Many aspects of viral RNA replication of hepatitis C virus (HCV) are still poorly understood. The process of RNA synthesis is driven by the RNA-dependent RNA polymerase (RdRp) NS5B. Most mechanistic studies on NS5B so far were performed with in vitro systems using isolated recombinant polymerase. In this study, we present a replicon model, which allows the intracellular assessment of noncanonical modes of initiation by the full HCV replicase. Our results add to the understanding of the biochemical processes underlying initiation of RNA synthesis by NS5B by the discovery of internal initiation in cellulo. Moreover, they validate observations made in vitro, showing that the viral polymerase acts very similarly in isolation and in complex with other viral and host proteins. Finally, these observations provide clues about the evolution of RdRps of positive-strand RNA viruses, which might contain the intrinsic ability to initiate internally.
KEYWORDS: NS5B, RNA polymerases, RNA synthesis, RNA-dependent RNA polymerase, RdRp, flaviviridae, hepatitis C virus, initiation, positive-strand RNA virus, subgenomic RNA
ABSTRACT
Initiation of RNA synthesis by the hepatitis C virus (HCV) RNA-dependent RNA polymerase (RdRp) NS5B has been extensively studied in vitro and in cellulo. Intracellular replication is thought to rely exclusively on terminal de novo initiation, as it conserves all genetic information of the genome. In vitro, however, additional modes of initiation have been observed. In this study, we aimed to clarify whether the intracellular environment allows for internal initiation of RNA replication by the HCV replicase. We used a dual luciferase replicon harboring a terminal and an internal copy of the viral genomic 5′ untranslated region, which was anticipated to support noncanonical initiation. Indeed, a shorter RNA species was detected by Northern blotting with low frequency, depending on the length and sequence composition upstream of the internal initiation site. By introducing mutations at either site, we furthermore established that internal and terminal initiation shared identical sequence requirements. Importantly, lethal point mutations at the terminal site resulted exclusively in truncated replicons. In contrast, the same mutations at the internal site abrogated internal initiation, suggesting a competitive selection of initiation sites, rather than recombination or template-switching events. In conclusion, our data indicate that the HCV replicase is capable of internal initiation in its natural environment, although functional replication likely requires only terminal initiation. Since many other positive-strand RNA viruses generate subgenomic messenger RNAs during their replication cycle, we surmise that their capability for internal initiation is a common and conserved feature of viral RdRps.
IMPORTANCE Many aspects of viral RNA replication of hepatitis C virus (HCV) are still poorly understood. The process of RNA synthesis is driven by the RNA-dependent RNA polymerase (RdRp) NS5B. Most mechanistic studies on NS5B so far were performed with in vitro systems using isolated recombinant polymerase. In this study, we present a replicon model, which allows the intracellular assessment of noncanonical modes of initiation by the full HCV replicase. Our results add to the understanding of the biochemical processes underlying initiation of RNA synthesis by NS5B by the discovery of internal initiation in cellulo. Moreover, they validate observations made in vitro, showing that the viral polymerase acts very similarly in isolation and in complex with other viral and host proteins. Finally, these observations provide clues about the evolution of RdRps of positive-strand RNA viruses, which might contain the intrinsic ability to initiate internally.
INTRODUCTION
Hepatitis C virus (HCV) is a single-stranded positive-sense RNA virus with a genome length of ∼9,600 nucleotides (nt). After entry and uncoating, it is released into the cytoplasm, where the single open reading frame (ORF) is translated via an internal ribosomal entry site (IRES) to produce a polyprotein. This is the precursor for three structural (core, E1, and E2) and seven nonstructural (NS) proteins (p7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B), which are processed by host and viral proteases. NS3 to NS5B constitute the viral replicase and are involved in the formation of the viral replication compartment (membranous web) and amplification of the viral genome (1). NS5B is the viral RNA-dependent RNA polymerase (RdRp) (2). It is supposed to initiate genome replication at the 3′ end of the viral RNA, a process which relies on primary sequence and structural elements (cis-acting replication elements [CREs]) within the positive-strand 3′ untranslated region [UTR; poly(U), X-tail (3, 4)] and the ORF (5BSL [5, 6]), as well as the negative strand 3′ end (7), in which the hairpin structures SL-I′ and SL-IIz′, control replication (8, 9). An asymmetric replication mechanism leads to an ∼10:1 positive-strand/negative-strand ratio in infected cells, likely due to differing initiation efficiencies at the respective initiation sites (10–12). Several studies have analyzed replication initiation at the biochemical and structural level. Although there is no evidence for strict sequence specificity, certain nucleotides are selected for by the polymerase. Some reports have found that a pyrimidine at the 3′-terminal position is essential for efficient initiation of RNA replication (13, 14) and a cytosine is strongly favored at position +2 (15). Accordingly, the 3′ end of the positive strand features a pangenotypically conserved uracil. In contrast, the 3′-terminal nucleotides of the negative strand are more variable. Most HCV isolates contain a UG/CG dinucleotide at this position. In addition, JFH-1 (genotype 2a [gt2a]) can support an additional G (introduced by in vitro transcription of viral genomes) at the 5′ positive end in cell culture, so the 3′-terminal sequence in the negative strand is converted to CU (15).
Moreover, it is well established that RNA structure also dictates initiation efficiency. For HCV NS5B the ideal initiation platform in vitro is composed of a short single-stranded region, followed by a stem-loop (12). It is not surprising that the 3′ end of the negative stand contains such a structure (SL-Iz′ and upstream single-stranded RNA [16]), and consequently positive-strand synthesis is highly efficient. Accordingly, significant alterations within these structures typically lead to severe impairment of positive- and negative-strand synthesis (3, 9, 17). The positive-strand 3′ terminus, on the other hand, is engaged in a stable stem-loop without overhang (3′ SL1), which drastically reduces the initiation efficiency of NS5B (12, 18). The structure of the enzyme explains this template selectivity. It folds in the classical conformation of RdRps, which is reminiscent of a right-hand with accordingly designated domains (thumb, palm, and finger domains). A flexible linker connects the catalytic subunit with a carboxy-terminal membrane anchor. Several groups have shown that NS5B can adopt a “closed” (preinitiation) and an “open” (initiation/elongation) form (19, 20). However, it has been reported that the structure fluctuates between open and closed conformations in solution (21). In the closed state, the envelopment of the catalytic center by the finger and palm domains offers just enough room to accommodate a single-stranded template and an initiating nucleotide (NTPi). The space is further constrained by the protrusion of the hydrophobic linker element and the flavivirus-specific beta-flap into the RNA binding groove. The latter was reported to ensure terminal initiation, since a mutant with deletions in the beta-flap was prone to internal initiation events (22). The first rate-limiting step of the initiation process is the generation of a dinucleotide primer from the NTPi (14, 20). The second one is the transition toward an open conformation, which requires significant rearrangements, including the displacement of the beta-flap from the catalytic center (20).
Several distinct modes of initiation have been observed for recombinant NS5B in vitro. The terminal de novo initiation utilizing a dinucleotide is the most physiologically relevant, since it allows the generation of a full copy of the genome. In addition, primer-dependent initiation, starting from a recessed 3′ end of double-stranded RNA (dsRNA), is possible. This can also lead to the generation of a copy-back product in vitro, where the terminal sequence of nascent RNA folds back into a hairpin structure and primes itself (2, 23). Alternatively, the synthesized strand can be used to switch templates under certain conditions (24, 25). Finally, another option is internal de novo initiation, which produces truncated copies of the template (17). Due to the lack of evidence for subgenomic mRNAs (sgRNAs) in the viral replication cycle, terminal de novo initiation is currently the exclusively proposed mechanism of initiation in case of HCV (reviewed in reference 1). However, other positive-strand RNA viruses are known to employ numerous different mechanisms in cellulo. For example, picornavirus polymerases first generate a protein primer by uridylation of a terminal protein at an internal CRE, which is then used to initiate a primer-dependent, terminal initiation of genome replication (reviewed in reference 26) and can also perform recombination by template switching to facilitate adaptation (27). Several members of the Nidovirus order even require a variety of template switching mechanisms for efficient propagation. For instance, coronaviruses and arteriviruses rely on sgRNAs to regulate gene expression and RNA amplification. By template switching from a conserved sequence element, they create common leader sequences for all sgRNAs, which facilitates specific recognition of viral templates (28–30). In contrast, alphaviruses generate their sgRNAs encoding the structural proteins most likely by internal initiation (31). Importantly, the catalytic core of RdRps of positive-strand RNA viruses is an evolutionarily conserved structure, which still is capable of featuring such divergent replication mechanisms (19, 32, 33).
In this study, we addressed the question whether alternative modes of initiation observed in vitro would also be possible in the context of the full HCV replicase in cells. Therefore, we made use of a bicistronic construct harboring two identical HCV 5′ UTRs. We found that this replicon design enables the selective amplification of a truncated RNA containing only the second cistron. This was abolished or enforced by altering the initiation signal at the internal or terminal UTR sequence, respectively. Via this mutational analysis, we could exclude template switching or other forms of recombination and suggest internal initiation as the source of the truncated products. In addition, we determined template requirements and frequency of these events. In conclusion, we propose that internal initiation of the HCV replicase can sporadically occur in cells and that general template requirements are similar to those of terminal initiation. Our data thereby suggest that the capability for internal initiation might be an evolutionarily conserved property of RdRps of positive-strand RNA viruses, even in cases where subgenomic messenger RNAs are not a regular component of the replication cycle.
RESULTS
Duplication of the HCV 5′ UTR results in truncated replicons.
The initiation process of the HCV RdRp NS5B is assumed to start at the respective 3′ termini of the positive and negative strands. It is termed terminal de novo initiation as it only requires an initiating nucleotide to start RNA synthesis. In cellulo, this has been the only observed functional initiation point. However, since alternative modes of initiation by HCV NS5B have been reported in vitro (e.g., internal initiation, copy-back) and numerous viruses employ a variety of initiation/elongation forms during their life cycle, we aimed to explore the capabilities of the HCV replicase to perform these functions in the cellular environment.
Since terminal de novo initiation is the only way to conserve genome integrity, other types of initiation may not be efficiently propagated and most likely elude detection. Therefore, to test for the possibility of the HCV replicase to initiate in a noncanonical fashion during intracellular replication, we made use of a reporter replicon with two identical HCV UTRs (terminal and internal) (Bi-RLuc-SG, Fig. 1A) (34). This design should allow for template switching mechanisms, using the provided homologous sequences, or internal initiation, since the internal UTR could potentially act as competing platform to recruit the replicase.
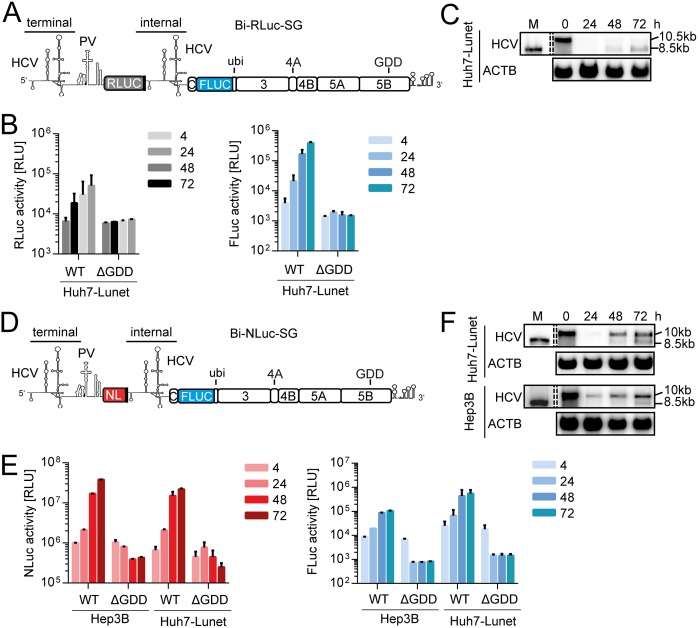
FIG 1.
