Recent studies using single-particle cryo-electron microscopy (cryoEM) revealed the mechanism underlying activation of viral fusion protein at the priming stage. However, characterizing the subsequent triggering stage underpinning transition from pre- to postfusion structures is difficult because single-particle cryoEM excludes unstable structures that appear as heterogeneous shapes. Therefore, population-based biochemical analysis is needed to capture features of unstable proteins. Here, we analyzed protease digestion products of a coronavirus fusion protein during activation; their sizes appear to be affected directly by the conformational state. We propose a model for the viral fusion protein in the intermediate state, which involves a compact structure and conformational changes that overcome steric hindrance within the three fusion protein subunits.
KEYWORDS: Western blotting, conformational change, coronavirus, intermediate, membrane fusion, spike
ABSTRACT
A fusion protein expressed on the surface of enveloped viruses mediates fusion of the viral and cellular membranes to facilitate virus infection. Pre- and postfusion structures of viral fusion proteins have been characterized, but conformational changes between them remain poorly understood. Here, we examined the intermediate conformation of the murine coronavirus fusion protein, called the spike protein, which must be cleaved by a cellular protease following receptor binding. Western blot analysis of protease digestion products revealed that two subunits (67 and 69 kDa) are produced from a single spike protein (180 kDa). These two subunits were considered to be by-products derived from conformational changes and were useful for probing the intermediate conformation of the spike protein. Interaction with a heptad repeat (HR) peptide revealed that these subunits adopt packed and unpacked conformations, respectively, and two-dimensional electrophoresis revealed a trimeric assembly. Based on biochemical observations, we propose an asymmetric trimer model for the intermediate structure of the spike protein. Receptor binding induces the membrane-binding potential of the trimer, in which at least one HR motif forms a packed-hairpin structure, while membrane fusion subunits are covered by the receptor-binding subunit, thereby preventing the spike protein from forming the typical homotrimeric prehairpin structure predicted by the current model of class I viral fusion protein. Subsequent proteolysis induces simultaneous packing of the remaining unpacked HRs upon assembly of three HRs at the central axis to generate a six-helix bundle. Our model proposes a key mechanism for membrane fusion of enveloped viruses.
IMPORTANCE Recent studies using single-particle cryo-electron microscopy (cryoEM) revealed the mechanism underlying activation of viral fusion protein at the priming stage. However, characterizing the subsequent triggering stage underpinning transition from pre- to postfusion structures is difficult because single-particle cryoEM excludes unstable structures that appear as heterogeneous shapes. Therefore, population-based biochemical analysis is needed to capture features of unstable proteins. Here, we analyzed protease digestion products of a coronavirus fusion protein during activation; their sizes appear to be affected directly by the conformational state. We propose a model for the viral fusion protein in the intermediate state, which involves a compact structure and conformational changes that overcome steric hindrance within the three fusion protein subunits.
INTRODUCTION
Class I viral fusion proteins drive the fusion of viral and cellular membranes to facilitate insertion of the viral genome into the host cytoplasm (1). Structural analyses, including cryo-electron microscopy (cryoEM) and X-ray crystallography, revealed pre- and postfusion structures, and biochemical analysis led to the proposal of a unified model for conformational changes of viral fusion proteins; receptor binding primes the formation of a membrane-embedded homotrimeric prehairpin structure that bridges viral and cellular membranes and then folds back on itself to form a hairpin structure, drawing the viral and cellular membranes into close proximity, resulting in lipid mixing (1–3). The postfusion form consists of a central N-terminal trimeric α-helical coiled coil (trimer of heptad repeat 1 [HR1]) surrounded by three C-terminal helices (HR2), generating a six-helix bundle (6HB) (1–3).
The molecular rearrangements occurring at the early stages of the fusion process underpinning the receptor-binding step in the fusion protein of human immunodeficiency virus 1, influenza virus, and coronavirus were revealed by high-resolution cryoEM (4–13). However, the dynamic rearrangements underpinning the transition from pre- to postfusion structures remain largely uncharacterized. During membrane fusion in influenza virus and herpes simplex virus, a V- or Y-shaped density was observed in the intermediate state by cryoEM (14, 15), and in the retroviral Env trimer, an asymmetric intermediate appeared to emanate from a single protomer in cryo-electron tomography experiments (16). For the Moloney murine leukemia virus Env protein, the sequential release of the surface subunit from the transmembrane subunit induces the formation of an asymmetric trimer (17). Adoption of an asymmetric conformation presumably overcomes steric hindrance within the three fusion protein subunits.
The coronavirus spike (S) glycoprotein is a class I viral fusion protein constructed of S1 and S2 subunits. The N-terminal S1 subunit is responsible for receptor binding, and the C-terminal membrane-anchored S2 subunit is important for virus-cell membrane fusion. Some coronaviruses, such as severe acute respiratory syndrome coronavirus (SARS-CoV), Middle East respiratory syndrome coronavirus (MERS-CoV), and mouse hepatitis virus type 2 (MHV-2), possess uncleaved 180-kDa S proteins. These viruses utilize cell surface or endosomal proteases (such as TMPRSS2, HAT, trypsin, elastase, or cathepsin L) to cleave S proteins during cell entry (2, 18–31). It remains controversial whether MERS-CoV S protein is processed by the cellular protease furin after internalization via endocytosis (32) because this finding was not supported by a recent study (33).
Our previous study showed that the S protein of MHV-2 requires a two-step conformational change process (26). The first step takes place after binding to a soluble form of the MHV receptor (CEACAM1a), upon which the metastable form of S protein is converted to a stable trimer possessing liposome-binding activity. The subsequent step is driven by protease digestion. From a single 180-kDa S protein, trypsin produces a 90-kDa subunit in the absence of receptor, but a 66-kDa subunit in the presence of receptor, and this 66-kDa species is thought to involve the formation of a 6HB structure (detected as a proteinase K-resistant 53-kDa subunit) (26). A similar result of the protease digestion pattern was also reported in SARS-CoV and MERS-CoV S proteins (4, 20). Therefore, the sizes of protease digestion products are thought to be directly affected by the conformational state of the coronavirus S protein. Studying the MHV-2 S protein could illuminate the conformational changes occurring in each step and may provide novel insight into viral class I fusion proteins.
RESULTS
Trypsin digestion generates three different sizes (90, 69, and 67 kDa) of S2 subunit.
A schematic diagram of the MHV-2 S protein and the binding sites of antibodies employed in this study is shown in Fig. 1A. Three anti-synthetic peptide antibodies, anti-S2a, anti-very highly conserved region (anti-VHCR), and anti-cytoplasmic tail (anti-CT), and a monoclonal antibody (MAb-10G) were used to detect the S2 subunits via Western blot analysis. To probe conformational changes of the MHV-2 S protein, an authentic virus rather than recombinant S protein was employed. Virus at 106 PFU in 10 μl of culture medium was treated with the soluble form of the MHV receptor (1 μM) at 37°C for 30 min, and 10 μg/ml trypsin was added, followed by incubation for 30 min. The main difference between previous (26) and present experiments is the concentration of trypsin (10 μg/ml trypsin was used instead of 1 μg/ml trypsin).
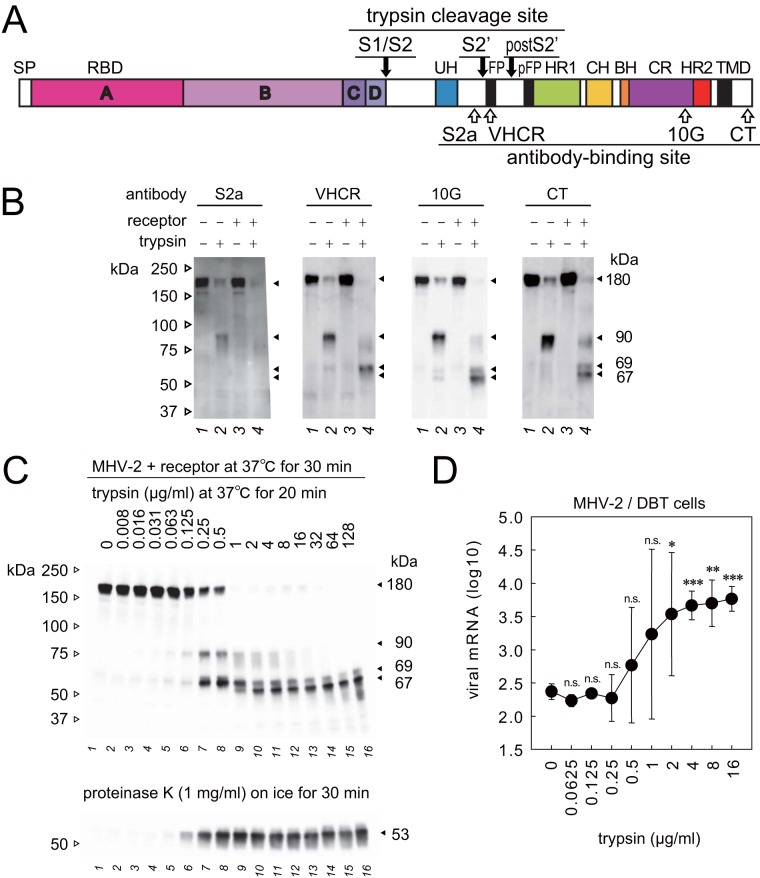
FIG 1.
Proteolytic activation of the MHV-2 spike (S) protein. (A) Schematic diagram of MHV-2 S protein organization. The letters A, B, C, and D indicate domains of the S1 subunit. SP, signal peptide; RBD, receptor-binding domain; UH, upstream helix; FP, fusion peptide; HR1/HR2, heptad repeats; CH, central helix; BH, β-hairpin; CR, connector region; TMD, transmembrane domain. The three trypsin cleavage sites are indicated by black arrows, and the four linear epitopes recognized by antibodies are indicated by white arrows. (B) Two-step conformational changes of S protein primed by receptor binding and triggered by trypsin. MHV-2 pretreated with a soluble form of the receptor (CEACAM1a, receptor) was incubated with trypsin (10 μg/ml). Samples were boiled and subjected to Western blot analysis using the indicated antibodies. (C) Trypsin concentration-dependent cleavage of S protein. MHV-2 pretreated with receptor was incubated with various concentrations of trypsin and then with proteinase K. Samples were boiled and subjected to Western blot analysis using MAb-10G antibody. (D) Trypsin concentration dependence of virus cell entry. MHV-2 was adsorbed onto DBT cells, and various concentrations of trypsin were added. After a 5 h of incubation, viral mRNA was quantified by real-time PCR (n = 6). Data were analyzed relative to the no trypsin control using two-tailed Student t tests. n.s., not significant; *, significant (P ≤ 0.05); **, highly significant (P ≤ 0.01); ***, very highly significant (P ≤ 0.001). Error bars indicate the SD.
