radonovich M, Dohoney K, et al. Gene expression profiling of ATL patients: compilation of disease-related genes and evidence for TCF4 involvement in BIRC5 gene expression and cell viability[J]. Blood, 2009, 113(17): 4016-26.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics. 2003;4(1):2. doi: 10.1186/1471-2105-4-2. [Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks[J]. BMC Bioinformatics, 2003, 4(1): 2.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.El Zawily A, Mcewen E, Toosi B, et al. The EphB6 receptor is overexpressed in pediatric T cell acute lymphoblastic leukemia and increases its sensitivity to doxorubicin treatment. Sci Rep. 2017;7(1):14767. doi: 10.1038/s41598-017-15200-3. [El Zawily A, Mcewen E, Toosi B, et al. The EphB6 receptor is overexpressed in pediatric T cell acute lymphoblastic leukemia and increases its sensitivity to doxorubicin treatment[J]. Sci Rep, 2017, 7(1): 14767.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu M, Breyssens H, Salter V, et al. Restoring p53 function in human melanoma cells by inhibiting MDM2 and cyclin B1/CDK1-phosphorylated nuclear iASPP. Cancer Cell. 2013;23(5):618–33. doi: 10.1016/j.ccr.2013.03.013. [Lu M, Breyssens H, Salter V, et al. Restoring p53 function in human melanoma cells by inhibiting MDM2 and cyclin B1/CDK1-phosphorylated nuclear iASPP[J]. Cancer Cell, 2013, 23(5): 618-33.] [DOI] [PubMed] [Google Scholar]
- 14.Wang Z, Wan L, Zhong J, et al. Cdc20: a potential novel therapeutic target for cancer treatment. Curr Pharm Des. 2013;19(18):3210–4. doi: 10.2174/1381612811319180005. [Wang Z, Wan L, Zhong J, et al. Cdc20: a potential novel therapeutic target for cancer treatment[J]. Curr Pharm Des, 2013, 19(18): 3210-4.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bie L, Zhao G, Cheng P, et al. The accuracy of survival time prediction for patients with glioma is improved by measuring mitotic spindle checkpoint gene expression. PLoS One. 2011;6(10):e25631. doi: 10.1371/journal.pone.0025631. [Bie L, Zhao G, Cheng P, et al. The accuracy of survival time prediction for patients with glioma is improved by measuring mitotic spindle checkpoint gene expression[J]. PLoS One, 2011, 6 (10): e25631.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shao C, Chien SJ, Farah E, et al. Plk1 phosphorylation of Numb leads to impaired DNA damage response. Oncogene. 2018;37(6):810–20. doi: 10.1038/onc.2017.379. [Shao C, Chien SJ, Farah E, et al. Plk1 phosphorylation of Numb leads to impaired DNA damage response[J]. Oncogene, 2018, 37 (6): 810-20.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Juríková M, Danihel L', Polák Š, et al. Ki67, PCNA, and MCM proteins: Markers of proliferation in the diagnosis of breast cancer. Acta Histochem. 2016;118(5):544–52. doi: 10.1016/j.acthis.2016.05.002. [Juríková M, Danihel L', Polák Š, et al. Ki67, PCNA, and MCM proteins: Markers of proliferation in the diagnosis of breast cancer [J]. Acta Histochem, 2016, 118(5): 544-52.] [DOI] [PubMed] [Google Scholar]
- 18.Park SW, Kang MR, Eom HS, et al. Somatic mutation of PIK3R1 gene is rare in common human cancers. Acta Oncol (Madr) 2010;49(1):128–30. doi: 10.3109/02841860903107890. [Park SW, Kang MR, Eom HS, et al. Somatic mutation of PIK3R1 gene is rare in common human cancers[J]. Acta Oncol (Madr), 2010, 49(1): 128-30.] [DOI] [PubMed] [Google Scholar]
- 19.Remke M, Pfister S, Kox C, et al. High-resolution genomic profiling of childhood T-ALL reveals frequent copy-number alterations affecting the TGF-beta and PI3K-AKT pathways and deletions at 6q15-16.1 as a genomic marker for unfavorable early treatment response. Blood. 2009;114(5):1053–62. doi: 10.1182/blood-2008-10-186536. [Remke M, Pfister S, Kox C, et al. High-resolution genomic profiling of childhood T-ALL reveals frequent copy-number alterations affecting the TGF-beta and PI3K-AKT pathways and deletions at 6q15-16.1 as a genomic marker for unfavorable early treatment response[J]. Blood, 2009, 114(5): 1053-62.] [DOI] [PubMed] [Google Scholar]
- 20.Scupoli MT, Donadelli M, Cioffi F, et al. Bone marrow stromal cells and the upregulation of interleukin-8 production in human Tcell acute lymphoblastic leukemia through the CXCL12/CXCR4 axis and the NF-kappaB and JNK/AP-1 pathways. Haematologica. 2008;93(4):524–32. doi: 10.3324/haematol.12098. [Scupoli MT, Donadelli M, Cioffi F, et al. Bone marrow stromal cells and the upregulation of interleukin-8 production in human Tcell acute lymphoblastic leukemia through the CXCL12/CXCR4 axis and the NF-kappaB and JNK/AP-1 pathways[J]. Haematologica, 2008, 93(4): 524-32.] [DOI] [PubMed] [Google Scholar]
- 21.Endo A, Tomizawa D, Aoki Y, et al. EWSR1/ELF5 induces acute myeloid leukemia by inhibiting p53/p21 pathway. Cancer Sci. 2016;107(12):1745–54. doi: 10.1111/cas.2016.107.issue-12. [Endo A, Tomizawa D, Aoki Y, et al. EWSR1/ELF5 induces acute myeloid leukemia by inhibiting p53/p21 pathway[J]. Cancer Sci, 2016, 107(12): 1745-54.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adamaki M, Lambrou GI, Athanasiadou AA, et al. HOXA9 and MEIS1 gene overexpression in the diagnosis of childhood acute leukemias: Significant correlation with relapse and overall survival. Leuk Res. 2015;39(8):874–82. doi: 10.1016/j.leukres.2015.04.012. [Adamaki M, Lambrou GI, Athanasiadou AA, et al. HOXA9 and MEIS1 gene overexpression in the diagnosis of childhood acute leukemias: Significant correlation with relapse and overall survival [J]. Leuk Res, 2015, 39(8): 874-82.] [DOI] [PubMed] [Google Scholar]