Loss of full-length bicistronic replicon over time. (A) Schematic representation of Bi-RLuc-SG (positive strand). The bicistronic replicon features two HCV 5′-UTR regions. The terminal UTR is connected by a polioviral IRES (PV), governing translation of a Renilla luciferase reporter gene (RLuc, gray). After a short spacer element, containing several stop codons, the second cistron features the internal UTR, a firefly luciferase (FLuc) gene, fused to the HCV nonstructural proteins, and the 3′ UTR via a ubiquitin linker. (B) Luciferase assay for Renilla (gray) and firefly (blue) activity at 4 to 72 h postelectroporation using Bi-RLuc-SG WT and ΔGDD in Hep3B cells. (C) Corresponding Northern blot analysis of Bi-RLuc-SG levels, using a probe against HCV positive strand RNA (HCV) or β-actin (ACTB). Lane M, In vitro transcript of monocistronic replicon as a size marker (the lane was cropped from the same gel). 10.5 or 10 kb, size of the full-length replicon; 8.5 kb, size of the truncated variant. (D) Schematic representation of Bi-NLuc-SG. All features are as described in panel A, with the RLuc replaced by a Nano luciferase reporter (NL, red). (E) Luciferase assay of Nano (red) and Firefly (blue) activity 4 to 72 h posttransfection in Hep3B and Huh7-Lunet cells. (F) Corresponding Northern blot analysis of Bi-NLuc-SG RNA levels in both cell lines. ΔGDD, replication deficient NS5B mutant. Bars for luciferase activity represent the relative light units (RLU) without normalization (means ± the SD) of three biological replicates, each measured in technical duplicates. Note that transfections in Hep3B cells were supplemented with 50 pmol of miR-122.
After transfection of this construct into Huh7-Lunet cells, we observed a strikingly low Renilla luciferase activity over the complete time course, rising barely above the level of the ΔGDD negative control (Fig. 1B, gray). While this phenotype might have been an issue of very low replication capacity, due to the excessive length of the construct (∼10.5 kb), the firefly luciferase contained within the second cistron was readily expressed (Fig. 1B, blue). Interestingly, Northern blot data revealed that the full-length replicon was not efficiently propagated over time, accompanied by the appearance of an RNA product of lower molecular mass (Fig. 1C, 10.5-kb band [full-length] and 8.5-kb band [truncated form]). Since the accumulation of this shortened RNA was independent of the full-length version, the product was presumed to be capable of autonomous replication. Its size furthermore matched that of a standard subgenomic reporter replicon, used as marker for RNA size (Fig. 1C, lane M, 8.5 kb). Since the second cistron encompasses such a replicon, containing all necessary elements for autonomous RNA replication, we assumed that this smaller product was likely generated by omission of the entire first cistron. We therefore performed a series of experiments to elucidate the underlying mode of action.
In an effort to optimize the reporter replicon further, we introduced a Nano luciferase instead of the Renilla gene (Bi-NLuc-SG, Fig. 1D). With this smaller reporter gene, generating a brighter luciferase signal, we aimed to increase overall replication and enable detection of the residual full-length genome. Moreover, we assessed the influence of the cellular background by performing the initial experiments with this replicon in Hep3B, as well as Huh7-Lunet, cells. Indeed, we were able to observe a strong increase of Nano luciferase (NLuc) reporter activity during the 72-h time course in both cell lines (Fig. 1E, red). The corresponding firefly luciferase levels were comparably enhanced (Fig. 1E, blue). Interestingly, Northern blot analyses of these samples also revealed a truncated variant, but with far lower abundance than full-length RNA (Fig. 1F). Since the results were similar in both cell types, we used Huh7-Lunet cells for the following experiments unless otherwise stated due to their superior permissiveness and the need for addition of exogenous microRNA-122 (miR-122) in Hep3B cells.
Impact of sequences upstream of the internal UTR copy on the abundance of truncated RNA species.
To examine the impact of the total replicon length on the abundance of truncated replicons in more depth, we designed variants of Bi-NLuc-SG, featuring nontranslated spacer regions of 500 or 750 nt, prior to the second cistron (Fig. 2A). The levels of Nano luciferase were markedly reduced for both mutants (Fig. 2B). Specifically, while the 500-nt spacer variant still displayed moderate NLuc activity upon replication, judged by comparison to the ΔGDD control, insertion of 750 nt abolished the NLuc reporter expression almost completely. Inversely, firefly activity was retained in both instances, arguing for replication of the truncated RNA independent of the presence of full-length template, albeit the maximum level was lower than that of the wild type (WT) (Fig. 2C).
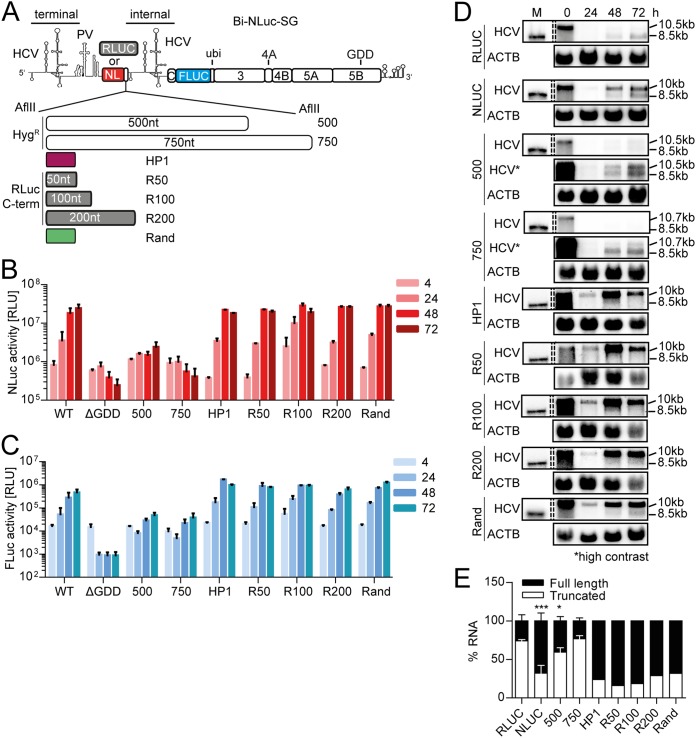
FIG 2.
Impact of spacer length and composition on the abundance of truncated replicons. (A) Schematic representation of the constructs used (positive strand). The nontranslated spacer region between NL and the internal UTR was elongated by insertions of 500 and 750 nt derived from the hygromycin resistance (Hygr) gene, RLuc C-terminal sequences, the transcriptional terminator of Turnip Crinkle virus (HP1), or a randomized control sequence (Rand). (B and C) Luciferase activity of Bi-NLuc-SG replicons harboring different spacer variants. NLuc activity is indicated in red (B), and FLuc activity is indicated in blue (C). Measurements were performed at 4 to 72 h, as indicated. (D) Northern blot analysis of total RNA extracted from transfected cells as in panels B and C at 0 to 72 h posttransfection, using a probe against HCV positive-strand RNA (HCV) or β-actin (ACTB). 10 to 10.7 kb, size of the full-length replicons; 8.5 kb, size of the truncated variant. (E) For Bi-RLuc-SG, Bi-NLuc-SG WT, 500, and 750 ratios of full-length versus truncated RNA were quantified at 72 h. ΔGDD, replication-deficient NS5B mutant. Bars for luciferase activity represent the RLU without normalization (means ± the SD) of three biological replicates for Bi-RLuc-SG, Bi-NLuc-SG WT, 500, and 750, each measured in technical duplicates. Two biological replicates were performed for HP1, R50-200, and Rand. Northern blots for Bi-RLuc-SG were performed in duplicate, and those for Bi-NLuc-SG and Hygr variants were performed in triplicates. All other mutants were blotted once. The statistical significance of differences in truncated RNA generation was determined for each construct against Bi-RLuc-SG. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
In accordance with the luciferase data, Northern blotting showed that the replication levels of both mutants were overall low, compared to the WT (Fig. 2D). In addition, quantification of the RNA species after 72 h revealed that the ratios of full-length to truncated RNA product were significantly altered in the mutants compared to the WT (Fig. 2D and E). The quantity of truncated product was around 50% for the 500-nt spacer and up to 80% in case of the 750-nt insertion, values very similar to those for the Renilla luciferase (RLuc) construct (Fig. 2E). However, this apparent shift in ratios was not due to more efficient formation of the truncated variants but rather to less efficient replication of the full-length genomes harboring insertions (Fig. 2D). Therefore, the longer templates were more easily outcompeted by the truncated form. Interestingly, the Bi-RLuc-SG, containing the Renilla luciferase instead of Nano luciferase generated truncated replicons with far higher efficiency than the Bi-NLuc-SG replicon containing the 500-nt insertion, despite a similar length of both constructs.
We therefore assessed the possibility that the gene encoding Renilla luciferase might contain specific cis-acting signals potentially facilitating the generation of truncated replicons. To address this hypothesis, we added 50 to 200 nt of the 3′ coding region of the Renilla luciferase gene as an additional spacer element to the Nano luciferase gene. As a further control for a potential impact of structured elements on truncation frequency, we added a strong stem-loop region located 5′ adjacent to the initiation site, which has been described as bona fide termination signal, enabling the transcription of sgRNA2 of Turnip Crinkle virus (HP1 [35]), compared to a 50-nt randomized sequence lacking predicted secondary structure (Rand, Fig. 2A to C). However, none of these insertions substantially affected NLuc activity (Fig. 2B and C) or the abundance of truncated replicon RNAs in Northern blot analyses (Fig. 2D and E). These data suggested that the increased number of truncated genomes observed for the Renilla constructs was likely not due to specific cis-acting sequences but rather due to the longer translation unit in the first cistron.
In summary, our data indicated a dual impact of the length of the first cistron on the abundance of truncated replicons: insertion of nontranslated spacer elements, irrespectively of secondary structures, rather reduced the replication rate of the full-length replicons, resulting in an apparent increase of truncated variants. In contrast, a longer translation unit in the first cistron stimulated the relative abundance of truncated genomes.
Mutational analysis of the 3′ terminus of the negative strand.
Apart from the standard terminal de novo initiation, we anticipated which forms of initiation would be able to produce truncated fragments (Fig. 3A). In case of internal initiation, both potential initiation sites would compete with binding of NS5B for terminal or internal initiation, respectively. Alternatively, various forms of template switching could generate truncated replicons. Template switching would essentially start with canonical terminal de novo initiation. After elongating a short stretch, the nascent strand could bind to downstream complementary sequences and cause NS5B to continue at this location.
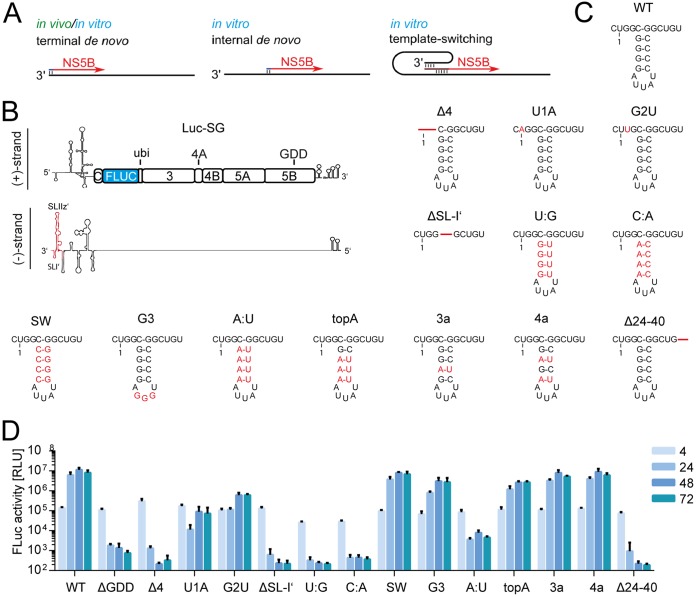
FIG 3.