As reported previously (26), trypsin-mediated cleavage of the 180-kDa S protein (Fig. 1B, 10G and CT, lane 1) generated a species of 90 kDa (Fig. 1B, 10G and CT, lane 2) in the absence of receptor. However, two different S2 subunit fragments (67 and 69 kDa) were observed following trypsin-mediated cleavage in the presence of receptor (Fig. 1B, 10G and CT, lane 4). The 67-kDa subunit was observed for the first time when a higher concentration of trypsin (10 μg/ml) was used in the present study. The sizes of these S protein subunits are revised from the previous study; the 200-kDa full-length S protein was revised to 180 kDa, the 80-kDa S2 subunit was revised to 90 kDa, and the 66-kDa S2 subunit was revised to 69 kDa (26).
The appearance of the 90-, 67-, 69-, and 53-kDa species was assessed at different trypsin concentrations. The 90- and 69-kDa species were detected following treatment with 0.25 or 0.5 μg/ml trypsin (Fig. 1C). In addition, a 67-kDa S2 subunit was observed when the concentration of trypsin was higher (≥1 μg/ml; Fig. 1C). Interestingly, both 67- and 69-kDa subunits were observed, even at the highest concentration of trypsin (128 μg/ml), which is ∼100-fold higher than needed to induce conformational changes of S protein according to a previous report (26), suggesting that a heterogeneous mix of S2 subunits (67 and 69 kDa) was stably produced from a single (180-kDa) S protein species. Treatment with 1 mg/ml proteinase K was carried out to probe the postfusion conformation that is thought to involve the formation of a 6HB structure (26). As shown in Fig. 1C, a proteinase K-resistant 53-kDa band was observed when the concentration of trypsin was ≥0.125 μg/ml (lane 6).
Quantification of virus cell entry triggered by trypsin.
The trypsin concentration required to induce virus cell entry was assessed. After virus adsorption for 30 min on ice, cells were treated with various concentrations of trypsin at 37°C for 30 min, and trypsin was then inactivated by addition of the serine protease inhibitor camostat mesylate. After 5 h of incubation at 37°C, cellular RNA was isolated, and real-time PCR was performed to quantify viral subgenomic mRNA7. The concentration of trypsin required to induce viral cell entry was ≥0.25 μg/ml (Fig. 1D), which corresponds approximately with the appearance of the 69- and 53-kDa fragments (Fig. 1C), but not the 67-kDa fragment.
Comparison of uncleaved and precleaved S proteins.
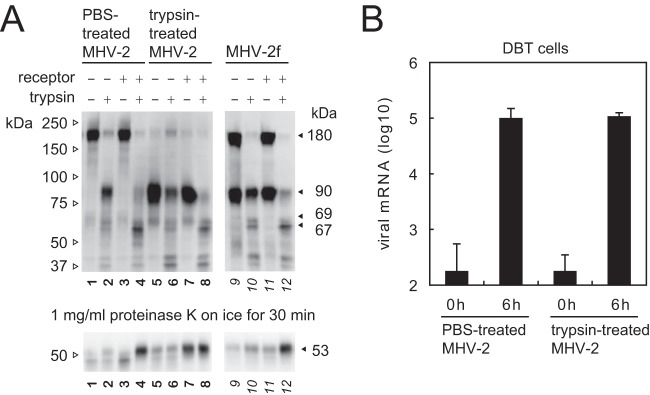
As reported previously, the MHV-2f variant of MHV-2 harbors an amino acid substitution at the S1/S2 cleavage site that is cleaved by furin during biogenesis; this variant is sufficient to induce cell-cell fusion without supplemental trypsin in the culture medium (29). To clarify whether cleavage at the S1/S2 site is sufficient for S protein priming prior to induction of conformational changes, we compared trypsin-treated MHV-2 and MHV-2f with MHV-2. To generate trypsin-treated MHV-2, the MHV-2 was exposed to 1 μg/ml trypsin (or phosphate-buffered saline [PBS] for the uncleaved virus control) for 1 h, followed by the addition of trypsin inhibitors (both soybean trypsin inhibitor [STI] and camostat). Next, the viruses were purified using a Sephadex G-75 column. Figure 2A shows that, as reported previously (29), half of the S protein of MHV-2f was cleaved (lane 9), whereas almost all of the S protein expressed by the trypsin-treated virus was cleaved (lane 5) (compare to PBS-treated MHV-2 S protein [lane 1]). The trypsin-treated virus was able to enter cells as efficiently as the PBS-treated virus (Fig. 2B). The viruses were then exposed to both receptor and trypsin to induce conformational changes. A higher concentration of trypsin (50 μg/ml) was needed to cleave the S protein, presumably due to residual trypsin inhibitors. Although background levels of the 67- and 69-kDa products were observed in the absence of receptor (lane 6) (presumably due to the use of preincubated viruses), clear production of a 67-kDa fragment was observed in the presence of receptor and trypsin (lane 8). Interestingly, induction of the proteinase K-resistant 53-kDa band from the trypsin-treated virus was observed in the presence of receptor, even in the absence of trypsin (lane 7); in contrast, the other two viruses (PBS-treated MHV-2 and MHV-2f) required both receptor and trypsin. This implies that the two types of cleaved S protein have different conformations: the precleaved S protein (cleaved by furin during biogenesis) needs additional cleavage after the receptor-binding step, whereas the trypsin-treated S protein (cleaved on virus particle) needs only receptor binding to trigger the conformational changes. Cleavage at the S2′ site (to generate the 69-kDa species) may trigger conformational changes, as reported previously (20, 30); however, cleavage at either the S1/S2 site or the S2′ site is presumably enough to activate the MHV-2 S protein. The 67- and 69-kDa species can be considered by-products of the S protein following conformational changes; therefore, these products were examined to probe the intermediate conformation(s) of the S protein.
FIG 2.
Proteolytic activation of the precleaved S protein. (A) Conformational changes in precleaved S proteins. To prepare the virus harboring a cleaved S protein, MHV-2 was treated with 1 μg/ml trypsin or PBS (uncleaved control) for 1 h, treated with trypsin inhibitors, and then purified on a Sephadex G-75 column. Conformational changes in PBS-treated MHV-2, trypsin-treated MHV-2, and MHV-2f harboring a precleaved S protein were examined as described for Fig. 1. Samples were boiled and subjected to Western blot analysis with MAb-10G antibody. (B) Cell entry by viruses. Viruses treated with PBS or trypsin were inoculated onto DBT cells, and cellular RNA was isolated after 0 or 6 h. Viral mRNA was quantified by real-time PCR (n = 4).
Characterization of a 67-kDa S2 subunit.
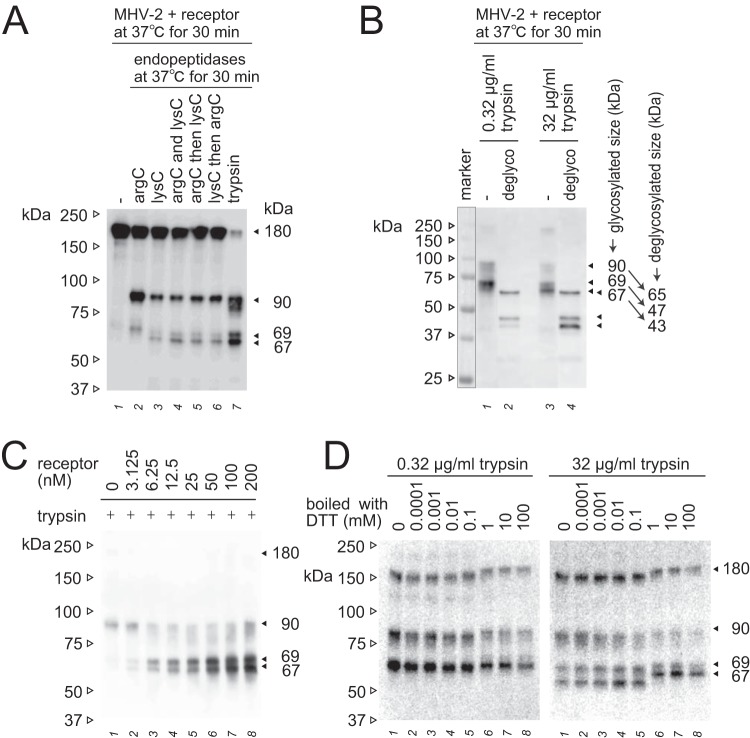
As identified in previous studies using MHV and SARS-CoV, the 90- and 69-kDa subunits are derived from cleavage at the S1/S2 site (R756) and the S2′ site (R907) (11, 19, 27, 28, 30, 31). To identify the trypsin cleavage site that produces the 67-kDa fragment, we attempted to gain indirect evidence because the 67-kDa subunit has not been successfully purified from the virus particle for amino-terminal sequencing. Both the 67- and 69-kDa subunits were detected by MAb-10G and anti-CT, but not by anti-S2a, even following increased exposure of blots, and only the 69-kDa subunit was detected by anti-VHCR (Fig. 1B). These results indicate that the 69-kDa subunit is a product of cleavage between S2a and VHCR epitopes at the S2′ site, as previously reported (11, 30), whereas the 67-kDa subunit is the result of cleavage at the C-terminal side of the VHCR epitope. Using the endoproteinases arg-C and lys-C to identify the specific arginine or lysine residue of the cleavage site, the 67-kDa subunit was found to be cleaved at a lysine residue (Fig. 3A).
FIG 3.
Additional characterization of the 67- and 69-kDa subunits. (A) Specific cleavage of the postS2′ site at arginine or lysine residues. Endopeptidase arg-C (20 μg/ml) and/or lys-C (20 μg/ml) were employed instead of trypsin to induce S protein conformational changes. (B) Deglycosylation of S2 subunits. S protein activated by receptor and trypsin was deglycosylated using a deglycosylation enzyme mix. Nonrelevant lanes on the same blot were sliced out in Adobe Photoshop to align the lanes shown. (C) Receptor concentration dependence. MHV-2 was treated with serially diluted soluble receptor and then with trypsin (10 μg/ml). (D) Effect of redox potential. The 67- and 69-kDa subunits induced by the treatment of receptor and trypsin (0.32 and 32 μg/ml) were boiled in sample buffer containing the indicated concentration of DTT. For all panels, after SDS-PAGE, Western blot analysis was carried out using MAb-10G antibody.
Next, deglycosylation of the S protein subunit was carried out using a commercial deglycosylation mix containing five enzymes that can completely remove N- and O-linked glycans from almost all glycoproteins except some plant and insect glycoproteins. The 90-, 69-, and 67-kDa bands were shifted to 65, 47, and 43 kDa, respectively, after treatment with 0.32 or 32 μg/ml trypsin (Fig. 3B). The 4-kDa size difference between 43- and 47-kDa deglycosylated bands indicates that the 67-kDa subunit is cleaved at K951, because the calculated molecular weight of the peptide fragment between S2′ (R907) and K951 is 4.8 kDa, whereas cleavage at the neighboring lysine K917 would yield a 1.3-kDa size difference and cleavage at K1008 would afford a 10.6-kDa size difference. The K951 cleavage site was therefore designated postS2′. Notably, the postS2′ cleavage site is in the 48th β-strand (β48), which forms an anti-parallel β-sheet with β47 in the prefusion form but not in the postfusion form of the S protein (10, 11). Cleavage at postS2′ is unlikely to contribute to S protein triggering because it is located at the C-terminal side of the fusion peptide (FP; Fig. 4A and B), and the appearance of the 67-kDa species does not correspond to the formation of the postfusion structure (53 kDa) or virus entry (Fig. 1C and D), as described above.
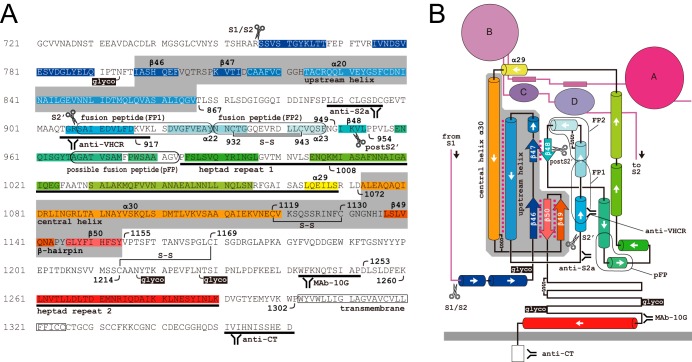
FIG 4.
Primary structure and topology of the MHV-2 S2 subunit. (A) Primary sequence of the S2 subunit. Gray areas indicate invariant motifs present in both pre- and postfusion structures. S-S, disulfide bridge; glyco, N-glycosylation site; scissor mark, trypsin cleavage site. The four antibody-binding sites and the functional motifs are depicted. (B) Topology of the prefusion form of S protein based on the cryoEM structure (10). Colors and labels correspond to those in panel A. Magenta dotted lines indicate molecular interactions between motifs.
Additional experiments were carried out to characterize the 67-kDa subunit. The receptor concentration was found to affect the total amount of 67- and 69-kDa species, but not the relative ratio between them (Fig. 3C), implying that two different cleavable features of the S2 subunit were induced in the receptor-binding step. Furthermore, inclusion of dithiothreitol (DTT) at concentrations of ≥1 mM in the sample buffer shifted the 67-kDa band to a greater extent than the 69- and 90-kDa bands during SDS-PAGE (Fig. 3D). Two cysteine residues at positions 1169 and 1214 (Fig. 4A and B), between the HR1 and HR2 motifs (in the connector region), were predicted to affect the conformation of the 67-kDa subunit. Meanwhile, cysteine residues 932 and 943, located upstream of HR1, and residues 1119 and 1130 within the invariant motifs may affect the smaller shift observed for the 69- and 90-kDa bands. The connector region, including the disulfide bond, presumably contributes to folding of the 67-kDa subunit, but not to that of the 69- and 90-kDa subunits.
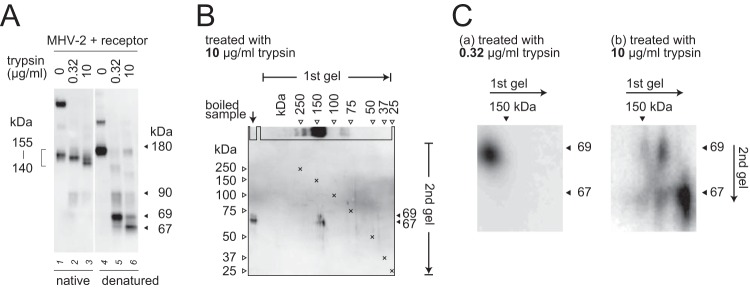
Two different sizes of S2 subunit are present in the trimer.
We next turned our attention to how two different sizes of S2 subunit (67 and 69 kDa) could be produced by trypsin digestion of a single 180-kDa S protein species, even though a single-sized (90-kDa) species is produced in the absence of receptor. We hypothesized that the S protein forms a heterogeneous trimer in the presence of receptor, so that each S protein in a trimer exposes a different cleavage site. In the first native SDS-PAGE step (gels were boiled before blotting so that the trimer could be detected by the antibody, as described in Materials and Methods), several different sizes of trimer were detected (Fig. 5A, lanes 2 and 3). In the second denaturing SDS-PAGE step, these trimer bands were separated into monomer bands (Fig. 5A, lanes 5 and 6). To examine the components of the trimer, we performed two-dimensional electrophoresis. After treatment with 10 μg/ml trypsin, both the 67- and 69-kDa bands, or the single 67-kDa band, were separated from three species of trimer at around 140 kDa (Fig. 5B and Cb), whereas only the 69-kDa band was detected following treatment with 0.32 μg/ml trypsin (Fig. 5Ca). These results suggest that a subset of S2 subunits can form a heterogeneous trimer comprising the 69- and 67-kDa species at a ratio of 3:0, 2:1, 1:2, or 0:3.
FIG 5.
Components of the S2 trimer after triggering. (A) Comparison of native and denatured S protein. MHV-2 treated with soluble receptor and trypsin was divided into two aliquots for unboiled (native) and boiled (denatured) treatments. After electrophoresis on a 3 to 10% gel, the gel was boiled in sample buffer at 105°C for 5 min in an autoclave and then transferred to a PVDF membrane. (B) Two-dimensional electrophoresis. The reaction mixture prepared as described for panel A was mixed with sample buffer containing molecular size markers, and two-dimensional electrophoresis was carried out. After the first electrophoresis step, the first gel (3 to 10%) was boiled in sample buffer, sliced along the markers, and laid onto the second gel (7.5%). After the second electrophoresis step, the gel was transferred to a PVDF membrane. X marks indicate overlaid molecular size markers. (C) Cropped and enlarged blots. Blots from two-dimensional electrophoresis and Western blot analysis corresponding to 0.32- or 10-μg/ml trypsin treatment were cropped at 67 and 69 kDa and enlarged by graphical manipulation software. For all panels, Western blot analysis was carried out using MAb-10G antibody.
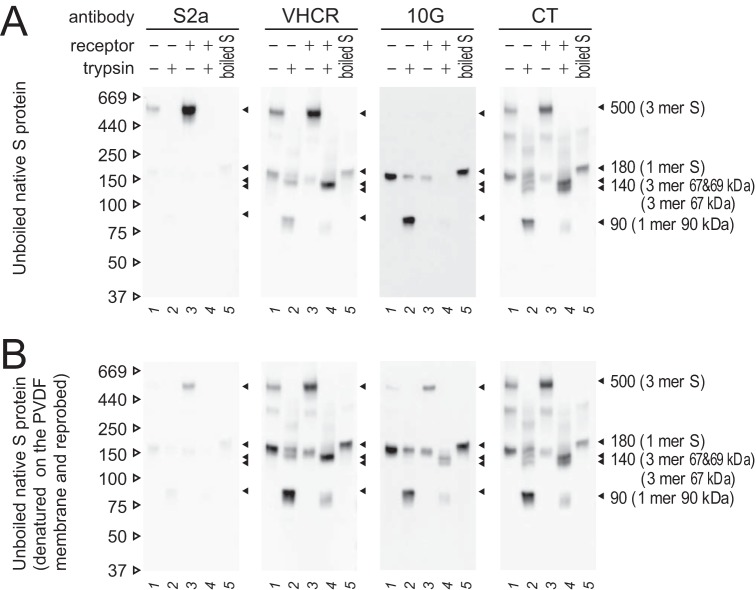
Exposed or buried epitope configurations in the S protein globule.
Further characterization of S2 subunits was performed to investigate exposed and buried configurations of epitopes, and the results are shown in Fig. 6. MHV-2 was treated with receptor and trypsin to induce conformational changes, as described above. S protein detection was performed using four anti-S2 antibodies recognizing linear epitopes. Following soaking in stripping buffer to denature the S protein on the polyvinylidene difluoride (PVDF) membrane, detection was then repeated with the same antibodies. When antibodies against the VHCR and CT epitopes were used, all bands detected in denatured blots (Fig. 6B) were the same as those observed in native blots (Fig. 6A), indicating that these epitopes are exposed on the S protein in both the native and denatured states. In the postfusion form of the S protein treated with receptor and trypsin, doublet bands at 140 kDa were observed using anti-CT antibody (Fig. 6A, CT, lane 4), while lower bands were not observed with the anti-VHCR antibody (Fig. 6A, VHCR, lane 4). This observation correlates with the appearance of 67- and 69-kDa bands in SDS-PAGE (Fig. 1B, CT and VHCR, lane 4), suggesting that the upper band of 140-kDa species is composed of 67- and 69-kDa subunits, whereas the lower band comprises three 67-kDa subunits. This is consistent with the results in Fig. 5Cb showing that the 67- and 69-kDa bands were separated from 140-kDa trimer bands.
FIG 6.
Exposed or buried configurations of epitopes in the S protein globule. (A) Native SDS-PAGE. Unboiled samples were subjected to Western blot analysis. (B) Denaturing and reprobing. PVDF membranes from panel A were soaked in stripping buffer for 5 min to denature the bound S protein and subjected to detection with the same antibodies a second time. For both panels, Western blot analysis was performed using the indicated antibodies.
Using anti-S2a antibody recognizing a linear epitope (Fig. 1B, S2a) (34), a 500-kDa band corresponding to a trimeric assembly was observed only after the receptor-binding step in native blots (Fig. 6A, S2a, lane 3), as observed in a previous study (26), but this was barely detectable in denatured blots (Fig. 6B, S2a, lane 3). We do not currently have an explanation for exposure of the S2a epitope, but this hydrophobic epitope is buried in the globule in the pre-fusion state (10) and is presumably exposed only in the native trimer after the receptor-binding step, before disappearing following cleavage by trypsin. When antibody recognizing the 10G epitope was used, monomeric S2 subunit species were detected in the native blot (Fig. 6A, 10G, lanes 3 and 4), but trimeric species were not observed, although they were detectable in the denatured blot (Fig. 6B, 10G, lanes 3 and 4). This indicates that the 10G epitope adopts a buried configuration in the trimer of pre- and postfusion forms. These observations provided clues that were used to predict the intermediate structure of the S2 subunit.