Firefly activity of mutated monocistronic constructs. (A) Overview of observed (in vivo and/or in vitro) modes of transcription initiation. Apart from terminal initiation, the mechanisms shown could explain the occurrence of truncated products. (B) Schematic of the Luc-SG positive (+) and negative (–) strands. Indicated in red is the region of the 3′ UTR of the negative strand relevant to the mutational analysis. (C) Representation of the wild-type sequence and changes introduced to the negative sense 3′ UTR; nucleotides deviating from JFH-1 consensus are indicated in red, deletions are indicated by red lines. Of note, due to the requirements of the T7 polymerase, requiring a terminal G, an additional 5′ G is present in all transfected in vitro transcripts. Therefore, the 3′ terminus of the negative strand contains a C (C-0), which is indicated. (D) Firefly luciferase activity of mutants of the 3′ UTR. Measurements were performed at 4 to 72 h. Bars for the luciferase activity represent the RLU without normalization (means ± the SD) of three biological replicates, each measured in technical duplicates.
To decide between these alternatives, we aimed to mutate the two initiation sites independently. Template switching mechanisms should be abrogated by mutations in the terminal initiation site, whereas internal initiation should be abolished by mutations in the internal initiation site. First, we sought to establish a set of mutations which reliably would inhibit initiation by NS5B and then to test these mutations for their impact on terminal or internal initiation mechanism. We chose mutations at the 3′ end of the negative strand, some of which were shown to abrogate replication in previous studies (Fig. 3C) focusing on the terminal single-stranded region, SL-I′ and SLII-z′ (Fig. 3B) and introduced these into a JFH-1 (gt2a) monocistronic reporter replicon (Luc-SG).
The large stem-loop SL-IIz′ was destroyed by deletion of 16 nt in its base (Fig. 3C, Δ24–40 [7]). In addition, the smaller stem, SL-I′, was altered either by deletion (Fig. 3C, ΔSL-I′ [7]), mutating the stem (Fig. 3C, SW, C:A, U:A, topA, 3a, and 4a [7, 9, 36]) or the loop (Fig. 3C and G [9]). Finally, the short stretch of terminal unpaired nucleotides was deleted (Fig. 3C, Δ4 [36]), or the terminal nucleotides changed (Fig. 3C, U1A and G2U [13, 15]). Since these mutations are adjacent to the viral IRES and the miR-122 binding sites, luciferase activity at 4 h after electroporation was used to exclude effects on initial translation, which was not substantially affected by any of the mutations (Fig. 3C and D). Reporter activity at later time points (24 to 72 h) was used to determine replication efficiency.
All deletions in the unpaired sequence, SL-I′ and SL-IIz′ efficiently suppressed replication (Fig. 3D), as reported previously (7). Changing the first or second nucleotide severely reduced replication levels but still allowed for some reporter activity (Fig. 3D). This might be the result of incomplete inhibition, due to initiation at C-0, or revertants, which could arise during replication. Changes in the loop of SL-I′ did not impair replication, neither did introduction of sequence variants derived from genotype 3a or 4a (Fig. 3D, G3, 3a, and 4a). Interestingly, a switch of the stem nucleotides had no effect, in contrast to the major impairment reported for genotype 1b (36). Altering larger portions of the stem, however, did have a negative impact on replication. When the top three base pairs were mutated to A:U, a 5-fold reduction of replication was observed. Mutation of four base pairs within the stem severely diminished viral propagation, irrespective of whether the structure was conserved (Fig. 3D, U:G and A:U) or not (Fig. 3D, C:A).
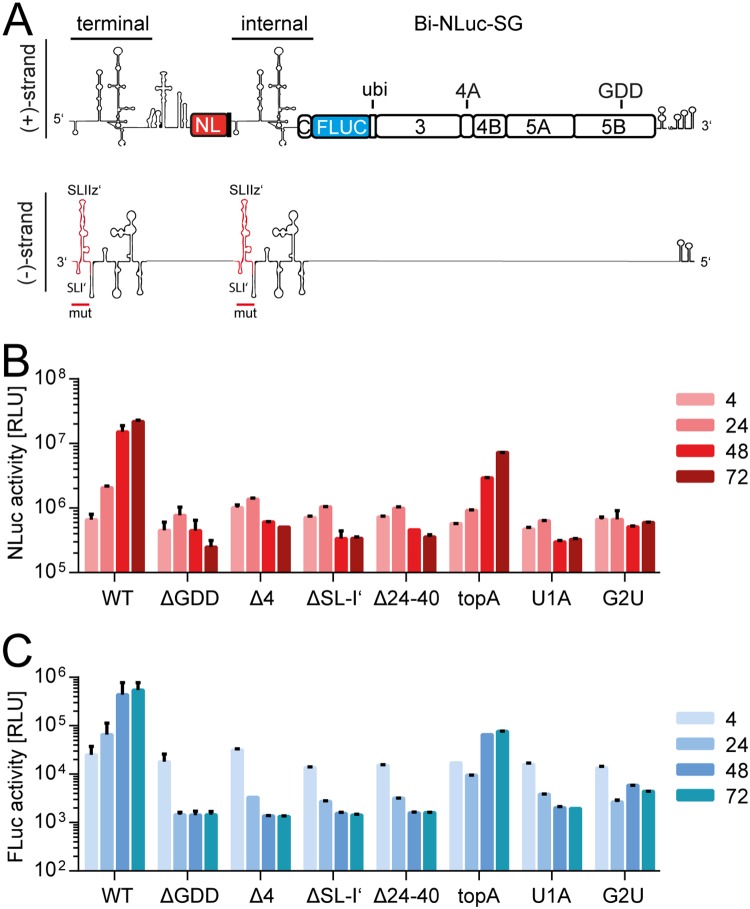
FIG 4.
Analysis of dual mutants. (A) Schematic of the Bi-NLuc-SG positive and negative strands. On the negative strand, red indicates the mutated terminal and internal site. (B and C) Measurement of Nano (red) and firefly (blue) luciferase activities of the mutants. ΔGDD, replication-deficient NS5B mutant Bars for luciferase activity represent the RLU without normalization (means ± the SD) of three biological replicates, each measured in technical duplicates. For a description of the mutations, refer to the legend for Fig. 3.
Several mutants were then selected for the screening in Bi-NLuc-SG, including mutations abolishing replication in Luc-SG (Δ24-40, Δ4, and ΔSL-I′), as well as some supporting various levels of residual replication (topA, U1A, and G2U). Next, both UTRs of Bi-NLuc-SG were modified (Fig. 4A, red outline). As expected, translation was not strongly affected by the mutations in the bicistronic context (Fig. 4B and C). However, most double mutants showed a loss-of-function phenotype for both reporters, only topA remained clearly replication competent (Fig. 4B and C). G2U showed a minimal rise of both reporters at later time points, whereas U1A was replication deficient, in line with the results from the Luc-SG experiments.
In summary, we could mostly confirm the already published effects of several alterations in the 3′ terminus of the viral negative strand for gt2a (7, 9, 13, 15, 36). We thereby obtained a set of mutations useable for subsequent experiments to define the mechanism underlying the observed truncated replicons.
Template requirements for RNA truncation.
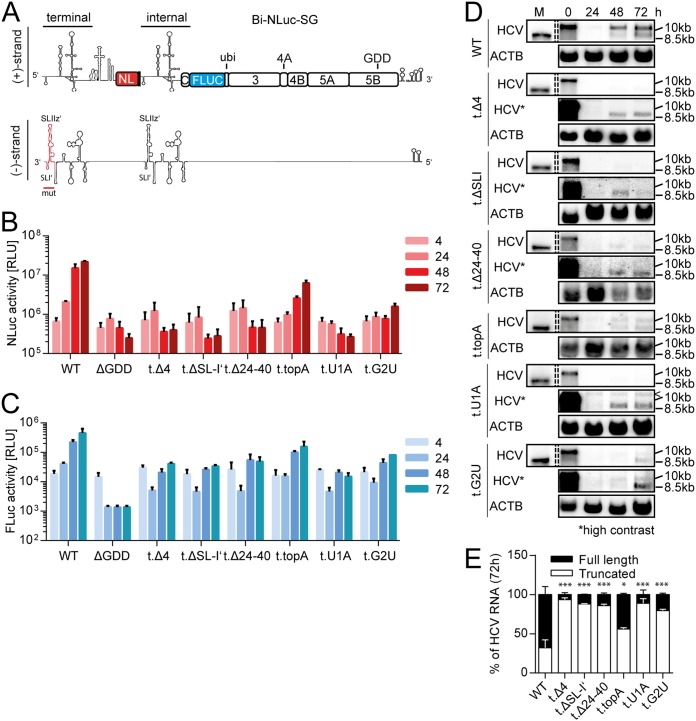
Next, we assessed the potential of the mutations to abrogate the formation of truncated replicons, when introduced only in the terminal or the internal initiation site of Bi-NLuc-SG. In case of template switching, we expected to see an increased rate of truncated replicons upon mutating the internal initiation site, since an intact sequence would be synthesized terminally and would thereby overcome the dysfunctional internal copy. In contrast, if the truncated replicons were due to internal initiation, the process should be blocked by mutations in the internal initiation site and promoted by mutations in the terminal copy of the UTR.
We first introduced mutations only in the terminal UTR to force NS5B toward internal initiation (Fig. 5A). As in Luc-SG, we were able to observe that all deletion mutants completely abolished terminal initiation from this site (Fig. 5B, Δ4, Δ24–40, and ΔSL-I′), as the Nano luciferase levels were at the level of the negative control. Also, the 3′-terminal point mutants, established to be strongly impaired in replication in the Luc-SG context, caused a massive decrease in NLuc activity, when introduced into Bi-NLuc-SG terminally (Fig. 5B, G2U and U1A). G2U and topA were again the only mutations to reach a 72-h value above the ΔGDD negative control (Fig. 5B). Therefore, in line with the data from Luc-SG (Fig. 3D), these mutations may still permit some NS5B initiation at the terminal site. Interestingly, firefly luciferase (FLuc) activity in all mutants with impaired replication capacity, as indicated by NLuc activity, showed a substantial increase compared to the negative control but remained lower than than for the WT (Fig. 5C, Δ4, ΔSL-I′, Δ24–40, topA, G2U, and U1A).
FIG 5.
Mutational analysis of the terminal UTR. (A) Schematic of the Bi-NLuc-SG positive and negative strands. The terminal 3′ UTR altered in the mutational analysis is indicated in red on the negative strand. (B and C) Measurement of Nano (B, red) and firefly (C, blue) luciferase activities of the mutants. (D) Northern blot analysis of total RNA extracted from transfected cells as described in panels B and C at 0 to 72 h posttransfection, using a probe against HCV positive-strand RNA (HCV) or β-actin (ACTB). 10.5 or 10 kb, size of the full-length replicon; 8.5 kb, size of the truncated variant. (E) The ratios of full-length versus truncated RNA were quantified. The “<” symbol indicates a background band. ΔGDD, replication-deficient NS5B mutant. Bars for luciferase activity represent the RLU without normalization (means ± the SD) of three biological replicates, each measured in technical duplicates. Northern blot analyses for quantification were performed three times for the WT and twice for each mutant. For a description of the mutations, refer to the legend of Fig. 3. The statistical significance of differences in truncated RNA generation was determined for each mutant against the WT. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Northern blot analysis of these mutants revealed that, as expected from the NLuc data, loss-of-function mutations at the terminal UTR only allowed for replication of the shorter fragment (Fig. 5D and E), reducing the full-length product to background levels. Only G2U, also showing residual Nano luciferase activity (Fig. 5B), retained a minor amount of full-length RNA (Fig. 5D and E, 1 and ∼20%). Again, the topA mutant showed an intermediate phenotype, where full-length replication was not dramatically impaired, but the ratio was slightly shifted toward the truncated RNA (Fig. 5D). These results clearly argued for internal initiation as the mechanism involved in the generation of the truncated replicons. Still, overall RNA levels in all mutants were much lower than in the WT (Fig. 5D), corresponding to the 1-log difference observed in FLuc activity between the WT and these mutants in the luciferase assay (Fig. 5C, Δ4, U1A, and G2U). This decreased level, and the plateau from 48 h onward observed in firefly luciferase activity, suggested that internal initiation was a rare event and only occurred in a small number of cells. Therefore, even if in these cells the resulting monocistronic replicon most likely replicated to very high levels, the bulk levels remained limited.