Timing of S protein cleavage.
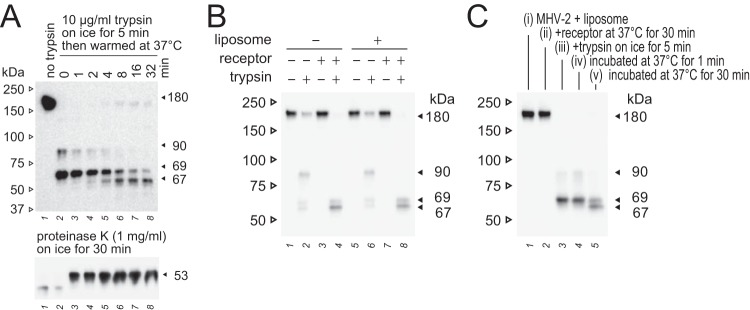
We next analyzed the timing of the appearance of 67-, 69-, and 53-kDa fragments during conformational changes of the S protein. To stop the reaction after different incubation times at 37°C, reaction tubes were quickly frozen in dry ice-methanol and immediately boiled in sample buffer containing the trypsin inhibitor. As shown in Fig. 7A, trypsin treatment on ice for 5 min following receptor treatment was sufficient to generate the 69-kDa but not the proteinase K-resistant 53-kDa species (lane 2). The 53-kDa band was induced after a 1-min incubation at 37°C (lane 3), and the 67-kDa band appeared and became prominent over 4 min (lane 5), indicating that the conformational changes that induce the 6HB structure are complete at 1 min after treatment with trypsin. In preliminary electron microscopy (EM) experiments (described below), conformational changes of the S protein were probed in the presence of liposomes. The appearance of S2 fragments (90, 69, and 67 kDa) and the timing of cleavage coinciding with their appearance were the same in the absence and presence of liposomes (Fig. 7B and C).
FIG 7.
Timing of the appearance of the 90-, 69-, 67-, and 53-kDa species. (A) Time course of 90-, 69-, 67-, and 53-kDa fragment generation. MHV-2 was treated with receptor and trypsin, and the reaction was stopped by freezing at the indicated time points. (B) Two-step conformational changes in the S protein primed by receptor binding and triggered by trypsin in the presence of liposomes. (C) Time course of the generation of 90-, 69-, and 67-kDa fragments in the presence of liposomes. MHV-2 was treated with receptor and trypsin in the presence of liposomes, and the reaction was stopped by freezing at the indicated time points. For all panels, the appearance of S2 subunits (90, 69, and 67 kDa) was detected by Western blot analysis using MAb-10G antibody.
Unpacked (accessible) and packed (occluded) HR conformations.
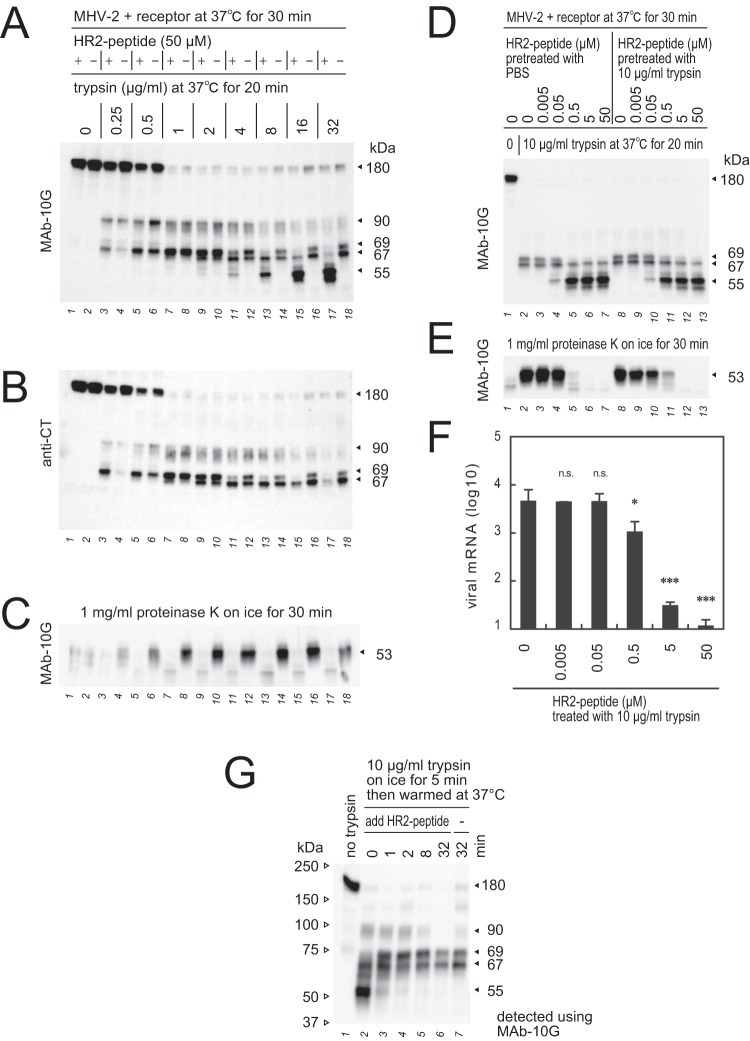
To characterize the conformational states of 67- and 69-kDa subunits after the receptor-binding step, HR2-mimicking peptide was used. This 39-amino-acid peptide derived from the HR2 region of the S2 subunit is water soluble and interferes with packing of the HR1/HR2 motif, thereby inhibiting virus infection (35). MHV-2 pretreated with soluble receptor was treated with HR2-mimicking peptide (50 μM) and various concentrations of trypsin. The 69-kDa but not the 67-kDa subunit disappeared following treatment with HR2-mimicking peptide, and the 55-kDa degradation product appeared after incubation in the presence of 8 μg/ml trypsin (Fig. 8A, lane 13). Presumably, protease-cleavable sites in the S2 subunit are vulnerable to scission by trypsin when the HR2-mimicking peptide is present due to the restriction of helical bundle formation by the HR motif; hence, the cleavage pattern was altered. This also indicates that the HR1/HR2 motif in the 67-kDa fragment forms a postfusion species in which the HR1 motif is occluded after the receptor-binding step.
FIG 8.
Interaction of HR2-mimicking peptide with the S2 subunit. (A) Effect of HR2-mimicking peptide (HR2-peptide) during S protein triggering. HR2-peptide (50 μM) was added to MHV-2 after the receptor-binding step, and reaction mixtures were treated with various concentrations of trypsin. (B) The polyvinylidene difluoride membrane from panel A was reprobed with anti-CT antibody. (C) Reaction mixtures from panel A were treated with proteinase K to generate the 53-kDa fragment. (D) Effect of trypsin on the HR2-peptide. HR2-peptide nontreated or pretreated with 10 μg/ml trypsin for 30 min was diluted and added to the reaction mixture containing MHV-2 and receptor, and reaction mixtures were treated with 10 μg/ml trypsin. (E) Reaction mixtures from panel D were treated with proteinase K to generate the 53-kDa fragment. (F) Blocking virus cell entry. MHV-2 was adsorbed onto DBT cells, and 10 μg/ml trypsin was added in the presence or absence of HR2-peptide. After 5 h of incubation, viral mRNA was quantified by real-time PCR (n = 6). Data were analyzed relative to the no-peptide control using two-tailed Student t tests, as described in the legend of Fig. 1. (G) Time course of HR1/HR2 motif packing. During S protein activation by receptor and trypsin, the reaction was stopped by freezing at the indicated time points, and HR2-peptide was added, followed by incubation for 20 min to facilitate the formation of 67- and 69-kDa fragments. For panels A to E and panel G, Western blot analysis was performed using the indicated antibodies.
Interestingly, HR2-mimicking peptide completely inhibited production of the proteinase K-resistant 53-kDa fragment (Fig. 8C, lanes 11 and 13), even in the presence of the packed 67-kDa species (Fig. 8A, lanes 11 and 13), suggesting that HR2-mimicking peptide interferes with 6HB formation. The trimer in the receptor-binding step is presumably constructed from both packed and unpacked HR1/HR2 motifs, and HR2-mimicking peptide interacts with an unpacked motif, and thereby interferes with 6HB formation. In the presence of 0.5 to 2 μg/ml trypsin, HR2-mimicking peptide did not appear to affect degradation of the 69-kDa fragment due to the low concentration of trypsin (Fig. 8A, lanes 5 to 10). In addition, trypsin degradation products in the presence of HR2-mimicking peptide were detected by MAb-10G antibody (Fig. 8A, lanes 13, 15, and 17) but not by anti-CT antibody (Fig. 8B, lanes 13, 15, and 17), indicating that the 10G epitope region is folded and consequently avoids cleavage by trypsin, whereas the C-terminal side of the 10G epitope, including the HR2 region (residues 1253 to 1302, Fig. 4A) is presumably unfolded and therefore degraded by trypsin. Although the mechanism remains unknown, HR2-mimicking peptide enhanced the production of the 69-kDa subunit following treatment with 0.25 μg/ml trypsin (Fig. 8B, lane 3).
Next, the concentration dependence of HR2-mimicking peptide was assessed. In the presence of ≥0.5 μM HR2-mimicking peptide, the 69- and 53-kDa bands disappeared (Fig. 8D and E, lanes 5 to 7), and the 55-kDa degradation products appeared (Fig. 8D, lanes 5 to 7). Pretreatment of HR2-mimicking peptide with 10 μg/ml trypsin did not affect the ability to cause the disappearance of the 69- and 53-kDa fragments (Fig. 8D and E, lanes 11 to 13), indicating that trypsin does not directly affect HR2-mimicking peptide. To determine whether the HR2-mimicking peptide actually blocks MHV-2 infection, real-time PCR-based virus entry assays were performed, as previously reported for SARS-CoV in which HR2-mimicking peptide blocks trypsin-mediated direct viral entry from the cell surface (36). In the presence of ≥0.5 μM HR2-mimicking peptide, virus entry was clearly blocked (Fig. 8F).
Timing of HR packing.
Next, the timing of HR1/HR2 motif packing in the S protein was analyzed. HR2-mimicking peptide was added to the S protein conformational change reaction at the indicated time points after stopping the reaction using dry ice-methanol, and mixtures were reincubated at 37°C for 20 min to facilitate production of the 67- and 69-kDa fragments (reincubation is required to visualize bands that reflect the structure present upon HR2-mimicking peptide addition). At each HR2-mimicking peptide addition time point, if the HR1/HR2 motif forms an unpacked structure, the HR2-mimicking peptide would be expected to bind to HR1 and trypsin would degrade the 69-kDa to produce the 55-kDa lacking the CT epitope as described above (Fig. 8A and B). As shown in Fig. 8G (lane 2), the 67-kDa subunit appeared, the 69-kDa subunit disappeared, and the 55-kDa degradation product appeared following treatment with HR2-mimicking peptide after the receptor-binding step, suggesting that both packed and unpacked HR1/HR2 motifs simultaneously occur in the reaction. After a 1-min incubation at 37°C, undegraded 69-kDa subunit, which does not interact with HR2-mimicking peptide, was observed (Fig. 8G, lane 3), suggesting that both the 67- and 69-kDa fragments form a packed structure at this time point.
Assembly of three S2 subunits at the center of the S protein trimer.
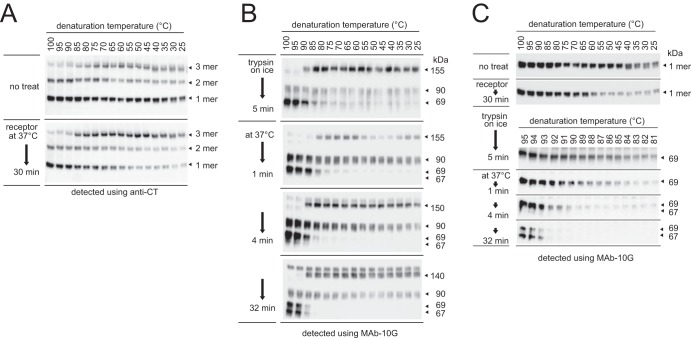
As shown in Fig. 5A and 6A, interactions between subunits in the S protein trimer were enhanced by receptor and trypsin treatment, and it remained stable in sample buffer containing 0.5% SDS during SDS-PAGE. To compare the strengths of intermolecular interactions within the trimer at each step during the conformational changes, reaction mixtures of virus treated with receptor and trypsin were frozen at the indicated time points using dry ice-methanol, mixed with sample buffer containing trypsin inhibitor and 0.5% SDS, and incubated at different temperatures between 25 and 100°C at intervals of 5°C. Both the trimer and the dissociated monomer were detected via Western blotting following soaking of the PVDF membrane in stripping buffer used for trimer detection by MAb-10G. As shown in Fig. 9A, the nontreated trimer in the prefusion state dissociated into the monomeric form at 25°C. After receptor binding, the dissociation temperature was increased to ∼60°C. The dissociation temperature of the trimer was further increased to 90°C after trypsin treatment and finally reached 95°C after 32 min (Fig. 9B).
FIG 9.
Timing of S2 subunit assembly. (A) Thermostability of the trimer in the receptor-binding step. The reaction analyzing conformational changes of S protein was stopped by freezing at the indicated time points after a 30-min treatment with receptor. (B) Thermostability of the trimer in the proteolysis step. Reaction mixtures prepared as described for panel A were treated with trypsin and stopped by freezing at the indicated time points. (C) More detailed analysis of S protein thermostability. The dissociation temperature of mixtures prepared as described for panel B was explored between 81 and 95°C at intervals of 1°C. For all panels, reaction mixtures were treated with sample buffer containing protease inhibitor and 0.5% SDS on ice and then incubated at the indicated temperature using a Veriti thermal cycler (Thermo Fisher). After electrophoresis and electroblotting, the PVDF membrane was soaked in stripping buffer for trimer detection by MAb-10G. Dissociated monomer bands from each step of the conformational changes were cropped and aligned to compare the dissociation temperature in panel C.
To explore the interactions of the three S2 subunits in more detail after the protease digestion step, experiments were performed between 81 and 95°C at intervals of 1°C, and the Western blots were cropped and aligned to compare the dissociated monomers (Fig. 9C). After trypsin treatment on ice, the dissociation temperature was ∼85°C, and this gradually increased to 94°C during incubation, corresponding with the appearance of the 67-kDa fragment. These results indicate that the three S2 subunits in the trimer partially assemble at the receptor-binding step, and their interaction is dramatically enhanced by trypsin treatment. The postS2′ site is finally cleaved after assembly.
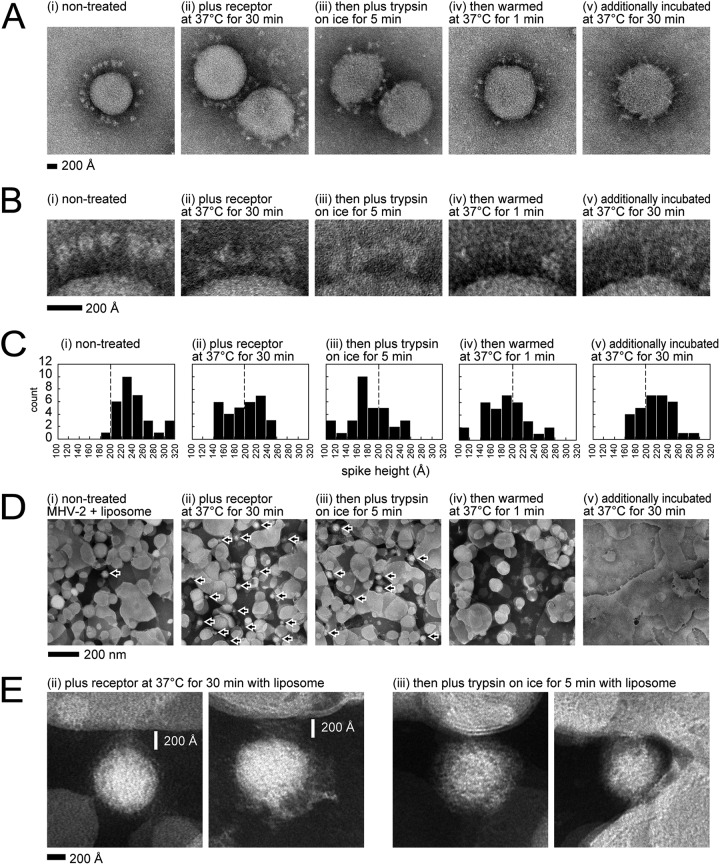
Negative-stain EM.
Each virus activation step described above was visualized by negative-stain EM (Fig. 10A), and enlarged views of images are shown in Fig. 10B. For nontreated virus, intact S protein globules were observed as uniform alignments on the viral membrane (Fig. 10Bi). The height (distance) from the viral membrane to the top of the S protein was measured and is presented as a histogram (Fig. 10C). The height of uniform nontreated S protein was ∼240 Å, but after receptor treatment, the shapes of S proteins became more variable, gaps between S protein globules and the viral membrane were reduced (Fig. 10Bii), and the heights of half of the S protein population were decreased to <200 Å (Fig. 10Cii). After trypsin treatment for 5 min on ice, the S1 subunits appeared to remain on the S2 subunit, their shapes became obscured, and the gaps were further reduced (Fig. 10Biii and Ciii). At 1 min after warming at 37°C, a few elongated cone-like structures were observed (Fig. 10Biv), as previously reported using recombinant S2 subunit (11). After additional incubation at 37°C for 30 min, almost all S protein globules disappeared, and many elongated cone-like structures appeared (Fig. 10Av and Bv).
FIG 10.
Visualization by negative-stain electron microscopy (EM). (A) Virus particles. MHV-2 treated with or without receptor (1 μM) and trypsin (10 μg/ml) to induce conformational changes in S protein was subjected to negative-stain EM. (B) Enlarged view of S protein on the viral particle. (C) Histogram of S protein height above the membrane. S proteins (n = 32) in five viral particles were measured, and the results are represented as a histogram. (D) Visualization of virus binding to liposomes. Liposomes were added to the reaction to induce conformational changes in the viral S protein. Viruses are indicated by arrows. (E) Enlarged view of virus binding to liposomes. The virus particles observed in panel D are enlarged.
The virus activation steps described above were then probed in the presence of liposomes. On EM grids, an excessive number of liposomes but very few viruses were observed in the absence of soluble receptor (Fig. 10Di), whereas many virus particles were observed on liposomes in the presence of soluble receptor (Fig. 10Dii). These results correspond to liposome flotation assays in a previous study in which virus binding to liposomes was induced by receptor (26). Virus particles disappeared at 1 min after trypsin treatment (Fig. 10Div), and fused liposomes were the dominant species observed at 30 min (Fig. 10Dv), suggesting that membrane fusion with liposomes was largely complete within 1 min, and liposomes then fused with each other using the remaining S protein. An enlarged view of virus-bound liposomes shown in Fig. 10Dii and iii is presented in Fig. 10E. The gaps between viruses and liposomes that are presumably bridged by S protein were ∼200 Å after the receptor-binding step (Fig. 10Eii), and this distance was reduced by trypsin treatment on ice (Fig. 10Eiii). A hazy density was evident between viruses and liposomes (Fig. 10E).
DISCUSSION
The S protein of the MHV-2 virion is uncleaved, similar to the S protein of MERS-CoV and SARS-CoV, which requires cellular protease following receptor binding to induce S1 dissociation from S2 and the subsequent conformational changes necessary for membrane fusion (26). In contrast, most MHV variants carry S proteins that are cleaved by cellular furin at the S1/S2 site during biogenesis, which was believed to require only receptor binding to induce membrane fusion (37, 38). However, previous studies of MHV and MERS-CoV suggest that the coronaviruses harboring precleaved S protein (cleaved by furin during biogenesis) at the S1/S2 site require further cellular proteases to facilitate cell entry (20, 30). Our results also suggest that the S protein cleaved during biogenesis by furin requires additional cleavage to induce conformational changes required to adopt a proteinase K-resistant conformation (Fig. 2A, lane 12). However, the trypsin-treated S protein cleaved on virus particles at the S1/S2 site does not require additional cleavage; it needs only receptor binding (Fig. 2A, lane 7). Therefore, these two types of S protein cleaved at the S1/S2 site are predicted to have different conformations. In the present study, using MHV-2, the 67- and 69-kDa products were considered to be by-products that reflect intermediate conformations of the S protein, although the importance of cleavage at the S2′ site was reported for coronavirus S proteins (20, 30).