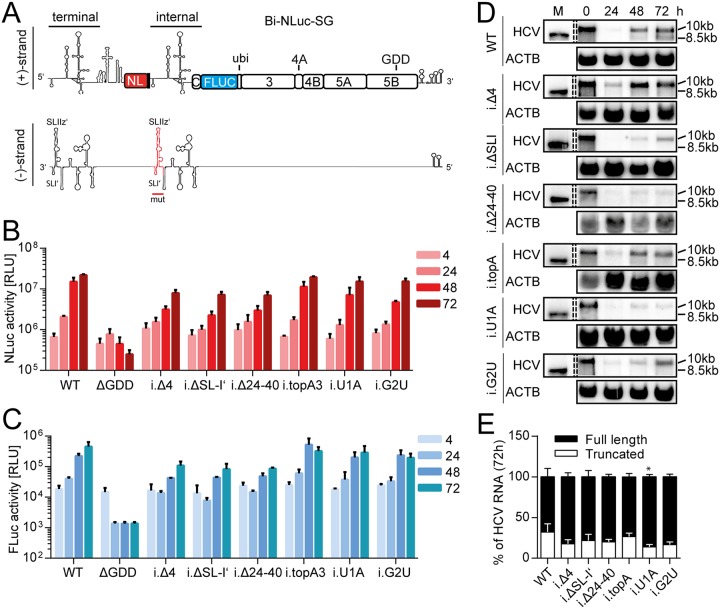
Next, we followed the complementing approach and generated mutants of the internal UTR copy of Bi-NLuc-SG (Fig. 6A). These mutations should prevent internal initiation and therefore suppress generation of truncated genomes (Fig. 6A to E). We indeed found high levels of either luciferase activity, regardless of the type of mutation inserted internally (Fig. 6B and C). For the deletion mutants, however, expression of both reporters was surprisingly lower than in the WT. It is not clear which mechanism is responsible for this phenotype. It might potentially be linked to miR-122 functions (34), since these deletions all affect one or both binding sites. Northern blot analysis of total RNA from internal mutants showed that all loss-of-function mutations did favor the full-length construct and lead to even less (∼10 to 15%) of the truncated form as Bi-NLuc-SG (∼30%, Fig. 6D and E). We also tested, whether this phenotype was due to structural rearrangements at the internal UTR, which might destabilize potential termination signals. However, especially the point mutants did not significantly alter the architecture of this region, as predicted by the RNAStructure online tool (data not shown).
FIG 6.
Mutational analysis of the internal UTR. (A) Schematic of the Bi-NLuc-SG positive and negative strands. The internal 3′ UTR altered in the mutational analysis is indicated in red on the negative strand. (B and C) Measurement of Nano (red) and firefly (blue) luciferase activities of the mutants. (D) Northern blot analysis of total RNA extracted from transfected cells as in panels B and C at 0 to 72 h posttransfection, using a probe against HCV positive-strand RNA (HCV) or β-actin (ACTB). 10 kb, size of the full-length replicon; 8.5 kb, size of the truncated variant. (E) The ratios of full-length versus truncated RNAs were quantified. ΔGDD, replication-deficient NS5B mutant. Bars for luciferase activity represent the RLU without normalization (means ± the SD) of three biological replicates, each measured in technical duplicates. Northern blot analyses for quantification were performed three times for the WT and twice for each mutant. For a description of the mutations, refer to the legend of Fig. 3. The statistical significance of differences in truncated RNA generation was determined for each mutant against the WT. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
The levels of truncated product generated by Bi-NLuc-SG were already very low; therefore, evaluation of the results was restricted by background signal and, in most cases, no statistically significant differences could be detected. Hence, we transferred the U1A point mutation inserted into either the terminal (t.U1A) or internal (i.U1A) UTR of the Bi-NLuc-SG 500 construct (Fig. 7A). Since the ratios of full-length and truncated products were approximately 50% each (Fig. 2D), we hoped to get a clearer outcome in terms of suppression or promotion of truncated replicons, respectively. While the overall replication capacity was expectedly rather low in this context, we could observe a significant reduction of NLuc activity for the terminal versus the internal mutant (Fig. 7B). The FLuc expression was comparable to wild-type Bi-Luc-SG 500 (Fig. 7C). Accordingly, albeit low overall RNA levels, the Northern blot-based quantification confirmed a marked shift of the ratio of full-length to truncated product (Fig. 7D and E). Specifically, a higher proportion of full-length was observed for the internal and more truncated RNA for the terminal mutant, corroborating the previous findings with Bi-NLuc-SG.
FIG 7.
Mutational analysis of the 500-nt spacer replicon. (A) Schematic representation of the 500-nt spacer replicon. The previously characterized U1A mutation was cloned into the terminal (500 t.U1A) or internal (500 i.U1A) UTR. (B and C) Measurement of Nano (B, red) and firefly (C, blue) luciferase activities for the mutants. (D) Northern blot analysis of total RNA extracted from transfected cells as in panels B and C at 0 to 72 h posttransfection, using a probe against HCV positive-strand RNA (HCV) or β-actin (ACTB). 10 kb, size of the full-length replicon; 8.5 kb, size of the truncated variant. (E) The ratios of full-length versus truncated RNA were quantified at the 72-h time point. ΔGDD, replication deficient NS5B mutant. Bars for luciferase activity represent the RLU without normalization (means ± the SD) of three biological replicates, each measured in technical duplicates. Northern blot analyses for quantification were performed three times for the 500-nt spacer replicon and twice for each mutant. The statistical significance of differences in truncated RNA generation was determined for each mutant against the WT. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
In summary, the disruption of the terminal UTR by initiation deficient mutations caused a higher proportion of truncated replicons, whereas mutations in the internal UTR suppressed this process. The marked effect of the tested mutations on the generation of truncated RNA levels argued against template switching or forms of homologous recombination but rather suggested internal initiation by NS5B as an underlying mechanism. Furthermore, these data indicated that the template requirements were similar for both terminal and internal initiation.
No impact of Xrn1 on the abundance of truncated replicons.
The fact that mutations of the terminal UTR stimulated the appearance of truncated replicons, whereas the same mutations in the internal copy abrogated the phenotype, would principally be in line with a mechanism implementing an exonucleolytic digest. A partial exonucleolytic digest by Xrn1 is common among flaviviruses, resulting in the generation of subgenomic flavivirus RNA (sfRNA) (reviewed in reference 37). Xrn1 also degrades HCV RNA, which is counteracted by the binding of miR-122 (38–40). We have further shown previously that miR-122 is capable of functional binding to the internal UTR copy (34). Therefore, Xrn1 might remove the first cistron and be stalled at the internal UTR, if miR-122 is bound (Fig. 8A). In this case, the generation of truncated replicons would be dependent on Xrn1 and should be abrogated in the absence of Xrn1. To address this hypothesis, we generated Hep3B cells with strongly reduced Xrn1 levels by CRISPR/Cas9 (Hep3BΔX1, Fig. 8B) and assessed the NLuc versus FLuc activity after transfection of Bi-NLuc-SG with or without a mutation in the terminal or internal copy of the HCV UTR (t.U1A and i.U1A, Fig. 8C and D) in Hep3B WT or Hep3BΔX1 cells. We found no substantial difference in NLuc or FLuc activities between the two Hep3B variants: the internal UTR mutation suppressed the occurrence of truncated replicons, indicated by high NLuc levels, whereas the terminal UTR mutants retained FLuc activity in the absence of detectable amounts of NLuc activity. However, if partial degradation by Xrn1 is the mechanism underlying the truncated replicons, their generation should be prevented or reduced upon knockdown of Xrn1 in case of terminally mutated replicons. We therefore concluded that Xrn1 is not involved in the generation of truncated HCV replicons.
FIG 8.
Examination of a potential Xrn1-based truncation mechanism. (A) Schematic of the experimental design to address a potential Xrn1-mediated mechanism of RNA truncation. Introduction of a mutant incapable of terminal initiation will lead to the loss of the original template. In the case of an Xrn1-mediated truncation mechanism, degradation of the first cistron and cessation of Xrn1 activity by internally bound miR-122 could create a replication-competent fragment. In contrast, in Xrn1-depleted cells the full-length RNA should not be truncated, and therefore replication would be inhibited entirely by the terminal mutation. (B) Western blot analysis of Xrn1 expression in naive or CRISPR/Cas9-treated Hep3B cells. (C and D) Measurement of Nano (C, red) and firefly (D, blue) luciferase activities for terminal (t.U1A) or internal (i.U1A) mutants in both cell types. ΔGDD, replication-deficient NS5B mutant. Bars for luciferase activity represent the RLU without normalization (means ± the SD) of three biological replicates, each measured in technical duplicates.
Quantification of internal initiation events on the single-cell level.
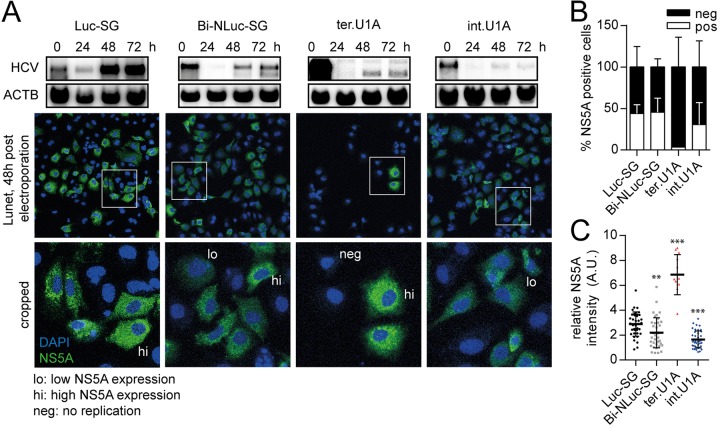
Because the previous experiments revealed that internal initiation was most likely the mechanism causing truncated replicons when identical terminal and internal initiation sites were available, we aimed to assess the frequency of this process on single cell level. We therefore performed experiments on Huh7-Lunet cells electroporated with a Luc-SG control, BI-NLuc-SG WT and point mutants harboring mutations in the terminal or internal UTR copy, respectively (Fig. 9A, t.U1A and i.U1A), and NS5A was used as a marker of HCV replication to assess the number of positive cells, as well as the replication level in individual cells. Interestingly, the percentage of NS5A positive cells was comparable between Luc-SG, BI-NLuc-SG, and i.U1A, whereas only a small fraction of cells was found to be HCV positive after transfection with t.U1A (Fig. 9A and B). However, almost all cells with replicating Luc-SG expressed high levels NS5A, correlating with the strong replication capability of the monocistronic replicon. In contrast, in case of BI-NLuc-SG high (hi) or low (lo) NS5A-expressing populations were found. Cells transfected with the i.U1A replicon, suppressing RNA truncations, instead all contained small amounts of NS5A, whereas t.U1A-transfected cells gave rise to a low number of positive cells, all with high NS5A levels (Fig. 9C), in line with the exclusive presence of the truncated replicon.
FIG 9.
Assessment of the frequency of internal initiation events. (A) Northern blots of the respective replicon are shown for estimation of the overall replication level. For t.U1A, a higher-contrast image is shown to visualize the faint bands at 48 and 72 h. Below are representative immunofluorescence images, with DAPI nuclear staining (blue) and detection of NS5A (green), 48 h after electroporation. The lower panel shows enlarged views of the indicated areas (white box), with examples of cells assigned to high (hi) and low (lo) NS5A-expressing groups or negative (neg) for HCV protein. (B) Relative amounts of NS5A-positive cells after electroporation with the indicated construct. (C) Intensity distribution of NS5A staining among HCV positive cells. ΔGDD, replication-deficient NS5B mutant. Bars represent means ± the SD of three biological replicates. In dot blots, each dot represents one cell. The statistical significance of differences in NS5A expression levels was determined for each construct compared to Luc-SG. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
These data suggest that internal initiation is a very rare event. However, once a truncated replicon is generated, it is further amplified with high efficiency, resulting in relatively high apparent replication levels upon bulk measurements.
DISCUSSION
In this study, we found that NS5B can create shorter RNA species from a bicistronic template containing two identical copies of the HCV UTR, one terminal and one internal. The truncated replicon matched the length of the second cistron. Our data overall suggest internal initiation of RNA synthesis by the viral replicase as an underlying mechanism.
In principle, several transcription dependent and independent mechanisms could cause the generation of truncated replicons. Transcription mediated events would occur during RNA amplification by the RdRp and could be caused by internal initiation or template switching. All available data on intracellular HCV genome replication by the viral polymerase NS5B so far indicate that RNA synthesis is exclusively initiated by a terminal de novo mechanism at the respective 3′ UTRs of the positive and negative strands. In contrast, analysis of recombinant purified NS5B in vitro revealed the capacity of the polymerase to inititate de novo RNA synthesis internally (17, 41) by primer-dependent copy-back initiation (22, 42, 43), as well as by template switching (24, 25). These data suggest that HCV NS5B can in principle generate truncated replicons by several mechanisms. Overall this process is reminiscent of the synthesis of subgenomic RNAs, which are produced by several subgroups of positive-strand RNA viruses (reviewed in reference 37).
A transcription-independent mechanism of heterologous recombination has been observed for bovine viral diarrhea virus (38). However, in this case an intact 5′ end would be fused to an internal sequence, which would not need to be functional. Therefore, such events should be unaffected by mutations of the internal initiation site and suppressed by mutations in the terminal UTR, which contradicts our observations. Cleavage by endo- or exonucleases at the initiation site would be another possibility, but since there is no evidence for sequence-specific RNases in eukaryotes, endonucleolytic cleavage with sufficient accuracy seems rather unlikely. We could further rule out exonucleolytic digest by Xrn1 as a potential mechanism, which generates truncated sfRNAs in flaviviruses (reviewed in reference 39). Therefore, our data clearly favor a transcription-based mechanism.
Several different mechanisms are used by positive-strand RNA viruses to generate subgenomic messenger RNAs on a regular basis, including discontinuous RNA synthesis/template switching or internal initiation of RNA synthesis (reviewed in reference 37). Synthesis of a set of subgenomic messengers is a regular feature of Coronaviridae (40) and Arteriviridae (44). All transcripts share a 5′ end but are generated by discontinuous synthesis of the negative strand (44–46), switching the template at homologous transcription regulating sequences (47). Concerning the ability of the HCV polymerase to promote such a mechanism, template switching by NS5B has indeed been observed under certain conditions in vitro (24, 25). In addition, NS5B has been shown to excessively produce short primers but dissociates from the template before progression into processive elongation (43, 48). This is due to the necessity of the polymerase to undergo a major structural change to switch from initiation to elongation, requiring an opening of the structure (reviewed in reference 1). Released RdRp-primer complexes could provide the basis for a template switching event, not requiring excessive homology. Generally, these mechanisms could explain the replication-competent subgenomic RNAs observed in vivo in HCV patients, containing partial core coding sequences followed by in-frame deletions up to the protease domain of NS2 (49, 50). This could also be a source of HCV intergenotypic recombinants found in some patients (51–54). Since template-switching products would feature the terminal 5′ UTR, mutations of the terminal UTR copy should abrogate the formation of truncated replicons. Vice versa, internal mutations would not affect template switching, particularly at the terminal nucleotides. However, all internal mutations abrogating initiation of RNA synthesis rather promoted the formation of shorter replicons. Therefore, we could exclude template switching as source for the truncated replicons we observed in our study.
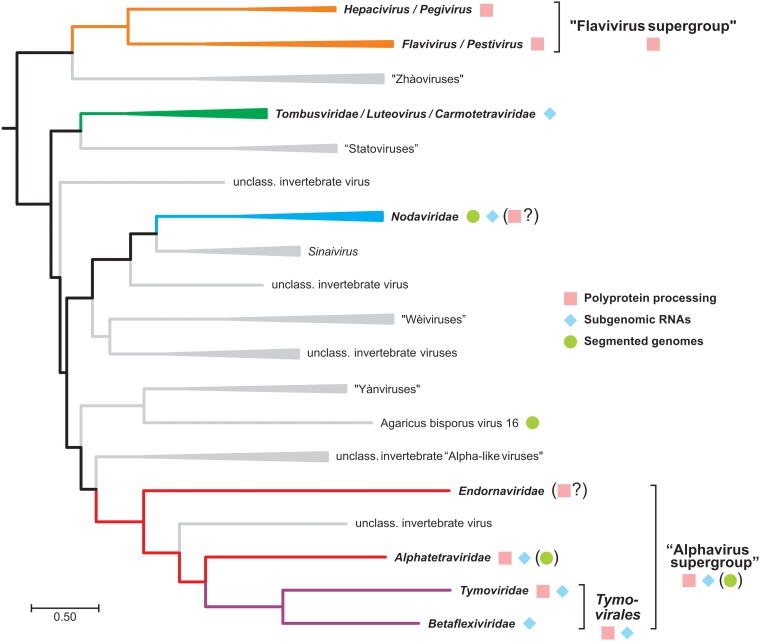
The production of replication-competent RNAs suggests that the complete 5′ UTR is precisely contained in the sgRNA. In addition, the marked effect of point and deletion mutations at the beginning of the second UTR (U1A, G2U, Δ4, and ΔSL-I′) speaks for a sequence- and structure-dependent positioning of the replicase at the start of the internal initiation site. Therefore, internal initiation of RNA synthesis is the only mechanism that fits to all of our data and represents our favorite mode of action for the generation of truncated replicons by HCV NS5B. Internal initiation of RNA syntheis is very common within the recently proposed branch 3 of the phylogenetic tree of RdRPs, which includes HCV and flaviviruses (Fig. 10) (55), and can be mediated either by internal initiation on the negative strand template or by premature termination of negative strand synthesis (reviewed in reference 37). Synthesis of subgenomic RNAs by premature termination of negative strand synthesis was proposed for nodaviruses (e.g., Flock House virus) and tombusviruses (e.g., Tomato bushy stunt virus) (reviewed in reference 56). Here, the polymerase is stalled at specific secondary structures in the positive strand during negative-strand synthesis, which are built by intermolecular (57) or intramolecular (58–60) RNA-RNA interactions. The polymerase then continues with positive-strand RNA synthesis and thereby generates a double-stranded replication intermediate of the subgenomic messenger, which can be amplified autonomously. In contrast, de novo initiation of RNA synthesis, starting at an internal promoter in the negative strand has been found for brome mosaic virus (61). The promoter consists of sequences and structures upstream and downstream of the initiation site (nt −95 to −20) (37, 62). A similar mechanism was proposed for alphaviruses (e.g., Sindbis virus and Togaviridae). Here, the minimal internal initiation site includes nt −19 to +5, but the activity is influenced by nt −98 to +14, relative to the initiation site (63).
FIG 10.
Rooted Bayesian phylogenetic tree of “branch 3” of the viral realm Riboviria. The tree is based on an alignment of 56 representative RdRp protein sequences as provided by Wolf et al. (55). Taxa in italics were formally approved by the ICTV (74; https://talk.ictvonline.org/taxonomy/). Colored groups with boldface labels represent viral taxa for which sufficient experimental data regarding the mode of replication have been obtained by methods of molecular virology. The “alphavirus supergroup” comprises, in addition to the depicted members, many important families of animal and plant pathogens, e.g., Togaviridae, Hepeviridae, Virgaviridae, Bromoviridae, and Closteroviridae.
Experimentally, both mechanisms are difficult to differentiate, and both in principle would fit our experimental results. In both cases, the production of subgenomic positive-strand RNA relies on an intact copy of the internal initiation site, which is in line with our data, since the generation of truncated replicons in our study was blocked by mutations of the internal UTR copy. Both mechanisms further require upstream elements, either in the positive or in the negative strand. Indeed, upstream sequences influenced sgRNA formation in our model, as the Renilla luciferase gene sequence showed a higher propensity to generate truncated products than nontranslated spacers, despite similar or even increased template length and distance between both UTR copies. Generally, the sequence preceding the internal UTR copy in our Bi-Luc-SG replicons was identical and comprised of a short spacer, devoid of strong secondary structures, as tested by RNA folding software (data not shown). Simply inserting parts of the Renilla coding sequence or even a transcription termination signal of Turnip Crinkle virus (35) did not increase the frequency of internal initiation. The most likely explanation for the difference between the RLuc and NLuc replicons therefore might be the length or nature of the translation unit, which might cause a clash between translating ribosomes in the first cistron and the replicase already engaged in negative-strand synthesis. A longer translation unit might therefore increase the probability of such events. However, such termination event would very likely not be precise, thereby formally anyhow requiring de novo initiation from an internal site.
The frequency of the synthesis of truncated subgenomic RNAs in our study is supposed to be very low and only visible due to the self-amplification capability of the truncated replicons. We furthermore forced this process by introduction of a second copy of the UTR within the HCV genome, which naturally does not occur. We therefore do not claim that the principle observation of internal initiation by the HCV polymerase has any direct relevance in the regular replication cycle but rather that this process indicates conserved features of viral RdRps. To further unravel the underlying mechanisms, an experimental model would be required, allowing a more direct measure of internal initiation efficiency in cells, to complement in vitro findings on ectopically expressed and purified NS5B, also in comparison to other RdRps. To this end, we tried to express the viral replicase and a positive- or a negative-strand template RNA containing two initiation sites from two separate plasmids; however, this approach did not give rise to any RNA synthesis driven by the replicase (data not shown). While such transcomplementation models have been successful for alphaviruses (64), it is still not clear how transcomplementation of the replicase can be achieved in case of HCV, but it seems to require NS5B translation in cis (65). We will therefore further modulate the sequence upstream of the internal initiation site in future experiments to better understand the determinants underlying internal initiation by HCV NS5B.
Generally, our knowledge on the regulation of translation and RNA synthesis is still scarce (reviewed in reference 1). It is neither clear, whether both happens simultaneously, nor is the nature of replication intermediates (RI) known. Some parts of the RIs seem to be double stranded, since they can be detected by dsRNA-specific antibodies in infected cells, which recognize stretches of >40 bp (66). In addition, HCV replication activates pattern recognition receptors specific for dsRNA (RIG-I [37, 62], MAVS [63], and TLR3 [67, 68]). However, parts of the RI might remain single stranded, particularly in regions encompassing strong secondary structures, as in case of the UTRs, thereby facilitating recognition of initiation sites by NS5B, only fitting single-stranded templates in its closed initiation state (69–71). A more comprehensive analysis of the mechanisms governing internal initiation in our model may therefore reveal valuable novel insights into general aspects of the regulation of HCV RNA synthesis and the nature of RIs.