Previous studies used cryoEM to examine conformational changes within the coronavirus S protein during the priming stage, which is induced by receptor binding; the results observed after receptor binding indicated an asymmetric trimer with opened and closed domains within the S1 subunit and a tightly assembled central helix in the S2 subunits (4, 6). However, no major differences in the HR region were observed. Although structural analysis using cryoEM is excellent for detecting stable protein structures, unstable structures such as the HR2 motifs in the S2 subunit cannot be analyzed using this technique, even if a large number of protein particles are captured; population-based biochemical analysis is needed to capture features of unstable proteins. Here, we predicted the dynamic rearrangements of S2 subunits underpinning the transition from pre- to postfusion structures based on Western blot analysis of the 67- and 69-kDa by-products that are induced following trypsin digestion of the S protein.
Our model of the S protein conformational changes was constructed based on previously reported pre- and postfusion structures (Protein Data Bank codes 3JCL and 6B3O, respectively). The S2 subunit of coronavirus features a topology similar to that of the paramyxovirus F protein, comprising a core β-sheet, an upstream helix, and a central helix, and these motifs are essentially identical in pre- and postfusion F protein structures (10, 39). These invariant MHV-2 S protein motifs are assumed not to change conformation and are displayed as a fixed globule represented by a gray column in Fig. 11. The flexible regions in the S2 subunit that undergo conformational changes are positioned in the FP region (residues 867 to 949) and the HR1 region (residues 954 to 1072), as shown in Fig. 4. In addition, the region downstream of the core β-sheet (residues 1155 to 1302) that includes the HR2 region also appears to be flexible (Fig. 4).
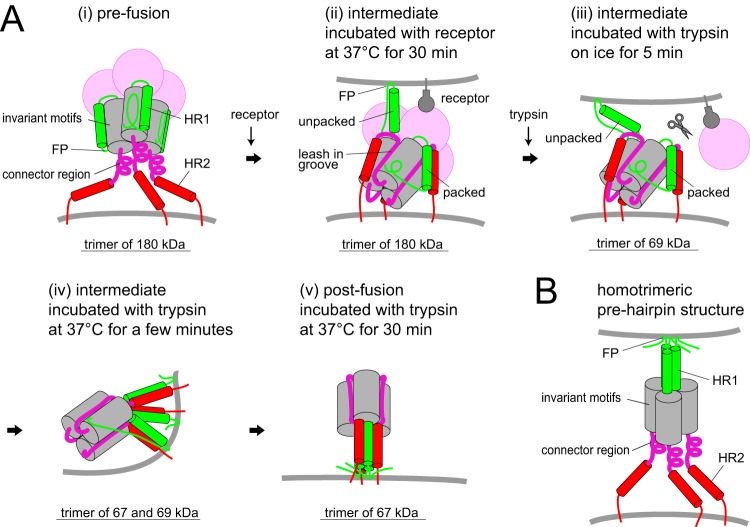
FIG 11.
Schematic diagram of S protein activation. (A) Conformational changes of the S protein trimer. (B) Homotrimeric prehairpin structure in the unified model of class I viral fusion protein assembly. FP, fusion peptide; HR1/HR2, heptad repeats.
In the present and previous studies, we observed eight features related to conformational changes that occur in the flexible regions of the S2 subunit. The first four features are induced by receptor binding. (i) During the first step, the S protein binds to the target membrane, as demonstrated by liposome-binding assays in a previous study (26) and negative-stain EM in this study (Fig. 10Dii). Because the FP region is located close to β48, at least one β48 per trimer must be dissociated from β47 in invariant motifs for the FP region to be free. It is important to remember that the exact location of the FP region of coronavirus S protein remains controversial. The FP region (residues 867 to 949) immediately follows an S2′ cleavage site (40), and another possible FP region (pFP; residues 967 to 983) is adjacent to the region upstream of HR1 (Fig. 4), as reported previously (41, 42). (ii) The metastable form of the S protein trimer is converted to a stable trimer (Fig. 6A, VHCR, lane 3), for which the dissociation temperature was increased from 25 to 60°C (Fig. 9C). During this stage, the movement of HR1 to form a trimeric α-helical coiled-coil on the distal side of the viral membrane is restricted because the S2 subunit is still covered by the S1 subunit in the uncleaved S protein (Fig. 10Aii). Therefore, regions at α29 and α30 (the central helix) are only capable of interacting within the trimer (Fig. 4B). This is consistent with a previous study in which the tightly packed central helix in the S2 subunit was detected by cryoEM after the receptor-binding step (4, 6). (iii) The 106 amino acids of the connector region (residues 1155 to 1260, Fig. 4A), including the 10G epitope and disulfide bond at positions 1169 and 1214, presumably form a folded structure because the 10G epitope in the trimer was undetectable in the native blot (Fig. 6A, 10G, lane 3) and was resistant to trypsin even in the presence of HR2-mimicking peptide (Fig. 8A). Reduction of the gap between the S protein globule and the viral membrane (∼200 Å) in EM images (Fig. 10Bii and Cii) may reflect a folded C-terminal side of the S2 subunit. In addition, the 10G region is likely fixed near helix α29 at the top of the S2 trimer to allow 6HB assembly in the following step (11). These observations indicate that, in the S protein structure appearing after the receptor-binding step, the core of the connector leash is packed into the groove of the invariant motif (Fig. 11Aii, purple lines), as seen in the postfusion structure (11). (iv) The HR1/HR2 motif forms the packed-hairpin structure observed as the 67-kDa subunit after trypsin treatment that is unable to interact with HR2-mimicking peptide (Fig. 8A, lane 13, and Fig. 8G, lane 2). Based on the steric configuration of the postfusion structure, HR1 of one S2 subunit must be packed with HR2 of another S2 subunit in the trimer to form a hairpin structure (11). In addition, unpacked HR1, observed as the 69-kDa band that was degraded following treatment with HR2-mimicking peptide and trypsin (Fig. 8G, lane 2), also appeared in this step. Interestingly, both 69- and 67-kDa subunits were present in the trimer, as revealed by two-dimensional electrophoresis (Fig. 5Cb), suggesting that a subset of S2 subunits form a heterogeneous trimer constructed from two different conformational states. We predict that the trimer includes at least one packed HR1/HR2 motif and extends at least one unpacked HR1 motif toward the target membrane to expose the FP region, and the S2 subunit is covered by the S1 subunit in the uncleaved S protein (Fig. 11Aii). These features suggest that the S protein cannot form the typical homotrimeric prehairpin structure predicted in the current model of class I viral fusion protein assembly (Fig. 11B).
The next four features of the conformational changes are induced by trypsin treatment, during which obscure-shaped S proteins were observed by EM (Fig. 10Biii). (v) The S1/S2 site is cleaved by trypsin to generate the 90-kDa subunit (Fig. 1C, lane 7, and Fig. 7A, lane 2), and the S2′ site is subsequently cleaved by trypsin to generate the 69-kDa subunit (Fig. 1C, lanes 7 to 16). Release of the S1 subunit from the S2 subunit presumably enhances interactions within three S2 subunits and triggers conformational changes in the HR1/HR2 motifs. (vi) All three HR1/HR2 motifs in the trimer appear to form a packed-hairpin structure at 1 min after warming at 37°C, because HR2-mimicking peptide cannot interact with them (Fig. 8G, lane 3). (vii) Simultaneously, the three HR1/HR2 motifs assemble at the center to form a 6HB core at 1 min after trypsin treatment (Fig. 7A, bottom, lane 3). At this stage, the dissociation temperature of the trimer was ∼90°C (Fig. 9C). (viii) The postS2′ cleavage site in β48 is finally cleaved by trypsin after assembly of the three HR motifs at the center, resulting in the appearance of the 67-kDa subunit (Fig. 9C bottom). The final 6HB structure is constructed from 67- and 69-kDa subunits or three 67-kDa subunits (Fig. 5Cb). Numerous elongated cone-like structures were observed by EM (Fig. 10Av).
As described above, at least one HR1/HR2 motif per trimer may form a packed-hairpin structure after the receptor-binding step and another exposes the FP region, as illustrated in Fig. 11Aii. This conformation presumably facilitates formation of an asymmetric trimer. In principle, the class I viral fusion protein trimer must break its rotational symmetry during activation to overcome the steric hindrance within the three fusion protein subunits (43, 44). When the different conformational states of the HR1/HR2 motif assemble in the trimer, the axis of the trimer is presumably tilted against the viral membrane (Fig. 11Avii), resulting in the breaking of rotational symmetry. This is followed by formation of 6HB and membrane fusion. Flexibility at the juxtamembrane region and the tilted orientation against the viral membrane required for membrane fusion were recently reported by a study examining influenza HA in detergent micelles analyzed by cryoEM (45). We anticipate that the mechanism suggested by Western blot analysis here will be confirmed by other approaches such as high-resolution cryoEM and/or in silico simulation.
MATERIALS AND METHODS
Virus, cells, soluble receptor, and HR2-mimicking peptide.
MHV-2 was propagated in DBT cells cultured in Dulbecco modified Eagle medium (DMEM) containing 10% tryptose phosphate broth (BD Difco) (46). Viruses were collected at 21 h postinfection and stored at –80°C. The soluble form of the MHV receptor (CEACAM1a) was produced using recombinant baculovirus and purified as previously described (47). HR2-mimicking peptide (DLSLDFEKLNVTLLDLTYEMNRIQDAIKKLNESYINLKE) was provided by B. J. Bosch (35) and dissolved in water at a concentration of 500 μM.
Proteases and protease inhibitors.
Trypsin (T8802; Sigma), proteinase K (166-21051; Wako, Japan), endoproteinase arg-C (P6056; Sigma), and endoproteinase lys-C (P3428; Sigma) were employed after being dissolved in PBS (pH 7.4). The inhibitors camostat (3193; Tocris Bioscience, UK), Pefabloc SC (11429868001; Roche, Switzerland), and E64d (330005; Calbiochem) were dissolved in dimethyl sulfoxide. Soybean trypsin inhibitor (STI; T-9128; Sigma) was dissolved in PBS.
Liposomes.
Lipids l-phosphatidylcholine (PC; egg; Avanti-Polar Lipids), l-phosphatidylethanolamine (PE; egg; Avanti-Polar Lipids), sphingomyelin (Sph; brain; Avanti-Polar Lipids), and cholesterol (Chol; Sigma) were mixed in a 1:1:1:1.5 molar PC:PE:Sph:Chol ratio, dried under N2 gas in a glass tube, and lyophilized overnight. After the addition of 1 ml of PBS (pH 7.2), the lipid suspension was vortexed and extruded 25 times through a 0.4-μm Nuclepore filter (GE Water & Process Technologies) using an Avanti Mini-Extruder. Liposomes were stored at 4°C and used within 1 week.