From an evolutionary standpoint, it is intriguing that even though the replication cycle of HCV likely relies exclusively on terminal de novo initiation, the capability of the polymerase for alternative modes of initiation is maintained, likely because the overall structure and functional motifs of RdRps are conserved among positive-strand RNA viruses and might have evolved from an ancestral reverse transcriptase (55). If the RdRp is inherently able to initiate internally, simple in-frame insertions creating a suitable platform for the replicase at a junction within the polyprotein could enable a novel regulatory step in the viral RNA life cycle resulting in the evolution of multipartite virus genomes. Whether this happens by insertion of a polymerase stop signal in the positive strand or by a subgenomic promoter in the negative strand might be less relevant. In line with this hypothesis, very different mechanisms are found within related supergroups of positive-strand RNA viruses. The Endornaviridae are the most striking example for such flexibility within the “alphavirus supergroup” (Fig. 10) (55), containing a dsRNA genome and lacking subgenomic RNAs. The “flavivirus supergroup” branches off very deep and yet has no known member regularly generating a subgenomic RNA (Fig. 10). Groups within branch 3 generating subgenomic RNAs by premature termination of negative strand synthesis (Nodaviridae and Tombusviridae) or by de novo initiation from a negative-strand promoter (most well-characterized members of the “alphavirus supergroup,” Bromoviridae, Togaviridae, and Hepeviridae [55]) are comparably distant from the “flavivirus supergroup.” Therefore, the evolutionary view also does not allow a clear judgement on the mechanism of the HCV RdRp in generating subgenomic transcripts, unless new viruses are found that might bridge those gaps in the future.
Conclusively, we have demonstrated that the HCV replicase is capable of internal initiation in cellulo and in the context of active replication, albeit with very low frequency. Moreover, we established that the template requirements for the target RNA are identical for terminal and internal initiation. Overall, our results suggest that the capability to initiate internally is a conserved feature of RdRps among positive-strand RNA viruses, even if a subgenomic RNA is not part of the regular replication cycle.
MATERIALS AND METHODS
Plasmids.
pFK-I341PIRLuc/I389FLuc-ubi/NS3-3′/JFH1 (Bi-RLuc-SG) and pFK-I341PINLuc/I389FLuc-ubi/NS3-3′/JFH1 (Bi-NLuc-SG) were described previously (34). These constructs contain two HCV 5′-UTR regions (terminal and internal), each followed by a luciferase reporter gene (RLuc, Renilla luciferase; NLuc, Nano luciferase; and FLuc, firefly luciferase), which are separated by a spacer region harboring three stop codons in each reading frame. Bi-RLuc-SG and Bi-NLuc-SG were created by insertion of the 341-nt 5′ UTR of HCV and the polio IRES from pFK-I341PILuc/NS3-3′/JFH1 and the Renilla luciferase gene from pCMV-Renilla or the Nano luciferase gene from pNL1.1.TK[NLuc/TK] Vector (Promega), respectively, into pFK-I389Luc-ubi/NS3-3′/JFH1 upstream of the HCV 5′ UTR.
To determine length-dependent effects of the first cistron on full-length RNA amplification, differently sized spacer fragments were generated by PCR from the hygromycin coding sequence of pWPI-hyg (excluding the start codon) and cloned into pFK-I341PINLuc/I389FLuc-ubi/NS3-3′/JFH1 with AflII (NEB). Correct orientation of the insert was confirmed by sequencing.
Mutants for initial evaluation of the impact of changes in the 3′(–) end on replication were designed, based on the pFK-I389Luc-ubi/NS3-3′/JFH1 (Luc-SG) plasmid, which harbors the sequence for a monocistronic HCV gt2a firefly luciferase replicon (72). All mutations were generated by standard PCR (Phusion Flash; Thermo Scientific) using mutagenesis primers and subsequently cloned into the vector using SbfI and XbaI (NEB).
The effects of these mutants on terminal versus internal initiation were tested by insertion into the bicistronic pFK-I341PINLuc/I389FLuc-ubi/NS3-3′/JFH1 vector (34). The first UTR (terminal) was mutated by PCR mutagenesis, and the fragment was inserted upstream of the spacer region via SbfI and AflII (NEB). Mutants of the internal UTR were created similarly, using AflII and XbaI.
pLenti-CRISPR-Xrn1 was produced by cloning a duplex of Xrn1-specific sgRNA oligonucleotides (gRNA miR-122 sense oligonucleotide [CACCGCCATAAAGAACACTTTCCT] and gRNA Xrn1 antisense oligonucleotide [AAACAGGAAAGTGTTCTTTATGGC]), designed with E-CRISP (www.e-crisp.de) into the pLenti-CRISPR plasmid (Addgene, catalog no. 52961) via BsmBI (NEB).
Sequenced clones were amplified in Escherichia coli DH5α, and plasmid DNA was extracted. More detailed information on the cloning procedures is available upon request.
Cell culture.
293T and human hepatoma cell lines Hep3B, Hep3BΔX1, and Huh7-Lunet cells were grown in Dulbecco modified Eagle medium (DMEM; Life Technologies) supplemented with 10% fetal calf serum (GE Healthcare/Sigma-Aldrich), 100 μg/ml penicillin, and 100 μg/ml streptomycin (Sigma-Aldrich) in a humidified incubator at 5% CO2 and 37°C.
Generation of Hep3BΔX1 cells by lentiviral transduction.
Lentiviruses were produced in HEK293T cells by CaPO4 transfection with pCMV-ΔR8.31 (HIV gag-pol), pMD2.G (VSV-G), and the pLenti-CRISPR-miR-122 vector in a 3:1:3 ratio, respectively. pCMV-ΔR8.31 and pMD.2G and were kindly provided by Didier Trono (University of Lausanne, Lausanne, Switzerland). Cell-free supernatants were harvested 48, 56, and 72 h posttransfection and used for transduction of Hep3B cells. Transduced cell pools were selected by supplementing the culture medium with 2 μg/ml puromycin. Relative Xrn1 expression was then tested by Western blotting using antibodies against Xrn1 (Santa Cruz, sc-165985) and Actin (Sigma-Aldrich, A5441) as loading controls.
In vitro transcription of HCV replicon RNA.
Plasmid DNA was linearized with MluI (NEB) and purified by phenol-chloroform extraction and ethanol precipitation prior to resuspension in RNase-free water. Then, 100-μl in vitro transcriptions from 5 μg of DNA were performed in rabbit reticulocyte lysate buffer (80 mM HEPES [pH 7.5], 12 mM MgCl2, 2 mM spermidine, 40 mM dithiothreitol [DTT]) supplemented with 3.125 mM nucleoside triphosphates, 1 U/μl RNasin (Promega), and 4 μl of recombinant T7 polymerase. In vitro transcriptions were incubated for 4 h or overnight at 37°C; 2 μl of additional T7 polymerase was added after 2 h. Template DNA was degraded by using 10 μl of RQ1 RNase-free DNase (Promega). RNA was phenol-chloroform extracted, precipitated with isopropanol at room temperature, and resuspended in RNase-free water, as previously described (34).
Electroporation.
Cells were trypsinized, pooled in DMEM, spun down at 700 × g for 5 min, washed with phosphate-buffered saline (PBS), and counted using a Neubauer counting chamber. Pellets were resuspended in Cytomix (120 mM KCl; 0.15 mM CaCl2; 10 mM K2HPO4/KH2PO4 [pH 7.6]; 25 mM HEPES; 2 mM EGTA; and 5 mM MgCl2, 2 mM ATP, and 5 mM glutathione) to a concentration of 1 × 107 cells/ml (Huh7-Lunet) or 1.5 × 107 cells/ml (Hep3B).
Electroporations for luciferase assays were performed using 5 μg of RNA with 200 μl of the cell suspension and carried out in a Bio-Rad GenePulser II with the corresponding 0.2-cm cuvettes at 0.975 μF and 166 mV. Then, 50 pmol of miR-122 duplex was added in the case of Hep3B cells. Electroporated cells were transferred into 6 ml of DMEM and seeded onto 6-well plates. After 4, 24, 48, and 72 h, the cells were washed with PBS and lysed using 350 μl of lysis buffer (1% Triton X-100, 25 mM glycyl glycine [pH 7.8], 5 mM MgSO4, 4 mM EGTA [pH 7.8], 10% glycerol, 1 mM DTT). Harvested cells were stored at –20°C.
For Northern blots, electroporations were carried out with three times 5 μg of RNA and 200 μl of cells in 0.2-cm cuvettes at 0.975 μF and 0.166 mV. The cells were pooled in Falcon tubes with 9 ml of DMEM; 6-ml portions of the suspension were then transferred to 6-cm dishes (3 ml for 24 h, 1.5 ml plus 1.5 ml of DMEM for 48 an 72 h), followed by growth at 37°C until harvest. The residual 3 ml was pelleted at 700 × g for 5 min and served as a 0-h control. All samples were harvested by washing with PBS and lysis with 750 μl of GITC (4 M guanidine thiocyanate, 25 mM sodium citrate, 0.5% sarcosyl, 0.1 M β-mercaptoethanol).
Luciferase assays.
The firefly luciferase activity of the monocistronic constructs was analyzed in a Lumat LB 9507 single tube reader (Berthold). Portions (100 μl) of lysed cells were added to 360 μl of assay buffer (25 mM glycyl glycine [pH 7.8], 15 mM KPO4 buffer [pH 7.8], 15 mM MgSO4, 4 mM EGTA [pH 7.8], 2 mM ATP, 1 mM DTT). Samples were measured for 20 s after injection of 200 μl of 1 mM d-luciferin (PJK) in 5 mM glycyl glycine.
For the bicistronic constructs, 40-μl portions of lysates were transferred to white-walled 96-well plates (Falcon) in duplicates. Measurement of the Nano and firefly luciferase activities was performed in a Mitras LB940 plate reader (Berthold) using the Nano-Glo dual-luciferase reporter assay system (Promega). Portions (40 μl) of lysate were supplemented with 40 μl of ONE-Glo EX reagent (Promega), followed by incubation for 10 min at room temperature before measurement of the firefly luciferase signal (10-s measurement). Subsequently, the FLuc activity was quenched by the addition of 40 μl of NanoDLR Stop&Glo buffer (Promega). The NLuc signal was measured after 10 min of incubation (1-s measurement).
All data from luciferase assays are reported as relative light units (RLU) without normalization.
Northern blots.
Total RNA extraction was performed by phenol-chloroform separation and isopropanol precipitation as described previously (73).
At least 10 μg of total RNA in 10 μl of H2O was supplemented with 4.1 μl of 100 mM NaPO4 (pH 7.0), 6 μl of 6 M glyoxal, and 20.5 μl of dimethyl sulfoxide. The samples were then denatured at 50°C for 1 h and cooled on ice, and 10.9 μl of loading buffer (0.25 mg/ml bromphenol blue, 0.25 mg/ml xylene cyanol, 10 mM NaPO4 [pH 7.0], 50% [vol/vol] glycerol) was added. Portions (25 μl) of the samples were run on a gel (1% agarose, 10 mM NaPO4 [pH 7.0]) in 10 mM NaPO4 buffer for 3 h at 100 V. The ion concentration of the buffer was kept constant by mixing using magnetic stir bars.
RNA was transferred to a HyBond N+ membrane (Amersham) by application of 20,000-Pa vacuum for 1 h, with the gel covered with 50 mM NaOH. The dried membrane was cross-linked using the auto-cross-link function of a UV Stratalinker 1800 (Stratagene) twice. Membranes were blocked in 10 ml hybridization buffer (5 × SSC; 5× Denhardt solution; 50% [wt/vol] formamide; 1% [wt/vol] sodium dodecyl sulfate) supplemented with 100 μl of heat-denatured salmon sperm DNA (100 μg/ml; denatured at 95°C prior to addition) at 68°C for at least 15 min.
32P-labeled HCV (nt 5979 to 6699) and β-actin antisense probes were synthesized in a 20-μl reaction mixture containing 1× transcription buffer (Promega), 10 mM DTT, 25 ng of template DNA (pFK1-389neo), a 0.5 mM AGU nucleotide mix, 5 μM cold CTP, 1 μM [32P]CTP, 0.5 μl of RNasin (Promega), and 1 μl of commercial RNA polymerase (T3 for HCV and T7 for β-actin; Promega). Reaction mixtures were incubated at 37°C for 1 h, and template DNA was degraded by the addition of 1 μl of DNase RQ (Promega), followed by incubation at 37°C for 15 min. Probes were purified by using a Nap-5 gel filtration column (Amersham) equilibrated in H2O. Hybridization was carried out overnight at 68°C using the respective probes in 10 ml of hybridization buffer.