Activation of MHV-2 spike (S) protein and Western blot analysis.
(i) Standard reaction. To evaluate conformational changes in the S protein occurring in the first step, a 10-μl solution of MHV-2 (107 PFU/100 μl) was mixed with 1.1 μl of soluble receptor (10 μM) and warmed at 37°C for 30 min. For conformational changes in the second step, 1.2 μl of trypsin (100 μg/ml) was added, followed by incubation at 37°C for 30 min. A 1/4 volume of sample buffer comprising 30% glycerol, 250 mM Tris (pH 6.8), 2.5% SDS, a small amount of bromophenol blue, 100 mM DTT, and 1 mM Pefabloc SC was added to the reaction, and the mixture was boiled at 100°C for 5 min. Samples were separated by SDS-PAGE on a 3 to 10% gradient or a 7.5% gel (e-PAGEL; ATTO, Japan), transferred to a PVDF membrane (Immobilon-P; Millipore), and soaked in ImmunoBlock (CTKN001; DS Pharma Biomedical, Japan) for 5 min. Western blot analysis was carried out using anti-S2 antibodies, a mouse monoclonal antibody recognizing the 10G epitope (MAb-10G), and rabbit anti-peptide antibodies recognizing the S2a region, the very highly conserved region (VHCR), and cytoplasmic tail (CT) epitopes (anti-S2a, anti-VHCR, and anti-CT, respectively), followed by horseradish peroxidase-conjugated anti-mouse (32430; Thermo) or anti-rabbit (sc-2054; Santa Cruz Biotech) IgG. Immunoreactive bands were visualized with an enhanced chemiluminescence kit (RPN2232; GE Healthcare) and a LAS-3000 instrument (Fuji, Japan). All experiments were repeated at least twice.
(ii) Reactions in the presence of liposomes. A 6-μl sample of liposomes was added, and the volume of receptor and trypsin was raised to achieve the target concentrations. Standard reactions were then carried out as described above.
(iii) Timing of HR packing. To stop the reaction at the indicated time points, reactions were quickly frozen in dry ice/methanol, and 1.4 μl of HR2-mimicking peptide (500 μM) was added. Samples were further incubated at 37°C for 20 min to facilitate the formation of 67- and 69-kDa fragments.
(iv) Generation of the proteinase K-resistant 53-kDa fragment. After assessing conformational changes of the S protein as described above, reaction mixtures were chilled on ice for 5 min and 1.5 μl of proteinase K (10 mg/ml) was added, followed by incubation on ice for 30 min.
(v) Deglycosylation of the S2 subunit. After assessing conformational changes of the S protein as described above, deglycosylation was carried out using protein deglycosylation mix (P6039S; NEB, UK) according to the manufacturer’s instructions.
(vi) SDS-PAGE of unboiled samples (native PAGE). After assessing conformational changes of the S protein as described above, sample buffer excluding DTT was added and unboiled mixtures were separated by SDS-PAGE (3 to 10% gradient or 7.5% gel; e-PAGEL).
(vii) Protein denaturation on PVDF membranes. After SDS-PAGE of unboiled samples and electrotransfer to a PVDF membrane, initial detection of native S protein was performed by Western blot analysis. The membrane was then soaked in stripping buffer (catalog no. 46428; Thermo Fisher) at room temperature for 5 min to denature the S protein, rinsed 10 times with rinse buffer (20845; Millipore), blocked with ImmunoBlock, and reprobed with anti-S2 antibody.
(viii) Two-dimensional SDS-PAGE. S protein was treated with receptor and trypsin as described above, mixed with sample buffer containing molecular size markers (1610373; Bio-Rad) without DTT, and separated by SDS-PAGE using a 3 to 10% gradient gel (first gel). After electrophoresis, the gel was wrapped in a heat-seal bag, soaked in sample buffer (0.5% SDS), boiled at 105°C for 5 min in an autoclave, sliced along the protein markers, placed onto a 7.5% gel (second gel), and subjected to electrophoresis and Western blot analysis.
Generating trypsin-treated MHV-2 harboring cleaved S protein.
A solution of MHV-2 (500 μl; 107 PFU/100 μl) was mixed with 5 μl of trypsin (100 μg/ml) or PBS (for uncleaved control) and incubated at 37°C for 1 h. Next, 5 μl of STI (10 mg/ml in PBS) and 5 μl of camostat (1 mM in PBS) were added, followed by incubation at room temperature for 10 min to inactivate trypsin. About 500 μl of virus solution was applied to a 2-ml bed volume of Sephadex G-75 (17005101; GE Healthcare) equilibrated with PBS (column size, 10 ml; 7311550; Bio-Rad). PBS (1.4 ml) was loaded onto the column, and eluent (200 μl fractions) was collected. Western blot analysis was carried out using MAb-10G to identify fractions containing MHV-2. Fractions 3 and 4 were used for the experiments shown in Fig. 2.
Virus cell entry assay.
DBT cells in a collagen-coated 96-well culture plate (4860-010; Iwaki, Japan) were treated with DMEM containing 10 μM E64d cathepsin inhibitor at 37°C for 30 min to block the endosomal virus entry pathway. Approximately 105 PFU of virus was used to infect 105 cells on ice. After a 30-min adsorption on ice, the virus was removed, and the cells were treated with various concentrations of trypsin. After a 30-min incubation, viral entry was stopped by adding DMEM containing 10 μM camostat and 10 μM E64d, and the samples were incubated at 37°C for 5 h. Cellular RNA was isolated from cells with the addition of 200 μl of Isogen (311-02501; Nippon Gene, Japan). Real-time PCR was performed to estimate the amount of newly synthesized viral mRNA7 as described below.
Quantitative estimation of viral mRNA by real-time PCR.
Real-time reverse transcription-PCR was performed to estimate the amount of MHV-2 mRNA7 as described previously. The target sequence was the MHV-2 N gene. Hybridization probes labeled with fluorescent dye, 5′-GCTCCTCTGGAAACCGCGCTGGTAATGG-3′ (3′-labeled with fluorescein isothiocyanate) and 5′-ATCCTCAAGAAGACCACTTGGGCTGACCAAACC-3′ (5′-labeled with LCRed640), were used to detect the amplified fragments. To amplify viral mRNA7, the oligonucleotides 5′-GTACGTACCCTTTCTACTC-3′ (MHV-2 leader) and 5′-CAAGAGTAATGGGGAACCA-3′ (MHV-2 mRNA7 reverse) were employed. PCR analysis involved reverse transcription at 61°C for 20 min, followed by PCR with an initial denaturation at 95°C for 30 s, followed by 40 cycles at 95°C for 5 s, 55°C for 15 s, and 72°C for 13 s. Reactions were performed using a LightCycler Nano instrument (Roche). The amount of virus in cells was calculated from the calibration curve.
Electron microscopy.
A 6-μl aliquot of UV-irradiated virus was absorbed onto glow-discharged 300-mesh heavy-duty carbon-coated Cu grids (Veco grids; Nisshin EM, Tokyo, Japan) for 2 min, and the excess was blotted onto filter paper (Whatman; GE Healthcare, Piscataway, NJ). Grids were then washed twice with Milli-Q water and negatively stained with 2% phosphotungstic acid. Data were collected using an HT7700 transmission electron microscope (Hitachi, Tokyo, Japan) operating at 100 kV electrons and magnification of ×30,000.
Statistical analysis.
Two-tailed Student t tests were used to analyze statistical significance. A P value of <0.05 was considered statistically significant (n.s., not significant; *, significant [P ≤ 0.05]; **, highly significant [P ≤ 0.01]; ***, very highly significant [P ≤ 0.001]). Error bars indicate standard deviations (SD).
ACKNOWLEDGMENTS
We thank Judith M. White (Virginia University) for valuable suggestions about this work, and we thank Fumihiro Taguchi (Chungnam National University) for providing an opportunity to work on this project. We also thank Noriyo Nagata (NIID) for technical suggestions and Berend-Jan Bosch and Peter J. M. Rottier (Utrecht University) for providing HR2-mimicking peptide.
This study was supported by a Grant-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (grant 17864517).
REFERENCES
- 1.White JM, Whittaker GR. 2016. Fusion of enveloped viruses in endosomes. Traffic 17:593–614. doi: 10.1111/tra.12389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Heald-Sargent T, Gallagher T. 2012. Ready, set, fuse! The coronavirus spike protein and acquisition of fusion competence. Viruses 4:557–580. doi: 10.3390/v4040557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harrison SC. 2015. Viral membrane fusion. Virology 479–480:498–507. doi: 10.1016/j.virol.2015.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walls AC, Xiong X, Park YJ, Tortorici MA, Snijder J, Quispe J, Cameroni E, Gopal R, Dai M, Lanzavecchia A, Zambon M, Rey FA, Corti D, Veesler D. 2019. Unexpected receptor functional mimicry elucidates activation of coronavirus fusion. Cell 176:1026–1039.e15. doi: 10.1016/j.cell.2018.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ozorowski G, Pallesen J, De Val N, Lyumkis D, Cottrell CA, Torres JL, Copps J, Stanfield RL, Cupo A, Pugach P, Moore JP, Wilson IA, Ward AB. 2017. Open and closed structures reveal allostery and pliability in the HIV-1 envelope spike. Nature 547:360–361. doi: 10.1038/nature23010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yuan Y, Cao D, Zhang Y, Ma J, Qi J, Wang Q, Lu G, Wu Y, Yan J, Shi Y, Zhang X, Gao GF. 2017. Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains. Nat Commun 8:1–9. doi: 10.1038/ncomms15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gui M, Song W, Zhou H, Xu J, Chen S, Xiang Y, Wang X. 2017. Cryo-electron microscopy structures of the SARS-CoV spike glycoprotein reveal a prerequisite conformational state for receptor binding. Cell Res 27:119–129. doi: 10.1038/cr.2016.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kirchdoerfer RN, Wang N, Pallesen J, Wrapp D, Turner HL, Cottrell CA, Corbett KS, Graham BS, McLellan JS, Ward AB. 2018. Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis. Sci Rep 8:15701. doi: 10.1038/s41598-018-36918-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee M, Yao H, Kwon B, Waring AJ, Ruchala P, Singh C, Hong M. 2018. Conformation and trimer association of the transmembrane domain of the parainfluenza virus fusion protein in lipid bilayers from solid-state NMR: insights into the sequence determinants of trimer structure and fusion activity. J Mol Biol 430:695–709. doi: 10.1016/j.jmb.2018.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walls AC, Tortorici MA, Bosch B-J, Frenz B, Rottier PJM, DiMaio F, Rey FA, Veesler D. 2016. Cryo-electron microscopy structure of a coronavirus spike glycoprotein trimer. Nature 531:114–117. doi: 10.1038/nature16988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walls AC, Tortorici MA, Snijder J, Xiong X, Bosch B-J, Rey FA, Veesler D. 2017. Tectonic conformational changes of a coronavirus spike glycoprotein promote membrane fusion. Proc Natl Acad Sci U S A 114:11157–11162. doi: 10.1073/pnas.1708727114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kirchdoerfer RN, Cottrell CA, Wang N, Pallesen J, Yassine HM, Turner HL, Corbett KS, Graham BS, McLellan JS, Ward AB. 2016. Prefusion structure of a human coronavirus spike protein. Nature 531:118–121. doi: 10.1038/nature17200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pallesen J, Wang N, Corbett KS, Wrapp D, Kirchdoerfer RN, Turner HL, Cottrell CA, Becker MM, Wang L, Shi W, Kong W-P, Andres EL, Kettenbach AN, Denison MR, Chappell JD, Graham BS, Ward AB, McLellan JS. 2017. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc Natl Acad Sci U S A 114:E7348–E7357. doi: 10.1073/pnas.1707304114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee KK. 2010. Architecture of a nascent viral fusion pore. EMBO J 29:1299–1311. doi: 10.1038/emboj.2010.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maurer UE, Sodeik B, Grünewald K. 2008. Native 3D intermediates of membrane fusion in herpes simplex virus 1 entry. Proc Natl Acad Sci U S A 105:10559–10564. doi: 10.1073/pnas.0801674105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Riedel C, Vasishtan D, Siebert CA, Whittle C, Lehmann MJ, Mothes W, Grünewald K. 2017. Native structure of a retroviral envelope protein and its conformational change upon interaction with the target cell. J Struct Biol 197:172–180. doi: 10.1016/j.jsb.2016.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sjöberg M, Löving R, Lindqvist B, Garoff H. 2017. Sequential activation of the three protomers in the Moloney murine leukemia virus Env. Proc Natl Acad Sci U S A 114:2723–2728. doi: 10.1073/pnas.1617264114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gierer S, Bertram S, Kaup F, Wrensch F, Heurich A, Krämer-Kühl A, Welsch K, Winkler M, Meyer B, Drosten C, Dittmer U, von Hahn T, Simmons G, Hofmann H, Pöhlmann S. 2013. The spike protein of the emerging betacoronavirus EMC uses a novel coronavirus receptor for entry, can be activated by TMPRSS2, and is targeted by neutralizing antibodies. J Virol 87:5502–5511. doi: 10.1128/JVI.00128-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Haan CAM, Stadler K, Godeke G, Bosch BJ, Rottier P. 2004. Cleavage inhibition of the murine coronavirus spike protein by a furin-like enzyme affects cell-cell but not virus-cell fusion. J Virol 78:6048–6054. doi: 10.1128/JVI.78.11.6048-6054.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park J-E, Li K, Barlan A, Fehr AR, Perlman S, McCray PB, Gallagher T. 2016. Proteolytic processing of Middle East respiratory syndrome coronavirus spikes expands virus tropism. Proc Natl Acad Sci U S A 113:12262–12267. doi: 10.1073/pnas.1608147113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reinke LM, Spiegel M, Plegge T, Hartleib A, Nehlmeier I, Gierer S, Hoffmann M, Hofmann-Winkler H, Winkler M, Pöhlmann S. 2017. Different residues in the SARS-CoV spike protein determine cleavage and activation by the host cell protease TMPRSS2. PLoS One 12:e0179177. doi: 10.1371/journal.pone.0179177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Belouzard S, Millet JK, Licitra BN, Whittaker GR. 2012. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses 4:1011–1033. doi: 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng Y, Shang J, Yang Y, Liu C, Wan Y, Geng Q, Wang M, Baric R, Li F. 2018. Lysosomal proteases are a determinant of coronavirus tropism. J Virol 92:e01504-18. doi: 10.1128/JVI.01504-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kawase M, Shirato K, Matsuyama S, Taguchi F. 2009. Protease-mediated entry via the endosome of human coronavirus 229E. J Virol 83:712–721. doi: 10.1128/JVI.01933-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simmons G, Reeves JD, Rennekamp AJ, Amberg SM, Piefer AJ, Bates P. 2004. Characterization of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) spike glycoprotein-mediated viral entry. Proc Natl Acad Sci U S A 101:4240–4245. doi: 10.1073/pnas.0306446101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsuyama S, Taguchi F. 2009. Two-step conformational changes in a coronavirus envelope glycoprotein mediated by receptor binding and proteolysis. J Virol 83:11133–11141. doi: 10.1128/JVI.00959-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Belouzard S, Chu VC, Whittaker GR. 2009. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc Natl Acad Sci U S A 106:5871–5876. doi: 10.1073/pnas.0809524106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Watanabe R, Matsuyama S, Shirato K, Maejima M, Fukushi S, Morikawa S, Taguchi F. 2008. Entry from the cell surface of severe acute respiratory syndrome coronavirus with cleaved S protein as revealed by pseudotype virus bearing cleaved S protein. J Virol 82:11985–11991. doi: 10.1128/JVI.01412-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamada YK, Takimoto K, Yabe M, Taguchi F. 1997. Acquired fusion activity of a murine coronavirus MHV-2 variant with mutations in the proteolytic cleavage site and the signal sequence of the S protein. Virology 227:215–219. doi: 10.1006/viro.1996.8313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burkard C, Verheije MH, Wicht O, van Kasteren SI, van Kuppeveld FJ, Haagmans BL, Pelkmans L, Rottier PJM, Bosch BJ, de Haan C. a M. 2014. Coronavirus cell entry occurs through the endo-/lysosomal pathway in a proteolysis-dependent manner. PLoS Pathog 10:e1004502. doi: 10.1371/journal.ppat.1004502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wicht O, Burkard C, de Haan CAM, van Kuppeveld FJM, Rottier PJM, Bosch BJ. 2014. Identification and characterization of a proteolytically primed form of the murine coronavirus spike proteins after fusion with the target cell. J Virol 88:4943–4952. doi: 10.1128/JVI.03451-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Millet JK, Whittaker GR. 2014. Host cell entry of Middle East respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein. Proc Natl Acad Sci U S A 111:15214–15219. doi: 10.1073/pnas.1407087111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matsuyama S, Shirato K, Kawase M, Terada Y, Kawachi K, Fukushi S, Kamitani W. 2018. Middle East respiratory syndrome coronavirus spike protein is not activated directly by cellular furin during viral entry into target cells. J Virol 92:1–12. doi: 10.1128/JVI.00683-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taguchi F, Shimazaki YK. 2000. Functional analysis of an epitope in the S2 subunit of the murine coronavirus spike protein: involvement in fusion activity. J Gen Virol 81:2867–2871. doi: 10.1099/0022-1317-81-12-2867. [DOI] [PubMed] [Google Scholar]
- 35.Bosch BJ, Rossen JWA, Bartelink W, Zuurveen SJ, de Haan CAM, Duquerroy S, Boucher CAB, Rottier P. 2008. Coronavirus escape from heptad repeat 2 (HR2)-derived peptide entry inhibition as a result of mutations in the HR1 domain of the spike fusion protein. J Virol 82:2580–2585. doi: 10.1128/JVI.02287-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ujike M, Nishikawa H, Otaka A, Yamamoto N, Yamamoto N, Matsuoka M, Kodama E, Fujii N, Taguchi F. 2008. Heptad repeat-derived peptides block protease-mediated direct entry from the cell surface of severe acute respiratory syndrome coronavirus but not entry via the endosomal pathway. J Virol 82:588–592. doi: 10.1128/JVI.01697-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sturman LS, Ricard CS, Holmes KV. 1990. Conformational change of the coronavirus peplomer glycoprotein at pH 8.0 and 37°C correlates with virus aggregation and virus-induced cell fusion. J Virol 64:3042–3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gallagher TM. 1997. A role for naturally occurring variation of the murine coronavirus spike protein in stabilizing association with the cellular receptor. J Virol 71:3129–3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McLellan JS, Yang Y, Graham BS, Kwong PD. 2011. Structure of respiratory syncytial virus fusion glycoprotein in the postfusion conformation reveals preservation of neutralizing epitopes. J Virol 85:7788–7796. doi: 10.1128/JVI.00555-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Madu IG, Roth SL, Belouzard S, Whittaker GR. 2009. Characterization of a highly conserved domain within the severe acute respiratory syndrome coronavirus spike protein S2 domain with characteristics of a viral fusion peptide. J Virol 83:7411–7421. doi: 10.1128/JVI.00079-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ou X, Zheng W, Shan Y, Mu Z, Dominguez SR, Holmes KV, Qian Z. 2016. Identification of the fusion peptide-containing region in betacoronavirus spike glycoproteins. J Virol 90:5586–5600. doi: 10.1128/JVI.00015-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luo Z, Weiss SR. 1998. Roles in cell-to-cell fusion of two conserved hydrophobic regions in the murine coronavirus spike protein. Virology 244:483–494. doi: 10.1006/viro.1998.9121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.von Messling V, Milosevic D, Devaux P, Cattaneo R. 2004. Canine distemper virus and measles virus fusion glycoprotein trimers: partial membrane-proximal ectodomain cleavage enhances function. J Virol 78:7894–7903. doi: 10.1128/JVI.78.15.7894-7903.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cohen FS, Melikyan GB. 2004. The energetics of membrane fusion from binding, through hemifusion, pore formation, and pore enlargement. J Membr Biol 199:1–14. doi: 10.1007/s00232-004-0669-8. [DOI] [PubMed] [Google Scholar]
- 45.Benton DJ, Nans A, Calder LJ, Turner J, Neu U, Lin YP, Ketelaars E, Kallewaard NL, Corti D, Lanzavecchia A, Gamblin SJ, Rosenthal PB, Skehel JJ. 2018. Influenza hemagglutinin membrane anchor. Proc Natl Acad Sci U S A 115:10112–10117. doi: 10.1073/pnas.1810927115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hirano N, Fujiwara K, Hino S, Matumoto M. 1974. Replication and plaque formation of mouse hepatitis virus (MHV-2) in mouse cell line DBT culture. Arch Gesamte Virusforsch 44:298–302. doi: 10.1007/BF01240618. [DOI] [PubMed] [Google Scholar]
- 47.Matsuyama S, Taguchi F. 2002. Receptor-induced conformational changes of murine coronavirus spike protein. J Virol 76:11819–11826. doi: 10.1128/jvi.76.23.11819-11826.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]