Membranes were washed twice in wash buffer I (2× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 0.1% SDS) and three times in wash buffer II (0.2 × SSC, 0.1% SDS) at 68°C for 15 min each. Bands were visualized after 2- to 48-h exposure of a phosphor imaging screen using a Molecular Imager FX scanner (Bio-Rad). Background subtraction and quantification of bands was performed by using QuantityOne software (Bio-Rad).
Immunofluorescence.
Huh7-Lunet cells were electroporated as stated above with Luc-SG, Bi-NLuc-SG, t.U1A, and i.U1A and then seeded on cover slides. After 48 h at 37°C, these slides were washed three times with PBS and fixed with 4% paraformaldehyde for 20 min at room temperature. After another three washes with PBS, the slides were permeabilized with 0.5% Triton X-100 in PBS for 5 min. The cells were then rinsed three times with PBS and stained using the 9E10 monoclonal antibody against NS5A (1:500 in PBS plus 3% [wt/vol] bovine serum albumin) for 1 h at room temperature. Unbound antibody was removed by three washes with PBS, followed by 1 h of incubation with goat anti-mouse Alexa 488-coupled secondary antibody. After a rinse with PBS, a 1:4,000 dilution of DAPI (4′,6′-diamidino-2-phenylindole) was applied for 1 min. The coverslips were mounted on a glass slide with Fluoromount G and then inspected on a Leica SP8 confocal microscope (×20 objective). NS5A expression in whole cells was imaged using z-stacks, and maximal intensity projections were quantified using Fiji.
Quantification and statistical analysis.
Where appropriate, a two-tailed Student t test was performed using Prism 6 software (GraphPad). All data are presented as means ± the standard deviations (SD). The sample size is specified in the corresponding figure legend. A P value of <0.05 was considered statistically significant. In graphs, statistical significance is indicated by asterisks (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
ACKNOWLEDGMENTS
We especially thank Rahel Klein and Ulrike Herian for excellent technical assistance. We are grateful to Charles M. Rice for antibody 9E10 and Takaji Wakita for the JFH-1 isolate.
This project was funded by grants from the Deutsche Forschungsgemeinschaft (LO 1556/4-1 and LO1556/4-2) to V.L.
REFERENCES
- 1.Lohmann V. 2013. Hepatitis C virus RNA replication. Curr Top Microbiol Immunol 369:167–198. doi: 10.1007/978-3-642-27340-7_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Behrens SE, Tomei L, De FR. 1996. Identification and properties of the RNA-dependent RNA polymerase of hepatitis C virus. EMBO J 15:12–22. doi: 10.1002/j.1460-2075.1996.tb00329.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Friebe P, Bartenschlager R. 2002. Genetic analysis of sequences in the 3′ nontranslated region of hepatitis C virus that are important for RNA replication. J Virol 76:5326–5338. doi: 10.1128/jvi.76.11.5326-5338.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.You S, Rice CM. 2008. 3′ RNA elements in hepatitis C virus replication: kissing partners and long poly(U). J Virol 82:184–195. doi: 10.1128/JVI.01796-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Diviney S, Tuplin A, Struthers M, Armstrong V, Elliott RM, Simmonds P, Evans DJ. 2008. A hepatitis C virus cis-acting replication element forms a long-range RNA-RNA interaction with upstream RNA sequences in NS5B. J Virol 82:9008–9022. doi: 10.1128/JVI.02326-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friebe P, Boudet J, Simorre JP, Bartenschlager R. 2005. Kissing-loop interaction in the 3′ end of the hepatitis C virus genome essential for RNA replication. J Virol 79:380–392. doi: 10.1128/JVI.79.1.380-392.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Friebe P, Lohmann V, Krieger N, Bartenschlager R. 2001. Sequences in the 5′ nontranslated region of hepatitis C virus required for RNA replication. J Virol 75:12047–12057. doi: 10.1128/JVI.75.24.12047-12057.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi ST, Lai MMC. 2006. HCV 5′ and 3′ UTR: when translation meets replication. In Tan SL. (ed), Hepatitis C viruses: genomes and molecular biology. Horizon Bioscience, Norfolk, United Kingdom: https://www.ncbi.nlm.nih.gov/books/NBK1624/. [Google Scholar]
- 9.Friebe P, Bartenschlager R. 2009. Role of RNA structures in genome terminal sequences of the hepatitis C virus for replication and assembly. J Virol 83:11989–11995. doi: 10.1128/JVI.01508-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang M, Williams O, Mittler J, Quintanilla A, Carithers RL Jr, Perkins J, Corey L, Gretch DR. 2003. Dynamics of hepatitis C virus replication in human liver. Am J Pathol 163:433–444. doi: 10.1016/S0002-9440(10)63673-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhong W, Ferrari E, Lesburg CA, Maag D, Ghosh SK, Cameron CE, Lau JY, Hong Z. 2000. Template/primer requirements and single nucleotide incorporation by hepatitis C virus nonstructural protein 5B polymerase. J Virol 74:9134–9143. doi: 10.1128/jvi.74.19.9134-9143.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kao CC, Yang X, Kline A, Wang QM, Barket D, Heinz BA. 2000. Template requirements for RNA synthesis by a recombinant hepatitis C virus RNA-dependent RNA polymerase. J Virol 74:11121–11128. doi: 10.1128/JVI.74.23.11121-11128.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cai Z, Liang TJ, Luo G. 2004. Effects of mutations of the initiation nucleotides on hepatitis C virus RNA replication in the cell. J Virol 78:3633–3643. doi: 10.1128/jvi.78.7.3633-3643.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferrari E, He Z, Palermo RE, Huang HC. 2008. Hepatitis C virus NS5B polymerase exhibits distinct nucleotide requirements for initiation and elongation. J Biol Chem 283:33893–33901. doi: 10.1074/jbc.M803094200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nandasoma U, McCormick C, Griffin S, Harris M. 2011. Nucleotide requirements at positions +1 to +4 for the initiation of hepatitis C virus positive-strand RNA synthesis. J Gen Virol 92:1082–1086. doi: 10.1099/vir.0.028423-0. [DOI] [PubMed] [Google Scholar]
- 16.Schuster C, Isel C, Imbert I, Ehresmann C, Marquet R, Kieny MP. 2002. Secondary structure of the 3′ terminus of hepatitis C virus minus-strand RNA. J Virol 76:8058–8068. doi: 10.1128/jvi.76.16.8058-8068.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Binder M, Quinkert D, Bochkarova O, Klein R, Kezmic N, Bartenschlager R, Lohmann V. 2007. Identification of determinants involved in initiation of hepatitis C virus RNA synthesis by using intergenotypic replicase chimeras. J Virol 81:5270–5283. doi: 10.1128/JVI.00032-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith RM, Walton CM, Wu CH, Wu GY. 2002. Secondary structure and hybridization accessibility of hepatitis C virus 3′-terminal sequences. J Virol 76:9563–9574. doi: 10.1128/jvi.76.19.9563-9574.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simister P, Schmitt M, Geitmann M, Wicht O, Danielson UH, Klein R, Bressanelli S, Lohmann V. 2009. Structural and functional analysis of hepatitis C virus strain JFH1 polymerase. J Virol 83:11926–11939. doi: 10.1128/JVI.01008-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Appleby TC, Perry JK, Murakami E, Barauskas O, Feng J, Cho A, Fox D III, Wetmore DR, McGrath ME, Ray AS, Sofia MJ, Swaminathan S, Edwards TE. 2015. Viral replication: structural basis for RNA replication by the hepatitis C virus polymerase. Science 347:771–775. doi: 10.1126/science.1259210. [DOI] [PubMed] [Google Scholar]
- 21.Davis BC, Thorpe IF. 2013. Molecular simulations illuminate the role of regulatory components of the RNA polymerase from the hepatitis C virus in influencing protein structure and dynamics. Biochemistry 52:4541–4552. doi: 10.1021/bi400251g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hong Z, Cameron CE, Walker MP, Castro C, Yao N, Lau JY, Zhong W. 2001. A novel mechanism to ensure terminal initiation by hepatitis C virus NS5B polymerase. Virology 285:6–11. doi: 10.1006/viro.2001.0948. [DOI] [PubMed] [Google Scholar]
- 23.Luo G, Hamatake RK, Mathis DM, Racela J, Rigat KL, Lemm J, Colonno RJ. 2000. De novo initiation of RNA synthesis by the RNA-dependent RNA polymerase (NS5B) of hepatitis C virus. J Virol 74:851–863. doi: 10.1128/jvi.74.2.851-863.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng CP, Panavas T, Luo G, Nagy PD. 2005. Heterologous RNA replication enhancer stimulates in vitro RNA synthesis and template-switching by the carmovirus, but not by the tombusvirus, RNA-dependent RNA polymerase: implication for modular evolution of RNA viruses. Virology 341:107–121. doi: 10.1016/j.virol.2005.06.042. [DOI] [PubMed] [Google Scholar]
- 25.Ranjith-Kumar CT, Sarisky RT, Gutshall L, Thomson M, Kao CC. 2004. De novo initiation pocket mutations have multiple effects on hepatitis C virus RNA-dependent RNA polymerase activities. J Virol 78:12207–12217. doi: 10.1128/JVI.78.22.12207-12217.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paul AV, Wimmer E. 2015. Initiation of protein-primed picornavirus RNA synthesis. Virus Res 206:12–26. doi: 10.1016/j.virusres.2014.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiao Y, Rouzine IM, Bianco S, Acevedo A, Goldstein EF, Farkov M, Brodsky L, Andino R. 2016. RNA recombination enhances adaptability and is required for virus spread and virulence. Cell Host Microbe 19:493–503. doi: 10.1016/j.chom.2016.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zuniga S, Cruz JL, Sola I, Mateos-Gomez PA, Palacio L, Enjuanes L. 2010. Coronavirus nucleocapsid protein facilitates template switching and is required for efficient transcription. J Virol 84:2169–2175. doi: 10.1128/JVI.02011-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zuniga S, Sola I, Alonso S, Enjuanes L. 2004. Sequence motifs involved in the regulation of discontinuous coronavirus subgenomic RNA synthesis. J Virol 78:980–994. doi: 10.1128/JVI.78.2.980-994.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van MG, Dobbe JC, Gultyaev AP, Luytjes W, Spaan WJ, Snijder EJ. 1999. Arterivirus discontinuous mRNA transcription is guided by base pairing between sense and antisense transcription-regulating sequences. Proc Natl Acad Sci U S A 96:12056–12061. doi: 10.1073/pnas.96.21.12056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pietila MK, Hellstrom K, Ahola T. 2017. Alphavirus polymerase and RNA replication. Virus Res 234:44–57. doi: 10.1016/j.virusres.2017.01.007. [DOI] [PubMed] [Google Scholar]
- 32.Posthuma CC, Te Velthuis AJW, Snijder EJ. 2017. Nidovirus RNA polymerases: complex enzymes handling exceptional RNA genomes. Virus Res 234:58–73. doi: 10.1016/j.virusres.2017.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen MW, Tan YB, Zheng J, Zhao Y, Lim BT, Cornvik T, Lescar J, Ng LFP, Luo D. 2017. Chikungunya virus nsP4 RNA-dependent RNA polymerase core domain displays detergent-sensitive primer extension and terminal adenylyltransferase activities. Antiviral Res 143:38–47. doi: 10.1016/j.antiviral.2017.04.001. [DOI] [PubMed] [Google Scholar]
- 34.Schult P, Roth H, Adams RL, Mas C, Imbert L, Orlik C, Ruggieri A, Pyle AM, Lohmann V. 2018. MicroRNA-122 amplifies hepatitis C virus translation by shaping the structure of the internal ribosomal entry site. Nat Commun 9:2613. doi: 10.1038/s41467-018-05053-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu B, Oliveri S, Mandic J, White KA. 2010. Evidence for a premature termination mechanism of subgenomic mRNA transcription in a carmovirus. J Virol 84:7904–7907. doi: 10.1128/JVI.00742-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luo G, Xin S, Cai Z. 2003. Role of the 5′-proximal stem-loop structure of the 5′ untranslated region in replication and translation of hepatitis C virus RNA. J Virol 77:3312–3318. doi: 10.1128/jvi.77.5.3312-3318.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sztuba-Solinska J, Stollar V, Bujarski JJ. 2011. Subgenomic messenger RNAs: mastering regulation of (+)-strand RNA virus life cycle. Virology 412:245–255. doi: 10.1016/j.virol.2011.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Becher P, Tautz N. 2011. RNA recombination in pestiviruses: cellular RNA sequences in viral genomes highlight the role of host factors for viral persistence and lethal disease. RNA Biol 8:216–224. doi: 10.4161/rna.8.2.14514. [DOI] [PubMed] [Google Scholar]
- 39.Slonchak A, Khromykh AA. 2018. Subgenomic flaviviral RNAs: what do we know after the first decade of research. Antiviral Res 159:13–25. doi: 10.1016/j.antiviral.2018.09.006. [DOI] [PubMed] [Google Scholar]
- 40.Sawicki SG, Sawicki DL. 2005. Coronavirus transcription: a perspective. Curr Top Microbiol Immunol 287:31–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ranjith-Kumar CT, Kao CC. 2005. Recombinant viral RdRps can initiate RNA synthesis from circular templates. RNA 12:303–312. doi: 10.1261/rna.2163106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lohmann V, Korner F, Herian U, Bartenschlager R. 1997. Biochemical properties of hepatitis C virus NS5B RNA-dependent RNA polymerase and identification of amino acid sequence motifs essential for enzymatic activity. J Virol 71:8416–8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhong W, Uss AS, Ferrari E, Lau JY, Hong Z. 2000. De novo initiation of RNA synthesis by hepatitis C virus nonstructural protein 5B polymerase. J Virol 74:2017–2022. doi: 10.1128/JVI.74.4.2017-2022.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pasternak AO, van den BE, Spaan WJ, Snijder EJ. 2001. Sequence requirements for RNA strand transfer during nidovirus discontinuous subgenomic RNA synthesis. EMBO J 20:7220–7228. doi: 10.1093/emboj/20.24.7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu HY, Brian DA. 2010. Subgenomic messenger RNA amplification in coronaviruses. Proc Natl Acad Sci U S A 107:12257–12262. doi: 10.1073/pnas.1000378107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sawicki D, Wang T, Sawicki S. 2001. The RNA structures engaged in replication and transcription of the A59 strain of mouse hepatitis virus. J Gen Virol 82:385–396. doi: 10.1099/0022-1317-82-2-385. [DOI] [PubMed] [Google Scholar]
- 47.Alonso S, Izeta A, Sola I, Enjuanes L. 2002. Transcription regulatory sequences and mRNA expression levels in the coronavirus transmissible gastroenteritis virus. J Virol 76:1293–1308. doi: 10.1128/JVI.76.3.1293-1308.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shim JH, Larson G, Wu JZ, Hong Z. 2002. Selection of 3′-template bases and initiating nucleotides by hepatitis C virus NS5B RNA-dependent RNA polymerase. J Virol 76:7030–7039. doi: 10.1128/jvi.76.14.7030-7039.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pacini L, Graziani R, Bartholomew L, De FR, Paonessa G. 2009. Naturally occurring hepatitis C virus subgenomic deletion mutants replicate efficiently in Huh-7 cells and are trans-packaged in vitro to generate infectious defective particles. J Virol 83:9079–9093. doi: 10.1128/JVI.00308-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cheroni C, Donnici L, Aghemo A, Balistreri F, Bianco A, Zanoni V, Pagani M, Soffredini R, D’Ambrosio R, Rumi MG, Colombo M, Abrignani S, Neddermann P, De Francesco R. 2015. Hepatitis C virus deletion mutants are found in individuals chronically infected with genotype 1 hepatitis C virus in association with age, high viral load and liver inflammatory activity. PLoS One 10:e0138546. doi: 10.1371/journal.pone.0138546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li YP, Ramirez S, Gottwein JM, Bukh J. 2011. Non-genotype-specific role of the hepatitis C virus 5′ untranslated region in virus production and in inhibition by interferon. Virology 421:222–234. doi: 10.1016/j.virol.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 52.Scheel TK, Galli A, Li YP, Mikkelsen LS, Gottwein JM, Bukh J. 2013. Productive homologous and non-homologous recombination of hepatitis C virus in cell culture. PLoS Pathog 9:e1003228. doi: 10.1371/journal.ppat.1003228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shi W, Carr MJ, Dunford L, Zhu C, Hall WW, Higgins DG. 2012. Identification of novel inter-genotypic recombinants of human hepatitis B viruses by large-scale phylogenetic analysis. Virology 427:51–59. doi: 10.1016/j.virol.2012.01.030. [DOI] [PubMed] [Google Scholar]
- 54.Kleine BM, Meyer D, Austermann-Busch S, Roman-Sosa G, Rumenapf T, Becher P. 2017. Nonreplicative RNA recombination of an animal plus-strand RNA virus in the absence of efficient translation of viral proteins. Genome Biol Evol 9:817–829. doi: 10.1093/gbe/evx046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wolf YI, Kazlauskas D, Iranzo J, Lucia-Sanz A, Kuhn JH, Krupovic M, Dolja VV, Koonin EV. 2018. Origins and evolution of the global RNA virome. mBio 9. doi: 10.1128/mBio.02329-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.White KA. 2002. The premature termination model: a possible third mechanism for subgenomic mRNA transcription in (+)-strand RNA viruses. Virology 304:147–154. doi: 10.1006/viro.2002.1732. [DOI] [PubMed] [Google Scholar]
- 57.Sit TL, Vaewhongs AA, Lommel SA. 1998. RNA-mediated trans-activation of transcription from a viral RNA. Science 281:829–832. doi: 10.1126/science.281.5378.829. [DOI] [PubMed] [Google Scholar]
- 58.Lindenbach BD, Sgro JY, Ahlquist P. 2002. Long-distance base pairing in flock house virus RNA1 regulates subgenomic RNA3 synthesis and RNA2 replication. J Virol 76:3905–3919. doi: 10.1128/jvi.76.8.3905-3919.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Choi IR, Ostrovsky M, Zhang G, White KA. 2001. Regulatory activity of distal and core RNA elements in tombusvirus subgenomic mRNA2 transcription. J Biol Chem 276:41761–41768. doi: 10.1074/jbc.M106727200. [DOI] [PubMed] [Google Scholar]
- 60.Choi IR, White KA. 2002. An RNA activator of subgenomic mRNA1 transcription in tomato bushy stunt virus. J Biol Chem 277:3760–3766. doi: 10.1074/jbc.M109067200. [DOI] [PubMed] [Google Scholar]
- 61.Koev G, Miller WA. 2000. A positive-strand RNA virus with three very different subgenomic RNA promoters. J Virol 74:5988–5996. doi: 10.1128/jvi.74.13.5988-5996.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wierzchoslawski R, Dzianott A, Bujarski J. 2004. Dissecting the requirement for subgenomic promoter sequences by RNA recombination of brome mosaic virus in vivo: evidence for functional separation of transcription and recombination. J Virol 78:8552–8564. doi: 10.1128/JVI.78.16.8552-8564.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wielgosz MM, Raju R, Huang HV. 2001. Sequence requirements for Sindbis virus subgenomic mRNA promoter function in cultured cells. J Virol 75:3509–3519. doi: 10.1128/JVI.75.8.3509-3519.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lemm JA, Rumenapf T, Strauss EG, Strauss JH, Rice CM. 1994. Polypeptide requirements for assembly of functional Sindbis virus replication complexes: a model for the temporal regulation of minus- and plus-strand RNA synthesis. EMBO J 13:2925–2934. doi: 10.1002/j.1460-2075.1994.tb06587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kazakov T, Yang F, Ramanathan HN, Kohlway A, Diamond MS, Lindenbach BD. 2015. Hepatitis C virus RNA replication depends on specific cis- and trans-acting activities of viral nonstructural proteins. PLoS Pathog 11:e1004817. doi: 10.1371/journal.ppat.1004817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Targett-Adams P, Boulant S, McLauchlan J. 2008. Visualization of double-stranded RNA in cells supporting hepatitis C virus RNA replication. J Virol 82:2182–2195. doi: 10.1128/JVI.01565-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schnell G, Loo YM, Marcotrigiano J, Gale M Jr. 2012. Uridine composition of the poly-U/UC tract of HCV RNA defines non-self-recognition by RIG-I. PLoS Pathog 8:e1002839. doi: 10.1371/journal.ppat.1002839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Grunvogel O, Colasanti O, Lee JY, Kloss V, Belouzard S, Reustle A, Esser-Nobis K, Hesebeck-Brinckmann J, Mutz P, Hoffmann K, Mehrabi A, Koschny R, Vondran FWR, Gotthardt D, Schnitzler P, Neumann-Haefelin C, Thimme R, Binder M, Bartenschlager R, Dubuisson J, Dalpke AH, Lohmann V. 2018. Secretion of hepatitis C virus replication intermediates reduces activation of Toll-like receptor 3 in hepatocytes. Gastroenterology 154:2237–2251. doi: 10.1053/j.gastro.2018.03.020. [DOI] [PubMed] [Google Scholar]
- 69.Caillet-Saguy C, Simister PC, Bressanelli S. 2011. An objective assessment of conformational variability in complexes of hepatitis C virus polymerase with non-nucleoside inhibitors. J Mol Biol 414:370–384. doi: 10.1016/j.jmb.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 70.Biswal BK, Cherney MM, Wang M, Chan L, Yannopoulos CG, Bilimoria D, Nicolas O, Bedard J, James MN. 2005. Crystal structures of the RNA-dependent RNA polymerase genotype 2a of hepatitis C virus reveal two conformations and suggest mechanisms of inhibition by non-nucleoside inhibitors. J Biol Chem 280:18202–18210. doi: 10.1074/jbc.M413410200. [DOI] [PubMed] [Google Scholar]
- 71.Labonte P, Axelrod V, Agarwal A, Aulabaugh A, Amin A, Mak P. 2002. Modulation of hepatitis C virus RNA-dependent RNA polymerase activity by structure-based site-directed mutagenesis. J Biol Chem 277:38838–38846. doi: 10.1074/jbc.M204657200. [DOI] [PubMed] [Google Scholar]
- 72.Kato N, Sugiyama K, Namba K, Dansako H, Nakamura T, Takami M, Naka K, Nozaki A, Shimotohno K. 2003. Establishment of a hepatitis C virus subgenomic replicon derived from human hepatocytes infected in vitro. Biochemistry Biophys Res Commun 306:756–766. doi: 10.1016/S0006-291X(03)01047-7. [DOI] [PubMed] [Google Scholar]
- 73.Chomczynski P, Sacchi N. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 162:156–159. doi: 10.1016/0003-2697(87)90021-2. [DOI] [PubMed] [Google Scholar]
- 74.Walker PJ, Siddell SG, Lefkowitz EJ, Mushegian AR, Dempsey DM, Dutilh BE, Harrach B, Harrison RL, Hendrickson RC, Junglen S, Knowles NJ, Kropinski AM, Krupovic M, Kuhn JH, Nibert M, Rubino L, Sabanadzovic S, Simmonds P, Varsani A, Zerbini FM, Davison AJ. 2019. Changes to virus taxonomy and the International Code of Virus Classification and Nomenclature ratified by the International Committee on Taxonomy of Viruses (2019). Arch Virol doi: 10.1007/s00705-019-04306-w. [DOI] [PubMed] [Google Scholar]